Igni18 from Ignicoccus hospitalis was identified to belong to the α/β-hydrolase superfamily and was cloned, purified and crystallized. The crystal belonged to space group R32 and contained one monomer in the asymmetric unit.
Keywords: Ignicoccus hospitalis, α/β-hydrolases, metallo-β-lactamases
Abstract
The hyperthermophilic crenarchaeon Ignicoccus hospitalis KIN4/I possesses at least 35 putative genes encoding enzymes that belong to the α/β-hydrolase superfamily. One of those genes, the metallo-hydrolase-encoding igni18, was cloned and heterologously expressed in Pichia pastoris. The enzyme produced was purified in its catalytically active form. The recombinant enzyme was successfully crystallized and the crystal diffracted to a resolution of 2.3 Å. The crystal belonged to space group R32, with unit-cell parameters a = b = 67.42, c = 253.77 Å, α = β = 90.0, γ = 120.0°. It is suggested that it contains one monomer of Igni18 within the asymmetric unit.
1. Introduction
Ignicoccus hospitalis KIN4/I is an anaerobic, hyperthermophilic crenarchaeon with an optimal growth temperature of 90°C (363 K). It was isolated from a submarine hydrothermal system at the Kolbeinsey Ridge in the north of Iceland (Paper et al., 2007 ▸). The archaeon belongs to the order Desulfurococcales and is an obligate chemolithoautotroph that grows by performing sulfur reduction using hydrogen as the electron donor. Among other uncommon metabolic pathways, I. hospitalis utilizes a rare autotrophic CO2-fixation pathway starting from acetyl-coenzyme A (Jahn et al., 2007 ▸). With 1.3 Mbp coding for 1444 proteins, it has one of the smallest genomes described for a free-living organism (Podar et al., 2008 ▸). It is also the only organism known to date that is capable of acting as a host for Nanoarchaeum equitans, which is so far the only cultivated member of the Nanoarchaeota (Paper et al., 2007 ▸; Huber et al., 2000 ▸). This is the only natural archaeon–archaeon interaction that has been described in vivo (Wrede et al., 2012 ▸), although it remains unclear whether it is a true mutualistic symbiosis or parasitism (Jahn et al., 2008 ▸). Owing to the small genome size of N. equitans (0.5 Mbp), I. hospitalis provides the biological macromolecules that it cannot synthesize owing to a lack of essential biosynthesis pathways such as those for lipids, amino acids and nucleotides (Waters et al., 2003 ▸) and even energetic precursors (Giannone et al., 2011 ▸, 2015 ▸).
Living at above 90°C requires adaptation of all enzymes in order to carry out the metabolic reactions that are necessary for existence. The amino-acid composition, especially a decrease in thermolabile residues such as asparagine and cysteine, hydrophobic interactions, aromatic interactions, ion pairs and increased salt-bridge networks, oligomerization and intersubunit interactions, packing and reduction of solvent-exposed surface area, flexibility of surface-exposed loops, and metal binding or substrate stabilization are some of the factors that have been attributed to confer protein stability at extreme temperatures (Unsworth et al., 2007 ▸; Kovacic et al., 2016 ▸).
Enzymes that belong to the α/β-hydrolase superfamily include metabolically important enzymes that carry out a large number of different hydrolysis and synthesis reactions. The metallo-β-lactamase (MBL) subfamily includes enzymes that hydrolyze thiol-ester, phosphodiester and sulfuric ester bonds, but also includes oxidoreductases. Many members of this subfamily are involved in mRNA maturation and DNA repair (Bebrone, 2007 ▸). The presence of MBL genes within the Eubacteria, Archaea and Eukaryota suggests a very ancient origin of this family (Garau et al., 2005 ▸). The first crystal structure of an archaeal metallo-β-lactamase was published in 2010 (ST1585 from Sulfolobus tokodaii; Shimada et al., 2010 ▸). MBL enzymes usually show a characteristic αβ/βα protein fold and are often dependent on Zn2+ ions (Meini et al., 2015 ▸).
To date, only four crystal structures of native I. hospitalis proteins are available: a cell-adhesion structural protein (PDB entry 3j1r; Yu et al., 2012 ▸), a membrane-associated octaheme cytochrome c (PDB entry 4q05; Parey et al., 2016 ▸), the type IV pilus-like filament protein Iho670 (PDB entry 5kyh; Braun et al., 2016 ▸) and a superoxide reductase (PDB entry 4bk8; Romão et al., 2018 ▸). Here, we present the recombinant production, purification and crystallization of an MBL-like domain-containing protein from I. hospitalis KIN4/I.
2. Materials and methods
2.1. Cloning and overexpression
The igni18 gene (NCBI accession No. IGNI_RS06455; start nucleotide No. 1115579, end nucleotide No. 1114878), which has been annotated as a metallo-hydrolase containing a metallo-β-lactamase fold (Podar et al., 2008 ▸), was amplified with Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific, Carlsbad, California, USA) from I. hospitalis KIN4/I genomic DNA kindly donated by Dr Harald Huber (University of Regensburg, Germany). The EasySelect Pichia Expression Kit (Invitrogen, Carlsbad, California, USA) was employed for cloning and expression. The oligonucleotides designed for the amplification of igni18 (Table 1 ▸) allowed the hydrolysis of the PCR product with EcoRI and NotI and cloning into the pPICZ-A expression plasmid (Invitrogen) using Escherichia coli DH5α. The start of the gene was modified to include a Kozak consensus sequence for yeast [5′-(G/A)NNATGG-3′; Romanos et al., 1992 ▸] and the stop codon was omitted for fusion with a C-terminal Myc epitope and His6 tag encoded by the vector sequence. These allow immunodetection and affinity-chromatographic purification of the produced fusion protein.
Table 1. Igni18 production information.
| Source organism | I. hospitalis KIN4/I |
| DNA source | I. hospitalis KIN4/I |
| Forward primer† (5′–3′) | CCGAGAATTC GACATGGCCACGGTTAAGCTGACCTAC |
| Reverse primer† (5′–3′) | AGCGGCCGCAAAATTCGAAGGTCACCGTCTCC |
| Cloning and expression vector | pPICZ-A |
| Cloning host | E. coli DH5α |
| Expression host | P. pastoris X-33 |
| Igni18_Myc_His6 amino-acid sequence | MATVKLTYFGHSAFHVEVDGVGIAIDPWITNPLSKTTLEDYLKNFKTDLVVITHAHEDHIGDALEIMRRTGAKFFSIHEIYVDLTQKGFQGIGANIGGPAKLDDVAPGLGIALTPATHSSYDKGVPTGAIIFKDGKALVYHAGDTGLFAEMQFIGELYAPKVALLPIGGHYTMDIEQALLATKLLRPEVVVPMHYNTFPPIRADPNEFKQKVESAGLAKVRVMEPGETVTFEFCGRQLGPEQKLISEEDLNSAVDHHHHHH |
Restriction sites are underlined and modifications are in bold.
The constructed pPICZ-A::igni18 expression vector purified from E. coli DH5α was linearized with MssI restriction endonuclease (ThermoFisher Scientific) prior to electroporation of Pichia pastoris X-33 competent host cells (Invitrogen) according to the manual. Insertion into the chromosome occurs via homologous recombination at the 5′ AOX1 (alcohol oxidase 1) region. Colonies carrying the construct appeared on YPD–agar plates containing 1 M sorbitol and 100 µg ml−1 zeocin (Invitrogen) after 3–5 d of incubation at 303 K. The clones were tested for multiple insertions of the construct by the ability to grow on YPD–agar containing 1 mg ml−1 zeocin. A total of eight multi-insertion clones were tested for expression, and the production of the protein fused with a His6 tag was verified by Western blotting using anti-His6-tag-specific antibodies as described in the manual (Invitrogen). The clone yielding the highest Igni18 production was selected. It showed the methanol-utilization slow (MutS) phenotype, in which one of the copies of pPICZ-A::igni18 replaced the original AOX1 locus, creating a mutant that can assimilate methanol only at slow rates. Fermentation was performed in 1.5 l buffered extra-YNB glycerol methanol (BYGM) auto-induction medium (Lee et al., 2017 ▸) in a fermenter (Minifors, INFORS AG, Bottmingen, Switzerland) for 45 h at 303 K. Glycerol is used as the preferable carbon source for initial growth. After 24 h, the glycerol is consumed and methanol induces the expression of the gene of interest. The cells were harvested by centrifugation and the cell pellet was stored at 193 K until further use. Macromolecule-production information is summarized in Table 1 ▸.
2.2. Protein purification and enzyme-activity assays
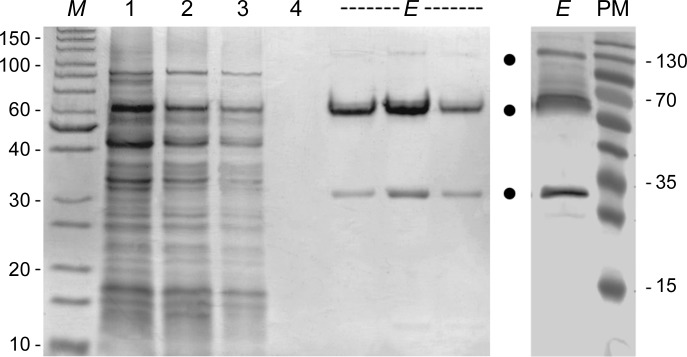
For immobilized metal ion-affinity chromatography (IMAC) purification of Igni18_Myc_His6, the cells were thawed on ice and suspended in 5 ml lysis buffer [10 mg ml−1 myristyl sulfobetaine (SB3-14), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.05 M NaH2PO4, 0.3 M NaCl pH 8.0] per gram. Cell disruption and partial purification were achieved by incubating the cells at 343 K for 1 h in the presence of the zwitterionic detergent SB3-14 (Zanna & Haeuw, 2007 ▸). Cell debris was removed from the crude cell extract by centrifugation at 15 000 rev min−1 for 30 min at 277 K (Sorvall RC6+ centrifuge, SS-34 rotor; Thermo Scientific, Braunschweig, Germany). The clear lysate was loaded onto a Protino Ni-TED 2000 Packed Column (Macherey-Nagel, Düren, Germany) and Igni18 was purified according to the manufacturer’s instructions. The buffer of the elution fractions was exchanged to 0.1 M potassium phosphate buffer pH 7.0 using an ultrafiltration unit with a pore size of 10 kDa (Vivaspin 20, Sartorius AG, Göttingen, Germany). The proteins were analyzed by polyacrylamide gel electrophoresis under denaturing conditions (SDS–PAGE) on 12%(w/v) gels stained with Coomassie Brilliant Blue G-250 (Laemmli, 1970 ▸) and by Western blotting (Fig. 1 ▸). Activity was assayed on para-nitrophenyl (pNP) esters with fatty-acid chain lengths ranging from two to 18 C atoms (Jaeger & Kovacic, 2014 ▸).
Figure 1.
Ni-TED affinity purification of Igni18_Myc_His6 (SDS–PAGE, left; Western blot, right). The samples were mixed with a loading dye containing 0.1 M dithiothreitol and were partially denatured at 368 K (95°C) for 10 min. Lane M, protein molecular-weight marker (PageRuler Unstained Protein Ladder, Thermo Scientific, Braunschweig, Germany; labeled in kDa); lane 1, lysate; lane 2, flowthrough; lane 3, initial column wash; lane 4, final column wash; lane E, elution fraction; lane PM, prestained protein molecular-weight marker (PageRuler Prestained Protein Ladder, Thermo Scientific, Braunschweig, Germany; labeled in kDa). Bands corresponding to monomeric Igni18 and oligomers are marked with dots and were confirmed by Western blot analysis using His6-specific antibodies.
2.3. Crystallization and preliminary X-ray analysis of Igni18
Crystallization trials were performed using the sitting-drop vapor-diffusion method at 293 and 285.15 K. To find an initial crystallization condition, various commercial kits from Qiagen (Hilden, Germany) and Molecular Dimensions (Suffolk, England) were used in MRC3 Swissci plates.
0.1 µl of homogenous recombinant Igni18 (at either 15 or 20 mg ml−1 in 0.1 M potassium phosphate buffer pH 7) was mixed with 0.1 µl reservoir solution and equilibrated against 40 µl reservoir solution using a pipetting robot (NT8, Formulatrix, Bedford, Massachusetts, USA).
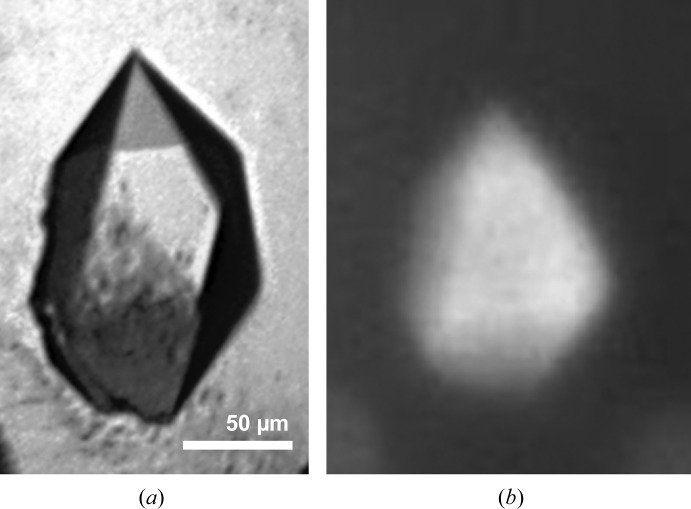
Crystals were grown at room temperature and reached final dimensions of around 130 × 90 × 80 µm after several months (Fig. 2 ▸ a). The crystallization condition consisted of 0.3 M magnesium nitrate hexahydrate, 0.1 M Tris pH 8, 22%(w/v) PEG 8000. To verify that the crystals contained protein, a fluorescence image was obtained using the intrinsic fluorescence of the tryptophan residue present in the Igni18 protein (Fig. 2 ▸ b). The crystal was cryoprotected by overlaying the drop with 2 µl mineral oil and subsequently flash-cooled in liquid nitrogen. Crystallization information is summarized in Table 2 ▸.
Figure 2.
Crystal of Igni18. Visible image (a) and UV image (b) of an Igni18 crystal obtained in 0.3 M magnesium nitrate hexahydrate, 0.1 M Tris pH 8, 22%(w/v) PEG 8000.
Table 2. Crystallization conditions.
| Method | Sitting-drop vapor diffusion |
| Plate type | MRC3, Swissci |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 20 |
| Buffer composition of protein solution | 0.1 M potassium phosphate pH 7 |
| Composition of reservoir solution | 0.3 M magnesium nitrate hexahydrate, 0.1 M Tris pH 8, 22%(w/v) PEG 8000 |
| Volume and ratio of drop | 200 nl, 1:1 |
| Volume of reservoir (µl) | 40 |
2.4. Data collection and processing
A data set was collected from a single Igni18 crystal on beamline ID30A-3 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France at 100 K. Initially, four frames were taken for characterization with 0.5° rotation at angles of 0°, 45°, 90° and 135°. The images were processed and a strategy was calculated using BEST (Bourenkov & Popov, 2010 ▸). A data set was then collected based on this strategy, resulting in the use of 0.05° rotation per frame with a total of 2500 frames (a total of 125° rotation). This data set was initially processed using the automated processing pipeline at the ID30A-3 beamline and was subsequently reprocessed using XDS (Kabsch, 2010 ▸), XSCALE and POINTLESS (Evans, 2006 ▸) to determine the space group. Data-collection and processing statistics are summarized in Table 3 ▸. To check for anomalous signal, the data were separately processed such that the Friedel pairs remained unmerged.
Table 3. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| X-ray source | ID30A-3, ESRF, Grenoble |
| Detector | EIGER 4M |
| Wavelength (Å) | 0.9677 |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 118.16 |
| Rotation range per image (°) | 0.05 |
| Total rotation range (°) | 125 |
| Exposure time (s) | 0.002 |
| Space group | R32 |
| a, b, c (Å) | 67.42, 67.42, 253.77 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 31.32–2.30 (2.382–2.300) |
| Total reflections | 70412 (7119) |
| Unique reflections | 10293 (1008) |
| R merge | 0.1004 (0.5445) |
| R meas | 0.1084 (0.5872) |
| R p.i.m. | 0.03987 (0.2152) |
| CC1/2 | 0.998 (0.885) |
| Wilson B factor (Å2) | 32.46 |
| 〈I/σ(I)〉 | 12.60 (3.37) |
| Completeness (%) | 99.78 (100.00) |
| Multiplicity | 6.8 (7.1) |
| Matthews coefficient (Å3 Da−1) | 2.17 |
| Solvent content (%) | 43.4 |
3. Results and discussion
A BLASTp search (Altschul et al., 2005 ▸) against the GenBank nonredundant database indicated that enzymes similar to Igni18 can only be found within the genus Ignicoccus with 78% identity. Furthermore, less similar enzymes with an identity of 52% or lower mostly belong to other members of the Crenarchaeota. The physiological function of this lipase and putative metallo-hydrolase in the respective hyperthermophilic organisms needs to be elucidated. To date, only about 61% of the protein-coding genes within I. hospitalis KIN4/I have bioinformatically predicted functions, implying that many pathways within this archaeon as well as its symbiont N. equitans are still unknown.
Initial attempts to produce the metallo-hydrolase Igni18 in different E. coli strains failed (data not shown); therefore, we used the yeast P. pastoris carrying multiple chromosomally integrated copies of the igni18 gene as an expression system. It seems that some of the problems regarding translation and post-translational modifications of the archaeal protein can be circumvented by using the eukaryotic P. pastoris compared with E. coli. After purification by affinity chromatography on Ni-TED agarose, the purity of Igni18_Myc_His6 was estimated to be greater than 90% and the yield was approximately 8 mg per litre of expression culture. The purified Igni18_Myc_His6 monomer has an apparent molecular weight of 30 kDa. which is in good agreement with the theoretical value of 28 614 Da (Fig. 1 ▸). The native protein seems to occur in an oligomeric state and this complex is highly stable, as expected from the nature of its native host; it can be denatured only partially with a reducing loading dye and heat incubation for 10 min at 368 K (95°C). Using a Western blot immunodetection method, the three bands could be identified as His6-tagged Igni18 (Fig. 1 ▸). Under semi-native running conditions, i.e. SDS–PAGE with nonreducing loading dye and without prior heat denaturation, only one single band could be observed on the gel at slightly below 130 kDa (data not shown). This suggests that native, soluble Igni18 predominantly exists in a multimeric form.
The enzyme activity of Igni18 was confirmed on pNP esters. Further assays are required with lipase-specific and MBL-specific substrates and need to be performed to reveal the precise biochemical function of Igni18. The optimal enzymatic activity of Igni18 was determined as 363 K (90°C), which corresponds to the optimal growth temperature of the host organism (Pérez-García et al., unpublished work).
The purified Igni18 was used to screen for suitable crystallization conditions using different commercial screening kits from Qiagen (Hilden, Germany) and Molecular Dimensions (Suffolk, England) and an NT8 pipetting robot (Formulatrix). Only a few Igni18 crystals appeared after several months in a condition consisting of 0.3 M magnesium nitrate hexahydrate, 0.1 M Tris pH 8, 22%(w/v) PEG 8000. Although the crystallization condition was further optimized by grid screening, only the crystal obtained in the initial screen displayed diffraction quality suitable for the collection of a data set.
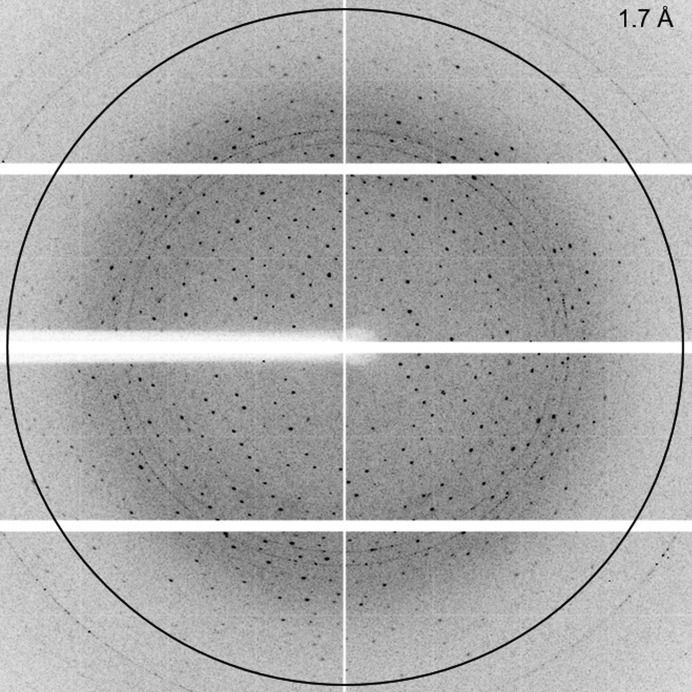
From this crystal, which diffracted to beyond 2.0 Å resolution (Fig. 3 ▸), a complete data set was collected on beamline ID30A-3 at the ESRF, Grenoble, France to a resolution of 2.3 Å. Here, we cut the data owing to incompleteness at higher resolutions. Preliminary X-ray diffraction analysis showed that the Igni18 crystal belonged to the hexagonal space group R32, with unit-cell parameters a = b = 67.42, c = 253.77 Å, α = β = 90.0, γ = 120.0°. Calculation of the unit-cell volume indicated the presence of one monomer in the asymmetric unit with a calculated VM of 2.17 Å3 Da−1 and a solvent content of 43.4% (Matthews, 1968 ▸).
Figure 3.
Diffraction image of an Igni18 crystal with a resolution of 1.7 Å at the edge of the detector (indicated by the black circle).
Interestingly, the anomalous signal after processing using XDS and XSCALE revealed the presence of an anomalous scatterer in the protein crystal, which was likely to result from endogenously bound metal within the Igni18 protein. Since Igni18 is a metallo-hydrolase, this suggests that this anomalous scatterer was bound to the protein directly during expression. Using the anomalous scattering analysis server (http://skuld.bmsc.washington.edu/scatter/), we analyzed which scatterer might be present. At the used wavelength of 0.9677 Å, we assume that manganese, iron, cobalt, nickel, copper and zinc might be possible candidates. They display scattering coefficients f′ or f′′ which range from 1.4 e for manganese to 3.9 e for iron. Since we did not add any of these ions to the buffers or the protein, we cannot precisely tell which one was bound by the Igni18 protein. By sequence homology, this protein belongs to the archaeal metallo-β-lactamase family, suggesting that it would be functional with zinc. However, we used a nickel column during purification, so this also might be the bound scatterer. Initial substructure analysis using AutoSol as part of the PHENIX suite (Adams et al., 2010 ▸) indicates that two atoms are bound within the asymmetric unit, suggesting, in combination with the Matthews coefficient, that one monomer has two anomalous scatterers bound. We used Auto-Rickshaw (Panjikar et al., 2009 ▸) to obtain the initial phases using the MR-SAD option, supplying only the Igni18 protein sequence as model input, which resulted in an initial model which is currently being refined. Although a monomer is present in the asymmetric unit, a trimeric Igni18 protein is likely to be built by the threefold axes given by the R32 symmetry. This suggests that despite the presence of only one monomer in the asymmetric unit the Igni18 protein crystallized as an oligomeric protein. The electron density is of good quality and allowed the entire sequence of Igni18 to be fitted unambiguously (see Supplementary Fig. S1).
Supplementary Material
Supplementary Figure S1.. DOI: 10.1107/S2053230X19002851/or5014sup1.pdf
Acknowledgments
X-ray diffraction measurements were performed on beamline ID30A-3 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. We are grateful to Guillaume Gotthard at the ESRF for providing assistance in using beamline ID30A-3. We thank Dr Harald Huber, University of Regensburg, Germany for providing us with genomic DNA of I. hospitalis. This project was partially funded by the Horizon2020 project INMARE (Grant agreement ID: 634486).
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schäffer, A. A. & Yu, Y.-K. (2005). FEBS J. 272, 5101–5109. [DOI] [PMC free article] [PubMed]
- Bebrone, C. (2007). Biochem. Pharmacol. 74, 1686–1701. [DOI] [PubMed]
- Bourenkov, G. P. & Popov, A. N. (2010). Acta Cryst. D66, 409–419. [DOI] [PMC free article] [PubMed]
- Braun, T., Vos, M. R., Kalisman, N., Sherman, N. E., Rachel, R., Wirth, R., Schoeder, G. F. & Egelman, E. H. (2016). Proc. Natl Acad. Sci. USA, 113, 10352–10357. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Garau, G., Di Guilmi, A. M. & Hall, B. G. (2005). Antimicrob. Agents Chemother. 49, 2778–2784. [DOI] [PMC free article] [PubMed]
- Giannone, R. J., Huber, H., Karpinets, T., Heimerl, T., Küper, U., Rachel, R., Keller, M., Hettich, R. L. & Podar, M. (2011). PLoS One, 6, e22942. [DOI] [PMC free article] [PubMed]
- Giannone, R. J., Wurch, L. L., Heimerl, T., Martin, S., Yang, Z., Huber, H., Rachel, R., Hettich, R. L. & Podar, M. (2015). ISME J. 9, 101–114. [DOI] [PMC free article] [PubMed]
- Huber, H., Burggraf, S., Mayer, T., Wyschkony, I., Rachel, R. & Stetter, K. O. (2000). Int. J. Syst. Evol. Microbiol. 50, 2093–2100. [DOI] [PubMed]
- Jaeger, K. E. & Kovacic, F. (2014). Methods Mol. Biol. 1149, 111–134. [DOI] [PubMed]
- Jahn, U., Gallenberger, M., Paper, W., Junglas, B., Eisenreich, W., Stetter, K. O., Rachel, R. & Huber, H. (2008). J. Bacteriol. 190, 1743–1750. [DOI] [PMC free article] [PubMed]
- Jahn, U., Huber, H., Eisenreich, W., Hügler, M. & Fuchs, G. (2007). J. Bacteriol. 189, 4108–4119. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kovacic, F., Mandrysch, A., Poojari, C., Strodel, B. & Jaeger, K. E. (2016). Protein Eng. Des. Sel. 29, 65–76. [DOI] [PMC free article] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Lee, J. Y., Chen, H., Liu, A., Alba, B. M. & Lim, A. C. (2017). Protein Expr. Purif. 137, 7–12. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Meini, M.-R., Llarrull, L. I. & Vila, A. J. (2015). FEBS Lett. 589, 3419–3432. [DOI] [PMC free article] [PubMed]
- Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S. & Tucker, P. A. (2009). Acta Cryst. D65, 1089–1097. [DOI] [PMC free article] [PubMed]
- Paper, W., Jahn, U., Hohn, M. J., Kronner, M., Näther, D. J., Burghardt, T., Rachel, R., Stetter, K. O. & Huber, H. (2007). Int. J. Syst. Evol. Microbiol. 57, 803–808. [DOI] [PubMed]
- Parey, K., Fielding, A. J., Sörgel, M., Rachel, R., Huber, H., Ziegler, C. & Rajendran, C. (2016). FEBS J. 283, 3807–3820. [DOI] [PubMed]
- Podar, M., Anderson, I., Makarova, K. S., Elkins, J. G., Ivanova, N., Wall, M. A., Lykidis, A., Mavromatis, K., Sun, H., Hudson, M. E., Chen, W., Deciu, C., Hutchison, D., Eads, J. R., Anderson, A., Fernandes, F., Szeto, E., Lapidus, A., Kyrpides, N. C., Saier, M. H. Jr, Richardson, P. M., Rachel, R., Huber, H., Eisen, J. A., Koonin, E. V., Keller, M. & Stetter, K. O. (2008). Genome Biol. 9, R158. [DOI] [PMC free article] [PubMed]
- Romanos, M. A., Scorer, C. A. & Clare, J. J. (1992). Yeast, 8, 423–488. [DOI] [PubMed]
- Romão, C. V., Matias, P. M., Sousa, C. M., Pinho, F. G., Pinto, A. F., Teixeira, M. & Bandeiras, T. M. (2018). Biochemistry, 57, 5271–5281. [DOI] [PubMed]
- Shimada, A., Ishikawa, H., Nakagawa, N., Kuramitsu, S. & Masui, R. (2010). Proteins, 78, 2399–2402. [DOI] [PubMed]
- Unsworth, L. D., van der Oost, J. & Koutsopoulos, S. (2007). FEBS J. 274, 4044–4056. [DOI] [PubMed]
- Waters, E., Hohn, M. J., Ahel, I., Graham, D. E., Adams, M. D., Barnstead, M., Beeson, K. Y., Bibbs, L., Bolanos, R., Keller, M., Kretz, K., Lin, X., Mathur, E., Ni, J., Podar, M., Richardson, T., Sutton, G. G., Simon, M., Söll, D., Stetter, K. O., Short, J. M. & Noordewier, M. (2003). Proc. Natl Acad. Sci. USA, 100, 12984–12988. [DOI] [PMC free article] [PubMed]
- Wrede, C., Dreier, A., Kokoschka, S. & Hoppert, M. (2012). Archaea, 2012, 596846. [DOI] [PMC free article] [PubMed]
- Yu, X., Goforth, C., Meyer, C., Rachel, R., Wirth, R., Schröder, G. F. & Egelman, E. H. (2012). J. Mol. Biol. 422, 274–281. [DOI] [PMC free article] [PubMed]
- Zanna, L. & Haeuw, J. F. (2007). J. Chromatogr. B, 846, 368–373. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1.. DOI: 10.1107/S2053230X19002851/or5014sup1.pdf