Abstract
Purpose
Elevated intraocular pressure (IOP) is an important risk factor for glaucoma. We constructed polygenic risk scores (PRSs) for IOP using the UK Biobank (UKB) data set to test whether the PRSs are associated with IOP and whether using them improves glaucoma prediction.
Methods
We conducted this study using 435,678 European participants from the UKB. We constructed weighted and unweighted PRSs using single nucleotide polymorphisms (SNPs) derived from the UKB data and previously reported IOP SNPs. We examined the associations of the PRSs with IOP and primary open-angle glaucoma (POAG) using linear and logistic regression, respectively. To quantify the discriminatory ability of the PRSs on POAG, we used the area under the receiver operating characteristic curve (AUC).
Results
The weighted PRS was significantly associated with IOP (P ∼ 10−200), after adjusting for age and sex. The PRS explained an additional 4% of variance in IOP. The weighted PRS was also significantly associated with POAG (P = 1.8 × 10−77). Subjects in the top quintile of the IOP PRS were 6.34 (95% confidence interval [CI]: 4.82–8.33; P = 2.1 × 10−57) times more likely to have POAG, compared to those in the bottom category. The weighted PRS improved the discriminatory power for POAG (AUC increased by 5%, P = 6.2 × 10−22) when added to the other covariates. The unweighted PRS exhibited similar results.
Conclusions
We determined that IOP PRSs are significantly associated with IOP and improve the prediction of POAG.
Translational Relevance
PRSs help reduce the burden of glaucoma by early detection of genetically susceptible individuals.
Keywords: genome-wide association study, polygenic risk score, intraocular pressure, primary open-angle glaucoma
Introduction
Glaucoma is a leading cause of irreversible blindness, affecting more than 70 million individuals worldwide.1 Primary open-angle glaucoma (POAG) is the most common form of glaucoma accounting for approximately 90% of all glaucoma cases. At present, since there is no cure for glaucoma, early detection is crucial. Elevated intraocular pressure (IOP) is one of the most significant risk factors for glaucoma. In addition, lowering IOP is the only proven therapeutic intervention that helps prevent the development and delay the progression of glaucoma.2–4 Therefore, identifying genetic factors that contribute to IOP will not only improve our understanding of its biological mechanisms, but may also aid in glaucoma prediction.
Polygenic risk scores (PRSs) examine the cumulative effect of genetic variants on a disease or trait by aggregating the individual genetic effects into a single measure. They provide a convenient way to quantify the overall genetic risk. In risk prediction, PRSs can outperform traditional risk factors, which supports the incorporation of PRSs into clinical practice.5 Furthermore, PRSs of endophenotypes can be used in risk prediction for disease.6 Aschard et al.7 reported a strong genetic correlation between IOP and POAG. Therefore, characterizing an individual's susceptibility to elevated IOP through PRSs should be an effective tool for glaucoma risk assessment. However, previous studies of IOP PRSs have yielded inconsistent findings. Ramdas et al.8 reported that their IOP PRSs were not significantly associated with IOP using a range of single nucleotide polymorphism (SNP) selection thresholds and only explained 0.2% additional variation in IOP in their European subjects (n = 10,972). Mabuchi et al.9 identified a significant association (P = 0.01) in a Japanese population (n = 762) using a PRS constructed from nine previously reported IOP genetic variants. Given the current inconclusive findings, using large-scale cohorts can help determine the utility of PRSs of IOP in glaucoma prediction.
Large-scale biobank repositories, such as the UK Biobank (UKB), are becoming available and have enabled identifying novel genetic variants associated with complex diseases/traits10 and more accurate estimation of SNP effect sizes. As such, in order to further elucidate the association between a cumulative measure of IOP-related genetic effects on IOP and prediction of glaucoma, we constructed and evaluated IOP PRSs using data from the UKB.
Materials and Methods
Ethics Statement
The UKB received approval from the North West Multi-centre Research Ethics Committee. Recruitment for the UKB was obtained by written consent. Our access to the resource was approved by UKB as complying with their Access Procedures and Ethics. We obtained fully de-identified data. Our research adheres to the tenets of the Declaration of Helsinki.
Study Sample, IOP Measurements, and Glaucoma
The following research was conducted using data from the UKB, an ongoing large prospective cohort study. Details regarding this cohort have been described elsewhere.11,12 Briefly, UKB recruited > 500,000 adult participants (40 to 70 years of age at enrollment) living in the United Kingdom who were registered with the National Health Service. Lifestyle, family, and medical information, as well as DNA samples, were collected. Ophthalmological data were also collected for a subset of study participants (∼118,000). For this study, we restricted our analysis to white participants and removed outliers with genetic ancestry at least six standard deviations from the means of the first two principle components.
IOP measurements were obtained using the Optical Response Analyzer (Reichert Corp., Philadelphia, PA). Both Goldman-correlated and corneal-compensated IOP measurements were collected. For this study, we used corneal-compensated IOP since it is less affected by corneal thickness.13,14 The average of the right and left eye IOP measurements was taken and used for downstream analysis. Study participants who received eye surgery within 4 weeks prior to the ocular assessment or those with possible eye infections did not receive IOP measurements. Furthermore, we excluded study participants with extreme values of IOP, that is, in the top and bottom 0.3 percentiles and outliers, including 673 subjects who had either eye surgery or used eye drop medications. We identified POAG using the International Classification of Diseases Tenth Revision (ICD-10) codes.
Genotyping, Imputation, and Quality Control
For this analysis, we used genetic data from the March 2018 data release. Details regarding the genetic data have been reported elsewhere.11,15 Briefly, a total of 488,377 study participants were genotyped on either the UK BiLEVE Axiom Array (807,411 markers; n = 49,950) or the UKB Axiom Array (825,927 markers; n = 438,427). The data were further imputed based on the 1000 Genomes Project, UK10K, and the Haplotype Reference Consortium reference panels. Quality control parameters were applied to both genetic markers and individual samples. After performing quality control, 92,693,895 genetic markers and 487,442 samples were included in the data release. We further excluded low quality (info score < 0.3) and rare (minor allele frequency < 0.5%) variants from the imputed data set, resulting in approximately 11.9 million variants for downstream analysis.
PRS Construction
Weighted and unweighted PRSs for IOP were constructed using SNPs derived from UKB data. For comparison purposes, we also constructed IOP PRSs using previously reported IOP SNPs. Risk alleles were coded as alleles associated with an increase in IOP. To avoid overfitting during the construction of the PRSs, we used independent data sets for training and testing. For evaluating the association between PRSs and IOP, we used a five-fold cross-validation. We randomly divided 110,964 white subjects with available IOP measurements into five groups (folds) of equal sizes. We then used a group for testing and the rest for training. This procedure was repeated five times. Each time, a different group was used as the testing set. For evaluating whether the IOP PRSs improve glaucoma prediction, we used the white subjects with IOP (n = 110,964) and without IOP (n = 324,713) as the training and testing data sets, respectively. To simplify the regression analyses, we excluded related subjects (inferred using KING16) from the testing data sets. To derive SNP effect sizes and select SNPs for downstream PRS calculation, we performed IOP genome-wide association studies (GWASs) on the training data sets using the BOLT-LMM software,17 adjusting for age, sex, array, and the first 10 principal components of genetic ancestry.10 We then selected independent SNPs using PLINK18,19 LD-based clumping with r2 < 0.3 and P < 5 × 10−5. After the clumping, 1256 SNPs on average (for the five cross-validation training data sets) and 1691 SNPs were retained for constructing IOP PRSs for the IOP testing and glaucoma testing data sets, respectively. For previously reported SNPs, effect estimates from the largest study were used if SNPs were reported in multiple studies. This resulted in 93 previously reported SNPs (Supplementary Table S1). The weighted PRSs were calculated by multiplying the risk allele by the corresponding effect estimate and summing these values together. We also constructed unweighted PRSs, which were calculated as the summation of the number of risk alleles, assuming each risk allele has the same effect.
Statistical Analysis
For testing the association between the PRSs and IOP, we carried out multiple linear regression analyses in the IOP testing data sets, adjusting for age and sex. Additional variance of IOP explained by the PRSs was also estimated. To investigate the association between the PRSs and POAG, we performed logistic regression analyses in the POAG testing data set. We also created quintiles of the IOP PRSs to compare the odds of POAG for study participants with higher PRSs to those with the lowest PRSs. We used the area under curve (AUC) of the receiver operating characteristic curve (ROC) to quantify the predictive ability of the PRSs on POAG. A significance cutoff of P ≤ 0.05 was used for all statistical analyses. R (v3.4.2)20 and SAS 9.4 (SAS, Inc., Cary, NC) were used for plotting and statistical analyses.
Results
Table 1 presents the characteristics of the study sample with IOP measurements. A total of 110,964 white European subjects were included in the IOP analyses. The mean (SD) of age was 58.2 (7.9) years and 53.4% of the participants were female. The average IOP (SD) was 16.0 (3.4; range: 7.0–39.0) mm Hg.
Table 1.
Characteristics of the Study Sample for IOP
| Sample Size |
Age, Mean (SD), y |
Female, % |
IOP, Mean (SD), mm Hg |
IOP, Range, mm Hg |
| 110,964 | 58.2 (7.9) | 53.4 | 16.0 (3.4) | 7.0–39.0 |
We performed multiple linear regression to examine the association between PRS and IOP, adjusting for age and sex. Table 2 shows the association results between the weighted PRS and IOP across five cross-validation runs. The weighted PRS was significantly associated with IOP with the P-values ranging from 3.5 × 10−209 to 1.0 × 10−190 in the IOP testing data sets, demonstrating that a higher PRS is associated with higher IOP. Moreover, the base model, consisting of age and sex, accounted for 3.3% of the variance in IOP. The inclusion of the weighted PRS in the model explained an additional 4% of variance in IOP, yielding a total of 7.3% variance explained. The unweighted PRS showed similar results and significance levels (Supplementary Table S2).
Table 2.
Association Between PRSs and IOP Across Five Cross-Validation Runs
| Model |
Number of SNPs Selected |
β |
SE |
P |
| Iteration 1 | 1253 | 0.2090 | 0.0070 | 1.0 × 10−190 |
| Iteration 2 | 1262 | 0.2180 | 0.0071 | 5.2 × 10−201 |
| Iteration 3 | 1242 | 0.2198 | 0.0072 | 3.2 × 10−202 |
| Iteration 4 | 1274 | 0.2178 | 0.0071 | 5.1 × 10−205 |
| Iteration 5 | 1250 | 0.2205 | 0.0071 | 3.5 × 10−209 |
We next assessed the association between the IOP PRSs and ICD-10 POAG. Table 3 summarizes the characteristics of POAG cases and controls from the POAG testing data set and logistic regression results. Out of the 324,713 participants in the POAG testing data set with ICD-10 codes, 896 study participants were identified as POAG cases and 248,871 were controls. The mean (SD) ages for cases and controls were 63.2 (5.7) years and 57.6 (8.0) years (P = 4.3 × 10−85), respectively. The proportion of females was 44.6% and 55.2% for cases and controls (P = 3.1 × 10−10). Cases exhibited higher systolic blood pressure (SBP) compared to controls, 143.5 (18.5) mm Hg to 138.2 (18.2) mm Hg (P = 2.7 × 10−17). The proportion of study participants with type 2 diabetes (T2D) was higher among cases than controls, 9.2% vs. 5.5% (P = 1.4 × 10−6). Significantly, cases had a higher weighted PRS compared to controls (P = 2.5 × 10−76), 211.2 (3.5) vs. 209.1 (3.4), respectively. Interestingly, in both univariate and multiple logistic regression analyses, the weighted PRS was much more significant, with the exception of age, than any other traditional risk factor. Adjusting for age and sex only, the weighted PRS was also significantly associated with POAG (P = 1.8 × 10−77). The unweighted PRS exhibited similar association results (data not shown).
Table 3.
Summary Statistics of POAG Testing Data Sets and Logistic Regression Results
| Cases (n = 896) |
Controls (n = 248,871) |
P (ULR) |
P (MLR) |
|
| Age, y | 63.2 (5.7) | 57.6 (8.0) | 4.3 × 10−85 | 3.4 × 10−70 |
| Female, % | 44.6 | 55.2 | 3.1 × 10−10 | 9.6 × 10−7 |
| BMI, kg/m2 | 27.4 (4.5) | 27.6 (4.7) | 1.6 × 10−1 | 1.1 × 10−4 |
| SBP, mm Hg | 143.5 (18.5) | 138.2 (18.2) | 2.7 × 10−17 | 6.5 × 10−2 |
| T2D, % | 9.2 | 5.5 | 1.4 × 10−6 | 1.1 × 10−2 |
| Weighted PRS | 211.2 (3.5) | 209.1 (3.4) | 2.5 × 10−76 | 2.2 × 10−75 |
ULR, univariate logistic regression; MLR, multiple logistic regression.
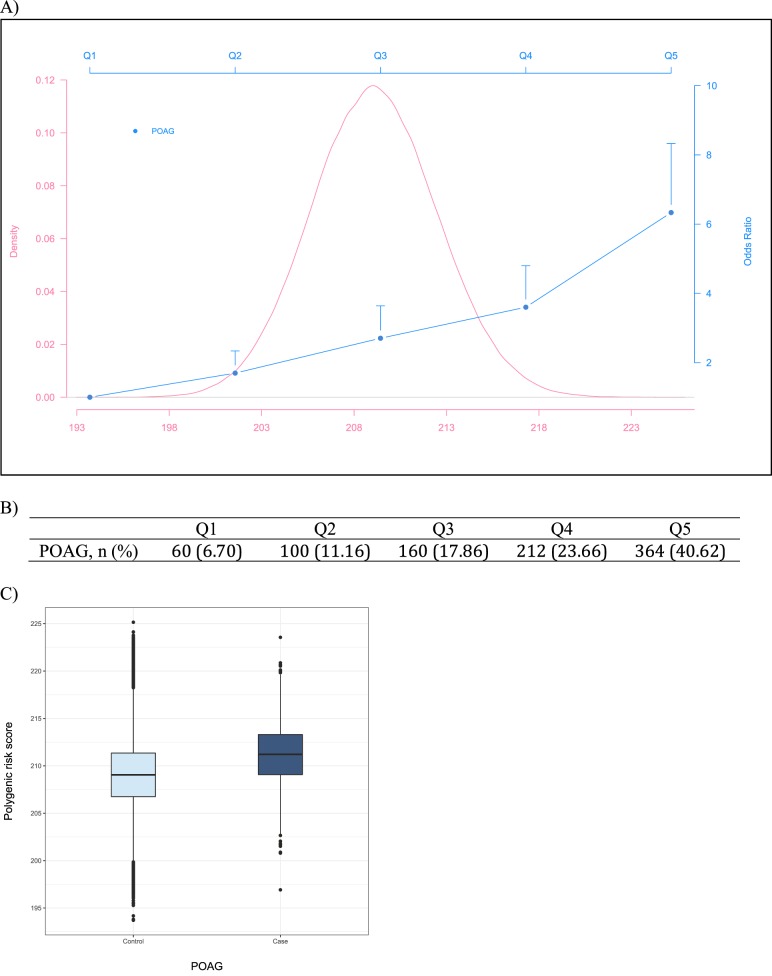
We further generated quintiles of the PRS to evaluate the relationship between the IOP PRS and POAG. Overall, there were significant, positive associations between the quintiles of the weighted IOP PRS with POAG (P = 2.6 × 10−62), after adjusting for age and sex. Figure 1 presents the association between the quintiles of the weighted IOP PRS and POAG. Compared to study participants in the lowest quintile, those in the second, third, fourth, and fifth quintiles of the IOP PRS have steadily increasing odds of POAG, with those in the highest quintile experiencing the greatest odds, odds ratio (OR) = 6.34 (95% confidence interval [CI]: 4.82–8.33; P = 2.1 × 10−57). Most POAG cases (40%) are in the highest quintile category of IOP PRS. The unweighted PRS showed similar results in POAG (OR = 5.37; 95% CI: 4.16–6.94; P = 1.8 × 10−55), when comparing study subjects in the highest quintile to those in the lowest (Supplementary Fig. S1).
Figure 1.
Distribution of the weighted IOP PRS and association with POAG. (A) Distribution of the weighted PRS and ORs of POAG comparing each of the four upper quintiles with the lowest quintile, adjusting for age and sex. The vertical lines represent the upper 95% CI for each OR. (B) Number of POAG cases (percentage) in each category. (C) Boxplots of PRS for POAG cases and controls.
We examined the discriminatory ability of the IOP PRSs on POAG using logistic regression and AUC. Figure 2 displays the ROC curves for models without and with the weighted IOP PRS on POAG. With only age and sex in the base model, the AUC for the model is 0.713 (95% CI: 0.698–0.729). When the weighted PRS from previously reported SNPs is added to the baseline model, there is a small increase in the AUC to 0.723 (95% CI: 0.708–0.739; P = 1.4 × 10−4). Adding the weighted PRS derived from UKB data yielded a significant increase in the AUC to 0.766 (95% CI: 0.751–0.781; P = 6.2 × 10−22). Interestingly, adding other traditional risk factors, that is, body mass index (BMI), SBP, and T2D, to the baseline model did not change the AUC much with only a 0.002 increase (P = 8.2 × 10−2). The unweighted IOP PRSs showed similar patterns of discriminatory ability on POAG (Supplementary Fig. S2).
Figure 2.
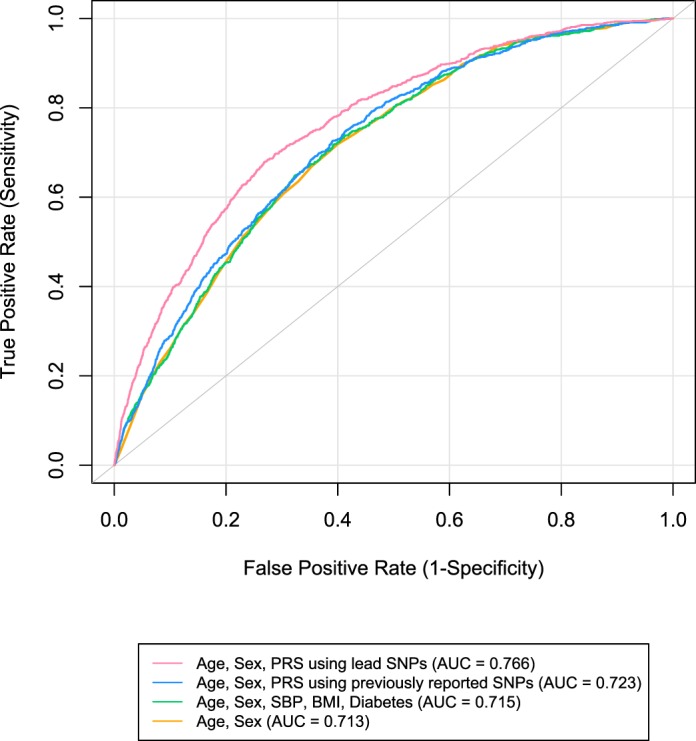
Receiver operating characteristic curves predicting POAG for weighted PRSs. The curves are based on logistic regression models adjusting for age and sex.
Discussion
In the present study, we constructed IOP PRSs, evaluated whether they are associated with IOP and POAG, and determined their discriminatory ability for POAG using independent training and testing data sets. We observed significant associations between the IOP PRSs and IOP, with increasing PRSs associated with higher IOP. Moreover, the PRSs explained an additional 4% of the variation in IOP. We also identified significant associations between the IOP PRSs and glaucoma, with study participants in the upper PRS quintiles experiencing greater odds of glaucoma compared to those in the lowest quintile. Additionally, the inclusion of the PRSs significantly improved the discriminatory ability for glaucoma. We observed similar results for the unweighted PRSs.
Glaucoma is a significant public health issue given the increasing number of individuals affected by the disease over the next several decades.21 Moreover, glaucoma is often regarded as the silent killer of sight. Tools that can identify individuals at a greater risk for the disease reduce its impact by preventing its development through early prevention and improved diagnosis. PRSs aggregate these individual variants identified through genetic association studies to estimate a cumulative measure of genetic risk, providing a better measurement of genetic predisposition for a specific health outcome like glaucoma. Hence, PRSs can serve as a promising clinical tool in early screening and identifying at-risk asymptomatic individuals for disease prevention. Early detection, which is crucial for glaucoma management, gives susceptible individuals the opportunity for earlier medical intervention.
We sought to construct and evaluate IOP PRSs for IOP and glaucoma through the inclusion of the large-scale UKB data set, a population-based cohort of over 500,000 participants,11 which can provide more definitive answers to the questions of whether IOP PRSs are associated with IOP and improve glaucoma prediction. We observed significant associations between the PRSs and IOP (P ∼ 10−200) and POAG (P = 1.8 × 10−77). Our findings showed substantially greater associations than previous studies reporting on the associations between IOP PRSs with IOP and POAG.9,22 Moreover, the magnitude of the association is much larger in the current report for study subjects in the highest PRS category, with a 3-fold greater odds of POAG in this study (OR = 6.34 [95% CI: 4.82–8.33]) compared to a multiethnic Asian study (OR = 2.00 [95% CI: 1.32–3.03]).22 We also observed a significant improvement in the AUC (5%) with the inclusion of the PRSs. The AUC with PRS is even higher than the combined effect of IOP- and vertical cup-to-disc ratio (VCDR)-PRSs (AUC increased by 3%) reported by Tham et al.22 in a Singapore multiethnic study (n = 6881). There are also a number of differences compared to two other recent studies, that is, Khawaja et al.23 and MacGregor et al.,24 that used UKB IOP and glaucoma data. A subtle yet important difference of our study is that we used a less stringent cutoff, that is, 5 × 10−5, instead of the genome-wide significance threshold, 5 × 10−8, to select SNPs for building PRSs. Given the polygenic nature of complex traits/diseases like IOP and glaucoma,10,23,24 the stringent 5 × 10−8 cutoff can miss many biologically relevant variants that do not reach genome-wide significance given the current sample sizes of GWASs. It is critical to include variants that do not reach GWAS significance in genetic risk prediction models, which was clearly shown in Khera et al.'s25 recent investigation of polygenic scores for several common diseases. For example, it required 6.6 million (assuming 0.1% of genome-wide SNPs are causal) and 5218 SNPs (P < 5 × 10−4 and r2 < 0.2) to reach optimal PRS performance for coronary artery disease and breast cancer, respectively.25 We tested a grid of P-value cutoffs for selecting SNPs, such as 0.01, 0.001, 10−4, 5 × 10−5, and 5 × 10−8.26 Balancing the prediction accuracy and easy interpretation, especially for nonstatisticians, we used 5 × 10−5 in this study. When we used GWAS-significant SNPs (P < 5 × 10−8) only for building PRSs, we got worse association results and worse prediction accuracy for IOP and POAG, respectively, compared to the results using the 5 × 10−5 cutoff (Supplementary Tables S3 and S4). This likely explained why we obtained a stronger discrimination accuracy in POAG (OR = 6.34) than MacGregor et al.24 (OR = 4.2) for the top IOP PRS quintile versus the bottom quintile comparison. Khawaja et al.23 used a regression-based model instead of PRS23 and got an AUC of 0.737 for glaucoma prediction using age, sex, IOP and POAG SNPs as covariates, while we obtained a better AUC = 0.766 using age, sex, and IOP PRS. The regression-based model, which includes each SNP as a covariate (rather than aggregating the genetic effects into a single PRS), can lead to model overfitting if too many SNPs are included as covariates. Overfitting issues are typically solved by shrinkage statistical methods, such as the LASSO and ridge regression.27 In addition to assess the discriminatory ability of IOP PRSs on predicting POAG, we tested their association with IOP, which can be useful to predict the risk of IOP elevation. Furthermore, our IOP PRSs showed a much stronger discriminatory ability for POAG than traditional risk factors such as T2D, BMI, and blood pressure (AUC difference: 5% vs. 0.1%). To our knowledge, this is the first time that IOP PRSs are reported to perform better than traditional risk factors in predicting POAG. Overall, our results demonstrated the utility of IOP PRSs to assess IOP elevation and glaucoma risk and its potential to serve as a clinically useful tool to reduce the occurrence of glaucoma.
PRSs have potential clinical utility in at least three intervention categories: (1) PRS-informed therapeutic intervention; (2) PRS-informed disease screening; and (3) PRS-informed life planning.28 First, PRSs are useful for identifying high-risk individuals for therapeutic interventions. PRSs also lead to the reclassification of individuals more accurately into appropriate disease risk categories. Recent studies suggest that individuals at top percentiles of PRSs of common diseases show high risk equivalent to monogenic mutations.25 Second, PRSs help improve disease screening and shift age-based criterion to a more refined PRS-augmented screening. PRSs also aid in the interpretation of testing results and in determining necessary treatment. Third, PRSs would be an effective tool for people to induce and maintain healthy behaviors. Interested readers of the clinical utility of PRSs can refer to the excellent review by Torkamani and colleagues28 for more details.
This study has many strengths, as well as several limitations. We used independent data sets for training and testing to avoid model overfitting. Moreover, we used the largest population-based cohort to date, that is, the UKB with half a million participants, which enabled us to obtain more stable effect size estimates and better risk predictions. We removed outliers based on principal component analysis (Supplementary Fig. S3). We further compared our IOP results, that is, effect sizes and P-values, with two recent large-scale IOP GWAS reports that used UKB data in their meta-analyses23,24 and found consistent results (Supplementary Fig. S4), demonstrating the robustness of our genetic association analyses. Our IOP PRSs performed much better than previous reports, including yielding stronger associations with IOP and better discriminatory abilities for POAG in independent testing data sets. An advantage of PRSs over traditional risk factors is that DNA information only needs to be collected and genotyped once. We also constructed unweighted PRSs, which performed similarly to weighted PRSs. With respect to the biological mechanisms, our results clearly demonstrated the polygenic nature of IOP genetics. Variants that do not reach GWAS significance also contribute to the prediction accuracy of IOP in addition to genome-wide significant variants. It is often challenging to interpret weaker signals in genetic analyses of complex traits/diseases.29 PRSs serve as an effective way to summarize the aggregative effect of the genetic variants. The pleiotropic nature and other biological mechanisms of the IOP loci have been documented in our previous GWASs of IOP,10 as well as others'.23,24 As for study limitations, we used ICD-10 POAG, which is not available for all UKB participants. Our study consisted of white participants only. The generalizability of these findings to other ethnic populations requires further investigation. We have only used IOP PRSs thus far. Glaucoma is a heterogeneous disease and some cases may not be due to elevated IOP. Including other PRSs, for example, glaucoma and vertical cup-disc ratio PRSs, in prediction models are likely to further improve prediction accuracy.
In conclusion, we constructed IOP PRSs using a larger number of SNPs than previous studies and observed significant associations between the PRSs and IOP. Greater PRSs were associated with higher IOP using the large-scale UKB cohort. The IOP PRSs were also significantly associated with glaucoma and increased the discriminatory ability for glaucoma. Moreover, the IOP PRSs outperformed traditional risk factors in predicting glaucoma, demonstrating the discriminatory power of aggregate genetic information as summarized in PRSs. Our findings highlight the potential far-reaching clinical utility of PRSs in reducing the global burden of devastating diseases like glaucoma by early detection of genetically susceptible individuals.
Supplementary Material
Acknowledgments
The authors thank the study participants from the UK Biobank and the staff who aided in data collection and processing.
Supported by National Institutes of Health (NIH; Bethesda, MD, USA) Grants R01EY022651, R01EY027315, RF1AG060472, and P30EY001792. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure: X.R. Gao, None; H. Huang, None; H. Kim, None
References
- 1.Weinreb RN, Leung CK, Crowston JG, et al. Primary open-angle glaucoma. Nat Rev Dis Primers. 2016;2:16067. doi: 10.1038/nrdp.2016.67. [DOI] [PubMed] [Google Scholar]
- 2.Collaborative Normal-Tension Glaucoma Study Group. Comparison of glaucomatous progression between untreated patients with normal-tension glaucoma and patients with therapeutically reduced intraocular pressures. Collaborative Normal-Tension Glaucoma Study Group. Am J Ophthalmol. 1998;126:487–497. doi: 10.1016/s0002-9394(98)00223-2. [DOI] [PubMed] [Google Scholar]
- 3.Kass MA, Heuer DK, Higginbotham EJ, et al. The Ocular Hypertension Treatment Study: a randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002;120:701–713. doi: 10.1001/archopht.120.6.701. discussion 829–730. [DOI] [PubMed] [Google Scholar]
- 4.Heijl A, Leske MC, Bengtsson B, et al. Reduction of intraocular pressure and glaucoma progression: results from the Early Manifest Glaucoma Trial. Arch Ophthalmol. 2002;120:1268–1279. doi: 10.1001/archopht.120.10.1268. [DOI] [PubMed] [Google Scholar]
- 5.Knowles JW, Ashley EA. Cardiovascular disease: the rise of the genetic risk score. PLoS Med. 2018;15:e1002546. doi: 10.1371/journal.pmed.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nannini DR, Kim H, Fan FD, Gao XY. Genetic risk score is associated with vertical cup-to-disc ratio and improves prediction of primary open-angle glaucoma in Latinos. Ophthalmology. 2018;125:815–821. doi: 10.1016/j.ophtha.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aschard H, Kang JH, Iglesias AI, et al. Genetic correlations between intraocular pressure, blood pressure and primary open-angle glaucoma: a multi-cohort analysis. Eur J Hum Genet. 2017;25:1261–1267. doi: 10.1038/ejhg.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramdas WD, Amin N, van Koolwijk LM, et al. Genetic architecture of open angle glaucoma and related determinants. J Med Genet. 2011;48:190–196. doi: 10.1136/jmg.2010.083337. [DOI] [PubMed] [Google Scholar]
- 9.Mabuchi F, Mabuchi N, Sakurada Y, et al. Additive effects of genetic variants associated with intraocular pressure in primary open-angle glaucoma. PLoS One. 2017;12:e0183709. doi: 10.1371/journal.pone.0183709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao XR, Huang H, Nannini DR, Fan F, Kim H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum Mol Genet. 2018;27:2205–2213. doi: 10.1093/hmg/ddy111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen NE, Sudlow C, Peakman T, Collins R, Biobank UK. UK biobank data: come and get it. Sci Transl Med. 2014;6:224ed224. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 13.Wasielica-Poslednik J, Berisha F, Aliyeva S, Pfeiffer N, Hoffmann EM. Reproducibility of ocular response analyzer measurements and their correlation with central corneal thickness. Graefes Arch Clin Exp Ophthalmol. 2010;248:1617–1622. doi: 10.1007/s00417-010-1471-1. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann M, Pitz S, Schmidtmann I, Pfeiffer N, Wasielica-Poslednik J. Tonographic effect of ocular response analyzer in comparison to Goldmann applanation tonometry. PLoS One. 2017;12:e0169438. doi: 10.1371/journal.pone.0169438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on ∼500,000 UK Biobank participants. Preprint at bioRxiv. 2017 doi: 10.1101/166298. [DOI]
- 16.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh PR, Tucker G, Bulik-Sullivan BK, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team. R: A language and environment for statistical computing. 2014 Available at: http://www.R-project.org/ Accessed December 1, 2017.
- 21.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Tham YC, Liao J, Vithana EN, et al. Aggregate effects of intraocular pressure and cup-to-disc ratio genetic variants on glaucoma in a multiethnic Asian population. Ophthalmology. 2015;122:1149–1157. doi: 10.1016/j.ophtha.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 23.Khawaja AP, Bailey JNC, Wareham NJ, et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50:778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacGregor S, Ong JS, An JY, et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50:1067–1071. doi: 10.1038/s41588-018-0176-y. [DOI] [PubMed] [Google Scholar]
- 25.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao XR, Fan F. Polygenic risk score is associated with intraocular pressure and improves glaucoma prediction in the UK Biobank cohort. Invest Ophthalmol Vis Sci. 2018;59:779. doi: 10.1167/tvst.8.2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, NY: Springer;; 2001. [Google Scholar]
- 28.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 29.Gao XR, Huang H, Kim H. Genome-wide association analyses identify 139 loci associated with macular thickness in the UK Biobank cohort. Hum Mol Genet. 2018 doi: 10.1093/hmg/ddy422. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 30.Choquet H, Thai KK, Yin J, et al. A large multi-ethnic genome-wide association study identifies novel genetic loci for intraocular pressure. Nat Commun. 2017;8:2108. doi: 10.1038/s41467-017-01913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hysi PG, Cheng CY, Springelkamp H, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46:1126–1130. doi: 10.1038/ng.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springelkamp H, Iglesias AI, Mishra A, et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017;26:438–453. doi: 10.1093/hmg/ddw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozel AB, Moroi SE, Reed DM, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014;133:41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blue Mountains Eye Study. Wellcome Trust Case Control Consortium 2 Genome-wide association study of intraocular pressure identifies the GLCCI1/ICA1 region as a glaucoma susceptibility locus. Hum Mol Genet. 2013;22:4653–4660. doi: 10.1093/hmg/ddt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nag A, Venturini C, Small KS, et al. A genome-wide association study of intra-ocular pressure suggests a novel association in the gene FAM125B in the TwinsUK cohort. Hum Mol Genet. 2014;23:3343–3348. doi: 10.1093/hmg/ddu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Springelkamp H, Iglesias AI, Cuellar-Partida G, et al. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum Mol Genet. 2015;24:2689–2699. doi: 10.1093/hmg/ddv027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.