Abstract
Type III protein secretion systems (T3SSs) or injectisomes are multi-protein nanomachines present in many gram-negative bacteria that have sustained long-standing close relationship with a eukaryotic host. These secretion systems have evolved to modulate host-cellular functions through the activity of the effector proteins they deliver. To reach their destination, T3SS effectors must cross the multi-barrier bacterial envelope and the eukaryotic cell membrane. Passage through the bacterial envelope is mediated by the needle complex, a central component of T3SSs that expands both the inner and outer membranes of gram-negative bacteria. A set of T3SS secreted proteins, known as translocators, form a channel in the eukaryotic plasma membrane through which the effector proteins are delivered to reach the host cell cytosol. While the effector proteins are tailored to the specific life style of the bacterium that encodes them, the injectisome is conserved among the different T3SSs. The central role of T3SSs in pathogenesis and their high degree of conservation make them a desirable target for the development of antimicrobial therapies against several important bacterial pathogens.
INTRODUCTION
Type III protein secretion systems (T3SSs) are multi-protein nanomachines present in many gram-negative bacteria with a close relationship with a eukaryotic host. The primary function of these machines is the delivery of bacterially encoded effector proteins into target eukaryotic cells(1–4), to modulate a myriad of cell biological processes for the benefit of the bacteria that encode them(5, 6). T3SSs are widespread in nature playing a central role in the pathogenic and symbiotic interactions between many bacteria and their hosts. Among the bacteria that encode T3SSs are many important human and plant pathogens. As the field has progressed, so has the amount of information available therefore precluding a comprehensive review of the literature. Therefore, in this chapter we will focus on the structural and architectural aspects of the type III system. To reflect current knowledge, and to help the reader better understand the structural organization of this machine, throughout this chapter, we will refer to the complete type III secretion machine as the injectisome, and we will describe in detail the different substructures that integrate it (i.e. the needle complex, the export apparatus, and the sorting platform). Readers are referred to other reviews for more specific aspects of the structure and function of these secretion machines(1–4).
The injectisome
The main structural element of the type III secretion system is the injectiome, a complex multi-protein structure composed of extracellular, envelope-associated and cytoplasmic elements or substructures. To facilitate their description, each of these elements will be discussed separately. However, it should be emphasized that the type III secretion machine is and operates as one single functional structural unit and that its separation into different sub-components, while useful, is somewhat arbitrary. Also, we note that the increasing knowledge on the structural organization of this system has rendered some previous descriptions of this system more confusing. Furthermore, the plethora of gene names has made comparison of systems across species somewhat challenging, thus, when referring to specific components of the injectisome, we will utilize a previously proposed universal nomenclature. Table 1 lists the specific names of proteins of the most studied T3SSs.
Table 1|.
Principal Components of Most Studied Type III Secretion Systems
| Universal Nomenclature |
Function | Yersinia |
Salmonella
SPI-1 |
Salmonella
SPI-2 |
E. coli
(EPEC/EHEC) |
Shigella | Chlamydia | P. aeruginose | P. syringae | Rhizobium | Flagellum |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Needle Complex (Flagellar Basal Body) | |||||||||||
| SctC | Secretin (OM ring) | YscC | InvG | SsaC | EscC | MxiD | CdsC | PscC | HrcC | RhcC1-RhcC2 | N/A |
| — | Secretin pilotin | YscW | InvH | — | — | MxiM | — | ExsB | HrpT | — | N/A |
| SctD | IM ring | YscD | PrgH | SsaD | EscD | MxiG | CdsD | PscD | HrpQ | Y4yQ | FliG |
| SctJ | IM ring | YscJ | PrgK | SsaJ | EscJ | MxiJ | CdsJ | PscJ | HrcJ | NolT | FliF |
| SctF | Needle subunit | YscF | PrgI | SsaG | EscF | MxiH | CdsF | PscF | HrpA | NopA-NopB | FlgE (Hook protein) |
| SctI | Inner rod subunit | YscI | PrgJ | SsaI | EscI | MxiI | — | PscI | HrpB | NolU | — |
| Export Apparatus (Inner Membrane Proteins) | |||||||||||
| SctU | Autoprotease/Su bstrate switching | YscU | SpaS | SsaU | EscU | Spa40 | CdsU | PscU | HrcU | RhcU | FlhB |
| SctV | Inner membrane channel | YscV | InvA | SsaV | EscV | MxiA | CdsV | PcrD | HrcV | Y4yR | FlhA |
| SctR | Inner membrane channel | YscR | SpaP | SsaR | EscR | Spa24 | CdsR | PscR | HrcR | RhcR | FliP |
| SctS | Inner membrane channel | YscS | SpaQ | SsaS | EscS | Spa9 | CdsD | PscS | HrcS | RhcS | FliQ |
| SctT | Inner membrane channel | YscT | SpaR | SsaT | EscT | Spa29 | CdsT | PscT | HrcT | RhcT | FliR |
| Sorting Platform (Flagellar C-ring) | |||||||||||
| SctQ | Core scafold | YscQ | SpaO | SsaQ | SepQ | Spa33 | CdsQ | PscQ | HrcQ | RhcQ | FliM-FliN |
| SctN | ATPase | YscN | InvC | SsaN | EscN | Spa47 | CdsN | PscN | HrcN | RhcN | FliI |
| SctL | Core scafold | YscL | OrgB | SsaK | EscL | MxiN | CdsL | PscL | HrpE | NolV | FliH |
| SctO | SctN/SctV linker | YscO | InvI | SsaO | EscO | Spa13 | CdsO | PscO | HrpO | Y4yJ | FliJ |
| SctK | Core scafold | YscK | OrgA | — | EscK | MxiK | — | PscK | HrpD | — | — |
| Regulatory proteins | |||||||||||
| SctP | Needle assembly regulator | YscP | InvJ | SsaP | EscP | Spa32 | CdsP | PscP | HrpP | — | FliK |
| SctW | Regulator of translocase secretion | YopN-TyeA | InvE | SsaL | SepL | MxiC | CopN | PopN | HrpJ | — | — |
| — | Initiation of needle assembly | ? | OrgC | ? | ? | MxiL | ? | ? | ? | ? | — |
| Translocators and tip complex protein | |||||||||||
| SctB | Translocon | YopD | SipC | SseD | EspB | IpaC | CopD | PopD | — | — | — |
| SctE | Translocon | YopB | SipB | SseC | EspD | IpaB | CopB | PopB | HrpK | NopX | — |
| SctA | Tip protein | LcrV | SipD | SseB | EspA | IpaD | CT584 | PcrV | — | — | — |
1). The needle complex
The needle complex (NC) is a major core element of the T3SS injectisome and its discovery in 1998 constituted a major breakthrough in the understanding of type III secretion machines(7). Up until then, T3SSs were simply a collection of genes required for protein secretion without a clear framework to explain how they may constitute a protein secretion machine. Since the first visualization and isolation from S. Typhimurium(7), it has been visualized in other bacteria showing a conserved architecture(8–10). It consists of a multi-ring base substructure embedded in the bacterial envelope and a needle-like extension protruding several nanometers from the bacterial surface (Figure 1). The needle is linked to the base through a substructure known as the inner rod, which docks into a socket-like structure within the NC composed of the export apparatus, which is thought to form a conduit to facilitate the passage of effector proteins through the bacterial inner membrane. At the distal side of the needle lies the needle tip complex, which senses the target host cell.
Figure 1.
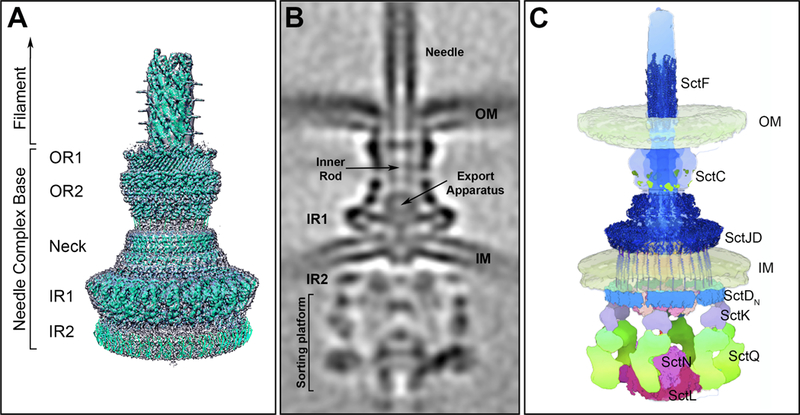
Salmonella Typhimurium SPI-1 encoded type III secretion system. (A) Surface view of the 3D reconstruction of the single particle cryo-EM map of the needle complex substructure with the atomic structures of the different needle complex components docked. OR1: outer ring 1; OR2: outer ring 2; IR1: inner ring 1; IR2: inner ring 2. (B) Central section of an overall cryo-ET structure of the complete injectisome in situ. Of note is the location of IR2 in the cytosolic side of the bacterial envelope. (C) Molecular model of the organization of the injectisome in situ with available atomic structures fitted into the model (figure adapted from DOI: 10.1016/j.cell.2018.01.034)
The base
The NC base is a multi-ring structure that spans the inner and outer membranes of the bacterial envelope. Despite its architectural complexity, the base of the NC is composed of a relatively small number of proteins. The inner rings (IR1 and IR2), located in the bacterial inner membrane and the cytoplasm, are composed of two proteins (SctJ and SctD form the IR1 while SctD alone forms the IR2), whereas the outer ring (OR) and the neck of the base are composed of a single protein (SctC), a member of the secretin family of outer membrane proteins. Single particle cryo-electron microscopy (cryo-EM) of isolated complexes has revealed a detailed view of the NC of the S. Typhimurium Pathogenicity Island 1 (SPI-1) encoded T3SS(11–13). This structure revealed that 15 SctC molecules form a double-walled stranded β-barrel complex in the outer membrane, that connects directly to 24 molecules of the inner ring components, amounting to a symmetry mismatch(12, 13). This mistmatch confers flexibility to the neck, which may be important during NC assembly. The IRs are composed of two concentric rings. A larger peripheral ring formed by SctD encircling a smaller internal ring formed by SctJ. This smaller ring is shielded on the sides by the SctD ring and on top by the SctC neck. Both SctD and SctJ share a similar although inverted topology, with a periplasmic and a cytoplasmic domain separated by a single transmembrane segment. The N-terminal domain of SctJ is lipidated and located in the periplasm and the C-terminal cytoplasmic domain is very short or absent in some homologues which also lack the transmembrane domain(14). In contrast, SctD has a longer N-terminal cytoplasmic domain that forms the IR2 and links the NC base to the sorting platform (see below). The soluble domains of some base components have been crystalized [i. e. the periplasmic domains of the outer membrane secretin EscC (SctC)(15), PrgH (SctD)(16) and EscJ (SctJ)(17)]. In all cases, the atomic structures showed a modular architecture of three topologically similar α/β domains. A comparison of the structures revealed a strong similarity between them despite the lack of detectable sequence identity. The fact that these three proteins arrange in a ring led to the hypothesis that this modular fold may represent a ring-building motif(15). However, this domain is also present in proteins that do not organize in rings and has been shown to be dispensable for ring formation(18). Therefore, the relationship between the presence of this domain and the ability to organize in rings remains unclear. The structure of the NC in situ determined by cryo electron tomography (cryo-ET) has shown that although the outer ring, neck, and inner rings of the in situ and isolated structures are virtually identical, there are some unique features revealed by the cryo-ET analysis(19) (Figure 1). The outer ring of the NC is inserted into the inner leaflet of the outer membrane resulting in an “inward pinch”. Furthermore, although the IR1, which is predicted to be located in the periplasmic space, completely overlap in the isolated and in in situ structures, the IR2, which is located in the cytoplasm, does not. In the in situ structure this ring is pushed further away from IR1 to accommodate the inner membrane, which separates both rings. Upon assembly of the sorting platform, the IR2 undergoes a significant conformation change to adopt a “6 patch” organization to accommodate the 6 pods of the sorting platform (see below)(19).
The needle
The NC base has a several nanometer-long needle extension that confers the NC its “syringe-like” appearance. The needle substructure is composed of a single, small protein subunit, SctF, polymerized in a helical fashion(20–23). In its native arrangement the length of the needle ranges between 30 and 70 nm, and its width from 10 to 13 nm. The atomic structure of the needle protein shows a α-helical hairpin arrangement with two α-helixes of similar size separated by a short loop most often containing two proline residues separated by two amino acids (the PXXP motif)(24–26). More recent studies involving cryo-EM(27) and solid state NMR(23) of recombinant and native needle polymers indicate that the needle has a right-handed helical organization consisting of ∼5.7 subunits per turn and a helical pitch of ∼24 Å. The entire assemble is traversed by a channel ∼25 Å in diameter. Although initially there were two incompatible atomic models for the needle polymer, several studies have now conclusively shown that the subunit orientation within the needle polymer places the extended N-terminus of the protomer facing the outside of the polymer and the C-terminus of the subunit facing the lumen. Residues that face the lumen of the needle are highly conserved and mostly polar, and analysis of their electrostatic potential reveals alternating positively- and negatively-charged regions. The implications of that observation are unclear but it is possible that such organization could play a role in the mechanisms of secretion through the needle channel. Within the filament, the individual proteins are stabilized by multiple inter- and intra-subunit contacts resulting in a rather rigid structure. Attention has been placed on a small kink (residues Val-20 to Asn-22 in the case of the S. Typhimurium SPI-1 needle protein) that interrupts the N-terminal α-helix and that it is not observed in the crystal structure of the soluble protomer. The implications of this observation has not been determined but it is conceivable that it could play a role in signal transduction during activation of the injectisome upon contact with target cells.
The inner rod
The inner rod is a substructure that links the needle to the NC base by docking to the export apparatus(11). Although by analogy with the flagellar system this substructure was originally referred to as a “rod”, cross-linking as well as stoichiometry studies suggest that this structure is most likely akin to a “washer” rather than a rod since it is predicted to have ~6 subunits(28, 29). Like the needle substructure, it is built from a single small protein subunit, SctI. The atomic structure of the inner rod of S. Typhimurium has been solved by CD and NMR spectroscopy(30). In its soluble monomeric form this protein is largely unfolded lacking tertiary structure. However, computational methods have suggested that the inner rod subunit shares a similar structure to that of the needle protein, an α-helical hairpin shape flanked by flexible regions. in silico modeling has also determined that the domains critical for filament assembly are well conserved between the needle and inner rod protein. However, the two subunits differ significantly at their N-terminus, and based on the needle filament structure, these differences would not allow the inner rod to elongate beyond one turn of the helix (~6 subunits)(28). This substructure has been implicated in substrate switching and needle length control(31, 32).
The needle tip complex and needle extension
The T3SS is inactive prior to contact with the eukaryotic host. In this inactive state, the needle filament is capped by a single protein that organizes in a tip complex(33, 34), or it is extended by another filament longer than the needle itself(35). The tip complex and the needle extension, play a role in sensing the environment preventing the premature unproductive secretion of effectors.
Based on their structure and biochemical properties the proteins that make up the tip complex can be divided in two groups: the SipD/IpaD (from Salmonella and Shigella respectively) and the LcrV/PcrV (from Yersinia and Pseudomonas). SipD/IpaD-like tip proteins are organized in an N-terminal α-helical hairpin, a long central coiled-coil domain, and a C-terminal region containing a mixture α/β domains(36, 37). The central coiled-coil domain is characteristic of all tip proteins although in LcrV/PcrV it is flanked by globular domains on the N- and C-termini, that give them a dumb-bell appearance(38). The central coiled-coil domain is important for the interaction of the tip protein with the needle filament and the needle protomer is expected to bind at multiple sites on this domain. The N-terminal α-helical hairpin folds independently and has been shown to act as a self-chaperone preventing the untimely oligomerization of the SipD/IpaD subunits in the bacterial cytoplasm. This self-chaperoning domain is absent in the LcrV/PcrV family of tip proteins, where a small cytoplasmic protein (LcrG/PcrG) functions as a chaperone instead(39).
Several crystal structures of different tip proteins have been solved, either alone(37, 38, 40), in combination with the needle protein (PrgI-SipD)(41), or in complex with bile salts(42, 43). What is needed however, is an atomic resolution view of the tip protein complex assembled at the needle tip. Presently there are low-resolution 3D reconstructions from electron micrographs of the Yersinia LcrV tip(44) and the Shigella IpaD tip(33). The Yersinia LcrV tip complex showed a well-defined structure characterized for the presence of a base (formed by the N-terminal globular domain), a neck (formed by the coiled-coil region), and a head (comprising the C-terminal globular domain). This precise organization could be accounted by a pentameric LcrV ring. The low-resolution structure of the IpaD tip complex is also compatible with a pentameric organization of the tip protein. This complex however does not show the characteristic morphology of the LcrV tip complex. It has been proposed that the Shigella tip complex may exhibit two different compositions, a homo-pentameric IpaD tip complex and a hetero-pentameric complex consisting on four IpaD molecules along with one IpaB molecule(45). Quantification of the relative abundance of the two different complexes was not feasible, therefore it is possible that the IpaD-IpaB complexes may represent a low proportion of injectisomes that have been activated prematurely while the pentameric IpaD structure represents the resting tip complex.
A significant modification in the tip complex occurs in several bacterial species including pathogenic E. coli strains (EPEC/EHEC). These injectisomes are characterized by the presence of a filamentous extension to the needle substructure, which in E. coli is formed by a single protein, EspA(35). Similar to the mechanism by which flagellin assembles into the flagellum, the EspA filament assembles by coiled-coil interactions between EspA subunits. However, electron micrographs of negative stained EspA filaments showed that they are distinct from flagella(46). The 3D reconstruction of the EspA filaments shows that they consist of a helical tube ~120 Å wide, containing a hollow central channel of ~25 Å in diameter with a continuous channel through which effector proteins are translocated. The EspA filament shows helical symmetry having 28 subunits in 5 turns for a 1 start helix (5.6 subunits/turn). A later study using cryo-EM of frozen-hydrated filaments has shown that the EspA filament displays heterogeneity in the structure(47) due to a fixed rotation between subunits, but a variable axial rise between adjacent subunits. How this variability in the structure relates to the function of the filaments has not yet been addressed.
The export apparatus
All T3SSs contain five conserved inner membrane proteins that are essential for their function, SctV, SctR, SctS, SctT, and SctU(48–51). Cryo-EM studies have correlated the presence of a defined density inside the inner membrane rings with the presence of the inner membrane proteins, indicating that at least a subset of these inner membrane proteins are located inside the NC structure creating a channel through which the secreted proteins can traverse the inner membrane(52). More recently, a cryo-EM structure of an in vitro assembled complex of 3 export apparatus components, SctR, SctS and SctT, from the homologous flagellar export apparatus was solved providing major insight into the organization of this substructure(53). The complex adopts a helical configuration and is organized with a stoichiometry 5:4:1 for FliP(SctR), FliQ(SctS), and FliR(SctT), respectively. Remarkably, despite the presence of several predicted transmembrane domains, none of these proteins adopt a canonical membrane protein configuration. Rather, the complex is arranged in a helical configuration in which a FliP/FliQ pair and an additional FliP molecule combine with one copy of FliR. FliR is structurally equivalent to the FliP/FliQ pair so the structure consists of six copies of a FliR-like element forming a single helical turn. The helical arrangement of these export apparatus components provides an optimal platform onto which the inner rod/needle filament can be effectively assembled. Two additional components of the export apparatus, SctV and SctU, whose location within the needle complex have not been precisely determined, are likely to play a more specialized role in type III secretion. SctV has a large cytoplasmic domain that crystalizes as a circular nonamer(18) and by cryo-ET can be seen as toroidal shape density immediately bellow the cytoplasmic side of the IR2 ring of the NC(19). It is possible that SctV may play a role in preparing the substrates for translocation through the export apparatus. It has also been suggested that the SctV family of proteins may work as a proton channel to energize the secretion process, although this activity has not been formally demonstrated(54). Another member of these inner membrane proteins, SctU, plays a role in substrate switching of the machine specificity form early substrates (i.e. needle complex components and regulators) to translocators and effectors. SctU, also known as the “switch protein”, is a protease that undergoes autocatalytic cleavage(55–62). This auto-cleaving event was proposed to be the signal that triggers substrate switching from early substrates (needle and inner rod proteins and other accessory proteins) to middle and late substrates (translocators and effectors). However, more recent experiments have demonstrated that the cleavage event per se does not provide a signal for substrate switching and that the cleavage may simply be required to provide SctU with the appropriate conformation for its secretion function(63).
2). The sorting platform
There are several conserved cytosolic proteins, SctQ, SctK, SctL, and SctN, that are required for type III secretion. They arrange in a complex that operates as a sorting platform to organize the secretion process through the T3SS establishing a hierarchy in the order in which substrates are secreted(64). The proteins that constitute the sorting platform are highly conserved among all T3SSs and have been shown to interact with one another(64–66). Recently, the structural organization of these proteins in situ was revealed by cryo-ET(19, 67)(Figure 1). The sorting platform exhibits a cage-like architecture, enclosed by 6 pod-like structures that emerge from the NC and converge into a 6-spoke wheel-like structure at its cytoplasmic side. This structure serves as scaffold to place the associated ATPase SctN and SctO in close apposition to the export apparatus. These studies have also revealed that SctQ accounts for most of the protein density associated with the pods and is linked to the wheel-like capping structure formed by SctL on one side and SctK, which links the sorting platform to the NC, on the other. The presence of the 6-pod structure stands in contrast with the appearance of a related structure found in the flagellar apparatus known as the C-ring, which forms a closed ring stably linked to the flagellar basal body(68). These structural differences may reflect the fundamentally different roles they play. There is evidence indicating that the sorting platform exhibits a dynamic behavior with cycles of assembly and disassembly although the specific role of this behavior in the function of T3SS or the mechanisms by which this behavior may be controlled are not currently known(69–71). It is possible that an alternatively translated product of the open reading frame encoding the core sorting platform components SctQ may be involved in this process(72–75). However, more research will be required to clarify these poorly understood aspects of type III secretion. The specific mechanisms by which the sorting platform may engage substrates is also poorly understood although it is expected that the associated ATPase, SctN, with the help of SctO may play a central role in the process of bringing the substrates to the ring formed by the cytoplasmic domain of SctV on the cytosolic side of the NC base.
Assembly of the injectisome
The assembly of the injectisome occurs in a step-wise manner (Figure 2). First, the export apparatus components SctR, SctS, and SctT, are inserted in the membrane(52) and subsequently form a pseudo hexameric assembly(53). Later, two additional membrane proteins, SctU snd SctV, are added to the export apparatus. The assembled export apparatus then serves as a template for the assembly of the inner rings of the NC(52). Nucleation of the inner rings around the export apparatus may also result in the “extraction” of the export apparatus components from the plasma membrane(53).
Figure 2.
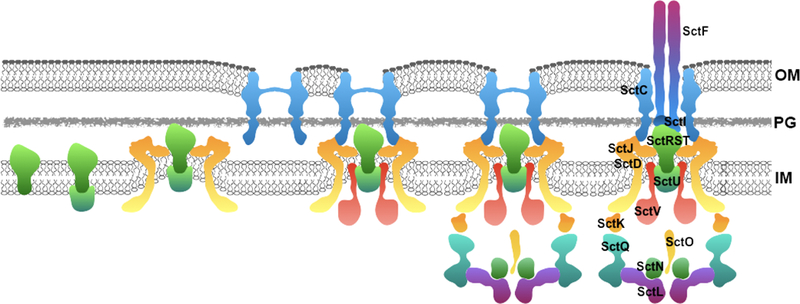
Model of the step-wise assembly of the injectisome. SctRST form a stable complex in the inner membrane (IM) to which SctU is recruited. This complex nucleates the assembly of the inner rings integrated by SctJ and SctD, which results in the extraction or “pulling” of the IM components from the bacterial plasma membrane. At the same time, the secretin is independently assembled into the outer ring and the two structures come together to form the needle complex base substructure to which SctV is subsequently recruited. Once the NC base is formed, the cytoplasmic sorting platform is recruited to the cytoplasmic side of the NC base and the system starts to function as a type III secretion machine dedicated to the delivery of early substrates such as the inner rod (SctI) and needle (SctF) subunits to complete the assembly of the entire injectisome.
The ORs and neck of the base are made of a single protein, SctC, belonging to the secretin family of outer membrane bacterial proteins(76). With the assistance of a “pilotin” SctC assembles into a pore in the outer membrane(77–80). Once the ORs and the IRs in association with export apparatus are independently assembled, they come together to form a complete NC base substructure. The cytoplasmic accessory proteins that form the sorting platform are recruited to the NC base that starts to function as a T3SS dedicated to the secretion of early substrates (22, 81). These early substrates include proteins that will create the needle, SctF, and the inner rod, SctI, as well as accessory regulatory proteins SctP, and the S. Typhimurium protein OrgC, which regulate the assembly process(82). Completion of the needle triggers substrate switching from early substrates (needle complex component and regulators of needle complex assembly) to translocators and effectors, and the injectisome is now ready to be activated by the contact with the host cell. An alternative outside-in assembly model has been proposed for Yersinia spp. although current evidence is not consistent with such model(83).
Activation of the type III secretion machine
A distinctive feature of T3SSs is that they require an activating signal to secrete and translocate effectors. Although the nature of the activating signal is poorly understood, in most cases it derives from the bacterial contact with target cells(84–86), although other agonists have also been described(87, 88). The activation step is presumably necessary to ensure that the effectors are not unproductively secreted into the extracellular environment prior to host cell contact. How cell contact triggers the activation of the T3SS is not known but the tip complex is predicted to be involved in the process. In support of this hypothesis it is possible to activate secretion in vitro by the addition of compounds such as bile salts or congo red that bind the tip complex and presumably induce conformational changes similar to those that may occur upon cell contact(42, 43, 89). These conformational changes are thought to be transduced to the secretion machine through the needle and inner rod structures. Consistent with this hypothesis, mutations in the needle and inner rod proteins have been identified that result in constitutive or altered secretion phenotypes(28, 90–92). Activation of the secretion machine leads to secretion of the translocators (SctB and SctE), which are deployed on the eukaryotic plasma membrane to form the translocation pore or translocon through which effector proteins directly reach the cytosol of the host cell (Figure 3). Recent cryo ET studies have provided insight into the organization of the translocon(93). These studies have observed a well-defined ‘bend’ on the target cell membrane in areas where the needle substructure makes contact with the host cell interface, reflecting the intimate association that is known to be required for optimal T3SS-mediated effector translocation(94). Notably, these studies showed the presence a distinct density within the region of the target host cell membrane in close apposition to the needle tip of the T3SS injectisome, which was correlated to the presence of the translocon. Although the structure was not detailed enough to provide insight into the stoichiometry and/or molecular organization of the translocon, it showed a structure ~13.5 nm in diameter and 8 nm in thickness, which was smaller than a structure (~60 nm in diameter) of the enteropathogenic E. coli translocons assembled from purified components on red blood cells(95). The reason for the different dimensions are unclear but they may reflect the differences between the in vivo and in vitro deployment of the translocases. For the translocation to be productive, the translocases must be engaged by the secretion machinery preceding the effectors, through mechanisms involving the sorting platform. Consistent with this notion, only the translocases can be detected in complex with the sorting platform prior to host cell contact, and it is only in the absence of the translocases that the effectors can be detected at this location(64).
Figure 3.
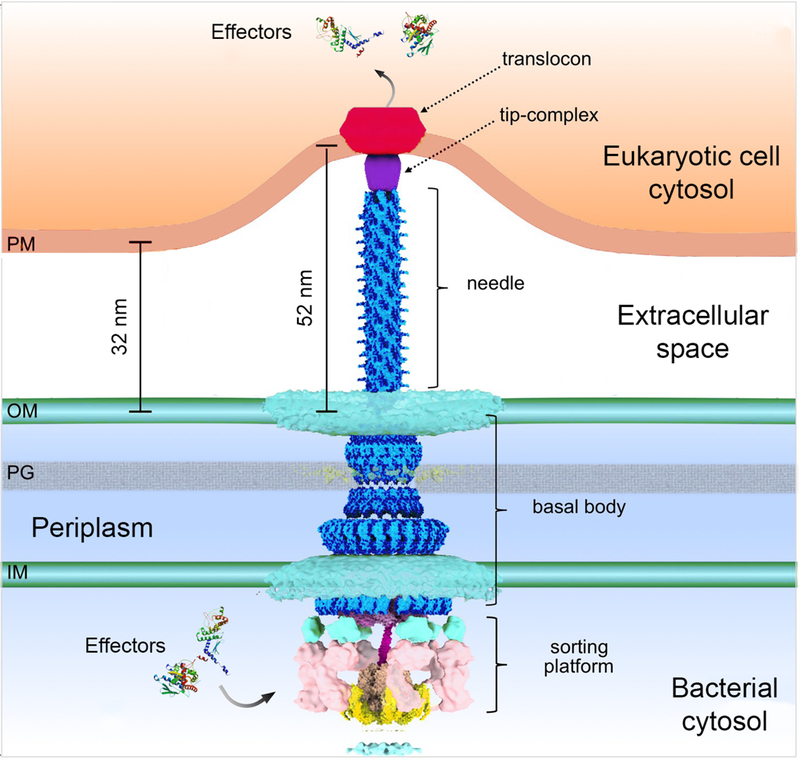
Model of the injectisome’s interaction with an eukaryotic host cell. Activation of the injectisome leads to secretion of the translocators that are deployed on the eukaryotic plasma membrane to form the translocon, which remains in contact with the needle to form a direct conduit between the bacterial and host cell cytosol that serves a passageway for the effector proteins (figure adapted from DOI: 10.7554/eLife.39514).
Concluding remarks
Type III secretion protein machines have sparked the interest of scientists for more than two decades. Although there has been remarkable advancements in the understating of the structure and function of these machines many knowledge gaps still remain. For example atomic information on some essential elements of the injectisome are still missing, the precise mechanism of substrate engagement by the secretion machine are poorly understood, and the mechanisms of eukaryotic cell sensing and signal transduction remain obscure. Addressing some of these fundamental questions most likely will require the development of novel experimental approaches to be able to study the function of these machine in live bacteria. The presence of these machines in many important bacterial pathogens has made them attractive targets for the development of next generation antimicrobials that can be deployed to prevent and combat many important infectious diseases.
REFERENCES
- 1.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. [DOI] [PubMed] [Google Scholar]
- 2.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Notti RQ, Stebbins CE. 2016. The Structure and Function of Type III Secretion Systems. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner S, Grin I, Malmsheimer S, Singh N, Torres-Vargas CE, Westerhausen S. 2018. Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol Lett 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks SW, Galan JE. 2013. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol 11:316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinaud L, Sansonetti PJ, Phalipon A. 2018. Host Cell Targeting by Enteropathogenic Bacteria T3SS Effectors. Trends Microbiol 26:266–283. [DOI] [PubMed] [Google Scholar]
- 7.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. [DOI] [PubMed] [Google Scholar]
- 8.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. 2001. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol 39:652–663. [DOI] [PubMed] [Google Scholar]
- 9.Daniell SJ, Takahashi N, Wilson R, Friedberg D, Rosenshine I, Booy FP, Shaw RK, Knutton S, Frankel G, Aizawa S. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell Microbiol 3:865–871. [DOI] [PubMed] [Google Scholar]
- 10.Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci U S A 98:11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schraidt O, Marlovits TC. 2011. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331:1192–1195. [DOI] [PubMed] [Google Scholar]
- 13.Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JR, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NC. 2016. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature doi: 10.1038/nature20576. [DOI] [PubMed]
- 14.Crepin VF, Prasannan S, Shaw RK, Wilson RK, Creasey E, Abe CM, Knutton S, Frankel G, Matthews S. 2005. Structural and functional studies of the enteropathogenic Escherichia coli type III needle complex protein EscJ. Mol Microbiol 55:1658–1670. [DOI] [PubMed] [Google Scholar]
- 15.Spreter T, Yip CK, Sanowar S, Andre I, Kimbrough TG, Vuckovic M, Pfuetzner RA, Deng W, Yu AC, Finlay BB, Baker D, Miller SI, Strynadka NC. 2009. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol 16:468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergeron JR, Worrall LJ, Sgourakis NG, DiMaio F, Pfuetzner RA, Felise HB, Vuckovic M, Yu AC, Miller SI, Baker D, Strynadka NC. 2013. A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog 9:e1003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NC. 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435:702–707. [DOI] [PubMed] [Google Scholar]
- 18.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Lara-Tejero M, Kong Q, Galan JE, Liu J. 2017. In Situ Molecular Architecture of the Salmonella Type III Secretion Machine. Cell 168:1065–1074 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem 278:17103–17107. [DOI] [PubMed] [Google Scholar]
- 21.Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH. 2010. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J Mol Biol 396:1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubori T, Sukhan A, Aizawa SI, Galan JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A 97:10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. 2012. Atomic model of the type III secretion system needle. Nature 486:276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. 2006. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci U S A 103:12529–12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, Tenenboim H, Goosmann C, Laube B, Thunemann AF, Zychlinsky A, Baldus M, Lange A, Griesinger C, Kolbe M. 2010. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol 17:788–792. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. 2006. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol 359:322–330. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Worrall LJ, Hong C, Vuckovic M, Atkinson CE, Caveney N, Yu Z, Strynadka NCJ. 2018. Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Commun 9:3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebre MD, Galan JE. 2014. The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc Natl Acad Sci U S A 111:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilkenat S, Franz-Wachtel M, Stierhof YD, Galan JE, Macek B, Wagner S. 2016. Determination of the Stoichiometry of the Complete Bacterial Type III Secretion Needle Complex Using a Combined Quantitative Proteomic Approach. Mol Cell Proteomics 15:1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong D, Lefebre M, Kaur K, McDowell MA, Gdowski C, Jo S, Wang Y, Benedict SH, Lea SM, Galan JE, De Guzman RN. 2012. The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J Biol Chem 287:25303–25311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637–640. [DOI] [PubMed] [Google Scholar]
- 32.Wood SE, Jin J, Lloyd SA. 2008. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol 190:4252–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epler CR, Dickenson NE, Bullitt E, Picking WL. 2012. Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol 420:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674–676. [DOI] [PubMed] [Google Scholar]
- 35.Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J 17:2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Wong Fat Sang N, Withers J, Gill R, Wood SP, Wood M, Fox GC, Cooper JB. 2006. High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J Mol Biol 363:125–136. [DOI] [PubMed] [Google Scholar]
- 37.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. 2007. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem 282:4035–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derewenda U, Mateja A, Devedjiev Y, Routzahn KM, Evdokimov AG, Derewenda ZS, Waugh DS. 2004. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 12:301–306. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhury S, de Azevedo Souza C, Plano GV, De Guzman RN. 2015. The LcrG Tip Chaperone Protein of the Yersinia pestis Type III Secretion System Is Partially Folded. J Mol Biol 427:3096–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaudhury S, Battaile KP, Lovell S, Plano GV, De Guzman RN. 2013. Structure of the Yersinia pestis tip protein LcrV refined to 1.65 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lunelli M, Hurwitz R, Lambers J, Kolbe M. 2011. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog 7:e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Nordhues BA, Zhong D, De Guzman RN. 2010. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry 49:4220–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. 2011. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci 20:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broz P, Mueller CA, Muller SA, Philippsen A, Sorg I, Engel A, Cornelis GR. 2007. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol 65:1311–1320. [DOI] [PubMed] [Google Scholar]
- 45.Cheung M, Shen DK, Makino F, Kato T, Roehrich AD, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ. 2015. Three-dimensional electron microscopy reconstruction and cysteinemediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol 95:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daniell SJ, Kocsis E, Morris E, Knutton S, Booy FP, Frankel G. 2003. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol Microbiol 49:301–308. [DOI] [PubMed] [Google Scholar]
- 47.Wang YA, Yu X, Yip C, Strynadka NC, Egelman EH. 2006. Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved approach to helical reconstruction. Structure 14:1189–1196. [DOI] [PubMed] [Google Scholar]
- 48.Allaoui A, Woestyn S, Sluiters C, Cornelis GR. 1994. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol 176:4534–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galan JE, Ginocchio C, Costeas P. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol 174:4338–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginocchio CC, Galan JE. 1995. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect Immun 63:729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groisman EA, Ochman H. 1993. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J 12:3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner S, Konigsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, Galan JE. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A 107:17745–17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhlen L, Abrusci P, Johnson S, Gault J, Deme J, Caesar J, Dietsche T, Mebrhatu MT, Ganief T, Macek B, Wagner S, Robinson CV, Lea SM. 2018. Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee PC, Rietsch A. 2015. Fueling type III secretion. Trends Microbiol 23:296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, Wolf-Watz H, Lloyd SA. 2003. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol 185:2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. 2002. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol 184:4500–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280:41236–41242. [DOI] [PubMed] [Google Scholar]
- 58.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127. [DOI] [PubMed] [Google Scholar]
- 59.Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, Lea SM. 2008. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol 69:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiesand U, Sorg I, Amstutz M, Wagner S, van den Heuvel J, Luhrs T, Cornelis GR, Heinz DW. 2009. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J Mol Biol 385:854–866. [DOI] [PubMed] [Google Scholar]
- 61.Lountos GT, Austin BP, Nallamsetty S, Waugh DS. 2009. Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci 18:467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bjornfot AC, Lavander M, Forsberg A, Wolf-Watz H. 2009. Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J Bacteriol 191:4259–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monjaras Feria JV, Lefebre MD, Stierhof YD, Galan JE, Wagner S. 2015. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. MBio 6:e01459–01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson MW, Plano GV. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol Lett 186:85–90. [DOI] [PubMed] [Google Scholar]
- 66.Spaeth KE, Chen YS, Valdivia RH. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A 112:1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas D, Morgan DG, DeRosier DJ. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J Bacteriol 183:6404–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diepold A, Kudryashev M, Delalez NJ, Berry RM, Armitage JP. 2015. Composition, formation, and regulation of the cytosolic c-ring, a dynamic component of the type III secretion injectisome. PLoS Biol 13:e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diepold A, Sezgin E, Huseyin M, Mortimer T, Eggeling C, Armitage JP. 2017. A dynamic and adaptive network of cytosolic interactions governs protein export by the T3SS injectisome. Nat Commun 8:15940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Lara-Tejero M, Bewersdorf J, Galan JE. 2017. Visualization and characterization of individual type III protein secretion machines in live bacteria. Proc Natl Acad Sci U S A 114:6098–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bzymek KP, Hamaoka BY, Ghosh P. 2012. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 51:1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDowell MA, Marcoux J, McVicker G, Johnson S, Fong YH, Stevens R, Bowman LA, Degiacomi MT, Yan J, Wise A, Friede ME, Benesch JL, Deane JE, Tang CM, Robinson CV, Lea SM. 2016. Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol 99:749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu XJ, Liu M, Matthews S, Holden DW. 2011. Tandem translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J Biol Chem 286:36098–36107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Notti RQ, Bhattacharya S, Lilic M, Stebbins CE. 2015. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun 6:7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Majewski DD, Worrall LJ, Strynadka NC. 2018. Secretins revealed: structural insights into the giant gated outer membrane portals of bacteria. Curr Opin Struct Biol 51:61–72. [DOI] [PubMed] [Google Scholar]
- 77.Daefler S, Russel M. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol Microbiol 28:1367–1380. [DOI] [PubMed] [Google Scholar]
- 78.Crago AM, Koronakis V. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol 30:47–56. [DOI] [PubMed] [Google Scholar]
- 79.Burghout P, Beckers F, de Wit E, van Boxtel R, Cornelis GR, Tommassen J, Koster M. 2004. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol 186:5366–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okon M, Moraes TF, Lario PI, Creagh AL, Haynes CA, Strynadka NC, McIntosh LP. 2008. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16:1544–1554. [DOI] [PubMed] [Google Scholar]
- 81.Magdalena J, Hachani A, Chamekh M, Jouihri N, Gounon P, Blocker A, Allaoui A. 2002. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J Bacteriol 184:3433–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kato J, Dey S, Soto JE, Butan C, Wilkinson MC, De Guzman RN, Galan JE. 2018. A protein secreted by the Salmonella type III secretion system controls needle filament assembly. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J 29:1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Menard R, Sansonetti P, Parsot C. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J 13:5293–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zierler MK, Galan JE. 1995. Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect Immun 63:4024–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mounier J, Bahrani FK, Sansonetti PJ. 1997. Secretion of Shigella flexneri Ipa invasins on contact with epithelial cells and subsequent entry of the bacterium into cells are growth stage dependent. Infect Immun 65:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bahrani FK, Sansonetti PJ, Parsot C. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun 65:4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. 2007. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75:2626–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. 2011. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry 50:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem 280:42929–42937. [DOI] [PubMed] [Google Scholar]
- 91.Cherradi Y, Schiavolin L, Moussa S, Meghraoui A, Meksem A, Biskri L, Azarkan M, Allaoui A, Botteaux A. 2013. Interplay between predicted inner-rod and gatekeeper in controlling substrate specificity of the type III secretion system. Mol Microbiol 87:1183–1199. [DOI] [PubMed] [Google Scholar]
- 92.Veenendaal AK, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ. 2007. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 63:1719–1730. [DOI] [PubMed] [Google Scholar]
- 93.Park D, Lara-Tejero M, Waxham MN, Li W, Hu B, Galan JE, Liu J. 2018. Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lara-Tejero M, Galan JE. 2009. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun 77:2635–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ide T, Laarmann S, Greune L, Schillers H, Oberleithner H, Schmidt MA. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol 3:669–679. [DOI] [PubMed] [Google Scholar]


