Abstract
Autophagy is an evolutionarily conserved lysosome/vacuole-dependent catabolic pathway in eukaryotes. Autophagy functions basally for cellular quality control and is induced to act as an alternative source of basic metabolites during nutrient-deprivation. These functions of autophagy are intimately connected to the regulation of metabolism, and the metabolic status of the cell in turn controls the nature and extent of autophagic induction. Here we highlight the co-regulation of autophagy and metabolism with a special focus on selective autophagy that, along with bulk autophagy, plays a central role in regulating and rewiring metabolic circuits. We outline the metabolic signals that activate these pathways, the mechanisms involved, and the downstream effects and implications while recognizing yet unanswered questions. We also discuss the role of autophagy in the development and maintenance of adipose tissue, an emerging player in systemic metabolic homeostasis, and describe what is currently known about the complex relationship between autophagy and cancer.
eTOC blurb
The integral regulatory interplay between autophagy and the cellular metabolic landscape is vital during development, essential for physiology, and crucial for preventing the occurrence of disease states. Klionsky and colleagues highlight the critical links between autophagy, especially selective autophagy, and metabolic homeostasis, and explores the mechanisms that govern these processes.
Introduction
Autophagy is a highly conserved eukaryotic pathway for maintaining cellular homeostasis through the degradation of superfluous and/or damaged intracellular materials. Autophagy can be either selective or non-selective. Non-selective autophagy describes the random engulfment and subsequent degradation of cytoplasmic material such as proteins and/or organelles (Dikic and Elazar, 2018). This process occurs continuously at a low, basal level facilitating the turnover and recycling of cytoplasmic contents but is also upregulated under conditions of nutrient deprivation. During starvation, degradation by non-selective autophagy provides simple macromolecules that can be utilized for essential anabolic synthesis. In addition, several forms of selective macroautophagy are now recognized, revealing a dynamic role of autophagy in cellular metabolism (Mizushima and Komatsu, 2011). Discriminant selection of autophagic cargo allows for the removal of dysfunctional/superfluous organelles as well as the generation of specific nutrients in response to environmental changes, thereby promoting cell survival and organismal health. Among other roles, selective autophagy allows the cell to adapt to lipid imbalance, glucose scarcity, amino acid deprivation, and iron shortage, and also facilitates cellular remodeling to accommodate major shifts in metabolism (Gatica et al., 2018).
Due to its diverse roles in maintaining metabolic homeostasis, autophagy plays a major role in general metabolic health and organismal development; autophagic imbalance has been linked to several mammalian pathologies including diabetes (Marasco and Linnemann, 2018), neurodegeneration (Frake et al., 2015) and cancer (Galluzzi et al., 2015). Autophagy-deficient mouse embryos die within a day of birth (Kuma et al., 2004), and adult mice induced to be autophagy-deficient die within 24 h of starvation due to hypoglycemia. Even when grown with sufficient food, these autophagy-deficient adults die in less than three months due to increased susceptibility to infection and neurodegeneration (Karsli-Uzunbas et al., 2014). Autophagy in mouse hypothalamic neurons regulates food intake and organismal energetics (Kaushik et al., 2011). Autophagy may also regulate circadian metabolic cycles by degrading core circadian proteins such as CRY1 (Toledo et al., 2018). A recent study in mice revealed the potential benefits of basal autophagy upregulation. Upregulated autophagy increases median lifespan by 12% and decreases susceptibility to age-related diseases such as cancer (Fernandez et al., 2018). These studies highlight a central role for autophagy in metabolic maintenance. This review will describe the various autophagic mechanisms that cells employ to combat metabolic perturbations and will touch on how these responses are important for systemic metabolism in health and disease.
Overview of autophagic mechanisms
The general mechanism of autophagy can be summarized as cargo deposition in the lysosome/vacuole, followed by cargo degradation by hydrolytic enzymes, and efflux of the resulting breakdown products into the cytosol. However, autophagy may be classified based on the mechanism of cargo entry:
Macroautophagy
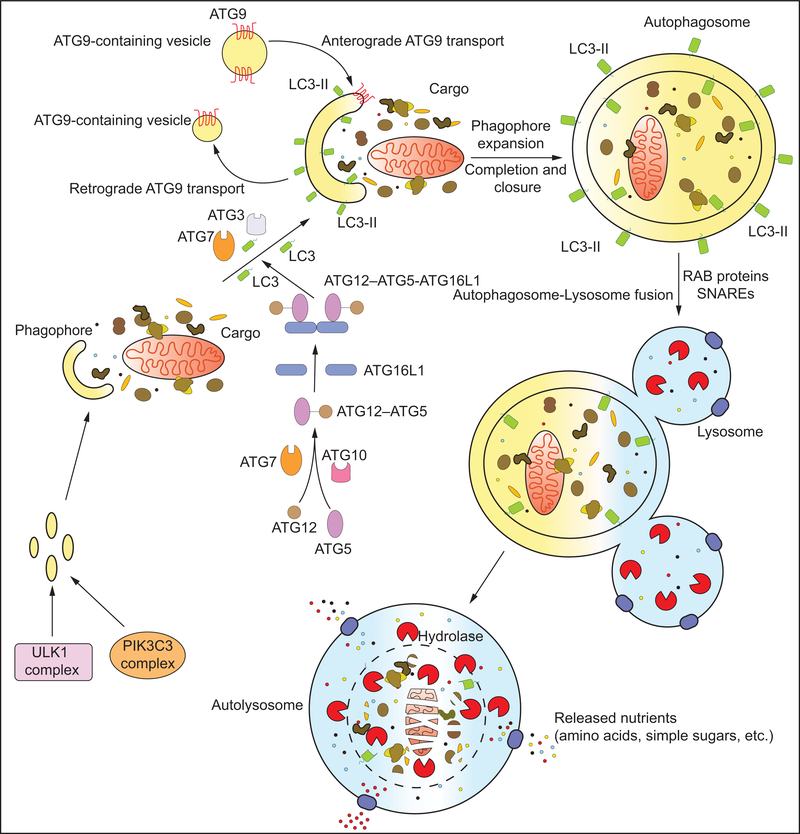
Macroautophagy (hereafter referred to as autophagy) begins with the initiation of the double-membrane phagophore by the ULK complex. The ULK complex is comprised of ULK1 (Atg1 in yeast) or ULK2, and several interacting proteins: ATG13, RB1CC1 and ATG101. ULK1 phosphorylates several components of the PIK3C3/VPS34 kinase complex that contains, in addition to PIK3C3/VPS34, PIK3R4/VPS15, BECN1, NRBF2 and other regulatory proteins such as ATG14, AMBRA1, SH3GLB1, RUBCN or UVRAG (Kihara et al., 2001; Itakura et al., 2008; Youle and van der Bliek, 2012). Phosphorylation of ATG14, BECN1, and/or AMBRA1 by ULK1 promotes PIK3C3/VPS34 activation and, in some cases, recruitment to the endoplasmic reticulum (ER) (Di Bartolomeo et al., 2010; Russell et al., 2013; Park et al., 2016a; Park et al., 2018). In mammals, activated PIK3C3/VPS34 produces local pools of phosphatidylinositol-3-phosphate (PtdIns3P) that define the region of phagophore initiation. One model suggests that PtdIns3P at the ER promotes the formation of omegasomes that act as sites of phagophore initiation (Ktistakis and Tooze, 2016). Subsequently, and possibly after detachment of the omegasome from the ER, membrane recruited from diverse sources including the ER, Golgi apparatus, plasma membrane and recycling endosomes (Axe et al., 2008; Knaevelsrud et al., 2013) feeds the expanding phagophore by a still poorly understood mechanism involving ATG9-containing vesicles. In contrast, a recent model proposes that phagophores evolve from RAB11A-enriched recycling endosomes. According to this model, RAB11A, along with PtdIns3P, plays a determining role in the recruitment of the early autophagy machinery. This suggests that recycling endosomes are primary platforms from which phagophores originate while the ER may contribute secondarily (Puri et al., 2018). A contribution of RAB11A-containing recycling endosomes to autophagosome formation in response to viral infection has also been suggested (Kuroki et al., 2018). In yeast, phagophore initiation occurs at the cytoplasmic phagophore assembly site (PAS). Here it is thought that tethering of Atg9-containing vesicles by the Atg1 kinase complex drives phagophore formation (Orsi et al., 2012).
Two essential ubiquitin-like conjugation systems drive autophagy. This machinery functions to covalently conjugate Atg8 (in yeast) and Atg8-family proteins (in mammals) to the phagophore membrane. The ubiquitin-like ATG12 protein is conjugated to ATG5 via the E1-like enzyme ATG7 and the E2-like enzyme ATG10. After processing by the protease, ATG4, Atg8-family proteins undergo conjugation to phosphatidylethanolamine (PE). This process, known as Atg8 lipidation, is mediated by the E1-like ATG7 and E2-like ATG3 enzymes, and the role of an E3-like ligase is filled by a complex between ATG12–ATG5 and ATG16L1 (Feng et al., 2014) (Figure 1). Mammalian Atg8-family proteins are split into two subfamilies: the MAP1LC3/LC3 family and the GABARAP family (Yu et al., 2018). Lipidation of Atg8/Atg8-family proteins allow for attachment to the phagophore where they recruit proteins containing an LC3-interacting region (LIR). Some LIR-containing proteins facilitate phagophore expansion and closure while others act as receptors, conveying cargo specificity to the growing phagophore. Several autophagy proteins are recruited to the expanding phagophore through PtdIns3P-interacting motifs such as FYVE and PX domains. One such protein is WIPI2 that binds ATG16L1, recruiting the ATG12–ATG5-ATG16L1 complex to the phagophore (Dooley et al., 2014). Phagophore expansion concludes in closure around the cargo at which point the vesicle is called an autophagosome. The autophagy protein machinery bound to the exterior membrane of the autophagosome dissociates prior to fusion with the lysosome. While the outer membrane of the autophagosome fuses with the lysosomal/vacuolar membrane, the inner membrane and enclosed contents are degraded by resident hydrolases to generate simple biomolecules such as glucose and amino acids that are released into the cytosol via lysosomal/vacuolar membrane transporters (Figure 1).
Figure 1: The molecular machinery of macroautophagy:
The hallmark of macroautophagy is the double membrane autophagosome that forms by the de novo assembly of membrane from various sources. The process begins with the formation of the phagophore, a process initiated by the ULK1 and VPS34 complexes. Expansion of the phagophore occurs via the continued recruitment of membrane vesicles by ATG9 as well as the conjugation of LC3 to the phagophore membrane (to form LC3-II). LC3 conjugation involves a two-step ubiquitin-like conjugation pathway involving ATG7, ATG10, ATG3, ATG12, ATG5 and ATG16 (refer to text for details). The phagophore expands around the cargo, finally closing to form a cargo-containing autophagosome. The autophagosome subsequently fuses with lysosome(s) by the concerted action of Rab and SNARE proteins to form the autolysosome. Lysosomal hydrolases degrade the inner autophagosomal membrane and the enclosed cargo. The breakdown products, simple macromolecules such as amino acids, are subsequently transported out in the cytoplasm by lysosomal transporters for reuse.
Several proteins involved in autophagy possess non-autophagy related functions as well (Cadwell and Debnath, 2018; Subramani and Malhotra, 2013). This includes the Atg8-family protein LC3, which modulates immune responses and inflammation by a process known as LC3-associated phagocytosis (Cunha et al., 2018; Martinez et al., 2015). ATG5, another essential autophagy protein promotes exosome formation by regulating the acidification of multivesicular bodies (MVBs) by uncoupling the V1V0-ATPase (Guo et al., 2017). However, direct metabolic roles for these functions have not been established yet and they will not be discussed further in this text.
Chaperone-Mediated Autophagy
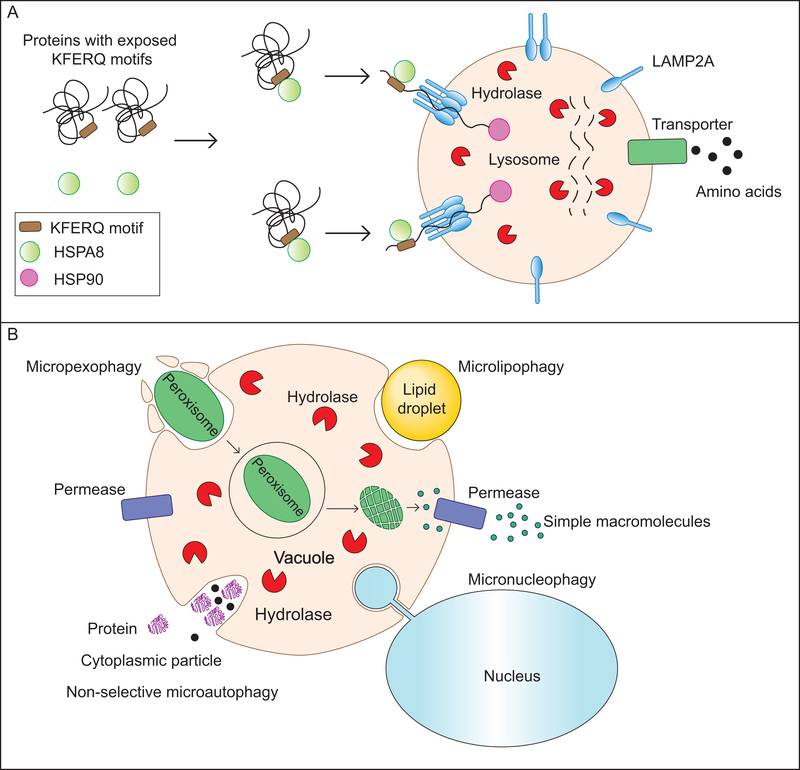
Chaperone-mediated autophagy (CMA) describes the HSPA8/Hsc70-dependent selective degradation of substrate proteins with an exposed KFERQ-like motif. Post-translational modifications (PTMs) allow for a great deal of diversification and regulation of the KFERQ-binding motif. For instance, a phosphorylated serine, threonine, or tyrosine can serve the role of a negatively charged amino acid in the binding motif (Kaushik and Cuervo, 2016). Similarly, acetylated lysine was recently shown to complete the CMA binding motif by acting as a pseudo-glutamine (Bonhoure et al., 2017). Additionally, PTMs at sites beyond the KFERQ motif can regulate recognition by determining whether the KFERQ motif is exposed. The dependence of some CMA motifs on PTM for completion allows the subset of CMA substrate proteins to change drastically in response to cellular conditions. Binding of the HSPA8 chaperone and associated co-chaperones to a substrate protein is followed by its lysosome targeting. Here, docking of the HSPA8-substrate complex to the lysosomal membrane is mediated by interaction with the cytoplasmic tail of the lysosomal transmembrane protein LAMP2A. At the time of HSPA8-substrate binding, LAMP2A is either monomeric or homodimeric, but soon after binding, HSPA8 dissociates and LAMP2A multimerizes to form a mature translocation complex. HSP90 (heat shock protein 90) stabilizes LAMP2A from within the lysosomal lumen, and interactions between GFAP (glial fibrillary acidic protein) and EEF1A/EF1α (eukaryotic translation elongation factor 1 alpha 1) regulate the rate of translocation complex assembly and disassembly (Bandyopadhyay et al., 2008; Bandyopadhyay et al., 2010). The substrate protein is unfolded and translocated into the lysosomal lumen via the multimeric LAMP2A complex where it is rapidly degraded by proteases (Figure 2A).
Figure 2: Other mechanisms of self-eating: Chaperone-mediated autophagy (CMA) and microautophagy:
(A) In yeast, microautophagy involves the sequestration of cargo by the protrusion/invagination of the vacuolar membrane followed by an inward scission leading to the formation of a cargo-containing lumenal vesicle. This vesicle is subsequently degraded by vacuolar hydrolases releasing simple breakdown products. Microautophagy can be non-selective, degrading cytosolic components randomly, or selective, specifically degrading lipid droplets (microlipophagy) or peroxisomes (macropexophagy). Another selective microautophagic process, not discussed in the text, is the piecemeal microautophagy of the nucleus (PMN) which degrades portions of the nucleus. (B) CMA is a lysosome-dependent protein degradation pathway that requires the cytosolic chaperone HSC70. Proteins with an exposed KFERQ or KFERQ-like motif are recognized and bound by HSC70. The complex then locates to the lysosomal membrane where the multimerization of LAMP2A allows the formation of a conduit for the delivery of the protein into the lysosomal lumen, a process facilitated by the lumenal chaperone HSP90. Lysosomal hydrolases break down the protein releasing amino acids which are transported into the cytosol.
Approximately 40% of proteins in the mammalian proteome contain a canonical KFERQ-like motif and several more contain PTM-inducible motifs (Kaushik and Cuervo, 2018), indicating that CMA may be a major intracellular protein degradation pathway. Indeed, this possibility is validated by studies showing that the selective blockage of CMA leads to the upregulation of other degradative pathways such as macroautophagy as well as increased proteasomal activity (Massey et al., 2006; Schneider et al., 2015). Conversely, cells upregulate CMA when macroautophagy is selectively inhibited, highlighting the concept that there is cross-talk between these pathways (Kaushik et al., 2008).
Microautophagy
Microautophagy describes the process in which autophagic cargo in the cytoplasm enters the lysosome (or endosome)/vacuole following a protrusion or invagination and inward pinching of the lysosomal (or endosomal)/vacuolar membrane. This results in the formation of a lumenal vesicle surrounding the cargo that is degraded along with its contents (Li et al., 2012). Microautophagy can be non-selective, incorporating cytoplasm randomly, or highly specific as is the case in micropexophagy, the selective degradation of peroxisomes by their direct sequestration into the lysosome (Oku and Sakai, 2016) (Figure 2B).
Cellular sensors integrate autophagy with cellular metabolic status
In mammals, starvation for 24–48 h induces autophagy in nearly all nucleated cells. However, circulating amino acid and glucose levels are relatively stable during this period due to the activity of homeostatic circuits involving the breakdown of systemic reserves (Galluzzi et al., 2014). Consequently, intracellular nutrient availability for most cells is highly dependent on factors influencing nutrient uptake from the extracellular milieu. Cellular nutrient uptake is modulated by a plethora of cytokines and hormones, with INS (insulin) and IGF1 (insulin like growth factor 1) being critical reporters of the fed state. GCG (glucagon) and epinephrine play a major role in conveying a fasting status through the GCG and ADRB/β-adrenergic receptors, respectively. Upon activation, these two guanosine-protein-coupled receptors (GPCRs) stimulate cAMP production. cAMP modulates autophagy via PRKA/PKA (protein kinase cAMP-dependent), MTOR (mechanistic target of rapamycin kinase), and the MAPK (mitogen-activated protein kinase) signaling cascade, although the exact mechanisms are yet to be defined (Lizaso et al., 2013; Wauson et al., 2014; Franco et al., 2017). While cAMP generally promotes autophagy through PRKA, studies indicate that the RAPGEF3/EPAC1 branch of cAMP signaling may reduce autophagosome biogenesis and autophagic flux in neurons (Williams et al., 2008) or during invasion by certain pathogens (Mestre and Colombo, 2012). Understandably given its major role in eukaryotic cell signaling, multiple MAPK pathways are intertwined with autophagy including the MAPK/JNK (Haberzettl and Hill, 2013), MAPK/ERK (Martinez-Lopez and Singh, 2014) and MAPK/p38 (He et al., 2018) pathways.
At the cellular level, starvation decreases the abundance of key nutrients such as glucose and amino acids, which eventually induces a decrease in downstream metabolites including TCA cycle intermediates. Importantly, a reduced supply of glucose and amino acids lowers the “energy charge” of the cell—the relative abundance of ATP in comparison to ADP and AMP. AMP kinase (AMPK) plays a major role in upregulating autophagy primarily, although not exclusively, in response to reduced energy charge. Another key regulator of autophagy, MTOR complex 1 (MTORC1), is highly responsive to intracellular amino acid levels (Figure 3).
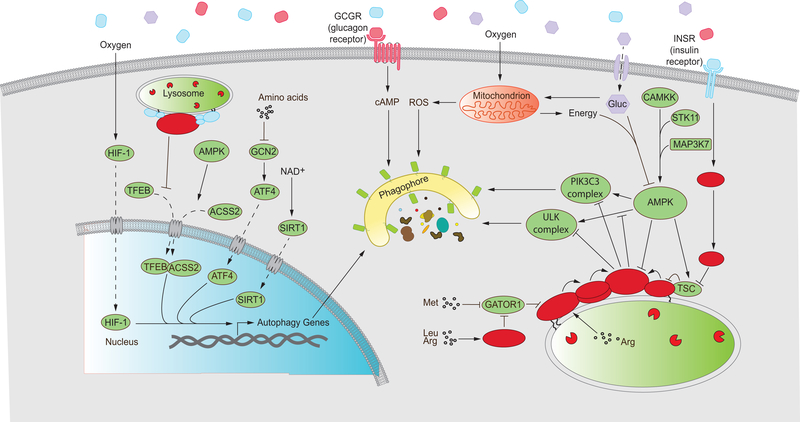
Figure 3: An intricate network of regulatory components and signaling pathways influence autophagy in response to cellular metabolic status.
Autophagy is regulated at multiple levels by cellular components that respond to specific or general metabolic cues. Several proteins such as ATF4, HIF-1, SIRT1 and TFEB modulate the expression of autophagy-related genes at the transcriptional level. These pathways are sensitive to the abundance of amino acids, oxygen availability, the reduction status of the cellular NAD pool and activation status of MTORC1 and AMPK. Expression of autophagy genes leads to autophagy induction, depicted as an expanding phagophore. Glucagon signals a fasted organismal status and upregulates autophagy through cAMP-dependent pathways. Glucose fuels oxidative phosphorylation in mitochondria, providing energy in the form of ATP but also generating ROS that indirectly upregulate autophagy. Low cellular energy charge activates AMPK in a process that requires upstream kinases such as CAMKK and STK11. AMPK promotes autophagy by activating the autophagy-initiating ULK1 and VPS34 complexes as well as inhibiting MTORC1 function. MTORC1 inhibits autophagy when recruited to the lysosome and activated. MTORC1 recruitment and activation occurs in response to the presence of both growth factors such as INS/insulin and an abundance of amino acids in the cytosol and lysosomal lumen. While INS signaling occurs through the PI3K-AKT-TSC axis, leading to the activation of the small GTPase RHEB, amino acid sufficiency is conveyed through the Ragulator complex that impinges on the small GTPases known as RRAGs. The RRAG complex represents a heterodimer between RRAGA (or B) and RRAGC (or D). Activated MTORC1 inhibits the ULK1 and VPS34 complexes to downregulate autophagy (Refer to text for details).
Solid arrows represent upregulation, blunt arrows represent repression and dashed arrows represent movement/transport. Gluc, glucose; Met, methionine; Arg, Arginine; Leu, leucine.
AMPK
AMPK is activated in response to energy charge and, to a lesser extent, nutrient status of the cell. Significant AMPK activation requires its phosphorylation by upstream kinases, principally STK11 (Woods et al., 2003). Binding of AMP stabilizes the phosphorylation status of AMPK and imparts an allosteric effect, both of which are required for full AMPK activation. ADP too can bind AMPK but only serves to preserve phosphorylation status (Xiao et al., 2007; Xiao et al., 2011; Chen et al., 2012). ATP can competitively bind AMPK, making this protein highly responsive to cellular energy availability. AMPK also responds to the glucose concentration independent of energy charge (Zhang et al., 2013; Zhang et al., 2014; Zhang et al., 2017). CAMKK (calcium/calmodulin dependent protein kinase kinase) activates AMPK by phosphorylating the same site as STK11; however, CAMKK activation is sensitive to intracellular Ca2+ concentration, thus coupling AMPK activation to extracellular signals that induce changes in cellular Ca2+ levels as well (Hoyer-Hansen et al., 2007). A novel and elegant mechanism of autophagy regulation involving AMPK has recently been proposed. During glucose starvation, AMPK phosphorylates ACSS2 (acetyl-CoA synthetase short chain family member 2) exposing a nuclear localization signal. Once imported to the nucleus, ACSS2 binds to TFEB and translocates to the promoter region of lysosome biosynthesis and autophagy genes. Here, ACSS2 locally generates acetyl-CoA that is used for histone H3 acetylation, enhancing gene expression and promoting autophagy (Li et al., 2017) (Figure 3).
A large role of AMPK in activating autophagy is the inhibition of MTORC1 through direct phosphorylation of its RPTOR subunit as well as via the activating phosphorylation of the MTORC1 inhibitor TSC2 (TSC complex subunit 2) (Inoki et al., 2006; Gwinn et al., 2008). Inhibition of MTORC1 simultaneously inhibits cellular anabolism and strongly induces autophagy. AMPK also phosphorylates BECN1 and PIK3C3/VPS34 subunits stimulating autophagic functions of PIK3C3/VPS34 kinase complexes and inhibiting non-autophagic functions, respectively (He et al., 2013; Kim et al., 2013). Finally, autophagy is upregulated by activating phosphorylation of ULK1 by AMPK (Figure 3). Other specialized AMPK signaling outputs have been reviewed elsewhere (Mihaylova and Shaw, 2011; Hardie et al., 2016). It is, however, interesting to note that increase in lifespan through dietary restriction in C. elegans occurs through AMPK activation highlighting the role of AMPK as a metabolic sensor (Weir et al., 2017).
MTORC1
The activity of MTORC1 is tied to cellular amino acid levels through several sensors that directly or indirectly modulate the activity of RRAG GTPases. RRAG complexes are responsible for recruiting MTORC1 to the lysosomal membrane, a necessary step in MTORC1 functionality (Sancak et al., 2008; Sancak et al., 2010). MTORC1-associated amino acid sensors include the SESN- (Chantranupong et al., 2014; Wolfson et al., 2016) and CASTOR- (Chantranupong et al., 2016) family proteins, which, in response to cytosolic leucine and arginine, respectively, modulate RRAG activity through the GTPase activating proteins GATOR1 and GATOR2 (Bar-Peled et al., 2013; Panchaud et al., 2013). MTORC1 may also sense leucine indirectly, in a celltype specific manner, through its metabolic product acetyl-CoA. Abundance of leucine leads to increased acetyl-CoA levels, which activate MTORC1 through acetylation of the RPTOR/RAPTOR regulatory subunit (Son et al., 2019). MTORC1 is activated by lysosomal lumenal arginine through an association between the Ragulator complex and SLC38A9 (Jung et al., 2015; Wyant et al., 2017). The protein SAMTOR is described as a link between MTORC1 activity and intracellular methionine levels (Gu et al., 2017). A role of FLCN-FNIP in amino acid signaling to MTORC1 is emerging but remains to be fully elucidated (Meng and Ferguson, 2018). Interestingly, RRAG activation is also sensitive to glucose starvation, tying MTORC1 activation to intracellular glucose levels (Efeyan et al., 2013). Additionally, HK2 (hexokinase 2), which regulates a rate-limiting step in glycolysis, inhibits MTORC1 through direct interaction in the absence of glucose (Roberts et al., 2014). MTORC1 activity is coupled to growth factors through a signaling cascade originating from INSR (insulin receptor). Ligand binding to INSR activates PIK3C (phosphatidylinositol-4,5-bisphosphate 3-kinase) that activates AKT/PKB. AKT phosphorylates the MTORC1 inhibitor TSC1/2, causing its dissociation from the lysosomal membrane-bound RHEB GTPase and subsequent MTORC1 activation (Gingras et al., 1998; Bhaskar and Hay, 2007; Menon et al., 2014) (Figure 3). A role of amino acids in TSC1/2 deactivation has also been proposed but is controversial (Demetriades et al., 2014).
MTORC1 acts to suppress autophagy in nutrient-replete conditions by several mechanisms. One such mechanism is the inhibitory phosphorylation of the ULK complex, which hinders ULK1 autophosphorylation and AMPK-dependent ULK1 phosphorylation (Kim et al., 2011). MTORC1 inhibits the PIK3C3/VPS34 kinase complex through phosphorylation of the regulatory subunits ATG14, AMBRA1, or UVRAG (Nazio et al., 2013; Yuan et al., 2013; Kim et al., 2015). MTORC1-mediated repression of autophagy also occurs via the phosphorylation of TFEB (transcription factor EB). Nuclear TFEB raises the catabolic capacity of the cell by upregulating both autophagy and lysosome biosynthesis (Figure 3). Phosphorylation of TFEB by MTORC1 leads to its disabling, cytosolic retention (Martina et al., 2012). In addition, phosphorylation of TFEB targets it for ubiquitination leading to proteasomal degradation (Sha et al., 2017).
Metabolite sensors
Glucose and amino acid depletion have indirect metabolic consequences that modulate autophagy. Amino acid starvation leads to the accumulation of uncharged tRNA that activates EIF2AK4/GCN2. EIF2AK4 phosphorylates EIF2S1/EIF2α (eukaryotic initiation factor 2 subunit alpha), reducing global translation but promoting the translation of ATF4 (activating transcription factor 4) that then transcriptionally activates numerous stress-responsive genes including some involved in autophagy (Deval et al., 2009; Ye et al., 2010; B’Chir et al., 2013) (Figure 3). Recently, GORASP2/GRASP55, a structural protein responsible for Golgi stacking and reassembly, was proposed as an intracellular glucose sensor. During glucose abundance, GORASP2 is O-GlcNAcylated, a PTM that is rapidly lost upon glucose starvation. De-O-GlcNAcylated GORASP2 is targeted to autophagosomes where it interacts with the lipidated form of LC3 (LC3-II) and subsequently with LAMP2 on the lysosomal membrane to promote autophagosome-lysosome fusion (Zhang et al., 2018).
Several groups have demonstrated that autophagy may be induced by the administration of free fatty acids (FFAs), both saturated and unsaturated. The mechanism of autophagy induction differs between these two classes with saturated FFAs activating PIK3C3/VPS34 kinase complexes through AMPK, MAPK8/JNK1, and EIF2AK2/PKR (Komiya et al., 2010; Shen et al., 2012; Niso-Santano et al., 2015). Although fatty acids are energy-rich nutrients, their abundance can be an indicator of starvation; an early response to starvation is the mobilization of fatty acids from intracellular stores. However, whether the intracellular generation of free fatty acids mediates the activation of autophagy by itself or in conjunction with other metabolites in vivo is yet to be established.
β-oxidation of FFAs in mitochondria feeds the TCA cycle by generating acetyl CoA. Under severe, prolonged starvation, free fatty acid stores dwindle, and acetyl CoA levels begin to drop due to consumption from several cellular processes. Without acetyl-CoA, acetylases lack an acetyl group donor, resulting in a shift in the proteome toward the deacetylated state. This favors both the transcriptional expression of pro-autophagic genes and the derepression of existing autophagic proteins (Eisenberg et al., 2014; Marino et al., 2014). Additionally, without acetyl-CoA to feed the TCA cycle, regeneration of NADH slows, shifting the cellular equilibrium toward the oxidized form, NAD+. Increased NAD+ levels activate SIRTs (sirtuins), a family of NAD+-dependent class III histone deacetylases. SIRTs induce autophagy through multiple mechanisms including the activation of FOXO (forkhead box protein O) transcription factors and core autophagy genes such as ATG5 and ATG7 (Lee et al., 2008; Hariharan et al., 2010) (Figure 3). Under some circumstances, cells combat diminishing acetyl-CoA levels by converting amino acids to TCA cycle intermediates such as α-ketoglutarate. This conversion, however, results in the production of ammonia which also induces autophagy, likely through the activation of AMPK and the unfolded protein response (Harder et al., 2014).
Hypoxia
Hypoxia may induce autophagy via several mechanisms. Initially, hypoxia results in reduced ATP production, and thus, reduced energy charge, activating AMPK. Another major link between hypoxia and autophagy is the transcription factor complex HIF-1. HIF1A/HIF1α, a critical HIF-1 subunit, is ubiquitinated under normoxia, resulting in its degradation by CMA. During oxygen deprivation, HIF1A is not ubiquitinated, allowing HIF-1 to mount a cellular hypoxic response (Hubbi et al., 2013; Ferreira et al., 2015). Once activated, HIF-1 promotes autophagosome assembly (Bellot et al., 2009; Zhao et al., 2012) (Figure 3). Additionally, HIF-1 appears to cross-talk with MTORC1 and MTORC2 (Hudson et al., 2002; Brugarolas et al., 2004). Hypoxia is also linked to autophagy though the generation of reactive oxygen species (ROS) (Scherz-Shouval et al., 2007; Chen et al., 2009).
Autophagy as a critical component of metabolic homeostasis
1. Autophagy promotes general nutrient/metabolite availability during starvation
In cultured cells, the withdrawal of serum or nutrients swiftly and potently induces bulk autophagy, which releases a wide range of metabolites. The degradation of glycogen releases glucose for glycolysis. The resulting pyruvate may be converted to acetyl-CoA and utilized for the TCA cycle and oxidative phosphorylation. Glucose may also be used for the generation of glycolytic intermediates that are substrates for anabolic synthesis. Alternatively, glucose-6-phosphate can be shunted into the oxidative pentose phosphate pathway to produce ribulose-5phosphate. Ribulose-5-phosphate is a precursor to the synthesis of nucleotides for DNA repair and replication, as well as NAD and NADP (Rabinowitz and White, 2010).
Amino acids produced from protein breakdown support the translation of a new set of proteins, thereby remodeling the proteome. Alternatively, amino acids may be converted into TCA cycle substrates via anaplerotic reactions and subsequently utilized for oxidative phosphorylation. For example, alanine is converted to pyruvate, glutamine to α-ketoglutarate, and aspartate to oxaloacetate. Triglycerides in lipid droplets (LDs) can be catabolized to release free fatty acids (FFAs) and glycerol. Glycerol is metabolized via glycolysis, whereas FFAs are metabolized in the mitochondria by β-oxidation to form acetyl-CoA that fuels the TCA cycle. The breakdown of ribosomal RNAs (rRNAs) provides a source of nucleotides. Organelles are also targeted for degradation non-selectively during starvation (Galluzzi et al., 2014).
The regulation of autophagy in organisms is more complex. Specific metabolic organs such as the liver and adipocytes through neuroendocrine circuits initially sense nutrient scarcity. During fed conditions, INS promotes glucose and lipid incorporation in the liver. Upon fasting, INS levels are reduced, and increased circulating GCG promotes the degradation of hepatic stores. The liver rapidly responds to starvation. Autophagy is upregulated in the liver following starvation, and predominantly degrades proteins to recycle amino acids for the first 6–8 h. If starvation persists, autophagy switches to the preferential degradation of lipids, and CMA is upregulated for the selective degradation of proteins. The mobilization of glucose also occurs in response to starvation in the liver (Madrigal-Matute and Cuervo, 2016).
Mammalian development is dependent on bulk autophagy. During the transition from the four-cell stage to the blastocyst, several developmental changes occur that require rapid protein synthesis. However, at this stage, the embryo is not supplied with maternal nutrition. To meet the increased demand for amino acids, autophagy is upregulated in the pre-implantation embryo. Mouse embryos that are genetically autophagy deficient (atg5−/− mice) and derived from an autophagy-deficient oocyte, thereby lacking maternal autophagy proteins/mRNA, do not survive this transition (Tsukamoto et al., 2008). Similar acute nutrient deprivation occurs immediately after birth. Neonatal pups deprived of placental nutrition suffer from hypoglycemia and hypolipidemia. Access to maternal milk, essential for survival during this period, is compromised in autophagy-deficient mice due to suckling defects. Neuron-specific restoration of Atg5 restores suckling behavior and survival in the atg5−/− mice, highlighting a role for autophagy in neuronal development (Yoshii et al., 2016). Autophagy also plays a prominent metabolic role during this period. In neonatal pups, autophagy degrades proteins and increases the circulating pool of amino acids that sustains developmental protein synthesis and supplements energy generated by the mobilization of storage carbohydrates and fats (Kuma et al., 2004).
2. Autophagy promotes the availability of specific nutrients/metabolites
The tremendous flexibility in the process of cargo capture and degradation allows bulk autophagy to make a wide range of metabolites available for cellular utilization. At the other end of the spectrum is selective autophagy that can mobilize specific metabolites as a response to specific cellular requirements. Selective autophagy regulates organellar and metabolic homeostasis by promoting the removal of dysfunctional/superfluous organelles downstream of metabolic cues. This involves the selective sequestration of specific cargo by a receptor that links cargo with LC3-II on the expanding phagophore. These receptors can be bona fide cargolocalized proteins or ubiquitin-binding proteins that also bind LC3-II (Figure 4A; Table 1). In this section, we explore important selective autophagy pathways, the mechanisms involved and the metabolic consequences.
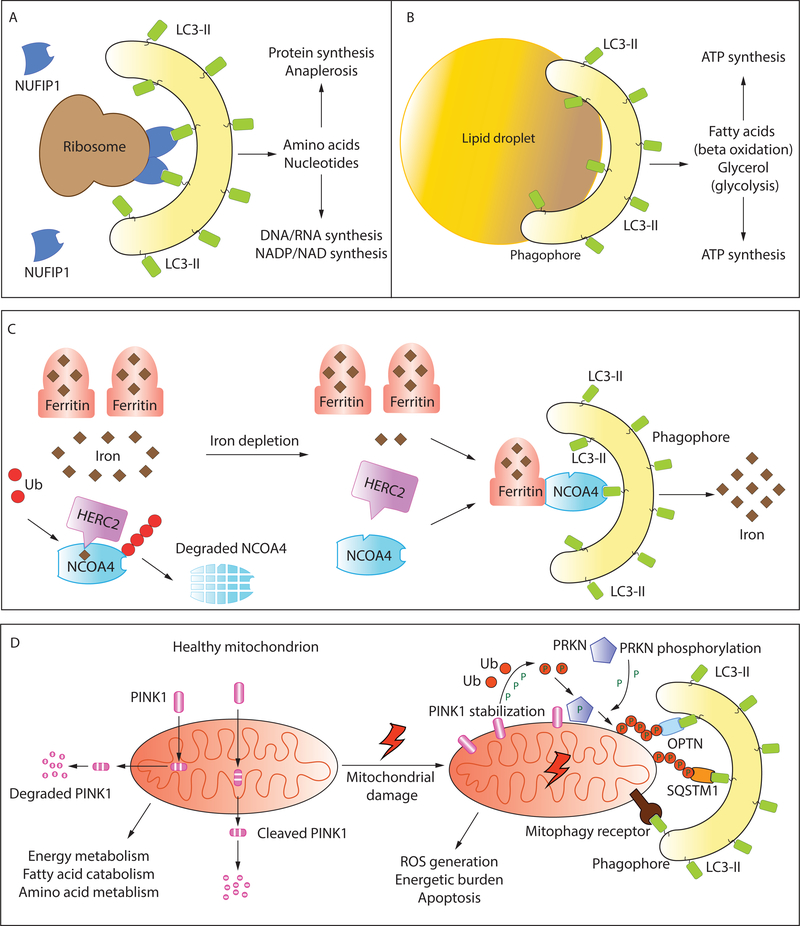
Figure 4: Selective autophagy as a modulator of metabolic homeostasis:
Selective autophagy removes dysfunctional/superfluous organelles downstream of metabolic cues. It also provides a source of raw material for several metabolic processes and pathways. Selective autophagy involves the sequestration of specific cargo by a LIR-containing receptor that links the cargo with LC3-II (see text for details). An example in (A) shows the selective targeting of ribosomes to the mitochondria by the ribophagy receptor NUFIP1. Designated receptors have not yet been identified for all types of selective autophagy. Selective uptake of lipid droplets (B) may simply occur by the formation and expansion of the phagophore on the surface of the droplet. (C) Ferritinophagy allows the iron-dependent regulation of Ferritin degradation. NCOA4 is the receptor that targets the iron-storing protein Ferritin to LC3-II. Under conditions of ironsufficiency, NCOA4 is ubiquitinated by HERC2 via an iron-dependent interaction, leading to NCOA4 degradation. When the cellular levels of free iron decline, this interaction is weakened allowing NCOA4 to target Ferritin to the phagophore. The degradation of Ferritin releases free iron. (D) Healthy mitochondria are the principal source of cellular ATP and regulate multiple metabolic circuits. Damaged mitochondria, that are detrimental, are removed by mitophagy. In the PINK1-PRKN dependent pathway of mitophagy, the kinase PINK1 which is imported and cleaved in healthy mitochondria, and subsequently targeted for cytosolic degradation, is stabilized on the OMM (outer mitochondrial membrane). PINK1 phosphorylates ubiquitin and the E3 ubiquitin ligase PRKN promoting large-scale ubiquitination of mitochondrial OMM proteins. Ubiquitinated proteins are recognized by ubiquitin-binding autophagy adaptors such as OPTN and SQSTM1 which also bind LC3-II, promoting mitochondrial degradation (refer to text for details). Mitophagy may also be orchestrated by OMM/IMM (inner mitochondrial membrane) proteins that directly bind LC3-II and function as mitophagy receptors (refer to text and Table 1 for details).
Table 1.
Receptors for Selective Autophagy
| Process | Organism | Receptor | Cargo | Metabolic trigger |
|---|---|---|---|---|
| Mitophagy | Yeast | Atg32 | Damaged/superfluous mitochondria | Hypoxia, mitochondrial damage, nonfermentable carbon source |
| Mammal | SQSTM1, OPTN, NBR1, BNIP3L (OMM), FUNDC1 (OMM), PHB2 (IMM) |
Damaged mitochondria (Ub), Superfluous mitochondria |
||
| Lipophagy | Yeast | - | Lipid droplet | Starvation, lipid overload |
| Mammal | - | Lipid droplet | ||
| Ribophagy | Yeast | - | Ribosome | Amino acid starvation, proteotoxicity, DNA damage |
| Mammal | NUFIP1 | Ribosome | ||
| Pexophagy | Yeast | Atg36, PpAtg30 | Peroxisome | Shift in carbon source |
| Mammal | SQSTM1, NBR1 | Peroxisome (Ub) | ||
| Glycophagy | Mammal | STBD1 | Glycogen | Starvation |
| Ferritinophagy | Mammal | NCOA4 | Ferritin | Iron insufficiency |
| Reticulophagy | Yeast | Atg39, Atg40 | Endoplasmic reticulum | ER stress recovery, starvation |
| Mammal | RETREG1/FAM134B, CCPG1 | Endoplasmic reticulum | ||
| Aggrephagy | Yeast | Cue5 | Protein aggregate (Ub) | Aggregate buildup/ubiquitination |
| Mammal | SQSTM1, NBR1, OPTN, TOLLIP, WDR81 | Protein aggregate (Ub) |
The table describes receptor proteins that orchestrate selective autophagy. Autophagy receptors have been highlighted from the yeast and mammalian systems. Some receptors are cargo-specific while others, such as ubiquitin (Ub)-binding receptors, are more general. The metabolic cues that most commonly trigger these mechanisms have also been listed. Pp, Pichia pastoris.
2.1. Ribophagy recycles superfluous protein synthetic machinery
Ribosomal RNA (rRNA) constitutes up to 80% of total RNA in a eukaryotic cell (Warner, 1999). The abundance of ribosomes, especially in actively growing cells, makes them a valuable pool of potentially mobilizable nucleic acids and amino acids. Selective autophagic degradation of ribosomes (ribophagy) was first described in yeast (Kraft et al., 2008; Ossareh-Nazari et al., 2010) but has since been found in mammals as well. Autophagic degradation of ribosomes also occurs as a part of other selective autophagic pathways such as mitophagy and lysophagy (An and Harper, 2018). The ribophagy receptor remained elusive in mammalian cells until recently, when NUFIP1 was identified as an autophagy receptor capable of binding both ribosomes and Atg8-family members (Wyant et al., 2018) (Figure 4A). Deletion of NUFIP1 exclusively prevents the normal decline in ribosomes under starvation conditions or upon MTORC1 inhibition; cells lacking functional NUFIP1 are also more susceptible to starvation-induced stress. Further, large fluctuations in ribosomal levels coincide with normal diurnal cycles in mice, suggesting a yet unappreciated role for targeted degradation of ribosomes in mammalian metabolism (Sinturel et al., 2017).
2.2. Lipid droplets are mobilized by the coordinated activation of lipolysis and lipophagy
Intracellular fats are stored in the form of LDs that are composed of a core of neutral lipid esters wrapped within a single layer of phospholipids and surrounded by a coat of structural proteins. Structural proteins, particularly PLINs (perilipins), not only shield the LD from the cytosol but also regulate the accessibility of lipogenic and lipolytic enzymes (Singh and Cuervo, 2012). LDs are dynamic metabolic stations (Greenberg et al., 2011) that interact and mediate lipid transfer with mitochondria (Rambold et al., 2015; Benador et al., 2018) and possibly other organelles such as the ER (Ozeki et al., 2005) and endosomes (Liu et al., 2007). Further, the hydrophobic nature of the LD allows it to bind and/or sequester proteins (Prevost et al., 2018). However, the most critical metabolic function of LDs is their role as mobilizable energy stores. LD catabolism can be initiated downstream of two distinct stimuli: nutrient deprivation and acute LD overload.
LDs may be catabolized by cytosolic lipases such as PNPLA2/ATGL and LIPE/HSL (lipase E, hormone sensitive type) (Zimmermann et al., 2004). LDs may also be degraded in the lysosome via an autophagy-dependent process known as lipophagy. Although a vacuole-dependent, autophagy-independent LD utilization mechanism has been proposed in yeast (Ouahoud et al., 2018), lipophagy is the major pathway for bulk LD-degradation in eukaryotes. Autophagy-incompetent cultured hepatocytes challenged with fatty acid overload exhibit increased triglyceride accumulation, as do hepatocytes in the autophagy-deficient mouse liver on a high-fat diet (Singh et al., 2009a). This provides a basis for metabolic disorders such as Wolman’s disease (Patrick and Lake, 1969) and cholesterol ester storage disease (CESD) (Burke and Schubert, 1972) that manifest due to deficiency of LIP (lipase, lysosomal acid type). Lipophagy is a critical metabolic pathway in neurons (Kaushik et al., 2011), brown adipose tissue (Martinez-Lopez et al., 2016) and macrophage foam cells (Ouimet et al., 2011; Lizaso et al., 2013). Basal levels of lipophagy are also required to prevent the excessive buildup of LDs (Lim et al., 2014). Although after sustained starvation there appears to be a specific sequestration of LDs within autophagosomes, an LD-specific autophagy receptor is yet to identified. Degradation of triglycerides and other lipids provides FFAs that can be metabolized through β-oxidation. TFEB, upregulated during nutrient deprivation (Settembre et al., 2013), is involved in the transcriptional upregulation of PPARGC1A (PPARG coactivator 1 alpha) and PPAR (peroxisome proliferator activated receptor) (Ghosh and Pahan, 2016), two master regulators of lipid catabolic processes, thereby connecting FFA generation to their subsequent utilization.
A proposed mechanism for selective incorporation suggests that nascent autophagosomes may form on the surface of the LD and then grow to sequester the LD partially, finally sealing off to form mature autophagosomes (Singh et al., 2009a; Singh and Cuervo, 2012) (Figure 4B). Interestingly, the lipidated form of LC3 was reported to be present on the LD surface (Shibata et al., 2009). Multiple RAB proteins also localize to the LD surface, some of which may play a role in regulating lipophagy. β-adrenergic stimulation promotes lipophagy in a RAB7-dependent manner (Lizaso et al., 2013). RAB7 may play an essential role during starvation-induced lipophagy by promoting the recruitment of lysosomes and multivesicular bodies (Schroeder et al., 2015). RAB10 colocalizes with autophagy proteins on the LD surface, and its ablation causes hepatocellular LD accumulation (Li et al., 2016).
The prevailing model of LD utilization suggests that LD catabolism occurs via a synergistic activation of lipolysis and lipophagy that promotes swift mobilization of lipid stores (Schulze et al., 2017). Both mechanisms require the removal of LD-associated PLINs by CMA. PLIN2 (perilipin 2) and PLIN3 are CMA substrates, and their degradation is upregulated after starvation, facilitated by the phosphorylation of PLIN2 by AMPK (Kaushik and Cuervo, 2015). The degradation of PLIN2 and PLIN3 allows both lipolytic enzymes and autophagy machinery access to the LD core. In the liver, PNPLA2 positively regulates lipophagy via the activation of SIRT1 (Lee et al., 2008; Sathyanarayan et al., 2017), indicating that lipolytic stimuli that activate PNPLA2 concomitantly promote lipophagy (Khan et al., 2015). PNPLA2 possesses a LIR motif and binds LC3 (Martinez-Lopez et al., 2016), an interaction critical for its LD localization.
In addition to macrolipophagy, the direct microautophagic uptake of LDs into the lysosome has also been proposed as a means of LD breakdown. In Saccharomyces cerevisiae, microlipophagy—the direct uptake of lipid droplets into the vacuole—is distinct from selective macroautophagic pathways (van Zutphen et al., 2014) and has been identified to function as a response pathway to chronic lipid imbalance (Vevea et al., 2015).
2.3. Ferritinophagy regulates iron availability
Iron, an essential micronutrient, is a cofactor for several enzymes and proteins. Iron-dependent heme synthesis in erythrocytes is critical for oxygen transport in mammals. Cytochromes utilize iron as a cofactor. Iron is also involved in the quenching of ROS as a part of antioxidative enzymes such as CAT (catalase) (Pantopoulos et al., 2012). Within the cell, iron is incorporated into the iron-sequestering protein ferritin (Zhao and Enns, 2012) Ferritin is a cage-like protein composed of multiple light (FTL) and heavy (FTH1) chain subunits surrounding a micelle of hydrated iron (Crichton, 1971; Lawson et al., 1991). The sequestration of iron is essential because free iron is prone to cycles of oxidation and reduction, producing detrimental ROS.
Equally important, however, is the regulated release of iron when needed. When the bioavailable iron level is low, it is replenished by ferritinophagy—the selective autophagic degradation of ferritin (Santana-Codina and Mancias, 2018). Lysosomal degradation of ferritin in response to iron depletion is autophagy-dependent in several cell types (Asano et al., 2011; Kishi-Itakura et al., 2014). The mechanism of selection, however, was unclear. Recently, NCOA4 (nuclear receptor coactivator 4) was identified as the cargo receptor that binds ferritin (Mancias et al., 2014), providing a basis for the selectivity. Inhibition of autophagy flux leads to an accumulation of NCOA4, confirming its identity as an autophagy substrate (Dowdle et al., 2014). NCOA4 binds the FTH1 subunit of ferritin (Mancias et al., 2015). Interestingly, although NCOA4 associates with multiple Atg8-family proteins in vitro, it does not possess a canonical LIR motif as seen with other autophagy receptors. It is possible that NCOA4 utilizes noncanonical LIR motifs (von Muhlinen et al., 2012). Recently, an ESCRT-dependent pathway that utilizes several autophagy proteins but not the Atg8-family, has also been proposed as a lysosomal targeting mechanism for the NCOA4-FTH1 complex (Goodwin et al., 2017).
The cellular level of NCOA4 is appropriately maintained to ensure regulated ferritinophagy. An iron-dependent interaction between NCOA4 and the E3 ubiquitin ligase HERC2 promotes the ubiquitination and degradation of NCOA4 when iron is abundant. When iron concentrations fall, NCOA4 is released and is available for binding and targeting ferritin for degradation (Mancias et al., 2015) (Figure 4C). The importance of NCOA4 in iron metabolism is highlighted by the massive accumulation of iron seen in several tissues of ncoa4−/− mice, especially splenic macrophages that function to reutilize iron from phagocytosed erythrocytes (Dowdle et al., 2014). ncoa4-null mice are also predisposed to anemia and sensitive to increased dietary intake of iron (Bellelli et al., 2016). Similarly, knockdown of ncoa4 leads to deficiencies in erythropoiesis in zebrafish (Mancias et al., 2015). NCOA4 also regulates the terminal differentiation of human erythroblasts (Gao et al., 2017). Further investigations will reveal other developmental and metabolic roles of this selective autophagy pathway.
2.4. Glycophagy works in concert with glycogenolysis to supply glucose
Glycogen, a branched polysaccharide, is an important contributor to glucose homeostasis. In mammals, excess circulating glucose is taken up by the liver and skeletal muscle and stored as glycogen. During periods of glucose scarcity, hormonally regulated glycogen degradation releases glucose. Glycogenolysis in skeletal muscle produces glucose that is predominantly utilized locally for sustaining muscle contraction. In contrast, glycogen breakdown in the liver, as a response to lowered blood glucose, leads to increased circulating glucose for systemic utilization (Mandl and Banhegyi, 2018). There are two principal pathways of glycogen catabolism: cytosolic glycogen undergoes a phosphorylytic degradation initiated by PYG (glycogen phosphorylase), whereas the glycogen present in the autophagic vacuole is hydrolyzed by the lysosomal enzyme GAA (glucosidase alpha, acid). The lysosomal targeting of glycogen is mediated selectively by glycophagy.
Glycophagy may rapidly provide glucose for immediate metabolic requirements while pathways such as gluconeogenesis are activated (Kuma et al., 2004; Kondomerkos et al., 2005). At the cellular level, glycophagy is regulated by the cAMP and MTOR pathways (Zhao et al., 2018). While not all glycophagy is selective, STBD1 (starch binding domain 1) has been identified as the receptor that selectively targets the glycogen particle for degradation. STBD1 binds to glycogen via a C-terminal glycan-binding domain and links it to the phagophore by its interaction with GABARAPL1 using an N-terminal LIR (Jiang et al., 2010; Jiang et al., 2011). The N terminus of STBD1 also contains a hydrophobic region that may independently mediate targeting to the phagophore.
In cardiac muscle, glycophagy is important for maintaining energy homeostasis. In the rodent heart, the pattern of STBD1 expression during fed and fasted states is sex specific as is the susceptibility to diabetic cardiomyopathy due to glycogen mishandling (Reichelt et al., 2013; Mellor et al., 2014). In Drosophila, autophagy is an efficient form of glycogen degradation in the skeletal muscle (Zirin et al., 2013). However, the specific role of glycophagy in most tissues in mammals is yet to be determined. The current consensus is that glycophagy works in concert with cytosolic glycogenolysis to orchestrate glucose metabolism. Unlike the phosphorylytic glycogenolysis, glycophagy produces non-phopshorylated glucose that can be utilized more rapidly. In mice, fast-twitch muscles that contain more glycogen deposits upregulate autophagy more than slow-twitch muscles that have lower glycogen supplies (Mizushima et al., 2004; Kaur and Debnath, 2015). The two pathways might also differ in terms of the glycogen substrate, and an attractive hypothesis is that glycophagy may preferentially target aberrantly branched glycogen particles for degradation (Mandl and Banhegyi, 2018). The importance of glycophagy is highlighted by the lysosomal storage disorder Pompe Disease that occurs due to the deficiency of GAA. The infantile disease presents as progressively lethal skeletal myopathy, respiratory and cardiac defects. The root cause lies in dysfunctional lysosomes where the degradation of glycogen is impaired, leading to energy deficiency in cardiac and skeletal muscle. Therapeutic intervention with the supplementary administration of recombinant human GAA has proven to be promising (Kishnani et al., 2007).
3. Autophagy maintains the metabolic circuit
3.1. Mitophagy and pexophagy influence aerobic metabolism
Mitochondria maintain cellular metabolism by providing ATP and regulating calcium availability. However, dysfunctional mitochondria generate ROS that not only damage cellular membranes and DNA but also lead to futile ATP consumption (Lemasters, 2014). Furthermore, severely damaged mitochondria release pro-apoptotic molecules that lead to cell death (Wang and Youle, 2009). Mitochondrial quality control is consequently a strictly regulated process. Mitochondrial maintenance is highly dynamic involving mitochondrial biogenesis, fusion, fission, and clearance (Mishra and Chan, 2016). In fact, a common mechanism to revive dysfunctional mitochondria involves fusion with healthy mitochondria (Nakada et al., 2001; Youle and van der Bliek, 2012). Mitochondrial stress also activates mitochondria-to-nucleus signaling that promotes cellular responses such as the ATFS-1-dependent mitochondrial unfolded protein response (UPRmt) (Zhao et al., 2002; Haynes et al., 2010; Nargund et al., 2012; Melber and Haynes, 2018) and the recently identified Pdr3-dependent mitochondrial compromised protein response (mitoCPR) (Weidberg and Amon, 2018). Mitochondria that are terminally damaged are removed through a process of selective autophagy called mitophagy. Mitophagy occurs at a low, basal level to continuously replace dysfunctional mitochondria, and a stronger mitophagy response may be evoked by increased mitochondrial insult. Metabolically active tissues use mitochondrial function extensively to meet their energy demands and have high basal levels of mitophagy (McWilliams et al., 2018) to facilitate mitochondrial turnover.
The mechanisms of mitophagy have been examined in several organisms. In S. cerevisiae, the outer mitochondrial membrane (OMM) protein Atg32 is the mitophagy receptor (Kanki et al., 2009). BNIP3L/NIX, an OMM protein acts as the mitophagy receptor in mitochondrial clearance during erythrocyte differentiation (Sandoval et al., 2008). Mitophagy is also induced under hypoxic conditions, where cells rely on anaerobic glycolysis, rendering mitochondria superfluous. FUNDC1 is a mitophagy receptor mediating hypoxia-dependent mitochondrial clearance (Liu et al., 2012). The inner mitochondrial membrane (IMM) protein, PHB2 (prohibitin 2), is a novel IMM-localized mitophagy receptor that is required for the clearance of paternal mitochondria in C. elegans (Wei et al., 2017).
The best-characterized pathway for mitophagy is the PINK1-PRKN pathway that responds to the loss of mitochondrial membrane potential. Membrane depolarization prevents the mitochondrial import of PINK1 and stabilizes it on the outer membrane (Kondapalli et al., 2012). PINK1 phosphorylates several substrates including ubiquitin and the E3 ligase PRKN which sets in motion a feed-forward loop that promotes large scale ubiquitination of mitochondrial membrane proteins (Koyano et al., 2014; Kane et al., 2014; Pickrell and Youle, 2015). Heavily ubiquitinated mitochondria are recognized by ubiquitin-binding autophagy receptors such as SQSTM1 and OPTN that also bind LC3, thereby linking mitochondria with phagophores (Geisler et al., 2010) (Figure 4D). However, most studies concerning the PINK1-PRKN pathway utilize the context of acute dissipation of mitochondrial membrane potential, precluding the identification of subtle pathways that are likely to be critical during pathophysiology (Gatica et al., 2018). A study showed that the phosphorylation of ubiquitin by PINK1 is sufficient to induce low-amplitude mitophagy, without the need for PRKN activity (Lazarou et al., 2015). Another PRKN-independent pathway for mitophagy that involves the recruitment of E3 ligase component RBX1 by SQSTM1, has been proposed to mitigate non-alcoholic fatty liver disease (Yamada et al., 2018). Additionally, mitophagy is independent of PINK1 in several metabolically active tissues in mice (McWilliams et al., 2018). A recent investigation concerning the in vivo relevance of PINK1 and PRKN has revealed that PINK1 and PRKN-dependent mitophagy might be critical in modulating TMEM173/STING-dependent innate immune responses to mitochondrial damage. The accumulation of mitochondrial damage leads to mitochondrial disruption which promotes inflammation. When subjected to acute or chronic mitochondrial stress, the levels of pro-inflammatory cytokines are significantly higher in mice lacking PINK1 or PRKN, indicating that these proteins likely play a critical role in limiting inflammation by mediating the timely removal of damaged mitochondria (Sliter et al., 2018).
The PINK1- and PRKN-dependent generation of mitochondria-derived vesicles (MDVs) removes localized, damaged portions of mitochondria (McLelland et al., 2016; Sugiura et al., 2014). Recently, piecemeal mitophagy, a process similar to MDV generation but with distinct cargo, was proposed to maintain basal mitochondrial homeostasis (Le Guerroue et al., 2017). Both mitophagy and MDV formation require mitochondrial fission that presumably performs two functions in this context: 1) It isolates portions of mitochondria that are damaged or disengages defective mitochondria from the mitochondrial reticular network; 2) it reduces the size of the cargo (mitochondria), promoting efficient sequestration. Consequently, mitochondrial damage and mitophagy are associated with reduced mitochondrial fusion and increased fission.
The peroxisome, involved in purine catabolism and the oxidation of fatty acids, is another important site for oxidative metabolism. The β-oxidation of very long chain FAs, branched chain FAs and the α-oxidation of phytanic acid exclusively occur in the peroxisome (Cho et al., 2018). The peroxisome produces ROS and reactive nitrogen species such as nitric oxide that are important regulators of cellular signal transduction pathways. Conversely, to quench these reactive species peroxisomes also produce antioxidant enzymes such as CAT (Bonekamp et al., 2009). Increased peroxisomal activity is promoted by increased peroxisomal protein synthesis by the transcriptional regulator PPARA (Pawlak et al., 2015). Conversely, peroxisomes can be selectively targeted for clearance by pexophagy when they are no longer beneficial.
Methylotrophic yeasts such as Pichia pastoris highlight pexophagy-mediated metabolic switching. The oxidation of methanol to formaldehyde, the first step in methanol metabolism, occurs exclusively in the peroxisome. P. pastoris maintains numerous peroxisomes when grown in methanol as the sole carbon source (van der Klei et al., 2006). When transferred from methanol to ethanol, peroxisomes are degraded by macropexophagy, whereas transfer to glucose results in micropexophagy (Tuttle and Dunn, 1995). In S. cerevisiae, the mechanism of selectivity for macropexophagy has been partly elucidated. ScAtg36 is the pexophagy receptor that links the peroxisomal membrane protein (PMP) Pex3 to both Atg11, the selective autophagy scaffold/adaptor protein, and Atg8 on the phagophore (Motley et al., 2012). In P. pastoris PpAtg30 is the selective receptor that interacts with PpPex3 and PpPex14 (Farre et al., 2008; Farre et al., 2013). As with mitophagy, macropexophagy is also promoted by peroxisomal fission (Mao et al., 2014).
In mammals, pexophagy occurs downstream of the ubiquitination of PMPs (Kim et al., 2008). Initially, PEX3 was identified as a ubiquitination substrate responsible for pexophagy induction but was subsequently found to be dispensable for pexophagy (Yamashita et al., 2014). Other PMPs, PEX5 and ABCD3/PMP70, are ubiquitinated by a mechanism involving the E3 ubiquitin ligase PEX2 and play important roles in pexophagy (Sargent et al., 2016; Zhang et al., 2015). Ubiquitinated proteins are recognized by SQSTM1 and NBR1, linking peroxisomes to the phagophore. Consequently, the depletion of SQSTM1 strongly inhibits pexophagy, whereas the exogenous expression of NBR1 strongly stimulates this process (Deosaran et al., 2013; Kim et al., 2008). PEX14 also mediates pexophagy under starvation conditions. Interestingly, PEX14 can directly interact with LC3 and, in complex with NBR1, promote peroxisome sequestration (Jiang et al., 2015). However, whether the ubiquitination of certain PMPs accelerates pexophagy or whether bulk ubiquitination of several PMPs acts as the ‘eat-me’ signal is not yet defined.
3.2. Autophagy regulates the levels of metabolic enzymes
Autophagy can influence energetics by directing the degradation of specific metabolic enzymes (Madrigal-Matute and Cuervo, 2016). Because of its selective nature, CMA is an important component of this regulatory mechanism. The regulation of the M2 splice isoform of the glycolytic enzyme PKM/PKM2 (pyruvate kinase M1/2) by CMA serves as an elegant example. M2 is the embryonic isoform of the enzyme while the M1 isoform is expressed ubiquitously in adult tissues. The preferential expression of M2 over M1 promotes rapid cell proliferation, a mechanism designed for the growth of embryonic cells, but also utilized by lung cancer cells (Christofk et al., 2008). M2 has a lower affinity for its substrate phosphoenolpyruvate than M1. M2 can also be acetylated under glucose sufficiency, which promotes its CMA-mediated degradation (Lv et al., 2011). Both factors combine to reduce the conversion of phosphoenolpyruvate to pyruvate, consequently reducing glycolytic flux. When abundant glucose is available, this mechanism allows for the accumulation of glycolytic intermediates for anabolic synthesis, a requirement of rapidly proliferating cells. Consistent with this, allografts of M1-expressing cells form smaller tumors than those of M2-expressing cells.
CMA regulates the cellular abundance of several metabolic enzymes and is a critical player in maintaining metabolic homeostasis (Kaushik and Cuervo, 2018). A study using tissue-specific lamp2a knockout in the mouse liver indicated that over 40% of CMA substrates are metabolic enzymes. These include a number of glycolytic enzymes as well as enzymes involved in triglyceride and steroid synthesis. Expectedly, the selective blockage of CMA in these mice leads to a drastic alteration in both lipid and carbohydrate metabolism and associated systemic changes such as reduced adipose tissue content, lowered body weight, increased energy expenditure and compromised responses to nutritional challenges such as starvation and lipid overload (Schneider et al., 2014). The loss of hepatic CMA also leads to a pronounced disruption of proteostasis with aging (Schneider et al., 2015). CMA may influence metabolic outcomes indirectly as well by regulating the levels of stress-responsive proteins such as HIF1A (Hubbi et al., 2013).
Identified in S. cerevisiae, the targeting of FAS (fatty acid synthase) for vacuolar degradation is a novel example of autophagy selectively degrading a single protein complex. FAS is a large enzymatic complex (Lomakin et al., 2007) that is preferentially delivered to the vacuole in an autophagy-dependent manner during nitrogen starvation. This requires interaction with Atg8 as well as the activity of Vac8 and Snx4/Atg24, two proteins involved in selective autophagy in yeast. FAS degradation during nitrogen starvation may serve to prevent the channeling of metabolic fuel for non-essential anabolic reactions because low FAS activity promotes cell viability (Shpilka et al., 2015) during starvation. Whether other protein complexes are also preferentially targeted by autophagy under similar or different conditions will be an interesting avenue for further exploration.
3.3. Autophagy is involved in proteostasis
The endoplasmic reticulum (ER) is the major cellular calcium store but also facilitates sterol synthesis and the folding and targeting of secretory pathway proteins. The ER houses chaperones, and the reducing environment allows disulfide bond formation (Bravo et al., 2013). The accumulation of unfolded proteins within the ER (Ron and Walter, 2007) causes ER stress. In haematopoietic cells, tunicamycin-induced ER stress engineers major metabolic alterations including glucose uptake and utilization followed by mitochondrial activation, which increase cellular oxygen consumption and overall ATP synthesis (Wang et al., 2011). ER stress is also associated with obesity, especially in the context of metabolic inflammation-induced dysfunction of the adipose tissue (Shan et al., 2017). Reduced protein secretion is another symptom of ER stress, altering the concentration of hormones and enzymes in circulation. ER stress is mitigated by the unfolded protein response that reduces general protein translation, upregulates proteasomal degradation, increases chaperone synthesis and promotes ER expansion (Araki and Nagata, 2011). Recovery from ER stress occurs via the removal of dilated ER subdomains by a process of selective autophagy known as reticulophagy (Smith et al., 2018)..
In yeast, Atg39 and Atg40 are the receptors for ER sequestration (Mochida et al., 2015), whereas in mammals RETREG1/FAM134B was the first identified reticulophagy receptor (Khaminets et al., 2015). These ER-resident proteins function similarly to known autophagy receptors and interact with Atg8-family proteins. RETREG1-dependent reticulophagy maintains the volume and structure of the ER, but the role of reticulophagy in recovery from ER stress was highlighted by the identification of a second reticulophagy receptor, CCPG1 (cell cycle progression 1) (Smith et al., 2018). Loss of RETREG1 causes sensory neuropathy in mice, whereas CCPG1 hypomorphic mice show impaired pancreatic proteostasis and exhibit a loss of polarization in the cells of the exocrine pancreas, underscoring the importance of reticulophagy. Sequestration of ER subdomains may also occur downstream of microbial infection and help resolve cellular stress (Moretti et al., 2017).
Aggregated proteins in the cytoplasm act as ATP sinks by consuming chaperone activity. Several proteins, such as amyloid-β and HTT are prone to aggregation, and these aggregates may promote apoptosis or necrosis (Stefani and Dobson, 2003). Aggrephagy—the selective degradation of protein aggregates—plays a pivotal role in removing toxic aggregates. The CUE domain-containing proteins Cue5 in yeast and its mammalian homolog TOLLIP, simultaneously bind to polyQ aggregates and Atg8-family proteins to promote aggregate-clearance (Lu et al., 2014a; Lu et al., 2014b). The ubiquitination of aggregated proteins plays an important role in their autophagy-dependent removal by recruiting autophagy receptors SQSTM1, NBR1 and OPTN (Kim et al., 2008; Pankiv et al., 2007; Kirkin et al., 2009). The SQSTM1-dependent degradation of aggregates also requires WDFY3/ALFY (Clausen et al., 2010), which acts as a scaffold for aggrephagy by binding lipids and proteins on the autophagosome (Filimonenko et al., 2010; Lystad et al., 2014). Another protein, WDR81, specifically interacts with LC3C and promotes aggrephagy (Liu et al., 2017b). Ubiquitin-mediated aggrephagy raises the question of substrate choice between autophagy and the ubiquitin-proteasome system. Several factors have been proposed to contribute to selectivity, including receptor oligomerization around the substrate, size of aggregates, the lysine residues used for linkage as well as the length and nature of the ubiquitin chain (Korolchuk et al., 2010; Lu et al., 2017; Verhoef et al., 2002).
Unlike conventionally secreted proteins, the yeast mating-factor is transported directly from the cytosol across the plasma membrane by an ABC transporter (Kuchler et al., 1989). The identification that IL1B/IL-1β, a mammalian cytokine, lacks a signal sequence (Rubartelli et al., 1990), initiated further interest in unconventional forms of protein secretion. An acyl-CoA binding protein known as AcbA in Dictyostelium and Acb1 in S. cerevisiae is secreted unconventionally (Duran et al., 2010; Manjithaya et al., 2010) with the secretion of this protein being dependent on autophagosome formation. However, these autophagosomes do not fuse with the lysosome/vacuole but rather with the plasma membrane. The secretion of leaderless peptides via autophagosomes is known as secretory autophagy (Ponpuak et al., 2015). The autophagymediated secretion of lysozyme occurs in intestinal Paneth cells in response to Salmonella infection. Secreted lysozyme confers protection from the invading pathogen (Bel et al., 2017). However, the metabolic consequences of autophagy-dependent secretion have not been clarified yet.
Autophagy influences metabolism during development and disease
In this section, we first highlight how autophagy influences systemic metabolism by regulating the development of adipose tissue. Adipose tissue works in concert with another primary metabolic modulator, the liver, to maintain metabolic homeostasis under conditions of nutrient deprivation. Autophagy is critical for the execution of hepatic functions—a subject of several excellent reviews (Ueno and Komatsu, 2017; Schneider et al., 2014; Madrigal-Matute and Cuervo, 2016). The dynamic and enigmatic role of autophagy in the pathogenesis and progression of cancer will be the focus of the second part of this section.
Autophagy in physiology: Adipogenesis and adipocyte maintenance
Adipocytes are specialized mammalian cells that preserve energy in the form of LDs and constitute the adipose tissue. Adipose tissue performs a range of metabolic, protective and endocrine functions and serves as a source of secreted factors such as TNF/TNFα and CFD/adipsin. Adipocytes can be white, brown or beige, with particular adipocytes serving specific functions (Rosen and Spiegelman, 2014; Zwick et al., 2018). Adipocyte differentiation is autophagy dependent. Autophagy is induced during adipogenesis in primary MEF cells, and the ablation of autophagy halts the differentiation program at an early stage. These undifferentiated cells show higher levels of apoptosis. Consequently, atg5−/− neonatal mouse pups show reduced subcutaneous fat deposits (Baerga et al., 2009).
Brown adipose tissue (BAT), constituted by mitochondria-rich, multilocular brown adipocytes, is primarily a heat-generating organ. Brown adipocytes express high levels of UCP1 (uncoupling protein 1) that uncouples mitochondrial electron transport from ATP synthesis. In these specialized adipocytes, LDs are metabolized to free fatty acids for β-oxidation and the ensuing mitochondrial electron transport builds up a proton gradient that is dissipated as heat (Fedorenko et al., 2012). BAT is, therefore, responsible for cold and diet-induced thermogenesis. Autophagy plays a critical role in the differentiation of brown adipocytes from MYF5+ progenitors. Autophagy inhibition in MYF5+ cells leads to impaired BAT differentiation and function in both pups and adult mice, highlighting the importance of autophagy in BAT differentiation during the entire lifespan. These mice also exhibit glucose intolerance, although defective skeletal muscle development contributes to that phenotype (Martinez-Lopez et al., 2013). A role for mitophagy in BAT maintenance has also recently been described. Mitophagy is induced in brown adipocytes during cold-induced thermogenesis in response to UCP1-mediated mitochondrial stress. This is coupled to mitochondrial biogenesis and serves a quality control function required for the preservation of BAT function (Lu et al., 2018b).
White adipose tissue (WAT), consisting of unilocular white adipocytes that contain few mitochondria, serves as the primary energy reserve in the body; LDs from white adipocytes are mobilized as fuel during nutrient deprivation. WAT is also an endocrine organ involved in the secretion of the appetite-regulating hormone LEP (leptin) (Kajimura, 2017), making it an important hub for metabolic regulation. Autophagy also plays an instrumental role in WAT differentiation. Adipocyte-specific atg7 knockout mice exhibit dramatically reduced body weight, as a direct consequence of reduced white adipose tissue mass. White adipocytes in the mutants are multilocular, show smaller lipid droplets, increased cytoplasm and a greater number of mitochondria. However, the mutants do not express markers of brown adipocytes, indicating that differentiation has not been rewired along a different fate. Consistent with the increase in mitochondria, these mice exhibit increased β-oxidation, reduced lipolysis, lower serum fatty acid levels and increased insulin sensitivity. Overall, these mice remain lean irrespective of diet (Zhang et al., 2009; Singh et al., 2009b). Mitochondrial abundance is a critical difference between white and brown adipocytes, and the ‘browning’ of WAT is associated with an increase in mitochondrial number. One of the factors that could promote this change is a reduction in mitophagy; recent report suggests that PRKN-mediated mitophagy is indeed downregulated during the process (Taylor and Gottlieb, 2017). Therefore, autophagy plays an instrumental role in maintaining the balance of WAT and BAT.
Beige adipocytes are an inducible form of thermogenic fat cells that reside within WAT. Brown and beige adipocytes share several morphological characteristics such as multilocular lipid droplets and numerous mitochondria but are developmentally distinct (Harms and Seale, 2013). Beige adipocytes express high levels of UCP1 and emerge upon thermogenic stimulation. However, upon withdrawal of stimulation, beige cells revert to a non-thermogenic, white-adipocyte like state and lose UCP1 expression. This reversion occurs without the appearance of an intermediate cell type and is prompted by autophagy-dependent mitochondrial clearance. The genetic deletion of Atg5 or Atg12 or the inhibition of lysosomal degradation using chloroquine in beige adipocytes promotes UCP1 retention and the maintenance of other beige-cell properties. Mitophagy induction during beige-to-white transition occurs through the cAMP-PRKA pathway (Altshuler-Keylin et al., 2016) and mitophagy in these cells is dependent on PRKN but not the UCP1-mediated loss of mitochondrial membrane potential (Lu et al., 2018a). Mice with prolonged maintenance of beige adipocytes exhibit decreased susceptibility to diet-induced obesity and insulin resistance (Altshuler-Keylin et al., 2016), indicating intriguing therapeutic avenues for these diseases.
Autophagy in pathology: The role of autophagy in cancer metabolism
As with normal cells, autophagy is an important regulator of metabolism in cancer cells (Kimmelman and White, 2017). The role of autophagy in cancer is dynamic and context dependent (Amaravadi et al., 2016). Inhibition of autophagy in mice by mosaic deletion of Atg5 or Atg7 promotes the development of liver neoplasms (Takamura et al., 2011; Inami et al., 2011). However, these neoplasms do not proceed to malignancy. In contrast, mice with monoallelic loss of BECN1, where autophagy is diminished but not absent, develop malignant tumors (Qu et al., 2003; Yue et al., 2003). Initiation of tumors in the case of partial BECN1 loss may not be solely due to decreased autophagy but also to secondary effects on tumor-suppressors such as TP53 (Liu et al., 2011); however, autophagy is critical for the maintenance of these tumors. These observations and others, coupled with infrequent mutations of core autophagy genes in human cancers (Lebovitz et al., 2015) indicate that autophagy may be important for tumor progression.
Autophagy in tumor-suppression: Helping cells protect themselves
In non-malignant cells autophagy is tumor-suppressive (Rybstein et al., 2018) and protects the cell from organellar dysfunction, protein-aggregation, redox imbalance, pathogens that possess transforming ability(Nakagawa et al., 2004) and genome destabilizers such as micronuclei and fragmented chromatin (Bartsch et al., 2017). Several genomic changes that compromise autophagy drive oncogenesis. The activation of the MTORC1 activating kinase AKT1 reduces autophagy and occurs frequently in cancers (Yi and Lauring, 2016). Oncogenic mutations in TP53/p53 that prevent its nuclear localization suppress autophagy because cytoplasmic TP53 inhibits ULK1 activation (Morselli et al., 2011). Mutations in U2AF1 (U2 small nuclear RNA auxiliary factor 1) that lead to aberrant ATG7 mRNA processing (Park et al., 2016b) are common in haematopoietic malignancies (Damm et al., 2012). The chromosomal translocation of BRD4 to the NUT locus causes an aggressive squamous cell carcinoma. BRD4 and the BRD4-NUT fusion protein were recently identified as transcriptional inhibitors of autophagy. Autophagy-deficient cells exhibit increased sensitivity to mitochondrial damage and ER stress, resulting in genomic instability and aneuploidy (Mathew et al., 2007) as well as reduced oncogene-induced senescence (Dou et al., 2015).
Autophagy drives tumor formation: Helping meet the metabolic needs of tumors
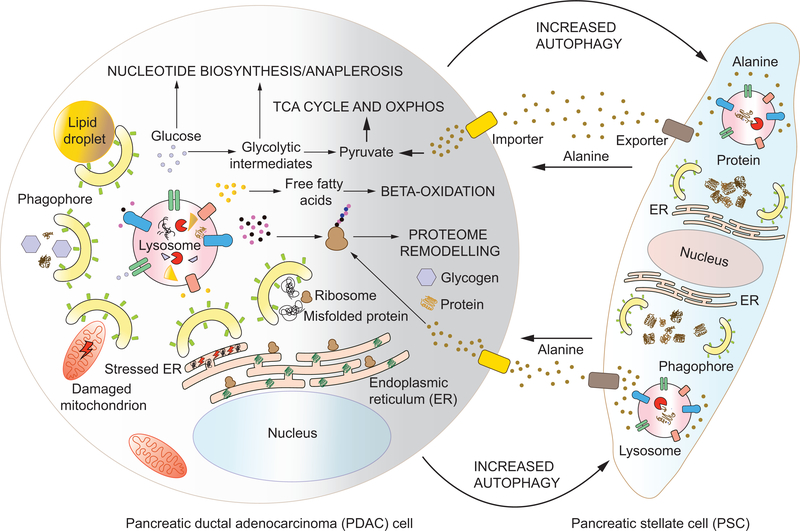
Once oncogenic transformation occurs, the role of autophagy switches, and tumors utilize autophagy as a cytoprotective mechanism (Rybstein et al., 2018). A large spectrum of tumors upregulate autophagy, a phenomenon associated with poor prognosis (Lazova et al., 2012). Autophagy fulfills the increased demands for energy and anabolism in rapidly proliferating cancer cells, producing simple biomolecules that can be used as energy sources or building blocks (Figure 5). Glycolytic flux is dependent on autophagy in genetically engineered mouse models (Wei et al., 2011; Lock et al., 2011). An acute systemic ablation of Atg7 revealed the importance of autophagy in physiological glucose homeostasis and lung tumor maintenance (Karsli-Uzunbas et al., 2014). Pancreatic ductal adenocarcinoma (PDAC), an aggressive cancer of the exocrine pancreas, exhibits MiT/TFE-family-dependent transcriptional upregulation of autophagy and lysosomal genes. The inhibition of TFE3 in PDAC decreases the pool of available metabolites, including lipids and nucleotides but particularly amino acids, highlighting the importance of autophagy in replenishing metabolic substrates (Perera et al., 2015). It is not surprising, therefore, that certain cancers such as pancreatic cancer show increased levels of basal autophagy (Yang et al., 2011). Glioblastomas and lung cancers also show a reliance on AMPK for maintenance of bioenergetics and tumor growth (Chhipa et al., 2018; Eichner et al., 2018). Additionally, AMPK-dependent upregulation of autophagy may be a mechanism of therapeutic resistance (Shteingauz et al., 2018). Indeed, inhibition of autophagy increases tumor sensitization to apoptosis (Fitzwalter et al., 2018).
Figure 5: Autophagy in tumor cells and the stroma sustains tumor progression:
Autophagy is a pro-tumorigenic pathway in transformed cells, helping them survive. Within cancer cells, autophagy removes detrimentally damaged mitochondria and helps relieve ER-stress. Autophagy is also responsible for the removal of toxic, misfolded proteins. The recycling of proteins, lipid droplets, glycogen and ribosomes by autophagy promotes energy metabolism and anabolic synthesis by providing substrates for metabolic pathways. In addition, certain cancer cells like pancreatic ductal adenocarcinoma (PDAC) cells induce autophagy in neighboring stromal cells, pancreatic stellate cells (PSCs) in the case of PDAC. The degradation of proteins by autophagy in PSCs promotes alanine production and secretion. PDACs import alanine, which may be channeled into protein synthesis or, more importantly, be converted to pyruvate. This allows an external source for pyruvate and subsequent mitochondrial energy production, thereby allowing PDACs to utilize glycolytic intermediates for nucleotide synthesis and anaplerotic reactions that fuel growth (see text for details).
The specific metabolic requirements characteristic of tumors can be met by selective autophagy (Guo et al., 2013; Strohecker et al., 2013). Malignant cells also use selective autophagy as a quality control pathway to limit organellar dysfunction that detrimentally affects tumor growth (White, 2015). Even in the hypoxic regions of tumors, mitochondrial oxidative phosphorylation generates a significant amount of ATP. The upregulation of oxidative phosphorylation may be a resistance mechanism to metformin in breast cancer (Lord et al., 2018). Conversely, damaged mitochondria promote apoptosis. Mitophagy removes damaged mitochondria, thereby regulating aerobic metabolism and preventing apoptotic signals and ROS-induced damage (Guo et al., 2011; Strohecker et al., 2013; Strohecker and White, 2014). Recently, mitophagy has been implicated in the maintenance of the cancer stem cell population in hepatocellular carcinoma (Liu et al., 2017a).
Autophagy in the tumor stroma: A helping hand
The metabolic role of autophagy within tumors includes not just its role in the transformed cells themselves but expands to the metabolic rewiring of the tumor microenvironment. The tumor microenvironment (stroma), including associated fibroblasts and immune cells acts as a major source of metabolic fuel for cancer cells (Gouirand et al., 2018). The role of the stroma in providing key metabolites such as glutamine (a carbon donor for nucleotide synthesis and other anaplerotic reactions), lactate (a substrate for the TCA cycle) and free fatty acids has been investigated (Yang et al., 2016; Sonveaux et al., 2008; Wen et al., 2017; Romero et al., 2015). Recent findings also indicate that the functions of the stroma, in concert with the mutated KRAS oncogene, may influence the epigenome and metabolome of the PDAC cells (Sherman et al., 2017).
PDAC forms a highly dense, nutrient-poor and oxygen-limiting tumor (Adamska et al., 2017). PDAC survival and growth is reliant on metabolic support from stroma-associated pancreatic stellate cells (PSCs) that provide PDACs with alanine. Alanine, secreted by the PSCs, is converted into pyruvate that can then enter the TCA cycle and oxidative phosphorylation (Sousa et al., 2016). This not only allows for increased ATP production within the PDAC cells but also makes more glucose available for amino acid or nucleotide biosynthesis. PDACs induce autophagy in PSCs leading to protein degradation that releases alanine as one of the end products (Figure 5). Consequently, autophagy-deficient PSCs do not secrete alanine. PDAC cells, cultured with conditioned medium from PSCs, grow more robustly and show higher levels of oxygen consumption than those cultured without conditioned medium, indicating the importance of the secreted alanine. However, conditioned medium from autophagy-deficient PSCs cannot promote growth of PDAC cells. Consistent with this finding, PSCs were found to have increased autophagy in the context of PDAC but not in acute pancreatitis, and the inhibition of autophagy in PSCs is detrimental to tumor growth in transplantation models (Endo et al., 2017).
Autophagy has also been implicated in promoting tumor growth by maintaining circulating arginine levels (Poillet-Perez et al., 2018). Although arginine is non-essential, arginine deficiency is detrimental to several tumor types that downregulate ASS1—an enzyme required for the de novo synthesis of arginine, using aspartate as a substrate. Downregulation of ASS1 is a metabolic switch adapted by these tumors to prevent the channeling of aspartate into arginine synthesis, instead using it for pyrimidine biosynthesis (Rabinovich et al., 2015), especially since aspartate is a limiting metabolite in hypoxic regions of tumors (Garcia-Bermudez et al., 2018). Conditional, systemic or liver-specific autophagy ablation leads to liver stress and increased release of the arginine-degrading enzyme ARG1 (arginase 1). Consequently, serum arginine levels are lowered leading to arginine deficiency and growth inhibition in tumors (Poillet-Perez et al., 2018).
Conclusion and perspectives
Macroautophagy, microautophagy and CMA regulate metabolic decisions and energy flux at multiple nodes, making self-digestion an integral part of cellular energetics. Consequently, there has been increasing interest in developing tools/drugs targeting these pathways to rectify pathological states that are directly or indirectly related to altered metabolism. However, this is not trivial given the ubiquitous role of autophagy in physiological homeostasis and the increasing recognition of non-autophagy related roles of autophagy-related proteins. The ideal therapeutic approach would specifically target autophagy-related functions of these components in a manner that allows for controlled changes in self-digestion. This will require further exploration of the basic autophagic mechanisms including answers to some of the outstanding questions highlighted in this text. One such intriguing area is the mechanism of mitophagy induction and progression in different mammalian cell types under physiological (not experimental) conditions, including the relevance of PINK1-PRKN and other parallel pathways. Another important issue to be mindful of is that a significant portion of our understanding of autophagy in mammals stems from studies in knockout mouse models where autophagy is completely abolished. This approach brings with it undesired off-target effects including possible unaccounted cellular damage and death. Instead, the use of hypomorphic alleles could provide a more accurate elucidation of autophagic mechanisms that would be instrumental in enabling the successful application of autophagy modulation as a therapeutic intervention for metabolism-related diseases.
ACKNOLWEDGEMENTS
This work was supported by NIH grant GM053396.
Footnotes
Declaration of Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adamska A, Domenichini A, and Falasca M (2017). Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, and Kajimura S (2016). Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metab 24, 402–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R, Kimmelman AC, and White E (2016). Recent insights into the function of autophagy in cancer. Genes Dev 30, 1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, and Harper JW (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, and Nagata K (2011). Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol 3, a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Komatsu M, Yamaguchi-Iwai Y, Ishikawa F, Mizushima N, and Iwai K (2011). Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol 31, 2040–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, and Ktistakis NT (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, and Bruhat A (2013). The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 41, 7683–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, and Jin S (2009). Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5, 1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U, Kaushik S, Varticovski L, and Cuervo AM (2008). The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 28, 5747–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, and Cuervo AM (2010). Identification of regulators of chaperone-mediated autophagy. Mol Cell 39, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, et al. (2017). Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26, 3960–3972. [DOI] [PubMed] [Google Scholar]
- Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, and Hooper LV (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli R, Federico G, Matte A, Colecchia D, Iolascon A, Chiariello M, Santoro M, De Franceschi L, and Carlomagno F (2016). NCOA4 Deficiency Impairs Systemic Iron Homeostasis. Cell Rep 14, 411–421. [DOI] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, and Mazure NM (2009). Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29, 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, Acin-Perez R, Shum M, Oliveira MF, Cinti S, et al. (2018). Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab 27, 869–885 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, and Hay N (2007). The two TORCs and Akt. Dev Cell 12, 487–502. [DOI] [PubMed] [Google Scholar]
- Bonekamp NA, Volkl A, Fahimi HD, and Schrader M (2009). Reactive oxygen species and peroxisomes: struggling for balance. Biofactors 35, 346–355. [DOI] [PubMed] [Google Scholar]
- Bonhoure A, Vallentin A, Martin M, Senff-Ribeiro A, Amson R, Telerman A, and Vidal M (2017). Acetylation of translationally controlled tumor protein promotes its degradation through chaperonemediated autophagy. Eur J Cell Biol 96, 83–98. [DOI] [PubMed] [Google Scholar]
- Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, et al. (2013). Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol 301, 215–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, and Kaelin WG Jr. (2004). Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18, 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JA, and Schubert WK (1972). Deficient activity of hepatic acid lipase in cholesterol ester storage disease. Science 176, 309–310. [DOI] [PubMed] [Google Scholar]
- Cadwell K, and Debnath J (2018). Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, and Sabatini DM (2014). The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang J, Zhang YY, Yan SF, Neumann D, Schlattner U, Wang ZX, and Wu JW (2012). AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nat Struct Mol Biol 19, 716–718. [DOI] [PubMed] [Google Scholar]
- Chen Y, Azad MB, and Gibson SB (2009). Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 16, 1040–1052. [DOI] [PubMed] [Google Scholar]
- Chhipa RR, Fan Q, Anderson J, Muraleedharan R, Huang Y, Ciraolo G, Chen X, Waclaw R, Chow LM, Khuchua Z, et al. (2018). AMP kinase promotes glioblastoma bioenergetics and tumour growth. Nat Cell Biol 20, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Kim YS, Jo DS, Choe SK, and Jo EK (2018). Pexophagy: Molecular Mechanisms and Implications for Health and Diseases. Mol Cells 41, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, and Cantley LC (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233. [DOI] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, et al. (2010). p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344. [DOI] [PubMed] [Google Scholar]
- Crichton RR (1971). Ferritin: structure, synthesis and function. N Engl J Med 284, 1413–1422. [DOI] [PubMed] [Google Scholar]
- Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, et al. (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, Della Valle V, Couronne L, Scourzic L, Chesnais V, et al. (2012). Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 119, 3211–3218. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, and Teleman AA (2014). Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126, 939–952. [DOI] [PubMed] [Google Scholar]
- Deval C, Chaveroux C, Maurin AC, Cherasse Y, Parry L, Carraro V, Milenkovic D, Ferrara M, Bruhat A, Jousse C, et al. (2009). Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J 276, 707–718. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364. [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, and Tooze SA (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12–5-16L1. Mol Cell 55, 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. (2015). Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al. (2014). Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Duran JM, Anjard C, Stefan C, Loomis WF, and Malhotra V (2010). Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, and Sabatini DM (2013). Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner LJ, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, Shokhirev MN, Leblanc M, Vera LI, Hutchins A, et al. (2018). Genetic Analysis Reveals AMPK Is Required to Support Tumor Growth in Murine Kras-Dependent Lung Cancer Models. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, et al. (2014). Nucleocytosolic depletion of the energy metabolite acetylcoenzyme a stimulates autophagy and prolongs lifespan. Cell Metab 19, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, Koikawa K, Okumura T, Sada M, Horioka K, et al. (2017). Autophagy Is Required for Activation of Pancreatic Stellate Cells, Associated With Pancreatic Cancer Progression and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology 152, 1492–1506 e1424. [DOI] [PubMed] [Google Scholar]
- Farre JC, Burkenroad A, Burnett SF, and Subramani S (2013). Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep 14, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, and Subramani S (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 14, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, and Kirichok Y (2012). Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151, 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, and Klionsky DJ (2014). The machinery of macroautophagy. Cell Res 24, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, et al. (2018). Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JV, Soares AR, Ramalho JS, Pereira P, and Girao H (2015). K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci Rep 5, 10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. (2010). The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 38, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzwalter BE, Towers CG, Sullivan KD, Andrysik Z, Hoh M, Ludwig M, O’Prey J, Ryan KM, Espinosa JM, Morgan MJ, et al. (2018). Autophagy Inhibition Mediates Apoptosis Sensitization in Cancer Therapy by Relieving FOXO3a Turnover. Dev Cell 44, 555–565 e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frake RA, Ricketts T, Menzies FM, and Rubinsztein DC (2015). Autophagy and neurodegeneration. J Clin Invest 125, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Martinez-Pinilla E, Navarro G, and Zamarbide M (2017). Potential of GPCRs to modulate MAPK and mTOR pathways in Alzheimer’s disease. Prog Neurobiol 149–150, 21–38. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. (2015). Autophagy in malignant transformation and cancer progression. EMBO J 34, 856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, and Kroemer G (2014). Metabolic control of autophagy. Cell 159, 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lee HY, Li W, Platt RJ, Barrasa MI, Ma Q, Elmes RR, Rosenfeld MG, and Lodish HF (2017). Thyroid hormone receptor beta and NCOA4 regulate terminal erythrocyte differentiation. Proc Natl Acad Sci U S A 114, 10107–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bermudez J, Baudrier L, La K, Zhu XG, Fidelin J, Sviderskiy VO, Papagiannakopoulos T, Molina H, Snuderl M, Lewis CA, et al. (2018). Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol 20, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, and Klionsky DJ (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, and Springer W (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12, 119–131. [DOI] [PubMed] [Google Scholar]
- Ghosh A, and Pahan K (2016). PPARalpha in lysosomal biogenesis: A perspective. Pharmacol Res 103, 144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, and Hay N (1998). 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev 12, 502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JM, Dowdle WE, DeJesus R, Wang Z, Bergman P, Kobylarz M, Lindeman A, Xavier RJ, McAllister G, Nyfeler B, et al. (2017). Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9. Cell Rep 20, 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouirand V, Guillaumond F, and Vasseur S (2018). Influence of the Tumor Microenvironment on Cancer Cells Metabolic Reprogramming. Front Oncol 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, and Mashek DG (2011). The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121, 2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, and Sabatini DM (2017). SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C, McCulloch D, et al. (2017). Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 43, 716–730 e717. [DOI] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27, 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, and Shaw RJ (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberzettl P, and Hill BG (2013). Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol 1, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder LM, Bunkenborg J, and Andersen JS (2014). Inducing autophagy: a comparative phosphoproteomic study of the cellular response to ammonia and rapamycin. Autophagy 10, 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, and Brunet A (2016). AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol 26, 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, and Sadoshima J (2010). Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res 107, 1470–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, and Seale P (2013). Brown and beige fat: development, function and therapeutic potential. Nat Med 19, 1252–1263. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, and Ron D (2010). The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 37, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Zhu H, Li H, Zou MH, and Xie Z (2013). Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes 62, 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, She H, Zhang T, Xu H, Cheng L, Yepes M, Zhao Y, and Mao Z (2018). p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J Cell Biol 217, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. (2007). Control of macroautophagy by calcium, calmodulindependent kinase kinase-beta, and Bcl-2. Mol Cell 25, 193–205. [DOI] [PubMed] [Google Scholar]
- Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, and Semenza GL (2013). Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J Biol Chem 288, 10703–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, and Abraham RT (2002). Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol 22, 7004–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. (2011). Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. (2006). TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, and Mizushima N (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19, 5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Hara-Kuge S, Yamashita S, and Fujiki Y (2015). Peroxin Pex14p is the key component for coordinated autophagic degradation of mammalian peroxisomes by direct binding to LC3-II. Genes Cells 20, 36–49. [DOI] [PubMed] [Google Scholar]
- Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, and Roach PJ (2010). Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem 285, 34960–34971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Wells CD, and Roach PJ (2011). Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun 413, 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Genau HM, and Behrends C (2015). Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol 35, 2479–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S (2017). Adipose tissue in 2016: Advances in the understanding of adipose tissue biology. Nat Rev Endocrinol 13, 69–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, and Youle RJ (2014). PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 205, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, and Klionsky DJ (2009). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. (2014). Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, and Debnath J (2015). Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 16, 461–472. [DOI] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2015). Degradation of lipid droplet-associated proteins by chaperonemediated autophagy facilitates lipolysis. Nat Cell Biol 17, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2016). AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 12, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2018). The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19, 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, and Cuervo AM (2008). Constitutive activation of chaperonemediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 19, 2179–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, and Singh R (2011). Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 14, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Khan SA, Sathyanarayan A, Mashek MT, Ong KT, Wollaston-Hayden EE, and Mashek DG (2015). ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1alpha/PPAR-alpha signaling. Diabetes 64, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, and Ohsumi Y (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, and Guan KL (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, and Guan KL (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, and Lippincott-Schwartz J (2008). Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 105, 20567–20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, and Kim DH (2015). mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 57, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC, and White E (2017). Autophagy and Tumor Metabolism. Cell Metab 25, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33, 505–516. [DOI] [PubMed] [Google Scholar]
- Kishi-Itakura C, Koyama-Honda I, Itakura E, and Mizushima N (2014). Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci 127, 4089–4102. [DOI] [PubMed] [Google Scholar]
- Kishnani PS, Corzo D, Nicolino M, Byrne B, Mandel H, Hwu WL, Leslie N, Levine J, Spencer C, McDonald M, et al. (2007). Recombinant human acid [alpha]-glucosidase: major clinical benefits in infantile-onset Pompe disease. Neurology 68, 99–109. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, et al. (2013). Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol 202, 331–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya K, Uchida T, Ueno T, Koike M, Abe H, Hirose T, Kawamori R, Uchiyama Y, Kominami E, Fujitani Y, et al. (2010). Free fatty acids stimulate autophagy in pancreatic beta-cells via JNK pathway. Biochem Biophys Res Commun 401, 561–567. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. (2012). PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2, 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondomerkos DJ, Kalamidas SA, Kotoulas OB, and Hann AC (2005). Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol 20, 689–696. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, and Rubinsztein DC (2010). Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584, 1393–1398. [DOI] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. (2014). Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, and Peter M (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10, 602–610. [DOI] [PubMed] [Google Scholar]
- Ktistakis NT, and Tooze SA (2016). Digesting the Expanding Mechanisms of Autophagy. Trends Cell Biol 26, 624–635. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, and Thorner J (1989). Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J 8, 3973–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, and Mizushima N (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Kuroki T, Osari S, Nagata K, and Kawaguchi A (2018). Influenza A Virus NS1 Protein Suppresses JNK1-Dependent Autophagosome Formation Mediated by Rab11a Recycling Endosomes. Front Microbiol 9, 3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, Cesareni G, et al. (1991). Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349, 541–544. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, and Pawelek JM (2012). Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res 18, 370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guerroue F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, and Behrends C (2017). Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell 68, 786–796 e786. [DOI] [PubMed] [Google Scholar]
- Lebovitz CB, Robertson AG, Goya R, Jones SJ, Morin RD, Marra MA, and Gorski SM (2015). Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy 11, 1668–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, and Finkel T (2008). A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 105, 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters JJ (2014). Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol 2, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WW, Li J, and Bao JK (2012). Microautophagy: lesser-known self-eating. Cell Mol Life Sci 69, 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, Li W, Lyu J, Rao G, Zhang X, et al. (2017). NucleusTranslocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell 66, 684–697 e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schulze RJ, Weller SG, Krueger EW, Schott MB, Zhang X, Casey CA, Liu J, Stockli J, James DE, et al. (2016). A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci Adv 2, e1601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, Ryu D, Koo SH, Kim HL, Kim J, et al. (2014). Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun 5, 4934. [DOI] [PubMed] [Google Scholar]
- Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. (2011). Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan ST, Cho C, Machida K, Chen D, and Ou JJ (2017a). Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol Cell 68, 281–292 e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14, 177–185. [DOI] [PubMed] [Google Scholar]
- Liu P, Bartz R, Zehmer JK, Ying YS, Zhu M, Serrero G, and Anderson RG (2007). Rab-regulated interaction of early endosomes with lipid droplets. Biochim Biophys Acta 1773, 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li Y, Wang X, Xing R, Liu K, Gan Q, Tang C, Gao Z, Jian Y, Luo S, et al. (2017b). The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J Cell Biol 216, 1301–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizaso A, Tan KT, and Lee YH (2013). beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, and Debnath J (2011). Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Xiong Y, and Steitz TA (2007). The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319–332. [DOI] [PubMed] [Google Scholar]
- Lord SR, Cheng WC, Liu D, Gaude E, Haider S, Metcalf T, Patel N, Teoh EJ, Gleeson F, Bradley K, et al. (2018). Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab 28, 679–688 e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, den Brave F, and Jentsch S (2017). Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol 19, 732–739. [DOI] [PubMed] [Google Scholar]
- Lu K, Psakhye I, and Jentsch S (2014a). Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563. [DOI] [PubMed] [Google Scholar]
- Lu K, Psakhye I, and Jentsch S (2014b). A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy 10, 2381–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Altshuler-Keylin S, Wang Q, Chen Y, Henrique Sponton C, Ikeda K, Maretich P, Yoneshiro T, and Kajimura S (2018a). Mitophagy controls beige adipocyte maintenance through a Parkindependent and UCP1-independent mechanism. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Fujioka H, Joshi D, Li Q, Sangwung P, Hsieh P, Zhu J, Torio J, Sweet D, Wang L, et al. (2018b). Mitophagy is required for brown adipose tissue mitochondrial homeostasis during cold challenge. Sci Rep 8, 8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. (2011). Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell 42, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, Kanegae Y, Kageyama S, Suzuki M, Saito I, Mizushima T, et al. (2014). Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep 15, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal-Matute J, and Cuervo AM (2016). Regulation of Liver Metabolism by Autophagy. Gastroenterology 150, 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Pontano Vaites L, Nissim S, Biancur DE, Kim AJ, Wang X, Liu Y, Goessling W, Kimmelman AC, and Harper JW (2015). Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Wang X, Gygi SP, Harper JW, and Kimmelman AC (2014). Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl J, and Banhegyi G (2018). The ER - Glycogen Particle - Phagophore Triangle: A Hub Connecting Glycogenolysis and Glycophagy? Pathol Oncol Res 24, 821–826. [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, and Subramani S (2010). Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Liu X, Feng Y, and Klionsky DJ (2014). The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 10, 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco MR, and Linnemann AK (2018). beta-Cell Autophagy in Diabetes Pathogenesis. Endocrinology 159, 2127–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. (2014). Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 53, 710–725. [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, and Puertollano R (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Athonvarangkul D, Sahu S, Coletto L, Zong H, Bastie CC, Pessin JE, Schwartz GJ, and Singh R (2013). Autophagy in Myf5+ progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep 14, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, and Singh R (2016). Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab 23, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, and Singh R (2014). ATGs: Scaffolds for MAPK/ERK signaling. Autophagy 10, 535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, et al. (2015). Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Massey AC, Kaushik S, Sovak G, Kiffin R, and Cuervo AM (2006). Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 103, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, and White E (2007). Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21, 1367–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Lee SA, McBride HM, and Fon EA (2016). Syntaxin-17 delivers PINK1/parkindependent mitochondrial vesicles to the endolysosomal system. J Cell Biol 214, 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, and Ganley IG (2018). Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab 27, 439–449 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, and Haynes CM (2018). UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res 28, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor KM, Varma U, Stapleton DI, and Delbridge LM (2014). Cardiomyocyte glycophagy is regulated by insulin and exposure to high extracellular glucose. Am J Physiol Heart Circ Physiol 306, H1240–1245. [DOI] [PubMed] [Google Scholar]
- Meng J, and Ferguson SM (2018). GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates Rag GTPase heterodimer nucleotide status in response to amino acids. J Cell Biol 217, 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, and Manning BD (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre MB, and Colombo MI (2012). cAMP and EPAC are key players in the regulation of the signal transduction pathway involved in the alpha-hemolysin autophagic response. PLoS Pathog 8, e1002664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, and Shaw RJ (2011). The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, and Chan DC (2016). Metabolic regulation of mitochondrial dynamics. J Cell Biol 212, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, and Komatsu M (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, and Ohsumi Y (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, and Nakatogawa H (2015). Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362. [DOI] [PubMed] [Google Scholar]
- Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, et al. (2017). STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 171, 809–823 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Shen S, Ruckenstuhl C, Bauer MA, Marino G, Galluzzi L, Criollo A, Michaud M, Maiuri MC, Chano T, et al. (2011). p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 10, 2763–2769. [DOI] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, and Hettema EH (2012). Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J 31, 2852–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, and Hayashi JI (2001). Intermitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med 7, 934–940. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, and Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. (2013). mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 15, 406–416. [DOI] [PubMed] [Google Scholar]
- Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Marino G, Cianfanelli V, BenYounes A, Troncoso R, Markaki M, Sica V, et al. (2015). Unsaturated fatty acids induce noncanonical autophagy. EMBO J 34, 1025–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, and Sakai Y (2016). Pexophagy in yeasts. Biochim Biophys Acta 1863, 992–998. [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, and Tooze SA (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, and Dargemont C (2010). Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11, 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouahoud S, Fiet MD, Martinez-Montanes F, Ejsing CS, Kuss O, Roden M, and Markgraf DF (2018). Lipid droplet consumption is functionally coupled to vacuole homeostasis independent of lipophagy. J Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, and Marcel YL (2011). Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab 13, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, and Fujimoto T (2005). Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci 118, 2601–2611. [DOI] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, and De Virgilio C (2013). Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 6, ra42. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, and Johansen T (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Pantopoulos K, Porwal SK, Tartakoff A, and Devireddy L (2012). Mechanisms of mammalian iron homeostasis. Biochemistry 51, 5705–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, et al. (2016a). The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 12, 547–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, et al. (2018). ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius CM, Ali AM, Zhu LJ, Mukherjee S, Raza A, et al. (2016b). U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3’ End Formation. Mol Cell 62, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick AD, and Lake BD (1969). Deficiency of an acid lipase in Wolman’s disease. Nature 222, 1067–1068. [DOI] [PubMed] [Google Scholar]
- Pawlak M, Lefebvre P, and Staels B (2015). Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 62, 720–733. [DOI] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, and Youle RJ (2015). The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, Su X, Maganti A, Jiang C, Lu W, et al. (2018). Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, and Deretic V (2015). Secretory autophagy. Curr Opin Cell Biol 35, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost C, Sharp ME, Kory N, Lin Q, Voth GA, Farese RV Jr., and Walther TC (2018). Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev Cell 44, 73–86 e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, Bento CF, Renna M, Menzies FM, and Rubinsztein DC (2018). The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev Cell 45, 114–131 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112, 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et al. (2015). Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, and White E (2010). Autophagy and metabolism. Science 330, 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, and Lippincott-Schwartz J (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32, 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt ME, Mellor KM, Curl CL, Stapleton D, and Delbridge LM (2013). Myocardial glycophagy - a specific glycogen handling response to metabolic stress is accentuated in the female heart. J Mol Cell Cardiol 65, 67–75. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Tan-Sah VP, Ding EY, Smith JM, and Miyamoto S (2014). Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell 53, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IL, Mukherjee A, Kenny HA, Litchfield LM, and Lengyel E (2015). Molecular pathways: trafficking of metabolic resources in the tumor microenvironment. Clin Cancer Res 21, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, and Walter P (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A, Cozzolino F, Talio M, and Sitia R (1990). A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J 9, 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, and Guan KL (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybstein MD, Bravo-San Pedro JM, Kroemer G, and Galluzzi L (2018). The autophagic network and cancer. Nat Cell Biol 20, 243–251. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, and Wang J (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana-Codina N, and Mancias JD (2018). The Role of NCOA4-Mediated Ferritinophagy in Health and Disease. Pharmaceuticals (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent G, van Zutphen T, Shatseva T, Zhang L, Di Giovanni V, Bandsma R, and Kim PK (2016). PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol 214, 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayan A, Mashek MT, and Mashek DG (2017). ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, and Elazar Z (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26, 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Suh Y, and Cuervo AM (2014). Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 20, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, Villarroya F, Santambrogio L, and Cuervo AM (2015). Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, and McNiven MA (2015). The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze RJ, Sathyanarayan A, and Mashek DG (2017). Breaking fat: The regulation and mechanisms of lipophagy. Biochim Biophys Acta Mol Cell Biol Lipids 1862, 1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. (2013). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y, Rao L, Settembre C, Ballabio A, and Eissa NT (2017). STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J 36, 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Wang X, Wu Y, Xu C, Xia Z, Dai J, Shao M, Zhao F, He S, Yang L, et al. (2017). The metabolic ER stress sensor IRE1alpha suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat Immunol 18, 519–529. [DOI] [PubMed] [Google Scholar]
- Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Marino G, Lachkar S, Senovilla L, et al. (2012). Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell 48, 667–680. [DOI] [PubMed] [Google Scholar]
- Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu S, Truitt ML, He N, Ding N, Liddle C, Atkins AR, et al. (2017). Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc Natl Acad Sci U S A 114, 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, and Uchiyama Y (2009). The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun 382, 419–423. [DOI] [PubMed] [Google Scholar]
- Shpilka T, Welter E, Borovsky N, Amar N, Shimron F, Peleg Y, and Elazar Z (2015). Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc Natl Acad Sci U S A 112, 1434–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteingauz A, Porat Y, Voloshin T, Schneiderman RS, Munster M, Zeevi E, Kaynan N, Gotlib K, Giladi M, Kirson ED, et al. (2018). AMPK-dependent autophagy upregulation serves as a survival mechanism in response to Tumor Treating Fields (TTFields). Cell Death Dis 9, 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, and Cuervo AM (2012). Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol 2012, 282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ (2009a). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, and Czaja MJ (2009b). Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 119, 3329–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, and Schibler U (2017). Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell 169, 651–663 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, et al. (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, and Wilkinson S (2018). CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 44, 217–232 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son SM, Park SJ, Lee H, Siddiqi F, Lee JE, Menzies FM, and Rubinsztein DC (2019). Leucine Signals to mTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metab 29, 192–201 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. (2008). Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 118, 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, and Dobson CM (2003). Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 81, 678–699. [DOI] [PubMed] [Google Scholar]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, and White E (2013). Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600Edriven lung tumors. Cancer Discov 3, 1272–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, and White E (2014). Targeting mitochondrial metabolism by inhibiting autophagy in BRAF-driven cancers. Cancer Discov 4, 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, and Malhotra V (2013). Non-autophagic roles of autophagy-related proteins. EMBO Rep 14, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, and McBride HM (2014). A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J 33, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, and Mizushima N (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D, and Gottlieb RA (2017). Parkin-mediated mitophagy is downregulated in browning of white adipose tissue. Obesity (Silver Spring) 25, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo M, Batista-Gonzalez A, Merheb E, Aoun ML, Tarabra E, Feng D, Sarparanta J, Merlo P, Botre F, Schwartz GJ, et al. (2018). Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab 28, 268–281 e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, and Mizushima N (2008). Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, and Dunn WA Jr. (1995). Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci 108 (Pt 1), 25–35. [DOI] [PubMed] [Google Scholar]
- Ueno T, and Komatsu M (2017). Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol 14, 170–184. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Yurimoto H, Sakai Y, and Veenhuis M (2006). The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta 1763, 1453–1462. [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, and Kohlwein SD (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 25, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef LG, Lindsten K, Masucci MG, and Dantuma NP (2002). Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet 11, 2689–2700. [DOI] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, and Pon LA (2015). Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell 35, 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, and Randow F (2012). LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 48, 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, and Youle RJ (2009). The role of mitochondria in apoptosis*. Annu Rev Genet 43, 95–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Eno CO, Altman BJ, Zhu Y, Zhao G, Olberding KE, Rathmell JC, and Li C (2011). ER stress modulates cellular metabolism. Biochem J 435, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Wauson EM, Dbouk HA, Ghosh AB, and Cobb MH (2014). G protein-coupled receptors and the regulation of autophagy. Trends Endocrinol Metab 25, 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, and Guan JL (2011). Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25, 1510–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chiang WC, Sumpter R Jr., Mishra P, and Levine B (2017). Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 168, 224–238 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, and Amon A (2018). MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD, and Mair WB (2017). Dietary Restriction and AMPK Increase Lifespan via Mitochondrial Network and Peroxisome Remodeling. Cell Metab 26, 884–896 e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YA, Xing X, Harris JW, Zaytseva YY, Mitov MI, Napier DL, Weiss HL, Mark Evers B, and Gao T (2017). Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell Death Dis 8, e2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E (2015). The role for autophagy in cancer. J Clin Invest 125, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, et al. (2008). Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol 4, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, and Sabatini DM (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, and Carling D (2003). LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13, 2004–2008. [DOI] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, and Sabatini DM (2017). mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 171, 642–654 e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, et al. (2007). Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449, 496–500. [DOI] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. (2011). Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, et al. (2018). Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in ParkinIndependent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease. Cell Metab 28, 588–604 e585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Abe K, Tatemichi Y, and Fujiki Y (2014). The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Achreja A, Yeung TL, Mangala LS, Jiang D, Han C, Baddour J, Marini JC, Ni J, Nakahara R, et al. (2016). Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab 24, 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. (2011). Pancreatic cancers require autophagy for tumor growth. Genes Dev 25, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, and Koumenis C (2010). The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29, 2082–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi KH, and Lauring J (2016). Recurrent AKT mutations in human cancers: functional consequences and effects on drug sensitivity. Oncotarget 7, 4241–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kuma A, Akashi T, Hara T, Yamamoto A, Kurikawa Y, Itakura E, Tsukamoto S, Shitara H, Eishi Y, et al. (2016). Systemic Analysis of Atg5-Null Mice Rescued from Neonatal Lethality by Transgenic ATG5 Expression in Neurons. Dev Cell 39, 116–130. [DOI] [PubMed] [Google Scholar]
- Youle RJ, and van der Bliek AM (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen Y, and Tooze SA (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HX, Russell RC, and Guan KL (2013). Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, and Heintz N (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 100, 15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng JW, Zhu M, et al. (2017). Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. (2014). The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20, 526–540. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 17, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang L, Lak B, Li J, Jokitalo E, and Wang Y (2018). GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev Cell 45, 245–261 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, and Jin S (2009). Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A 106, 19860–19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Guo H, Zhang CS, Lin SY, Yin Z, Peng Y, Luo H, Shi Y, Lian G, Zhang C, et al. (2013). AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab 18, 546–555. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tang M, Liu M, and Chen L (2018). Glycophagy: An emerging target in pathology. Clin Chim Acta 484, 298–303. [DOI] [PubMed] [Google Scholar]
- Zhao N, and Enns CA (2012). Iron transport machinery of human cells: players and their interactions. Curr Top Membr 69, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, and Hoogenraad NJ (2002). A mitochondrial specific stress response in mammalian cells. EMBO J 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen G, Zhang W, Xu N, Zhu JY, Jia J, Sun ZJ, Wang YN, and Zhao YF (2012). Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1alpha/BNIP3 signaling pathway. J Cell Physiol 227, 639–648. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al. (2004). Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386. [DOI] [PubMed] [Google Scholar]
- Zirin J, Nieuwenhuis J, and Perrimon N (2013). Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 11, e1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick RK, Guerrero-Juarez CF, Horsley V, and Plikus MV (2018). Anatomical, Physiological, and Functional Diversity of Adipose Tissue. Cell Metab 27, 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]