The production of nonglycosylated human furin ectodomain via the BacMam expression system is reported. The protein is equally active compared with the glycosylated protein and is readily crystallized; X-ray diffraction was used to solve a high-resolution structure.
Keywords: furin, nonglycosylated, apo, serine proteinase, hydrolase, BacMam
Abstract
Furin, also called proprotein convertase subtilisin/kexin 3 (PCSK3), is a calcium-dependent serine endoprotease that processes a wide variety of proproteins involved in cell function and homeostasis. Dysregulation of furin has been implicated in numerous disease states, including cancer and fibrosis. Mammalian cell expression of the furin ectodomain typically produces a highly glycosylated, heterogeneous protein, which can make crystallographic studies difficult. Here, the expression and purification of nonglycosylated human furin using the BacMam technology and site-directed mutagenesis of the glycosylation sites is reported. Nonglycosylated furin produced using this system retains full proteolytic activity indistinguishable from that of the glycosylated protein. Importantly, the nonglycosylated furin protein reliably forms extremely durable apo crystals that diffract to high resolution. These crystals can be soaked with a wide variety of inhibitors to enable a structure-guided drug-discovery campaign.
1. Introduction
The proprotein convertase subtilisin/kexin (PCSK) family of enzymes play a crucial role in processing and trafficking of a wide variety of precursor proteins such as hormones, receptors and enzymes (Seidah & Prat, 2012 ▸). These enzymes regulate the spatial and temporal production of the active form of the proproteins and play a critical role in cellular function and homeostasis. Furin (PCSK3) was the first human enzyme to be reported with a sequence relationship to the bacterial subtilisin and yeast kexin enzymes (Thomas, 2002 ▸; van de Ven et al., 1990 ▸). Thus, it is a widely studied enzyme and much is understood about its role in converting proproteins to functional proteins. Furin has also been widely studied owing to its association with many disease states, including cancer, metabolic diseases and fibrotic disorders, as well as bacterial and viral infections (Becker et al., 2012 ▸; Couture et al., 2015 ▸).
A truncated preparation of heterogeneous glycosylated mouse furin (∼60 kDa) produced in Chinese hamster ovary (CHO) cells was used to determine the first crystal structure of furin in complex with the decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone inhibitor at 2.6 Å resolution (Henrich et al., 2003 ▸). Subsequently, structures of human furin with two different inhibitors were reported at 2.3 and 2.7 Å resolution (Dahms et al., 2014 ▸). In this case, uniformly glycosylated, soluble furin protein was prepared from an engineered human embryonic kidney cell line (HEK293S Gnt−), requiring a three-part purification scheme including customized inhibitor-affinity chromatography. Most recently, using the same expression and purification process, Dahms and coworkers reported higher resolution structures of glycosylated furin at 1.8–2.0 Å resolution in unliganded and ligand-bound functional forms (Dahms et al., 2016 ▸, 2018 ▸). Our attempts at preparing reliable diffraction-quality crystals of glycosylated human furin were unsuccessful. Consequently, we chose to produce nonglycosylated furin, hypothesizing that such a change would improve crystallization and diffraction. Here, we describe BacMam transduction of mammalian cells to produce ample quantities of active, homogeneous protein containing aspartic acid substitutions at two known N-linked glycosylation sites, Asn387 and Asn440 (N387D/N440D), that can be used for biochemical and high-throughput crystallographic studies (Ames et al., 2007 ▸; Scott et al., 2007 ▸; Dukkipati et al., 2008 ▸). This nonglycosylated form of the furin ectodomain has similar enzymatic properties to the native protein and readily crystallizes in the apo form, providing an excellent system for small-molecule soaks.
2. Materials and methods
2.1. Macromolecule production
The BacMam technology is used under license from MGH Research Ventures and Licensing. The Gateway vector pENTR/D was from Life Technologies. Ni-Sepharose High Performance and Superdex 75 resins were from GE Healthcare. All other reagents were from Sigma unless otherwise noted. Cells for protein expression were a GlaxoSmithKline (GSK) proprietary CHO-Gam-E1A cell line (CHO-GE), a derivative of CHO K1 cells that expresses GAM1 and Ad5 E1A proteins. Clones were selected based on their ability to be efficiently transduced in suspension by BacMam-GFP virus (J. P. Condreay, W. S. Clay and T. A. Kost, unpublished data). Harvest filters were 5.0 µm MicroCap Single-Use depth filters from ErtelAlsop and 0.2 µm PES filters from Meissner. The plasmid ‘furin aa1–574’ insert was created in a series of three reactions, beginning with untagged furin cDNA plasmid and the primers ‘furin_H3_M1_F’ (5′-AGA CCC AAG CTT GCC ACC ATG GAG CTG AGG CCC-3′) and ‘furin_A574_R1’ (5′-CCT GGA AGT ACA GGT TCT CAC CGG CGG TGC CAT AGA GTA CGA G-3′) (Table 1 ▸). This 1762 bp amplicon was used in a second PCR to add a TEV cleavage site and a FLAG tag to the 3′ end using the primer pair ‘H3_Koz_Met_F’ (5′-AGA CCC AAG CTT GCC ACC ATG-3′) and ‘TEV_FLAG_R’ (5′-ACC CTT GTC ATC GTC GTC CTT GTA GTC GCC CTG GAA GTA CAG GTT CTC ACC-3′). Finally, the second PCR product was used as template with the primers ‘H3_Koz_Met_F’ and ‘FLAG_6H_Xba_R2’ (5′-CAG GCT CTA GAC TAT TAG TGA TGG TGG TGA TGG TGA CCC TTG TCA TCG TCG TCC TTG-3′) to add a His6 tag. This amplicon was ligated into the HindIII–XbaI sites of the pFastBacMam-1 vector.
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| DNA source | Lung and brain cDNA libraries |
| Forward primer | AGA CCC AAG CTT GCC ACC ATG GAG CTG AGG CCC |
| Reverse primer | CCT GGA AGT ACA GGT TCT CAC CGG CGG TGC CAT AGA GTA CGA G |
| Cloning vector | pENTR/D-TOPO |
| Expression vector | pFastBacMam-1, a modified version of pFastBac1 (Life Technologies) in which the CMV IE promoter has been substituted for the baculovirus polyhedrin promoter (Condreay et al., 1999 ▸) |
| Expression host | CHO-Gam-E1A cell line (CHO-GE) |
| Complete amino-acid sequence of the construct produced | DVYQEPTDPKFPQQWYLSGVTQRDLNVKAAWAQGYTGHGIVVSILDDGIEKNHPDLAGNYDPGASFDVNDQDPDPQPRYTQMNDNRHGTRCAGEVAAVANNGVCGVGVAYNARIGGVRMLDGEVTDAVEARSLGLNPNHIHIYSASWGPEDDGKTVDGPARLAEEAFFRGVSQGRGGLGSIFVWASGNGGREHDSCNCDGYTNSIYTLSISSATQFGNVPWYSEACSSTLATTYSSGNQNEKQIVTTDLRQKCTESHTGTSASAPLAAGIIALTLEANKDLTWRDMQHLVVQTSKPAHLNANDWATNGVGRKVSHSYGYGLLDAGAMVALAQDWTTVAPQRKCIIDILTEPKDIGKRLEVRKTVTACLGEPNHITRLEHAQARLTLSYNRRGDLAIHLVSPMGTRSTLLAARPHDYSADGFNDWAFMTTHSWDEDPSGEWVLEIENTSEANNYGTLTKFTLVLYGTAGENLYFQGDYKDDDDKGHHHHHH |
A glycosylation-site double mutant was generated by using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Stratagene) on plasmid template ‘furin aa1–574’. Plasmid ‘furin aa1–574 N387D/N440D’ was created with the primer pair ‘DM3192’ (5′-CTC ACC CTG GAG GCC AAT AAG GAC CTC ACA TGG CGG GAC ATG CAA-3′) and ‘DM3193’ (5′-GCC ATG GTG GCC CTG GCC CAG GAT TGG ACC ACA GTG GCC CCC CAG-3′). BacMam viruses for transductions were made from the shuttle plasmids furin aa1–574 and furin aa1–574 N387D/N440D.
BacMam viruses were amplified in Sf9 cells using Hyclone SFX medium containing 5% heat-inactivated FBS using standard baculovirus generation protocols. The BacMam viruses were not concentrated prior to use and the titer was determined using the BaculoTiter Assay Kit and Detection Reagents Protocol (Invitrogen). CHO-GE cells were grown in suspension and routinely passaged three times per week using vented Corning shake flasks in the GSK proprietary CHO cell medium MR1-4 supplemented with 300 µg ml−1 hygromycin and 500 µg ml−1 G418 (Life Technologies). The growth parameters were 110 rev min−1, 37°C and 5% CO2. Production was performed in shake flasks or in Wave Bags up to the 25 l scale (Kadwell & Overton, 2016 ▸). One day prior to transduction, the cells were seeded at 0.5 × 106 cells ml−1 in MR1-4 medium without selection. In Wave Bags, the growth parameters were 37°C, 20 rocks per minute, angle 10° and air flow 0.5 l min−1. On the day of transduction, BacMam virus was added at a multiplicity of transduction (MOT) of 10. At the time of transduction, sodium butyrate was added to achieve a final concentration of 5 mM and the temperature was reduced to 31°C. 48–72 h post-transduction, the conditioned medium was harvested by filtration using the filters noted above. At large scale, the conditioned medium was concentrated from 25 l to ∼1–2 l and washed three times with 10 l chilled 10 mM Tris–HCl, 100 mM NaCl, 0.5 mM CaCl2 pH 7.4 using tangential-flow filtration (Millipore Pellicon) with a 10 kDa cutoff membrane.
The entire protein-purification protocol was conducted at 4°C. The protocol was nearly identical for both mature furin proteins: aa108–574-TEV-FLAG-His6 and aa108–574-TEV-FLAG-His6 N387D/N440D. For conditioned medium, an equal volume of 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 0.5 mM CaCl2 was added. A typical final volume was ∼4 l. Imidazole was added to the diluted medium to 5 mM final concentration using a 1 M imidazole pH 7.5 stock solution. A 150 ml volume of prewashed Ni-Sepharose (GE Healthcare) 50% slurry was added to the medium. The solution was stirred slowly with a raised stir bar overnight at 4°C. The mixture was loaded onto an XK50 column (GE Healthcare) with a slow drip. After the solution had finished loading, the column was washed extensively with 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 0.5 mM CaCl2, 5 mM imidazole. The protein was eluted with a gradient over about 1 l (about ten column volumes) to a final buffer solution consisting of 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM CaCl2, 100 mM imidazole using a flow rate of 2 ml min−1 and 10 ml collection tubes. SDS–PAGE was used to analyze the fractions for furin content and fractions from 25% to 75% were typically pooled. The solution was concentrated to about 30 ml using an Amicon Ultra 30K cutoff device and filtered with a 0.22 µm filter (Costar μStar LB). The protein was loaded in 10 ml runs onto a Superdex 75 HiLoad 26/60 column (GE Healthcare) equilibrated with 10 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2 using a 2.5 ml min−1 flow rate and collected using 2 ml fractions. Furin typically eluted in the 170 ml range as a single peak. Fractions in this region were analyzed for purity with SDS–PAGE and appropriate fractions were pooled. Note that the FLAG tag was not required for purification to homogeneity (>95%) and the TEV cleavage site for tag removal did not need to be utilized for successful crystallization.
For enzymatic characterization, the respective furin protein was diluted into 20 mM HEPES pH 7.5, 1 mM CaCl2, 2 mM CHAPS, 0.1 mg ml−1 BSA buffer at a twofold concentration for assay [200 pM in Fig. 2(a) or various concentrations in Fig. 2(b)]. The FAM-TAMRA [5-FAM, carboxyfluorescein; 5-TAMRA, carboxytetramethylrhodamine; 5-FAM-Gln-Arg-Val-Arg-Arg-Ala-Val-Gly-Ile-Asp-Lys(5-TAMRA)-OH; custom synthesis from AnaSpec Inc.] substrate peptide was diluted to the twofold concentration used in the assay (2 µM). The furin solution (5 µl) was added to 384-well plates (Greiner low volume, No. 784076). The reaction was initiated by rapid addition of the substrate-peptide solution (5 µl) to the furin solution. Fluorescence was monitored using an Envision (PerkinElmer) with 485 nm excitation and 535 nm emission with an FITC top mirror using 30 s intervals. Initial rates were analyzed with linear regression (Microsoft Excel). No fluorescence change was observed in all enzyme-free controls (data not shown).
2.2. Crystallization
Apo furin (aa108–574-TEV-FLAG-His6 N387D/N440D) in 10 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2 was spin-concentrated to 6 mg ml−1 and used for vapor-diffusion crystallization. Sitting drops were set up with 400 nl protein solution and 400 nl crystallization reagent [11–13% polyethylene glycol (PEG) 8000, 0.11–0.16 M potassium dihydrogen phosphate, 0.1 M HEPES pH 7.5] and incubated at 4°C (Table 2 ▸). For X-ray diffraction studies, crystals were harvested and cryopreserved using crystallization reagent amended with 30% ethylene glycol. The crystals were stable at 4°C for over one year, demonstrating consistent morphology and X-ray diffraction quality.
Table 2. Crystallization.
| Method | Sitting-drop vapor diffusion |
| Plate type | MRC 2-drop |
| Temperature (°C) | 4 |
| Protein concentration (mg ml−1) | 6 |
| Buffer composition of protein solution | 150 mM NaCl, 5 mM CaCl2, 10 mM HEPES pH 7.5 |
| Composition of reservoir solution | 11–13% PEG 8000, 0.11–0.16 M potassium dihydrogen phosphate, 0.1 M HEPES pH 7.5 |
| Volume and ratio of drop | 800 nl, 1:1 |
| Volume of reservoir (µl) | 60 |
2.3. Data collection and processing
A single apo crystal at 100 K was used to collect X-ray diffraction data to 1.89 Å resolution using a Rigaku FR-E generator equipped with a Saturn 944+ detector (Table 3 ▸). HKL-2000 was used to index, integrate and scale the data (Otwinowski & Minor, 1997 ▸). The initial structure was determined by molecular replacement using internally determined human furin coordinates that were derived from the published mouse structure (PDB entry 1p8j; Henrich et al., 2003 ▸). The reported space group contains one molecule in the crystallographic asymmetric unit.
Table 3. Data collection and processing.
| Diffraction source | Rigaku FR-E SuperBright rotating anode |
| Wavelength (Å) | 1.54 |
| Temperature (K) | 100 |
| Detector | Rigaku Saturn 944+ CCD |
| Crystal-to-detector distance (mm) | 43 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 30 |
| Space group | C2 |
| a, b, c (Å) | 95.98, 66.60, 88.44 |
| α, β, γ (°) | 90.00, 122.41, 90.00 |
| Mosaicity (°) | 0.45 |
| Resolution range (Å) | 50.00–1.89 (1.96–1.89) |
| Total No. of reflections | 135597 |
| No. of unique reflections | (3678) |
| Completeness (%) | 99.8 (98.8) |
| Multiplicity | 3.6 (3.1) |
| 〈I/σ(I)〉 | 13.7 |
| Overall B factor from Wilson plot (Å2) | 11.830 |
2.4. Structure solution and refinement
The starting model was iteratively built with Coot (Emsley et al., 2010 ▸) and refined with REFMAC (Murshudov et al., 2011 ▸) or PHENIX (Adams et al., 2010 ▸) using all hydrogens and maximum-likelihood restraints. The crystallographic data and final refinement statistics are summarized in Table 4 ▸. The reported interatomic distances are between heavy atoms unless specified and were measured with Coot or PyMOL (http://www.schrodinger.com). The deposited coordinates for the apo furin structure (PDB entry 4z2a; http://www.rcsb.org) have a MolProbity (Chen et al., 2010 ▸) score of 1.40 (97th percentile) and a clashscore of 1.33 (99th percentile). Structure figures were generated using PyMOL.
Table 4. Structure refinement.
| Resolution range (Å) | 29.581–1.890 (1.976–1.890) |
| Completeness (%) | 99.7 |
| σ Cutoff | F > 1.340σ(F) |
| No. of reflections, working set | 36491 (4486) |
| No. of reflections, test set | 1173 (129) |
| Final R cryst | 0.148 (0.174) |
| Final R free | 0.183 (0.210) |
| No. of non-H atoms | |
| Protein | 3524 |
| Ion | 14 |
| Ligand | 4 |
| Water | 408 |
| Total | 3950 |
| R.m.s. deviations | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.260 |
| Average B factors (Å2) | |
| Protein | 16.6 |
| Ion | 24.2 |
| Ligand | 23.1 |
| Water | 23.2 |
| Ramachandran plot | |
| Favored regions (%) | 96.3 |
| Additionally allowed (%) | 3.2 |
| Outliers (%) | 0.2 |
3. Results and discussion
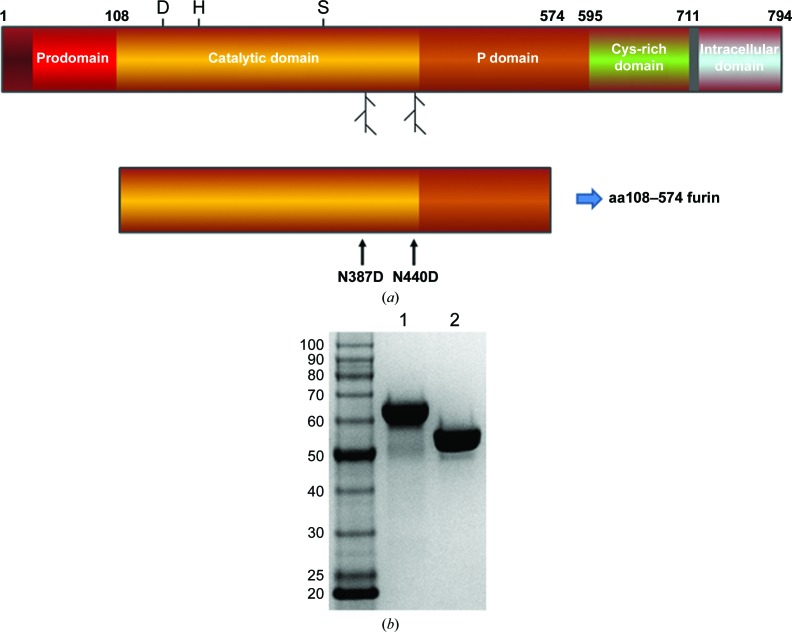
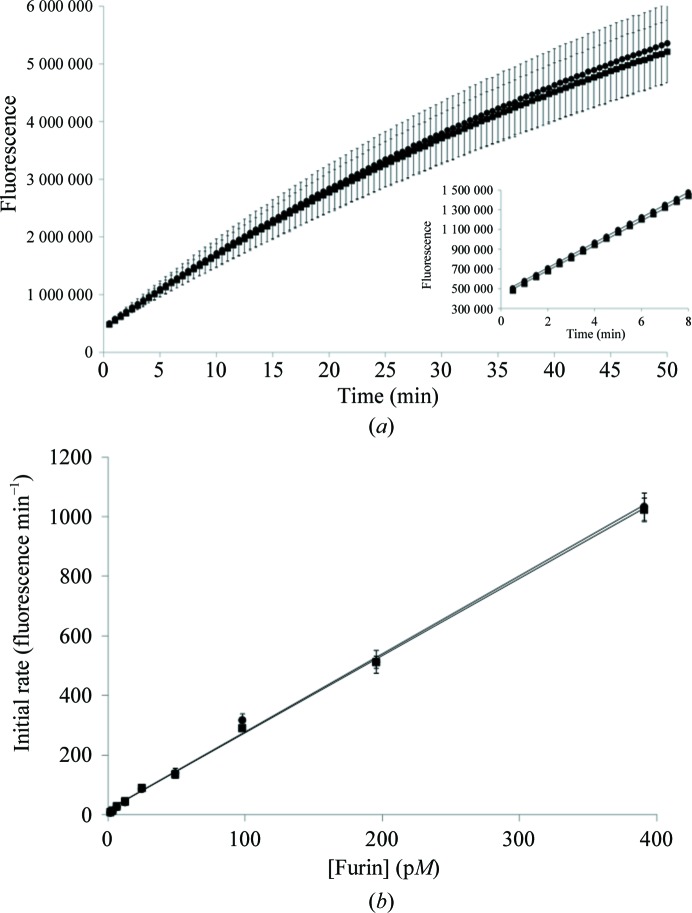
Both the glycosylated and nonglycosylated furin proteins were secreted as endoproteolytically cleaved products (Creemers et al., 1995 ▸) and were purified to homogeneity (Fig. 1 ▸). For the prodomain-processed, glycosylated aa108–574-TEV-FLAG-His6 furin protein, the experimental mass-spectrometric data typically showed multiple mass species in the range 60 774–58 879 Da, compared with the expected mass of 53 449 Da, owing to about 5–7 kDa from glycosylation adducts. The exact amount of glycosylation varied from preparation to preparation. The experimental mass of the prodomain-processed, nonglycosylated aa108–574-TEV-FLAG-His6 N387D/N440D furin protein was typically 53 445.5 Da, compared with the expected 53 451.8 Da, where the difference of approximately 6 Da is owing to the expected three disulfide bonds. As noted in Section 2, we added the TEV protease site (ENLYFQ↓G) to the expression constructs in case affinity-tag removal was necessary for crystallographic studies. However, digestion with TEV protease for the removal of the His6 and FLAG tags was not necessary for crystallization. We observed no evidence of glycosylation with the N387D/N440D double mutant even though it is reported that furin contains a potential additional glycosylation site at Asn553 (UniProtKB P09958; http://www.uniprot.org/uniprot/P09958). Typical protein yields were approximately 2–3.5 and 0.5–1.5 mg furin protein per litre of conditioned medium for the glycosylated aa108–574-TEV-FLAG-His6 furin and the nonglycosylated aa108–574-TEV-FLAG-His6 N387D/N440D furin, respectively. The activity of the glycosylated and nonglycosylated aa108–574 furin proteins was assessed using an internally quenched consensus substrate peptide (as described in Section 2). The progress curves for both enzymes were identical (Fig. 2 ▸ a). An enzyme-dilution test was completed for both glycosylated and nonglycosylated furin. Both enzymes displayed ideal enzymatic behavior over a wide range of concentrations (Fig. 2 ▸ b). Overall, these results show that N-linked glycosylation is not essential for furin ectodomain production using the BacMam technology and for enzymatic function (Roth, 2002 ▸).
Figure 1.
Furin ectodomain constructs and protein purification. (a) Schematic of the human furin protein along with the ectodomain expression construct discussed here. The location of the catalytic triad and the two N-linked glycosylation positions are noted on the full-length schematic. The amino-acid mutant positions are noted on the construct schematic. (b) Example of a Coomassie-stained SDS–PAGE gel showing the prodomain-processed, glycosylated aa108–574-TEV-FLAG-His6 furin protein (lane 1) and the prodomain-processed, nonglycosylated aa108–574-Tev-FLAG-His6 N387D/N440D furin protein (lane 2).
Figure 2.
Characterization of glycosylated and nonglycosylated furin protein. Glycosylated aa108–574-Tev-FLAG-His6 furin (circles) and nonglycosylated aa108–574-TEV-FLAG-His6 N387D/N440D furin (squares). (a) Comparison of furin activity progress curves for glycosylated and nonglycosylated furin using the FAM-TAMRA substrate peptide as noted in Section 2. (b) Comparison of enzymatic rate versus protein concentration.
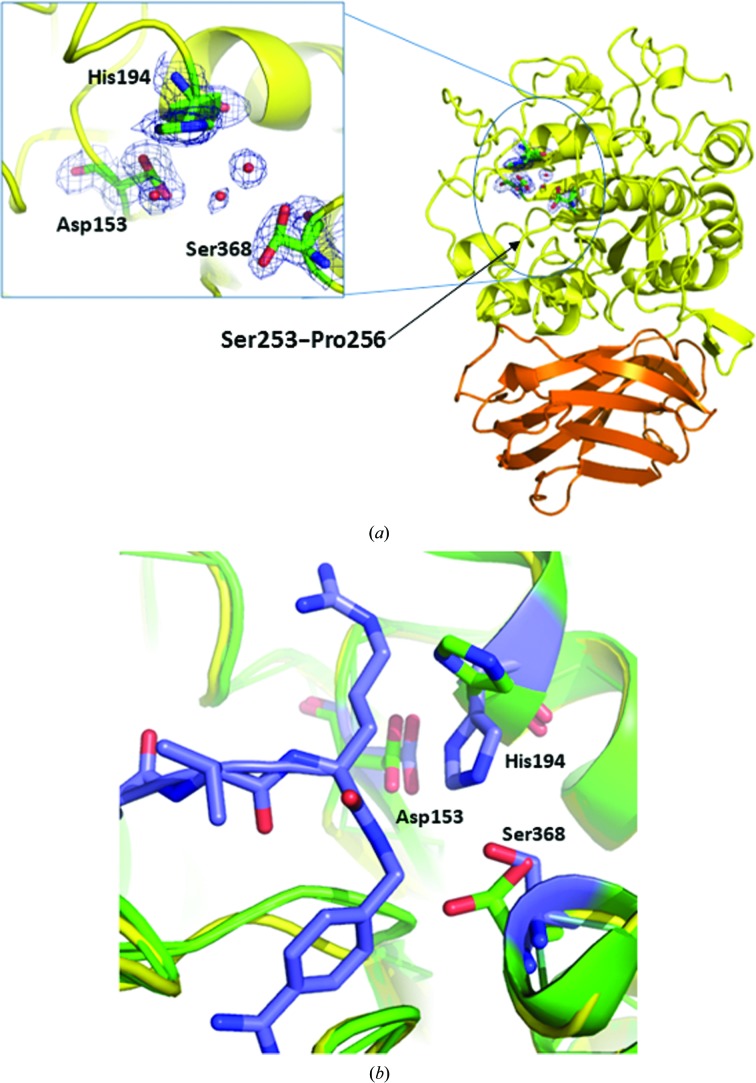
The global fold of the nonglycosylated apo human furin structure compares very favorably with the glycosylated forms of human and mouse furin bound to inhibitors (Fig. 3 ▸). This suggests that the nonglycosylated protein is fit for the purpose of inhibitor discovery and characterization studies. The catalytic domain of our human furin structure compares almost identically with the glycosylated human furin structures bound to peptidomimetic benzamidine inhibitors (PDB entries 4omc and 4omd; Dahms et al., 2014 ▸). The high sequence identity of 99.6% and structural homology yields an r.m.s.d. of 0.45 Å when the A chains are superimposed using the Coot SSM function. We note that the side chain of the catalytic triad residue His194 adopts a different rotamer to that in the liganded human glycosylated structures, but the side chains of Asp153 and Asn295 have similar conformations. The Ser368 side chain is observed as two rotamers in the apo structure, while in the glycosylated human structures it is found as a single rotamer. Finally, we observe that in the absence of the peptidomimetic inhibitor the Ser253–Pro256 strand containing Trp254 moves slightly away from the substrate cleft.
Figure 3.
Ribbon representation of the crystal structure of apo furin. (a) The catalytic domain is shown in yellow and the P domain is shown in orange. The inset is an enlarged view of the active-site triad with 2F o − F c electron density. Ser368 shows two rotamers fitted to the electron density. Ordered waters are rendered as red spheres. The black arrow points to the Ser253–Pro256 strand, as noted in Section 3. (b) Superposition of apo (green) with liganded (purple) furin (PDB entry 4omd). Apo furin shows differing rotamers for His154 and Ser368.
Likewise, the catalytic domain of our human furin structure compares almost identically to the glycosylated mouse furin structure bound to a peptidomimetic chloromethylketone inhibitor (PDB entry 1p8j; Henrich et al., 2003 ▸). The high sequence identity of 97.4% and structural homology yields an r.m.s.d. of 0.47 Å when the A chains are superimposed using the Coot SSM function. Similar to above, the side chain of the catalytic triad residue His194 adopts a different rotamer to that in the liganded mouse structure, but the Asp153 and Asn295 side chains are identical. The Ser368 side chain is observed as two rotamers in the apo structure, while in the mouse structure it contributes to forming the tetrahedral hemiketal. Finally, as noted above with human furin, we observe that in the absence of the chloromethylketone inhibitor the Ser253–Pro256 strand containing Trp254 is also slightly further away from the substrate cleft.
Our apo, nonglycosylated furin structure superimposes with the recently reported apo, glycosylated structure of Dahms et al. (2016 ▸) with an r.m.s.d. of 0.39 Å. Only a few locations in the sequence show differences of greater than 1.0 Å (the N-terminal residue at amino acid 110 and residues 125, 126, 188, 189, 440 and 558) and are at surface sites that are likely to be affected by crystal packing. One significant difference between the two reported apo structures is at the catalytic residue His154, where rotamer differences are obvious. We have performed numerous inhibitor soaks using furin apo crystals and these efforts will be the subject of subsequent reports. It should be noted that another recent report also shows that apo furin crystals are suitable for soaking small-molecule inhibitors (Dahms et al., 2017 ▸). In our experience, small-molecule inhibitors with inhibition constants in the high-micromolar range or more potent have yielded liganded structures (to be published at a later date).
Overall, we have shown that the BacMam method is capable of producing native furin ectodomain and nonglycosylated furin double-mutant protein from mammalian cells. Removal of the glycosylation sites reduced the protein yield, but did not cause problems with protein purification. Biochemical characterization studies show that glycosylation does not perturb the enzymatic activity of the furin ectodomain. Finally, our observation that nonglycosylated furin readily crystallizes as an apo protein will greatly enable future small-molecule inhibitor studies and drug-discovery efforts for both furin and related PCSKs.
Supplementary Material
PDB reference: human furin, 4z2a
Acknowledgments
We thank Dr Tom Meek for early ideas and support of this work. We also thank Catherine Simmons for early contributions and Wendy White for mass spectrometry. Finally, we thank Drs Allen Oliff, Jeff Axten, Melisa Ho and Sanjay Kumar for helpful discussions and support.
Funding Statement
This work was funded by GlaxoSmithKline grant .
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Ames, R. S., Kost, T. A. & Condreay, J. P. (2007). Exp. Opin. Drug. Discov. 2, 1669–1681. [DOI] [PubMed]
- Becker, G. L., Lu, Y., Hardes, K., Strehlow, B., Levesque, C., Lindberg, I., Sandvig, K., Bakowsky, U., Day, R., Garten, W. & Steinmetzer, T. (2012). J. Biol. Chem. 287, 21992–22003. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Condreay, J. P., Witherspoon, S. M., Clay, W. C. & Kost, T. A. (1999). Proc. Natl Acad. Sci. USA, 96, 127–132. [DOI] [PMC free article] [PubMed]
- Couture, F., Kwiatkowska, A., Dory, Y. L. & Day, R. (2015). Exp. Opin. Ther. Pat. 25, 379–396. [DOI] [PubMed]
- Creemers, J. W. M., Vey, M., Schäfer, W., Ayoubi, T. A. Y., Roebroek, A. J. M., Klenk, H.-D., Garten, W. & Van de Ven, W. J. M. (1995). J. Biol. Chem. 270, 2695–2702. [DOI] [PubMed]
- Dahms, S. O., Arciniega, M., Steinmetzer, T., Huber, R. & Than, M. E. (2016). Proc. Natl Acad. Sci. USA, 113, 11196–11201. [DOI] [PMC free article] [PubMed]
- Dahms, S. O., Hardes, K., Becker, G. L., Steinmetzer, T., Brandstetter, H. & Than, M. E. (2014). ACS Chem. Biol. 9, 1113–1118. [DOI] [PMC free article] [PubMed]
- Dahms, S. O., Hardes, K., Steinmetzer, T. & Than, M. E. (2018). Biochemistry, 57, 925–934. [DOI] [PubMed]
- Dahms, S. O., Jiao, G.-S. & Than, M. E. (2017). ACS Chem. Biol. 12, 1211–1216. [DOI] [PubMed]
- Dukkipati, A., Park, H. H., Waghray, D., Fischer, S. & Garcia, K. C. (2008). Protein Expr. Purif. 62, 160–170. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Henrich, S., Cameron, A., Bourenkov, G. P., Kiefersauer, R., Huber, R., Lindberg, I., Bode, W. & Than, M. E. (2003). Nature Struct. Mol. Biol. 10, 520–526. [DOI] [PubMed]
- Kadwell, S. H. & Overton, L. K. (2016). Baculovirus and Insect Cell Expression Protocols, edited by D. W. Murhammer, pp. 263–284. New York: Humana Press.
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Roth, J. (2002). Chem. Rev. 102, 285–303. [DOI] [PubMed]
- Scott, M. J., Modha, S. S., Rhodes, A. D., Broadway, N. M., Hardwicke, P. I., Zhao, H. J., Kennedy-Wilson, K. M., Sweitzer, S. M. & Martin, S. L. (2007). Protein Expr. Purif. 52, 104–116. [DOI] [PubMed]
- Seidah, N. G. & Prat, A. (2012). Nature Rev. Drug Discov. 11, 367–383. [DOI] [PubMed]
- Thomas, G. (2002). Nature Rev. Mol. Cell Biol. 3, 753–766. [DOI] [PMC free article] [PubMed]
- Ven, W. J. M. van de, Voorberg, J., Fontijn, R., Pannekoek, H., van den Ouweland, A. M. W., van Duijnhoven, H. L. P., Roebroek, A. J. M. & Siezen, R. J. (1990). Mol. Biol. Rep. 14, 265–275. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human furin, 4z2a