Abstract
Rapidly improving sequencing technology coupled with computational developments in sequence assembly are making reference-quality genome assembly economical. Hundreds of vertebrate genome assemblies are now publicly available, and projects are being proposed to sequence thousands of additional species in the next few years. Such dense sampling of the tree of life should give an unprecedented new understanding of evolution and allow a detailed determination of the events that led to the wealth of biodiversity around us. To gain this knowledge, these new genomes must be compared through genome alignment (at the sequence level) and comparative annotation (at the gene level). However, different alignment and annotation methods have different characteristics; before starting a comparative genomics analysis, it is important to understand the nature of, and biases and limitations inherent in, the chosen methods. This review is intended to act as a technical but high-level overview of the field that should provide this understanding. We briefly survey the state of the genome alignment and comparative annotation fields and potential future directions for these fields in a new, large-scale era of comparative genomics.
Keywords: genome alignment, genome annotation, comparative genomics
Introduction
Alignment is possibly the most fundamental problem in genomics. The alignment problem is to establish a mapping between the letters of a set of sequences that approximates some relation that the user is interested in. In comparative genomics, we are generally interested in the homology relation—that is, does the lineage of two bases coalesce at a single base in a single organism at some (recognizably recent) point in time? In typical real-world comparative genomics, there is no clear proof of homology, as we have absolutely no access to the true history of every base in a set of sequences. However, we can use our knowledge of molecular evolution to construct very good approximations to the homology relation. The potential for using sequence similarity to approximate homology was recognized and applied very early on, starting with the pioneering work of Needleman & Wunsch (1) on optimal pairwise global alignment. The pairwise global alignment work was quickly specialized to perform local alignment, which calculates the optimal alignment of subsequences rather than sequences, by Smith & Waterman (2).
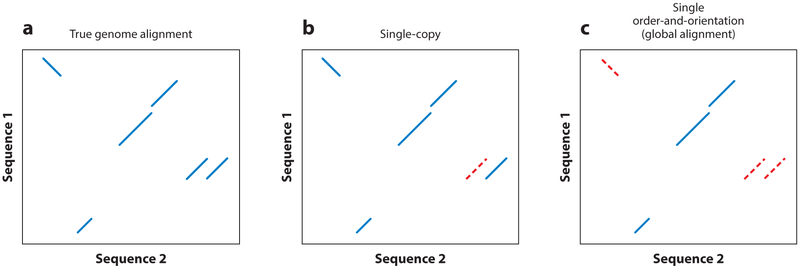
The traditional dynamic-programming algorithms require O(nm) (i.e., roughly proportional to the product of n and m) time and space, where n and m are the lengths of the two sequences; obviously, as n and m grow to genome scale, the problem becomes too expensive to solve in practice. Another consideration is how genome rearrangements complicate the alignment problem. Smith–Waterman and Needleman–Wunsch both produce alignments that have fixed order and orientation; that is, insertions, deletions, and substitutions are the only allowed edit operations. When looking within short or well-conserved sequences, like genes, this requirement is usually fulfilled. But at large evolutionary distances and looking within a sufficiently large window, genomes almost always contain more complex rearrangements with respect to each other—inversions, transpositions, and duplications all cause breaks in order and orientation that cannot be captured under constant order and orientation (see Figure 1).
Figure 1.
An example of how different heuristics affect a genome alignment. All panels are dotplots: A line with positive slope indicates an alignment from the positive strand of sequence 1 to the positive strand of sequence 2, and a negative slope indicates an alignment from the positive strand of sequence 1 to the negative strand of sequence 2. Solid blue lines represent alignments, and red dashed lines represent where alignments have been missed. (a) The true alignment between the two sequences. (b) The same alignment if a single-copy aligner perfectly recovered the true alignment, except for the ignored duplication. (c) The same alignment according to a global or approximately global aligner: No edit operations except insertions, deletions, and substitutions are allowed, so substantial alignment is missing.
As long DNA sequences became available, it was soon recognized that Needleman–Wunsch or Smith–Waterman alignments were far too slow to be useful for megabase-scale sequences, much less chromosome-scale sequences. The impractical running time of global alignment drove the development of several tools (3–5) that produce an approximately optimal global alignment through the use of high-confidence anchors in a single order and orientation, which are then used to partition the alignment into smaller problems that can be more efficiently solved. These anchors provided a very efficient and reliable way to break up the alignment problem but relied on a constant order and orientation, which excludes any possibility of noticing rearrangements.
What is genome alignment?
One obvious possible solution to the problems of rearrangement and duplication is to use a fast, approximate local alignment algorithm and simply use the collection of all local alignments that it finds as the whole-genome alignment. However, naively applying a local alignment approach has its own problems. Local alignments, when applied at genome-wide scale, have both too-low sensitivity and too-low specificity to be useful at substantial evolutionary distances. That is, local alignments will miss a homologous sequence that, by chance, happened to be further diverged than the sensitivity of the aligner could detect. They will also capture spurious alignments that can obscure more useful data. Even when they correctly identify homologous regions, the end user is more often interested in orthology rather than homology: Ancient duplications may share similar sequence but often do not share similar function. For our purposes in this article, we call any alignment that allows rearrangements (i.e., does not have fixed order and orientation) and attempts to determine orthology rather than just homology (even if restricted to a single copy) a whole-genome alignment (or for short, a genome alignment). Most whole-genome alignment methods are based on local alignments but do some filtering and postprocessing to construct a useful end product (6). Genome alignment tools offer more than simply a collection of local alignments—they must make decisions about where homology begins and mere similarity ends, and additionally they must make decisions about what is orthologous and not merely homologous. The size of the problem in whole-genome alignment of large genomes (e.g., mammalians) causes alignments to take too long to be practical, forcing efficiency considerations to be taken into account. At the same time, they must handle genome rearrangements—global aligners cannot properly align genomes that are diverged by even a few millions of years, because the collinearity restriction of global alignment causes so many homologies to be missed.
Determining orthology and the single-copy heuristic.
Choosing the single best target alignment for each region (based on alignment score or percent identity), which we call the single-copy strategy, is a common, if overly simplistic, way (7, 8) to deal with the problems that duplications cause. It is simplistic because the best-fit strategy will not always find a correct ortholog, and indeed even a reciprocally best-fit is not enough to guarantee finding an ortholog (9). Perhaps more importantly, choosing a single best sequence ignores lineage-specific duplications. When lineage-specific duplication occurs, a gene outside that lineage will have multiple orthologs in the lineage and should be aligned to multiple copies (10). Single-copy alignments implicitly assume that orthology is a one-to-one relationship. However, in nature, orthology can often be a one-to-many relationship (10). When that assumption of one-to-one orthology is violated, single-copy alignments can be very misleading.
Multiple Alignment
Often it is necessary to consider the alignment between a set of more than two sequences, which we call multiple alignment. A multiple alignment is defined as an equivalence relation ~ on a set of sequences S = {s1, s2,…,} such that for two bases b1 ∈ s1 ∈ S and b2 ∈ s2 ∈ S, b1 ~ b2 if they are considered to be aligned to each other. Here ~ is the alignment relation: the aligner’s estimate of orthology or homology between bases. The alignment is partitioned into columns by the equivalence classes of ~; i.e., every base is related to all bases in its column, and no two bases in different columns are related. Unfortunately, even simple formulations of the multiple alignment problem are significantly more difficult than their pairwise alignment equivalent and known to be NP-hard (11). Heuristics must be employed to efficiently solve the multiple alignment problem. Progressive alignment is the most popular strategy for approximate multiple alignment (12). Progressive alignment uses as an additional input a guide tree relating the input sequences. The most closely related sequences are aligned first, then the resulting alignment is itself aligned to other sequences or alignments, following the structure of the guide tree. Often consensus sequences are used as a method of aligning alignments.
Reference-Free Alignment
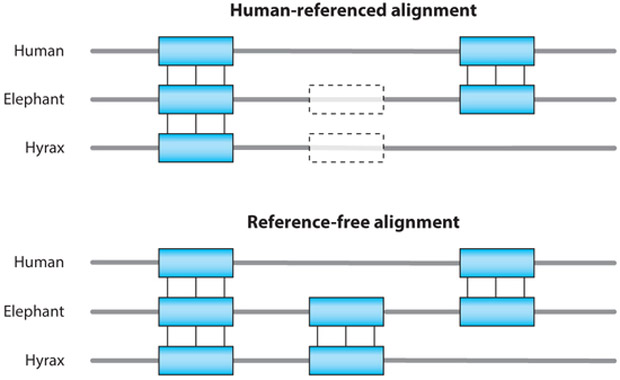
Because the multiple alignment problem is so difficult, a common heuristic is to use a single reference genome to base the alignment on. All other sequences in the multiple alignment are simply aligned to this genome in a pairwise fashion, then the several pairwise alignments are combined to form a reference-biased multiple alignment. This approach performs very well when viewed from the reference genome, but information relating genomes distant from the reference is lost. See Figure 2 for an illustration of this effect. In the mid- to late-2000s, the first methods for reference-free multiple genome alignment allowing multiple copies began to appear [notably the Enredo-Pecan-Ortheus (EPO) pipeline (13) and the A-Bruijn aligner (14)]. The EPO pipeline especially began to see wide use as part of the Ensembl genome browser (15). Although impressive, these pipelines left significant room for improvement, especially with regard to finding small-scale order-and-orientation-breaking rearrangements (13).
Figure 2.
A diagram showing the difference between a reference-biased and a reference-free multiple alignment. In a human-biased multiple alignment, any large regions that are deleted in human, or inserted somewhere else in the tree, cannot be aligned.
Genome histories.
Alignments are conventionally described as a set of columns, each containing a set of bases that are all related to each other by some alignment relation ~. Usually this relation represents orthology rather than homology. However, in that case, this model falls apart when considering reference-free alignments that can allow a single site in one genome to be aligned to multiple sites in another genome. The orthology relation is not transitively closed (10), so it is impossible in the general case to create a set of columns containing bases that are all orthologous to each other. The only way to represent a reference-free, multicopy, orthologous multiple genome alignment is by associating the alignment with phylogenetic trees, which are inferred (even if implicitly) during the alignment process. These trees must be reconciled (16) with the species tree so that the duplication and speciation events are distinguished, to enable the determination of orthology relationships. We term these types of alignments genome histories to reflect that they require a different representation than typical alignments (which can be represented by a collection of only blocks or columns).
A genome history {S, ~, Tc, ts, L} consists of a set of genomes S; a multiple alignment ~ relating the bases of those genomes; a reconciled tree called a column tree t ∈ Tc for each column in that alignment; a species tree ts; and, optionally, a set of links L between columns, indicating the ordering of the ancestral chromosomes. The columns of the genome history reflect the homology rather than the orthology relation. Because homology is transitive, the homology-based alignment can be represented by columns. The set of trees (hereafter referred to as column trees) indicate the evolutionary history of the bases in each column. Where there are duplications, gains, or losses, the column tree t ∈ Tc will differ from the species tree ts. Though the genome history representation we present here is not the only possible representation, any other representation (such as a collection of all pairwise orthology relationships) can be transformed into this one.
A genome history can be used to define both orthology and paralogy relations. The orthology relation, which we symbolize by ~o, uses the column trees of the genome history to determine which of the homologous bases in a column are also orthologous to each other. The orthologous bases are those homologous bases whose lineage coalesces in a speciation event in the reconciled column tree (10). The paralogy relation ~p simply relates homologous bases that are not orthologs.
A genome history can be projected onto any genome to create a more conventional referenced multiple alignment. These projected, reference-based alignments are collections of columns, each containing exactly one reference base, in which every base in the column is orthologous to the reference base but not necessarily orthologous to every other base in the column. These projected alignments are useful because they can be represented in conventional formats like the Multiple Alignment Format (MAF) and used as input to existing analysis tools.
Local Alignment Tools
Because genome alignment tools usually rely heavily on local alignments of some form, local alignment tools play a large role in genome alignments. Because finding all-against-all optimal local alignments has prohibitive time and memory requirements, approximate local aligners in the vein of BLAST (basic local alignment search tool) (17) are used almost exclusively. These aligners typically look for short sections of exact matches called seeds [which may sometimes include positions that are allowed to vary, to increase sensitivity (18)] and then extend the alignment away from those seeds. Local aligners used for genome alignment are often different than read aligners like BWA (Burrows–Wheeler Aligner) (19). Though they use the same basic ideas, local alignment between genomes generally involves much more evolutionary distance than read aligners, which are generally optimized for aligning reads to a reference genome that is near-identical to the sample. BLAT (BLAST-like alignment tool) (20) is a popular, fast local alignment tool that is useful at short evolutionary distances, though it can handle longer evolutionary distances with its “translated BLAT” translated-protein versus translated-protein mode. BLASTZ (7) and its successor LASTZ (21) are local aligners tuned to be more sensitive than normal BLAST, using PatternHunter-esque spaced seeds (18), while also allowing transitions for increased sensitivity. LAST (22) is a similar aligner, which can potentially use much smaller seeds than other aligners, without spending time going through uninteresting, highly repetitive alignments, because it extends partial matches until a low enough multiplicity is reached using an efficient substring index.
MashMap2 (23, 24), an approximate local aligner, is much faster than all the local aligners described above. However, currently it generates not a base-level alignment but an approximate correspondence between long regions combined with a similarity score. While base-level alignment is required for many tasks, approximate local aligners prove very useful for studying large-scale phenomena, such as chromosomal rearrangements.
Genome Alignment Methods
Most genome aligners, at a high level, work in two stages: filtering, in which a large number of local alignments are generated and filtered down to remove spurious false-positive alignments and identify homologous, rearrangement-free regions [locally collinear blocks in the terminology used by Mauve (25)], and refinement, in which the homologous regions undergo alignment with a collinear aligner. (Some aligners keep a subset of the original local alignments as anchors to be included in the final alignment, whereas others throw away all the original local alignments and align the rearrangement-free regions from scratch.) The filtering step can take many different forms, but many involve constructing a graph representation of the alignment and using various heuristics to simplify the graph (for a review, see Reference 26). A summary of popular or historically significant genome alignment tools is given in Table 1 (for pairwise alignment) and Table 2 (for multiple alignment).
Table 1.
Pairwise genome alignment tools
| Program | Year (Reference) | Description |
|---|---|---|
| MUMmer | 1999 (4, 117) | Fast aligner relying on maximal unique matches from a query sequence to a reference sequence; recent versions remove the colinearity restriction of the first version and improve the speed |
| Chains and nets | 2003 (29) | Combines fragmented local alignments into larger, high-scoring chains, which are arranged into hierarchical nets representing rearrangements |
| Shuffle-LAGAN | 2003 (27) | A glocal (global + local) aligner that is less restrictive than global alignment but still enforces monotonicity of the blocks relative to one sequence |
Table 2.
Popular and/or historically important multiple genome alignment tools
| Program | Year (Reference) | Reference-bias | Single-copy | Description |
|---|---|---|---|---|
| TBA | 2004 (8) | ✓ | Collinear multiple aligner (using MULTIZ internally) that produces a collection of partially ordered threaded blocksets | |
| Mugsy | 2011 (32) | Uses a graph-based method to segment the alignment problem into locally collinear blocks: small subregions with no local rearrangements, which are fed into a collinear multiple aligner | ||
| MULTIZ (autoMZ) | 2004 (8) | ✓ | ✓ | Multiple alignment based on pairwise alignment from every genome to a single reference |
| ABA | 2004 (14) | Aligner based on the concept of A-Bruijn graphs | ||
| EPO | 2008 (13, 37) | * | Graph-based aligner that allows duplications and optionally produces ancestral reconstructions | |
| VISTA-LAGAN (SuperMap) | 2009 (36) | Progressive aligner based on Shuffle-LAGAN (27) | ||
| Mauve | 2004 (25) | ✓ | Finds maximal unique matches present in every input species, then attempts to remove small matches that cause rearrangements that disrupt collinearity | |
| progressiveMauve | 2010 (31) | ✓ | Progressive aligner that attempts to remove anchors causing small rearrangements by optimizing a breakpoint-weighted score | |
| Cactus | 2011 (40) | Graph-based aligner that attempts to remove anchors representing small rearrangements |
Although the core method behind EPO is reference free, as currently applied its anchor generation is reference biased.
Pairwise genome alignment tools.
We now briefly survey some of the most significant pairwise genome alignment tools.
MUMmer.
MUMmer (4) is an extremely fast pairwise alignment tool, which is able to align the human and chimp genomes within less than 4 h. It achieves this speed by using a suffix-tree data structure to find all maximal unique matches between the two input genomes. Optionally, the nucmer script included in the package can perform gapped extension between these matches to generate a more complete alignment. MUMmer is an efficient package for aligning very similar genomes, though as a trade-off for its impressive speed, its sensitivity, especially with the default settings, is somewhat lower than slower aligners like LASTZ.
Shuffle-LAGAN.
Shuffle-LAGAN (27) is a pairwise genome alignment tool that aims to draw a compromise between the drawbacks of global and local alignment, using a method the authors call glocal alignment. The method works by first performing an all-against-all local alignment of the two genomes using CHAOS (28), then finding a maximal-scoring 1-monotonic map, which groups a subset of local alignments into chains, each of which contains local alignments with only a single order and orientation. This map is restricted so that the chains must be nondecreasing with respect to a single reference genome, while they can be in an arbitrary order in the other genome to represent rearrangements. This allows homology to be detected despite rearrangements, though it will not be able to detect duplications in the nonreference genome. The alignment is then further refined by discarding the local alignments within the chains and instead realigning the region bounded by each chain with the approximate global aligner LAGAN (5).
Chaining and netting.
Chaining (29) is a powerful technique for making sense of pairwise local alignments. Chains are simply maximal-scoring combinations of local alignments that maintain a single order and orientation. Chaining provides a good way of filtering out spurious alignments, which are likely to form short, low-scoring chains. However, the set of chains can often include distant paralogs or spurious sequence, which makes it difficult to understand the rearrangements that have taken place between the two input genomes. Netting (29) is a related technique that makes rearrangements relative to a reference genome much easier to find. In essence, netting finds the best-scoring set of chains that covers the bases of the reference genome only once. This makes it very easy to find high-confidence rearrangements like transpositions, inversions, and deletions but removes any duplications in the target genome, instead choosing a single copy to align to. Chaining and netting is very fast, but because the process fundamentally relies on local alignments as input, the overall process of generating chained and netted alignments mostly depends on the speed of the local aligner used.
Multiple genome alignment tools.
The following multiple genome alignment tools are some of the most historically important or popular in the field.
Mauve/progressiveMauve.
Mauve (25) is a reference-free multiple genome aligner that works by first finding all blocks that contain maximal unique matches from every species to use as anchors. To remove spurious matches, small matches that cause rearrangements are removed until the alignment can be partitioned into locally collinear blocks that are all above a certain size. These blocks are then further refined to attempt to create alignment problems small enough that they can be handled using a conventional collinear multiple aligner, in this case CLUSTAL W (30). The collection of these multiple alignments forms the final genome alignment.
The original version of Mauve performed poorly in large regions that were present in some but not all genomes, because only blocks containing sequence from every genome were used as anchors. progressiveMauve (31) was developed to relax this restriction. It builds a phylogenetic tree from the input sequences and then uses that tree as a guide to progressively apply an algorithm similar to the original Mauve at each internal node. Because of this progressive decomposition, its runtime scales linearly in the number of genomes.
Mugsy.
Mugsy (32) is a reference-free multiple genome aligner that uses a graph-based algorithm to segment a large collection of local alignments into smaller, rearrangement-free subproblems called locally collinear blocks, which can be fed into a conventional nongenome multiple aligner. Mugsy first generates all-against-all pairwise alignments using MUMmer (4) and then constructs a graph representation of the local alignment relationships. This graph is used to segment the large alignment problem into smaller locally collinear subproblems, which are then aligned using a specialized version of T-Coffee (33).
MULTIZ/TBA.
MULTIZ (8) is a reference-biased multiple genome alignment tool originally developed as part of the TBA (threaded blockset aligner) (8) program. Because TBA is restricted to producing multiple alignments that have only a single order and orientation (though an unpublished version exists that removes that restriction), MULTIZ sees much wider use than TBA itself. It is the tool currently used to generate the multiple alignments on the UCSC Genome Browser (34).
MULTIZ, in effect, is a method of aligning alignments. To produce MULTIZ alignments in practice, usually pairwise alignments from a given reference to all other species are generated using a local alignment tool, sometimes postprocessed using chains and nets, and then the autoMZ command is used to progressively align these pairwise alignments using a guide tree.
ABA.
The A-Bruijn alignment method (ABA) (14) uses A-Bruijn graphs (introduced in 35) to filter a collection of local alignments, removing inconsistencies and small rearrangements using simplification operations on the graph. The method is in principle reference free if the input alignments are generated in an unbiased way. Though the method was mostly applied to protein alignment (where individual domains are often duplicated and shuffled during evolution), it was also shown to be capable of aligning small chloroplast genomes more completely than TBA.
VISTA-LAGAN.
VISTA-LAGAN, also known as SuperMap (36), is a reference-free multiple alignment tool built on the Shuffle-LAGAN (27) pairwise alignment algorithm. Unlike Shuffle-LAGAN, VISTA-LAGAN can detect duplications in any genome, not just a reference genome. VISTA-LAGAN progressively aligns each pair of genomes, creating an ancestral ordering of the alignment blocks at each step (which is not intended to be an accurate ancestral reconstruction) to continue the alignment to further outgroup genomes.
EPO.
The EPO pipeline (13, 37) is a reference-free multiple alignment pipeline that, unlike TBA, can handle rearrangements. It is in wide use, being one of the main multiple genome alignments available on the Ensembl genome browser (38). The process begins with a relatively sparse set of anchor points that are known homologies within a set of genomes. The Enredo algorithm builds a sequence graph from these anchors and, through various operations, attempts to remove homologies that are likely to be spurious or uninteresting. The Pecan algorithm then fills in the gaps between the sparse anchors selected by the Enredo algorithm. The Ortheus algorithm (37) is then optionally run to generate ancestral sequences for all blocks, creating a genome history. Although EPO is in principle reference free, the method that is currently used to generate its anchors is reference biased (13).
Cactus.
Cactus uses an overall strategy similar in principle to the anchoring approach described above. The notion of a cactus graph (39) is used to create a filtered, high-confidence set of anchors. The unaligned space between anchors is then aligned using a sensitive pair hidden Markov model (HMM) to create a final multiple alignment. The first step of the Cactus process is to take small, uncertain local alignments captured by LASTZ (21) [which is similar to BLAST (17)] and combine them naively to create a multiple alignment. Given the typical evolutionary distances involved, LASTZ is tuned to be very sensitive but not very precise. The low precision means that the local alignments may be spurious (for example, when a small seed happened to match, and be extended, in a region that is not truly homologous). The local alignments may also conflict—that is, several alignments may disagree on how to align a particular region. These inconsistencies and spurious alignments will manifest as tiny rearrangements—breaks in order and orientation—in the alignment. By using the Cactus Alignment Filter (CAF) algorithm defined in Reference 40, these small rearrangements, which are unlikely to be biological, in the multiple alignment are discovered and removed, producing an alignment that contains only rearrangements longer than a certain length. After this process, the cactus graph contains anchors that are very likely to represent true regions of homology but will have unaligned regions of homology between the anchors, which local alignment was not sensitive enough to pick up, or which were deleted in the CAF process. The Base Alignment Refinement (BAR) process (40) fills in these unaligned but homologous regions. Because its runtime is dominated by the all-against-all local alignment process, Cactus scales quadratically in the number of input genomes, assuming they are all the same length.
ProgressiveCactus.
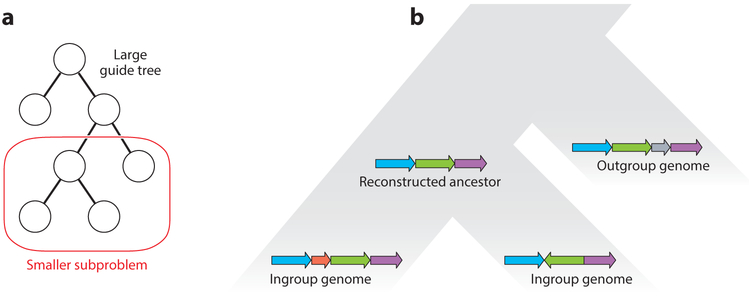
The version of Cactus published in 2011 (40) was highly effective at aligning a small number of genomes in the tens to hundreds of megabases (41), but because it scaled quadratically with the total size of all genomes in the alignment problem, it could not efficiently create alignments of hundreds of vertebrate-sized genomes. The recently developed progressiveCactus, a progressive-alignment extension to the original Cactus algorithm, scales linearly with the number of genomes, allowing efficient alignments of hundreds of genomes. The progressiveCactus process works as follows (see Figure 3). First, the problem is decomposed into several subproblems using an input guide tree. There is one subproblem per internal node in the guide tree. Each subproblem involves aligning several genomes using the traditional Cactus process: the ingroup (children of the internal node) and outgroup (nondescendants of the internal node) genomes. This alignment subproblem is then used to infer a reference assembly that contains only the blocks that should be present in the ancestral genome. All blocks containing only a single ingroup sequence are deemed to be insertions in that lineage and removed from the ancestor. Similarly, all blocks composed of only outgroups represent deletions in the branch above the ancestor and are removed. The filtered blocks are arranged into sequences according to an algorithm that attempts to maximize the consistency between the order and orientation of all the sequences in the alignment (42). The base-level sequence for these blocks is then generated by finding the maximum-likelihood base for each column using the guide tree. This assembly is a reconstruction of the ancestral genome at that node, which functions as a consensus sequence for the ingroups below it. The reference assembly is then fed as input into subproblems further up the guide tree.
Figure 3.
An example of how progressive genome alignment works, focused on aligners like VISTA-LAGAN (SuperMap) (36) and progressiveCactus (40), which reconstruct ancestral genomes as input for further alignment steps. (a) A large guide tree (usually the species tree), which may include many species, is divided up into smaller local alignment problems of a few genomes each. (b) A diagram of what occurs within each subproblem. Each subproblem is focused on reconstructing a single ancestral genome, which is then used as input for subproblems further up the tree. Ingroup genomes (children of the ancestor in question) and, optionally, outgroup genomes (nondescendants of the ancestor) are aligned together. A plausible ancestral reconstruction is generated for use in later subproblems.
Alignment Formats
The fact that genome alignments include the potential for rearrangements and duplications makes representation in collinear alignment formats like aligned-FASTA impossible, because they represent alignments as only a series of insert, delete, and substitution operations. The most popular format for genome alignments currently is MAF. MAF is capable of representing referenced multiple alignments with rearrangements; however, because MAF is a column/block-oriented format, it is impossible to represent complex orthology relationships in a reference-free way (see section titled Genome Histories) without extending the format.
The Hierarchical Alignment Format (HAL) (43) was designed to be an efficiently accessible format representing a genome history, including any available ancestral reconstructions. HAL allows projection from this genome history onto any reference genome (including ancestors), creating a multiple genome alignment showing what is orthologous (related by ~o) to every base in that genome. This projection can be output in a traditional format like MAF or simply used on demand to visualize the alignment (44) or as part of downstream analysis pipelines.
COMPARATIVE ANNOTATION
Introduction
Genome annotation is the process of finding functional elements in a genome assembly. Generally, these take the form of protein coding genes, but they can also include noncoding transcripts (45), chromatin configuration (46), DNase hypersensitivity (47), CpG islands (48), and population variation (49, 50).
The task of automatically annotating genome assemblies has been considered since the first full-length genomes were released in the mid-1990s (51–53). This task is often divided into two categories: ab initio prediction, or the computational prediction of exon-intron structure using statistical models, and sequence alignment–based approaches, which map any expressed sequence tag (EST), complementary DNA (cDNA), or protein sequences onto an assembled sequence to discover transcripts (38). Some annotation pipelines combine both sources of transcript prediction to generate a final annotation set (54, 55).
Recent improvements in sequencing technologies, including long-read (56) and linked-read technology (57), have provided the ability to produce high-quality genome assemblies at prices that make genome assembly an affordable experiment for labs across the world. This has led to the formation of consortia that aim to produce genome assemblies on a wide scale, such as the Vertebrate Genome Project (58).
This rapid increase in the availability of high-quality genome assemblies necessitates the introduction of automated methods that can scale and can leverage the improved phylogenetic information that such an array of assemblies can provide. For example, the 200 Mammals Project is specifically designed to allow for the calculation of base-level conservation across mammalian evolutionary history. This discriminatory power can be leveraged downstream of the assemblies to improve whole-genome alignments as well as annotations, and it provides a framework for annotating genes that allows for gain/loss of genes throughout evolution, instead of the current models that rely heavily on annotating relationships relative to mouse and human.
In this new era of genome assembly, consideration must be given not just to assembly and alignment but also to annotation. Annotation is central to the question of how to use this explosion of genome assemblies, and high-quality annotation sets with orthology mappings across species will enable a wide range of comparative genomics analyses.
Sequence-Based Comparative Annotation
In conjunction with the release of the first mouse draft assembly (59), multiple different tools were created to try and leverage comparative information to human to look for genes, including TWINSCAN (60), SGP (61), and SLAM (62). Table 3 provides an overview of comparative annotation tools, including those written in the years following the mouse genome assembly. These tools provide probabilistic frameworks that combine established single-genome gene prediction approaches (63, 64) with informant data obtained through genomic alignments to improve gene predictions. Notably, all of these tools work only on pairwise alignments and cannot use information extrinsic to this alignment and the underlying input sequences.
Table 3.
Overview of comparative annotation tools
| Program | Year (Reference) | Description |
|---|---|---|
| ROSETTA | 2000 (110) | Uses pairwise genomic alignments to find regions of homology; incorporates a splice junction and exon length model |
| SGP−1/−2 | 2001 (61) | Uses pairwise genomic alignments to find syntenic loci; evaluates a coding and splice model in these loci |
| TWINSCAN | 2003 (60) | Uses local alignments between a target genome and a reference (informant) genome to identify regions of conservation |
| SLAM | 2003 (62) | Treats two alignments in a symmetric way, predicting pairs of transcripts |
| EvoGene | 2003 (111) | Phylogenetic HMM that performs ab initio prediction of genes across a multiple-sequence alignment (more than two genomes), making use of phylogenetic information |
| ExoniPhy | 2004 (112) | Phylogenetic HMM that performs ab initio predictions across a multiple-sequence alignment |
| DOGFISH | 2006 (113) | Two-step program that combines a classifier that scores potential splice sites using a multiple-sequence alignment and an ab initio gene predictor that makes use of the scores from the classifier to predict gene structures |
| N-SCAN | 2006 (65) | Extends the TWINSCAN model to N genomes |
| CONTRAST | 2007 (68) | Uses a combination of SVM and CRF predictors, providing a big boost over traditional HMMs |
| DOUBLESCAN | 2002 (118) | Uses a pair HMM to simultaneously predict gene structures and conservation in two aligned sequences |
Abbreviation: CRF, conditional random field; HMM, hidden Markov model; SVM, support vector machine.
As more vertebrate genomes were sequenced, the need for comparative gene predictors that could use more than one informant species arose. Some of the previous tools were reengineered, as is the case with N-SCAN (65, 66). However, N-SCAN predicted only 35% of human genes correctly, and using a multiple sequence alignment was no more accurate than using a high-quality pairwise alignment (67). In contrast, CONTRAST (68) remarkably was able to accurately predict 65% of human genes using 11 informant genomes. Prior to CONTRAST, practically all gene prediction tools relied on HMM, a generative model, whereas CONTRAST relied on a discriminative support vector machine (SVM) model. The SVM is used to model coding regions, whereas an additional model called a conditional random field (CRF) is used to model the gene structure itself. A CRF can be considered a generalization of a HMM (69).
For the most part, after the initial mouse genome project, none of these tools have been used on full vertebrate genomes. There are two reasons for this. First, these tools require very careful parameter training, which must be performed on every genome in the alignment. Second, these tools require evaluating all pairwise comparisons leading to running times quadratic in the number of genomes. Combined with the overall lower efficacy of comparative prediction versus transcriptome and proteome sequence alignment approaches, this has led the field of comparative gene finding to languish for the past 10 years.
Transcriptome Evidence-Based Comparative Annotation
None of the annotation programs described above were capable of incorporating extrinsic information, instead relying entirely on sequence composition. In species with sufficient transcriptome data, mapping these data to the assembly generally performs far better at gene finding than the de novo approaches outlined above (70, 71).
In the early 2000s, projects like the Mammalian Gene Collection (72) were generating full-length cDNA sequences for model organisms. These full-length transcripts, in addition to ESTs, were being stored in databases like GenBank (73), supplemented by submissions from labs around the world. Tools were developed to incorporate alignments of these sequences in gene prediction, including N-SCAN-EST (74), GenomeWise (75), and AUGUSTUS (70, 76, 77) (Table 4).
Table 4.
Overview of gene prediction tools that incorporate transcriptome data
| Program | Year (Reference) | Description |
|---|---|---|
| GeneWise | 2004 (75) | HMM-based gene prediction tool using extrinsic evidence; MAKER2 can make use of it |
| N-SCAN-EST | 2006 (74) | HMM-based gene prediction tool that makes use of EST and genomic alignments, incorporating phylogenetic information |
| AUGUSTUS | 2004 (76, 77) | CRF-based gene prediction tool with many modes; features are still being added; can perform ab initio gene prediction as well as incorporate extrinsic evidence; has the ability to provide nonlinear weights to various types of evidence |
| EVM | 2008 (114, 115) | A chooser algorithm that combines previously predicted gene sets with extrinsic information to construct consensus gene sets |
| PASA | 2003 (116) | Uses alignments of cDNA, EST, or RNA-seq to predict gene structures, including alternative splice events |
| MAKER2 | 2008 (55, 78) | An all-in-one pipeline that runs programs including AUGUSTUS and GeneWise with extrinsic information such as RNA-seq or protein sequences to both predict annotations and construct a gene set |
Abbreviation: cDNA, complementary DNA; CRF, conditional random field; EST, expressed sequence tag; HMM, hidden Markov model; RNA-seq, RNA sequencing.
Although these tools were developed for annotating single genomes with extrinsic information from the same genome, they can be applied in a comparative fashion. Many species of interest have limited transcriptome data available but are closely related to well-annotated species. Examples include mouse versus rat and human versus other great apes. Alignment of related transcript sequences is used in the gene builds produced by both Ensembl (38) and RefSeq (54).
Another approach is to use alignments of protein sequences instead of transcript sequences, which are more robust across long phylogenetic distances. The popular annotation pipeline MAKER2 (55) provides such functionality and recommends providing protein sequences from at least two related genomes (78).
For more distantly related species, approaches that generate profiles of proteins and protein domains may be used. Databases such as InterPro (79) store precomputed models of protein sequences and motifs that are conserved across long periods of evolutionary history. GeneWise (75) can perform gene prediction using a profile HMM like those stored in InterPro. Additionally, AUGUSTUS-PPX (80) is an extension of the AUGUSTUS annotation program that models protein families and combines them with the existing ab initio model.
Transcript Projection
Transcript projection uses sequence alignments to project the coordinates of an existing annotation in one genome to another genome. This powerful approach leverages high-quality annotations in well-studied organisms to annotate diverse transcripts in related genomes. Many genes and isoforms are expressed in specific tissue types (81), at specific developmental time points, or only in response to specific environmental conditions (82). Gathering the data for a new species to fully annotate their transcriptome is thus prohibitively expensive. Transcript projection methods allow researchers to bypass this by making use of the high-quality information in well-studied organisms (83, 84). Additionally, traditional ab initio gene-finding models rely heavily on the signature of protein coding genes, which limits these models’ ability to predict untranslated region (UTR) sequences; noncoding RNAs, such as long noncoding RNAs; and pseudogenes. Transcript projection methods can be combined with any available extrinsic information from either the genome in question or related genomes and provide the highest-quality annotation as a result (70). See Table 5 for an overview of transcript projection methods.
Table 5.
Overview of transcript projection tools
| Program | Year (Reference) | Description |
|---|---|---|
| Projector | 2004 (85) | Similar to DOUBLESCAN but extends the model to make use of annotation information on one sequence to inform the other; works better than GENEWISE over long branch lengths |
| AIR | 2005 (86) | Integrates multiple forms of extrinsic evidence to perform alternative splice junction prediction |
| transMap | 2007 (70, 87) | Uses whole-genome alignments to project existing annotations from one genome to one or more other genomes |
| CESAR | 2016 (84) | Uses a HMM to adjust splice sites in whole-genome alignments, improving transcript projections |
Abbreviations: AIR, Annotation Integrated Resource; CESAR, Coding Exon-Structure Aware Realigner; HMM, hidden Markov model.
The first tool to perform transcript projection was Projector (85), which uses a pair HMM and models exon-intron structure through a pairwise alignment, similar to how tools like TWINSCAN work. However, Projector can make use of the known gene information in one sequence in the alignment to restrict the probability paths to those that match the known gene. A subsequent tool, Annotation Integrated Resource (AIR) (86), introduced the concept of a splice graph, a directed acyclic graphic structure that represents exons as vertices and introns as edges, in which isoforms of a gene are paths through this graph. AIR projects transcripts from a reference genome through a syntenic alignment to score the paths in the graph, reducing the large number of biologically improbable combinations.
transMap.
transMap (70, 87), first developed in conjunction with improvements to AUGUSTUS to model extrinsic information (77), relies on whole-genome alignments to project existing annotations from one to the other genome in the alignment. This process is purely arithmetic, but it has proven to be immensely helpful at providing extrinsic information to guide AUGUSTUS and improve on purely sequence-based prediction. This simplicity means that transMap runs in linear time with the number of genomes and transcripts being projected. Compared with methods that incorporate EST alignments, transMap provides both full-length transcript information and isoform information. transMap provided the biggest benefit to specificity in AUGUSTUS predictions in all cases except those in which the existing cDNA repertoire for the species in question exceeded the quality of the reference (70).
CESAR.
Coding Exon-Structure Aware Realigner (CESAR) (84) is a tool that projects exons through a whole-genome alignment, handling splice site shifts that are a common feature of evolutionary change. CESAR is a straightforward HMM that takes as input the linear alignment of a single exon to other genomes with a small amount of flanking intronic sequence and outputs a realigned region that accounts for exon frame and evolutionary change. CESAR was able to achieve a nearly 89% accuracy at realigning splice sites, leading the number of frameshifts seen when mapping human genes to mouse to drop from 2.7% to 0.3%. These spurious frameshifts must be addressed when working with transcript projection methods. CESAR 2.0 (88) made multiple improvements, most noticeably in runtime. Mapping of all human protein coding genes to mouse was recorded as taking seven hours on a single core of a desktop computer.
Comparative AUGUSTUS
AUGUSTUS recently added a novel objective function parameterization option, called Comparative AUGUSTUS or AugustusCGP, that makes use of whole-genome alignments to predict coding genes simultaneously in every genome in the alignment (89). With recent updates, training the AugustusCGP model is straightforward and integrated in the AUGUSTUS binary. In contrast to previous comparative gene-finding tools, AugustusCGP runs linearly in the number of genomes, making the possibility of annotating dozens of genomes computationally tractable. Currently, AugustusCGP relies on a referenced multiple genome alignment format called MAF, and as such it cannot annotate genes that are not present in the reference.
AugustusCGP makes use of the phylogenetic information inherent in the whole-genome alignment to look for evidence of negative selection. This is done by calculating the ratio of synonymous and nonsynonymous substitutions in candidate exons. AugustusCGP can incorporate extrinsic evidence, including RNA sequencing (RNA-seq), Iso-Seq, and cDNA alignments, loading these into a database that allows for predictions to be parallelized and scaled. AugustusCGP can also incorporate evidence from annotation sets on one or more genomes in the alignment, which can help guide comparative annotation efforts by providing strong hints on where genes are expected to be found. Making use of as many of these forms of extrinsic evidence as possible is recommended, as it greatly improves the accuracy and specificity of the model. AugustusCGP, along with all genefinding tools, can often produce false positives. For the Mouse Genomes Project (90), as well as the default in the Comparative Annotation Toolkit (CAT) pipeline (83), we find that filtering for predictions with at least two splices reduces the false-positive rate to an acceptable level. However, this can remove interesting unspliced genes like olfactory receptors (91).
GENOME ANNOTATION PIPELINES
The most commonly used annotation pipeline for individual researchers is MAKER2 (55). MAKER2 was designed to enable researchers to annotate genome assemblies they produce and not rely on institutional pipelines. This makes MAKER2 amenable to annotating nonmodel and nonvertebrate species, including prokaryotes. MAKER2 combines multiple forms of extrinsic evidence with ab initio prediction and provides a fully automated end-to-end annotation pipeline. MAKER2 can make use of multiple gene prediction tools, including GeneWise, GeneMark, SNAP, and AUGUSTUS. MAKER2 also performs repeat masking of the assembly.
However, MAKER2 has drawbacks. It is technically challenging to run the pipeline, and it relies on difficult-to-use parallel computing paradigms. MAKER2 also requires that all extrinsic evidence exist in the form of sequences, and as such requires that RNA-seq data undergo de novo assembly before use. MAKER2 does not attempt to track orthology relationships or anything about the genes predicted and so requires subsequent processing of the gene models to determine gene family and protein domain information. MAKER2 also does not annotate UTR sequences or noncoding genes, which can present challenges when using the annotations to quantify expression.
Most vertebrate annotation sets available right now are produced by large institutional pipelines at either Ensembl (Ensembl Gene Build) or the National Center for Biotechnology Information (RefSeq). These institutional efforts are important for the larger size and complexity of vertebrate genomes, but as computing power becomes cheaper, the need to outsource these efforts is diminishing. Turnaround for annotation by RefSeq often takes months. Both of these pipelines do track orthology relationships and assign gene common names where applicable.
This process of tracking orthology relationships will become increasingly important as the number of assembled vertebrate genomes continues to grow. A systematic framework that can track complex orthology relationships is required. Ensembl has put effort into this with the Compara browser (92).
Comparative Annotation Toolkit (CAT)
CAT is a comparative annotation pipeline that combines a variety of parameterizations of AUGUSTUS, including Comparative AUGUSTUS, with transMap projections through whole-genome progressiveCactus alignments to produce an annotation set on every genome in a Cactus alignment (83). CAT is an attempt to synthesize all of the possible methods of genome annotation, relying on transcript projection, transcriptome and proteome alignments, simultaneous gene finding, and single-genome gene finding with full-length cDNA reads. CAT leverages high-quality gene sets like those produced by GENCODE on mouse and human to project annotations to other genomes, augmented with predictions that add species specificity and detect gene family expansion and collapse.
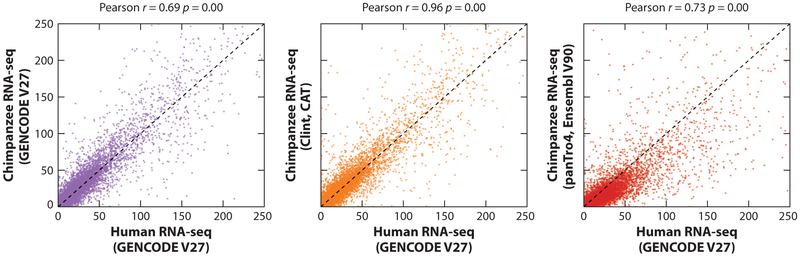
Recent work showed that CAT was capable of leveraging GENCODE to reannotate the rat genome, improving on the existing RefSeq and Ensembl annotations (83). CAT was also applied to the great apes, reannotating the existing great ape assemblies as well as annotating the new PacBio-derived great ape assemblies. In all cases, CAT provided the highest isoform concordance compared with the Ensembl annotations when compared with an Iso-Seq data set generated from induced pluripotent stem cells from each of the great ape species. Isoform concordance for the chimpanzee annotation of the new PacBio assembly was 82.1%, the same as it was for human GRCh38 using GENCODE V27. In testing these new annotation sets by using them to quantify RNA-seq, an average 9,518 more expressed genes were found in the great ape species than when the Ensembl annotations were used. These annotation sets greatly improved the ability to perform cross-species RNA-seq expression estimates, with Pearson r = 0.96 seen when comparing CAT annotation of chimpanzee to human, compared with r = 0.69 when mapping the same RNA-seq to human directly and r = 0.73 when using common gene names in Ensembl V90 to perform cross-species comparison (Figure 4).
Figure 4.
Comparing RNA sequencing (RNA-seq) expression quantification across different species with Comparative Annotation Toolkit (CAT). Kallisto (109) protein-coding gene-level expression for chimpanzee induced pluripotent stem cell (iPSC) RNA-seq is compared with human across all of the chimpanzee annotation and assembly combinations as well as when mapped directly to human. In all cases, the x-axis is the transcripts per million of human iPSC data mapped to GRCh38 annotated with GENCODE V27. The highest correlation (Pearson r = 0.96) is seen when comparing Clint (panTro6) annotated with CAT to GRCh38. The value p is the p-value of observing the Pearson correlation.
Noncoding Annotation
Comparative annotation of noncoding genes has been considered since the explosion of comparative genomics tools in the mouse genome project era (93). MAKER2, along with most standard annotation tools like AUGUSTUS, cannot detect noncoding transcripts owing to the inherent difficulty in detecting these without the strong statistical signal that protein coding transcripts provide. Tools like transMap can leverage noncoding annotations curated in high-quality genomes to transfer these annotations to new genomes. However, important spliced noncoding transcripts like long intergenic noncoding RNAs are inherently less conserved, especially at the sequence level (94–96). This can present challenges to using whole-genome alignments to annotate such transcripts. It also can be difficult to determine whether a predicted noncoding RNA is actually expressed in a species without finding it in an extrinsic data set. An opposite problem also exists—it is not difficult to find a nonconserved open reading frame in a noncoding transcript, and tools like AUGUSTUS will predict coding transcripts at noncoding loci, particularly when provided extrinsic evidence that contains expression of these loci. Tools like PhyloCSF (97) can help diagnose these regions by leveraging whole-genome alignments to evaluate conservation of coding signal, but they are not foolproof, and the authors recommend manual curation. GENCODE is using the PhyloCSF model to help improve manual annotation efforts.
Personal Human Genomes and Intraspecies Annotation
The rapidly decreasing price of genome assembly has opened the possibility of assembly of multiple individuals of a single species (98, 99). This is particularly interesting in the case of humans, where personalized diploid assembly (57, 100) may prove useful in understanding complex loci and could lead to deeper understanding of disease phenotypes. One step to understanding these personalized de novo assemblies of humans is to annotate them and evaluate them for deleted, duplicated, or rearranged gene content. Rearranged gene content in particular can be difficult to detect with standard short-read resequencing methods.
DISCUSSION
The Future of Whole-Genome Alignment
The average evolutionary distance between sequenced species is getting much shorter as more genomes are sequenced. In the next several years, thousands of genomes will be released owing to the efforts of projects like the Vertebrate Genomes Project/Genome 10K (101), Bat 1K, 200 Mammals, Insect 5K (102), and the Earth Biogenome Project. By necessity, many comparative genomics projects will focus on alignment between hundreds to thousands of closely related genomes, instead of tens of distantly related genomes. Evaluation of simulated data has shown that although whole-genome aligners vary drastically in accuracy over long evolutionary distances [F-scores ranging from 0.12 to 0.80 in a mammal-wide simulated alignment (41)], they all perform extremely well over closer evolutionary distances [F-scores ranging from 0.97 to 0.99 in a primate-wide alignment (41)]. In some ways, then, genome alignment will become easier because finding homologies is simpler with less evolutionary distance. However, in other ways, it will become more difficult. Creating an alignment of thousands of large genomes is a unique challenge, one that no aligner is currently prepared for. In addition, because the rate of new assemblies being generated will increase, with new assemblies projected to come every few days or weeks rather than months, maintaining alignments at community comparative genomics resources like Ensembl Compara (15) or the UCSC Genome Browser (34) will necessitate adding new genomes to existing resources piecemeal, rather than regenerating alignments from scratch.
As large sequencing projects produce hundreds to thousands more assemblies in the coming years, reference bias in multiple-genome alignments may become more of a problem. Though reference-biased alignments will serve the genome that they are referenced on well (usually a popular genome like human or mouse), it will certainly be cost prohibitive to generate a full alignment referenced on all, or even most, new assemblies. This can be a disadvantage to researchers working on nonmodel organisms, who may not have the resources to run a full alignment referenced on their genome. A reference-free alignment would be more easily shared as a resource useful to many different communities researching many different species. However, reference-free alignment is a substantially more difficult problem than reference-biased alignment. In principle, a reference-free alignment should be equally good for every included genome. However, it may be the case that for any given genome, the quality of a reference-free alignment may be worse than if a new reference-biased alignment were generated referenced on that genome. To live up to their potential, reference-free aligners should aim to equal the quality of reference-biased aligners on reference genomes.
The unique challenges facing genome alignment are twofold: Compared with global alignment, the challenge is to capture rearrangements; compared with local alignment, the challenge is to detect orthology rather than mere homology. The Alignathon (41) showed that modern genome aligners generally capture homologies well in the presence of rearrangements. However, orthology detection in modern genome aligners is still very simplistic and not very accurate. Many aligners still operate under a single-copy restriction, which, although a useful simplification of the alignment problem, obscures crucial aspects of genome evolution. Others, like Cactus (40) or EPO (13), can support multiple orthology in theory but in practice use simple heuristics that can often lead to aligning paralogs or missing alignment to orthologs.
Determining orthology accurately, efficiently, and on a genome-wide scale is possibly the most difficult unsolved problem in genome alignment. The problem can be framed as building phylogenetic trees for every column in the genome history, after which orthology relationships among the column’s bases can be easily established using a reconciliation algorithm (16). Simply applying maximum-likelihood methods such as RAxML (103) will not be sufficient for multiple reasons. First, these methods require near-prohibitive amounts of computer power to apply genome wide in large alignments: Building trees from even a relatively small set of 2,000 1,000-bp alignment regions among 48 avian species can take more than 100 CPU days to compute (104). Second, and more importantly, with large numbers of genomes, the size of regions with the same duplication content can become smaller and smaller, leading to less and less phylogenetic information available for any given region, which could increase the chance of errors. It may be helpful to incorporate syntenic information to try to improve the accuracy of finding orthology relationships genome wide, though it seems that there are still unanswered questions about how best to solve that problem (105). One advantage of using syntenic information to establish orthology is that it may enable tracking of orthologous loci [sometimes called toporthologs or positional orthologs (106)] in addition to tracking of orthologous sequences. Keeping track of orthologous loci may be useful to better track gene conversion events, which will cause discrepancies between the toporthology and orthology of a given region.
This review has focused on interspecies comparison, but the future must include more convergence in thinking, models, and reasoning about both inter- and intraspecies variation. A key, relevant development in modeling population variation is the genome graph (107, 108), which represents variation by encoding individual genomes as paths through a graph structure representing the combined genome alignment. This process allows for variation to be comprehensively captured and reduces the necessity of depending on a linear reference that does not accurately represent haplotypes present in the population. Extending the genome alignment process to handle graph-to-graph alignment, rather than merely sequence-to-sequence alignment, will bring together the fields of comparative and population genomics by enabling the integration of the analysis of inter- and intraspecies variation. Such graphs also naturally fit with methods that model uncertainty about ancestral genomes, and therefore some of the software developed for genome graphs might be useful in modeling ancestral genome reconstructions.
The Future of Comparative Annotation
In the coming new era of comparative genomics in which high-quality genomes of many species or many individuals of the same species are plentiful, comparative annotation will play a central role in helping to synthesize useful information out of the deluge of data. All of the comparative annotation paradigms described above are insufficient for the annotation of true many-to-many orthology.
As the number of assembled genomes grows, it will become possible to track these relationships in new ways. New approaches and tools must be developed to assess these relationships. Cactus provides a method for generating alignments amenable to annotating complex orthology relationships, but currently no comparative annotation tool can approach this. Comparative AUGUSTUS, for example, still relies on an alignment file format that is referenced on a specific genome. We see the possibility of a future where these problems are sidestepped through iterative use of pipelines like CAT. Gene predictions can be performed on many genomes, perhaps picking representatives evenly spread throughout the tree of life that have high-quality assemblies and a decent variety of extrinsic evidence like RNA-seq available. These predictions can then be mapped via transMap to other genomes in the alignment and then assessed, combined, and collapsed in a process similar to the consensus-finding step that CAT currently performs.
ACKNOWLEDGMENTS
The authors were supported by the National Institutes of Health (3U54HG007990, 1U01HL137183, and 5U41HG007234) and the W.M. Keck Foundation (DT06172015). We thank the reviewers for their help and excellent suggestions.
Footnotes
DISCLOSURE STATEMENT
I.T.F. is an employee of 10x Genomics. M.D. is part of the GENCODE consortium, which makes use of comparative genomic software, and is involved in the development of the University of California, Santa Cruz, comparative genomes suite of tools. The other authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443–53 [DOI] [PubMed] [Google Scholar]
- 2.Smith T, Waterman M. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195–97 [DOI] [PubMed] [Google Scholar]
- 3.Bray N, Dubchak I, Pachter L. 2003. AVID: a global alignment program. Genome Res. 13:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLOS Comput. Biol. 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, et al. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batzoglou S 2005. The many faces of sequence alignment. Brief. Bioinform. 6:6–22 [DOI] [PubMed] [Google Scholar]
- 7.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, et al. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13:103–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M, Kent WJ Riemer C, Elnitski L, Smith AFA, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson T 2007. Reciprocal best hits are not a logically sufficient condition for orthology. arXiv:0706.0117 [q-bio.GN] [Google Scholar]
- 10.Koonin EV. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309–38 [DOI] [PubMed] [Google Scholar]
- 11.Jiang T, Wang L. 1994. On the complexity of multiple sequence alignment. J. Comput. Biol. 1:337–48 [DOI] [PubMed] [Google Scholar]
- 12.Feng DF, Doolittle RF. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25:351–60 [DOI] [PubMed] [Google Scholar]
- 13.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. 2008. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 18:1814–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raphael B, Zhi D, Tang H, Pevzner P. 2004. A novel method for multiple alignment of sequences with repeated and shuffled elements. Genome Res. 14:2336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, et al. 2017. Ensembl 2017. Nucleic Acids Res. 45:D635–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zmasek CM, Eddy SR. 2001. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics 17:821–28 [DOI] [PubMed] [Google Scholar]
- 17.Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–10 [DOI] [PubMed] [Google Scholar]
- 18.Ma B, Tromp J, Li M. 2002. PatternHunter: faster and more sensitive homology search. Bioinformatics 18:440–45 [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris R 2007. Improved pairwise alignment of genomic DNA. PhD thesis, Coll. Eng., Pa. State Univ., University Park, PA [Google Scholar]
- 22.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21:487–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain C, Koren S, Dilthey A, Phillippy AM, Aluru S. 2018. A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics 34:i748–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain C, Dilthey A, Koren S, Aluru S, Phillippy AM. 2018. A fast approximation algorithm for mapping long reads to large reference databases. J. Comput. Biol. 25:766–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehr B, Trappe K, Holtgrewe M, Reinert K. 2014. Genome alignment with graph data structures: a comparison. BMC Bioinform. 15:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brudno M, Malde S, Poliakov A, Do CB, Couronne O, et al. 2003. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19:i54–i62 [DOI] [PubMed] [Google Scholar]
- 28.Brudno M, Morgenstern B. 2002. Fast and sensitive alignment of large genomic sequences. In Proceedings of the IEEE Computer Society Bioinformatics Conference, pp. 138–47. New York: IEEE; [PubMed] [Google Scholar]
- 29.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. 2003. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. PNAS 100:11484–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling AE, Mau B, Perna NT. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLOS ONE 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notredame C, Higgins D, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–17 [DOI] [PubMed] [Google Scholar]
- 34.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, et al. 2017. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46:D762–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pevzner PA, Tang H, Tesler G. 2004. De novo repeat classification and fragment assembly. Genome Res. 14:1786–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubchak I, Poliakov A, Kislyuk A, Brudno M. 2009. Multiple whole-genome alignments without a reference organism. Genome Res. 19:682–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, et al. 2008. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 18:1829–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, et al. 2016. The Ensembl gene annotation system. Database 2016:baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paten B, Diekhans M, Earl D, John JS, Ma J, et al. 2011. Cactus graphs for genome comparisons. J. Comput. Biol. 18:469–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paten B, Earl D, Nguyen N, Diekhans M, Zerbino D, Haussler D. 2011. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 21:1512–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earl D, Nguyen N, Hickey G, Harris RS, Fitzgerald S, et al. 2014. Alignathon: a competitive assessment of whole-genome alignment methods. Genome Res. 24:2077–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen N, Hickey G, Zerbino DR, Raney B, Earl D, et al. 2015. Building a pan-genome reference for a population. J. Comput. Biol. 22:387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hickey G, Paten B, Earl D, Zerbino D, Haussler D. 2013. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29:1341–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen N, Hickey G, Raney BJ, Armstrong J, Clawson H, et al. 2014. Comparative assembly hubs: web-accessible browsers for comparative genomics. Bioinformatics 30:3293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, et al. 2012. GENCODE: the reference human genome annotation for the encode project. Genome Res. 22:1760–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, et al. 2013. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152:642–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ENCODE Proj. Consort. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:636–40 [DOI] [PubMed] [Google Scholar]
- 48.Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev. 25:1010–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29:308–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.1000 Genomes Proj. Consort. 2010. A map of human genome variation from population-scale sequencing. Nature 467:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letovsky SI, Cottingham RW, Porter CJ, Li PW. 1998. GDB: The Human Genome Database. Nucleic Acids Res. 26:94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulp D, Haussler D, Reese MG, Eeckman FH. 1996. A generalized hidden Markov model for the recognition of human genes in DNA. Proc. Int. Conf. Intelligent Syst. Mol. Biol. 4:134–42 [PubMed] [Google Scholar]
- 54.Pruitt KD, Tatusova T, Maglott DR. 2006. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantarel BL, Korf I, Robb SM, Parra G, Ross E, et al. 2008. Maker: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon D, Huddleston J, Chaisson MJ, Hill CM, Kronenberg ZN, et al. 2016. Long-read sequence assembly of the gorilla genome. Science 352:aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weisenfeld NI, Kumar V, Shah P, Church D, Jaffe DB. 2016. Direct determination of diploid genome sequences. Genome Res. 27(5):757–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haussler D, O’Brien SJ, Ryder OA, Barker FK, Clamp M, et al. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J. Hered. 100:659–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterston RH, Pachter L. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–62 [DOI] [PubMed] [Google Scholar]
- 60.Flicek P, Keibler E, Hu P, Korf I, Brent MR. 2003. Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map. Genome Res. 13:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiehe T, Gebauer-Jung S, Mitchell-Olds T, Guigó R. 2001. SGP-1: prediction and validation of homologous genes based on sequence alignments. Genome Res. 11:1574–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexandersson M, Cawley S, Pachter L. 2003. SLAM: cross-species gene finding and alignment with a generalized pair hidden Markov model. Genome Res. 13:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeh RF, Lim LP, Burge CB. 2001. Computational inference of homologous gene structures in the human genome. Genome Res. 11:803–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gelfand MS, Mironov AA, Pevzner PA. 1996. Gene recognition via spliced sequence alignment. PNAS 93:9061–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross SS, Brent MR. 2006. Using multiple alignments to improve gene prediction. J. Comput. Biol. 13:379–93 [DOI] [PubMed] [Google Scholar]
- 66.van Baren MJ, Koebbe BC, Brent MR. 2007. Using N-SCAN or TWINSCAN to predict gene structures in genomic DNA sequences. Curr. Protoc. Bioinform. 20:4.8.1–4.8.16 [DOI] [PubMed] [Google Scholar]
- 67.Flicek P 2007. Gene prediction: compare and CONTRAST. Genome Biol. 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gross SS, Do CB, Sirota M, Batzoglou S. 2007. CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction. Genome Biol. 8:R269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lafferty J, McCallum A, Pereira FC. 2001. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Dep. Pap., Dep. Comput. Inf. Sci., Univ. Pa., Philadelphia [Google Scholar]
- 70.Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–44 [DOI] [PubMed] [Google Scholar]
- 71.Hoff K, Stanke M. 2015. Current methods for automated annotation of protein-coding genes. Curr. Opin. Insect Sci. 7:8–14 [DOI] [PubMed] [Google Scholar]
- 72.Mamm. Gene Collect. Progr. Team. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. PNAS 99:16899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. 2000. GenBank. Nucleic Acids Res. 28:15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei C, Brent MR. 2006. Using ESTs to improve the accuracy of de novo gene prediction. BMC Bioinform. 7:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Birney E, Clamp M, Durbin R. 2004. GeneWise and genomewise. Genome Res. 14:988–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. Augustus: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32:W309–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yandell M, Ence D. 2012. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13:329–42 [DOI] [PubMed] [Google Scholar]
- 79.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–48 [DOI] [PubMed] [Google Scholar]
- 80.Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27:757–63 [DOI] [PubMed] [Google Scholar]
- 81.Consort GTEx. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, et al. 2011. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. mBio 2:e00198–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fiddes IT, Armstrong J, Diekhans M, Nachtweide S, Kronenberg ZN, et al. 2018. Comparative Annotation Toolkit (CAT)—simultaneous clade and personal genome annotation. Genome Res. 28:1029–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma V, Elghafari A, Hiller M. 2016. Coding exon-structure aware realigner (CESAR) utilizes genome alignments for accurate comparative gene annotation. Nucleic Acids Res. 44:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meyer IM, Durbin R. 2004. Gene structure conservation aids similarity based gene prediction. Nucleic Acids Res. 32:776–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Florea L, Di Francesco V, Miller J, Turner R, Yao A, et al. 2005. Gene and alternative splicing annotation with AIR. Genome Res. 15:54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D. 2007. Comparative genomics search for losses of long-established genes on the human lineage. PLOS Comput. Biol. 3:e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma V, Schwede P, Hiller M. 2017. CESAR 2.0 substantially improves speed and accuracy of comparative gene annotation. Bioinformatics 33:3985–87 [DOI] [PubMed] [Google Scholar]
- 89.König S, Romoth L, Gerischer L, Stanke M. 2016. Simultaneous gene finding in multiple genomes. Bioinformatics 32:3388–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lilue J, Doran AG, Fiddes IT, Abrudan M, Armstrong J, et al. 2018. Multiple laboratory mouse reference genomes define strain specific haplotypes and novel functional loci. bioRxiv 235838 10.1101/235838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sosinsky A, Glusman G, Lancet D. 2000. The genomic structure of human olfactory receptor genes. Genomics 70:49–61 [DOI] [PubMed] [Google Scholar]
- 92.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19:327–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rivas E, Eddy SR. 2001. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinform. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Pérez-Lluch S, Abad A, et al. 2017. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 49:1731–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diederichs S 2014. The four dimensions of noncoding RNA conservation. Trends Genet. 30:121–23 [DOI] [PubMed] [Google Scholar]
- 96.Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27:i275–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517:608–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jain M, Koren S, Miga KH, Quick J, Rand AC, et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36:338–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Korlach J, Gedman G, Kingan SB, Chin CS, Howard JT, et al. 2017. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience 6:gix085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koepfli KP, Paten B, Genome 10K Community Sci., O’Brien SJ. 2015. The Genome 10K project: a way forward. Annu. Rev. Anim. Biosci. 3:57–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, et al. 2011. Creating a buzz about insect genomes. Science 331:1386. [DOI] [PubMed] [Google Scholar]
- 103.Stamatakis A 2014. RaxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mirarab S, Bayzid MS, Boussau B, Warnow T. 2014. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 346:1250463. [DOI] [PubMed] [Google Scholar]
- 105.Chauve C, El-Mabrouk N, Guéguen L, Semeria M, Tannier E. 2013. Duplication, rearrangement and reconciliation: a follow-up 13 years later In Models and Algorithms for Genome Evolution, Vol. 19, ed. Chauve C, El-Mabrouk N, Tannier E, pp. 47–62. London: Springer [Google Scholar]
- 106.Dewey CN. 2011. Positional orthology: putting genomic evolutionary relationships into context. Brief. Bioinform. 12:401–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paten B, Novak AM, Eizenga JM, Garrison E. 2017. Genome graphs and the evolution of genome inference. Genome Res. 27:665–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marschall T, Marz M, Abeel T, Dijkstra L, Dutilh BE, et al. 2018. Computational pan-genomics: status, promises and challenges. Brief. Bioinform. 19:118–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bray N, Pimentel H, Melsted P, Pachter L. 2015. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–27 [DOI] [PubMed] [Google Scholar]
- 110.Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. 2000. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 10:950–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pedersen JS, Hein J. 2003. Gene finding with a hidden Markov model of genome structure and evolution. Bioinformatics 19:219–27 [DOI] [PubMed] [Google Scholar]
- 112.Siepel A, Haussler D. 2004. Computational identification of evolutionarily conserved exons. In Proceedings of the Eighth Annual International Conference on Research in Computational Molecular Biology, ed.Gusfield D, Bourne P, Istrail S, Pevzner P, Waterman M, pp. 177–86. New York: Assoc. Comput. Mach. [Google Scholar]
- 113.Carter D, Durbin R. 2006. Vertebrate gene finding from multiple-species alignments using a two-level strategy. Genome Biol. 7(Suppl. 1):S6.1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8:1494–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Delcher AL, Kasif S, Fleischmann RD, Peterson J, White O, Salzberg SL. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meyer IMM, Durbin R. 2002. Comparative ab initio prediction of gene structures using pair HMMs. Bioinformatics 18:1309–18 [DOI] [PubMed] [Google Scholar]