Abstract
The metabolic oligosaccharide engineering (MOE) strategy using unnatural sialic acids has recently enabled the visualization of the sialome in living systems. However, MOE only reports on global sialylation and dissected information regarding subsets of sialoside is missing. Here we describe the synthesis and utilization of sialic acids modified with a sydnone reporter for the metabolic labeling of sialoconjugates. We showed that the positioning of the reporter on the sugar significantly altered its metabolic fate. Further in vitro enzymatic assays revealed that the 9-modified neuraminic acid is preferentially accepted by the sialyltransferase ST6Gal-I over ST3Gal-IV, leading to the favored incorporation of the reporter into linkage specific α2,6-N-linked sialoproteins. This sydnone-sugar raises the possibility of investigating the roles of specific sialosides.
Keywords: chemical reporter, click chemistry, metabolic incorporation, sialic acids, sydnone
Graphical Abstract
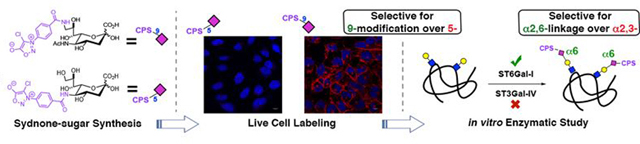
In search of selectivity: Incorporation of a clickable sydnone reporter onto sialic acid had a profound impact on its metabolic fate. While the 5-modified sugar was not utilized by cells, the 9-modified analog was strongly displayed on cell-surface sialosides. Further studies revealed that the 9-modified sialic acid is preferentially accepted by the sialyltransferase ST6Gal-I over ST3Gal-IV, leading to the incorporation of the reporter into linkage specific α2,6-N-linked sialoproteins.
Sialic acids are negatively charged carbohydrates that are usually found at the non-reducing end of complex glycans decorating cell-surface proteins and lipids.[1] They are therefore ideally placed for interacting with the cell’s exogenous environment and consequently for mediating physiological and pathological events including cell adhesion, host-pathogen interactions and cancer progression.[2] In mammals, the vast array of existing sialoconjugates are biosynthesized in a non-templated manner by 20 sialytransferases (STs) that catalyze the transfer of the same donor substrate (cytidine-5′-monophospho-N-acyl-neuraminic acid (CMP-Neu5Ac)) to various specific acceptor glycoconjugates.[3]
By exploiting the donor promiscuity of STs, the metabolic oligosaccharide engineering (MOE) strategy has emerged as a fundamental technology for interrogating the sialome in living systems.[4] In this two-step approach, a bioorthogonal chemical handle is first introduced into sialoconjugates by feeding cells a sialic acid modified with a reporter, which can then be selectively ligated to a complementary bioorthogonal probe for direct visualization or affinity capture. The inconspicuous organic azide has elegantly been employed as a versatile chemical reporter for the tagging of cell-surface sialoconjugates[5] that can be detected in live cells and living animals under strict metal-free conditions either by Staudinger ligation,[6] or by rapid 1,3-dipolar cycloaddition with tailored cyclooctynes (SPAAC).[7, 8] Terminal[9] and strained alkenes[10] have recently been utilized for tagging sialoconjugates in living cells, however they often suffer from poor reactivity with the complementary tetrazine probe, which has so far precluded their application in living animals.
Despite the many attractive features of employing azido-sugars for the MOE, azides can be reduced into amines in biological systems, by the action of thiols.[11] Controversially, this may lead to the accumulation of secondary byproducts with unknown metabolic fate and biological effects. To overcome this limitation, we envisaged that sialic acids fitted with a sydnone moiety, an aromatic mesoionic 1,3-dipole, would be valuable unnatural sugars for addition to the MOE toolbox. We and others have recently shown that sydnones can react in a metal-free strain-promoted click fashion with cyclooctyne reagents for the in vitro bioconjugation of purified proteins.[12–16]
Herein, we report on the preparation and utilization of sialic acids bearing a sydnone reporter as novel metabolic precursors for the labeling of sialoconjugates in living cells (Figure 1). We observed that while 5-modified neuraminic acid with sydnone (Neu5SydCl) was not metabolically incorporated into sialosides, a sialic acid analog having the sydnone moiety at its 9-position (Neu9NSydCl) was well tolerated by the biosynthetic machinery of the cell, leading to robust display of the reporter on cell-surface sialoconjugates. Interestingly, further in vitro enzymatic studies revealed that unlike CMP-Neu5Ac, CMP-Neu9NSydCl is not a universal donor substrate of all the STs involved in the synthesis of N-glycans, ultimately leading to the favored incorporation of the reporter into linkage specific α2,6-N-linked sialoproteins, a beneficial outcome, since MOE, while useful for reporting on global sialylation, is usually unable to differentiate among the various types of sialosides.[17–19]
Figure 1.
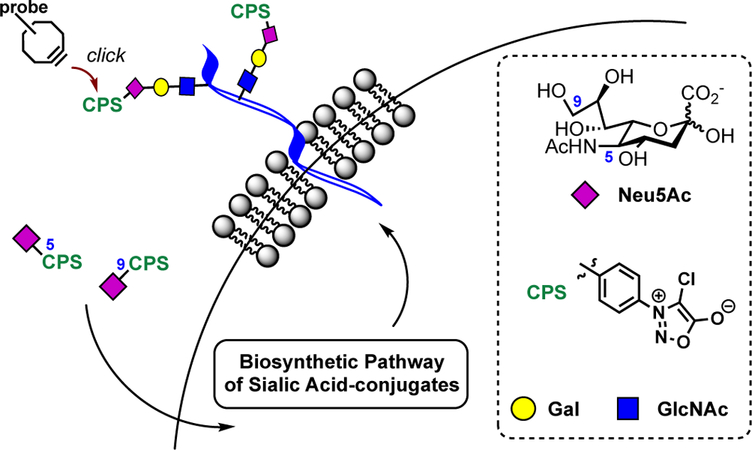
Chemical remodeling of cell-surface sialoconjugates using sydnone-modified sialic acids.
Performing organic reactions in living systems remains a formidable chemical challenge. Both the chemical reporter and the associated probe must not only exhibit stringent biostability but also fast and exquisite reactivity with each other. Consequently, we first examined the stability of chlorosydnones under simulated physiological conditions in the presence of glutathione (GSH). GSH is a cellular thiol-containing tripeptide present in millimolar concentrations in the cytosol of mammalian cells. We focus our attention on chlorosydnone since we have previously shown that introduction of a chlorine atom on the sydnone ring significantly enhanced its reactivity.[15, 16] In this context, we incubated an aqueous solution of N-chlorosydnone-benzoyl mannosamine 1 (ManNSydCl), and for reference N-azidoacetyl mannosamine, having an anomeric p-methoxyphenyl group (ManNAz-PMP 2, Scheme S1 for synthetic details), supplemented with 100-fold GSH at 37 °C, and monitored their respective stability over time by HPLC (Figures S1-S2). Gratifyingly, both α- and β-anomers of 1 were found highly stable, as no decomposition was observed for over 72 h. Conversely, incubating the azide 2, quickly led to the appearance of a new HPLC peak with shorter retention time, that was identified as the expected reduced product by NMR spectroscopy and mass spectrometry, highlighting the greater biostability of the sydnone reporter.
Next, to identify the most reactive cyclooctynyl-probe with the chlorosydnone reporter, we determined the second-order rate constants of cycloadditions of three representative cyclooctynes commonly used in bioconjugation, MeO-DIBO,[20] MeO-DIBAC[21] and BCN[22], with 4-chloro-3-phenylsydnone (3) by 1H-NMR spectroscopy in CD3OD at 25 °C (Figures S3-S6). While 3 reacted slowly with MeO-DIBO (k = 0.06 M−1 s−1), its rate constant with MeO-DIBAC (k = 0.30 M−1 s−1) was on par with the cycloaddition of DIBAC with azide. Interestingly, 3 reacted notably faster with the less sterically hindered BCN (k = 0.57 M−1 s−1), at a suitable rate constant for the labeling of complex glycans in living cells.
To validate the physiological applicability of our system, we next turned our attention to employing the chlorosydnone reporter for the labeling of sialosides in living cells. Accordingly, we synthesized two unnatural neuraminic acids modified either at their 5- or 9-position with a chlorosydnone. Neu5SydCl (4) was prepared enzymatically from ManNSydCl (1) in 88% yield using recombinant Sialic acid Aldolase from E. coli, in the presence of sodium pyruvate (Scheme 1A). On the other hand, incorporation of the chlorosydnone onto the 9-position of sialic acid was achieved by chemical modification of 9-azido neuraminic acid 5 (Neu9Az) (Scheme 1B). Briefly, Neu9Az[23] was reduced into the amine 6 by hydrogenation over Pd/C which was subsequently used for the incorporation of the chlorosydnone using classical N-hydroxysuccinimide coupling protocol, providing the desired Neu9NSydCl (7). Following a similar approach, we also prepared the azido-analog Neu9NAz (8).
Scheme 1.
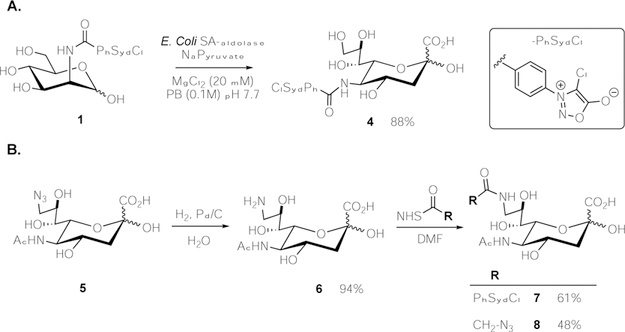
Preparation of 5- and 9-modified neuraminic acids. A. Enzymatic synthesis of Neu5SydCl (4); B. Chemical synthesis of Neu9NSydCl (7) and Neu9NAz (8).
Next, U2OS cells were incubated for 48 h with 2 mM of neuraminic acids, Neu5Ac (negative control), Neu9NAz (8) (positive control), and both sydnone modified compounds Neu5SydCl (4) and Neu9NSydCl (7). Importantly, even at high concentration, none of the unnatural sugars exhibited any particular cytotoxicity (Figures S9-S10). Labeled cells were then reacted with BCN-biotin conjugate (50 μM) for 1 h and stained with Streptavidin-AlexaFluor568 for analysis by confocal fluorescence microscopy (Figure 2A). Robust cell surface staining was observed when Neu9NSydCl (7) was employed, demonstrating the successful metabolic labeling of chlorosydnone into sialosides of living cells. To note, while employing BCN has previously been reported to be unspecific due to cross-reactivity with endogenous thiols,[24] we did not observe any substantial unspecific labeling by flow cytometry (Figure 2B and Figure S8), perhaps because our protocol only reports on cell-surface modification. Flow cytometry analysis also revealed that the cell-surface incorporation of Neu9NSydCl (7) was as efficient as the azido-analog 8 (Figure 2B). In marked contrast, performing the MOE with Neu5SydCl (4) resulted in no cell membrane staining (Figure 2), indicating that positioning of the chemical reporter on the sialic acid scaffold is of prime importance for a successful MOE. To ensure that this noted difference in cell metabolization was specific to the reporter and not cell-type dependent, we also performed the MOE on human lung carcinoma (A549) cells. Fluorescence microscopy only showed cell-surface staining with Neu9NSydCl (7) (Figure S7), suggesting that the cellular discrimination between the 5- and 9-modified sialic acid with the chlorosydnone is an intrinsic feature of sialoconjugates production.
Figure 2.
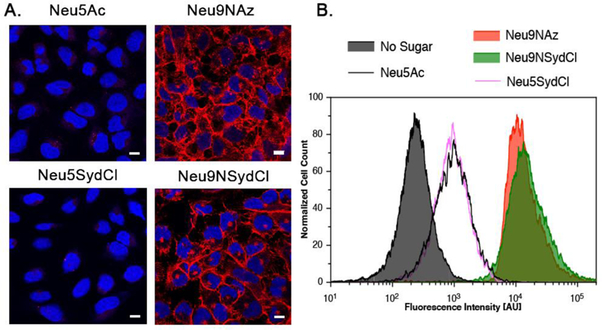
Metabolic glycan labeling of living U2OS cells with modified neuraminic acids. A. Fluorescence microscope images of U2OS cells cultured with Neu5Ac, Neu9NAz (8), Neu5SydCl (4) and Neu9NSydCl (7), treated with BCN-biotin and stained with Streptavidin-AlexaFluor568 (Red) and DAPI (Blue), Scale bar: 10 μm. B. Flow cytometry analysis of U2OS cells cultured respectively with No Sugar, Neu5Ac, Neu9NAz (8), Neu5SydCl (4) and Neu9NSydCl (7). All cells were treated with BCN-biotin and Streptavidin-AlexaFluor568.
Intrigued by the non-metabolization of Neu5SydCl (4) by cells, we decided to investigate which biosynthetic enzyme may be responsible for blocking its incorporation into sialosides. To be integrated into complex glycans, cytosolic sialic acids must first be activated in the nucleus into their CMP-analogs by the action of CMP-sialic acid synthetase (CSS), followed by their transport to the Golgi where they are utilized by sialyltransferases (STs).[25] To investigate the substrate tolerance of CSS, we devised an in vitro enzymatic-1H-NMR coupled assay using bacterial CSS from N. meningitidis (NmCSS). We selected NmCSS over its mammalian analog due to its commercial availability. Favorably, murine CSS and NmCSS have been shown to share a similar overall active-site architecture involved in substrate binding.[26] Accordingly, Neu5Ac and its unnatural analogs Neu5SydCl (4), Neu9NSydCl (7) and Neu9NAz (8) were individually incubated at 37 °C with NmCSS and CTP (Figure 3A), and the crude mixture was analyzed by 1H-NMR spectroscopy to determine the formation of the respective CMP-sialic acids (Figure 3B and Figures S11-S14). The chemical shifts of the protons present on the C3-carbon atom of sialic acid (dashed lines in Figure 3) provided distinct peaks changes between 1.6 and 2.6 ppm upon CMP-activation, ideal to detect product formation. After 5 h, while over 90% of Neu5Ac was converted into CMP-Neu5Ac, modification at the 9-position with either an azido (Neu9NAz) or a chlorosydnone (Neu9NSydCl) moiety only slightly interfered with the CMP-activation with a product conversion of 75% and 50%, respectively. However, having the chlorosydnone on the 5-position of sialic acid seemed to prevent NmCSS from generating the desired CMP-Neu5SydCl, as no product was detected, even upon prolonged reaction time (up to 24 h, Figure S13). Collectively, these results suggest that Neu5SydCl (4) is not recognized by CSS as an appropriate substrate and is therefore not activated into its CMP-analog, which precludes its incorporation into cell-surface glycans.
Figure 3.
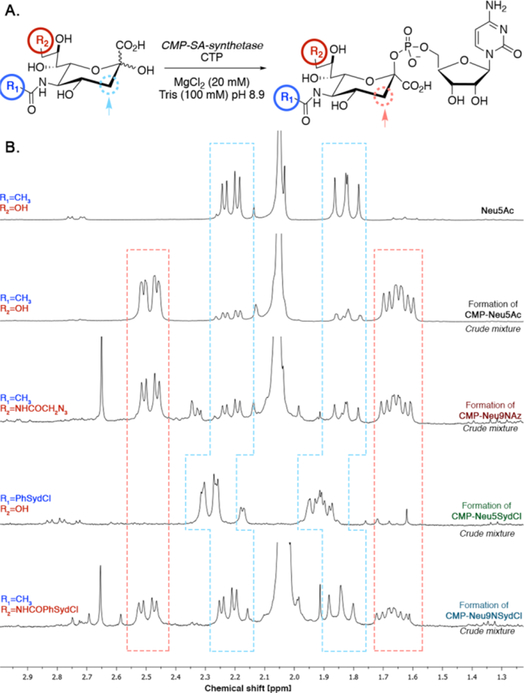
Enzymatic-1H-NMR coupled assay for the detection of CMP-sialic acids formation. A. Enzymatic formation of CMP-sialic acid using NmCSS. B. 1H-NMR spectra of the corresponding crude reaction mixtures for CMP-Neu5Ac, -Neu9NAz, -Neu5SydCl and -Neu9NSydCl. Protons on the C3-carbon atom of the starting sialic acid (dashed blue line) and the CMP-sialic acid product (dashed orange line).
Having established that the presence of the sydnone reporter on sialic acid could have dramatic impact on enzymes substrate recognition, we sought to examine the promiscuity of STs involved in the biosynthesis of N-linked sialoproteins towards CMP-Neu9NSydCl (9). We chose to investigate N-glycan structures because they are the most prevalent modifications found in vertebrate glycoproteins.[27] Sialylation of N-glycoproteins is mainly controlled by ST6Gal-I and ST3Gal-IV that adds a sialic acid residue to N-acetyllactosamine (LacNAc) termini of N-glycans in an α2,6- and α2,3-linkage, respectively.
To determine the tolerance of human ST6Gal-I and ST3Gal-IV in terms of accepting CMP-sialic acid analogs, we measured the kinetic parameters of both enzymes for the transfer of CMP-Neu5Ac and its unnatural analogs CMP-Neu9NSydCl (9) and CMP-Neu9Az (10) to LacNAc-butyl (11), as a model acceptor (Table 1 and Figures S15-S16). ST6Gal-I seemed to be promiscuous in regards to utilizing CMP-sialic acid analogs with a slight preference for sydnone 9, exhibiting kcat/KM values of similar order. In contrast, the activity of ST3Gal-IV was clearly more impacted by the presence of the reporters. ST3Gal-IV displayed a 10-fold decrease in kcat/KM values for sydnone 9 (kcat/KM = 1.7 ± 1.4 mM−1 min−1, the lack of recognition of 9 by ST3Gal-IV prevented Vmax to be reached (Figure S16) and therefore the KM(app) and kcat(app) values should be considered estimates) compared to azide 10 and the natural CMP-Neu5Ac substrate, suggesting that in a physiological context, while CMP-Neu9Az (10) would similarly be employed by both enzymes, CMP-Neu9NSydCl (9) would be preferentially utilized by ST6Gal-I over ST3Gal-IV.
Table 1.
Kinetic constants of the sialylation of LacNAc-butyl (11) by human ST6Gal-I or ST3Gal-IV in the presence of CMP-Neu5Ac, -Neu9Az (10) or -Neu9NSydCl (9)a
| ST6Gal-I | ST3Gal-IV | |||||
|---|---|---|---|---|---|---|
| Donor | KM(app) [μM] | kcat(app) [min−1] | kcat/KM [mM−1.min−1] | KM(app) [μM] | kcat(app) [min−1] | kcat/KM [mM−1.min−1] |
| CMP-Neu5Ac | 119 ± 16 | 11 ± 0.4 | 92 ± 25 | 300 ± 66 | 1.4 ± 0.1 | 4.7 ± 1.5 |
| CMP-Neu9Az (10) | 385 ± 88 | 18 ± 1.7 | 47 ± 19 | 237 ± 59 | 3.4 ± 0.3 | 14.3 ± 5.1 |
| CMP-Neu9SNSydCl (9) | 86 ± 21 | 10 ± 0.7 | 116 ± 33 | 3057 ± 2720 | 5.2 ± 3.7 | 1.7 ± 1.4 |
Data obtained using fixed enzyme and LacNAc-butyl concentrations while varying the concentrations of the CMP-sialic acid donors.
To further validate the preferential incorporation of Neu9NSydCl (7) into α2,6-N-sialoproteins in a more complex system, we incubated asialofetuin, a model glycoprotein displaying LacNAc terminated N-glycans, with human ST6Gal-I or ST3Gal-IV in the presence of either CMP-Neu5Ac or its unnatural analogs chlorosydnone 9 and azide 10 overnight at 37 °C in cacodylate buffer (50 mM, pH 7.5). The labeled proteins were then treated with BCN-FITC conjugate at 37 °C for 3 h to reveal the presence of the reporters by in gel-fluorescence imaging (Figure 4). A robust fluorescent band, corresponding to enzymatically sialylated asialofetuin, was clearly detected when both azido- and sydnone-modified CMP-sialic acids were employed in the presence of ST6Gal-I, confirming the promiscuity of this enzyme. In sharp contrast, when ST3Gal-IV was utilized, fluorescent labeling was only observed in the presence of azide 10, highlighting the selective incorporation of Neu9NSydCl (7) into α2,6-N-sialoproteins in a more complex biochemical context.
Figure 4.
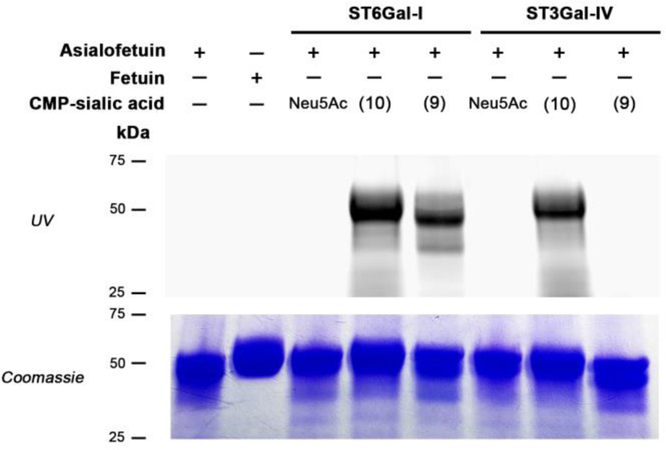
In-gel visualization of asialofetuin (2.4 mg/mL) incubated with human ST6Gal-I (19.5 μg/mL) or ST3Gal-IV (19.5 μg/mL) in the presence of CMP-Neu5Ac (1.2 mg/mL) or its unnatural analogs CMP-Neu9Az (10) (1.2 mg/mL) or CMP-Neu9NSydCl (9) (1.7 mg/mL), or no reagent (−). The presence of the chemical reporter was detected by fluorescence imaging (incubation with BCN-FITC (top row; λexc: trans UV)) and by Coomassie Blue stain to reveal total protein content (bottom row).
In summary, we have demonstrated that chlorosydnones are valuable chemical reporters for the MOE of living cells. In particular, they not only proved to react positively fast with strained cyclooctynes but also showed high stability in the presence of thiols. In addition, we showed that unlike azide, the chlorosydnone reporter is not an innocent bystander and its presence on sialic acid was not unnoticed by the biosynthetic pathway of sialoconjugates. For instance, while employing Neu9NSydCl led to robust cell-surface labeling, Neu5SydCl was not metabolized by cells and we identified CMP-sialic acid synthetase as the enzymatic roadblock. Furthermore, CMP-Neu9NSydCl was shown to be preferentially accepted by the sialyltransferase ST6Gal-I over ST3Gal-IV, leading to the favored incorporation of the sydnone reporter into linkage specific α2,6-N-linked sialoproteins. Neu9NSydCl raises therefore the possibility to selectively track and capture a subset of sialosides, a desirable outcome, since MOE with existing azido-sugars can only report on global sialylation. Further studies are currently being performed in our laboratory to explore the impact of chlorosydnone on a larger panel of human sialyltransferases.
Supplementary Material
Acknowledgments
Financial support from CNRS (ATIP-Avenir program to F.F.) is gratefully acknowledged. This study has also received financial support from the French State in the frame of the “Investments for the Future” Programme IdEx Bordeaux (ANR-10-IDEX-03–02 to F.F.) and from the Laboratory of Excellence TRAIL (ANR-10-LABX-57 to F.F.), EraNet Neuron TRAINS, and US National Institutes of Health grants (P41GM103390, P01GM107012, and R01GM130915 to K.W.M.).
References
- [1].Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, Essentials of Glycobiology, 2nd ed., Cold Spring Harbor Laboratory Press, 2009. [PubMed] [Google Scholar]
- [2].Varki A, Nature 2007, 446, 1023–1029. [DOI] [PubMed] [Google Scholar]
- [3].Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P, Biochimie 2001, 83, 727–737. [DOI] [PubMed] [Google Scholar]
- [4].Wratil PR, Horstkorte R, Reutter W, Angew. Chem. Int. Ed 2016, 55, 9482–9512. [DOI] [PubMed] [Google Scholar]
- [5].Saxon E, Bertozzi CR, Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- [6].Prescher JA, Dube DH, Bertozzi CR, Nature 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- [7].Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Friscourt F, Boons GJ, in Click Chemistry in Glycoscience: New Developments and Strategies, 2013, pp. 211–233. [Google Scholar]
- [9].Niederwieser A, Spate AK, Nguyen LD, Jungst C, Reutter W, Wittmann V, Angew. Chem. Int. Ed 2013, 52, 4265–4268. [DOI] [PubMed] [Google Scholar]
- [10].Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA, J. Am. Chem. Soc 2012, 134, 18638–18643. [DOI] [PubMed] [Google Scholar]
- [11].Handlon AL, Oppenheimer NJ, Pharm. Res 1988, 5, 297–299. [DOI] [PubMed] [Google Scholar]
- [12].Wallace S, Chin JW, Chem. Sci 2014, 5, 1742–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Narayanam MK, Liang Y, Houk KN, Murphy JM, Chem. Sci 2016, 7, 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bernard S, Audisio D, Riomet M, Bregant S, Sallustrau A, Plougastel L, Decuypere E, Gabillet S, Kumar RA, Elyian J, Trinh MN, Koniev O, Wagner A, Kolodych S, Taran F, Angew. Chem. Int. Ed 2017, 56, 15612–15616. [DOI] [PubMed] [Google Scholar]
- [15].Favre C, de Cremoux L, Badaut J, Friscourt F, J. Org. Chem 2018, 83, 2058–2066. [DOI] [PubMed] [Google Scholar]
- [16].Favre C, Friscourt F, Org. Lett 2018, 20, 4213–4217. [DOI] [PubMed] [Google Scholar]
- [17].Moller H, Bohrsch V, Bentrop J, Bender J, Hinderlich S, Hackenberger CPR, Angew. Chem. Int. Ed 2012, 51, 5986–5990. [DOI] [PubMed] [Google Scholar]
- [18].Mbua NE, Li XR, Flanagan-Steet HR, Meng L, Aoki K, Moremen KW, Wolfert MA, Steet R, Boons GJ, Angew. Chem. Int. Ed 2013, 52, 13012–13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nischan N, Kohler JJ, Glycobiology 2016, 26, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Friscourt F, Ledin PA, Mbua NE, Flanagan-Steet HR, Wolfert MA, Steet R, Boons GJ, J. Am. Chem. Soc 2012, 134, 5381–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Debets MF, Prins JS, Merkx D, van Berkel SS, van Delft FL, van Hest JCM, Rutjes F, Org. Biomol. Chem 2014, 12, 5031–5037. [DOI] [PubMed] [Google Scholar]
- [22].Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes F, van Hest JCM, Lefeber DJ, Friedl P, van Delft FL, Angew. Chem. Int. Ed 2010, 49, 9422–9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Han S, Collins BE, Bengtson P, Paulson JC, Nat. Chem. Biol 2005, 1, 93–97. [DOI] [PubMed] [Google Scholar]
- [24].van Geel R, Pruijn GJM, van Delft FL, Boelens WC, Bioconjugate Chem 2012, 23, 392–398. [DOI] [PubMed] [Google Scholar]
- [25].Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VDP, Yarema KJ, Glycobiology 2009, 19, 1382–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Krapp S, Munster-Kuhnel AK, Kaiser JT, Huber R, Tiralongo J, Gerardy-Schahn R, Jacob U, J. Mol. Biol 2003, 334, 625–637. [DOI] [PubMed] [Google Scholar]
- [27].Moremen KW, Tiemeyer M, Nairn AV, Nat. Rev. Mol. Cell Biol 2012, 13, 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


