CdOppA and CdAppA are putative extracellular peptide-binding proteins serving cognate ABC transporters in Clostridium difficile. Using purified proteins, no evidence was found for (i) peptide binding to either protein in Thermofluor experiments or (ii) the presence of peptides in the crystal structure of CdAppA. Further sequence analysis suggests that CdOppA is a nickel-transporter protein and that CdAppA may be involved in the transport of a restricted set of peptides.
Keywords: Clostridium difficile, peptide transport, OppA, AppA, sporulation
Abstract
Peptides play an important signalling role in Bacillus subtilis, where their uptake by one of two ABC-type oligopeptide transporters, Opp and App, is required for efficient sporulation. Homologues of these transporters in Clostridium difficile have been characterized, but their role, and hence that of peptides, in regulating sporulation in this organism is less clear. Here, the oligopeptide-binding receptor proteins for these transporters, CdAppA and CdOppA, have been purified and partially characterized, and the crystal structure of CdAppA has been determined in an open unliganded form. Peptide binding to either protein could not be observed in Thermofluor assays with a set of ten peptides of varying lengths and compositions. Re-examination of the protein sequences together with structure comparisons prompts the proposal that CdAppA is not a versatile peptide-binding protein but instead may bind a restricted set of peptides. Meanwhile, CdOppA is likely to be the receptor protein for a nickel-uptake system.
1. Introduction
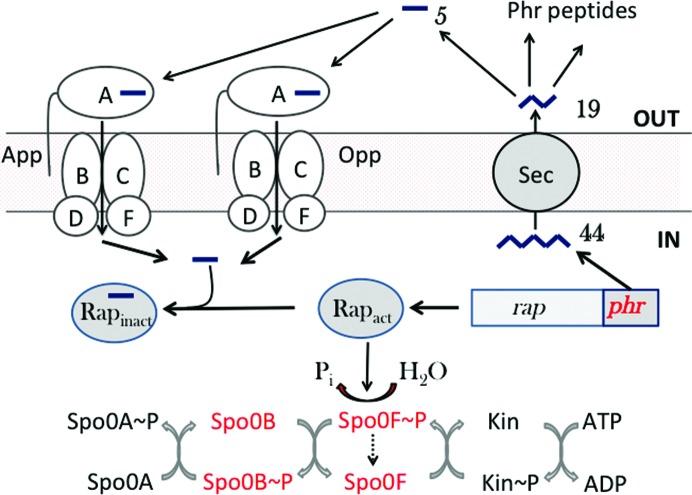
The formation of dormant spores is the ultimate response of Bacillus subtilis to starvation. As a time- and resource-intensive process, sporulation is under elaborate regulation. At the heart of this regulatory system is an expanded two-component system termed the phosphorelay (Burbulys et al., 1991 ▸), which is fed by multiple sensor kinases that integrate environmental and metabolic signals. This leads to the phosphorylation of the master regulator of sporulation, Spo0A. In opposition, the system is drained by Rap phosphatases such as RapA and RapE, which dephosphorylate the phosphorelay component Spo0F phosphate (Spo0F~P), delaying the accumulation of Spo0A phosphate (Spo0A~P) and the entry into sporulation (Fig. 1 ▸). For sporulation to proceed, the Rap phosphatases must be inhibited by specific peptides imported into the cell by one of two oligopeptide permeases, Opp or App. The relevant substrates are derived from Phr (phosphatase regulator) polypeptides that are secreted by the bacteria, processed extracellularly by mechanisms yet to be characterized, and later reimported as oligopeptides (Perego, 1997 ▸; Perego & Brannigan, 2001 ▸).
Figure 1.
Peptide signalling in sporulation in B. subtilis. The sporulation phosphorelay is shown at the bottom. One of up to five sensor kinases (Kin) autophosphorylates and relays a phosphoryl group via Spo0F and Spo0B to Spo0A. A short polypeptide (44) encoded by a phr gene situated downstream of a cognate rap phosphatase gene is exported from the cell by the Sec system, with the removal of a secretion signal peptide, to give a 19-mer (19). Outside the cell, under circumstances that favour sporulation, environmental proteases cleave the peptide, producing short oligopeptides such as the pentapeptide (5) shown, which may be imported into the cell by either App or Opp. The imported peptides bind to the cognate Rap phosphatase and prevent it from dephosphorylating Spo0F phosphate (Spo0F~P). This allows increased flux through the sporulation phosphorelay and the accumulation of Spo0A~P so that sporulation may commence. Counterparts of the components shown in red have yet to be identified in C. difficile.
The opp and app genes encode ATP-binding-cassette (ABC) uptake systems for oligopeptides and their role is complemented in B. subtilis by a third ABC transporter, the dipeptide permease Dpp. The specificity of ABC transporters is in a large part determined by an extracellular (periplasmic and membrane-anchored in Gram-negative and Gram-positive bacteria, respectively) solute-binding protein which captures the ligand and delivers it to a set of cognate membrane components for transport (Wilkinson & Verschueren, 2003 ▸; Maqbool et al., 2015 ▸). It is likely that Opp and App have overlapping specificities, with Opp responsible for the uptake of peptides of 2–5 residues in length and App handling longer peptides. The former assertion is supported by the possession of an RxGWxxD motif shared with the well characterized OppA from Salmonella typhimurium (StOppA), which binds dimer to pentamer peptides, with the highest affinity for tripeptides and tetrapeptides, and where the conserved Arg and Asp are involved in binding to the α-carboxylate and the α-amino groups of peptides, respectively (Tame et al., 1994 ▸; Sleigh et al., 1997 ▸). The latter assertion is supported by peptide-binding studies and the crystal structure of the receptor component of App, AppA, which revealed a bound nonapeptide (Levdikov et al., 2005 ▸; Picon & van Wely, 2001 ▸).
The interest here is in the putative peptide transporters of Clostridium difficile and their contribution to the regulation of spore formation in this pathogen. C. difficile infection (CDI) is a major health concern as a principal cause of hospital-acquired antibiotic-associated diarrhoea. Spores of C. difficile are agents of CDI transmission. Spo0A is conserved in C. difficile and at least one sporulation sensor kinase has been identified (Underwood et al., 2009 ▸; Pettit et al., 2014 ▸). Curiously, the phosphorelay that is characteristic of sporulation in B. subtilis is condensed to a conventional two-component system in C. difficile. The role of Opp and App has been investigated in C. difficile sporulation. The opp genes are organized into what appears to be a single transcription unit (oppBCADF), while the app genes are in two divergently transcribed operons (appABC and appDF), where the oppBC and appBC genes encode integral membrane permeases and the oppDF and appDF genes encode cytoplasmic ATPases (Edwards et al., 2014 ▸). In contrast to B. subtilis, the effect of opp/app deletion is to increase the frequency of sporulation, suggesting that peptide transport plays a very different role in spore formation in C. difficile (Edwards et al., 2014 ▸). Examination of the sequences of the solute-binding proteins CdAppA and CdOppA suggested to us that the structural basis of peptide recognition was likely to differ from that seen in other peptide-transporter proteins. To address this question, we set out to determine the crystal structures of CdAppA and CdOppA and to define their peptide-binding profiles. Here, we report the structure of CdAppA in the unliganded form. The absence of bound peptide in the crystals, the observations from a series of unsuccessful peptide-binding experiments and a re-examination of the sequences lead us to conclude that Opp is a probable nickel transporter and that App is a peptide transporter with narrow specificity.
2. Materials and methods
2.1. Protein production and purification
For biochemical and structural studies, we sought to overproduce soluble forms of CdAppA (locus tag CD630_26720) and CdOppA (locus tag CD630_08550). In C. difficile both proteins are secreted, becoming myristoylated on Cys1 of the mature protein and anchored in the cell membrane (Charlton et al., 2015 ▸). Sequence analysis and structure prediction suggested that polypeptides encompassing residues Gly31–Glu498 of CdAppA and Ser2–Glu502 of CdOppA would be soluble and compact. The corresponding coding sequence of CdAppA was amplified from C. difficile 630 chromosomal DNA using the primers F_CdAppA and R_CdAppA (Table 1 ▸) and Q5 High Fidelity DNA Polymerase (NEB), and the ∼1.5 kb amplification product was cloned into pET-YSBLLIC3C (Fogg & Wilkinson, 2008 ▸) using the NEBuilder HiFi DNA Assembly Cloning Kit to generate the expression plasmid pET-CdAppA, the sequence of which was verified. For CdOppA production, an Escherichia coli codon-optimized coding sequence was purchased from GenScript. The primers F_CdOppA and R_CdOppA (Table 1 ▸) were then used for amplification and cloning into pET-YSBLLIC3C as described above, generating pET-CdOppA. In these plasmids, the coding sequence of the peptide-binding protein is fused to a sequence encoding a human rhinovirus (HRV) 3C protease-cleavable hexahistidine tag. Recombinant protein was produced from E. coli BL21(DE3) cells harbouring either pET-CdAppA or pET-CdOppA, and the respective proteins were initially purified by two steps of nickel-chelation chromatography separated by treatment with HRV 3C protease (purified in-house). The latter procedure involved the digestion of the recombinant tagged protein at a 1:100 (protease:substrate protein) mass ratio during overnight dialysis at 277 K to remove the imidazole. Whereas the CdOppA fusion was readily cleaved by the protease, HRV 3C protease treatment of the CdAppA fusion failed to remove the tag. Subsequently, size-exclusion chromatography fractionation yielded proteins (∼20 mg per litre) which were judged to be 95% homogeneous from Coomassie-stained polyacrylamide gels. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| CdAppA | |
| Source organism | C. difficile |
| DNA source | C. difficile 630 chromosomal DNA |
| Forward primer F_CdAppA† | ttctgttccagggaccagcaGGAGCTTTTGCAAACGTTAAAG |
| Reverse primer R_CdAppA† | atatgtgaggagaaggcgcgTTATTCTACATACAGTTTAGACCAA |
| Expression vector | pET-YSBLLIC3C |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MGSHHHHHHSSGLEVLFQGPAGAFANVKEDSLASNIVYAPLYTYEKGNLVNYLAEKVDFKDSKELTIKLKSNLKWHDGKPITAEDVLFTFNTVLDEKQNSPSRQYLLVGEKPVKVEKIDDLTVKITLPTASESFLYGISKISPIPKHVFEGESNIAKSEKNNNPVGSGAFKFKEWKKGESIVFEKNADYFGGEPKADSIALKIIPNEASQEAALNNGEISLMKTSAEGYEKAKSNSNLQTYTYSEERLNYIVFNQNISNMANKEVRQALSYALNRNEMIESAYGKEGSVPAKSILVPEADFYTEEGVEGYDQDTNKAKDLLDKSGVKIDKLKIGYNTGRFGHKNYALVAQQELKKIGIEAEIVPYESKAFFNILFSNSTECDMYVNGYAWGLEPNPYRGMFETGQYCNQTKYSNAEIDALWEKGFTELNKEKREEIYKQIQQDISKDAPIYTIDYEQNLMAAQKNLKGIKDAKPSPAILFEDWSKLYVE |
| CdOppA | |
| Source organism | C. difficile |
| DNA source | Synthetic coding sequence codon-optimized for E. coli expression |
| Forward primer F_CdOppA† | ttctgttccagggaccagcaAGCAGCGGTGGCGACAAG |
| Reverse primer R_CdOppA† | atatgtgaggagaaggcgcgTTACTCAATGGTCCAATCCGCG |
| Expression vector | pET-YSBLLIC3C |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MGSHHHHHHSSGLEVLFQGPASSGGDKDKKADTPKDGKVLVYGSNDYTSINPALYEHGEINSLIFNGLTAHDENNKVVPCLAKDWKFDEATNTYTFNLRDDVKWHDGEKFTANDVKFTIETIMNPDNASEIASNYEDITKIDVVNDNTIKITLKAPNTAMLDYLTVGVLPKHALEGKDIATDEFNQKPIGTGPFKLEKWDKGQSITLVKNSDYFVKEPGLDKVVFKIVPDDKAKAMQLKSGELDLAQITPKDMSNFEKDEKNFKVNIMKTADYRGILYNFNSKFFKDKKAKGLPNALSYAIDRKAIVDSVLLGHGVPAYSPLQMGPYNNPDIEKFEYNPEKAKQEIEKLGWKLGSDGIYEKEGTKLAFEITAGESDQVRVDMAKICAQQLKEIGVDAKAVVVTETDWANQDAHLIGWGSPFDPDDHTYKVFGTDKGANYSAYSNPTIDKILQKARETEDKDEKLKLYKQFQVEMTKDMPYTFIAYIDAIYVGKPNIKGLTPDTVLGHHGVGIFWNIADWTIE |
The upper-case sequence is complementary to the template DNA and the lower-case flanking sequence is complementary to the vector DNA for use in HiFi cloning.
2.2. Crystallization
Protein concentrations were determined using an Epoch Microplate Spectrophotometer and using the extinction coefficient at 280 nm calculated from the sequence. Crystallization experiments were set up as sitting drops in 96-well plates using Hydra 96 and Mosquito liquid-handling systems to dispense the well and drop solutions, respectively. A variety of commercially available crystallization screens were trialled. Suitably diffracting crystals of CdAppA were grown from 0.2 M sodium iodide, 0.1 M bis-Tris propane, 20% PEG 3350 pH 6.5 in a drop consisting of 150 nl mother liquor and 150 nl protein solution at 20 mg ml−1. Diffracting crystals of CdOppA could be not be obtained despite exhaustive trials. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting drop |
| Plate type | 96-well MRC/Wilden |
| Temperature (K) | 291 |
| Protein concentration (mg ml−1) | 20 |
| Buffer composition of protein solution | 50 mM Tris–HCl pH 8.0, 150 mM NaCl |
| Composition of reservoir solution | 0.2 M sodium iodide, 0.1 M bis-Tris propane, 20% PEG 3350 |
| Volume and ratio of drop | 150 nl, 1:1 |
| Volume of reservoir (µl) | 100 |
2.3. Data collection and processing
A single CdAppA crystal was captured in a nylon loop and cooled in liquid nitrogen prior to diffraction data collection on beamline I04 at Diamond Light Source (DLS). Diffraction data extending to 2 Å spacing were processed using xia2 (Winter, 2010 ▸). The crystals belonged to space group P212121, with one molecule in the asymmetric unit. Data-collection and processing statistics are summarized in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | I04, DLS |
| Wavelength (Å) | 0.9795 |
| Temperature (K) | 100 |
| Detector | Dectris PILATUS3 S6M |
| Crystal-to-detector distance (mm) | 341 |
| Rotation range per image (°) | 0.1 |
| Total rotation range (°) | 220 |
| Exposure time per image (s) | 0.04 |
| Space group | P212121 |
| a, b, c (Å) | 45.6, 106.1, 109.5 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.4 |
| Resolution range (Å) | 54.76–2.06 (2.11–2.06) |
| Total No. of reflections | 261521 (18017) |
| No. of unique reflections | 33617 (2440) |
| Completeness (%) | 99.9 (99.9) |
| Multiplicity | 7.8 (7.4) |
| 〈I/σ(I)〉 | 17.9 (2.0) |
| R r.i.m. | 0.102 (1.26) |
| Overall B factor from Wilson plot (Å2) | 27.75 |
2.4. Structure solution and refinement
The structure of CdAppA was solved by molecular replacement using MrBUMP (Keegan & Winn, 2008 ▸) in the CCP4i2 interface (Potterton et al., 2018 ▸). MrBUMP performs a homology search of a subset of structures in the PDB and creates a set of search models from the template structures that are used for molecular replacement. The best solution from MrBUMP was derived from CtaP from Listeria monocytogenes (PDB entry 5isu; Center for Structural Genomics of Infectious Diseases, unpublished work), which has 41% sequence identity to CdAppA. The resulting model was refined in REFMAC5 (Murshudov et al., 1997 ▸, 2011 ▸) and automatic model building was then performed using Buccaneer (Cowtan, 2006 ▸) before iterative rounds of manual model building in Coot (Emsley et al., 2010 ▸) and refinement in REFMAC5. Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 54.82–2.00 (2.05–2.00) |
| Completeness (%) | 100 |
| σ Cutoff | None |
| No. of reflections, working set | 34874 (2524) |
| No. of reflections, test set | 1823 (139) |
| Final R cryst (%) | 20.9 (33.0) |
| Final R free (%) | 27.2 (34.0) |
| No. of non-H atoms | |
| Protein | 3755 |
| Ion | 1 |
| Water | 224 |
| Total | 3980 |
| R.m.s. deviations | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.47 |
| Average B factors (Å2) | |
| Overall | 38.32 |
| Protein | 37.53 |
| Ion | 45.07 |
| Water | 39.26 |
| Ramachandran plot | |
| Favoured regions (%) | 95.81 |
| Additionally allowed (%) | 3.77 |
| Outliers (%) | 0.21 |
3. Results and discussion
3.1. Structure description
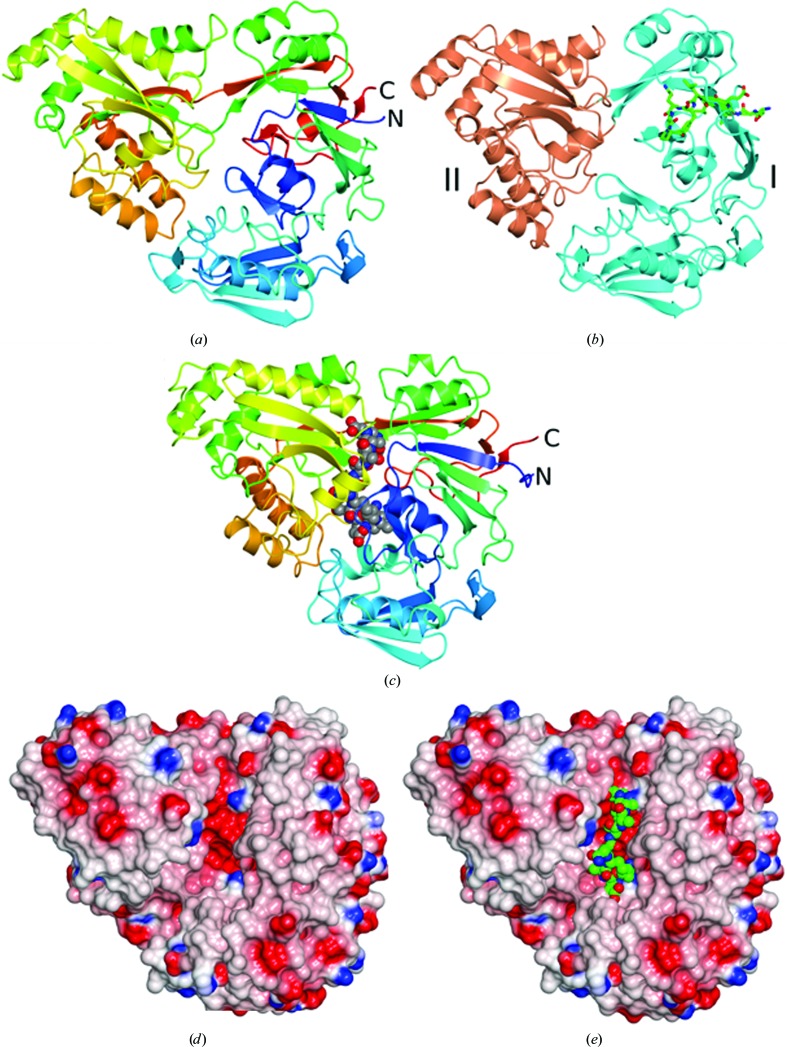
Residues Gly31–Glu498 are well defined in the electron-density maps, with additional density observed corresponding to 11 residues of the fusion tag (Ser-Gly-Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro-Ala). The overall structure of CdAppA is shown in Fig. 2 ▸(a). It closely resembles other cluster C substrate-binding proteins in the PDB (Berntsson et al., 2010 ▸). The closest structural matches are an unliganded form of L. monocytogenes CtaP (PDB entry 5isu), an unliganded B. anthracis extracellular solute-binding protein of unknown specificity (PDB entry 5u4o; Center for Structural Genomics of Infectious Diseases, unpublished work), an unliganded form of the Campylobacter jejuni nickel-binding protein NikZ (PDB entry 4oet; Lebrette et al., 2014 ▸) and AppA from B. subtilis bound to a nonapeptide (PDB entry 1xoc; Levdikov et al., 2005 ▸). Overlaying these structures with CdAppA gives r.m.s.d. values in the range 2.1–4.0 Å over 450–473 residues. Extracellular solute-binding proteins comprise two lobes, with ligand binding accompanied by closure of the lobes around the substrate according to a mechanism that has been likened to a Venus fly trap (Mao et al., 1982 ▸; Wilkinson & Verschueren, 2003 ▸). In the CdAppA structure, residues 31–253 and 466–498 constitute lobe I, with residues 254–465 constituting lobe II (Fig. 2 ▸ b). Unlike the structure of BsAppA (PDB entry 1xoc), which is of a closed liganded form (Fig. 2 ▸ c), the CdAppA structure is of an open unliganded form. In the open form of CdAppA there is a prominent groove (Fig. 2 ▸ d) which is the expected site of ligand binding, as illustrated in Fig. 2 ▸(e), where the nonapeptide ligand from BsAppA is displayed in the context of the CdAppA structure. Splitting the structure of CdAppA into its two lobes and superposing these individually with BsAppA gives lower r.m.s.d. values of 2.0 Å over 245 residues for lobe I and 1.4 Å over 204 residues for lobe II. Comparison with the BsAppA–nonapeptide complex structure suggests hinge opening by approximately 30° in the unliganded CdAppA (Figs. 2 ▸ a and 2 ▸ c).
Figure 2.
Structure of CdAppA. (a, c) Ribbon rendering of the polypeptide chain colour ramped from the N-terminus (blue) to the C-terminus (red) for CdAppA (a) and BsAppA (c). In (c) the nonapeptide ligand is shown as spheres coloured by atom type with carbon in grey, oxygen in red and nitrogen in blue. (b) CdAppA in ribbon format with lobe I in cyan and lobe II in coral and with the residues derived from the purification tag drawn as cylinders coloured by atom type (carbon, grey; oxygen, red; nitrogen, blue). (d) Electrostatic surface rendering of CdAppA showing the prominent groove situated between the two lobes. (e) Surface rendering of CdAppA as in (d) with the nonapeptide ligand from the BsAppA structure displayed following superposition of the protein chains using the SSM superpose routine in CCP4mg, which was also used to render these images (McNicholas et al., 2011 ▸).
In retrospect, it is clear that the CdAppA expression construct was truncated too severely at the amino-terminus such that the sequence that would form the first β-strand of the β-sheet in lobe I is missing. This strand, β1, which is integral to the seven-stranded β-sheet, is instead formed by residues from the HRV 3C cleavage-recognition element of the purification tag (Fig. 2 ▸ b). This interesting structural compensation results in the partial burial of the HRV 3C cleavage-recognition sequence and explains the failure of the protease to cleave off the purification tag.
No residual electron density was observed between the two lobes in the region of the structure that would be expected to form the binding pocket. The altered N-terminus is not expected to affect ligand binding, although this possibility cannot be excluded. Salt bridges between the α-amino and α-carboxylate groups of the substrate peptides and the side chains of Asp/Glu or Arg residues are recurring features of many peptide-binding proteins. As shown in Fig. 2 ▸ and Supplementary Fig. S1, the RxGWxxD motif of StOppA appears at a structurally equivalent site to V394NGYAWG in CdAppA. Thus, both the Asp and the Arg side chains that form ion pairs with the α-amino and α-carboxylate of peptide ligands in StOppA are absent. The Asp residue of this motif is also conserved in DppA and MppA from E. coli, which bind dipeptides and murein tripeptide, respectively, but the Arg is absent as the peptide C-termini are handled differently in these proteins (Dunten & Mowbray, 1995 ▸; Maqbool et al., 2011 ▸; Bhatt et al., 2018 ▸). The Asp154 and Arg373 residues of BsAppA which make ion-pairing interactions with the nonapeptide ligand appear as Ser148 and His351, respectively, in CdAppA (Supplementary Fig. S1). The absence of conservation of these signature peptide-binding residues in CdAppA suggests a mode of binding that is divergent from that seen in previous peptide-binding protein structures. An examination of the residues lining the groove between the two lobes that normally forms the substrate-binding site suggests Glu490 and Arg348 of CdAppA as potential substrate-anchoring residues (Supplementary Fig. S1).
3.2. Peptide-binding assays
The observation of an open unliganded structure in the crystals of CdAppA was initially surprising. ABC transporter substrate-binding proteins invariably co-purify and crystallize with cognate ligands, and crystal structures can reveal the specificity of transporters of hitherto uncertain function (Maqbool et al., 2011 ▸; Müller et al., 2005 ▸). Achieving crystals of unliganded solute-binding proteins often requires the application of partial unfolding–refolding regimes (Lanfermeijer et al., 1999 ▸). The absence of bound peptide in CdAppA crystals therefore suggests that the recombinant protein has not encountered its cognate ligand(s) during expression in E. coli and subsequent purification. Since peptides are readily available and were observed in the crystal structures of StOppA, BsAppA and E. coli MppA, CdAppA is an outlier, suggesting that it may not be a conventional peptide-binding protein.
To explore this further, we carried out peptide-binding assays of CdAppA and CdOppA. For these experiments, we removed any endogenous ligand by (i) loading the tagged protein onto a nickel-chelation column, (ii) partially unfolding the immobilized protein by washing with 2 M guanidinium hydrochloride (GdnHCl), (iii) refolding the protein through stepwise removal of the GdnHCl and (iv) elution from the column. To investigate peptide binding, we used differential scanning fluorimetry (Thermofluor) to measure changes in thermal denaturation upon the addition of potential ligands. These experiments were carried out in 50 mM Tris–HCl pH 8.0 buffer containing 150 mM NaCl. CdAppA has a melting temperature (T m) of 45°C, which is similar to that of BsOppA (50°C), which was studied in parallel (Adam Hughes, unpublished observations), but significantly lower than that of CdOppA (65°C). This suggests the possibility that the non-native N-terminal sequence destabilizes the CdAppA fold.
Ten peptides were explored in these experiments, including the tetrapeptides KKKK, DDDD, VAPG and SNSS, the pentapeptides ARNQT and SRNVT, the heptapeptide GRGDSPK, the octapeptide DYKDDDDK and the decapeptide RGDSPASSKL. We found no evidence of ligand binding in experiments using 10–20 µM of either protein (CdOppA or CdAppA) at peptide concentrations of up to 2 mM. In contrast, all of the tetrapeptides and pentapeptides tested bound tightly to OppA from B. subtilis (A. Hughes, unpublished observations). These observations suggest that neither CdAppA nor CdOppA are general peptide-binding proteins.
This conclusion is consistent with the lack of conservation of the signature residues involved in peptide binding in the broad-specificity OppA-type and AppA-type peptide-binding proteins. It should be noted that (i) other modes of peptide binding are possible, as exhibited by OppA from Lactococcus lactis, which also lacks these signature residues (Berntsson et al., 2011 ▸) and (ii) our peptide-binding experiments were performed with a restricted set of peptides. That said, the failure to observe peptide binding is perhaps consistent with the finding that wild-type C. difficile is unable to grow on minimal media containing peptides as the sole source of amino acids (Edwards et al., 2014 ▸).
3.3. CdAppA may have restricted substrate specificity
The closest structural homologue of CdAppA is CtaP (PDB entry 5isu) from L. monocytogenes, which is annotated as an oligopeptide-binding protein. It was named cysteine transport-associated protein (CtaP) following a study which showed that CtaP is required for bacterial growth in the presence of low concentrations of cysteine (Xayarath et al., 2009 ▸), and it was suggested that CtaP was involved in the uptake of free cysteine. A later study proposed that CtaP is involved in the uptake of a lipoprotein-derived peptide pheromone that enhances the escape of L. monocytogenes from host-cell vacuoles (Xayarath et al., 2015 ▸). Peptide pheromone-uptake systems have been described elsewhere, most notably in Enterococcus faecalis, where PrgZ regulates bacterial conjugation. In contrast to the E. faecalis OppA protein, which has a broad substrate specificity similar to the sequence-independent peptide-binding OppAs of E. coli and S. typhimurium, PrgZ has a narrower specificity and a much higher affinity for pheromone peptides (Berntsson et al., 2012 ▸). The crystal structure of CtaP (PDB entry 5isu) is of an unliganded form and provides few clues regarding substrate preference. The structural resemblance of CtaP and CdAppA suggests that CdAppA may also function in the transport of a restricted set of peptides, perhaps accounting for our failure to observe ligand binding with a general set of peptides.
3.4. CdOppA is a putative nickel-binding protein
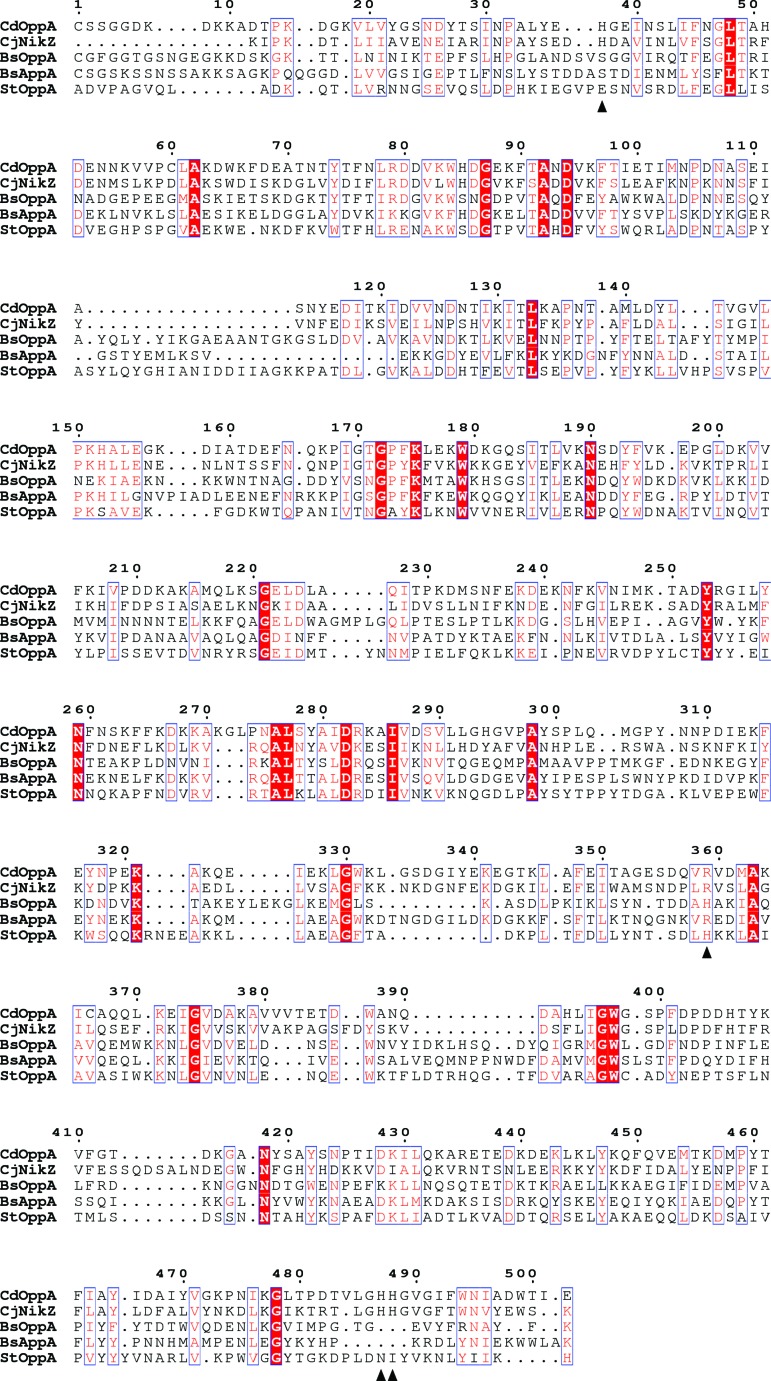
The absence of evidence for peptide binding in CdOppA prompted us to perform a BLAST search of the PDB using the CdOppA sequence. This identified NikZ, a nickel-binding protein from C. jejuni, as the closest match, with 39% sequence identity and 93% coverage. NikZ binds a nickel histidine chelate (Lebrette et al., 2014 ▸), with three histidine side chains from the protein forming coordinate bonds to the nickel ion and an arginine residue forming a two-pronged salt bridge to the chelating histidine carboxylate. These residues are conserved as His37, His487, His488 and Arg359 in CdOppA; in contrast, none of the histidines is conserved in bona fide OppAs of known structure (Fig. 3 ▸). At the position corresponding to Arg359 in CdOppA, a histidine or arginine is present in several OppAs, and in StOppA this histidine is known to be involved in binding the carboxylate of tetrapeptide ligands (Tame et al., 1995 ▸). Supporting the notion that CdOppA is a receptor for nickel import, the C. difficile genome encodes other components of the minimum machinery for nickel utilization (Zeer-Wanklyn & Zamble, 2017 ▸), namely a putative nickel response regulator (NikR), the three subunits (UreABC) of a nickel-dependent urease, and accessory proteins involved in nickel-ion loading (UreG) and the prevention of nickel-mediated toxicity (HypB).
Figure 3.
CdOppA as a putative nickel-binding protein. Alignment of the sequence of CdOppA with the sequences of the nickel-binding protein NikZ from C. jejuni, BsOppA, BsAppA and StOppA. Invariant and conserved residues are indicated by a red background and blue boxes, respectively. The positions of the nickel-chelating and histidine-binding residues of NikZ are denoted by black triangles below the sequence. The conservation of these residues selectively in CdOppA is apparent. The RxGWxxD motif is present in BsOppA and StOppA at residues corresponding to 394–400 in the sequence of CdOppA.
In relation to peptide regulation of sporulation, neither of the two rap phosphatase genes in C. difficile is followed by a phr coding sequence. Coupled with the absence of spo0F, the phosphorylated gene product of which is the substrate of RapA and RapE in B. subtilis, it is fair to conclude from the present discussion that sporulation in C. difficile is not regulated in an analogous manner by extracellular peptides.
Supplementary Material
PDB reference: C. difficile AppA, 6i3g
Supplementary Figure S1.. DOI: 10.1107/S2053230X1900178X/dp5121sup1.pdf
Acknowledgments
We thank Diamond Light Source for beamline access to I04 (Proposal No. MX-13587) that contributed to the results presented here. We thank Simon Cutting (RHUL) for providing C. difficile chromosomal DNA.
Funding Statement
This work was funded by Biotechnology and Biological Sciences Research Council grant BB/MO11151 to Adam M. Hughes.
References
- Berntsson, R. P.-A., Schuurman-Wolters, G. K., Dunny, G., Slotboom, D.-J. & Poolman, B. (2012). J. Biol. Chem. 287, 37165–37170. [DOI] [PMC free article] [PubMed]
- Berntsson, R. P.-A.., Smits, S. H. J., Schmitt, L., Slotboom, D.-J. & Poolman, B. (2010). FEBS Lett. 584, 2606–2617. [DOI] [PubMed]
- Berntsson, R. P.-A., Thunnissen, A.-M. W. H., Poolman, B. & Slotboom, D.-J. (2011). J. Bacteriol. 193, 4254–4256. [DOI] [PMC free article] [PubMed]
- Bhatt, F., Patel, V. & Jeffery, C. J. (2018). Biology (Basel), 7, 30. [DOI] [PMC free article] [PubMed]
- Burbulys, D., Trach, K. A. & Hoch, J. A. (1991). Cell, 64, 545–552. [DOI] [PubMed]
- Charlton, T. M., Kovacs-Simon, A., Michell, S. L., Fairweather, N. F. & Tate, E. W. (2015). Chem. Biol. 22, 1562–1573. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Dunten, P. & Mowbray, S. L. (1995). Protein Sci. 4, 2327–2334. [DOI] [PMC free article] [PubMed]
- Edwards, A. N., Nawrocki, K. L. & McBride, S. M. (2014). Infect. Immun. 82, 4276–4291. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fogg, M. J. & Wilkinson, A. J. (2008). Biochem. Soc. Trans. 36, 771–775. [DOI] [PubMed]
- Keegan, R. M. & Winn, M. D. (2008). Acta Cryst. D64, 119–124. [DOI] [PMC free article] [PubMed]
- Lanfermeijer, F. C., Picon, A., Konings, W. N. & Poolman, B. (1999). Biochemistry, 38, 14440–14450. [DOI] [PubMed]
- Lebrette, H., Brochier-Armanet, C., Zambelli, B., de Reuse, H., Borezée-Durant, E., Ciurli, S. & Cavazza, C. (2014). Structure, 22, 1421–1432. [DOI] [PubMed]
- Levdikov, V. M., Blagova, E., Brannigan, J. A., Wright, L., Vagin, A. A. & Wilkinson, A. J. (2005). J. Mol. Biol. 345, 879–892. [DOI] [PubMed]
- Mao, B., Pear, M. R., McCammon, J. A. & Quiocho, F. A. (1982). J. Biol. Chem. 257, 1131–1133. [PubMed]
- Maqbool, A., Horler, R. S., Muller, A., Wilkinson, A. J., Wilson, K. S. & Thomas, G. H. (2015). Biochem. Soc. Trans. 43, 1011–1017. [DOI] [PubMed]
- Maqbool, A., Levdikov, V. M., Blagova, E. V., Hervé, M., Horler, R. S., Wilkinson, A. J. & Thomas, G. H. (2011). J. Biol. Chem. 286, 31512–31521. [DOI] [PMC free article] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Müller, A., Thomas, G. H., Horler, R., Brannigan, J. A., Blagova, E., Levdikov, V. M., Fogg, M. J., Wilson, K. S. & Wilkinson, A. J. (2005). Mol. Microbiol. 57, 143–155. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Perego, M. (1997). Proc. Natl Acad. Sci. USA, 94, 8612–8617. [DOI] [PMC free article] [PubMed]
- Perego, M. & Brannigan, J. A. (2001). Peptides, 22, 1541–1547. [DOI] [PubMed]
- Pettit, L. J., Browne, H. P., Yu, L., Smits, W. K., Fagan, R. P., Barquist, L., Martin, M. J., Goulding, D., Duncan, S. H., Flint, H. J., Dougan, G., Choudhary, J. S. & Lawley, T. D. (2014). BMC Genomics, 15, 160. [DOI] [PMC free article] [PubMed]
- Picon, A. & van Wely, K. H. M. (2001). Mol. Biol. Today, 2, 21–25.
- Potterton, L., Agirre, J., Ballard, C., Cowtan, K., Dodson, E., Evans, P. R., Jenkins, H. T., Keegan, R., Krissinel, E., Stevenson, K., Lebedev, A., McNicholas, S. J., Nicholls, R. A., Noble, M., Pannu, N. S., Roth, C., Sheldrick, G., Skubak, P., Turkenburg, J., Uski, V., von Delft, F., Waterman, D., Wilson, K., Winn, M. & Wojdyr, M. (2018). Acta Cryst. D74, 68–84. [DOI] [PMC free article] [PubMed]
- Sleigh, S. H., Tame, J. R. H., Dodson, E. J. & Wilkinson, A. J. (1997). Biochemistry, 36, 9747–9758. [DOI] [PubMed]
- Tame, J. R. H., Dodson, E. J., Murshudov, G., Higgins, C. F. & Wilkinson, A. J. (1995). Structure, 3, 1395–1406. [DOI] [PubMed]
- Tame, J. R. H., Murshudov, G. N., Dodson, E. J., Neil, T. K., Dodson, G. G., Higgins, C. F. & Wilkinson, A. J. (1994). Science, 264, 1578–1581. [DOI] [PubMed]
- Underwood, S., Guan, S., Vijayasubhash, V., Baines, S. D., Graham, L., Lewis, R. J., Wilcox, M. H. & Stephenson, K. (2009). J. Bacteriol. 191, 7296–7305. [DOI] [PMC free article] [PubMed]
- Wilkinson, A. J. & Verschueren, K. H. G. (2003). ABC Proteins: From Bacteria to Man, 1st ed., edited by I. B. Holland, S. P. C. Cole, K. Kuchler & C. F. Higgins, pp. 187–207. London: Academic Press.
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Xayarath, B., Alonzo, F. III & Freitag, N. E. (2015). PLoS Pathog. 11, e1004707. [DOI] [PMC free article] [PubMed]
- Xayarath, B., Marquis, H., Port, G. C. & Freitag, N. E. (2009). Mol. Microbiol. 74, 956–973. [DOI] [PMC free article] [PubMed]
- Zeer-Wanklyn, C. J. & Zamble, D. B. (2017). Curr. Opin. Chem. Biol. 37, 80–88. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C. difficile AppA, 6i3g
Supplementary Figure S1.. DOI: 10.1107/S2053230X1900178X/dp5121sup1.pdf