Abstract
The potential of CRISPR–Cas9-based therapeutic genome editing is hampered by difficulties in the control of the in vivo activity of CRISPR–Cas9. To minimize any genotoxicity, precise activation of CRISPR–Cas9 in the target tissue is desirable. Here, we show that, by complexing magnetic nanoparticles (MNPs) with recombinant baculoviral vectors (BVs), CRISPR–Cas9-mediated genome editing can be activated locally in vivo via a magnetic field. BV was chosen for in vivo gene delivery because of its large loading capacity and its ability to locally overcome systemic inactivation by the complement system. We demonstrate that a locally applied magnetic field can enhance the cellular entry of MNP-BVs, thereby avoiding BV inactivation and causing a transient transgene expression in the target tissue. Because BVs are inactivated elsewhere, gene delivery and in vivo genome editing via MNP-BVs are tissue-specific.
The CRISPR–Cas9 system is a revolutionary genome-editing technology that can efficiently modify target genes in mammalian cells1. Preclinical studies have shown that the CRISPR–Cas9 system provides unprecedented opportunities for treating a variety of genetic diseases and infectious diseases2–7. Although in vitro genome editing of cultured cells has many clinical applications, for potentially curing a wide range of diseases including muscular dystrophy, cystic fibrosis and cancer8,9 it is necessary to perform in vivo genome editing. The CRISPR–Cas9 system targets a short stretch of DNA via the hybridization of a complementary guide RNA (gRNA) and the binding of the Cas9 protein, which recognizes a protospacer adjacent motif (PAM)1. The gRNAs can hybridize to DNA sequences containing base mismatches and DNA and RNA bulges compared with the intended target sequence10. Consequently, the CRISPR–Cas9 system can have off-target activities, causing gene mutations, deletions, insertions or translocations, which may lead to tumorigenic or other deleterious events10–13. A major challenge for clinical applications of CRISPR–Cas9-based in vivo genome editing is, therefore, to selectively activate the CRISPR–Cas9 system in the desired tissue or organ in order to maximize therapeutic efficacy and minimize genotoxicity.
To improve the specificity of the CRISPR–Cas9 system, many tools have been developed for identifying potential gRNA off-target sites14, and the Cas9 nucleases have been designed with controllable nuclease activities15,16. For example, the Cas9 nucleases have been fragmented into nonfunctional units, which can dimerize to form active nucleases upon blue light radiation16. Cas9 can also be delivered as inducible transgenes that can only be translated in the presence of a chemical cue, e.g. doxycycline15. However, in in vivo applications, optical signals cannot penetrate deeply into the body owing to the strong absorption and scattering of light by biological tissues. The chemically-regulated Cas9 expression relies on the biodistribution of transgenes. Alternatively, in vivo genome editing can be controlled through targeted delivery of the CRISPR–Cas9 system. In particular, the viral vectors with tissue tropism, e.g., the adeno-associated viral vector (AAV)17, are being explored for tissue-specific genome editing in vivo3,13,18. However, most viral vectors for in vivo gene delivery are derived from the viruses originated from human or other mammals4. It is difficult to control the systemic dissemination and replication of these viral vectors, which increases the risk of genotoxicity19.
Recent studies have shown that magnetic nanomaterials can be used to alter molecular or cellular processes in vivo mechanically or thermally in response to a magnetic field20–23. Compared with chemical or optical signals, a magnetic field with a strength of 1.5–3.0 Tesla, such as that in a typical magnetic resonance imaging scanner, has no evident adverse effect to the human body and the magnetic field is not attenuated by the tissue24. Here we show the development of a magnetic responsive gene delivery system consisting of a baculoviral vector complexed with magnetic iron oxide nanoparticles (MNP-BV) that enables spatial control of in vivo genome editing.
The baculoviral vector (BV) is derived from a cylindrical insect virus (Autographa californica multicapsid nucleopolyhedrovirus, AcMNPV, 30~60 nm in diameter and 250~300 nm in length)25. The large size of BV allows an extraordinary DNA packing capacity (> 38 kb) compared to most other viruses, thus enabling the integration of multiple gene expression cassettes into a single viral vector26. Although BV lacks the ability to replicate in mammalian cells, it can transduce many types of mammalian cells with high efficiency and low cytotoxicity, providing a robust and transient gene expression25–29. However, in vivo applications of BV have been hurdled by its inactivation by the complement system27. Previous studies have shown that BV injected systemically can trigger the classical pathway of the complement system and lead to a significant reduction of viral transduction30. BV injected locally into muscle or tumor can induce moderate transduction due to reduced exposure to the complement system31,32. It has been shown that BV can transduce tissues that are naturally immune privileged, such as brain, eye or testis33,34. The complement induced BV inactivation can also be circumvented to some extent by protecting BV with a surface coating or treating the animal with agents that inhibit the complement system30,35,36. In this study, we show that the serum inactivation of BV can be utilized as an “off” switch to limit systemic activities of BV, and an external magnetic field can serve as an “on” switch for tissue-specific genome editing by promoting margination and cell entry of the MNP-BV complex locally. This hybrid nanoparticle-viral vector system provides a unique delivery vehicle that provides spatial control of CRISPR–Cas9-mediated in vivo genome editing.
Results and Discussion
Construction of the MNP-BV vector
Recombinant BV was produced, purified and concentrated according to published protocols (see Online Methods). The titer of various BVs was determined by a qPCR assay and a plaque assay (Supplementary Figure S1). Magnetic iron oxide nanoparticles (MNPs) that can bind to BV were synthesized in three steps. First, magnetite nanocrystals were synthesized through thermodecomposition of iron acetylacetonate in benzyl ether37. As-synthesized nanocrystals were 15.5±1.1 nm in diameter and had a saturation magnetization of 87.2 emu/g, similar to that of bulk magnetite (Supplementary Figures S2a, b). Water-dispersible MNPs were generated by coating the nanocrystals with copolymers of phospholipid and polyethylene glycol (PEG) using a dual solvent exchange method38. Coated MNPs (MNP-PEG) were then conjugated with TAT peptide (GRKKRRQRRRPQ)39, a positively charged peptide used to facilitate the interaction between MNPs and BV surface. TAT peptide conjugation to MNPs was confirmed by zeta potential measurements, gel shifting assay and DNA retardation assay (Supplementary Table S1 and Supplementary Figures S2c, d). TAT-conjugated MNPs (MNP-TAT) can disperse in aqueous buffers with negligible magnetic interactions, but upon exposure to a magnetic field, they migrate against the field gradient as nanomagnets. In most of the experiments reported in this study, the magnetic field was generated by using NdFeB block magnets with a residual induction of 1.48 Tesla (Supplementary Figure S3).
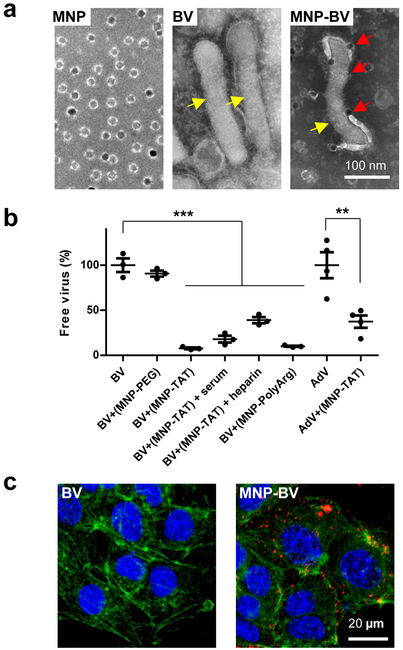
When MNP-TAT was mixed with BV in phosphate buffered saline (PBS), multiple MNP-TAT could attach to a single BV to form the MNP-BV hybrid (Figure 1a). We found that, when mixed with MNP-TAT, more than 90% of BVs could be pulled down by a magnetic plate, while mixing with MNP-PEG (i.e., without TAT) had a negligible effect on the dispersion of BVs (Figure 1b). When MNP-TAT was pre-incubated with heparin, a negatively charged molecule, the pull-down efficiency was reduced to 60% (Figure 1b). Further pull-down assays were performed respectively using MNPs conjugated with positively charged peptide poly-arginine (MNP-polyArg) instead of TAT, and mixed with adenoviruses (AdV) instead of BV, revealing similar trends (Figure 1b). Our results suggest that the interaction between BV and MNP-TAT is largely due to the nonspecific electrostatic interactions between the positively charged peptide and negatively charged viral surface40. Importantly, when serum was added to the sample, the pull-down efficiency of BV mixed with MNP-TAT was only reduced by 10% (Figure 1b), suggesting that BV and MNP-TAT could remain associated during systemic circulation.
Figure 1. Nanomagnets improve endocytosis of BV.
(a) TEM images of MNPs, BV, and MNP-BV hybrids via TAT. The samples were negatively stained with phosphotungstic acid. In the left panel, the white corona surrounding the dark cores is the coating layer of MNPs. The right panel shows multiple MNPs (red arrows) may be associated with a single BV through TAT. (b) Association between MNP and BV. BV or adenovirus (AdV) were mixed with unconjugated MNP (MNP-PEG), MNP conjugated with TAT peptide (MNP-TAT), and MNP conjugated with (Arg)12 (MNP-polyArg) and incubated on a magnetic plate under designated conditions (see Methods). The copies of free viruses were quantified by qPCR. Data represent mean ± SEM. **p=0.0038 based on one-tailed Student’s t-test, ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test. (c) MNPs under the external magnetic field enhanced endocytosis of BV. Cells incubated with BV alone (left panel) or MNP-BV (right panel) under a magnetic field were stained and examined with fluorescence microscopy. Blue, nuclei; Green, actin fibers; Red, BV stained with an anti-vp39 antibody.
Nanomagnets improve cell entry and transduction efficiency of BV in vitro
We further investigated the effect of nanomagnets on the interactions of BV with cultured Hepa 1–6 cells that have high BV infectibility (Supplementary Figure S4) 41. The cells were incubated with BV or MNP-BV for 10 minutes, much shorter than that used in a typical in vitro viral transduction assay. Cells incubated with BV alone had negligible intracellular BV as examined by immunostaining with the anti-vp39 antibody (Figure 1c) 41–43. In contrast, under a magnetic field, a large number of MNP-BV complexes entered the cells after a short period (10 minutes) of incubation (Figure 1c, Supplementary Videos V1, V2). TEM images of cell cross-sections show co-existence of MNPs and BV in the lysosomes (Supplementary Figure S5), suggesting concurrent cellular internalization of MNPs and BV.
To examine the effect of nanomagnets on BV-induced transgene expression in vitro, we constructed BV-LUC and BV-eGFP, BV vectors that contain the luciferase and eGFP cassette respectively (Figure 2a). BV-eGFP was mixed with MNP-TAT, and the mixture was incubated with Hepa 1–6 cells under a magnetic field for 30 minutes, resulting in a much larger number of eGFP-positive cells compared with having BV-eGFP alone, BV-eGFP under magnetic field (MF) but without MNPs, or BV-eGFP with MNPs but without applied magnetic field (Figures 2b, d). When BV-eGFP was mixed with MNPs labeled with a fluorophore, DiI (ex/em = 549/565 nm), MNPs could be observed in perinuclear vesicles in the cells that had a strong eGFP expression, indicating that transgene expression occurred in the presence of MNPs (Figure 2c). The BV transduction efficiency was determined by quantifying eGFP fluorescence and luciferase activity in the cells incubated with BV-eGFP and BV-LUC respectively (Figures 2d, e). The luciferase activity shown in Figure 2e was normalized with that from cells incubated with BV-LUC alone for 4 hours, resulting in a high level of luminescence (Supplementary Figure S4c).
Figure 2. Nanomagnets improve BV-mediated transgene expression in vitro.
(a) Expression vectors carrying the cassettes for luciferase (BV-LUC) and eGFP (BV-eGFP) respectively. (b) Fluorescence images of BV-mediated eGFP expression. Hepa 1–6 cells were incubated with BV or MNP-BV respectively for 30 minutes with or without a magnetic field directing to the cell surface. (c) Colocalization of eGFP (green) and MNPs (red) in the cells treated with MNP-BV-eGFP. Blue, cell nucleus staining. (d) BV-mediated eGFP expression. (e) BV-mediated luciferase expression. The luciferase activity was normalized with that from the cells incubated with BV-LUC alone for 4 hours (Supplementary Figure S4c). The transgene expression of BV was enhanced by the combination of attached MNPs and an applied magnetic field, while the effect of attaching MNPs or applying the magnetic field alone was negligible. Data represent mean ± SEM. ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test.
Clearly, having BV mixed with MNPs or under the magnetic field alone did not affect the transgene expression (Figures 2d, e). Having BV mixed with MNPs and applying a magnetic field could increase the transgene expression by 5 folds compared with that of BV alone (Figure 2e). We found that with both MNPs and an applied magnetic field, BV-eGFP and BV-LUC could efficiently transduce many other cell types such as HeLa, U87, and HUVEC (Supplementary Figure S6). The increase in the transgene expression due to nanomagnets is larger in cells with lower BV infectability such as HeLa cells (Supplementary Figure S7). At the MOI used in the in vitro experiments, MNP-BV had a low effect on cell viability, similar to a lipofectamine-based transfection (Supplementary Figure S8) 26. However, MNP-BV could have a moderate toxicity when a large amount of MNP is used with high BV MOI.
The results shown in Figure 2 were obtained with an MNP to BV ratio of ~104:1 in the MNP-BV mixture, so the vast majority of MNPs were not attached to BV. Using MNPs without the TAT peptide conjugated, we found that nanomagnets alone could enhance the cellular uptake of BV as well as BV-LUC induced transgene expression (Supplementary Figure S9). It has been shown that an important pathway of cellular uptake of BV is the macropinocytosis mediated by the baculovirus envelope glycoprotein, gp-64, which depends on actin polymerization in the cells44. Consistently, we found that the cells treated with cytochalasin D, an actin depolymerization agent, showed disrupted actin filament structure and reduced BV uptake compared to control (untreated) cells (Supplementary Figure S9). However, subsequent use of MNPs together with the magnetic field could partially restore actin filament formation and BV uptake. Our results suggest that the increase in the cellular uptake of MNP-BV complexes is partly attributable to magnetic force-induced changes in actin filaments21,45.
Nanomagnets can help overcome serum inactivation of BV in vitro
To determine if MNPs can protect BV from serum inactivation similar to that of polymer coating or ligand displaying25,27,28, we performed BV transduction in a culture medium with 50% of adult mouse serum (AMS), which contains the complement system to inactivate BV. When the cells were incubated with BV alone, BV transduction was suppressed by AMS as indicated by the low percentage of eGFP-positive cells and negligible luciferase expression (Figures 3a, b). Neither MNPs nor the magnetic field alone could rescue BV-eGFP or BV-LUC transduction. In contrast, when associated with MNPs and under an applied magnetic field, BV-eGFP had high transduction efficiency and BV-LUC induced a high level of luciferase expression in Hepa 1–6 cells (Figures 3a, b), suggesting that the inactivation of BV by AMS was diminished. When MNP-BV-LUC was used under a magnetic field, the level of transgene expression in the presence of AMS was even higher than that induced by BV-LUC alone without AMS (Figures 2e and 3b).
Figure 3. Nanomagnets help BV overcome serum inactivation in vitro.
(a) and (b) In vitro activation of transgene expression against adult mouse serum (AMS). Hepa 1–6 cells were incubated with BV-eGFP (a) or BV-LUC (b) for 30 minutes in a culture medium containing 50% AMS. The luciferase activity was normalized with that from cells incubated with BV-LUC alone for 4 hours without AMS (Supplementary Figure S4c). Data represent mean ± SEM. ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test. (c) Copies of BV genome and transgene mRNA in transduced cells. Hepa 1–6 cells were incubated with premixed BV-LUC and MNPs under designated conditions. Copies of BV genome and luciferase mRNA were quantified by qPCR. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test. (d) Nanomagnet induced BV transduction in a static cell culture. The cells cultured in a chamber slide were incubated with MNP-BV-eGFP in the culture medium containing 50% of AMS for 30 minutes, while the left half of the chamber was placed on a block magnet. Most eGFP-positive cells localized in the area on top of the magnet. (e) Nanomagnet-induced BV transduction in an artificial vessel. HUVECs were seeded in a silicone tubing (i.d. = 0.3 mm). MNP-BV-eGFP was infused in culture medium containing 50% of AMS with a syringe pump for 10 minutes while the tubing was aligned with the edge of a bar magnet. Red arrows mark the range of the magnet. The dark streak in the bright field image was MNP-BV-eGFP captured by the magnetic force. After infusion, the cells were incubated with fresh culture medium and GFP expression was examined after 24 hours. GFP expression was only found in the HUVECs next to the magnet.
To further elucidate the mechanisms underlying the complement evasion, we quantified the BV genome copy and the transgene expression in BV transduced cells (Figure 3c). AMS dramatically decreased the endocytosis of BV as indicated by the much reduced BV genome copies in cultured cells. MNP-TAT alone did not have a significant effect on the BV endocytosis nor the transgene expression. Under an applied magnetic field, the endocytosis of BV complexed with MNP-TAT increased by 20 folds in the normal medium and by more than 4 folds in the presence of AMS. These results suggest that BV inactivation was mainly due to inhibition of BV cell entry by either enzymatic degradation of BV or masking of the BV surface. It is likely that with MNP-BVs the physical forces generated by MNPs under the magnetic field induce a rapid cellular uptake of BV, which may outpace the cascade involved in serum-medicated BV inactivation. Even when the MNP-BVs were mixed with AMS for 30 minutes before applying the magnetic field, the level of BV internalization was not reduced, confirming the high stability of the MNP-BV complex in the serum found in our pull-down study (Figure 1b). However, having MNPs complexed with BV reduced the level of luciferase mRNA level in Hepa 1–6 cells, possibly due to interference with the endosomal escape of BV. The combined effects of MNPs and the magnetic field on overcoming the BV evasion from the complement system are depicted schematically in Supplementary Figure S10. Our results suggest that MNPs together with the magnetic field can enhance the endocytosis of BV through enrichment of BVs on the cell surface, and the increase in the interaction of BV with cell membrane via positively charged TAT peptide. It is also possible that magnetic force exerted on MNPs on the cell surface and in the intracellular vesicles may induce changes in actin filaments, similar to what we reported previously,23,46 which may further increase BV endocytosis. This, however, does not rule out other mechanisms, such as microtubule and heparin sulfate mediated endocytosis.
We investigated if serum inactivation and magnetic activation could be combined to provide spatial control of BV transduction. Cells seeded in a chamber slide were incubated with MNP-BV-eGFP in the presence of AMS; only half of the chamber was placed on a block magnet. We found that after 12 hours post-transduction, most eGFP-positive cells were in the area above the magnet (Figure 3d). The level of transgene expression increased with the MNP to BV ratio and the strength and the duration of the applied magnetic field (Supplementary Figure S11). When a mixture of BV and MNP-TAT were infused through a silicone tubing at physiologically relevant flow rates, BV could be captured by a block magnet placed next to the tubing (Supplementary Figure S12). This suggests that the block magnet can increase the margination of the MNP-BV complex circulating in the blood vessels and enhance the contact between MNP-BV and the vascular endothelium. In addition, an artificial vein was created by growing a layer of endothelial cells in the silicone tubing. The MNP-BV-eGFP vector in culture medium containing AMS was infused into the tubing at a flow rate of 7 mm/s. A section of the tubing was placed along a block magnet during the infusion. After overnight incubation, we found that only the cells in the tubing next to the magnet showed eGFP fluorescence (Figure 3e), further demonstrating the ability to provide spatial control of BV transduction.
We integrated the cassettes encoding eGFP, the Streptococcus pyogenes (Sp) Cas9, and the gRNA targeting mouse VEGFR2 gene into one plasmid for BV packaging (BV-CRISPR), thanks to BV’s large DNA loading capacity (> 38 kb) (Figure 4a). When delivered as a plasmid or via the BV-CRISPR vector respectively into Hepa 1–6 cells, the CRISPR–Cas9 system had cutting efficiencies of 9–30% of the mouse VEGFR2 gene (Supplementary Figure S13). When Hepa 1–6 cells were incubated with the MNP-BV vector carrying CRISPR–Cas9 (MNP-BV-CRISPR) in the medium containing 50% AMS, both the eGFP expression and CRISPR–Cas9 induced gene modification rate increased with the strength of the applied magnetic field (Figure 4b). Without applying a magnetic field to overcome BV inactivation by AMS, there was no eGFP expression or site-specific VEGFR2 gene modification in Hepa 1–6 cells (Figure 4b).
Figure 4. On- and off- switch of genome editing by the magnetic field and the complement system.
(a) BV-CRISPR expression vectors. eGFP expression cassette, guide RNA and cas9 expression cassette can all be integrated into one virus. eGFP was used for identifying transduced cells. (b) Magnetic field-triggered mouse VEGFR2 gene editing in vitro. The cells were incubated with MNP-BV (MOI = 100) in culture medium containing 50% of AMS for 30 minutes in the presence of a magnetic field at difference strength. Transgene expression was examined by eGFP fluorescence at 24 hours after transduction. There was no transgene expression in the cells without the magnetic treatment. The level of transgene expression increased with the strength of the magnetic field. After 72 hours, the cells were harvested and examined with the T7E1 assay. Consistent with the trend in eGFP expression, the CRISPR-mediated VEGFR2 disruption correlated with the magnetic field strength.
In vivo BV transduction and genome editing via local injection
It has been shown that conventional viral vectors (such as adenoviral vectors) injected to tumor tissue locally (intratumoral injection) can disseminate through systemic circulation and induce a high level of transgene expression in the liver19. In contrast, in vivo transduction of BV via local injection is most effective in immune privileged tissues lack of the complement system33,34; otherwise BV transduction is suppressed, presumably caused by a partial exposure to the complement system due to local bleeding and in situ secretion of the complement factors. We performed intratumoral injection of BV and found that BV-LUC could induce a moderate transgene expression in subcutaneous tumors (Figures 5a, b). If the tumor was injected with MNP-BV-LUC and subjected to a magnetic field for 1 hour following injection, the transgene expression was increased markedly (Figures 5a, b). Ex vivo imaging of dissected tumors and vital organs showed that the transgene expression was limited to injected tumor tissue (Figures 5c, d). Local BV transduction and transgene expression can also be achieved in the mouse hind limb following intramuscular injection (Supplementary Figure S14).
Figure 5. Magnetic field-enhanced transgene expression and genome editing in subcutaneous tumors.
The hybrid vehicle with luciferase expressing cassette (MNP-BV-LUC) was injected directly into a subcutaneous tumor. The tumor was then placed between two block magnets for 1 hour. (a) and (b) Bioluminescence analysis of transgene expression in the tumor at 24 hours post transduction. Data represent mean ± SEM. *p = 0.03 based on one-tailed Student’s paired t-test. (c) and (d) Bioluminescence analysis of transgene expression in the dissected tumors and vital organs. Data represent mean ± SEM. **p = 0.004 based on one-tailed Mann Whitney test. In a parallel study, the mice were injected with MNP-BV-CRISPR. (e) Flow chart of genome editing analysis of CRISPR–Cas9 targeted cells from the mouse tumor. (f) Analysis of in vivo mouse VEGFR2 gene editing using T7E1 assay. (g) Analysis of genome editing in the tumor and vital organs. n = 3 per group. In the tumor, multiple samples were collected at the regions near the block magnet. Note that the samples without detectable editing rate, i.e. < 0.1%, are plotted as having 0.05% editing rate.
To examine MNP-BV-CRISPR induced genome editing, we injected the mice intratumorally with MNP-BV-CRISPR and placed the tumor under a magnetic field. Following the workflow illustrated in Figure 5e, at 24 hours post MNP-BV-CRISPR delivery, the eGFP positive cells were harvested from the mouse tumors and T7E1 assays were performed to quantify the gene modification rate. We found that the MNP-BV-CRISPR induced site-specific gene modification in transduced cells with a ~30% indel rate (Figure 5f). In a parallel study, at 96 hours post-transduction, the mouse tumors and vital organs were homogenized and the modifications in the mouse VEGFR2 gene were analyzed by next-generation sequencing (NGS). We found that gene modification was only detectable in the tumor (Figure 5g). The indel rates (0 to 4.7%) varied among the samples collected from different parts of the tumor as a result of the heterogeneous distribution of the magnetic field and MNP-BV-CRISPR following direction injection. A representative mutation pattern detected by NGS of mouse VEGFR2 locus is summarized in Supplementary Figure S15. Local injection of MNP-BV coupled with a magnetic field, therefore, provides a means for efficient local gene delivery and tissue-specific genome editing, overcoming the challenge of non-specific dissemination.
In vivo activation of BV transduction and local genome editing via systemic delivery
It is well known that viral vectors delivered via systemic injection can reach vascular endothelium and in the case of fenestrated or leaky vessels, the stromal cells and parenchymal cells behind the vessel wall. Although BV administrated intravenously can circulate throughout the body, circulating BV can trigger the classical complement cascade that leads to BV inactivation30. As shown in our in vitro BV transduction study, a magnetic field can drive MNP-BV toward cell surface and enhance its cellular uptake with faster kinetics, which overcomes BV inactivation by the complement system. We expect that magnetic activation of BV can enable selective in vivo genome editing in the tissue exposed to the applied magnetic field.
We first tested the nanomagnet-based approach in vivo with the mouse liver which can be readily influenced by a block magnet. MNP-BV-LUC was administrated systemically through tail vein injection, and the mouse liver was positioned on top of a block magnet for 1 hour (Figure 6a). The transgene expression was evaluated by examining the luciferase activity with live animal imaging. Consistent with the results from our in vitro studies, the mice treated with MNP-BV-LUC and subjected to a magnetic field showed strong luminescence in the liver, whereas there was no luminescence in the mice treated with BV-LUC alone, or with MNP-BV-LUC but without applying a magnetic field (Figures 6b, c). Ex vivo examination confirmed that the high luciferase expression was only in the liver tissue exposed to the magnetic field; other vital organs including heart, lung, spleen, and kidney did not show luminescence signal (Figures 6d, e). The level of luciferase expression in the liver also increased with the strength of the magnetic field (Supplementary Figure S16). Importantly, the luciferase expression in mouse liver lasted less than 48 hours (Figures 6f, g), indicating that nanomegnet-induced transgene expression is transient. The MNP-BV-LUC did not induce significant acute liver damage (Supplementary Figures S17 and S18).
Figure 6. Magnetic field enables liver-specific transgene expression and genome editing in vivo via systemic injection.
(a) Schematic diagram of MNP-BV-mediated transgene delivery in liver. MNP-BV-LUC was administrated to the mouse through i.v. injection. A block magnet is pressed against the mouse liver to trigger local transgene expression. The contour plot indicated the magnetic force applied to individual MNPs at a distance of 1 mm from the top of the magnet. (b) and (c) Bioluminescence analysis of transgene expression. Nude mice were injected with PBS (WT), BV alone, MNP-BV, and MNP-BV followed with MF treatment for 1 hour. In the positive control, C3 knockout mice were injected with BV alone. In all groups, the dosage of the virus was 109 PFU virus per mouse. After 24 hours post-injection, the mice were imaged using an IVIS™ small animal live imaging system. (c) Plots the bioluminescence value in an ROI enclose the liver. Data represent mean ± SEM. ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test. Note that the magnetic field triggered high transgene expression in mice injected with MNP-BV, while without the magnetic treatment, the signal is negligible due to serum inactivation. (d) and (e) Biodistribution of transgene expression. In the (MNP-BV)+MF group, the organs were isolated 24 hours after injection, and bioluminescence of vital organs was measured ex vivo. As shown in the inset, the liver showed a high level of transgene expression, while bioluminescence was undetectable in the lung, kidney, spleen, and heart. All luminescence activity was normalized to the peak value in the plot. Data represent mean ± SEM. ***p < 0.001 based on one-way ANOVA and post-hoc Dunnett test. (f) and (g) In vivo transgene expression at 24 hours and 48 hours post-injection. Data represent mean ± SEM. *p = 0.018 based on one-tailed Student’s t-test. Additionally, MNP-BV-CRISPR was administrated to the mice under the magnetic field and VEGFR2 gene editing was analyzed. (h) Analysis of in vivo mouse VEGFR2 gene editing using T7E1 assay. At 24 hours post MNP-BV-CRISPR delivery, eGFP positive cells were selected from mouse liver tissue and T7E1 assays were performed to quantify the gene modification rate. (i) Analysis of genome editing in vital organs. n = 4 per group. Multiple samples were collected in the mouse liver at the regions near the block magnet, and gene editing was quantified using NGS. Note that the samples without detectable editing rate, i.e. < 0.1%, are plotted as having 0.05% editing rate.
The MNP-BV transduction activated by a magnetic field was further examined in the spleen, where a small block magnet was placed next to the spleen following the tail vein injection of MNP-BV (Supplementary Figure S19a). At 24 hours post-transduction, ex vivo imaging of dissected organs showed that the luciferase activity in the spleen was significantly higher than that in the heart, lung, and kidney (Supplementary Figures S19b, c), suggesting that the nanomagnets induced transgene expression in vivo can be switched on locally with a good tissue specificity. Since the liver was exposed to the applied magnetic field as well, luciferase activity also increased in the liver.
For in vivo genome editing, the mice were injected with MNP-BV-CRISPR and subjected to a magnetic field targeting mouse liver similar to that shown in Figure 6a. Following the workflow illustrated in Figure 5e, at 24 hours post MNP-BV-CRISPR delivery, the eGFP positive cells were harvested from mouse liver tissue and T7E1 assays were performed to quantify the gene modification rate. We found that the nanomagnets induced site-specific gene modification in the transduced mouse liver cells with a ~50% indel rate (Figure 6h). A representative pattern of the indels at the VEGFR2 target locus is shown in Supplementary Figure S20. Our NGS analysis suggests that ~96% of mutations (3N+1, 3N+2) may lead to a frameshift. In a parallel experiment, mouse organs including liver, heart, lung, spleen and kidney were harvested 4 days post MNP-BV-CRISPR delivery, and the genomic DNA was extracted for NGS analysis. Site-specific gene modification was detected in the liver but not in the off-target organs (Figure 6i). As expected, without selecting BV-transduced cells that are eGFP positive, the gene editing rates in the liver tissue samples collected somewhat randomly was much lower than that with selection, since the magnetic-field activated BV transduction is localized, and the liver samples for NGS were chosen by guessing where the magnetic field was effectively applied. Nevertheless, the NGS results shown in Figure 6i confirmed that there was efficient magnetic-field-driven genome editing with high tissue-specificity.
To determine if spatial control of genome editing can be realized in another tissue, we also performed genome editing in the tumor-carrying mice following systemic injection of MNP-BV-CRISPR. The tumor was placed between two block magnets for 1 hour following injection, and the analysis of gene editing in the tumor was performed in eGFP positive cells selected with the same workflow illustrated in Figure 5e. Gene modifications at the VEGFR2 locus in the transduced cells in the tumor were quantified by both the T7E1 assay and NGS analysis. Indels due to specific gene editing were detected in the tumor albeit at a lower frequency compared to those induced by intratumoral injection (Supplementary Figure S21). The decrease in the gene editing efficiency is likely due to the limited amount of MNP-BV penetrating into the tumor stroma47.
Taken together, our results provide strong evidence of efficient magnetic-field-driven BV transduction and transgene expression in different organs, and of spatially controlled genome editing in different tissues such as the liver and the tumor. Specifically, our in vivo studies demonstrate that the nuclease activity of the CRISPR–Cas9 system packaged in MNP-BV can be induced in the target tissues/organ on-demand by the applied magnetic field, and both direct injection and systemic injection of MNP-BV enable site-specific genome editing. The MNP-BV based in vivo delivery system takes advantage of the ability of nanomagnets to overcome BV inactivation locally, thus enabling spatial control of in vivo gene delivery and genome editing. Our studies also provide insight into the mechanism of binding between BV and TAT-modified magnetic nanoparticles, and how the MNP-BV system prevents complement inactivation of BV.
Effective transduction of the MNP-BV complex in vivo is ‘triggered’ by the applied magnetic field, which serves as an activator of transgene expression and genome editing. BV activation and gene-editing efficiency depend on the strength and spatial distribution of the magnetic field, the distribution of MNP-BV in the tissue, and the accessibility of the target gene in somatic cells. In this proof-of-concept study, although the block magnets used for both in vitro and in vivo experiments could generate a strong magnetic field (1.48 Tesla), the resulting magnetic force applied to the MNPs attached to BV varies with the location (distance and angle relative to the block magnet) of the MNP-BV complex. Due to the size of the magnet used (see Supplementary Figure S19) and the challenge in positioning the block magnet very close to the mouse spleen, under systemic delivery, it was difficult to have BV transduction only in the spleen, not in the liver (which is next to the spleen and has a much larger size). Further, when delivered systemically, it is likely that MNP-BVs could only be transduced in the vascular endothelium in the tumor and the few layers of stroma cells behind the leak vessel, resulting in a much reduced gene editing rate compared with that of local injection. With a large magnetic device consisting of a set of coils optimally positioned, a strong magnetic field gradient could be applied anywhere in the body, thus overcoming these limitations. Efficient gene editing in specific cells in the target tissue can be achieved by optimizing the route of administration. Owing to the large DNA loading capacity of BV, the MNP-BV based delivery system also has the potential to facilitate multiplexed genome editing in vivo.
Methods
Production of baculoviral vector
Recombinant baculoviral vectors (BV), including BV-LUC, BV-eGFP, and BV-CRISPR, were constructed using the Bac-to-Bac baculovirus expression system (Thermo Fisher). Briefly, the expression cassette of CMV-Luciferase, EF1α-eGFP or EF1α-eGFP-U6-sgRNA-CBh-Cas9 was inserted into the pFastBac vector and transformed into DH10Bac competent cells. The recombinant bacmids containing virus genome and the expression cassette were extracted using the PureLink™ HiPure Plasmid Miniprep Kit (Thermo Fisher). The recombinant bacmids were transduced into Spodoptera frugiperda (Sf9) insect cells using Cellfectin II (Thermo Fisher). The insect cell culture medium containing the budded viruses was centrifuged and filtered to remove cell debris using the Bottle Top Filters (0.45 μm, Thermo Scientific). The collected recombinant BV was amplified in Sf9 for two more passages. The passage 3 was used for in vitro and in vivo studies. To concentrate and purify the viral particles, BV was centrifuged at 18,600 g (JLA-10.500 rotor, Beckman Coulter) for 150 min at 4°C. The virus pellets were dispersed with 1 mL of phosphate-buffered saline (PBS). Collected BV was centrifuged on a sucrose cushion (25% sucrose with 5 mM NaCl and 10 mM EDTA) at 80,000 g (55.2 Ti rotor, Beckman Coulter) for 75 min at 4°C. The virus pellets were dispersed with 0.5 mL of PBS and stored at 4°C. The titer of BV stocks was determined using both the plaque assay and the qPCR titration assay (Supplementary Figure S1)
Synthesis of magnetic iron oxide nanoparticles
Magnetic iron oxide nanoparticles (MNPs) were synthesized according to previously published protocols 37,38. In brief, magnetite nanocrystals were synthesized through thermodecomposition of Fe(acac)3 (Sigma) in benzyl ether using oleic acid (Sigma) and oleylamine (Sigma) as the capping molecules 37. As-synthesized nanocrystals were coated with DSPE-mPEG2000 (Avanti lipids) and DSPE-PEG-maleimide (Avanti lipids) at a molar ratio of 9:1 using a dual solvent exchange method 38. To conjugate peptides to the surface of MNPs, freshly coated MNPs were mixed with cys-TAT peptides (CGYGRKKRRQRRR, Genscript) or (Arg)12-cys (RRRRRRRRRRRRC, Genscript) at a molar ratio of 1:400 in PBS and incubated overnight. Unconjugated peptides were removed by washing the nanoparticles with deionized water in centrifugal filter tubes (cutoff MW = 100k Da). The physical properties of the MNPs were characterized using transmitted electron microscopy (TEM), dynamic light scattering (DLS) (Mobius, Wyatt) and SQUID (MPMS, Quantum Design).
In vitro magnetic pull-down study
BV-LUC (2×108 PFU) or AdV-LUC (2×108 PFU, Vector Biolabs) were mixed with 20 µg Fe of unconjugated MNPs (MNP-PEG), MNPs conjugated with TAT (MNP-TAT) or MNPs conjugated with (Arg)12 (MNP-polyArg) in PBS for 20 minutes. In the heparin group, MNP-TAT was first mixed with heparin (10 units) for 20 minutes before mixing with BV. Then the mixtures were added to 500 µL culture medium in 8-well chamber slides. In the serum group, the medium contained 50% adult mouse serum (AMS). The chamber slides were incubated on a magnetic plate for 10 minutes with gentle shaking. In the end, 50 µL supernatant was collected and copies of free viruses were quantified by qPCR (BacPAK qPCR Titration Kit, Clontech).
Cell culture
Mouse hepatoma cells (Hepa 1–6), human cervical adenocarcinoma cells (HeLa), and human glioblastoma cells (U87) were purchased from ATCC. Human umbilical vein endothelial cells (HUVEC) were purchased from Lonza. All cells were cultured according to the standard protocols from the distributors without further authentication. The cells were tested for mycoplasma routinely.
In vitro BV transduction
In vitro BV transduction was performed with either BV-eGFP, BV-LUC or BV-CRISPR. eGFP fluorescence was used to evaluate transduction efficiency or spatial distribution of BV transduction. Luciferase activity was used to examine the level of transgene expression. In a typical in vitro BV transduction experiment, the cells were seeded in a chamber slide. Before BV transduction, 2×106 PFU BV was mixed with 4 µg Fe of MNPs for 20 minutes. Then the cells were incubated with the mixture for 30 minutes with or without the magnet. In each group, the cells were transduced with BV at an MOI of 100 PFU per cell unless otherwise specified. After transduction, the cells were incubated with fresh medium. At 24 hours post-transduction, the cells transduced by BV-eGFP were examined for eGFP fluorescence by flowcytometry or fluorescence microscopy. For the cells transduced by BV-LUC, luciferase activity was measured using an in vitro luciferase kit (ONE-Glo™ Luciferase Assay System, Promega) in a microplate reader. The cells transduced with BV-CRISPR were cultured for 72 hours and the genomic DNA was extracted and analyzed with a T7E1 assay for gene editing efficiency as described below.
Cytotoxicity study
Hepa 1–6 cells were cultured in 96-well plates and incubated with BV at designated MOIs with or without MNPs for 12 hours. After treatment, the cells were incubated in fresh medium for 3 days and cell viability was evaluated by the MTT assay.
Immunofluorescence staining
The cells were seeded in chamber slides and incubated with BV or MNP-BV under designated conditions. After treatment, the cells were fixed in 4% paraformaldehyde for 20 minutes, permeabilized with PBS containing 0.1% Triton for 3 minutes and blocked with 5% BSA for 1 hour at room temperature. BV was detected by incubating the cells with an antibody against vp39 (kindly provided by Prof. Loy Volkman and Dr. Taro Ohkawa) overnight at 4°C followed by an Alexa Fluor 647 goat anti-mouse IgG antibody (Abcam) 48. After that, the cells were stained with Alexa Fluor 488 phalloidin (Thermo Fisher) and Hoechst 33342 (Thermo Fisher). The images were acquired with a confocal microscope (Nikon A1 Confocal Microscope).
Genomic DNA extraction, PCR amplification, and T7E1 assay
Genomic DNA was extracted from the cell or tissue samples using the Quick-DNA Miniprep Plus Kit (Zymo Research). The amplicon containing the CRISPR cutting site was amplified by PCR using Q5 High-Fidelity 2X Master Mix, forward primer: CCCCCATTCGCTAGTGTGTA, and reverse primer: AGCACGGAGTGATTGATGCC with the following PCR condition: 98°C for 30 s, 34 × (98°C for 10 s, 63°C for 30 s, 72°C for 30 s), 72°C for 2 min, hold at 4°C. The PCR products were analyzed by gel electrophoresis in 1% agarose and purified with MinElute PCR Purification Kit (Qiagen). Mismatched duplex DNA was obtained by denaturation and renaturation of 1 μg of purified PCR product. The renatured DNA was digested with T7 endonuclease I (New England BioLabs) at 37°C for 1 hour and analyzed by gel electrophoresis in 2% agarose. The rate of gene modification was calculated using the following equation.
Next Generation Sequencing (NGS) for indels in sorted cells and tissue homogenate
The genomic region containing the cutting site in mouse VEGFR2 gene was amplified by PCR using Q5 High-Fidelity 2X Master Mix, forward primer: TCTACAGTCCGACGATCATGAAAGAACACCCAAGGGAGG, and reverse primer: GACGTGTGCTCTTCCGATCGGGACGGAGAAGGAGTCTGT with the following PCR condition: 98°C for 30 s, 34 × (98°C for 10 s, 63°C for 30 s, 72°C for 10 s), 72°C for 2 min, hold at 4°C. The second round of PCR was used to attach the Illumina P5 adapters and sample-specific barcodes to the target amplicons. PCR products were pooled and purified through gel extraction (QIAquick Gel Extraction Kit). The concentrations of purified DNA samples were determined using Qubit Fluorometer (Thermo Fisher). Sequencing libraries were sequenced with the Illumina MiSeq Personal Sequencer. The indels were analyzed using the custom script available at https://github.com/piyuranjan/NucleaseIndelActivityScript.
Magnetically activated BV transduction in vivo
All animal studies were approved by the Institutional Animal Care and Use Committee at Rice University. Athymic nude mice (4–5 weeks old female) were purchased from Charles River. C3 knockout mice were purchased from the Jackson Laboratory. The mice were randomly allocated to the experimental groups without blinding. In the mouse tumor model, 3 × 106 Hepa 1–6 cells were dispersed in 100 μL of PBS containing 5 mg/mL matrigel and injected into the right flank of the mice subcutaneously. The mice were used in the experiments when the tumor size reached 6–8 mm in diameter.
To investigate MNP-BV-induced transduction via local injection, BV-LUC (2×107 PFU) or MNP-BV-LUC (2×107 PFU BV mixed with 20 µg of Fe MNP) dispersed in 40 µL sterile PBS was injected intratumorally. The tumor was placed between 2 parallel N52 grade NdFeB block magnet (L×W×H = 1″×1/2″×1/2″) (K&J Magnetics) for one hour under anesthesia. At 24 hours post-injection, each mouse was injected with in vivo luciferase substrate (Promega) i.p. and imaged using an IVIS Kinetic III live imaging system (Perkin Elmer). After that, the mice were immediately euthanized and the tumor and vital organs were dissected for ex vivo imaging.
To investigate MNP-BV induced transduction via systemic injection, the mice were injected with BV-LUC (109 PFU) or MNP-BV-LUC (109 PFU BV mixed with 0.1 mg of Fe MNP) dispersed in 200 µL sterile PBS through the tail vein. To activate BV transduction in the liver, the mice were placed on an N52 grade NdFeB block magnet (L×W×H = 1″×1/2″×1/2″) for one hour under anesthesia (Figure 6a). At 24 hours post-transduction, each mouse was injected with in vivo luciferase substrate (Promega) i.p. and the luciferase activity was examined using the IVIS imaging system. After that, the mice were euthanized immediately and vital organs were dissected and imaged ex vivo.
Magnetically activated in vivo genome editing
Tumor-bearing mice were used to test genome editing in tumor via direct injection. The mice were injected with MNP-BV-CRISPR as described above. One group of mice was euthanized on day 1 post-injection. Tumors were harvested, minced under sterile conditions, and digested in Hank’s Balanced Salt Solution (HBSS) containing 300 U/mL collagenase (Stem Cell Technology), 100 U/mL hyaluronidase (Stem Cell Technology), and 1 mg/mL DNase I (Stem Cell Technology), for 1 hour at 37°C with periodic shaking. Undigested tissue fragments were further treated with 0.25% Trypsin-EDTA (Gibco) for 5 to 10 min. The isolated cells were collected through a cell sieve (100 μm, Greiner Bio-One). The red blood cells were removed with the red blood cell lysing buffer (Sigma-Aldrich). The remaining cells were sorted for GFP using BD FACSMelody Cell Sorter. The cutting efficiency of the target gene was examined in sorted cells with the T7E1 assay. The induced indel patterns were evaluated by next-generation sequencing (NGS). Another group of mice was euthanized on day 4 and the gene editing frequency in the tumor and the vital organs were evaluated by NGS in the gene segments amplified directly from tissue homogenate.
To test genome editing via systemic injection, the mice were injected with MNP-BV-CRISPR and subjected to the magnetic treatment. Editing of the target gene was examined as described above. In the group for the T7E1 assay, the liver was harvested and the liver cells were isolated using a Liver Dissociation Kit (Miltenyi Biotec).
In vivo toxicity of MNP-BV
To examine the in vivo toxicity of MNP-BV, vital organs and blood were harvested from treated mice after 10 days post-injection. The organs were fixed in 10% formalin solution overnight and embedded in paraffin. Histology evaluation was performed in tissue sections stained with hematoxylin and eosin. Alanine transaminase (ALT) and aspartate aminotransferase (AST) levels in the blood were measured using the ALT ELISA Kit (Biocompare) and AST Colorimetric Kit (Biovision) respectively, according to the manufacturers’ instructions.
Statistics
GraphPad Prism (GraphPad Software) was used for all the calculations. Data were analyzed using one-tailed Student’s t-tests, Mann-Whitney test and one-way ANOVA and post-hoc Dunnett test. The difference with p < 0.05 was considered statistically significant (* denotes p < 0.05; ** denotes p < 0.01; *** denotes p < 0.001; and n.s. denotes p > 0.05).
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information. The raw datasets are available from the corresponding author upon reasonable request.
The custom script used to analyze the indels in the NGS data is available at https://github.com/piyuranjan/NucleaseIndelActivityScript.
Supplementary Material
Acknowledgment
We thank Professor Loy Volkman and Dr. Taro Ohkawa for kindly providing the anti-vp39 antibody, and Tim Davis, Lin Hong, and Anirban Ray for their assistance. This work was supported by the National Institutes of Health through a Nanomedicine Development Center Award (PN2EY018244 to G.B.) and the Cancer Prevention and Research Institute of Texas (RR140081 and RR170721 to G.B.).
Footnotes
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information are available online at www.nature.com/reprints.
Competing interests
H. Z., S. T., and G. B. filed a US patent application based on the results presented in this paper.
References
- 1.Sander JD & Joung JK CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 32, 347–355, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 339, 819–823, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 32, 551–553, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swiech L et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 33, 102–106, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox DB, Platt RJ & Zhang F Therapeutic genome editing: prospects and challenges. Nat Med. 21, 121–131, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao HK et al. Use of the CRISPR–Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 6, 6413, (2015). [DOI] [PubMed] [Google Scholar]
- 7.Dever DP et al. CRISPR–Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 539, 384–389, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson CE et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 351, 403–407, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabebordbar M et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 351, 407–411, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YN et al. CRISPR–Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42, 7473–7485, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cradick TJ, Fine EJ, Antico CJ & Bao G CRISPR–Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 41, 9584–9592, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 31, 822–826, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 31, 827–832, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CM, Cradick TJ, Fine EJ & Bao G Nuclease Target Site Selection for Maximizing On-target Activity and Minimizing Off-target Effects in Genome Editing. Mol Ther. 24, 475–487, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dow LE et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 33, 390–394, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nihongaki Y, Kawano F, Nakajima T & Sato M Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 33, 755–760, (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zincarelli C, Soltys S, Rengo G & Rabinowitz JE Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol Ther. 16, 1073–1080, (2008). [DOI] [PubMed] [Google Scholar]
- 18.Yin H et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 34, 328–333, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y et al. Systemic dissemination of viral vectors during intratumoral injection. Mol Cancer Ther. 2, 1233–1242, (2003). [PubMed] [Google Scholar]
- 20.Stanley SA, Sauer J, Kane RS, Dordick JS & Friedman JM Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat Med. 21, 92–98, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannix RJ et al. Nanomagnetic actuation of receptor-mediated signal transduction. Nat Nanotechnol. 3, 36–40, (2008). [DOI] [PubMed] [Google Scholar]
- 22.Wheeler MA et al. Genetically targeted magnetic control of the nervous system. Nat Neurosci. 19, 756–761, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu Y et al. Magnetic forces enable controlled drug delivery by disrupting endothelial cell-cell junctions. Nat Commun. 8, 15594, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sammet S Magnetic resonance safety. Abdom Radiol. 41, 444–451, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Airenne KJ et al. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol Ther. 21, 739–749, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansouri M et al. Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat Commun. 7, 11529, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CY, Lin CY, Chen GY & Hu YC Baculovirus as a gene delivery vector: recent understandings of molecular alterations in transduced cells and latest applications. Biotechnol Adv. 29, 618–631, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kost TA, Condreay JP & Jarvis DL Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 23, 567–575, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindriksen S et al. Baculoviral delivery of CRISPR–Cas9 facilitates efficient genome editing in human cells. PLoS One. 12, e0179514, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann C & Strauss M Baculovirus-mediated gene transfer in the presence of human serum or blood facilitated by inhibition of the complement system. Gene Ther. 5, 531–536, (1998). [DOI] [PubMed] [Google Scholar]
- 31.Strauss R et al. Baculovirus-based vaccination vectors allow for efficient induction of immune responses against plasmodium falciparum circumsporozoite protein. Mol Ther. 15, 193–202, (2007). [DOI] [PubMed] [Google Scholar]
- 32.Swift SL et al. Evaluating baculovirus as a vector for human prostate cancer gene therapy. PLoS One. 8, e65557, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C et al. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol Ther. 17, 2058–2066, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haeseleer F, Imanishi Y, Saperstein DA & Palczewski K Gene transfer mediated by recombinant baculovirus into mouse eye. Invest Ophthalmol Vis Sci. 42, 3294–3300, (2001). [PMC free article] [PubMed] [Google Scholar]
- 35.Kaikkonen MU, Maatta AI, Yla-Herttuala S & Airenne KJ Screening of complement inhibitors: shielded baculoviruses increase the safety and efficacy of gene delivery. Mol Ther. 18, 987–992, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raty JK et al. Enhanced gene delivery by avidin-displaying baculovirus. Mol Ther. 9, 282–291, (2004). [DOI] [PubMed] [Google Scholar]
- 37.Sun S et al. Monodisperse MFe2O4 (M = Fe, Co, Mn) nanoparticles. J Am Chem Soc. 126, 273–279, (2004). [DOI] [PubMed] [Google Scholar]
- 38.Tong S, Hou S, Ren B, Zheng Z & Bao G Self-assembly of phospholipid-PEG coating on nanoparticles through dual solvent exchange. Nano Lett. 11, 3720–3726, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torchilin VP Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Adv Drug Deliv Rev. 60, 548–558, (2008). [DOI] [PubMed] [Google Scholar]
- 40.Small DA & Moore NF Measurement of surface charge of baculovirus polyhedra. Appl Environ Microbiol. 53, 598–602, (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyce FM & Bucher NLR Baculovirus-mediated gene transfer into mammalian cells. P Natl Acad Sci USA. 93, 2348–2352, (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohkawa T, Volkman LE & Welch MD Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol. 190, 187–195, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matilainen H et al. Baculovirus entry into human hepatoma cells. J Virol. 79, 15452–15459, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kataoka C et al. Baculovirus GP64-mediated entry into mammalian cells. J Virol. 86, 2610–2620, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romet-Lemonne G & Jegou A Mechanotransduction down to individual actin filaments. Eur J Cell Biol. 92, 333–338, (2013). [DOI] [PubMed] [Google Scholar]
- 46.Shen H, Tong S, Bao G & Wang B Structural responses of cells to intracellular magnetic force induced by superparamagnetic iron oxide nanoparticles. Phys Chem Chem Phys. 16, 1914–1920, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan F et al. Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size. Cancer Res. 55, 3752–3756, (1995). [PubMed] [Google Scholar]
- 48.Danquah JO, Botchway S, Jeshtadi A & King LA Direct interaction of baculovirus capsid proteins VP39 and EXON0 with kinesin-1 in insect cells determined by fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy. J Virol. 86, 844–853, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information. The raw datasets are available from the corresponding author upon reasonable request.
The custom script used to analyze the indels in the NGS data is available at https://github.com/piyuranjan/NucleaseIndelActivityScript.