Abstract
There is increasing evidence that genomic instability is a prerequisite for cancer progression. Here we show that SIM2s, a member of the bHLH/PAS family of transcription factors, regulates DNA damage repair through enhancement of homologous recombination, and prevents epithelial mesenchymal transitions (EMT) in an ATM dependent manner. Mechanistically, we found that SIM2s interacts with ATM and is stabilized through ATM-dependent phosphorylation in response to ionizing radiation (IR). Once stabilized, SIM2s interacts with BRCA1 and supports RAD51 recruitment to the site of DNA damage. Loss of SIM2s through the introduction of shSIM2 or the mutation of SIM2s at one of the predicted ATM phosphorylation sites (S115) reduces homologous recombination efficiency through disruption of RAD51 recruitment, resulting in genomic instability and induction of EMT. The EMT induced by the mutation of S115 is characterized by a decrease in E-cadherin and an induction of the basal marker, K14, resulting in increased invasion and metastasis. Together, these results identify a novel player in the DNA damage repair pathway and provides a link in DCIS progression to IDC through loss of SIM2s, increased genomic instability, EMT and metastasis.
INTRODUCTION
Breast cancer is one of the most common cancers in women, affecting approximately one in eight women during their lifetime. Most, if not all, invasive breast cancers have a pre-invasive stage defined as ductal carcinoma in situ (DCIS). DCIS consists of a heterogeneous group of diseases characterized by a neoplastic mammary lesion that is confined to the ductal-lobular system of the breast14. Due to increasing adoption of screening mammography, the diagnosis of DCIS has risen from less than 1% of breast cancers to 15–25% and accounted for over 54,944 new cases of DCIS in 201314, 36, 51; yet, as few as 20% of diagnosed DCIS progress to invasive ductal carcinoma (IDC)48, 54. Given the significant morbidity associated with adjuvant therapy, many clinicians are currently reluctant to recommend radiation or tamoxifen therapy for DCIS patients20, 21, 32, 48, 54. Therefore, there is a critical need to find therapeutic and intervention strategies to prevent breast cancer progression in those DCIS patients at high risk for invasive breast cancer.
DCIS is characterized by aberrant epithelial cell growth into the ductal lumen, and increased severity is categorized by elevated levels of calcification and necrosis70. Currently, a number of markers, including human epidermal growth factor receptor 2 (HER2), p53, and ER, are used to gauge early disease severity11. Unfortunately, the genetic profile between late-stage DCIS and IDC are considered indistinguishable, which makes these markers unusable as indicators for DCIS progression to IDC38. There is increasing evidence that progression from early ductal hyperplasia to the onset of IDC is a result of escalating levels of genomic instability, which culminate in the accrual of detrimental mutations in tumor suppressing genes38. These aberrant mutations can vary from single-nucleotide polymorphisms (SNPs) to the translocation, duplication, fragmentation, or deletion of vast chromosomal regions5, 23, 34, 64. Aneuploidy and gene silencing resulting from these mutations can significantly deplete tumor-suppressing factors and lead to the increased expression of pro-proliferative factors, which further enhance breast cancer progression.
DNA double-stranded breaks (DSB) can lead to chromosomal alterations if left unrepaired and are generally repaired via the error-prone non-homologous end-joining (NHEJ) pathway. Interestingly, it is mutations within the homologous recombination (HR) DDR pathway that have been implicated in familial breast cancer. Studies show a strong correlation between hereditary mutations in BRCA1/BRCA2 and increased incidence of high-grade DCIS and progression from DCIS to IDC75. A pivotal mediator of the HR pathway is Ataxia-telangiectasia mutated (ATM)30. Although ATM has been found to be partially dispensable during NHEJ, it is crucial for accessing DSBs in heterochromatin26. Repair within these DNA structures happens many orders of magnitude slower than elsewhere in the genome, taking hours instead of minutes. Even though heterochromatin only accounts for a fraction of the DNA within a cell, DNA damage that occurs here can only be repaired in the presence of ATM and can persist for days in the absence of ATM26, 45, 50.
Although previously only associated with familial breast cancer (5–10% of total breast cancer cases), a growing body of evidence shows that BRCA misregulation and mutations are more abundant in sporadic cancers than previously thought, possibly being present in as many as 82% of ovarian cancers9, 31,61. Loss of these crucial DDR factors has been associated with a distinct tumor profile, including loss of ER, progesterone receptor (PR), and HER2, providing a strong association between loss of BRCA and highly invasive triple negative breast cancers (TNBC)9, 67. Interestingly, the loss of HR in these tumors presents an effective treatment option with the induction of synthetic lethality through use of PARP (poly (ADP-ribose) polymerases) inhibitors as one of the most promising treatments.
Previously, our lab has shown that the bHLH-PAS transcription factor family member single-minded 2s (SIM2s; short isoform expressed from SIM2) plays a role in mammary gland development, is down-regulated in primary breast, and that loss of SIM2s expression is associated with an epithelial mesenchymal transition (EMT) both in normal breast and breast cancer cell lines27, 37, 39, 44, 57, 58, 73. We have used the DCIS.com progression model to demonstrate that re-expression of SIM2s inhibits growth and metastasis and promotes a more luminal-like phenotype; whereas down-regulation of SIM2s leads to an increase in invasive potential58. This system allows for the formation of DCIS structures that spontaneously progress into IDC when injected intraductally or into the flanks of mice4. Although research has been conducted into the role of SIM2s in different cancers, the mechanism behind SIM2s activation has remained elusive1, 29, 57. Here, for the first time, we show a novel role for SIM2s activation in response to DNA damage.
RESULTS
SIM2s expression is lost during progression to IDC
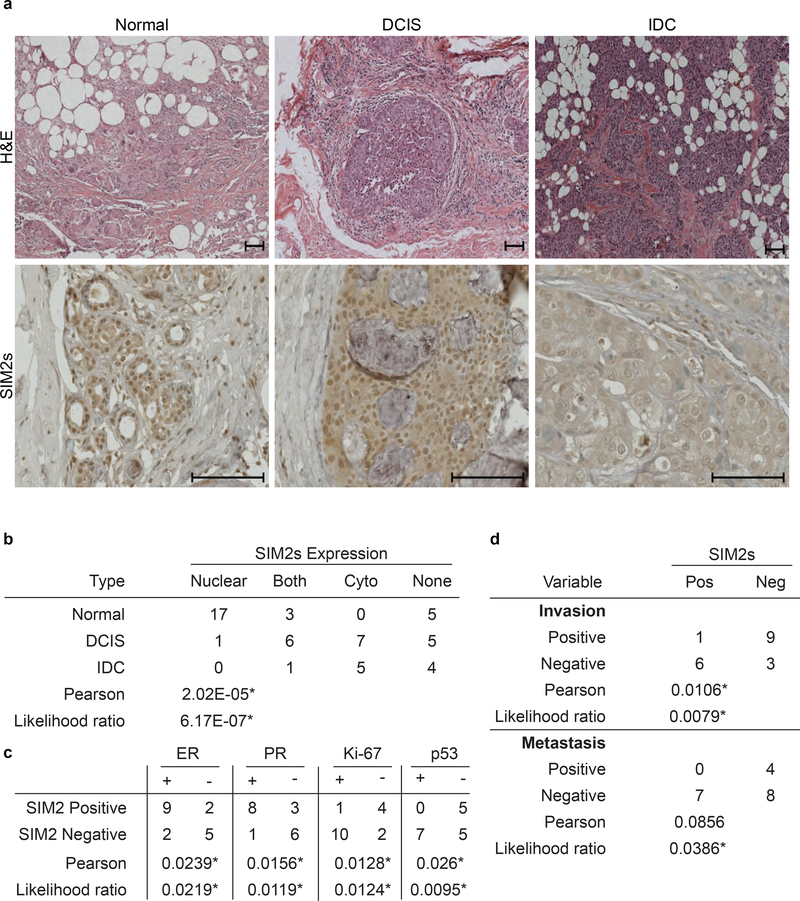
To investigate the expression of SIM2s during progression from normal breast tissue through DCIS to IDC, we analyzed human tissue microarrays containing normal breast, DCIS, DCIS with local invasion, and IDC for differences in SIM2s by IHC in 27 patients diagnosed with DCIS or IDC. The results showed high, nuclear expression of SIM2s in normal tissue. Although SIM2s expression remained high in DCIS, a shift towards cytoplasmic localization was apparent. Interestingly, SIM2s loss was apparent in sections containing IDC (Fig. 1a). Statistical analysis of SIM2s staining revealed a significant correlation between SIM2s expression and the state of breast disease (Normal, DCIS, or IDC) (p<6.7E−7) (Fig. 1b). SIM2s expression significantly correlated with ER (p<0.0239) and PR (p<0.0156) expression in DCIS samples, but not HER2 (p<0.1377), indicating a relationship between SIM2s and luminal stage breast cancer. SIM2s expression was also significantly correlated with p53 expression (p<0.0095) and inversely related to Ki67 expression (p<0.0124) in DCIS samples (Fig. 1c). Importantly, loss of SIM2s correlated with increased micro-invasion and metastasis, supporting a role for SIM2s in inhibiting breast cancer progression (Fig. 1d; p<0.0079).
Figure 1: Loss of SIM2s correlates with increased metastasis and invasion.
(a) Histological staining of human normal, DCIS, and IDC samples for H&E and SIM2s. Samples were imaged at 25× magnification. Scale bars, 100μm. (b) Statistical analysis of breast type with SIM2s expression. Tissue microarrays were analyzed categorically to compare the location of SIM2s staining versus the breast type. (c) Statistical analysis of SIM2s status correlated with ER, PR, Ki-67, and p53 status in DCIS specific pathology reports (n=18). (d) Prognosis of micro invasion and/or metastasis was compared with binomial SIM2s staining. Likelihood ratio and Pearson’s chi-squared tests were performed to test correlations. *p-value<0.05.
SIM2s regulation of the DDR pathway
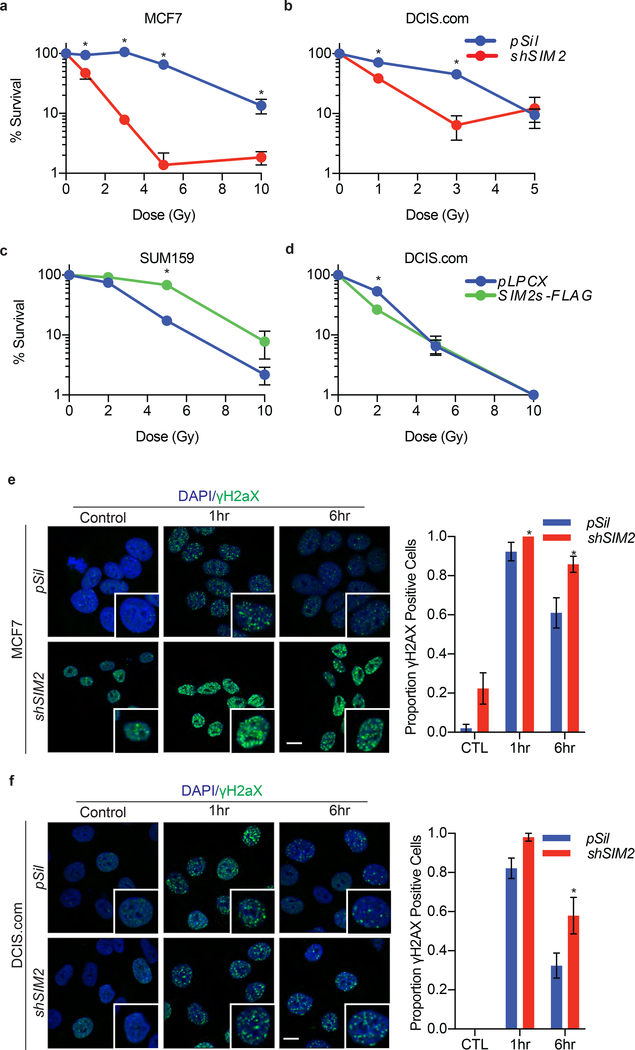
To determine the impact of SIM2s on DDR, we utilized breast cancer cell lines that express differing levels of endogenous SIM2s with SUM159 (low), MCF7 (high), and MCF10-DCIS.com (DCIS.com-medium)l37, 58. We modified SIM2s levels by over-expression (SIM2s-FLAG) or with a previously validated shSIM239. DNA damage was induced with IR and colony formation assays were performed28, 37, 39, 58. The results show that shSIM2 sensitized cells to IR (Fig. 2a, b), whereas over-expression of SIM2s had a protective effect in SUM159 cells (Fig. 2c). Interestingly, there was no significant change in DCIS.com-SIM2s-FLAG cells, possibly due to the presence of endogenous SIM2s in control cells (Fig. 2d).
Figure 2: SIM2s increases cell survivability by reducing DNA damage.
(a-d) Clonogenic survival assays of (a) MCF7 and (b) DCIS.com cells containing shSIM2 or control were treated with the indicated doses of irradiation. Cells were then plated and allowed to grow for 7 days before being counted. (c) SUM159 and (d) DCIS.com cells over-expressing SIM2s-FLAG or control were treated with the indicated doses of IR and sampled as before. (e) MCF7 and (f) DCIS.com cells containing shSIM2 or control were treated with 2GYs IR or left untreated and fixed at the indicated time points. Cells were then immunostained for γH2AX, and cells containing more than 10 foci were counted as positive (n=50). Scale bars, 10μm. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
To investigate this protective effect of SIM2s, we examined how loss of SIM2s affected genomic stability. As DNA damage is detected, ATM phosphorylates histone H2AX (γH2AX), which then forms foci at the site of DNA damage10, 71. We found that MCF7-shSIM2 cells treated with 2GYs IR had an increase in basal γH2AX foci compared to control cells (p<0.00815) and recovered slower from DNA damage compared to control cells (p<0.00163; Fig. 2e). Although the DCIS.com-shSIM2 lacked the initial basal elevation of γH2AX foci, they did have significantly more γH2AX foci 6 hours after treatment with 2GYs IR (p<0.00143; Fig. 2f). The delayed resolution of γH2AX foci has been previously associated with the ATM-dependent DDR within heterochromatin structures, where repair is both temporally and spatially separated from other DDR in order to prevent inappropriate cross-over events26, 45, 50.
SIM2s is phosphorylated by ATM and stabilized in response to IR
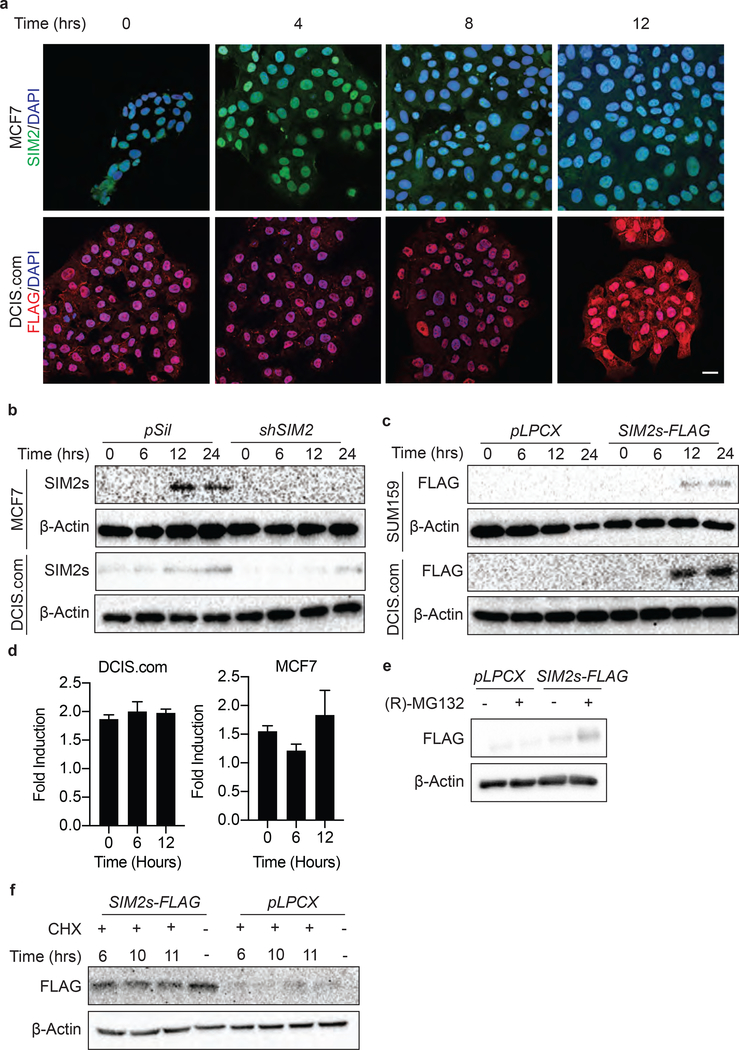
The bHLH-PAS family of transcription factors plays an important role in development and in response to environmental cues; however, mechanisms that activate SIM2s have remained elusive18, 74. To better understand the role SIM2s may play in DDR, we assessed SIM2s levels in DCIS.com-SIM2s-FLAG and MCF7 cells treated with IR. IF analysis showed an elevated level of nuclear SIM2s-FLAG after treatment with 2GYs IR (Fig. 3a). SIM2s activation was further confirmed by western blot analysis in MCF7 and DCIS.com treated with 2GYs IR (Fig. 3b). The efficiency of our previously validated shSIM2 construct was further confirmed in MCF7 and DCIS.com cells, with no SIM2s detected 12 hours after 2GYs IR treatment (Fig. 3b)39. SUM159-SIM2s-FLAG and DCIS.com-SIM2s-FLAG cells also showed SIM2s stabilization after treatment with 2GYs IR (Fig. 3c).
Figure 3: SIM2s is stabilized with IR treatment.
(a) WT MCF7 cells or DCIS.com cells over-expressing SIM2s-FLAG were treated with 2GYs of IR and fixed at the indicated time points before immunostaining for the presence of FLAG. Scale bar, 20μm. (b) Efficiency of SIM2s knockdown by shSIM2 was confirmed in MCF7 and DCIS.com cells via western blot. Stabilization of endogenous SIM2s 12 hours after IR treatment was further confirmed via western blot. (c) The FLAG epitope was confirmed to not interfere with SIM2s stabilization in the DCIS.com and SUM159 cells over-expressing SIM2s-FLAG. (d) DCIS.com and MCF7 cells were treated with 2GYs of IR, and RNA was harvested at the indicated time points. RT-qPCR analysis of SIM2s indicates no change in SIM2s mRNA after treatment with 2GYs IR. (e) Western blot analysis of DCIS.com SIM2s-FLAG and pLPCX control cells treated for 2 hours with 10μM (R)-MG132 or DMSO. (f) Confirmation of SIM2s stabilization at the protein level was confirmed in DCIS.com cells over-expressing SIM2s-FLAG. Cells were treated with 2GYs IR and then dosed with 50μg/mL of the translation inhibitor, cycloheximide (CHX), at the indicated time points before being harvested 12 hours post-IR. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
We next sought to determine if the increase in SIM2s levels was due to an up-regulation of SIM2s gene expression. Interestingly, in both normal MCF7 and DCIS.com cells, we observed no change in SIM2s levels after treatment with 2GYs IR (Fig. 3d). SIM2s is ubiquitinated by the E3 ubiquitin ligase, Parkin, leading to its rapid proteasome-dependent degradation47. Proteasomal inhibition with 10μM (R)-MG132 for 2 hours inhibits this pathway and resulted in increased SIM2s levels (Fig. 3e). This led us to theorize that SIM2s could be stabilized post-translationally. To test this, we treated DCIS.com-SIM2s-FLAG cells with 50μg/mL cycloheximide (CHX) to inhibit translation at the indicated time points after treatment with 2GYs of IR and harvested all samples 12 hours after IR treatment. No significant change in SIM2s levels was observed, supporting our hypothesis that SIM2s is stabilized post-translationally (Fig. 3f).
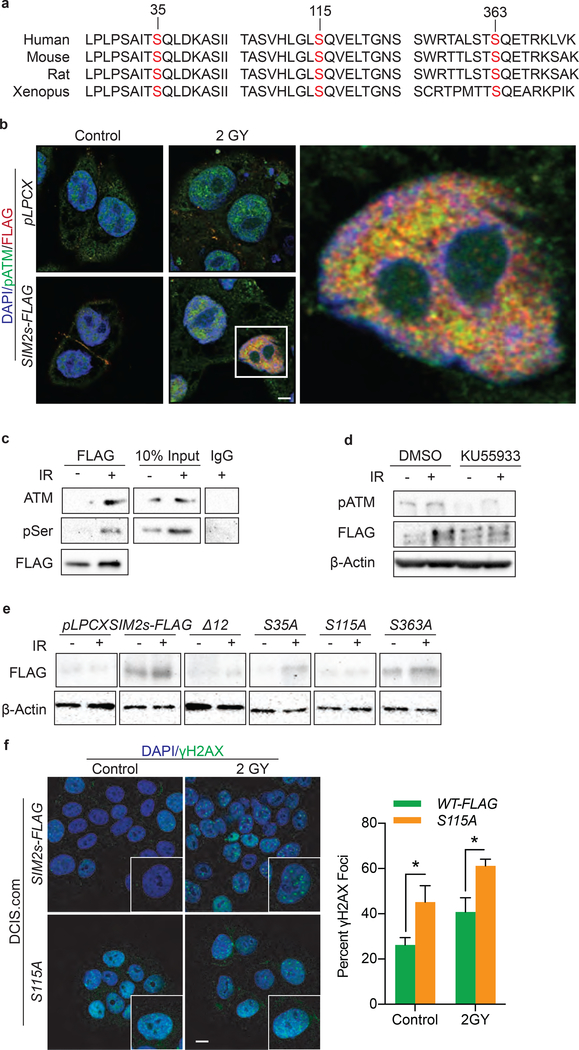
To investigate the possibility that SIM2s is stabilized by a post-translational modification in response to IR, we analyzed SIM2s for phosphorylation sites of known DDR kinases and identified 12 total ATM consensus sites, three of which are highly conserved across Mus Musculus, Homo Sapiens, and Xenopus laevis (S35, S115, S363) (Fig. 4a)7. To determine if ATM interacts with and phosphorylates SIM2s, DCIS.com-SIM2s-FLAG cells were treated with 2GYs of IR and assessed for nuclear co-localization of FLAG and pATM via IF (Fig. 4b). This interaction was confirmed through immunoprecipitation of FLAG in DCIS.com-SIM2s-FLAG cells treated with 2GYs IR followed by immunoblotting for ATM (Fig. 4c). Additionally, an increase in phospho-serine residues at the molecular weight of SIM2s was overserved after treatment with 2GYs IR (Fig. 4c). To determine if ATM is necessary for IR-induced SIM2s stabilization, we pretreated SUM159-SIM2s-FLAG cells with KU55933, a selective ATM inhibitor, for 2 hours prior to treatment with 2GYs of IR. Western blot analysis showed a significant reduction in SIM2s stabilization in KU55933 treated cells in response to IR (Fig. 4d). These findings suggest that SIM2s is stabilized in response to IR through phosphorylation.
Figure 4: S115 is necessary for SIM2s stabilization.
(a) SIM2s was analyzed for ATM consensus sequences. Three highly conserved sites were found and confirmed with NetPhos 3.15. (b) Co-localization of SIM2s and pATM 12 hours after IR treatment was confirmed in DCIS.com cells. Scale bar, 10μm. (c) Immunoprecipitation of SIM2s-FLAG 12 hours after treatment with 2GYs IR in DCIS.com cells by a FLAG specific antibody shows SIM2s interacts with ATM in response to IR. In addition, an increase in pSer is seen in response to IR at the molecular weight of SIM2s-FLAG. (d) Treatment with 10μM of KU55933 inhibited SIM2s stabilization in response to 2GYs IR. (e) Serine to alanine point mutations were generated in SIM2s-FLAG at S35, S115, S363, or at all predicted ATM consensus sites (Δ12) and then stably transfected into DCIS.com cells. Mutants were analyzed for SIM2s stabilization 12 hours post-treatment with 2GYs IR, with only the S115A mutant not responding to IR. (f) DCIS.com cells over-expressing SIM2s-FLAG or SIM2s-S115A were treated with 2GYs IR and fixed 6 hours later before being immunostained for the presence of γH2AX foci. Cells containing 10 or more γH2AX foci were counted as positive (n=50). Scale bar, 10μm. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
We next generated a DCIS.com-SIM2s-FLAG cell line with serine to alanine mutations at all the predicted ATM consensus sites (S35, S115, S203, S216, S309, S343, S352, S361, S363, S392, S393, S426; SIM2sΔ12). To test if the abrogation of these consensus sites effected SIM2s stabilization in response to IR, we treated SIM2sΔ12 cells with 2GY IR and isolated protein 12 hours later. Interestingly, there was no increase in SIM2s levels with these 12 sites mutated (Fig. 4e).
To narrow down which serine residue is necessary for SIM2s stabilization we generated DCIS.com-SIM2s-FLAG cells lines containing point mutations at the three highly conserved ATM-consensus sites: serine 35 (S35A), 115 (S115A), or 363 (S363A). Utilizing these cells lines, we sought to determine if one of these mutations inhibited SIM2s stabilization. Immunoblot analysis found that S115A no longer showed SIM2s stabilization 12 hours after treatment with 2GYs IR (Fig. 4e). As inefficient transduction of S115A could have led to these findings, we verified SIM2s RNA levels in these cells via RT-qPCR and found a significant increase in SIM2s RNA when compared to the pLPCX control cells (Supplemental Figure 3). The interaction of SIM2s and ATM, suggests that SIM2s may be acting downstream of ATM; however, western blot analysis revealed that loss of SIM2s does not affect p53 activation (data not shown).
The loss of stabilization observed with S115A led us to predict that cells containing this mutation would behave similarly to cells containing shSIM2 and have impaired DDR. Comparing DCIS.com-SIM2s-FLAG to S115A revealed that cells containing S115A had significantly more γH2AX foci 6 hours after 2GYs IR than cells over-expressing SIM2s-FLAG (Fig. 4f).
Loss of SIM2s impairs homologous recombination
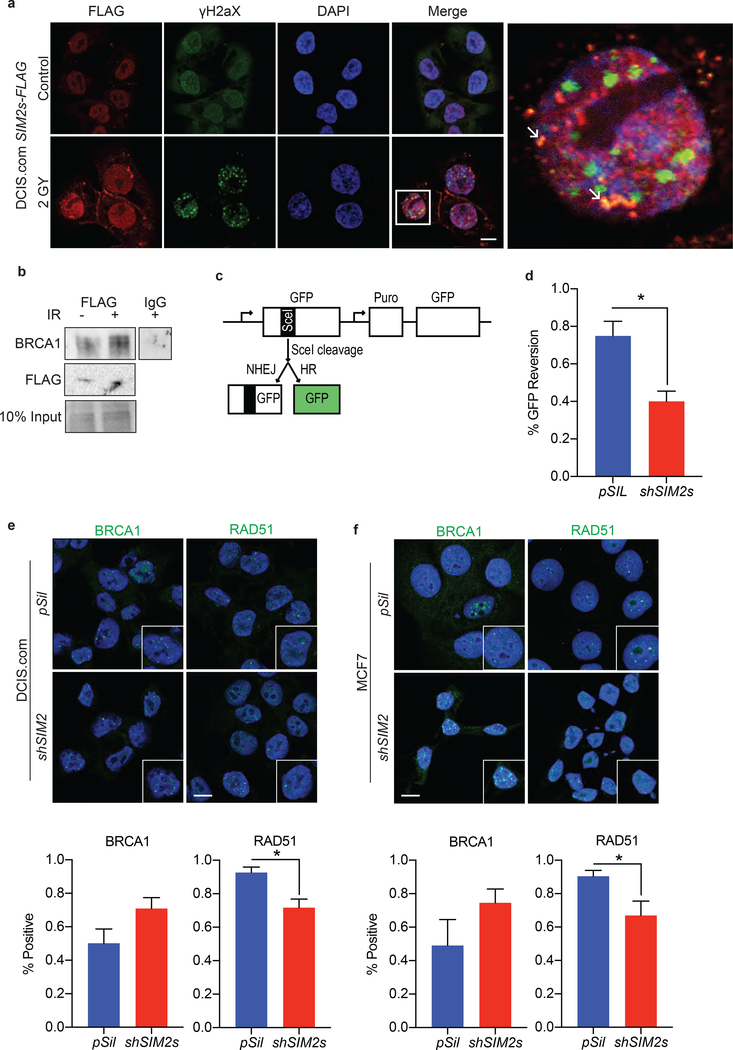
As SIM2s is a transcription factor we sought to determine if SIM2s regulates DDR gene expression. Interestingly, loss of SIM2s did not have a negative effect on ATM, TP53, 53BP1, BRCA1, BRCA2, or RAD51 levels 6 hours after treatment with 2GYs IR (Supplemental Figure 1). This finding led us to hypothesize that SIM2s directly interacts with proteins involved in DDR. Utilizing IF, we observed that SIM2s co-localizes with γH2AX in response to IR (Fig. 5a). The localization of this interaction to the nuclear periphery is consistent with a role for SIM2s in aiding ATM repair of damaged heterochromatin via HR53. To further test this, we immunoprecipitated FLAG from IR treated DCIS.com-SIM2s-FLAG 12 hours after treatment and found that IR enhanced SIM2s interaction with BRCA1 (Fig. 5b).
Figure 5: SIM2s is involved in HR and is necessary for RAD51 recruitment.
(a) DCIS.com cells over-expressing SIM2s-FLAG were treated with 2GYs IR and probed for γH2AX and FLAG 12 hours later. Arrows indicate sites of co-localization. Scale bar, 10μm. (b) The interaction of SIM2s with BRCA1 was further assessed by immunoprecipitation of SIM2s with a FLAG specific antibody from DCIS.com SIM2s-FLAG cell lysate 12 hours after 2GYs IR or mock treatment. (c) Diagram of drGFP locus reporter plasmid structure. (d) The effect that loss of SIM2s has on HR was assessed using the drGFP reporter system in conjunction with a plasmid encoding SceI. The presence of GFP was analyzed via flow cytometry (10,000 events) and represented as the percentage of cells expressing GFP. (e) DCIS.com or (f) MCF7 cells containing a scrambled vector or shSIM2 were treated with 2GYs of IR and probed for BRCA1 or RAD51 6 hours later (n=50). Cells containing 10 or more foci were counted as positive. Scale bars, 10μm. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
To investigate the effect of SIM2s on HR, we utilized a GFP reporter system to assay the efficiency of HR in cells containing shSIM249. This system contains an expressed non-functional GFP with a SceI cleavage site and an unexpressed functional GFP. Upon treatment with SceI, cells capable of HR use the functional GFP template for repair. In this way, HR efficacy can be assessed by quantification of GFP activity in cells following transduction of cells containing drGFP with SceI (Fig. 5c). Here, we employed this system to investigate whether depletion of SIM2s inhibits HR. Our results showed a significant decrease in GFP reversion in cells containing shSIM2 compared to controls, suggesting that loss of SIM2s impairs HR (p<0.022; Fig. 5d).
The dynamics that determine which DDR pathway is chosen have been well studied and depend upon the location of the break, the stage of the cell cycle, and the break type42. In instances where BRCA1 is lost, cells are no longer able to remove p53BP1 and a dramatic increase in p53BP1 foci is observed12. Interestingly, we observed no change in p53BP1 foci in cells containing shSIM2 (Supplemental Figure 2). This finding suggests that SIM2s may be acting downstream of the NHEJ/HR fate determining steps. In line with this hypothesis, we observed that loss of SIM2s leads to a significant decrease in RAD51 recruitment in DCIS.com (p<0.012) and MCF7 (p<0.003) cells after treatment with IR, whereas there was no change in BRCA1 recruitment (Fig. 5e, f, Supplemental Figure 4). Together, these data show that loss of SIM2s impairs HR by decreasing the efficiency of RAD51 loading in response to IR.
Loss of SIM2s sensitizes cells to treatment with PARP inhibitors
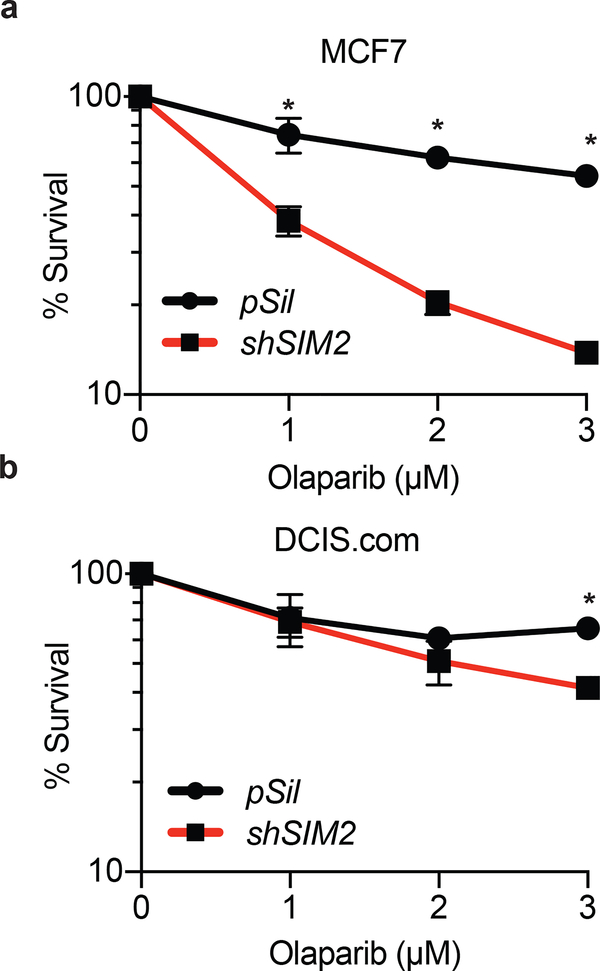
Recent studies have shown that loss of factors crucial for HR, including BRCA1, BRCA2, and RAD51, sensitizes cells to PARP inhibitors in pre-clinical and clinical trials8, 16, 19, 68, 72. PARPs are necessary in early DDR to poly-ADP-ribosylate target histones, leading to the destabilization of chromatin structure and exposure of damaged DNA for invasion of DDR machinery56. This combination of loss of both single-stranded break repair systems as well as impaired DSB repair leads to the accrual of DNA damage that eventually overwhelms and kills the cell. As such, we hypothesized that loss of SIM2s would sensitize cells to treatment with PARP inhibitors. Consequently, we dosed shSIM2 containing cells with the indicated doses of Olaparib, a potent PARP1/PARP2 inhibitor, and assayed for differences in proliferation72. Loss of SIM2s significantly reduced cell survival in a dose-dependent manner, further supporting a role for SIM2s in HR (Fig. 6, Supplemental Figure 4).
Figure 6: Loss of SIM2s sensitizes cells to synthetic lethal treatments.
Percent cell survival in (a) MCF7 and (b) DCIS.com cells containing shSIM2 or control with increasing doses of the PARP inhibitor Olaparib. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
SIM2s mutation at S115 leads to EMT
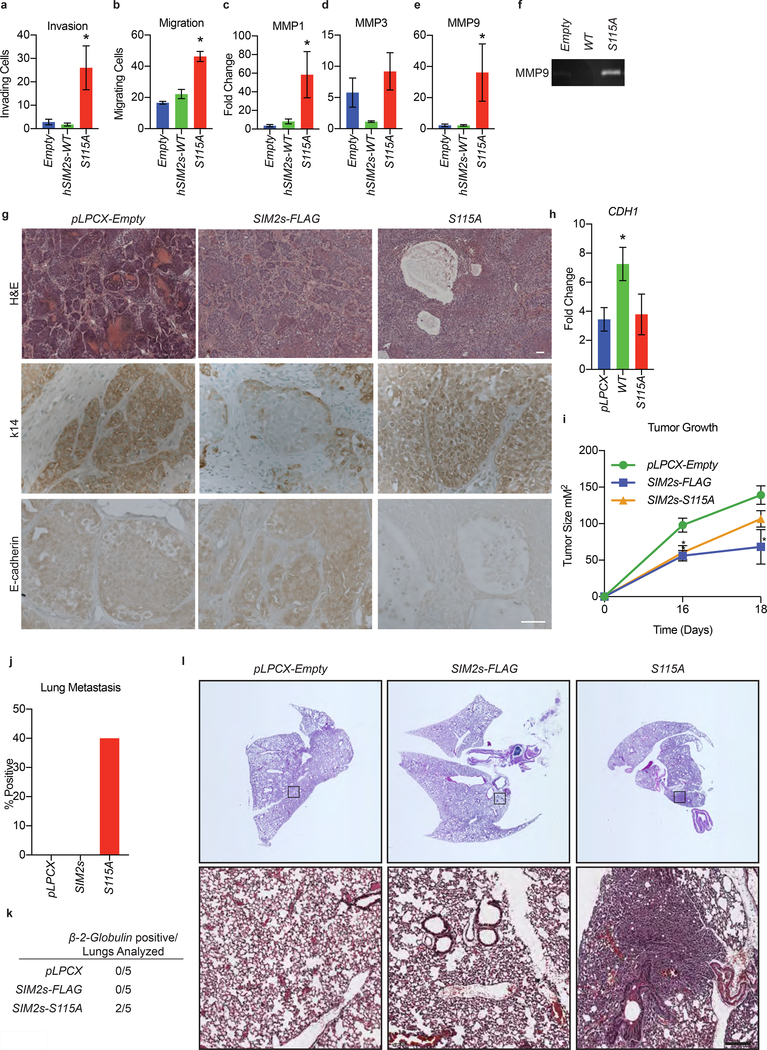
We have previously shown that SIM2s is an inhibitor of EMT in normal breast and breast cancer cell lines and in the mouse mammary gland, which are further supported by our current findings (Fig. 1)27, 37, 39. As we have identified a role for SIM2s in the recruitment of RAD51, we sought to determine if the S115A mutation impairs the EMT inhibitory function of SIM2s. Using Boyden chamber assays, we found that the S115A mutant resulted in a significant increase in the ability of DCIS.com cells to not only migrate (p<0.0009), but also to invade (p<0.0485), through an extra-cellular matrix (Fig. 7a, b). We have shown that the invasive phenotype observed with loss of SIM2s can be attributed to an increase in matrix metalloprotease (MMP) activity37, 39. Here, we found that S115A cells have an increase in MMP1 (p<0.006) and MMP9 (p<0.05) expression (Fig. 7c-e). This also correlated with an increase in MMP9 activity in these cells, as demonstrated by gelatin zymography (Fig. 7f).
Figure 7: Mutation of SIM2s at S115 promotes basal marker expression and tumor metastasis.
(a) DCIS.com cells containing the indicated constructs were plated in Boyden chamber inserts containing Matrigel and measured for invasive potential. (b) DCIS.com cells containing the indicated plasmids were plated in Boyden chamber inserts and measured for migratory potential. (c-e) DCIS.com cells containing the indicated constructs were harvested for RNA and analyzed for the expression of (c) MMP1, (d) MMP3, and (e) MMP9 via RT-qPCR. (f) Matrix-metalloprotease activity was confirmed via gelatin zymograph in DCIS.com cells containing SIM2s-FLAG, S115A, or empty vector. (g) H&E staining of tumors isolated from the indicated mice. Scale bar, 100μm. Tumor sections were stained for the indicated basal and luminal markers. Scale bar, 50μm. (h) RNA was isolated from the largest tumor from each mouse and analyzed for CDH1 expression via RT-qPCR. (i) Seventy-five thousand DCIS.com cells containing SIM2s-FLAG, SIM2s-S115A, or empty vector were injected into the flanks of nude mice, and tumor growth was measured. (j) Percent and (k) enumeration of mice positive for lung metastasis in the indicated tumors was measured by RT-qPCR analysis using a human specific β−2-globulin primer that does not cross-react with mouse. (l) Lungs were sectioned before being H&E stained. Top: slide scanned whole lung sections. Bottom: lung sections under 10X magnification. Scale bar, 200μm. Values indicate the mean ± SE with n=3 unless otherwise stated. Student’s t-test was performed to test significance. *p-value<0.05.
To determine if these characteristics translated into an increased invasive potential in vivo, control, DCIS.com-SIM2s-FLAG, or S115A were injected into the flanks of nude mice and monitored for tumor growth. Histological analysis of tumors revealed the SIM2s-FLAG tumors formed distinct lobular structures, whereas the S115A tumors exhibited a more invasive phenotype with increased areas of necrosis (Fig. 7g). Staining of sections from SIM2s-FLAG tumors revealed a single layer of keratin 14 (K14) positive cells, whereas S115A tumors stained positive for K14 with no defined organization (Fig. 7g). Further, we found that S115A tumors contained decreased E-cadherin compared to SIM2s-FLAG, suggesting induction of EMT (Fig. 7g). RT-qPCR analysis of RNA isolated from tumors showed a significant increase of CDH1 in SIM2s-FLAG (p<0.02) but not in S115A (Fig. 7h) tumors.
Analysis of tumor growth, showed decreased tumor growth in both SIM2s-FLAG (p<0.0001) and S115A (p<0.002) tumors, as compared to controls (Fig. 7i). This finding is consistent with our previous studies, which showed that DCIS.com-shSIM2 xenografts leads to increased invasion and metastasis, independent of tumor growth58.
As previous studies have shown that loss of SIM2s leads to an increase in lung metastasis in xenografts, we hypothesized that S115A tumors would have increased lung metastasis58. As such, RNA was isolated from mouse lungs, and RT-qPCR analysis was performed with human-specific β−2-globulin primers58. We found an increase in human specific β−2-globulin positive lungs in mice that were injected with S115A cells compared to either the SIM2s-FLAG or control groups (Fig. 7j, k). To further validate these findings, we stained lung sections gathered from these mice with H&E. In two of the five mice that were injected with S115A, dense masses of cells, with defined boundaries could be identified in the lung tissues (Fig. 7l). Together, these findings link SIM2s-dependent DDR and HR to inhibition of EMT and DCIS progression.
Discussion
It is well recognized that genomic instability is a crucial factor in breast cancer progression41. A great deal of research has focused on key components of the DDR pathway, including BRCA1, BRCA2, ATM, and 53BP124, 25, 76. Possessing a mutated BRCA allele increases an individual’s lifetime breast cancer risk by more than 10-fold, raising the risk from 0.08 to 0.8246, 61. A growing body of evidence shows that BRCA1 or BRCA2 mutations are highly prevalent in not only familial breast cancer, but also in sporadic tumors9, 31. Of note, these tumors are typically highly invasive and non-responsive, with as many as 68% of these tumors being TNBC 43.
In this study, we show that loss of SIM2s leads to increased genomic instability and correlates with progression from DCIS to IDC (Fig. 1, 2). Of interest is the finding that loss of SIM2s impairs HR by impairing RAD51 loading in response to IR, abrogating a major DDR pathway (Fig. 5). Failure of a cell to initiate this crucial step has been linked with an increased incidence of SNPs, chromosomal abnormalities5, 23, 34, 35, 64. These findings parallel previous studies demonstrating that the loss of BRCA1 results in increased γH2AX foci and sensitivity to mutagenic agents (Fig. 2, 6)55, 65, 77, 78. Previous studies have also suggested that BRCA mutant carriers exhibit higher-grade DCIS cases than would be expected in sporadic tumors75. This correlation between defects in HR machinery and more severe tumorigenesis may explain the negative correlation between SIM2s expression and progression to IDC (Fig. 1).
Importantly, loss of SIM2s sensitizes cells to synthetic lethality treatments (Fig. 6). This strategy for targeted tumor treatment is already being employed with varying levels of success for individuals presenting BRCA1 and BRCA2 deficiencies8, 19. Preliminary results from studies with PARP inhibitors in combinational therapies have shown a 51% pathological complete response (up from 26% in control individuals) in TNBC individuals52.
The regulation of orphan bHLH-PAS family members, including SIM2s, has remained elusive3, 6, 15. Here we have shown a novel mechanism for the stabilization of SIM2s in response to IR (Fig. 3). The discovery of a new factor involved in ATM signaling pathways could have clinical implications as improper ATM signaling has been suggested to result in a 2–5× increase in breast cancer risk, which can be seen in individuals with Ataxia-telangiectasia50, 62, 63.
ATM has previously been identified as a necessary component in the repair of heterochromatic DNA DSBs26. Interestingly, the repair of heterochromatic DNA via HR is both temporally and spatially separated from NHEJ, as HR repair sites are moved to the nuclear periphery prior to repair42, 66. This is thought to occur in order to maintain genomic fidelity by preventing improper recombinational events in highly repetitive sequences66. Our finding that there is a delayed response in γH2AX resolution with loss of SIM2s is consistent with this temporal shift and suggests that SIM2s is involved in HR (Fig. 2).
EMT, typified by the activation of molecular pathways that promote loss of epithelial character, is known to play an important role in breast cancer invasion and metastatic progression59. During cancer progression, these processes are thought to promote the breakdown of the basement membrane, leading to increased invasion into the surrounding stroma and metastasis33, 40. Previously, it had been assumed that EMT and DDR were not mechanistically linked. More recently evidence has suggested that EMT-inducing transcription factors, including SLUG and TWIST, play a role in cancer progression by promoting EMT and overriding DDR through inhibition of CDKN1A (p21)2. Interestingly, loss of SIM2s correlates with a more metastatic phenotype (Fig. 1, 7). The S115A point mutation is sufficient to imbue mesenchymal characteristics on what would normally be a highly differentiated xenograft (Fig. 7). This transition includes an increase in the basal marker K14 and a decrease in E-cadherin. S115A tumors also exhibit large necrotic centers, a condition associated with group 2 and 3 (intermediate and high-grade) lesions on the Van Duys classification scale (Fig. 7)60. Further, the S115A mutation impairs DDR, showing a significant decrease in γH2AX resolution after treatment with IR (Fig. 4). With the dual role of this SNP both in impairing DDR and transitioning cells to a more mesenchymal phenotype, SIM2s could bridge the gap between DDR and EMT.
Here we provide evidence that SIM2s impedes DCIS progression. We also provide a mechanism for SIM2s activation in DDR and regulation of genomic stability. As DCIS is a non-obligate precursor to IDC, the identification of new diagnostic markers that correlate with an increased risk for progression to IDC is critically important13. Given the strong correlation between loss of WT SIM2s and metastatic potential, SIM2s is an ideal candidate for genetic screenings.
METHODS
Cell Culture.
MCF7 and SUM159 cells were from American Type Culture Collection (ATCC) and were maintained in accordance with their guidelines. MCF10-DCIS.com cells were generously donated by Dr. Dan Medina (Baylor College of Medicine, Houston, TX, USA).
Generation of cell lines.
Point mutations in the SIM2 gene were generated via long cDNA synthesis (Invitrogen). Plasmids were amplified using Subcloning Efficiency™ DH5α™ competent cells (Life Technologies). Plasmid DNA was isolated using the HiPure Plasmid Maxiprep kit (Life Technologies) or the ZymoPURE Plasmid DNA Isolation Kit (Zymo Research). Viral transduction was then preformed as previously described39. Puromycin selection (2μg/mL) was started the following day and maintained for a week.
shSIM2 sequences.
The shSIM2 was generated by inserting 5’ - GAT CCG GTC GTT CTT TCT TCG AAT TTC AAG AGA ATT CGA AGA AAG AAC GAC CTC TTT TTT GGA AA-3’ into pSilencer U6-retro 5.1 shRNA vector (Ambion) as previously described39. The specificity of this shSIM2 was confirmed by generating a second shSIM2 by inserting 5’-GAT CCG GTC ACC ACC AAA TAC TAC TTC AAG AGA-3’ into pSilencer U6-retro 5.1 shRNA vector (Ambion) as previously described73.
drGFP homologous recombination assay.
Plasmids were amplified and harvested as above. Eight micrograms of drGFP (Addgene) and pCBA-SCEI (Addgene) were mixed with GeneJuice (EMD Millipore) in Opti-MEM (Life Technologies) and incubated at room temperature for 15 minutes. The mixture was added to HEK293 cells and incubated at 37°C for 2 hours. Cells were trypsinized and pelleted prior to resuspension in PBS. The fraction of GFP reversion was assessed by flow cytometry (BD Acurri C6). Ten thousand events were counted.
RNA isolation and real-time qPCR (RT-qPCR).
As previously described58. Primers can be found in Supplemental Table 1.
Clonogenic survival assay.
Cells were plated at equal numbers on 60mm tissue culture dishes and irradiated using a RS 2000 (Rad Source) at indicated doses. Cells were immediately washed with PBS, trypsinized (Life Technology), pelleted at 200 × g (Eppendorf 5710R), and resuspended in growth media. Viable cells were counted using a Cellometer Auto 1000 (Nexcelom Bioscience LLC) prior to plating 500 cells/plate on 10cm tissue culture dishes. Cells were incubated at 37°C and 5% CO2 for 7 days and then washed with PBS before being fixed with 4% paraformaldehyde (Santa Cruz). Cells were stained with 0.01% crystal violet (Sigma-Aldrich) and washed twice with H2O. Dishes were imaged using a ChemiDoc MP (Bio-Rad), and colonies were counted using ImageJ software. Percent survival was calculated as previously described 22.
Survival Assay.
10,000 cells were plated into 60mm plates and dosed with the indicated concentrations of Olaparib (Cayman Chemical Company). Cells were incubated at 37°C and 5% CO2 for 4 days before being trypsinized and counted on a Cellometer Auto 1000 (Nexcelom Bioscience LLC).
Immunoblotting and zymography.
Cells were washed with cold PBS and lysed in high salt buffer (50mM HEPES, 500mM NaCl, 1.5mM MgCl2, 1mM EDTA, 10% glycerol, 1% Triton X-100, pH 7.5) supplemented with 1mM Na3VO4 (Sigma) and 1mM complete ULTRA tablets mini EDTA-free Easy pack (Roche). Protein concentration was determined using the DC Protein Assay (Bio-Rad) with BSA as a standard. Immunoblotting and zymography were then conducted as previously described39. Antibodies can be found in supplemental table 2. Blots were imaged on a ChemiDoc MP (Bio-Rad) after incubating in ProSignal Pico ECL Spray (Genesee Scientific) for 3 minutes.
ATM inhibition.
Indicated cells were pre-treated with 10μM KU55933 (Cayman Chemical Company) for 2 hours before being dosed with 2GYs ionizing radiation (IR). Protein was harvested from plates as described above and analyzed via immunoblotting.
Co-immunoprecipitation.
Cells were lysed in high salt buffer 12 hours after treatment. 2g of protein lysate were mixed with 200μl of anti-FLAG M2 beads (Sigma, M8823) or IgG control beads (Cell Signaling, 5873S) after equilibrating the beads, IP was conducted according to the manufacturer’s instructions β-mercaptoethanol was added prior to boiling for an additional 3 minutes and immunoblotting.
Immunofluorescent (IF) staining of cells.
IF was conducted as previously described39. Antibody concentrations can be found in supplemental table 2. All images were taken using a Zeiss 780 confocal microscope.
Xenografts.
4–6-month-old, female Nu/Nu mice were purchased from Jackson Laboratory and singly housed. Sample size was determined by previous studies using this cell line and mouse model systems. We have previously found that (n=5) is sufficient to provide a significant difference in tumor growth and metastasis58. The indicated cells were mixed 1:1 with Matrigel (Corning) and kept on ice. Seventy-five thousand cells were then injected into each flank of nude mice. Mice were monitored daily for tumor growth and were euthanized once tumors reached a critical size, in accordance with IACUC procedures. During all subsequent experiments, scientists were blinded, and only made aware of the groups after data had been collected.
Immunostaining tissue sections.
IHC was performed as previously described 57
Tissue Micrographs.
Tissue micrographs containing DCIS and DCIS with localized IDC were provided in collaboration with Dr. Fariba Behbod (University of Kansas Medical Center, Kansas City, KS). ER, PR, HER2, p53, and Ki67 status of patients was known. Following placement in preservation media (LiforCell, Lifeblood Medical, Inc.), biopsy tissue was stored at 4°C until processing, as previously described17, 69.
Cell migration and invasion.
25,000 cells suspended in serum-free media were plated in FluoroBlok Cell Culture Inserts (Corning) that either contained Matrigel (invasion) or did not (migration). Invasion and migration assays were then conducted as previously described39.
Statistical Analysis.
To address scientific rigor, all experiments in cell lines and xenografts were conducted in biological triplicates at a minimum, technical duplicates, and repeated three times. Normal distribution was confirmed before conducting two-tailed Student’s t-test. Likelihood ratio and Pearson statistical test were used for goodness of fit comparisons. Significance was considered at p<0.05.
Study Approval.
Animal studies were approved by the Texas A&M University Laboratory Animal Care Committee. Patients gave written informed consent for participation in this University of Kansas Medical Center Institutional Review Board approved study, which allowed collection of de-identified surgical tissue for research.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank the Histology Core Facility at Texas A&MUniversity College of Veterinary Medicine & Biomedical Sciences for tissue preparation and H&E staining and the University of Kansas Medical Center for providing the human samples, tissue microarray, and pathology information. Confocal microscopy was performed in the Texas A&M University College of Veterinary Medicine & Biomedical Sciences Image Analysis Laboratory, supported by NIH-NCRR (1S10RR22532–01) grant.
We acknowledge support from the University of Kansas (KU) Cancer Center’s Biospecimen Repository Core Facility staff for helping obtain human specimens. The authors also acknowledge support from the KU Cancer Center’s Cancer Center Support Grant (P30 CA168524).
FUNDING
National Cancer Institute R21CA190941 (WP); R01HD083952 (CO-PI WP, MR); R21CA185460 (FB)
Department of Defense (DOD-CDMRP) W81XWH-11–1-0158 (KCS)
American Association for Cancer Research (AACR) – 2014 Breast Cancer Research Foundation – Translational Breast Cancer research (FB)
Footnotes
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
SUPPLEMENTARY DATA
Supplementary Data are available at Oncogene online.
REFERENCES
- 1.Aleman MJ, DeYoung MP, Tress M, Keating P, Perry GW, Narayanan R. Inhibition of Single Minded 2 gene expression mediates tumor-selective apoptosis and differentiation in human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America 2005; 102: 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer cell 2008; 14: 79–89. [DOI] [PubMed] [Google Scholar]
- 3.Bardos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta 2005; 1755: 107–120. [DOI] [PubMed] [Google Scholar]
- 4.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res 2009; 11: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 2006; 45: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 6.Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH-PAS proteins in cancer. Nat Rev Cancer 2013; 13: 827–841. [DOI] [PubMed] [Google Scholar]
- 7.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004; 4: 1633–1649. [DOI] [PubMed] [Google Scholar]
- 8.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434: 913–917. [DOI] [PubMed] [Google Scholar]
- 9.Burgess M, Puhalla S. BRCA 1/2-Mutation Related and Sporadic Breast and Ovarian Cancers: More Alike than Different. Front Oncol 2014; 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001; 276: 42462–42467. [DOI] [PubMed] [Google Scholar]
- 11.Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. The New England journal of medicine 2004; 350: 1430–1441. [DOI] [PubMed] [Google Scholar]
- 12.Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci 2012; 125: 3529–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowell CF, Weigelt B, Sakr RA, Ng CK, Hicks J, King TA et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol 2013; 7: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowell CF, Weigelt B, Sakr RA, Ng CK, Hicks J, King TA et al. Progression from ductal carcinoma in situ to invasive breast cancer: Revisited. Molecular oncology 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 2011; 124: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y, Yamaguchi H, Wei Y, Hsu JL, Wang HL, Hsu YH et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med 2016; 22: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsarraj HS, Valdez KE, Hong Y, Grimm SL, Ricci LR, Fan F et al. NEMO, a Transcriptional Target of Estrogen and Progesterone, Is Linked to Tumor Suppressor PML in Breast Cancer. Cancer Res 2017; 77: 3802–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein DJ, Martinu L, Michaud JL, Losos KM, Fan C, Joyner AL. Members of the bHLH-PAS family regulate Shh transcription in forebrain regions of the mouse CNS. Development 2000; 127: 4701–4709. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434: 917–921. [DOI] [PubMed] [Google Scholar]
- 20.Fisher B, Dignam J, Wolmark N, Wickerham DL, Fisher ER, Mamounas E et al. Tamoxifen in treatment of intraductal breast cancer: National Surgical Adjuvant Breast and Bowel Project B-24 randomised controlled trial. Lancet 1999; 353: 1993–2000. [DOI] [PubMed] [Google Scholar]
- 21.Fisher ER, Dignam J, Tan-Chiu E, Costantino J, Fisher B, Paik S et al. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) eight-year update of Protocol B-17: intraductal carcinoma. Cancer 1999; 86: 429–438. [DOI] [PubMed] [Google Scholar]
- 22.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1: 2315–2319. [DOI] [PubMed] [Google Scholar]
- 23.Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 2006; 6: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ 2006; 13: 1003–1016. [DOI] [PubMed] [Google Scholar]
- 25.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S et al. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J Biol Chem 2004; 279: 15402–15410. [DOI] [PubMed] [Google Scholar]
- 26.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular cell 2008; 31: 167–177. [DOI] [PubMed] [Google Scholar]
- 27.Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA et al. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA et al. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene 2009; 28: 1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halvorsen OJ, Rostad K, Oyan AM, Puntervoll H, Bo TH, Stordrange L et al. Increased expression of SIM2-s protein is a novel marker of aggressive prostate cancer. Clin Cancer Res 2007; 13: 892–897. [DOI] [PubMed] [Google Scholar]
- 30.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics 2010; 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst 2002; 94: 1396–1406. [DOI] [PubMed] [Google Scholar]
- 32.Houghton J, George WD, Cuzick J, Duggan C, Fentiman IS, Spittle M et al. Radiotherapy and tamoxifen in women with completely excised ductal carcinoma in situ of the breast in the UK, Australia, and New Zealand: randomised controlled trial. Lancet 2003; 362: 95–102. [DOI] [PubMed] [Google Scholar]
- 33.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol 2005; 17: 548–558. [DOI] [PubMed] [Google Scholar]
- 34.Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR et al. Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 2005; 65: 7612–7621. [DOI] [PubMed] [Google Scholar]
- 35.Kim TM, Ko JH, Hu L, Kim SA, Bishop AJ, Vijg J et al. RAD51 mutants cause replication defects and chromosomal instability. Molecular and cellular biology 2012; 32: 3663–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar G, Redick M, Dixon GD. New techniques for mammography screening: advantages and limitations. Missouri medicine 2005; 102: 138–141. [PubMed] [Google Scholar]
- 37.Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis 2007; 28: 259–266. [DOI] [PubMed] [Google Scholar]
- 38.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol 2010; 4: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Molecular and cellular biology 2008; 28: 1936–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. The Journal of cell biology 2006; 172: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loeb LA. A mutator phenotype in cancer. Cancer Res 2001; 61: 3230–3239. [PubMed] [Google Scholar]
- 42.Lorkovic ZJ, Berger F. Heterochromatin and DNA damage repair: Use different histone variants and relax. Nucleus 2017; 8: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomarkers Prev 2012; 21: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metz RP, Kwak HI, Gustafson T, Laffin B, Porter WW. Differential transcriptional regulation by mouse single-minded 2s. J Biol Chem 2006; 281: 10839–10848. [DOI] [PubMed] [Google Scholar]
- 45.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J 2000; 19: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman B, Austin MA, Lee M, King MC. Inheritance of human breast cancer: evidence for autosomal dominant transmission in high-risk families. Proceedings of the National Academy of Sciences of the United States of America 1988; 85: 3044–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okui M, Yamaki A, Takayanagi A, Kudoh J, Shimizu N, Shimizu Y. Transcription factor single-minded 2 (SIM2) is ubiquitinated by the RING-IBR-RING-type E3 ubiquitin ligases. Exp Cell Res 2005; 309: 220–228. [DOI] [PubMed] [Google Scholar]
- 48.Page DL, Dupont WD, Rogers LW, Jensen RA, Schuyler PA. Continued local recurrence of carcinoma 15–25 years after a diagnosis of low grade ductal carcinoma in situ of the breast treated only by biopsy. Cancer 1995; 76: 1197–1200. [DOI] [PubMed] [Google Scholar]
- 49.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999; 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 2006; 38: 873–875. [DOI] [PubMed] [Google Scholar]
- 51.Rudloff U, Jacks LM, Goldberg JI, Wynveen CA, Brogi E, Patil S et al. Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010; 28: 3762–3769. [DOI] [PubMed] [Google Scholar]
- 52.Rugo HS, Olopade OI, DeMichele A, Yau C, van ‘t Veer LJ, Buxton MB et al. Adaptive Randomization of Veliparib-Carboplatin Treatment in Breast Cancer. The New England journal of medicine 2016; 375: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nature cell biology 2015; 17: 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders ME, Schuyler PA, Dupont WD, Page DL. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer 2005; 103: 2481–2484. [DOI] [PubMed] [Google Scholar]
- 55.Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Molecular cell 2014; 54: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nature reviews Molecular cell biology 2006; 7: 517–528. [DOI] [PubMed] [Google Scholar]
- 57.Scribner KC, Wellberg EA, Metz RP, Porter WW. Singleminded-2s (Sim2s) promotes delayed involution of the mouse mammary gland through suppression of Stat3 and NFkappaB. Mol Endocrinol 2011; 25: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scribner KC, Behbod F, Porter WW. Regulation of DCIS to invasive breast cancer progression by Singleminded-2s (SIM2s). Oncogene 2013; 32: 2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev 2003; 120: 1351–1383. [DOI] [PubMed] [Google Scholar]
- 60.Silverstein MJ, Poller DN, Waisman JR, Colburn WJ, Barth A, Gierson ED et al. Prognostic classification of breast ductal carcinoma-in-situ. Lancet 1995; 345: 1154–1157. [DOI] [PubMed] [Google Scholar]
- 61.Society AC. Cancer Facts & Figures 2018. American Cancer Society: Atlanta, 2018. [Google Scholar]
- 62.Swift M, Reitnauer PJ, Morrell D, Chase CL. Breast and other cancers in families with ataxia-telangiectasia. The New England journal of medicine 1987; 316: 1289–1294. [DOI] [PubMed] [Google Scholar]
- 63.Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst 2005; 97: 813–822. [DOI] [PubMed] [Google Scholar]
- 64.Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R et al. Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res 1997; 57: 1222–1227. [PubMed] [Google Scholar]
- 65.Trenz K, Schutz P, Speit G. Radiosensitivity of lymphoblastoid cell lines with a heterozygous BRCA1 mutation is not detected by the comet assay and pulsed field gel electrophoresis. Mutagenesis 2005; 20: 131–137. [DOI] [PubMed] [Google Scholar]
- 66.Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A et al. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Molecular cell 2016; 63: 293–305. [DOI] [PubMed] [Google Scholar]
- 67.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 2004; 4: 814–819. [DOI] [PubMed] [Google Scholar]
- 68.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376: 235–244. [DOI] [PubMed] [Google Scholar]
- 69.Valdez KE, Fan F, Smith W, Allred DC, Medina D, Behbod F. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. J Pathol 2011; 225: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Cleef A, Altintas S, Huizing M, Papadimitriou K, Van Dam P, Tjalma W. Current view on ductal carcinoma in situ and importance of the margin thresholds: A review. Facts, views & vision in ObGyn 2014; 6: 210–218. [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H, Adhikari S, Butler BE, Pandita TK, Mitra S, Hegde ML. A Perspective on Chromosomal Double Strand Break Markers in Mammalian Cells. Jacobs J Radiat Oncol 2014; 1. [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Ding Q, Fujimori H, Motegi A, Miki Y, Masutani M. Loss of CtIP disturbs homologous recombination repair and sensitizes breast cancer cells to PARP inhibitors. Oncotarget 2016; 7: 7701–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wellberg E, Metz RP, Parker C, Porter WW. The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation. Development 2010; 137: 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J Biol Chem 2002; 277: 10236–10243. [DOI] [PubMed] [Google Scholar]
- 75.Yang RL, Mick R, Lee K, Graves HL, Nathanson KL, Domchek SM et al. DCIS in BRCA1 and BRCA2 mutation carriers: prevalence, phenotype, and expression of oncodrivers C-MET and HER3. J Transl Med 2015; 13: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci 2004; 95: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zamborszky J, Szikriszt B, Gervai JZ, Pipek O, Poti A, Krzystanek M et al. Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou C, Smith JL, Liu J. Role of BRCA1 in cellular resistance to paclitaxel and ionizing radiation in an ovarian cancer cell line carrying a defective BRCA1. Oncogene 2003; 22: 2396–2404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.