Summary
Primed epiblast stem cells (EpiSCs) can be reverted to a pluripotent embryonic stem cell (ESC)-like state by expression of single reprogramming factor. We used CRISPR activation to perform a genome-scale, reprogramming screen in EpiSCs and identified 142 candidate genes. Our screen validated a total of 50 genes, previously not known to contribute to reprogramming, of which we chose Sall1 for further investigation. We show that Sall1 augments reprogramming of mouse EpiSCs and embryonic fibroblasts and that these induced pluripotent stem cells are indeed fully pluripotent including formation of chimeric mice. We also demonstrate that Sall1 synergizes with Nanog in reprogramming and that overexpression in ESCs delays their conversion back to EpiSCs. Lastly, using RNA sequencing, we identify and validate Klf5 and Fam189a2 as new downstream targets of Sall1 and Nanog. In summary, our work demonstrates the power of using CRISPR technology in understanding molecular mechanisms that mediate complex cellular processes such as reprogramming.
Keywords: CRISPR/Cas9, CRISPR activation, epiblast stem cells, reprogramming, genome-wide screen, activation screen, gain-of-function, CRISPR screen, reprogramming pathways, induced pluripotent stem cells
Graphical Abstract
Highlights
-
•
Genome-scale CRISPRa screen in mouse EpiSCs identifies novel reprogramming factors
-
•
50 novel genes, including Sall1 and Fam189a2, identified to mediate reprogramming
-
•
Sall1 synergizes with Nanog to increase reprogramming efficiency in EpiSCs and MEFs
-
•
RNA-seq provides insight into downstream pathways of Sall1 and Nanog-mediated reprogramming
In this study, Metzakopian and colleagues use CRISPR activation to perform a genome-scale reprogramming screen on mouse epiblast stem cells identifying 50 novel candidates. In addition, the authors provide evidence of Sall1 being a potent reprogramming gene, capable of synergizing with Nanog. Lastly, using RNA-seq, the authors provide insight into potential downstream targets of Sall1 and Nanog.
Introduction
The ability of pluripotent stem cells (PSCs) to self-renew and their potential to differentiate into multiple cell types makes them useful for clinical applications (Martello and Smith, 2014). PSCs can either be derived from early embryos or be induced (iPSCs) by reprogramming somatic cells with Yamanaka factors, i.e., Oct4, Sox2, c-Myc, and Klf4 among other transcription factors, mRNAs, microRNAs, and small molecules (Hou et al., 2013, Sandmaier and Telugu, 2015, Takahashi and Yamanaka, 2006, Warren et al., 2010). During early mouse embryo development, at least two types of PSCs can be derived, naive embryonic stem cells (ESCs) from the inner mass of the blastocyst and primed post-implantation epiblast stem cells (EpiSCs) (Nichols and Smith, 2009, Tesar et al., 2007). While both have the potential to differentiate into multiple lineages, only ESCs can contribute extensively to chimeras, showing unbiased developmental potential. Both ESCs and EpiSCs express major pluripotent transcription factors such as Oct4 and Sox2 at similar levels. In EpiSCs, however, reduced expression of pluripotency-associated factors such as Rex1 and Klf4 and elevated levels of early differentiation markers such as Fgf5, Gata6, and Otx2 indicate their restricted developmental potential. Interestingly, EpiSCs cultured in fully defined ESC medium (with inhibition of MAPK and GSK3 and supplementation with LIF; hereafter 2i/LIF medium) can be reprogrammed into ESCs by overexpressing only a single gene––such as Nanog, Klf4, or Nr5a2 (Guo and Smith, 2010)––making them an ideal model system for genetic screens.
Recently, CRISPR/Cas9 has gained importance by achieving simple, precise, and rapid editing of the genome, enabling large-scale experiments such as genetic screening. While the RNA-programmable (single guide RNA [sgRNA]) endonuclease Cas9 is used to induce double-strand breaks in defined genomic locations, its catalytically dead variant (dCas9) can be fused with transcriptional activators and directed toward promoter regions to increase gene expression (CRISPR activation, CRISPRa) (Doudna and Charpentier, 2014, Gaj et al., 2013).
Genome-wide screening is a powerful unbiased approach to discover genes and pathways that underlie biological processes. To date, identification of key transcription factors and epigenetic modifiers within naive and primed PSCs has been investigated by employing either gain-of-function (GoF) screens using cDNA libraries and PiggyBac transposons or loss-of-function screens using RNA interference (Gayle et al., 2015, Hu et al., 2009, Pritsker et al., 2006).
Here, we describe the development and application of a genome-scale CRISPRa screen to identify genes that contribute to mouse EpiSC reprogramming. We show that our screening approach not only detects established reprogramming factors such as Oct4 and Nanog, but also identifies previously unreported candidate genes capable of reprogramming. We focus on the role of Sall1, a transcription factor belonging to the Spalt-like gene family, which has been implicated in cellular reprogramming in a number of studies but has not been sufficiently investigated (Basta et al., 2017, Gaspar-Maia et al., 2013, Mansour et al., 2012). Our work substantiates Sall1 as a potent reprogramming gene candidate by demonstrating its ability to reprogram EpiSCs and mouse embryonic fibroblasts (MEFs) to iPSCs. In addition, we show that Sall1 may exert its functions by interacting synergistically with Nanog to reprogram cells to ground state pluripotency.
Results
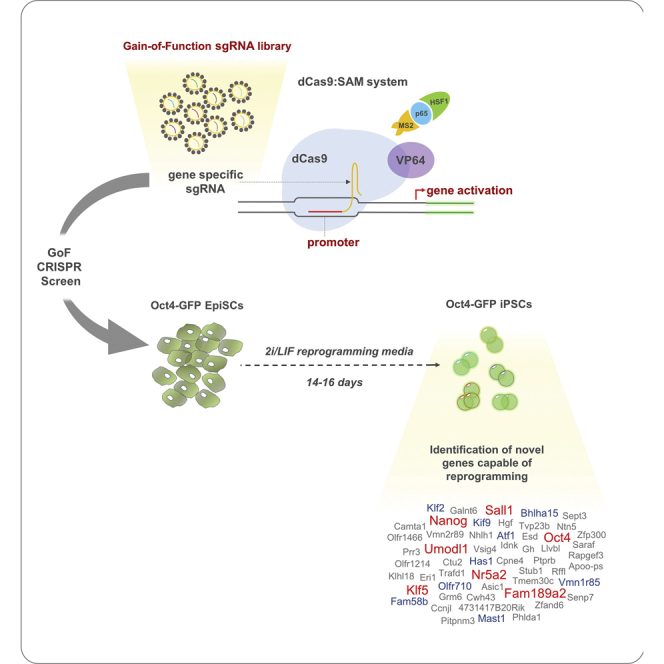
GoF CRISPRa Screen Identifies Reprogramming Genes
Initially, we sought to determine the optimal Cas9 transactivation system, as several variants have been published (Chavez et al., 2016, Konermann et al., 2015, Tanenbaum et al., 2014). To that end, we created PiggyBac-transposable (Yusa et al., 2011) expression vectors with a Blasticidin-mCherry cassette for four different dCas9-CRISPRa systems: dCas:VP160, dCas9:SunTag, dCas9:VPR, and dCas9:SAM (Figure S1).
Furthermore, we designed a versatile sgRNA expression construct (pKLV-PB-U6-gRNA-PGK-Puro-T2A-TagBFP) (Metzakopian et al., 2017) with a selectable and a fluorescent marker (puromycin and BFP, Figure S1), which can be stably integrated into target genomes as lentivirus or via PiggyBac-mediated transposition (Yusa et al., 2011).
We directed single sgRNAs guides against the promoter region of Ascl1 and Neurog2, genes with low baseline expression, in HEK293 cells. After stable integration of dCas9-CRISPRa and the sgRNA vectors via transposition and antibiotic selection, qRT-PCR revealed that dCas9:SAM achieved the highest overexpression of both target genes and thus was chosen for all subsequent experiments (Figure 1A).
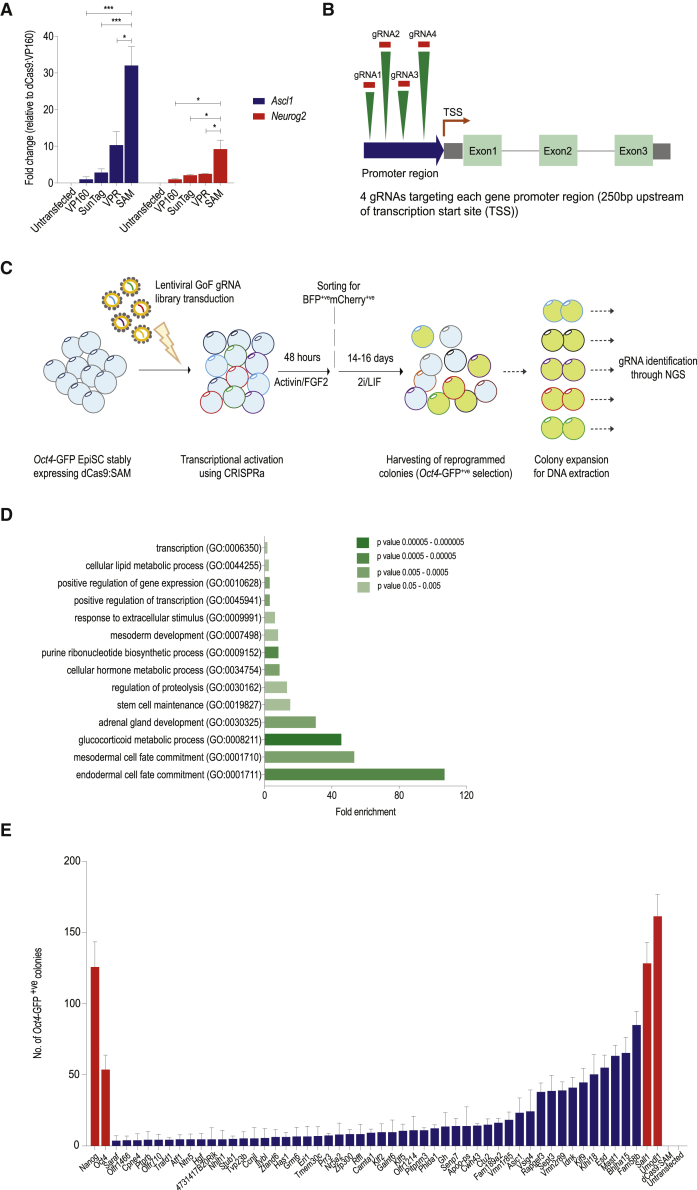
Figure 1.
GoF EpiSC Reprogramming Screening with CRISPRa and sgRNA Library
(A) Activation of Ascl1 and Neurog2 in HEK293 cells. Cells were transfected with one sgRNA per target and four different dCas9 versions. qRT-PCR normalized to Gapdh, fold change relative to dCas9:VP160 (mean of experimental triplicates ± SD, ∗p < 0.05; ∗∗∗p < 0.001).
(B) sgRNA design targeting gene promoters in the murine genome.
(C) Screening strategy in Oct4-GFP EpiSCs stably expressing dCas9:SAM, lentiviral transduction (MOI = 0.3) of the sgRNA library. Reprogramming in 2i/LIF for 14–16 days, after sorting for transduced cells. NGS identified candidate sgRNAs in Oct4-GFP+ve iPSC colonies.
(D) GOTOOLBox analysis of 142 genes identified in GoF screening. Pathways with fold change compared with reference; colors indicate p values.
(E) Validation of 54 genes including Nanog and Oct4 in dCas9:SAM-Oct4-GFP EpiSCs with single sgRNAs (Oct4-GFP+ve iPSC colonies, mean of 3 independent experiments ± SD).
See also Figures S1 and S2 and Tables S5, S6, and S7.
To perform a genome-scale activation screen, we designed a pooled library of 87,863 sgRNAs targeting a 250-bp region upstream of the transcription start site (TSS) of 19,994 genes with an average of 4 guides each (Figure 1B; Table S5).
We decided to use EpiSC derived from Oct4-GFP reporter transgenic mice as they have been used for this purpose before (Yang et al., 2010). Characteristically for EpiSCs these cells already exhibit a baseline Oct4 (and therefore GFP) expression. However, only cells successfully reprogrammed to the naive pluripotent state are able to maintain and increase Oct4 expression upon plating in aforementioned 2i/LIF medium. Thus, successfully reprogrammed Oct4-GFP EpiSCs can be identified by their strong GFP expression (Figure S2A) and the characteristic ESC-like morphology, and grow as distinct colonies, whereas EpiSCs failing to reprogram either detach and die or differentiate.
We stably integrated dCas9:SAM into Oct4-GFP EpiSCs via PiggyBac transposition and then transduced 100 × 106 dCas9:SAM-expressing EpiSCs with our library at a MOI of 0.3 (Figure S2B). Two days later, we used fluorescence-activated cell sorting 10 × 106 to successfully transduce cells by BFP expression, giving a library coverage of around 114-fold. These BFP+ve cells were seeded in 2i/LIF medium to select for reprogramming cells. After 14–16 days of culture in 2i/LIF, 480 GFP+ve colonies were harvested for expansion (Figure 1C). Next-generation sequencing revealed 146 sgRNAs targeting 142 different genes (Table S6). These included known reprogramming factors Nanog (Mitsui et al., 2003), Klf2 (Qiu et al., 2015), and Nr5a2 (Guo and Smith 2010), confirming the specificity of the screen.
GOTERM analysis (Castro et al., 2011) on these 142 genes identified an enrichment in pathways related to transcriptional activation, expression of various transcription factors and enrichment toward stem cell maintenance (Figure 1D; Table S6).
To validate these candidate genes individually, we chose the highest performing sgRNA for each from the library, including Nanog as a positive control and again transduced dCas9:SAM-expressing Oct4-GFP EpiSCs. We expected the validation rate to be no higher than 50%, as small-scale single colony sub-sampling showed an average of two sgRNAs present in most colonies (data not shown), where one sgRNA presumably acts as the driver responsible for reprogramming, while the other is co-amplified as a passenger. As before, GFP+ve ESC-like colonies could be observed for Oct4, Nanog, and 52 of the candidate genes, resulting in a 36% validation rate (Table S7). The efficiency of reprogramming was gene dependent ranging from 5 to 165 colonies per 1 × 106 cells transfected (Figure 1E). Among the genes with the highest colony counts were positive controls Nanog and Oct4, as well as transcription factors Klf2 and Nr5a2 with a known role in reprogramming, confirming the validity of our CRISPRa approach.
Gene Dosage Is Critical for Oct4-Mediated Reprogramming
We observed that CRISPRa-mediated induction of the pluripotency marker Oct4 produced a significant number of ESC-like colonies, contradicting previous studies showing that cDNA-mediated Oct4 expression is inefficient in EpiSC reprogramming (Guo and Smith, 2010, Niwa et al., 2000). Indeed, we were unable to generate any iPSC colonies in our EpiSCs with Oct4 cDNA, while CRISPRa achieved robust reprogramming (Figure 2B).
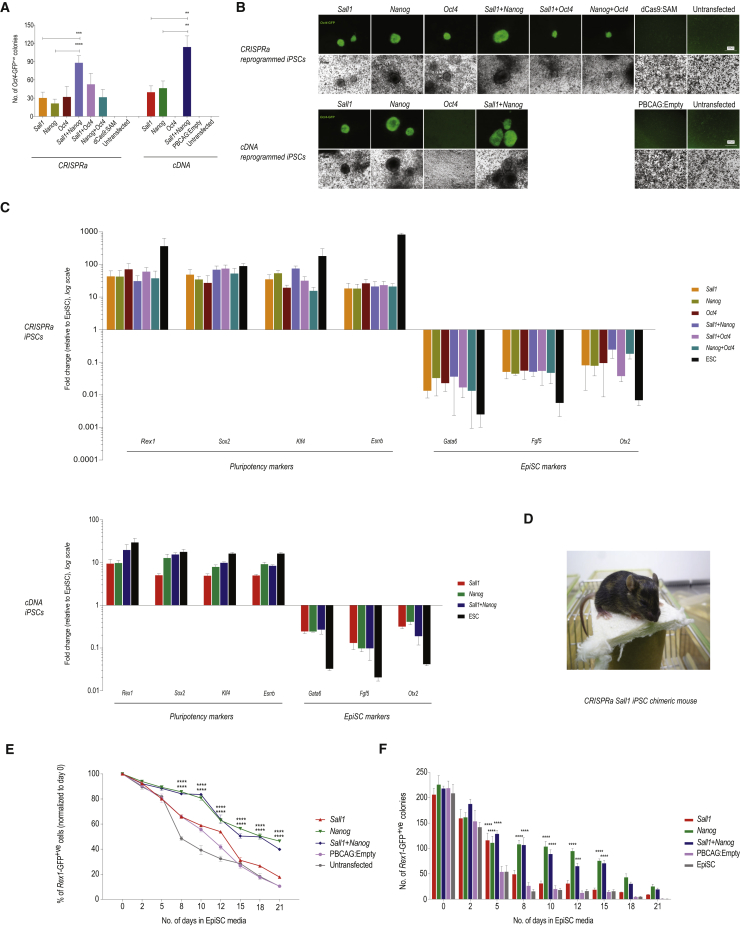
Figure 2.
Sall1 and Nanog Reprogram EpiSCs and Influence ESC Differentiation
(A) Reprogramming efficiencies of Sall1, Nanog, and Oct4 in Oct4-GFP EpiSCs stably transfected with CRISPRa or cDNA (Oct4-GFP+ve colonies, mean of 3 independent experiments ± SD, ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
(B) Morphology of Oct4-GFP+ve colonies at day 20 in 2i/LIF is similar to ESC colonies. No colonies were observed in untransfected or mock transfected EpiSCs.
(C) qRT-PCR expression profiles of pluripotency markers and EpiSC markers in iPSC colonies normalized to Gapdh and relative to EpiSCs (mean of 3 independent experiments ± SD).
(D) Chimeric mouse produced with CRISPRa Sall1-induced PSCs injected into C57B/6 blastocyst.
(E) Flow cytometric analysis of Rex1-GFP+ve cells cultured in EpiSC medium at the timepoints indicated. Cells were stably transfected with Sall1 or Nanog cDNA, or empty vector and cultured in EpiSC medium (mean of 3 independent experiments ± SD, ∗∗∗∗p < 0.0001 versus PBCAG:Empty).
(F) Number of Rex1-GFP+ve ESC colonies recovered after ESCs were converted in EpiSC medium at indicated timepoints (600 cells plated at time point zero; mean of 3 independent experiments ± SD, ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 versus PBCAG:Empty).
We speculated that gene dosage might be the underlying issue and compared endogenous Oct4 induction in EpiSCs with exogenous overexpression in more detail: compared with ESCs, CRISPRa-mediated induction of Oct4 mRNA achieved roughly half the physiological expression level, while exogenous Oct4 cDNA slightly surpassed it (Figure S2C). Doxycycline (Dox) titration of tet-inducible Oct4 resulted in amounts comparable with Oct4 cDNA and only very low concentrations of Dox gave levels similar to CRISPRa. Nevertheless, all cDNA-mediated overexpression conditions still failed to reprogram. On the protein level, all Oct4 cDNA conditions produced disproportionally higher amounts than expected from the mRNA levels (Figure S2D, top panel). CRISPRa, on the other hand, achieved Oct4 protein expression similar to that in ESCs (Figure S2D, bottom panel). We suspect that differences in mRNA stability are the cause, as CRISPRa-driven endogenous mRNA should be physiologically regulated, while exogenous mRNA could be more stable due to differing polyadenylation.
The importance of physiological expression levels agrees with our observation that, although our screening library contained an average of four sgRNAs per gene, almost all candidate genes from our screen were derived from only one specific sgRNA per target. Indeed, sgRNAs showed vastly different activities in a distribution that suggests a dependency on the distance of the sgRNA to the TSS (Figure S2E). This is also supported by a recent report (Liu et al., 2018a), which shows proof-of-principle MEF reprogramming using CRISPRa. In their experiments, only sgRNAs targeting the Oct4 promoter in very specific locations (−71 and −127 bp from TSS) achieved activation sufficient for reprogramming, while in our experiments an sgRNA −101 bp from the TSS was successful.
Sall1 Facilitates EpiSC Reprogramming in Cooperation with Nanog
Umodl1 and Sall1 were the two most potent validated candidates from our screen. We confirmed that Umodl1 upregulates Lifr, Essrb, Nanog, and Sox2, and downregulated Tgfbr1 as would be expected in iPSC reprogramming when medium was switched from EpiSC to 2i/LIF (Figure S2F). Sall1, a member of the Spalt-family of transcription factors, has been reported to cooperate with Nanog to promote the maintenance of ESC state (Karantzali et al., 2011, Novo et al., 2016) and to play an important role in reprogramming and ESC differentiation (Basta et al., 2017, Mansour et al., 2012). However, the downstream targets of Sall1 involved in reprogramming have not been sufficiently explored. Having found that Sall1 is also able to independently reprogram EpiSCs, we set to investigate the underlying mechanisms.
First, we asked whether Sall1 and Nanog also act synergistically in EpiSC reprograming by overexpressing them individually and in combination in Oct4-GFP EpiSCs. We performed these experiments both with CRISPRa as well as cDNA-mediated overexpression, and also verified that the observed activity of Sall1-specific sgRNA was not due to cross-reactivity with Sall4, a known pluripotency factor (Figure S2G).
Three days after transfection, qRT-PCR showed a 2.5- to 3.5-fold increase in expression of Sall1, Nanog, and Oct4 mediated by CRISPRa and a 10- to 20-fold increase in expression through cDNA (Figure S3A). Co-expression of Sall1 and Nanog resulted in a marked increase in Oct4-GFP+ve ESC-like colony numbers (Figures 2A and 2B). As above, Oct4 induction via CRISPRa successfully reprogrammed EpiSCs (but not cDNA overexpression), without showing significant synergy with either Sall1 or Nanog. Pluripotency markers examined by qRT-PCR (Rex1, Sox2, Klf4, and Essrb) were markedly increased; concordantly, EpiSC markers Gata6, Fgf5, and Otx2 showed decreased expression (Figure 2C). Sall1 reprogrammed EpiSCs (MF1 and C57BL/6 background) contributed significantly to chimeras when injected into C57BL/6 blastocysts (Figures 2D and S3E).
To exclude the possibility that the baseline GFP expression of the Oct4-GFP reporter EpiSCs might skew the correct identification of successfully reprogrammed EpiSCs, we repeated these experiments with Nanog-GFP reporter EpiSCs (Yang et al., 2010), which show strong GFP expression on successfully entering the naive ESC state, but virtually none in the primed EpiSC state (Guo and Smith, 2010). Both gene induction using CRISPRa and overexpression via cDNA confirmed the reprogramming capability of Sall1 alone and in synergy with Nanog (Figures S3B–S3D). Notably, colony formation assays in 2i/LIF recapitulated the results obtained with Oct4-GFP EpiSCs and the reprogramming kinetics as measured in time course experiments were comparable between the two reporter cell lines (Figure S3F).
Sall1 and Nanog Delay Differentiation of ESCs into EpiSCs
ESCs readily differentiate into EpiSCs in culture medium containing the EpiSC self-renewal factors Activin and fibroblast growth factor 2 (FGF2) (Guo et al., 2009). To investigate whether higher levels of Sall1 and Nanog can delay this conversion we generated stable cDNA transfectants in Rex1-GFP ESCs (Wang et al., 2011). We cultured the cells in EpiSC media and quantified the Rex1-GFP+ve population as a measure of cells remaining in the ESC ground state in a 21-day time course. Nanog and Sall1+Nanog maintained a significantly higher proportion of GFP+ve cells than Sall1 (Figure 2E). The expression of naive pluripotency and EpiSC markers analyzed by qRT-PCR followed a similar pattern (Figures S3G–S3I), although Sall1 delayed upregulation of differentiation markers Fgf5 and Otx2. Concordantly, when plated in 2i/LIF medium, Nanog and Sall1+Nanog overexpressing cells retained the ability to form ESC colonies through most of the time course, and Sall1 preserved colony formation capacity until after 6 days (Figure 2F). While Sall1 might not have the same capacity as Nanog to keep the ESC ground state, it may confer a longer “formative state” (Smith, 2017) during conversion.
Sall1 Promotes MEF Reprogramming and Works Synergistically with Nanog
To test whether Sall1 enhances somatic cell reprogramming, we stably transfected Oct4-GFP reporter MEFs (Oct4-GFP-MEFs) with the Yamanaka factors (CAG4F, Figure S1), dCas9:SAM and sgRNAs against Sall1 and/or Nanog. In ESC media, Sall1 sgRNA transfected MEFs produced a significantly higher number of Oct4-GFP+ve and alkaline phosphatase-positive (AP+ve) colonies (Figures 3A and S4A) with ESC-like morphology (Figure S4B) than CAG4F alone, mirroring the results obtained from EpiSC reprogramming, including synergy between Sall1 and Nanog.
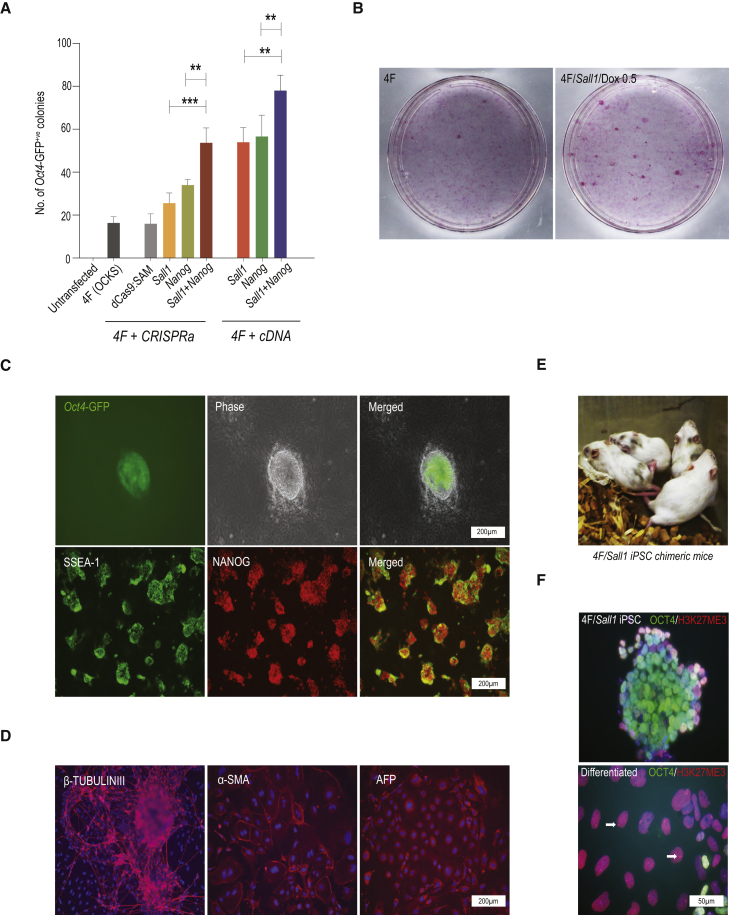
Figure 3.
CRISPRa Gene Induction and cDNA-Mediated Overexpression of Sall1, Nanog Reprogrammed MEF to iPSCs
(A) (4F + CRISPRa) MEFs stably transfected with CAG4F and gRNAs against Sall1/Nanog/Sall1+Nanog in ESC medium (Oct4-GFP+ve colonies after 18 days; mean of 3 independent experiments ± SD, ∗∗p < 0.01; ∗∗∗p < 0.001) (4F + cDNA) MEFs stably transfected with TRE4F, TRENanog, and TRESall1 (all co-transfected with PBEF-1αTet3G), induced with 0.5 μg/mL Dox for 12 days and counted on day 18 (mean of 3 independent experiments ± SD, ∗∗p < 0.01).
(B) Alkaline phosphatase-positive (AP+ve)-stained ESC colonies reprogrammed from MEFs by 4F alone and in combination with Sall1 (induced with Dox at 0.5 μg/mL).
(C) iPSCs reprogrammed from C57B/6J MEF with 4F/Sall1. Oct4-GFP expression and ESC-like morphology (upper panel), immunofluorescent staining for pluripotency markers SSEA-1 and NANOG (lower panel).
(D) In vitro differentiation of C57B/6 MEF reprogrammed iPSCs with 4F/Sall1; neuronal differentiation in N2B27 (immunofluorescence for β-tubulin III+); mesoderm and endoderm differentiation in M10 (alpha smooth muscle actin [α-SMA] and alpha fetoprotein [AFP] antibody staining).
(E) Chimeric mice produced with 4F/Sall1-iPSCs injected into CD1 blastocysts.
(F) Inactivation of X chromosomes in female 4F/Sall1-iPSCs (co-immunostaining for H3K27me3 and Oct4. Arrows indicate H3K27me3 foci).
To examine the dynamics of MEF reprogramming, we co-transfected Oct4-GFP-MEFs with tet-inducible Sall1 (TRESall1, Figure S1; Table S2) and CAG4F, and induced expression with three concentrations of Dox (0.1, 0.5, and 1.0 μg/mL) to find a suitable concentration to mediate reprogramming in ESC medium. After 18 days, Dox concentrations of 0.5 or 1.0 μg/mL resulted in a significant 2- to 3-fold increase in Oct4-GFP+ve and AP+ve colonies (Figures 3B and S4C) and we chose 0.5 μg/mL Dox for all subsequent experiments. To determine the active window for Sall1, we induced expression at 0, 2, 4, 6, 8, 10, and 12 days of reprogramming and found that only activation during the first 4 days resulted in higher reprogramming efficiency (Figure S4D).
As Nanog has been reported to promote MEF reprogramming (Theunissen et al., 2011) we tested for synergy with Sall1 by transfecting MEFs with tet-inducible Yamanaka factors, Nanog and Sall1 (TRE4F, TRENanog, and TRESall1). Indeed, co-expression of Sall1+Nanog/4F led to a 1.5-fold increase in colony number compared with either factor alone (Figures 3A and S4A).
The Sall1-iPSCs derived from these experiments were morphologically similar to ESCs with a compact dome-like structure and Oct4-GFP expression. Immunofluorescent staining of these iPSCs showed protein expression of the ESC-markers SSEA-1 and NANOG (Figure 3C). In differentiation medium (DMEM/10% fetal calf serum [FCS]) or N2B27 medium (Ying et al., 2003), these iPSCs exited ground state pluripotency and differentiated into mesoderm, endoderm, and ectoderm lineages as confirmed by immunofluorescent staining for expression of smooth muscle actin, alpha fetoprotein, and β-tubulin III (Figure 3D). In addition, when we injected these iPSCs into blastocysts, live chimeras were born (Figure 3E), confirming the pluripotency of these Sall1-iPSCs.
Female mESCs have two activated X chromosomes when maintained at ground state (Lessing et al., 2013) and randomly inactivate one of them once they undergo differentiation. Staining with anti-H3K27me3 antibody detects this event as foci on the inactivated X chromosome (Silva et al., 2008). We derived iPSCs from female MEFs by co-transfecting with 4F/Sall1 as before and then differentiated them in DMEM/10% FCS for 5 days. Loss of Oct4 expression demonstrated successful differentiation and the presence of H3K27me3 foci indicated X chromosome silencing. In contrast iPSC cultured in 2i/LIF strongly expressed Oct4 protein and lacked any H3K27me3 foci (Figure 3F).
Together, this demonstrates that Sall1 can enhance 4F-driven somatic cell reprogramming and that 4F/Sall1 reprogrammed iPSCs are naive and pluripotent.
Yamanaka factors Oct4, Sox2, and Klf4 are essential and sufficient for reprogramming, albeit at a lower efficiency than in conjunction with c-Myc; all three can be replaced by other transcription factors or small molecules such as Gata3 (Shu et al., 2013) or valproic acid (Biswas and Jiang, 2016, Huangfu et al., 2008). However, in co-transfection experiments, Sall1 was unable to substitute for any of the factors in MEFs (Figures S4E–S4G).
RNA Sequencing Identifies Potential Mechanisms of Cellular Reprogramming Mediated by Sall1 and Nanog
We performed RNA sequencing (RNA-seq) for Oct4-GFP EpiSCs overexpressing Sall1 and/or Nanog via cDNA for 24 h. Our analysis identified 372 genes differentially expressed specific to Sall1-transfected cells, and 307 genes specific to Nanog. We observed a large overlap of 568 genes (45%) between both sets (Figure 4A; Table S8) and GOTERM analysis (Castro et al., 2011) revealed that they are involved in a number of developmental processes and signaling cascades (Figure 4B; Table S8).
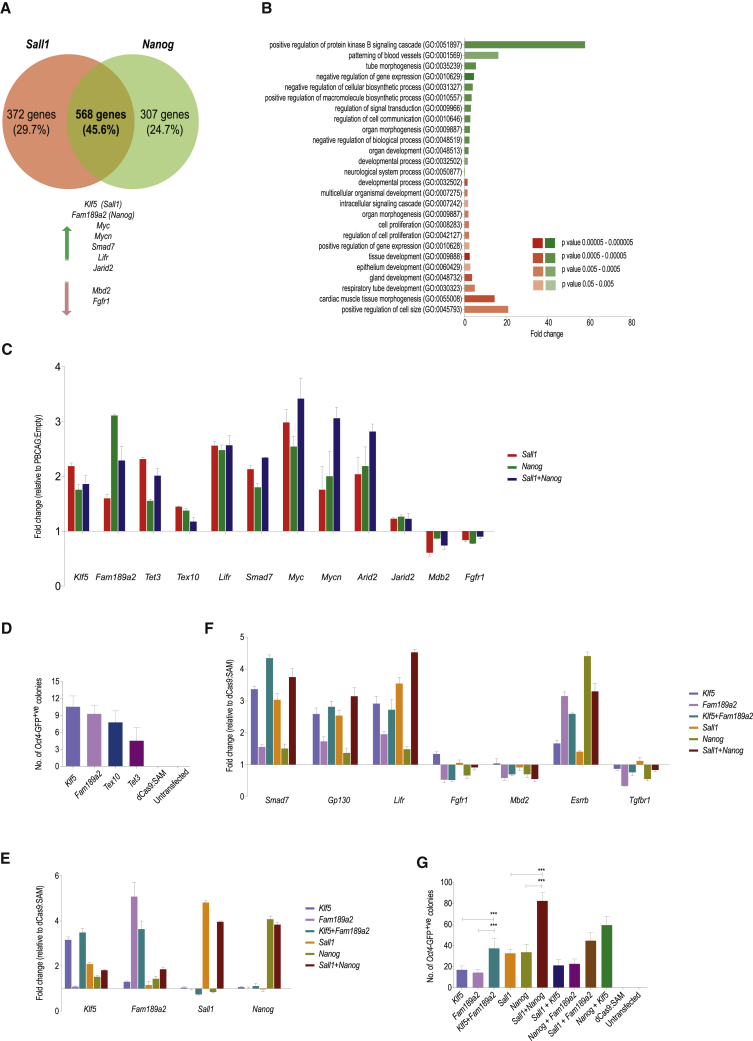
Figure 4.
RNA-Seq Identifies Potential Mechanisms of Reprogramming Mediated by Sall1 and Nanog
(A) Venn diagram of genes being differentially expressed in Sall1 and Nanog overexpressing cells (cutoff padj < 0.001). Upregulated (green arrow) and downregulated (red arrow) genes for further experiments were chosen from the overlap between Sall1 and Nanog, except Klf5 and Fam189a2.
(B) GOTOOLBox analysis of common regulated genes (fold change compared with reference, colors indicate p values).
(C) qRT-PCR validations of RNA-seq (24 h after transfection, normalized to Gapdh and relative to PBCAG:Empty; mean of 3 independent experiments ± SD).
(D) Reprogramming of Oct4-GFP EpiSCs via CRISPRa-mediated gene induction of Klf5, Fam189a2, Tex10, and Tet3 (Oct4-GFP+ colonies after 2i/LIF for 20 days; mean of 3 independent experiments ± SD).
(E) qRT-PCR for Klf5, Fam189a2, Sall1, and Nanog after CRISPRa-mediated induction of Klf5 and Fam189a2 (flow-sorted for sgRNA expression after 24 h, normalized to Gapdh and relative to dCas9:SAM; mean of 3 independent experiments ± SD).
(F) qRT-PCR expression levels of key regulators in JAK/STAT3 and TGF-β signaling (flow-sorted for sgRNA expression 24 h after changing to 2i/LIF media, 48 h after transfection, normalized to Gapdh and relative to dCas9:SAM; mean of 3 independent experiments ± SD).
(G) Reprogramming of Oct4-GFP EpiSCs via CRISPRa-mediated gene induction of Klf5, Fam189a2, Sall1, and Nanog (Oct4-GFP+ve colonies after 20 days in 2i/LIF; error bars represent mean of 3 independent experiments ± SD).
Among those commonly regulated genes were Myc, Mycn, Tet3, Tex10, Jarid2, Fgfr1, Mbd2, Lifr, and Smad7 (Figure 4A) which have previously been implicated in the promotion of cellular reprogramming or inhibition of ESC differentiation (Li et al., 2016, Iseki et al., 2016, Bagci and Fisher, 2013, Fidalgo et al., 2016, Jinek et al., 2012, Niwa et al., 1998, Hall et al., 2009, Festuccia et al., 2017). Upregulation of Lifr and downregulation of Fgfr1 is expected in EpiSC reprogramming and validates our RNA-seq and qRT-PCR data (Figure 4C). Furthermore, we found 215 genes that were only regulated when Sall1 and Nanog were overexpressed together (Figure S4H; Table S8), such as Dnmt3c and Hdac9, reported to be involved in the epigenetic regulation of male germ cell maintenance (Barau et al., 2016) and muscle differentiation (Mihaylova and Shaw, 2013), respectively; as well as a modest upregulation of Utf1, a transcription factor known to synergize with the Yamanaka factors in reprogramming (Zhao et al., 2008).
We had already independently identified the genes Klf5 and Fam189a2 in our GoF screen (Figure 1E; Table S5) and RNA-seq showed them to be potentially regulated by Sall1 and Nanog, respectively. We validated the RNA-seq results with qRT-PCR and found a good correlation between both methods (Figures 4C and S4I). While Klf5 narrowly failed the stringent p value cutoff for the RNA-seq results in Nanog overexpressing cells, qRT-PCR indicated that Klf5 may be regulated by Nanog as well, albeit to a lesser extent than by Sall1. Fam189a2 on the other hand seemed to be regulated significantly stronger by Nanog than Sall1. When we co-expressed Sall1 and Nanog, we did not observe a significant synergistic increase of expression for these downstream targets (Figure 4C); we did, however, for the genes Myc, Mycn (Chappell and Dalton 2013), and Arid2, all of which have been shown to play a role in reprogramming and chromatin remodeling (Awe and Byrne, 2013, Singhal et al., 2010).
We used CRISPRa to induce expression of Klf5, Fam189a2, Tex10, and Tet3 in Oct4-GFP EpiSCs and found that all were able to augment reprogramming into iPSCs (Figure 4D). Reprogramming by Fam189a2 occurred in 10 days, while Klf5, Tex10, and Tet3 required between 14 and 20 days. In all cases, the number of reprogrammed colonies was significantly lower compared with Sall1 or Nanog (Figure 4G), which may indicate that multiple downstream targets of Sall1 and Nanog participate in reprogramming.
We tested the regulatory relationship between Sall1+Nanog and Klf5+Fam189a2 by transfecting EpiSCs with CRISPRa for Klf5 and Fam189a2. Transcription increased significantly for Klf5 and Fam189a2, but not for Sall1 and Nanog, indicating Klf5 and Fam189a2 are downstream targets (Figure 4E). qRT-PCR for some of the key genes differentially regulated in the RNA-seq data showed that both Sall1 and Klf5 upregulated Smad7, Gp130, and Lifr, suggesting the repression of transforming growth factor β (TGF-β) signaling and activation of Jak/Stat3 signaling. Nanog and Fam189a2 on the other hand downregulated Fgfr1, Tgfr1, and Mbd2 and upregulated Esrrb expression, indicating the repression of FGF and TGF-β signaling, inhibition of epigenetic repression and promotion of self-renewal and pluripotency (Figure 4F). Functionally, co-activation of both Klf5 and Fam189a2 generated significantly more Oct4-GFP+ve colonies than either gene alone. As expected, co-activation of either Sall1 and its downstream target Klf5 or Nanog and its downstream target Fam189a2 showed no synergistic effects in Oct4-GFP+ve colony production, whereas co-activation of either Sall1 and Fam189a2 or Nanog and Klf5 did, although less than Sall1 and Nanog co-activation. These results suggest that Klf5 and Fam189a2 are situated downstream of Sall1 and Nanog, respectively, and can synergize as well (Figure 4G).
Discussion
To date few CRISPR activation screens have been performed (Bester et al., 2018, Heaton et al., 2017, Liu et al., 2018b) using previously established GoF libraries (Kampmann, 2018, Konermann et al., 2015). However, none of them targeted stem cell reprogramming and, while some recent publications have used CRISPRa in this field of research, they have been restricted to a few genes to demonstrate proof-of-concept (Guo et al., 2017, Liu et al., 2018a, Weltner et al., 2018).
Our present study shows that a genome-scale CRISPRa screen, in conjunction with an experimental model such as EpiSCs, in which a single overexpressed gene may mediate reprogramming to pluripotency, is a powerful tool for gene discovery. We identified 142 candidate reprogramming factors, among them Nanog, known to reprogram EpiSCs to iPSCs (Silva et al., 2009), validating our screen. Similarly, we found the Yamanaka factor Oct4 (Takahashi and Yamanaka, 2006), which is critical for the maintenance of ESCs and differentiation (Niwa et al., 2000). Curiously, CRISPRa-induced Oct4 readily and robustly reprogrammed EpiSCs into iPSCs, while overexpression via cDNA failed in our experiments and those of others (Guo and Smith, 2010). We reason that gene dosage is one critical aspect to explain this behavior and that excessive levels of Oct4 can be detrimental to pluripotency, which tallies with previous studies suggesting that artificially reduced Oct4 levels maintain ESCs in a robust pluripotent state, whereas wild-type levels enable differentiation (Gao et al., 2013, Karwacki-Neisius et al., 2013, Radzisheuskaya et al., 2013).
The important implication for CRISPRa-mediated screens is that tiled sgRNAs in regulatory regions of genes can, as we and others show (Liu et al., 2018a), provide a variety of expression levels unachievable with exogenous cDNA, giving a higher probability of matching the physiological gene dosage. Conceivably, the choice of the CRISPRa system may well influence the outcome of a screen and repeating our screen with a different CRISPRa system at lower activation efficiencies than SAM could produce a non-redundant list of candidate genes.
While the positional aspect of sgRNA efficiency certainly serves to explain why most of our candidate genes were only identified by a single sgRNA in our screen, we also acknowledge that reprogramming is inherently a very inefficient process and, thus, a very large initial cell number may be required to cover a genome-wide library deeply enough to give a sufficient number of cells a chance to gain pluripotency. While we performed our GoF screen with 10 × 106 library-transduced cells (library coverage 114×), a deeper coverage or a more focused library promises to uncover reprogramming candidates the present screen might have missed.
Our screen identified Sall1, a member of the Spalt-like gene family, as a potent EpiSC reprogramming factor. Sall1 and Sall4 have been implicated in the establishment of pluripotency (Gaspar-Maia et al., 2013) in studies showing that the action of demethylase Utx on Sall1 and Sall4 is required to enable MEF reprogramming (Mansour et al., 2012). Furthermore, it has been demonstrated that Sall4 activates Oct4 expression while Sall1 is a direct binding partner of Nanog (Karantzali et al., 2011, Zhang et al., 2006) and has been suggested to be required in Nanog-mediated open heterochromatin maintenance within ESCs and EpiSCs (Novo et al., 2016). So far, it is unclear whether Sall1 plays an active role in EpiSC reprogramming. In our work, we demonstrate that endogenous as well as exogenous Sall1 can reprogram EpiSCs, and that Sall1 synergizes with Nanog in reprogramming EpiSCs and MEFs. However, Sall1 cannot replace Oct4, Sox2, or Klf4 in MEF reprogramming, suggesting that it is unable to initiate the reprogramming machinery in more differentiated cells. One of its roles may be in facilitating epigenetic modification and nucleosome remodeling, e.g., through interaction with Nanog and the deacetylase complex (NurD) (Basta et al., 2017).
Unlike Nanog, the ability of Sall1 to reprogram EpiSCs is insufficient to keep ESCs in the naive pluripotent state, only marginally delaying loss of pluripotency in differentiation experiments. However, it slowed expression of EpiSC markers Fgf5 and Otx2, and preserved the ability to generate ESC-like colonies. Sall1 inhibited Otx2 expression in embryoid body differentiation of ESCs, and a formative pluripotent phase between naive and primed states was postulated when cells lost naive pluripotency markers and gained post-implantation markers such as Otx2 and Oct6 among others (Karantzali et al., 2011, Smith, 2017). Considering that, even after 21 days in differentiation medium, some Sall1 overexpressing cells still formed ESC-like colonies in 2i/LIF, these cells may be stalled in a formative state.
We used RNA-seq to identify downstream targets of Sall1 and Nanog in EpiSCs and found genes previously implicated in pluripotency or stem cell maintenance. Esrrb, a downstream target of Nanog, plays an important role in maintaining ESCs pluripotency and reprogramming by interacting with the core pluripotency network via Sox2 (Adachi et al., 2013). Tex10 was recently reported to be a pluripotency factor and partner of Sox2, capable of promoting MEF reprogramming (Ding et al., 2015), a role we further extended to EpiSC reprogramming. Tet3 is a member of the ten-eleven translocation (Tet) protein family, which regulate DNA methylation. Tet1 and Tet2 have already been implicated in somatic reprogramming and Tet2 has been reported to promote EpiSCs to a naive state (Bagci and Fisher, 2013, Fidalgo et al., 2016). Here, we show that Tet3 can mediate EpiSC reprogramming as well.
The Kruppel-like factor family proteins Klf2, 4, and 5 are also pluripotency factors and both Klf2 and Klf4 have been shown to facilitate reprogramming (Jeon et al., 2016). The potential of Klf5 however is unclear as it has been reported to be incapable of reprogramming EpiSCs in a study by Hall et al. (2009), while Jeon et al. (2016) and recently Azami et al. (2018) both were able to derive iPSCs from EpiSCs by cDNA-mediated Klf5 overexpression. We identified Klf5 in our GoF screen and confirmed its ability to reprogram EpiSCs via CRISPRa transcriptional activation. These incongruent observations may reflect a Goldilocks effect similar to our observations with Oct4, highlighting the utility of different overexpression approaches to discover new pluripotency factors. LIF-dependent activation of Jak/Stat3 and its role in ESC self-renewal and reprogramming has been widely studied to date (Tang and Tian, 2013, Yu et al., 2017). Overexpression of Klf5 via cDNA may compensate for the absence of LIF in maintaining ESC pluripotency and thereby be capable of reprogramming EpiSCs via LIF-independent pathways (Ema et al., 2008, Jeon et al., 2016). Besides Klf5, our data also indicate that Sall1 positively and negatively regulates the Jak/Stat3 and TGF-β pathways, respectively (via Gp130, Lif receptor, and Smad7), together providing insights into the role of Sall1 in EpiSC reprogramming.
Fam189a2 was identified as a new target of Nanog in EpiSC reprogramming and our data showed that both Nanog and Fam189a2 downregulate Tgfbr1 and upregulate Esrrb expression. We postulate that the observed synergy between Sall1 and Nanog as well as their downstream effectors Klf5 and Fam189a2 is partially due to the combined activation of Jak/Stat3, suppression of TGF-β signaling and upregulation of pluripotent genes such as Esrrb.
In conclusion, using a genome-scale CRISPR activation screen in the well-established EpiSCs reprogramming model, we identify known and previously unknown genes that can mediate cellular reprogramming in EpiSCs. We demonstrate that the transcription factor Sall1 can effectively reprogram EpiSCs and MEFs, and provide new insights into the role of Sall1 in promoting and maintaining pluripotency. Other reprogramming candidates such as transcription factors Atf1 and Bhlha15, kinases Idnk and Has1, several olfactory receptor genes (Olfr), and others with less known functions such as Umodl1 and Prr3 deserve further in-depth investigation. Our studies demonstrate the strengths of CRISPR activation screens in the identification of factors that were previously not reported in molecular reprogramming and in illuminating biological pathways.
Experimental Procedures
GoF gRNA Library Design
In brief, the GoF library targeted the region of up to 250 bp upstream of the TSS of each protein-coding gene. Up to 4 guides of 19 bp length were selected per gene. Guide sequences with off-target sites exhibiting fewer than 3 mismatches over their 19 bp length were omitted from the design.
A selection algorithm was designed to spread high-quality guides across the target region. To this end, the region upstream of the TSS was divided into quarters of roughly equal length. Starting with the quarter closest to the TSS the algorithm looped over quarters picking the best guide, by quality score, in each if available, and adding it to the library until no more guide fitting the constraints could be found, or the target number of five guides per genes was reached. A constraint for the GC content of less than 55% was applied in the first loop and then relaxed to 70%.
GoF Reprogramming Screen
The GoF sgRNA library was synthesized by Custom Array, and the oligo pools were cloned into the lentiviral sgRNA expression plasmid via Gibson assembly as described by Shalem et al. (2014), with minor modifications.
In brief, Oct4-GFP EpiSC cells stably expressing dCas9:SAM were first generated and were expanded to 100 × 106 cells for lentiviral transduction of the GoF library. Library transduction was carried out at an MOI of 0.3. After 2 days, 10 × 106 BFP+ve Oct4-GFP EpiSCs were sorted by flow cytometry and plated in 2i/LIF in order to allow selection for reprogrammed cells. After 14–16 days in 2i/LIF, the individual reprogrammed colonies, verified by Oct4-GFP fluorescence, were picked and transferred to 96-well plates for colony expansion and genomic DNA extraction. PCR amplification on the genomic DNA, across the stably integrated sgRNA, was performed using primers described previously (Koike-Yusa et al., 2014) and NGS was used to identify the sgRNA sequences.
All experimental procedures are detailed out in the Supplemental Information.
Author Contributions
J.Y. initiated and designed the project, performed GoF screening and EpiSC, MEF reprogramming with cDNA, analyzed the data, and wrote the manuscript. S.S.R. performed EpiSC reprogramming, ESC conversion, MEF reprogramming with gRNA, RNA-seq, analyzed the data, and wrote the manuscript. M.J.F. performed EpiSC reprogramming and RNA-seq data analysis, and wrote the manuscript. G.L. and X.Z. performed microinjection and analyzed chimera data. H.P. designed the GoF sgRNA library and performed RNA-seq data analysis. D.A.G. performed experiments to determine various dCas9-mediated overexpression levels. P.L. and A.B. financially supported the project and interpreted the data. E.M. initiated, designed, and supervised the project, performed GoF screening, validation, analyzed and interpreted data, and wrote the manuscript.
Acknowledgments
We thank Caroline Sinclair, Michael Woods, Evelyn Grau, and Brendan Doe from the research support facility (RSF) at the Sanger Institute for providing blastocysts. We also thank Prof. Austin Smith and Dr. Ge Guo from Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute for the reagents. This work was funded by the Wellcome Trust (WT098051). J.Y. is funded by the National Science Foundation of China (NSFC) (31871491). E.M. is funded by the UK Dementia Research Institute (RG86445) and Parkinson's Disease UK (F-1501).
Published: March 21, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.02.010.
Accession Numbers
The accession number for the data reported in this paper is ArrayExpress: E-MTAB-7692.
Supplemental Information
References
- Adachi K., Nikaido I., Ohta H., Ohtsuka S., Ura H., Kadota M., Wakayama T., Ueda H.R., Niwa H. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol. Cell. 2013;52:380–392. doi: 10.1016/j.molcel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Awe J.P., Byrne J.A. Identifying candidate oocyte reprogramming factors using cross-species global transcriptional analysis. Cell Reprogram. 2013;15:126–133. doi: 10.1089/cell.2012.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azami T., Matsumoto K., Jeon H., Waku T., Muratani M., Niwa H., Takahashi S., Ema M. Klf5 suppresses ERK signaling in mouse pluripotent stem cells. PLoS One. 2018;13:e0207321. doi: 10.1371/journal.pone.0207321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagci H., Fisher A.G. DNA demethylation in pluripotency and reprogramming: the role of tet proteins and cell division. Cell Stem Cell. 2013;13:265–269. doi: 10.1016/j.stem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., Herault Y., Guillou F., Bourc'his D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909–912. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- Basta J.M., Robbins L., Denner D.R., Kolar G.R., Rauchman M. A Sall1-NuRD interaction regulates multipotent nephron progenitors and is required for loop of Henle formation. Development. 2017;144:3080–3094. doi: 10.1242/dev.148692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester A.C., Lee J.D., Chavez A., Lee Y.R., Nachmani D., Vora S., Victor J., Sauvageau M., Monteleone E., Rinn J.L. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–664.e20. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Jiang P. Chemically induced reprogramming of somatic cells to pluripotent stem cells and neural cells. Int. J. Mol. Sci. 2016;17:226. doi: 10.3390/ijms17020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro D.S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L.G., Hunt C. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J., Dalton S. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb. Perspect. Med. 2013;3:a014381. doi: 10.1101/cshperspect.a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Huang X., Shao N., Zhou H., Lee D.F., Faiola F., Fidalgo M., Guallar D., Saunders A., Shliaha P.V. Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Ema M., Mori D., Niwa H., Hasegawa Y., Yamanaka Y., Hitoshi S., Mimura J., Kawabe Y., Hosoya T., Morita M. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Festuccia N., Owens N., Navarro P. Esrrb, an estrogen-related receptor involved in early development, pluripotency, and reprogramming. FEBS Lett. 2017 doi: 10.1002/1873-3468.12826. [DOI] [PubMed] [Google Scholar]
- Fidalgo M., Huang X., Guallar D., Sanchez-Priego C., Valdes V.J., Saunders A., Ding J., Wu W.S., Clavel C., Wang J. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016;19:355–369. doi: 10.1016/j.stem.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yang J., Tsang J.C., Ooi J., Wu D., Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Reports. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Qadeer Z.A., Hasson D., Ratnakumar K., Leu N.A., Leroy G., Liu S., Costanzi C., Valle-Garcia D., Schaniel C. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle S., Pan Y., Landrette S., Xu T. piggyBac insertional mutagenesis screen identifies a role for nuclear RHOA in human ES cell differentiation. Stem Cell Reports. 2015;4:926–938. doi: 10.1016/j.stemcr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Ma D., Huang R., Ming J., Ye M., Kee K., Xie Z., Na J. An inducible CRISPR-ON system for controllable gene activation in human pluripotent stem cells. Protein Cell. 2017;8:379–393. doi: 10.1007/s13238-016-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Heaton B.E., Kennedy E.M., Dumm R.E., Harding A.T., Sacco M.T., Sachs D., Heaton N.S. A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep. 2017;20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., Elledge S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Iseki H., Nakachi Y., Hishida T., Yamashita-Sugahara Y., Hirasaki M., Ueda A., Tanimoto Y., Iijima S., Sugiyama F., Yagami K. Combined overexpression of JARID2, PRDM14, ESRRB, and SALL4A dramatically improves efficiency and kinetics of reprogramming to induced pluripotent stem cells. Stem Cells. 2016;34:322–333. doi: 10.1002/stem.2243. [DOI] [PubMed] [Google Scholar]
- Jeon H., Waku T., Azami T., Khoa le T.P., Yanagisawa J., Takahashi S., Ema M. Comprehensive identification of Kruppel-like factor family members contributing to the self-renewal of mouse embryonic stem cells and cellular reprogramming. PLoS One. 2016;11:e0150715. doi: 10.1371/journal.pone.0150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2018;13:406–416. doi: 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E., Lekakis V., Ioannou M., Hadjimichael C., Papamatheakis J., Kretsovali A. Sall1 regulates embryonic stem cell differentiation in association with nanog. J. Biol. Chem. 2011;286:1037–1045. doi: 10.1074/jbc.M110.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacki-Neisius V., Goke J., Osorno R., Halbritter F., Ng J.H., Weisse A.Y., Wong F.C., Gagliardi A., Mullin N.P., Festuccia N. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H., Li Y., Tan E.P., Velasco-Herrera Mdel C., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing D., Anguera M.C., Lee J.T. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- Li Q., Lex R.K., Chung H., Giovanetti S.M., Ji Z., Ji H., Person M.D., Kim J., Vokes S.A. The pluripotency factor NANOG binds to GLI proteins and represses hedgehog-mediated transcription. J. Biol. Chem. 2016;291:7171–7182. doi: 10.1074/jbc.M116.714857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Chen M., Liu Y., Qi L.S., Ding S. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 2018;22:252–261.e4. doi: 10.1016/j.stem.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Liu Y., Yu C., Daley T.P., Wang F., Cao W.S., Bhate S., Lin X., Still C., 2nd, Liu H., Zhao D. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell. 2018;23:758–771.e8. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Gafni O., Weinberger L., Zviran A., Ayyash M., Rais Y., Krupalnik V., Zerbib M., Amann-Zalcenstein D., Maza I. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Metzakopian E., Strong A., Iyer V., Hodgkins A., Tzelepis K., Antunes L., Friedrich M.J., Kang Q., Davidson T., Lamberth J. Enhancing the genome editing toolbox: genome wide CRISPR arrayed libraries. Sci. Rep. 2017;7:2244. doi: 10.1038/s41598-017-01766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova M.M., Shaw R.J. Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol. Metab. 2013;24:48–57. doi: 10.1016/j.tem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Novo C.L., Tang C., Ahmed K., Djuric U., Fussner E., Mullin N.P., Morgan N.P., Hayre J., Sienerth A.R., Elderkin S. The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev. 2016;30:1101–1115. doi: 10.1101/gad.275685.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsker M., Ford N.R., Jenq H.T., Lemischka I.R. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D., Ye S., Ruiz B., Zhou X., Liu D., Zhang Q., Ying Q.L. Klf2 and Tfcp2l1, two Wnt/beta-catenin targets, act synergistically to induce and maintain naive pluripotency. Stem Cell Reports. 2015;5:314–322. doi: 10.1016/j.stemcr.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A., Chia Gle B., dos Santos R.L., Theunissen T.W., Castro L.F., Nichols J., Silva J.C. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat. Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmaier S.E., Telugu B.P. MicroRNA-mediated reprogramming of somatic cells into induced pluripotent stem cells. Methods Mol. Biol. 2015;1330:29–36. doi: 10.1007/978-1-4939-2848-4_3. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W., Tang X., Yang H., Shen L., Zuo X. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N., Graumann J., Wu G., Arauzo-Bravo M.J., Han D.W., Greber B., Gentile L., Mann M., Scholer H.R. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Gilbert L.A., Qi L.S., Weissman J.S., Vale R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Tian X.C. JAK-STAT3 and somatic cell reprogramming. JAKSTAT. 2013;2:e24935. doi: 10.4161/jkst.24935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L.S., Rad R., Guo G., Zhang S. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. U S A. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltner J., Balboa D., Katayama S., Bespalov M., Krjutskov K., Jouhilahti E.M., Trokovic R., Kere J., Otonkoski T. Human pluripotent reprogramming with CRISPR activators. Nat. Commun. 2018;9:2643. doi: 10.1038/s41467-018-05067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., van Oosten A.L., Theunissen T.W., Guo G., Silva J.C., Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Yu Y., Gu S., Li W., Sun C., Chen F., Xiao M., Wang L., Xu D., Li Y., Ding C. Smad7 enables STAT3 activation and promotes pluripotency independent of TGF-beta signaling. Proc. Natl. Acad. Sci. U S A. 2017;114:10113–10118. doi: 10.1073/pnas.1705755114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tam W.L., Tong G.Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yin X., Qin H., Zhu F., Liu H., Yang W., Zhang Q., Xiang C., Hou P., Song Z. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.