Abstract
To date, a large number of long non-coding RNAs (lncRNAs) have been recently discovered through functional genomics studies. Importantly, lncRNAs have been shown, in many cases, to function as master regulators for gene expression and thus, they can play a critical role in various biological functions and disease processes including cancer. Although the lncRNA-mediated gene expression involves various mechanisms, such as regulation of transcription, translation, protein modification, and the formation of RNA-protein or protein-protein complexes, in this review we discuss the latest developments primarily in important cell signaling pathways regulated by lncRNAs in cancer.
Introduction
It is well known that although a large number of RNA species are transcribed from the human genome, protein-coding sequences account for a very small fraction of total transcripts (Birney et al 2007, Kapranov et al 2007). The rest of transcripts are non-coding RNAs (ncRNAs) and they lack the coding potential except for those capable of producing functional small peptides (Anderson et al 2015, Nelson et al 2016). ncRNAs were initially considered as transcriptional noises (Gibb et al 2011), however, it has become increasingly apparent that they may play a critical role in diverse cellular processes from normal development to disease processes (Esteller 2011, Fatica and Bozzoni 2014, Wang et al 2013a, Wilusz 2016). There are two major classes of regulatory non-coding RNAs based on their size, 1) small non-coding RNAs with less than 200 nt in length, represented by microRNAs and 2) long non-coding RNAs (lncRNAs) with larger than 200 nt in length. As a well-characterized group of ncRNAs, microRNAs cause gene silencing by mRNA degradation or translation repression via the RNA interference pathway (Hayes et al 2014). On the other hand, gene expression regulated by lncRNAs is more complex, often involving multiple mechanisms (Rinn and Chang 2012).
As a matter of fact, lncRNAs were discovered a long time ago although the term ‘lncRNA’ had not been coined until recently. For example, XIST and H19 were reported in 1980s and 1990s through screening of cDNA libraries (Bartolomei et al 1991, Brown et al 1992). To date, there are overwhelming numbers of lncRNAs reported in different databases such as non-code database (http://www.noncode.org), and LNCipedia (http://www.lncipedia.org). The number of lncRNAs continues growing (Blythe et al 2016).
There are several ways to classify lncRNAs, one of which is based on the genomic level organization. In this regard, there are at least four groups of lncRNAs based on their relative positions to protein-coding genes: 1) intergenic lncRNAs, i.e., lincRNAs, that are transcribed from DNA sequences between two protein-coding genes; 2) intronic lncRNAs are those that are generated from introns of protein-coding genes; 3) overlapping lncRNAs are defined as transcripts overlapping known protein coding genes; and 4) antisense lncRNAs that are transcribed in an opposite direction to a protein-coding gene (Katayama et al 2005, Marques and Ponting 2014, Mattick and Rinn 2015, Ulitsky and Bartel 2013). Regardless of their genomic organization, it appears that most, if not all, of these lncRNAs can regulate gene expression and thus, they could be major players in cancer biology.
Regulation of gene expression by lncRNAs
Various mechanisms have been implicated in the lncRNA-mediated gene regulation, which can be attributed to their ability to interact with DNA, RNA, or protein. For example, lncRNAs may serve as signals to promote transcription, or as decoys to repress transcription, or as epigenetic regulators, or as scaffolds to interact with various protein partners to form ribonucleoprotein complexes (Mercer et al 2009, Ulitsky and Bartel 2013, Wang and Chang 2011). Based on levels of gene expression, lncRNA-mediated gene expression may take place at transcriptional and/or posttranscriptional levels.
At the transcriptional level, lncRNAs may participate in direct transcription by interacting with transcriptional complex or DNA elements such as promoters involved in transcription. More broadly, lncRNAs may be involved in modulation of chromatin structures by recruiting chromatin modifying enzymes, leading to expression or repression of a large number of genes. For instance, lncRNAs can interact with DNA binding proteins to preclude the access of these proteins to DNA recognition elements such that transcription is either induced or repressed depending on the nature of targeted proteins. In this regard, GAS5 has been shown to compete with glucocorticoid response element (GRE) on the DNA by occupying the DNA-binding domain of the glucocorticoid receptor (GR) and prevent the access of GR to target DNA (Kino et al 2010). Similarly, PANDA, a p53 inducible lncRNA, interacts with transcription factor NF-YA to titrate NF-YA away from the target gene containing chromatin (Baldassarre and Masotti 2012). There are many reports that lncRNAs can recruit chromatin modifying enzymes such that they impact their expression at the epigenetic level (Meller et al 2015). A notable example is HOTAIR which plays a critical role in cancer metastasis through the interaction with the polycomb repressive complex 2 (PRC2) (Gupta et al 2010). In this case, HOTAIR alters histone (H3K27) methylation pattern and gene expression to promote tumor cell invasion. On the other hand, suppression of HOTAIR causes reduction of cell invasion. This is primarily attributed to the ability of HOTAIR to serve as a scaffold for selected histone modification enzymes, impacting expression of a specific set of genes (Tsai et al 2010).
Posttranscriptional regulation primarily involves mRNA stability, splicing and modifications; it can also include translational regulation and protein stability, and subcellular localization. It is well known that certain mRNAs have a very short half-life, especially for early genes such as c-Jun and c-Myc. However, their stability can be significantly increased in cancer cells. In the case of c-Myc, mRNA turnover is a major mechanism of posttranscriptional regulation in part through an AU rich element (ARE) in the 3′-untranslated region (3′-UTR) and ARE binding factors such as AUF1 because ARE binding by AUF1 is associated with the mRNA stability of a target gene (DeMaria and Brewer 1996). In support of this notion, our recent study suggests that the interaction of Linc-RoR with hnRNP I and/or AUF1 impacts the c-Myc stability (Huang et al 2016). In this way, Linc-RoR is involved in the selective regulation of mRNA stability of specific genes such as c-Myc.
Alternative splicing is an important mechanism for genetic diversity and thus, it can impact cancer initiation and progression. It is known that the vast majority of eukaryotic genes can be expressed as various alternative splice variants (Pan et al 2008). However, regulation of gene splicing is often complex; splicing factors are not the only factors required for gene splicing. Moreover, these factors often work together to select correct splice sites at a given condition. Recent studies suggest that lncRNAs are also involved in the regulation of alternative splicing. LncRNAs may cooperate with heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNP A1 (Kashima et al 2007) and hnRNP C (Zarnack et al 2013) to facilitate their binding to exonic splicing silencer (ESS) or intronic splicing silencer (ISS) elements for alternative splicing. On the other hand, MALAT1 can regulate alternative splicing of many genes by interaction with the serine–arginine (SR) proteins and influencing their subnuclear localization (Tripathi et al 2010). Our recent studies indicate that PCGEM1 and BC200 regulate alternative splicing of androgen receptor (AR) and Bcl-x, respectively, through interaction with various splicing factors (Singh et al 2016, Zhang et al 2016b).
Translational regulation provides another mechanism for lncRNA-mediated gene expression. In this regard, lincRNA-p21 serves as an inhibitor of JunB and β-catenin translation in a HuR dependent manner (Yoon et al 2012). The mRNA UTRs often serve a regulatory element for translational control. For instance, lncRNA antisense (Uchl1) promotes UCHL1 protein synthesis by enhancing the interaction of the 5′ overlapping sense protein-coding mRNA with active polysomes (Carrieri et al 2012). Several reports indicate that p53 translation can be regulated through its 5′ or 3′ UTR (Halaby and Yang 2007) (Grover et al 2008). In particular, we show that Linc-RoR is able to suppress p53 translation by interacting with the phosphorylated hnRNP I, and blocking its interaction with p53 5′-UTR (Zhang et al 2013a).
Of interest, different lncRNAs may share the same target. A well-known target is Ezh2 which can be targeted by multiple lncRNAs. For instance, HOTAIR interacts with Ezh2 to suppress a number of genes such as JAM2 whereas it induces genes such as SNAIL (Gupta et al 2010). Several other lncRNAs can also target Ezh2. For example, suppression of MALAT1 increases E-cadherin expression, whereas it reduces β-catenin expression in an Ezh2 dependent manner (Hirata et al 2015). Furthermore, ANCR regulates the stability of Ezh2, leading to suppression of invasion and metastasis of breast cancer (Li et al 2017). These findings further support the complexity of lncRNA-mediated gene regulation. However, it remains to be determined as to whether these lncRNAs simultaneously or selectively target Ezh2 in different cellular contents, and Ezh2 is essential to regulation of gene expression by these lncRNAs. Thus, systematic characterization of this regulatory network may provide a better answer to this question.
Cell signaling pathways
Cell signaling plays a pivotal role in a variety of cellular processes in response to intracellular or extracellular stimuli. Numerous signaling pathways have been identified in the cell, and they are often critical to a cascade of biological reactions and gene expression. A given signal pathway often consists of a number of signaling molecules. Although these signaling molecules may not be directly involved in transcription, they can ultimately impact gene expression because these signaling molecules can regulate activity of transcription factors directly or indirectly. Emerging evidence suggests that lncRNAs are involved in various pathways. Due to limited space, we selectively discuss those we believe the most important to cancer, including p53, NF-κB, PI3K/AKT and Notch.
LncRNA and p53 regulatory network
p53 is a well-known tumor suppressor and is frequently mutated or deleted in cancer. p53 can regulate a large set of genes involved cell cycle, apoptosis and DNA repair. Therefore, the cellular p53 level is delicately balanced. Regulation of p53 level takes place primarily at translation, and posttranslational modifications and protein stability. Increasing evidence suggests that in addition to protein factors, lncRNAs also play a role in this p53 regulatory network (Zhang et al 2014). For instance, a large number of lncRNAs have been recently shown to serve as p53 effectors, participating in downstream events in a p53-dependent manner. Of great interest, among those p53-regulated lnRNAs, some members of this group of lncRNAs may also regulate p53 expression or its stability through an auto-regulatory feedback loop.
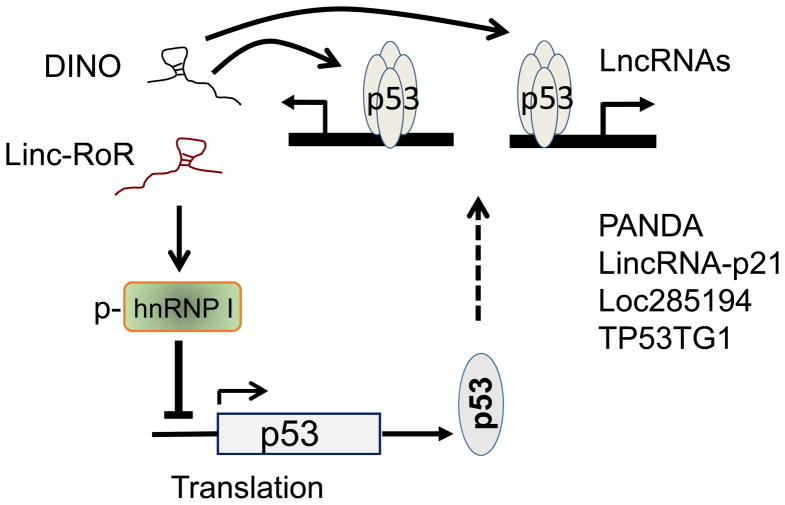
The p53 protein level is usually low under normal physiological conditions. However, upon DNA damage, p53 is substantially increased primarily due to the increased stability. Protein modifications such as phosphorylation and acetylation play an important role in such a way that the major p53 inhibitor MDM2 is no longer able to direct p53 to degradation. The role of lncRNAs in regulation of p53 stability has been demonstrated by a recent study (Schmitt et al 2016). DINO (Damage Induced Noncoding) is induced by DNA damage in a p53-dependent manner. In this regard, DINO is required for the p53-mediatd phenotypes, including cell cycle arrest and apoptosis in response to DNA damage. In particular, DINO expression alone is sufficient to activate damage signaling and cell cycle arrest (Schmitt et al 2016), suggesting that DINO can mimic the function of DNA damage-induced p53. Mechanistically, DINO physically interacts with p53 to stabilize it; this stabilized p53 in turn transactivates the downstream targets including DINO itself. Thus, this is a positive feed-back loop such that the DNA damage-induced p53 is greatly amplified (Fig. 1).
Fig. 1. p53 and lncRNA auto-feedback regulatory network.
Both DINO and Linc-RoR are p53 transcriptional targets. While DINO stabilizes p53, Linc-RoR inhibits p53 translation.
However, once p53 is induced, the signal has to be turned down or shut off, otherwise the cell may also suffer from p53-induced damage. It is well known that MDM2 is a major player to keep p53 level low under normal conditions. After acute DNA damage, the cell may also need a mechanism to rapidly turn p53 off. However, MDM2 alone may be not sufficient where other factors such as Linc-RoR can provide an additional mechanism because Linc-RoR can turn down p53 translation (Zhang et al 2013a).
Our study indicates that Linc-RoR can be induced by p53 in response to DNA damaging agents. Consistent with this finding, Linc-RoR promoter carries several conserved p53-binding sites and p53 interacts with these sites, as determined by ChIP assays (Zhang et al 2013a). Of interest, Linc-RoR is also able to suppress p53 (see below) to keep p53 in check (Fig. 1).
This Linc-RoR-mediated repression of p53 involves hnRNP, an RNA binding protein with multiple functions. The interaction of hnRNP I with the 5′-UTR of p53 mRNA promotes p53 translation (Grover et al 2008). Our study indicates that Linc-RoR can also interact with hnRNP I. Once bound by Linc-RoR, hnRNP I is no longer able to stimulate p53 translation (Zhang et al 2013a). Of interest, although hnRNP I is an abundant nuclear protein, a small fraction (<5%) is phosphorylated (p-hnRNP I) and is detected in the cytoplasm. Further study indicates that only p-hnRNP I is able to interact with Linc-RoR. Therefore, this forms a competition between Linc-RoR and the 5′-UTR of p53 mRNA for hnRNP I, ultimately impacting p53 translation (Zhang et al 2013a). In addition, miR-145 serves as a direct target of p53 and thus, DNA damage induces p53-mediated miR-145 expression (Sachdeva et al 2009), which in turns is able to target Linc-RoR through CeRNA mechanisms (Wang et al 2013b). In this way, the elevated miR-145 also helps keep Linc-RoR level low. Evidently, these mechanisms can work together to control the cellular p53 level in response to environmental cues.
To date, a large number of p53-regulated lncRNAs have been reported and they can impact various cellular processes. Most of them are direct transcriptional targets of p53, such as lincRNA-p21 functioning as a repressor in the p53 pathway (Huarte et al 2010). Similarly, PANDA is involved in p53-regulated cell cycle progression and apoptosis (Hung et al 2011). Our study indicates that Loc285194 serves as a p53 effector, suppressing tumor cell growth (Liu et al 2013).
Some of these p53 effector lncRNAs are involved in stemness while others play a role in DNA repair. For example, LncPRESS1 was identified as a p53-regulated lncRNA by RNA-seq and ChIP-seq. LncPRESS1 is highly expressed in human embryonic stem cells and it is repressed by p53 during differentiation (Jain et al 2016). On the other hand, TP53TG1 is tumor suppressor and plays a role in the DNA damage response. Importantly, TP53TG1 is able to interact with YBX1 to prevent its nuclear localization such that the YBX1-mediated activation of oncogenes is suppressed (Diaz-Lagares et al 2016). Similarly, p53 regulated LINP1 promotes the interaction of Ku80 with DNA-PKcs, and thus, enhances DNA repair (Zhang et al 2016a).
Finally, a specific group of p53-regulated lncRNAs are enhancer RNAs (p53-eRNAs). These eRNAs are called p53-eRNAs because they are expressed in a p53-dependent manner (Melo et al 2013). Given the nature of p53-mediated expression, the p53-eRNA target genes are also under control by p53, although these target genes may not carry classical p53-binding sites. Evidently, through this mechanism the repertoire of p53 regulated genes are significantly increased, and at the same time, p53 may be able to selectively turn on a group of genes. Finally, a recent study suggests that at least some of these eRNAs can be controlled by lncRNAs. For instance, LED (LncRNA activator of Enhancer Domains) is involved in regulation of p53 eRNA expression and it is required for p53-induced cell cycle arrest (Leveille et al 2015). Therefore, these findings add another layer of complexity in the p53-regulatory system.
LncRNA-mediated NF-κB signaling
NF-κB is a ubiquitously expressed pleiotropic transcription factor present in almost all cell types and NF-κB pathway plays a critical role in many biological processes such as inflammation, cell growth, and cancer metastasis (Peppicelli et al 2013, Shi et al 2000, Xu and Fidler 2000). Under normal conditions, NF-κB stays in the cytoplasm as a heterotrimeric complex consisting of the subunits p50, p65, and the inhibitory subunit IκBα. In response to inducing stimuli such as TNF-α, IκBα undergoes phosphorylation, ubiquitination and proteolytic degradation. The p65/p50 subunits are translocated into the nucleus where they regulate expression of a specific set of target genes (Lu and Stark 2015). Aberrant regulation of NF-κB and its downstream targets often leads to inflammation, drug/radiation resistance, and cancer metastasis (Aggarwal 2004).
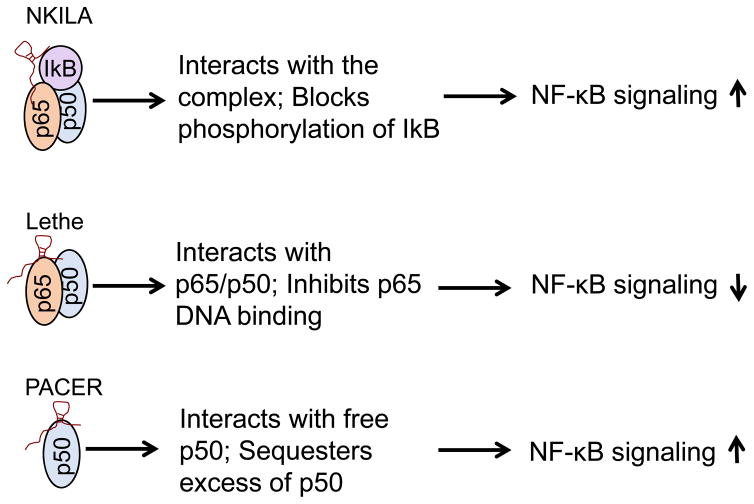
Several studies have implicated a number of lncRNAs in NF-κB signaling directly or indirectly (Krawczyk and Emerson 2014, Liu et al 2015, Rapicavoli et al 2013), but we primarily focus on those that have direct effect on the NF-κB complex where lncRNAs serve as a scaffold. In the first case, NF-κB Interacting LncRNA (NKILA) interacts with the NF-κB/IκB complex in such a way that NKILA prevents phosphorylation of IκB by IKK (Liu et al 2015). NKILA is primarily present in the cytoplasm, where it can inhibit both basal and cytokine-stimulated NF-κB activation. The underlying mechanism may involve the ability of NKILA to form a stable heterotrimeric complex (Fig. 2), which may explain why a low level of NKILA is associated with breast cancer metastasis and poor patient prognosis (Liu et al 2015). Furthermore, NKILA is upregulated by NF-κB, forming a negative feedback loop. Therefore, this example illustrates that lncRNAs can function as a scaffold to recruit various proteins in signaling pathways.
Fig. 2. Regulation of NF-κB signaling by lncRNAs.
Both NKILA and PACER serve as an NF-κB promoter. On the other hand, Lethe suppresses NF-κB activity.
Another example of NF-κB-associated lncRNAs is Lethe which can be induced by NF-κB-mediated proinflammatory cytokines (Rapicavoli et al 2013). In particular, Lethe suppresses NF-κB activity by physical interaction with the p65 subunit. Since Lethe appears to be negatively regulated by p65, this is another example of lncRNA-mediated negative feedback regulatory system for NF-κB (Fig. 2). However, the cancer relevance of Lethe remains to be determined yet.
It is well known that p65 and p50 can form heterodimers or homodimers; the heterodimeric p65-p50 complex appears to be the most abundant, serving as a transcriptional activation complex. On the other hand, p50-p50 homodimers often play an inhibitory role in transcription. A nuclear lncRNA PACER (p50-Associated COX-2 Extragenic RNA) was identified to be able to interact with free p50. Both COX2 and PACER can be induced by PMA. On the other hand, PACER can regulate COX2 transcription because PACER knockdown is required for COX2 expression (Krawczyk and Emerson 2014). The mechanism may involve sequestration of free p50 such that the repressive p50-p50 homodimers are decreased and the active p65-p50 heterodimers are increased in response to stimuli such as LPS. Therefore, PACER functions in cis to enhance NF-κB-dependent COX2 transcription (Fig. 2). Further evidence suggests that PACER can be induced by IL-1β, leading to a high level of COX2 (Pearson et al 2016). Given that the heterodimeric p65-p50 complex regulates a large number of genes, it would be interesting to determine how PACER specifically impacts COX2 expression. Similar to PACER, lincRNA-COX2 is required for the transcription of NF-κB-regulated late-primary inflammatory response genes stimulated by LPS (Hu et al 2016).
There are still several other lncRNAs associated with NF-κB pathway, primarily through in an indirect manner or as NF-κB effectors. For instance, HOTAIR decreases IκBα transcription through the polycomb repressive complex 2 (PRC2) in ovarian cancer (Ozes et al 2016). Since IκBα serves as an inhibitor for NF-κB, a decreased level of IκBα means that activation of NF-κB can last for a longer time, leading to increased expression of NF-κB-regulated genes. Furthermore, HOTAIR can activate NF-κB possibly through cAMP dependent protein kinase (PKA), or nuclear kinases such as MSK1 and MSK2 (Wu et al 2016). Finally, nuclear MALAT1 was reported to interact with p65/p50 heterodimer such that expression of TNFα and IL-6 is suppressed (Zhao et al 2016). However, this observation seems to contradict to what have been reported regarding the function of MALAT1 and NF-κB. Thus, further evidence is needed to define the role of MALAT1 in this aspect.
LncRNAs as AKT regulators
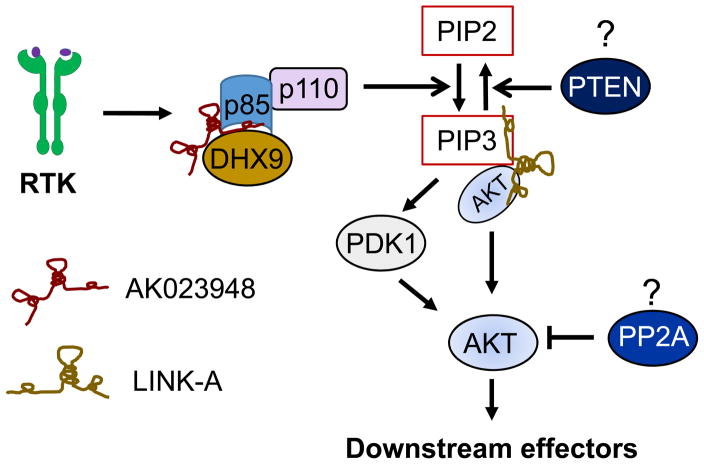
Overwhelming evidence supports the role of AKT in cancer (Thorpe et al 2015). At the top of this pathway, growth factors first interact with receptor tyrosine kinase (RTK), generating the second messenger PIP3 through PI3K, which subsequently activates critical downstream targets such as AKT and mammalian target of rapamycin (mTOR). Regulation of AKT activity often involves vast arrays of players. Two well-known AKT regulators are PI3K and PTEN. PI3K is a kinase that converts PIP2 to PIP3, serving as a positive AKT regulator, whereas PTEN is a phosphatase that reverse PIP3 to PIP2, acting as a negative AKT regulator. In addition, other factors such as PP2A or microRNAs can also regulate AKT activity. Our recent study suggests that lncRNA AK023948 is directly involved in AKT pathway (Koirala et al 2017). For example, knockdown or knockout of AK023948 suppresses AKT activity; re-expression of AK023948 in the KO cells (i.e., rescue experiment) restores the AKT activity. Thus, AK023948 is a positive regulator for AKT. Functionally, AK023948 interacts with ATP dependent RNA helicase A (RHA/DHX9) and the regulatory subunit of PI3K, p85.
Despite the importance of p85 in AKT pathway and cancer, information about the regulation of p85 is limited. This regulatory subunit consists of two major isoforms, p85α and p85β, however, evidence indicates that p85β plays an oncogenic role whereas p85α functions as a tumor suppressor (Taniguchi et al 2010). Consistent with these findings, p85β is a primary target for AK023948 in breast cancer cells. Mechanistically, both AK023948 and DHX9 are critical to the p85 stability. The interaction of AK023948 with DHX9 and p85 helps to stabilize p85 and thus, promote AKT activity (Fig. 3).
Fig. 3. Regulation of AKT activity by lncRNAs.
AK023948 promotes AKT activity by stabilizing p85, a regulatory subunit of PI3K; LINK-A facilitates the interaction of AKT and PIP3, stimulating AKT activity. It remains to be determined whether lncRNAs can interact with PTEN or PP2A and thus impact AKT activity.
In addition to PI3K, other components of ATK pathway can also be subject to regulation by lncRNAs. For instance, a recent report suggests that LINK-A (long intergenic non-coding RNA for kinase activation) can regulate AKT activity, but through a different mechanism. LINK-A directly interacts with the AKT pleckstrin homology domain and phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to promote AKT activity. LINK-A is required for AKT-PIP3 interaction and consequent enzymatic activation (Lin et al 2017). Evidently, LINK-A is a lipid-binding lncRNA (Fig. 3). The PIP3-binding motif of LINK-A is critical to resistance to AKT inhibitors. Thus, through targeting various components of the AKT pathway, lncRNAs play a very unique role and dysregulation of these lncRNAs may ultimately contribute to the increased AKT activity in cancer.
There are a few other lncRNAs implicated in regulation of AKT activity; they may exert their functions through a direct or indirect way. For example, FER1L4 inhibits AKT activity by promoting PTEN expression (Qiao and Li 2016). Similarly, BC087858 was reported to be involved in the PI3K/AKT and MEK/ERK pathways and epithelial-mesenchymal transition (EMT) through upregulation of ZEB1 and Snail (Pan et al 2016). In osteosarcoma cells MALAT1 knockdown significantly suppresses pAKT level whereas ectopic expression of MALAT1 promotes tumor growth and metastasis by activating PI3K/AKT signaling cascade (Xu et al 2015). This activation of AKT may be through a post-transcriptional regulation mechanism (Dong et al 2015). However, further evidence is still needed to define their role in AKT pathway. In particular, more studies are required to provide detailed mechanisms of how these lncRNAs impact AKT pathway.
The significance of AKT in cancer is further supported by the findings that NF-κB serves an important AKT effector. Thus, AKT-associated lncRNAs may also affect NF-κB activity to certain degree, further highlighting the importance of these AKT-associated lncRNAs. In addition, it is well known that phosphatases such as PTEN and PP2A can negatively regulate AKT activity. Thus, it is conceivable that any lncRNA associated with these phosphatases will also have an impact on AKT activity. However, to date little is known regarding the potential involvement of lncRNAs in regulation of these phosphatases. Evidently, identification of such potential lncRNAs would provide more comprehensive insight into regulation of AKT pathway.
Notch-associated lncRNAs
The Notch signaling pathway is also a highly conserved cell signaling system in most multicellular organisms, and is critical to diverse cellular processes such as stemness, cell proliferation, differentiation, and cell death. The role of Notch signaling in stem cells and malignancy is well documented. Of interest, a number of lncRNAs have been shown to be regulated by Notch; among them is LUNAR1 that is essential for T-ALL growth by maintaining high expression of IGF1R mRNA through a cis-activation mechanism (Trimarchi et al 2014). On the other hand, Notch1 activation can specifically induce expression of TUG1 (taurine upregulated gene 1) which in turn promotes self-renewal of glioma stem cells (Katsushima et al 2016), highlighting the importance of Notch-lncRNA axis in the regulation of self-renewal of glioma cells. Finally, NALT (NOTCH1 Associated LncRNA In T-Cell Acute Lymphoblastic Leukemia 1) is upregulated in bone marrow of T ALL specimens. While suppression of NALT by RNAi or suppression of Notch by GSI inhibitors impairs, ectopic expression of NICD promotes cell proliferation or xenograft tumor growth (Wang et al 2015). This NALT-mediated activation of Notch signaling may be through transcription activation by NALT although further studies are needed to confirm this possibility.
Other signaling pathways
Although there are many other signaling pathways that lncRNAs can play role in corresponding signaling pathways, due to space limit, we will just discuss two here, Wnt/β-catenin and HIF1α.
Aberrant activation of Wnt/β-catenin signaling has been implicated in several types of cancer because Wnt/β-catenin signaling controls a number of oncogenic genes, especially c-Myc, leading to cell proliferation and tumorigenesis. In addition to Linc-RoR controlling the stability of c-Myc through AUF (Huang et al 2015), a recent study identifies lncRNA MYU as a direct target of c-Myc. The interaction of MYU with hnRNP K can stabilize CDK6 expression and thereby promote the G1-S transition of the cell cycle (Kawasaki et al 2016). Similarly, the ultraconserved lncRNA uc.158 serves as a cancer-specific mediator of Wnt/β-catenin pathway in liver cancer (Carotenuto et al 2016). Given the important role of the Wnt/β-catenin signaling in liver pathophysiology, this may explain at least in part why dysregulation of this pathway promotes the development and progression of different cancers, including hepatobiliary tumors.
As an important feature of lncRNAs in serving as a scaffold, lncRNAs can interact with different kinases in cytoplasm that are often critical to cell signaling. For instance, BRK and LRRK2 are required for HIF1α signaling. This same LINK-A, which was recently shown to facilitate the recruitment of PIP3 by AKT (Lin et al 2017), can also interact with BRK and LRRK2. Importantly, LINK-A is required for BRK to interact with the EGFR:GPNMB complex and it promotes BRK kinase activity. The BRK-dependent HIF1α phosphorylation at Tyr565 interferes with its hydroxylation at Pro564, leading to HIF1α stabilization even under normoxic conditions (Lin et al 2016).
Role of lncRNAs in cancer
Given the role of cell signaling pathways in cancer initiation, progression and metastasis, lncRNAs involved in these pathways can impact all aspects of tumorigenesis. As a result, lncRNAs can play an oncogenic or a tumor suppressive role.
For instance, a large number of lncRNAs can promote tumor growth and metastasis. PVT1 is required for a high level of c-Myc protein; PVT1 RNA and c-Myc protein expression is highly correlated in primary human tumors. Suppression of PVT1 inhibits c-Myc-driven tumorigenic potency (Tseng et al 2014). MALAT1 was first identified in lung cancer MALAT1 (Ji et al 2003) and is one of the most studied lncRNAs in cancer. For example, MALAT1 can promote tumorigenesis through Wnt/β-catenin pathway, EMT, PI3K/AKT pathway, ERK/MAPK pathway and angiogenesis (Liu et al 2017). PCAT-1 functions as a prostate-specific regulator of cell proliferation and it has been implicated in a subset of prostate cancer patients (Prensner et al 2011). PCGEM1 is a prostate cancer specific lncRNA (Srikantan et al 2000); it can promote cell proliferation (Petrovics et al 2004) and reduce apoptosis induced by anticancer drugs (Fu et al 2006). HOTAIR is upregulated in breast tumors and its expression is strongly associated with breast cancer metastasis and patient survival (Gupta et al 2010). Finally, LINK-A expression and activation of LINK-A-dependent signaling pathway correlate with triple-negative breast cancer (TNBC), and they together promote breast cancer glycolysis reprogramming and tumorigenesis (Lin et al 2016). Moreover, LINK-A-dependent AKT hyperactivation can lead to tumorigenesis and resistance to AKT inhibitors. (Lin et al 2017).
Our own studies suggest that Linc-RoR and UCA1 are upregulated in breast cancer and suppression of their expression inhibits tumor growth (Huang et al 2014, Huang et al 2015); AK023948 is upregulated in breast cancer and it promotes tumor cell growth (Koirala et al 2017). On the other hand, Loc285194 and GAS5 may function as tumor suppressors in breast cancer (Liu et al 2013, Zhang et al 2013b). These are just a few of lncRNA examples important to tumorigenesis. Although the detailed molecular mechanism of lncRNA-mediated tumorigenesis still remains to be elucidated yet, evidently lncRNA-mediated cell signaling is an important contributing factor.
Concluding remarks and future directions
The signaling pathways discussed above were discovered a long time ago; and key players in these pathways are well characterized and described in most textbooks. We thought that we knew everything about them. However, the fact is that we still have a lot to learn. Although lncRNAs do not appear to be as important as signaling molecules themselves to certain degree, it is evident that lncRNAs are critical to this regulatory network by providing a background support to make the signaling more smoothly or making signaling more flexible in response to environmental cues.
The most important feature of lncRNA-mediated regulation of cell signaling is that lncRNAs are able to function as a scaffold through which lncRNAs are capable of interacting with various signaling molecules. In this case, lncRNAs may act as various aptamers. Depending on the type and/or number of their binding partners, lncRNAs can often exert multiple functions, especially by directly interacting with signaling molecules. Given the complexity of cell signaling, a critical challenge is how to identify lncRNAs carrying functional domains (or aptamers) that interact with corresponding binding partners. Currently available technologies such as RNA precipitation and RNA immunoprecipitation (RIP) or cross-linking immunoprecipitation (CLIP) are important for identification of such binding partners. Especially, RNA precipitation combined with high throughput mass-spectrometry and RIP/CLIP combined with deep sequencing provide critical research tools in exploring lncRNA functions in cell signaling. Moreover, functional screening from RNAi libraries or CRISPR-based libraries are more advanced technologies for identification of their functions specific to a particular signaling pathway. Together, systematic characterization of lncRNA-mediated cell signaling would be possible.
The most exciting experience we have been through in past years with regard to lncRNA-mediated cell signaling is probably the finding that classic signaling molecules and other factors including lncRNAs are often interconnected and cooperate to exert cellular functions. Therefore, it is important to keep in mind that lncRNAs alone may not be sufficient to drive cell signaling; likewise, classic signaling molecules may not work by themselves or they alone may not work efficiently. Given that lncRNA research is still at the early stage, we expect that there will be many more lncRNAs associated with various signaling pathways to be identified in near future. Further characterization of these lncRNAs will provide new insight into how lncRNAs and signaling molecules work together, impacting various aspects of tumorigenesis. As a result, these lncRNAs may serve as cancer biomarkers or therapeutic targets.
Acknowledgments
This work was supported by NIH grant R01 CA154989 (YM).
Abbreviations
- 3′-UTR
3′-untranslated region
- ARE
AU rich element
- DINO
Damage Induced Noncoding
- EMT
epithelial-mesenchymal transition
- GAS5
growth arrest-specific 5
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- hnRNP
heterogeneous nuclear ribonucleoprotein
- lncRNA
long ncRNA
- LED
LncRNA activator of Enhancer Domains
- LINK-A
long intergenic non-coding RNA for kinase activation
- LINP1
lncRNA in nonhomologous end joining pathway 1
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- PRC2
polycomb repressive complex 2
- RTK
receptor tyrosine kinase
- RISC
RNA-induced silencing complex
- SAM
synergistic activation mediator
- TUG1
taurine upregulated gene 1
References
- Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre A, Masotti A. Long non-coding RNAs and p53 regulation. Int J Mol Sci. 2012;13:16708–16717. doi: 10.3390/ijms131216708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe AJ, Fox AH, Bond CS. The ins and outs of lncRNA structure: How, why and what comes next? Biochimica et biophysica acta. 2016;1859:46–58. doi: 10.1016/j.bbagrm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Carotenuto P, Fassan M, Pandolfo R, Lampis A, Vicentini C, Cascione L, et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2016 doi: 10.1136/gutjnl-2016-312278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. The Journal of biological chemistry. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, Soler M, Setien F, Goyal A, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA and cell biology. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–2198. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaby MJ, Yang DQ. p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene. 2007;395:1–7. doi: 10.1016/j.gene.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in molecular medicine. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer research. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol. 2016;196:2799–2808. doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell death & disease. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic acids research. 2015 doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, Ding X, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic acids research. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Molecular cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature reviews Genetics. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Human molecular genetics. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nature communications. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Komiya M, Matsumura K, Negishi L, Suda S, Okuno M, et al. MYU, a Target lncRNA for Wnt/c-Myc Signaling, Mediates Induction of CDK6 to Promote Cell Cycle Progression. Cell reports. 2016;16:2554–2564. doi: 10.1016/j.celrep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun. 2017;8:14422. doi: 10.1038/ncomms14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nature communications. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell death and differentiation. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nature cell biology. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nature cell biology. 2017;19:238–251. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front Biosci (Landmark Ed) 2017;22:66–80. doi: 10.2741/4472. [DOI] [PubMed] [Google Scholar]
- Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic acids research. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Stark GR. NF-kappaB: Regulation by Methylation. Cancer research. 2015;75:3692–3695. doi: 10.1158/0008-5472.CAN-15-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Intergenic lncRNAs and the evolution of gene expression. Curr Opin Genet Dev. 2014;27:48–53. doi: 10.1016/j.gde.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- Meller VH, Joshi SS, Deshpande N. Modulation of Chromatin by Noncoding RNA. Annual review of genetics. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET, et al. Long Intergenic Noncoding RNAs Mediate the Human Chondrocyte Inflammatory Response and Are Differentially Expressed in Osteoarthritis Cartilage. Arthritis Rheumatol. 2016;68:845–856. doi: 10.1002/art.39520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppicelli S, Bianchini F, Contena C, Tombaccini D, Calorini L. Acidic pH via NF-kappaB favours VEGF-C expression in human melanoma cells. Clinical & experimental metastasis. 2013;30:957–967. doi: 10.1007/s10585-013-9595-4. [DOI] [PubMed] [Google Scholar]
- Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochemical and biophysical research communications. 2016;478:507–512. doi: 10.1016/j.bbrc.2016.06.160. [DOI] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nature genetics. 2016;48:1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Le X, Wang B, Xiong Q, Abbruzzese JL, Xie K. Regulation of interleukin-8 expression by cellular pH in human pancreatic adenocarcinoma cells. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2000;20:1023–1028. doi: 10.1089/10799900050198471. [DOI] [PubMed] [Google Scholar]
- Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, et al. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell death & disease. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Aleman JO, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer research. 2010;70:5305–5315. doi: 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nature reviews Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen L, Chen B, Li X, Kang J, Fan K, et al. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis. 2013a;4:e765. doi: 10.1038/cddis.2013.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Developmental cell. 2013b;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu P, Lin R, Rong L, Xue Y, Fang Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Scientific reports. 2015;5:13749. doi: 10.1038/srep13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu J, Li W, Liu G, Li Z. LncRNA-HOTAIR promotes TNF-alpha production in cardiomyocytes of LPS-induced sepsis mice by activating NF-kappaB pathway. Biochemical and biophysical research communications. 2016;471:240–246. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- Xu L, Fidler IJ. Acidic pH-induced elevation in interleukin 8 expression by human ovarian carcinoma cells. Cancer research. 2000;60:4610–4616. [PubMed] [Google Scholar]
- Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell research. 2013a;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. Journal of molecular cell biology. 2014;6:181–191. doi: 10.1093/jmcb/mju013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nature structural & molecular biology. 2016a;23:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell death and differentiation. 2013b;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou N, Huang J, Ho TT, Zhu Z, Qiu Z, et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget. 2016b;7:15481–15491. doi: 10.18632/oncotarget.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS letters. 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]