Abstract
The Hippo pathway controls organ size and tissue homeostasis, and its dysregulation often contributes to tumorigenesis. Extensive studies have shown that the Hippo pathway inhibits cell proliferation, and survival in a cell-autonomous manner. We examined the function of the Hippo pathway kinases LATS1/2 (large tumor suppressor 1 and 2) in cancer cells. As expected, loss of LATS1/2 promotes cancer cell growth in most cell lines. Surprisingly, however, LATS1/2 deletion inhibits the growth of murine MC38 colon cancer cells, especially under detachment conditions. This growth inhibitory effect caused by LATS1/2 deletion is due to uncontrolled activation of Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ), the key downstream transcriptional coactivators inhibited by LATS1/2. We identified Wnt inducible signaling pathway protein 2 (Wisp2) and coiled-coil domain containing 80 (Ccdc80) as direct targets of YAP/TAZ. Their expression is selectively induced by LATS1/2 deletion in MC38 cells. Furthermore, deletion of WISP2 and CCDC80 prevents the growth inhibitory effect of LATS1/2 loss in MC38 cells. Our study demonstrates that the function of LATS1/2 in cell growth is cell context dependent, suggesting that LATS1/2 inhibition can be a therapeutic approach for some cancer types.
Keywords: Hippo, LATS, YAP, WISP2, MC38
Introduction
The Hippo pathway was initially identified through genetic mosaic screenings in Drosophila melanogaster as a key contributor of organ size determination1. Core components of the Hippo pathway are highly conserved in evolution and the mammalian Hippo pathway also restricts tissue growth by controlling cell proliferation and apoptosis2–4. Aberrant expression or dysregulation of the Hippo pathway has been linked to human cancers5–7. Upstream kinases of the mammalian Hippo pathway include mammalian STE20-like protein kinase 1/2 (MST1 and MST2),mitogen-activation protein kinase kinase kinase kinase (MAP4Ks) family members, and large tumor suppressor 1/2 (LATS1/2). Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ; also known as WWTR1) are key effectors of the Hippo signaling pathway8. YAP/TAZ are mainly inhibited by LATS1/2-dependent phosphorylation, which leads to cytoplasmic localization and ubiquitin-mediated degradation9, 10. When activated, the unphosphorylated YAP/TAZ translocate into the nucleus and bind to the TEA domain (TEAD) family of transcription factors (TEAD1–4) to induce expression of genes that control many different biological processes11–14.
Previous studies have convincingly established that the Hippo pathway acts as a tumor suppressor by inhibiting cell proliferation and differentiation, as well as stimulating cell death5. Many studies in mouse models show that loss of Hippo signaling or overexpression of YAP is sufficient to promote tumor formation15. For example, MST1/2 deficiency in the liver results in hepatocellular carcinoma and YAP overexpression induces liver tumors9, 16–20. Loss-of function mutations of the Hippo pathway upstream component NF2 contributes to schwannoma and meningioma with high activation of YAP/TAZ21, 22. Furthermore, deletion of LATS2 is associated with malignant mesothelioma23, 24. Although most of the studies support the tumor suppressor model of the Hippo pathway, dual functions of the Hippo pathway in cancer biology has been suggested as YAP may have a tumor suppressive function in hematological cancers25, 26 and lung squamous cell carcinoma27.
In this study, we examined the effect of LATS1/2 deletion in 8 mouse cancer cell lines and found that in most cell lines LATS1/2 deletion moderately or dramatically enhanced anchorage independent cell growth. Interestingly, MC38 colon cancer cells are addicted to LATS1/2 as cell growth inversely correlates with the degree of LATS1/2 deletion. Cells with complete LATS1/2 deletion display severe growth retardation and senescent phenotypes. The phenotypes of LATS1/2 deletion in MC38 cells are exacerbated under detachment conditions. Our study demonstrates cell type dependent functions of LATS1/2 in controlling cancer cell growth and the underlying mechanism.
Results
LATS1/2 deletion shows cell type-dependent effect in cell growth.
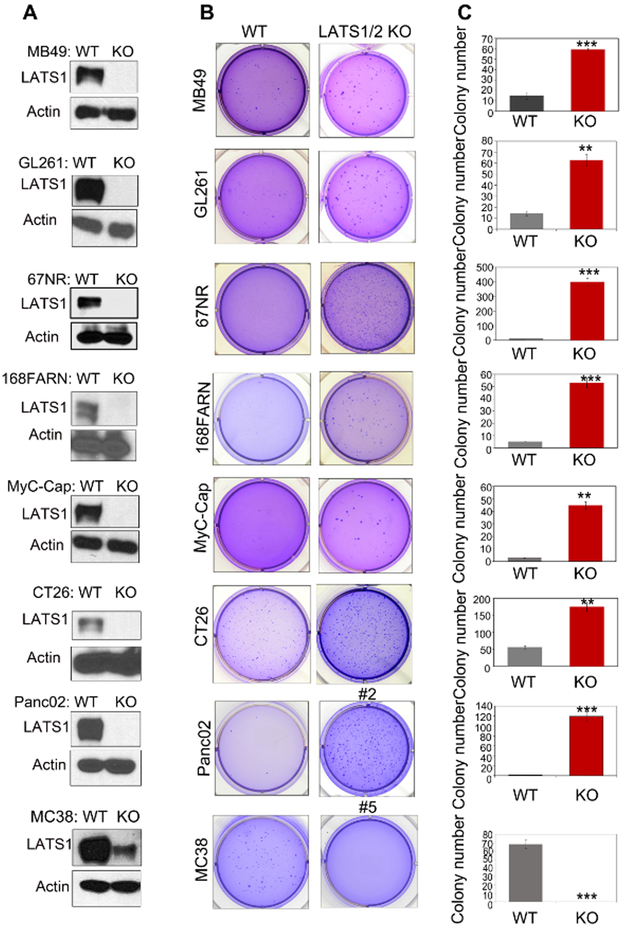
We had previously observed that deletion of LATS1/2 in mouse cancer cell lines B16, SCC7, and 4T1 enhanced immunogenicity of the cancer cells, leading to a tumor growth suppression of the LATS1/2 knockout (KO) cells in syngeneic immune competent mice28. In an effort to expand this discovery, we deleted LATS1/2 in many mouse cancer cell lines, including MC38 and CT26 colon cancer cells, Panc02 pancreatic cancer cells, MB49 bladder cancer cells, GL261 glioma cancer cells, MyC-CaP prostate cancer cells, and 168FARN and 67NR breast cancer cells, using CRISPR/Cas9 technology29. LATS1/2 KO cells were readily generated with the exception of MC38. By evaluating DNA sequence and protein expression of LATS1/2, we successfully obtained complete LATS1/2 KO clones for each of the above mentioned cell lines (Fig. 1a). It is noteworthy that it was very challenging to obtain complete LATS1/2 KO clone in MC38 cells. We analyzed more than 100 clones and obtained many partial LATS1/2 deletion clones, but only had a few LATS1/2 complete KO cells (Supplementary Figs. 1a and b), which grew extremely poorly (see data in Fig. 2). These observations suggest a very surprising and interesting possibility that LATS1/2 are required for growth and/or survival of MC38 cells.
Fig1. Deletion of LATS1/2 enhances anchorage-independent growth of cancer cells with the exception of MC38.
a: LATS1/2 deleted cells were subjected to immunoblot analysis with antibodies to LATS1 and Actin.
b: LATS1/2 deletion promotes anchorage-independent growth of MB49, GL261, 67NR, 168FARN, MyC-CaP, CT26 and Panc02 cells, but inhibits MC38 (clone #5) in vitro. Soft-agar colony-formation assay was performed and the colonies were stained with crystal violet for quantification.
c: Quantification of the results presented in B. Data are mean±SD from three independent experiments. Student’s t-test was applied. ** denotes p<0.01, *** denotes p<0.001.
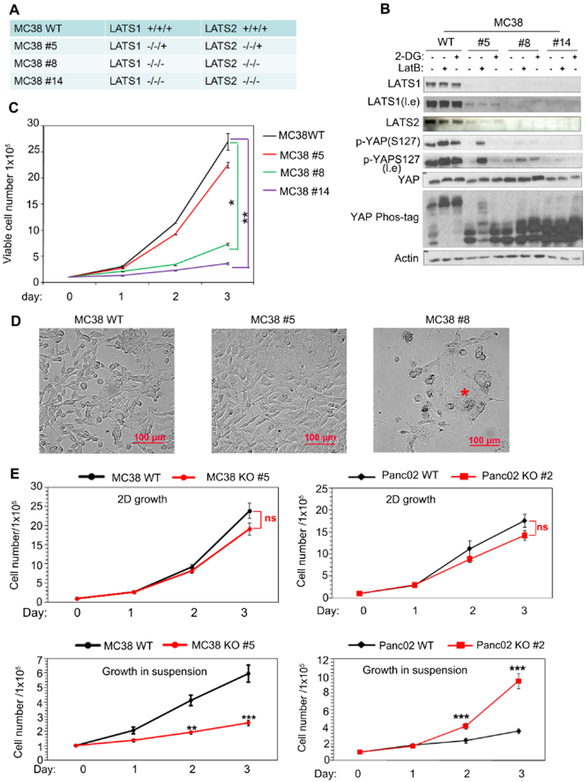
Fig 2. LATS1/2 deletion has opposite effects on cell growth in different cell lines.
a: Genomic sequencing confirms deletion of LATS1/2 in MC38 clones.
b: Immunoblot showing various degrees of LATS1/2 deletion in MC38 cells. Three independent clones (#5, #8 and #14) are shown. Cells were treated with 2-DG (2-deoxyglucose) or LatB (latrunculin B) to induce YAP phosphorylation, which was detected by a phosphor-YAP antibody as well as a phos-tag gel. l.e. denotes longer exposure.
c: LATS1/2 deletion inhibits MC38 cell growth in a dosage-dependent manner. MC38 WT and LATS1/2 deletion cells (1×105) were plated in 6-well culture dishes and viable cells were counted after trypan blue staining. Three replicates were included in this experiment. The error bars represent s.d. *p<0.05, ** p<0.01; two-way ANOVA test.
d: LATS1/2 deletion causes morphological changes in MC38 cells. * marks enlarged senescent cells. Scale bar, 100 μm.
e: LATS1/2 deletion has opposite effects in MC38 and Panc02 cells in 2D (attachment) and suspension (detachment) conditions. Cells (1×105) were plated in 6-well culture dishes and cell number was determined with trypan blue staining. Three replicates were included. The error bars represent s.d. ns (p>0.05); *** p<0.001. Student’s t-test was applied.
LATS1/2 are well known as tumor suppressors. We examined anchorage independent growth of WT and LATS1/2 KO cells by a soft agar assay. LATS1/2 deletion strongly increased anchorage independent growth in pancreatic cancer Panc02, prostate cancer MyC-CaP, breast cancer 168FARN and 67NR cells. LATS1/2 deletion also increased growth of colon cancer CT26, glioma GL261, and bladder cancer MB49 (Figs. 1b and c). These results are consistent with the tumor suppressor function of LATS1/2. In contrast, partial deletion of LATS1/2 in MC38 clone #5, which could grow under normal two-dimensional (2D) culture, strongly inhibited anchorage independent cell growth. This result contradicts the current notion of LATS1/2 as tumor suppressors, and indicates that LATS1/2 show a cell type dependent effect in cell growth.
We tested whether reducing LATS1/2 expression levels could inhibit MC38 cell growth under regular cell culture conditions. MC38 has three alleles of both LATS1 and LATS2. We compared the LATS1/2 partial deletion clone #5 and complete deletion clone #8 and #14 (Fig. 2a). Either energy starvation by 2-DG (2-deoxyglucose) or disruption of F-actin by LatB (Latrunculin B) induced YAP phosphorylation in wild type MC38 as determined by both phospho-YAP antibody and phos-tag gel (Fig. 2b). These observations suggest that the Hippo pathway is operational in MC38 cells. Partial deletion of LATS1/2 (clone #5) showed a significant reduction in YAP phosphorylation while complete LATS1/2 deletion (clone #8 and 14) abolished YAP phosphorylation upon treatment with either 2-DG or LatB (Fig. 2b). We also found that LATS1/2 were pivotal for MC38 cell growth as complete LATS1/2 deletion dramatically reduced cell growth (Fig. 2c). Microscopic examinations showed that MC38 clone #8 cells were larger in size with flattened morphology (Fig. 2d), reminiscent of cellular senescence. These results indicate that LATS1/2 have a positive role for MC38 cell growth in a gene dosage dependent manner.
We compared the cell growth of WT and LATS1/2 KO cells under 2D conditions as well as under anchorage-independent conditions (in cell suspension). Because LATS1/2 complete KO MC38 cells grow extremely poorly and are hard to manipulate, we focused on the LATS1/2 partial deletion clone #5. Both LATS1/2 KO Panc02 cells and MC38 clone #5 showed cell growth similar to the corresponding WT cells on a 2D surface. Notably, Panc02 LATS1/2 KO cells showed a significantly stronger growth potential than WT cells in suspension (Fig. 2e). In contrast, LATS1/2 partial deletion inhibited MC38 clone #5 growth in soft agar (Fig. 1b) and in suspension (Fig. 2e). Together, the above observations indicate that LATS1/2 deficiency has different effects on cell growth in different cancer cell types and this effect is exacerbated by detachment. Therefore, the outcome of LATS1/2 deletion is strongly influenced by culture conditions, such as anchorage.
YAP/TAZ regulation by LATS1/2 is comparable between MC38 and Panc02 cells.
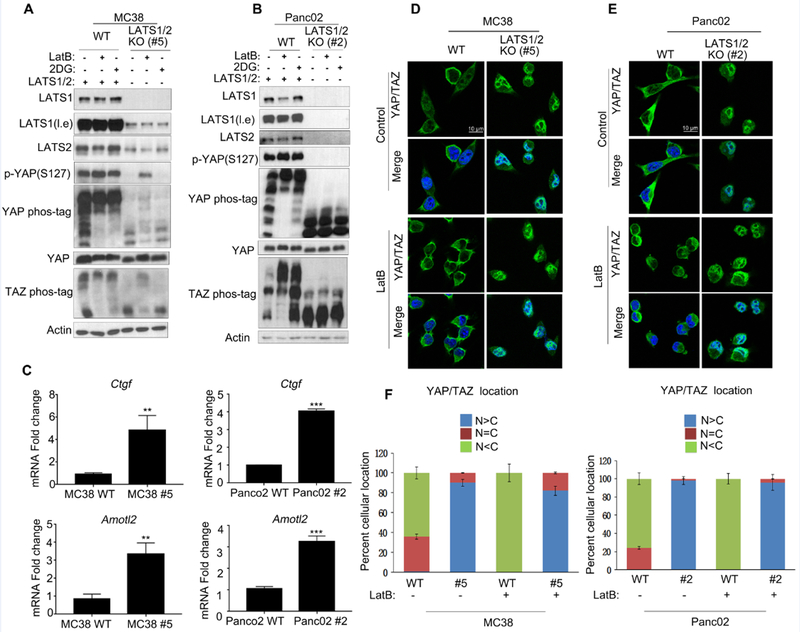
YAP/TAZ are the major downstream effectors of the Hippo pathway. LATS1/2 directly phosphorylate YAP/TAZ on multiple serine residues, resulting in YAP/TAZ cytoplasmic localization and inactivation. To test whether YAP/TAZ are similarly regulated by LATS1/2 in MC38 and Panc02 cells, we examined YAP/TAZ phosphorylation with a phospho-YAP antibody or by mobility shift on a phos-tag gel. We found that YAP/TAZ phosphorylation was induced by energy stress with 2-DG or actin depolymerization with LatB (Fig. 3a and b) in both WT MC38 and Panc02 cells. LATS1/2 deletion abolished YAP/TAZ phosphorylation in Panc02 while partial LATS1/2 deletion in MC38 clone #5 reduced YAP/TAZ phosphorylation (Figs. 3a and b). Consistent with the functional importance of YAP/TAZ phosphorylation, LATS1/2 deletion also increased the expression of YAP/TAZ target genes Ctgf and Amotl2 in both MC38 and Panc02 cells (Fig. 3c). We further observed that LatB treatment induced YAP/TAZ cytoplasmic localization in WT MC38 and Panc02 cells, but not the LATS1/2 deletion cells (Figs. 3d, e and f). Therefore, LATS1/2 similarly regulate YAP/TAZ phosphorylation, subcellular location, and transcriptional activity in both MC38 and Panc02.
Fig 3. LATS1/2 inhibit YAP/TAZ function in both MC38 and Panc02 cells.
a: Partial deletion of LATS1/2 in MC38 diminishes YAP/TAZ phosphorylation in response to actin polymerization inhibitor LatB and energy stress by 2-DG treatment. Wild-type and LATS1/2 partial knockout MC38 clone #5 were seeded on six-well plates and treated with LatB and 2-DG for 1 hour. Cells were collected for determination of YAP/TAZ phosphorylation. l.e., long exposure of films.
b: LATS1/2 deletion abolishes YAP/TAZ phosphorylation in Panc02 cells. Experiments are similar to panel A.
c: LATS1/2 deletion increases the expression of YAP/TAZ target genes Ctgf and Amotl2. mRNA levels of Ctgf and Amotl2 expression were measured by quantitative real-time PCR. Results were from triplicated experiments. The error bars represent s.d. ** p<0.01; *** p<0.001. Student’s t-test was applied.
d-e: LATS1/2 deletion induces constitutive nuclear YAP/TAZ localization. LatB-treated or non-treated MC38 or Panc02 cells were subjected to immunostaining with the YAP/TAZ antibody (green) along with DAPI for DNA (blue). Scale bar, 10 μm.
f: Quantification of data from panels D-E. Ten randomly chosen fields for each treatment were examined. Typically, 100 cells were counted in each field. N and C denote nuclear and cytoplasmic.
Uncontrolled YAP/TAZ activity mediates the growth inhibitory effect of LATS1/2 deletion in MC38 cells
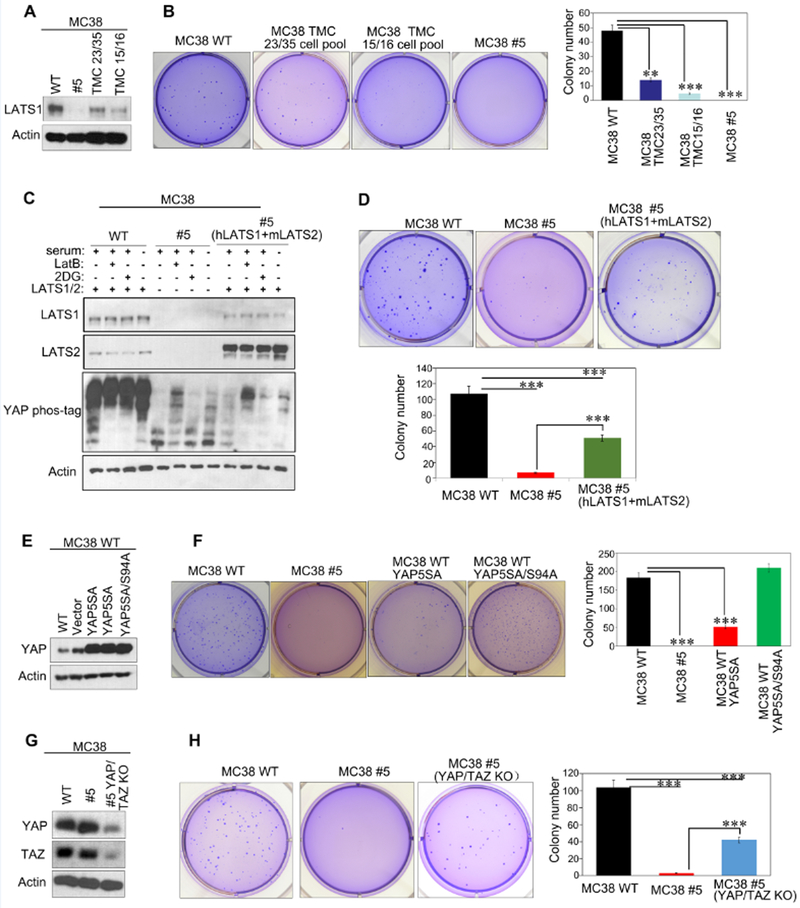
To avoid potential off-target effect, we generated MC38 LATS1/2 knockout cell pools using two independent CRISPR guide sequences (TMC15/16 and TMC23/35) (Fig. 4a). We found that LATS1/2 deficient cell pools decreased colony formation in soft agar. The LATS1/2 knockout efficiency correlated with the reduction in colony formation (Fig. 4b). Next, we re-expressed human LATS1 and mouse LATS2 in MC38 clone #5. Western blotting results showed that LATS1/2 re-expression significantly restored YAP phosphorylation in response to 2-DG treatment or serum starvation (Fig. 4c). Importantly, re-expression of LATS1/2 partially but significantly rescued colony formation of the MC38 clone #5 in soft agar (Fig. 4d). These results further support that LATS1/2 deletion inhibits MC38 cell growth.
Fig 4. YAP/TAZ mediate the growth inhibitory effect of LATS1/2 deletion in MC38.
a: Immunoblots showing CRISPR-mediated deletion of LATS1/2 in MC38 cells. MC38 WT, cell pools generated by two independent CRISPR guide sequences (TMC23/35 and TMC15/16) and cell clone #5 were subjected to immunoblot analysis.
b: LATS1/2 deletion decreases anchorage-independent growth of MC38. Cells described in panel A were subjected to soft-agar colony-formation assay, and the colonies were stained with crystal violet for quantification. Data are mean±SD from 3 independent experiments. **p<0.01; *** p<0.001; one-way ANOVA test.
c: Re-expression of LATS1/2 rescues YAP phosphorylation in MC38 clone #5. MC38 #5 cells infected with retroviruses encoding human LATS1 and mouse LATS2 or with control empty retrovirus were treated with serum starvation, LatB or 2-DG, and then analyzed for YAP phosphorylation by immunoblot.
d: LATS1/2 re-expression restores anchorage independent growth of MC38 clone #5. Indicated cells were subjected to soft-agar colony-formation assay, and the colonies were stained with crystal violet for quantification. Data are mean±SD from 3 independent experiments. *** p<0.001; one-way ANOVA test.
e: Expression of YAP5SA or YAP5SA/S94A in MC38 WT cells. MC38 WT cells stably expressing control vector, YAP5SA or YAP5SA/S94A were subjected to immunoblot with indicated antibodies.
f: Expression of the active mutant YAP5SA inhibits anchorage-independent growth of MC38 cells. Soft-agar colony-formation assay of cells in panel E was performed and the colonies were stained with crystal violet for quantification. Date are mean±SD from 3 independent experiments. *** p<0.001; one-way ANOVA test.
g: CRISPR-mediated deletion of YAP/TAZ in MC38 clone #5. Cell lysates (MC38 WT, clone #5, and cell pool of YAP/TAZ deletion in MC38 clone #5) were subjected to immunoblot analysis to show the reduction of YAP/TAZ proteins.
h: YAP/TAZ deletion increases anchorage-independent growth of MC38 clone #5. Indicated cells were subjected to soft-agar colony-formation assay, and the colonies were stained with crystal violet for quantification. Date are mean±SD from 3 independent experiments. *** p<0.001; one-way ANOVA test.
YAP/TAZ are the major physiological targets of LATS1/2 and activation of YAP/TAZ is generally believed to promote cell growth and tumorigenesis. Our observation suggests two possibilities: the function of YAP/TAZ in MC38 is growth inhibitory; or other LATS1/2 downstream targets other than YAP/TAZ may mediate the cell growth inhibition in MC38 cells. To this end, we generated MC38 cells stably expressing YAP5SA (a constitutively active mutant that is not inhibited by LATS1/2) or YAP5SA/S94A (this is a functionally inactive mutant of YAP5SA because it cannot bind to the downstream transcription factor TEAD)14 (Fig. 4e and Supplementary Fig. 1c). Overexpression of YAP5SA, but not the inactive YAP5SA/S94A, inhibited cell growth of MC38 cells under soft agar, 2D surface, and suspension conditions (Fig. 4f and Supplementary Fig. 1d). These data suggest that MC38 cells are rather unique, in which active YAP has growth inhibitory effect. Next, we investigated the functional significance of YAP/TAZ in mediating the growth inhibitory effects caused by LATS1/2 knockout. We generated YAP/TAZ knockout pool of MC38 clone #5. The reduction of YAP/TAZ proteins was confirmed by Western blotting (Fig. 4g). As expected, expressions of YAP target genes Ctgf and Amotl2 were reduced by YAP/TAZ deletion in MC38 clone #5 (Supplementary Fig. 1e). Importantly, we observed that deletion of YAP/TAZ in MC38 clone #5 increased colony formation (Fig. 4h). Together, our data support a model that high YAP/TAZ activity is responsible for the growth suppressing effect caused by LATS1/2 knockout in MC38 cells.
LATS1/2 deletion in MC38 cells suppresses proliferation and promotes apoptosis in suspension culture
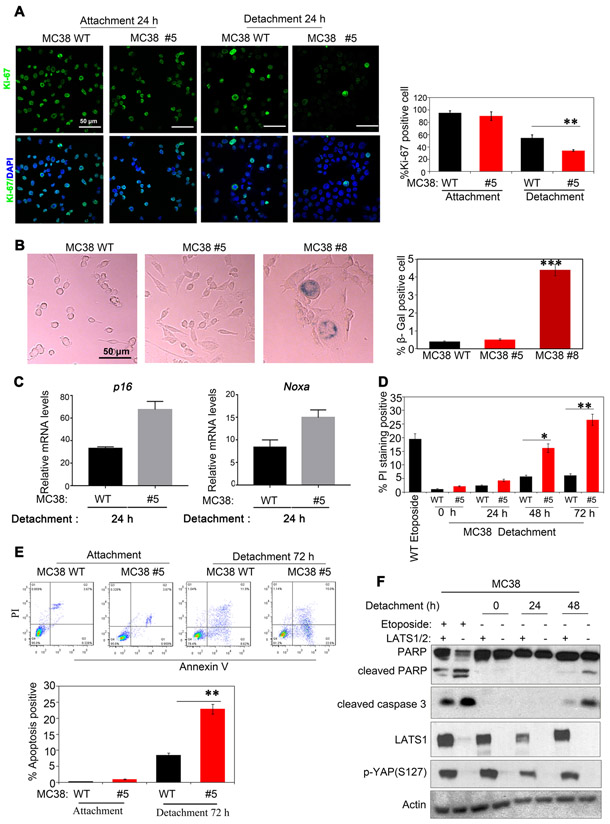
To uncover the mechanism of growth inhibition by LATS1/2 loss in MC38 cells, we examined MC38 clone #5 on 2D surfaces and in cell suspension. We observed that LATS1/2 deficiency significantly reduced proliferation of MC38 cells in suspension culture as determined by Ki-67 staining (Fig. 5a), consistent with the results in Figure 2. However, more complete LATS1/2 deletion, as in clone #8, severely retarded cell growth even under attachment and cells showed an enlarged morphology, similar to senescent cells. Staining with the senescence marker SA-β-gal revealed that clone #8 cells are positive for SA-β-gal activity (Fig. 5b). These data indicate that LATS1/2 deletion results in cellular senescence in MC38 cells. p53 and p16/pRB axes are major senescence-triggering pathways 30. We found that p16 and the p53 target gene Noxa were increased in MC38 clone #5 (Fig. 5c), suggesting that knockout of LATS1/2 induces cellular senescence in MC38 cells.
Fig 5. LATS1/2 deletion in MC38 represses cell proliferation and promotes cell apoptosis in suspension culture.
a: Cell proliferation is reduced by LATS1/2 deletion. Cells harvested from attachment or detachment conditions were subjected to immunostaining with Ki-67 for proliferation (green) and DAPI for DNA (blue). ** p<0.01; one-way ANOVA test. Quantification is shown in the right panel.
b: Loss of LATS1/2 promotes senescence in MC38 cells. MC38 WT, LATS1/2 partial deletion clone #5, and complete LATS1/2 deletion clone #8 were cultured and stained for the senescence marker SA-β-gal. Quantification is shown in the right panel. Data are mean±SD from 3 independent experiments. *** p<0.001; one-way ANOVA test.
c: LATS1/2 deletion increases senescence related gene expression in MC38 cells. Noxa and p16 mRNA expressions were determined by Q-PCR.
d: LATS1/2 deletion increases cell death. MC38 WT and clone #5 were cultured in suspension for the indicated times. PI (propidium iodine) staining followed by FACS detects cell death. Etoposide treatment overnight (50 μM) was included as a positive control. Data are mean±SD from 3 independent experiments. * p<0.05; ** p<0.01. one-way ANOVA test.
e: LATS1/2 deletion promotes apoptosis. MC38 WT and clone #5 were cultured for 72 h in suspension. 0.5 × 105 cells were stained with PE Annexin V and analyzed by FACS within 1 h. Data are mean±SD from 3 independent experiments. ** p<0.01, one-way ANOVA test.
f: LATS1/2 deletion increased apoptosis markers of cleaved caspase 3 and PARP. MC38 WT and LATS1/2 deletion clone #5 were cultured in suspension for 24 h or 48 h. Etoposide treatment overnight (50 μM) was used as a positive control. Western blot was performed with indicated antibodies.
The effect of LATS1/2 on cell death was also determined using PI staining. A significant increase of PI positive cells was found in MC38 clone #5 cultured in suspension (Fig. 5d). Furthermore, we examined apoptosis using PE-Annexin V apoptosis kit (Fig.5e) as well as Western blotting for apoptosis markers, cleaved PARP and cleaved caspase 3 (Fig. 5f). Apoptosis was increased in MC38 clone #5 upon suspension culture (Figs. 5e and 5f). Taken together, we propose that LATS1/2 deletion in MC38 cells represses cell proliferation as well as increases senescence and apoptosis, especially under detachment condition.
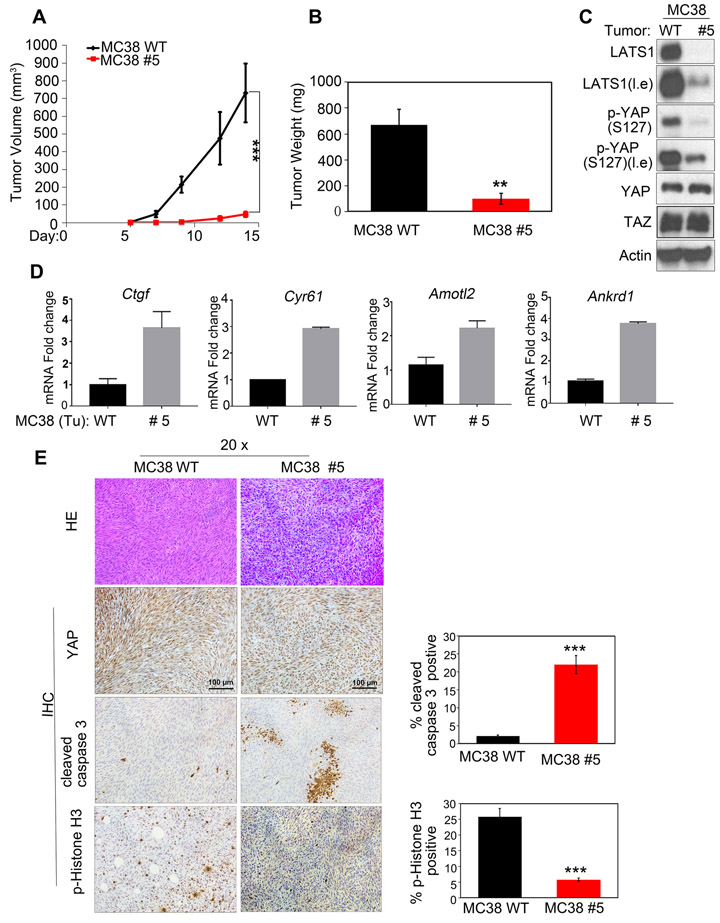
LATS1/2-deficiency inhibits tumor growth in vivo.
To evaluate the role of LATS1/2 in MC38 tumor growth, we performed xenograft experiments in nude mice. LATS1/2 deficient cells grew tumors slower than the control MC38 WT group (Figs. 6a and b). Western blotting analysis confirmed the reduction of LATS1 protein and YAP S127 phosphorylation in tumors from the MC38 clone #5 (Fig. 6c). Consistently, expression of YAP target genes (Ctgf, Cyr61, Amotl2, and Ankrd1) was elevated in the LATS1/2 deficient tumor samples (Fig. 6d). In addition, we found an increase of cleaved caspase 3, a marker for apoptosis, and a decrease of phospho-histone H3, a marker for proliferation, in LATS1/2 deficient tumors (Fig. 6e). Collectively, our results show that LATS1/2 deletion inhibits MC38 cell growth in vitro and in vivo.
Fig 6. Loss of LATS1/2 inhibits MC38 tumor growth in xenografted mice.
a-b: Deletion of LATS1/2 in MC38 cells inhibits tumor growth in vivo. MC38 WT and LATS1/2 deletion clone #5 (1×106 cells for each injection) were implanted subcutaneously into nude mice. Tumor volume was monitored and tumor weight determined at the end of the experiments. ** p<0.01, *** p<0.001. The p value of tumor volume was determined using two-way ANOVA test. The p value of tumor weight was Student’s t-test.
c: YAP phosphorylation is decreased in the LATS1/2 deleted tumors. Tumor samples (panel B) were harvested and then subjected to immunoblot analysis with indicated antibodies.
d: Increased expression of YAP/TAZ target genes in the LATS1/2 deleted tumors. RNA was isolated from tumor samples (panel B) and analyzed by RT-PCR for YAP/TAZ target genes. Normalized data are expressed relative to the corresponding value for WT tumors and are mean±SD; n=3 tumors for each group.
e: Loss of LATS1/2 decreases cell proliferation and increases apoptosis in MC38 tumors. Immunohistochemistry staining for YAP, cleaved caspase3, and p-Histone H3 in representative tumor tissue. Scale bar, 100 µm. Data are mean±SD from 5 independent fields. *** p<0.001; Student’s t-test was applied.
The YAP/TEAD target genes WISP2 and CCDC80 function downstream of LATS1/2 in MC38 cell growth regulation.
It has recently been reported that YAP hyperactivation induces reactive oxygen species (ROS) accumulation and suppresses lung squamous cell carcinoma growth 27. This led us to investigate if LATS1/2 deletion increased ROS levels in MC38 cells. We found that ROS levels were not significantly elevated in MC38 clone #5 (Supplementary Figs. 2a and b). These results suggest that ROS is unlikely to be the key factor responsible for the growth inhibition in MC38 clone #5.
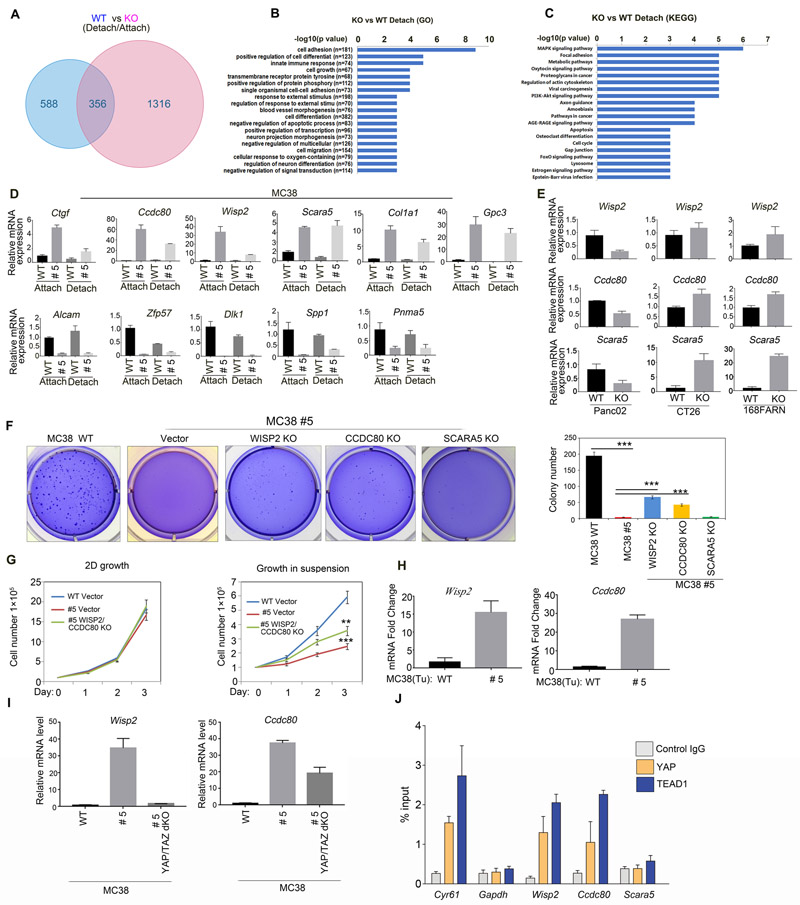
Activation of the transcription co-activators YAP/TAZ is the best-known consequence of LATS1/2 deletion. To further explore the mechanism by which LATS1/2 regulate cell growth in MC38, we performed a genome-wide RNA-seq analysis of MC38 WT and LATS1/2 deleted clone #5 under attachment and detachment conditions. Comparison of cells in attachment and detachment culture conditions revealed 1672 genes (fold change ≥2; p value< 0.05) were changed in MC38 clone #5 and, 944 genes were changed in WT cells. 356 of these genes were common to both groups (Fig. 7a). Interestingly, MC38 clone #5 showed a more dramatic transcription alteration in response to detachment. Pathway analysis showed that the genes altered by detachment may play roles in cell growth, apoptosis and cancer development. These genes are known to be involved in cell adhesion, differentiation, growth, MAPK and PI3K-AKT signaling pathways (Figs. 7b and c, Supplementary Figs. 2c and d). We confirmed the altered expression of selected genes, including the upregulated Ctgf, Wisp2, Ccdc80, Scara5, Col1a1, Gpc3 and the downregulated Alcam, Zfp57, Spp1, Pnma5 and Dlk1 (Fig. 7d).
Fig 7. The YAP target genes Wisp2 and Ccdc80 contribute to growth inhibition by LATS1/2 deletion in MC38 cells.
a: Genome-wide RNA-seq analysis of MC38 WT and clone #5. Genes changed under between detachment and attachment conditions.
b: Top 20 enriched gene Ontology (GO) terms. KO vs WT under detachment.
c: Top 20 enriched KEGG analysis. KO vs WT under detachment. Fold>1.5, p<0.05.
d: Effects of LATS1/2 deletion on gene expression in MC38 cells. Total RNA extracted from MC38 WT and clone #5 was subjected to RT-PCR analysis for the indicated genes. Data are mean±SD from triplicated experiments.
e: Effects of LATS1/2 deletion on expression of Wisp2, Ccdc80 and Scara5 in Panc02, CT26, and 168FARN cells. Experiments were similar to panel D.
f: Deletion of WISP2, CCDC80 restores anchorage-independent growth of MC38 clone #5. Soft agar assay was performed and the colonies were stained with crystal violet for quantification, which is shown in the right panel. Data are mean±SD from 3 independent experiments. one-way ANOVA test.***p < 0.001.
g: WISP2 and CCDC80 deletion rescues cell growth defects in MC38 LATS1/2 KO cells in suspension conditions. Cells (1×105) were plated in 6-well culture dishes and cell number was determined with trypan blue staining. Three replicates were included. The error bars represent s.d. ** p<0.01; *** p<0.001. Student’s t-test was applied.
h: Increased expression of Wisp2 and Ccdc80 in the LATS1/2 deleted tumors tissue. RNA was isolated from mouse tumor samples and analyzed by RT-PCR for Wisp2 and Ccdc80 genes. Normalized data are expressed relative to the corresponding value for WT tumors and are mean±SD; n=3 tumors for each group.
i:YAP/TAZ deletion reduces the expression of Wisp2 and Ccdc80. Total RNA extracted from indicated cells were subjected to real time PCR analysis for the indicated genes. Data are means ± SD of from triplicated experiments.
j:YAP and TEAD1 bind to the promoters of Wisp2 and Ccdc80. Chromatin immunoprecipitation (ChIP) with an antibody against YAP or TEAD1 or control IgG was performed. The precipitated DNA was quantitated by real-time PCR with primers specific for putative TEAD binding elements in promoters. Gapdh was included as a negative control. Data are means ±SD of quadruplicates.
LATS1/2 deletion specifically inhibits anchorage independent growth in MC38 cells, but not in Panc02 or other cell lines (Fig.1). We hypothesized that growth-related genes that are selectively affected by LATS1/2 deletion in MC38 but not in other cell lines may contribute to the unique LATS-deletion-caused growth inhibitory effect in MC38. We found that Wisp2, Ccdc80 and Scara5 were highly expressed in MC38 clone #5, but not in Panc02 LATS1/2 KO cells. Actually, these three genes were repressed by LATS1/2 deletion in Panc02 cells (Fig. 7e). In contrast, other genes, such as Ctgf, Gpc3, Alcam and Spp1, were similarly altered by LATS1/2 deletion in both MC38 and Panc02 cells (Fig. 7d and Supplementary Fig. 2e). Furthermore, Wisp2 and Ccdc80 were only weakly altered by LATS1/2 deletion in CT26 and 168FARN cells whereas Scara5 was strongly increased (Fig. 7e). Thus, WISP2 and CCDC80 are selectively induced by LATS1/2 deletion in MC38 cells.
We examined the significance of WISP2, CCDC80 and SCARA5 in anchorage independent growth of MC38 clone #5. Three independent CRISPR guide sequences were tested for each candidate gene. We found that deletion of WISP2, CCDC80 partially restored the anchorage independent growth of MC38 clone #5 (Fig.7f). Deletion of Wisp2 produced more significant effect. In contrast, deletion of SCARA5 had little effect (Fig. 7f). We also found that deletion of WISP2 and CCDC80 in clone #5 partially, but significantly, rescued cell growth defects in suspension (Fig.7g and Supplementary Fig. 2f). Furthermore, Q-PCR results showed that Wisp2 and Ccdc80 also were up-regulated in LATS1/2 KO xenografts tumor tissue (Fig.7h). Together, our data show that elevated expressions of WISP2 and CCDC80 in MC38 clone #5 contribute to the inhibition of anchorage independent growth.
Next, we tested whether YAP/TAZ were required for WISP2 and CCDC80 induction in MC38. We found that YAP/TAZ knockout reduced the expression of both WISP2 and CCDC80 (Fig. 7i). To test whether Wisp2 and Ccdc80 are direct target genes of YAP, we performed chromatin immunoprecipitation with a YAP or TEAD antibody. PCR primers were designed to amplify the most proximal DNase-hypersensitive open chromatin region from transcription start site of each gene (primer sequences in supplemental methods). All amplified regions had putative TEAD binding motif CATTCC within 200 bps except for Gapdh, which has no putative TEAD motif in its DNase-hypersensitive region. Cyr61, a well-characterized direct target of YAP-TEAD, was included as a positive control. We observed that both YAP and TEAD bind to the putative TEAD binding elements in the promoters of Wisp2 and Ccdc80, but not Scara5 (Fig. 7j), suggesting that Wisp2 and Ccdc80 are direct targets of YAP and TEAD. The above data support a model in which LATS deletion in MC38 results in YAP/TAZ activation, induction of downstream target genes including Wisp2 and Ccdc80, and inhibition of anchorage independent growth. This growth inhibitory effect is specific to MC38 possibly because WISP2 and CCDC80 are selectively induced by LATS1/2 deletion in MC38 cells.
Discussion
In this report, we show that LATS1/2 similarly inhibit YAP/TAZ in all cancer cell lines as indicated by the increased phosphorylation and cytoplasmic localization of YAP/TAZ. However, the effect of LATS1/2 deletion on cell growth is cell type dependent. LATS1/2 deletion promotes cell growth (particularly anchorage independent growth) in most cancer cell lines, but inhibits MC38 colon cancer cell growth. Moreover, the outcome of LATS1/2 deletion is cell culture condition dependent. For example, LATS1/2 deletion has a minor effect on Pan02 under 2D culture, but strongly promotes growth both in soft agar and suspension (Fig.1 and Fig. 2e). Likewise, LATS1/2 partial deletion in MC38 inhibits anchorage independent growth, but has a minor effect on 2D culture. The more complete LATS1/2 knockout MC38 clones (such as #8 and #14) grow very slowly and appear to be senescent even under the attachment condition (Fig.2c and 2d). This indicates that LATS1/2 are pivotal for MC38 cell growth. Partial deletion of LATS1/2 in MC38 cells (#5 clone: LATS1-/−/+;LATS2-/−/+) can maintain cell growth in the attachment condition, but their growth is severely retarded in detachment condition or soft agar. The precise mechanism for why this happens is unclear. One possibility is that LATS1/2 deletion creates stress that can be exacerbated under anchorage independent conditions. This is in line with previous studies demonstrating important functions of the Hippo pathway in sensing extracellular stiffness31. In addition, re-expression of LATS1/2 as well as deletion of YAP/TAZ in MC38 LATS1/2 KO clone #5 showed only a partial rescue of anchorage independent cell growth. One possible reason for the partial rescue of cell growth in LATS1/2 re-expression is that we could not fully restore LATS1/2 functions as indicated by the partial rescue of YAP phosphorylation (Fig. 4C). One caveat of the YAP/TAZ deletion experiment is that we use YAP/TAZ KO pool cells, but not a clone with complete YAP/TAZ ablation. Naturally, the degrees of YAP/TAZ reduction vary in individual cells. These factors may explain the partial rescue phenotypes.
Generally, LATS1/2 are considered to be tumor suppressors and YAP/TAZ function as oncoproteins. However, YAP has been suggested to be a tumor suppressor in cancers32. In addition, YAP suppresses lung squamous cell carcinoma through ROS accumulation27. Our study establishes that LATS1/2 are not universal tumor suppressors, and their function in cell growth is cell type dependent and attachment dependent. LATS1/2 deletion inhibits MC38 by increasing senescence and apoptosis as well as reducing proliferation.
Mechanistic investigations reveal that WISP2 and CCDC80 induction by LATS1/2 deletion in MC38 contributes to inhibition of anchorage independent growth. We show that WISP2 and CCDC80 are direct YAP/TEAD target genes. WISP2 is a CCN family protein, which comprises six members (CCN1–6) including CYR61, CTGF, NOV, WISP1, WISP2, WISP3 33. Previous studies show that CYR61, CTGF, NOV, and WISP1 have tumor promoting effects, while WISP2 and WISP3 exert tumor suppressor functions. WISP2 inhibits cell growth, migration and invasion of breast cancer cells and gastric cancer cells 34, 35. CCDC80 expression is frequently down-regulated in primary colorectal cancers and human thyroid carcinomas 36, 37. In addition, CCDC80 can suppress anchorage independent growth and sensitize cells to anoikis and CD95-induced apoptosis 38. Our data reveal that WISP2 and CCDC80 are downstream of the Hippo pathway and act as key effectors in mediating the function of LATS1/2 and YAP/TAZ in modulating MC38 cell growth. It should be noted that high expression of WISP2 and CCDC80 occurs in MC38 clone #5 under attachment conditions (Fig.7d), which does not displays a strong growth inhibition (Fig.2e). Therefore, the function of WISP2 and CCDC80 in cell growth could be condition dependent, such as cell attachment.
VGLL4 is known to suppress YAP-mediated transcription by interacting with TEADs, and it is thought to be crucial in the regulation of both Hippo and Wnt signaling outputs in human colorectal cancer39. We perform RT-PCR for Vgll4 in both MC38 (WT and LATS1/2 KO) and CT26 (WT and LATS1/2 KO) under 2D and suspension conditions. The expression of Vgll4 slightly decreased in MC38 clone # 5 and increased in CT26 clone #16 under 2D and suspension conditions (Supplemental Fig.3C), indicating that Vgll4 is unlikely the key factor. We knockdown Vgll4 in MC38 and CT26 WT cells and find that Ctgf and Cyr61 expression decreased both in MC38 and CT26 (Supplemental Fig.3D). These results show that Vgll4 potentiates YAP-mediated transcription in MC38 WT and CT26 WT cells, but Vgll4 expression is different between MC38 LATS1/2 KO cells and CT26 LATS1/2 KO cells.
Extensive efforts are currently being made to develop YAP/TEAD inhibitors for cancer treatment. Given the possible cell growth inhibitory effect of YAP in some cancer cells, it might be important to determine the function of YAP as oncogene or tumor suppressor in a specific cancer before using YAP/TEAD inhibitors for therapy. Given the strong requirement of LATS1/2 for MC38 cell growth, one may speculate that LATS1/2 inhibition could have anti-growth effect in certain cancer types, suggesting LATS1/2 as potential cancer targets.
Materials and Methods
Cell culture and CRISPR-mediated gene deletion
MC38 and CT26 colon cancer cells, Panc02 pancreatic cells, 168FARN and 67NR breast cancer cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin solution (Gibco) at 37°C in a humidified 5% CO2 atmosphere. MB49 bladder cancer cells, GL261 glioma cancer cells, MyC-CaP prostate cancer cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin solution (Gibco). Panc02, CT26 and MyC-CaP cells were available from ATCC. MC38 cells were available from Kerafast, Int. GL261 cells were from DSMZ. MB49 cells were from MERCK. 67NR and 168FARN cells were from Dr.Yang, Jing lab. The cells were tested for mycoplasma and they are free of mycoplasma contamination.
LATS1/2 deficient cells were created using the CRISPR genome editing technology. The CRISPR plasmids were transfected into MC38, Panc02, CT26, 168FARN, 67NR, MB49, GL261 and MyC-CaP cells. 24 h after transfection, cells were treated with puromycin (2–10 µg/ml) for 3 days and then sorted into 96-well plates with a single cell in each well. Colonies were grown up, clones were screened by Western blot analysis with antibodies against LATS1 and LATS2. The guide sequences used in this experiment (IDT, USA) are provided in supplemental methods.
Retroviral infection
MC38 cells stably expressing empty vector, YAP5SA or YAP5SA/S94A were generated by retroviral infection. For expression virus, 293 phoenix retrovirus packing cells were transfected with pQCXIH empty vector (or pBABE empty), pQCXIH-YAP5SA, pQCXIH-YAP5SA/S94A, pQCXIH-LATS1, or pBABE-LATS2 vectors. Forty-eight hours after transfection, retroviral supernatant was supplemented with 5 μg/ml polybrene, filtered through a 0.45 μm filter, and used to infect the target cells. Forty-eight hours after infection, cells were selected with either 200 μg/ml hygromycin (for pQCXIH constructs) or 10 μg/ml puromycin (for pBABE constructs) in culture medium.
Soft agar colony formation assay
Each 6-well plate was coated with 1.5 ml of bottom agar (0.5% Difco agar noble in normal culture medium). Cells were suspended in 1.5 ml of 0.35% agar containing cells in culture medium and poured onto culture plates. The final cell concentration was (2.5–5) ×103 cells/well. The plates were placed in a 5% CO2 humidified incubator at 37°C. Colonies were counted 2 to 3 weeks after plating.
SA-β-gal staining
SA-β-gal (senescence-associated β-galactosidase) was done in a 12 well plate using a staining kit (Cell Signaling Technology). Cells were washed with PBS, then fixed and stained following the manufacturer’s instructions. For each sample SA-β-gal positive and total cell numbers were counted in 5 different microscopic fields (> 200 cells per field).
Annexin V apoptosis detection
MC38 WT and LATS1/2 deficient cells were cultured in suspension for 24 h, 48 h and 72 h. Cells were washed twice with cold PBS and then resuspended in Annexin V Binding Buffer at a concentration of 1×106 cells/ml, 1×105 cells were transferred to 5 ml culture tube, 5 ul of PE Annexin V (Annexin V conjugated to fluorochromes including Phycoerythrin (PE)) and 7-AAD were added to the 5mL culture tube. Cells were gently vortexed and incubated for 15 min at 25℃ in the dark, 400 ul of 1×Binding Buffer was then added to each tube. Annexin V staining was analyzed by flow cytometry within 1 h.
Intracellular ROS detection
Intracellular ROS detection was performed following manufacturer’s instructions (DCFDA Cellular ROS Detection Assay Kit, ab113851). Briefly, MC38 WT and LATS1/2 deficient cells were seeded in 6-well plates. Cells were incubated with the DCFH-DA probe for 30 minutes, harvested, and subjected to FACS analysis.
ChIP (chromatin immunoprecipitation)
LATS1/2 KO MC38 cells were cross-linked with 1% formaldehyde for 15 min, and chromatin DNA was digested with micrococcal nuclease (MNase). Immunoprecipitation reactions were carried out with chromatin extracts overnight at 4°C. Precipitated DNA was quantitated by real-time PCR analysis. All ChIP signals were normalized to the input.
Animal experiments
8–9-week-old female nude mice were provided by the animal care program at University of California, San Diego. All animal protocols were in accordance with the NIH Guide for the Care and Use of Laboratory Animals. To assess cancer cell proliferation in vivo, MC38 WT and LATS1/2 deficient cells (1×106) were subcutaneously transplanted into both back flanks of 8–9-week-old female nude mice. Eight mice were assigned to each group. Tumor height and width were measured with a caliper every 2–3 days to calculate tumor volume (= width2 × height ×0.523). For subcutaneous tumour growth, the maximum single tumour cannot exceed 1.5 cm in diameter in mice according to the guidelines provided by the animal care program at University of California, San Diego, and no experiments in this study generated tumour burden over this limit. Three weeks later primary tumor masses were collected from nude mice, fixed in 4% paraformaldehyde, and embedded in paraffin. For the xenograft studies, the mice were randomly divided into different groups according to ID number. No blinding was carried out. We did not exclude any data.
Tissue staining
Primary tumor paraffin sections were deparaffinized and rehydrated with xylene and alcohol gradients, and then incubated in 0.3% H2O2. After antigen retrieval using 10 mM sodium citrate (pH 6.0), Sections were incubated with anti-YAP, anti-p-Histone H3 anti-cleaved caspase 3 (1:200), at room temperature for 1 h, followed by a reaction with biotin-labeled secondary antibodies for 30 min. Staining was performed using Vectastain ABC kits and 3, 3′-diaminobenzidine (DAB) peroxidase substrate kits (Vector Laboratories, Burlingame, CA, USA).
Immunofluorescence analysis
Cells were fixed for 10 min at room temperature with 4% paraformaldehyde in PBS, permeabilized with 0.3% Triton X-100 in PBS, incubated with the blocking buffer (PBST containing 5% bovine serum albumin), and subsequently probed with anti-YAP/TAZ, anti-Ki-67 (1:200) and Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes). Slides were mounted using VectaShield with 4′, 6-diamidino-2-phenylindole (DAPI, Vector Laboratories). Digital images were acquired using Laser scanning confocal microscope with 6–100× magnification.
Western blot analysis
Total proteins from the cell extracts were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). After probing with primary antibodies, membranes were washed in Tris-buffered saline containing 0.05% Tween-20 (TBST) and incubated with a horse-radish peroxidase-linked secondary antibody. Finally, Western signals were detected using HRP substrate luminal reagent (Millipore). The primary antibodies used are provided in the supplemental methods.
RNA extraction and real-time RT–PCR analysis
Total RNA was extracted using RNeasy Plus Mini Kit (QIAGEN), according to the manufacturer’s instructions. Real-time PCR analysis was performed using KAPA SYBR FAST qPCR kit (Kapa Biosystems, USA) and an Applied 7300 Real-Time PCR System. Relative mRNA levels were determined by normalizing to the endogenous Gapdh mRNA.
RNA interference
Cells were transfected with different siRNAs using Lipofectamine RNAiMAX Reagent (ThermoFisher Scientic). Briefly, cells were seeded in six-well plates and transfected with siRNA and Lipofectamine RNAiMAX Reagent, which were each incubated separately in Opti-MEM for 5 min, mixed together for 10 min at room temperature, and then the mixture was applied to the cells plated in 1 ml of medium (final siRNA concentration, 80 nM). The siRNA sequences used in this experiment (GenePharma, Shanghai, China) were as follows:
sicon (siNC): UUCUCCGAACGUGUCACGUTT
siTcf4: GGGACAUGCAUGGAAUCAUTT
siVgll4-1: GCCAAAGCACUAGGUGACATT
siVgll4-2: CCUCAUGGAGCAGCCUCUUTT
Statistical analysis.
The experiments shown in Figs.6a and 6b are representative of 2 independent experiments performed with similar results. All in vitro assays were performed in triplicates. Microsoft Excel was used for t-tests, Groups were compared by two-tailed t-tests or ANOVA using GraphPad Prism statistical programs (GraphPad Prism, San Diego, CA). p values were noted in the figure legends. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Zhipeng Meng, Rui Chen, Shenghong Ma, Zhengming Wu, Kimberly C, Lin and Min Luo for technical assistance; JiaYu Chen and Xiangdong Fu for help in bioinformatics analysis. This work was supported by grants from the National Institutes of Health (CA196878, CA217642, and GM51586) to K.L.G. This study was supported by grants from National Natural Science Foundation of China (31871402, 81402162), and the Natural Science Foundation of Zhejiang Province (LY17H160060) to W.W.P.
Footnotes
Disclosure of Potential Conflicts of Interest
Dr. Kun-Liang Guan is a co-founder and has an equity interest in Vivace Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010; 19: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews Drug discovery 2014; 13: 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015; 163: 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry ER, Camargo FD. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 2013; 25: 247–253. [DOI] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer 2013; 13: 246–257. [DOI] [PubMed] [Google Scholar]
- 6.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nature reviews Cancer 2015; 15: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer cell 2016; 29: 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev 2016; 30: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007; 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 2007; 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Current biology : CB 2008; 18: 435–441. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 2008; 14: 388–398. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 2008; 14: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, Ye X, Yu J, Li L, Li W, Li S et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008; 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Liu P, Zhou X, Wang T, Feng X, Sun YP et al. Endothelin Promotes Colorectal Tumorigenesis by Activating YAP/TAZ. Cancer research 2017; 77: 2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell 2009; 16: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A 2010; 107: 1431–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 2010; 107: 1437–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. Journal of hepatology 2015; 63: 1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Current biology : CB 2007; 17: 2054–2060. [DOI] [PubMed] [Google Scholar]
- 21.Oh JE, Ohta T, Satomi K, Foll M, Durand G, McKay J et al. Alterations in the NF2/LATS1/LATS2/YAP Pathway in Schwannomas. J Neuropathol Exp Neurol 2015; 74: 952–959. [DOI] [PubMed] [Google Scholar]
- 22.Baia GS, Caballero OL, Orr BA, Lal A, Ho JS, Cowdrey C et al. Yes-associated protein 1 is activated and functions as an oncogene in meningiomas. Molecular cancer research : MCR 2012; 10: 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nature genetics 2016; 48: 407–416. [DOI] [PubMed] [Google Scholar]
- 24.Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukui T et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer research 2011; 71: 873–883. [DOI] [PubMed] [Google Scholar]
- 25.Barry ER MT, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 2013; 493: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med 2014; 20: 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Zhang W, Pan Y, Gao Y, Deng L, Li F et al. YAP Suppresses Lung Squamous Cell Carcinoma Progression via Deregulation of the DNp63-GPX2 Axis and ROS Accumulation. Cancer research 2017; 77: 5769–5781. [DOI] [PubMed] [Google Scholar]
- 28.Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 2016; 167: 1525–1539.e1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nature reviews Cancer 2015; 15: 397–408. [DOI] [PubMed] [Google Scholar]
- 31.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M et al. Role of YAP/TAZ in mechanotransduction. Nature 2011; 474: 179–183. [DOI] [PubMed] [Google Scholar]
- 32.Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell death and differentiation 2008; 15: 1752–1759. [DOI] [PubMed] [Google Scholar]
- 33.Barreto SC, Ray A, Ag Edgar P. Biological characteristics of CCN proteins in tumor development. Journal of BUON : official journal of the Balkan Union of Oncology 2016; 21: 1359–1367. [PubMed] [Google Scholar]
- 34.Dhar G, Banerjee S, Dhar K, Tawfik O, Mayo MS, Vanveldhuizen PJ et al. Gain of oncogenic function of p53 mutants induces invasive phenotypes in human breast cancer cells by silencing CCN5/WISP-2. Cancer research 2008; 68: 4580–4587. [DOI] [PubMed] [Google Scholar]
- 35.Ji J, Jia S, Jia Y, Ji K, Hargest R, Jiang WG. WISP-2 in human gastric cancer and its potential metastatic suppressor role in gastric cancer cells mediated by JNK and PLC-gamma pathways. British journal of cancer 2015; 113: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferraro A, Schepis F, Leone V, Federico A, Borbone E, Pallante P et al. Tumor suppressor role of the CL2/DRO1/CCDC80 gene in thyroid carcinogenesis. The Journal of clinical endocrinology and metabolism 2013; 98: 2834–2843. [DOI] [PubMed] [Google Scholar]
- 37.Herbst A, Bayer C, Wypior C, Kolligs FT. DRO1 sensitizes colorectal cancer cells to receptor-mediated apoptosis. Oncology letters 2011; 2: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bommer GT, Jager C, Durr EM, Baehs S, Eichhorst ST, Brabletz T et al. DRO1, a gene down-regulated by oncogenes, mediates growth inhibition in colon and pancreatic cancer cells. The Journal of biological chemistry 2005; 280: 7962–7975. [DOI] [PubMed] [Google Scholar]
- 39.Jiao SLC, Hao Q, Miao H, Zhang L, Li L, Zhou Z. VGLL4 targets a TCF4–TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nature communications 2017; 8 14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.