Abstract
Ataxia telangiectasia mutated and RAD3 related (ATR) protein kinase plays critical roles in ensuring DNA replication, DNA repair, and cell cycle control in response to replication stress, making ATR inhibition a promising therapeutic strategy for cancer treatment. To identify genes whose loss makes tumor cells hypersensitive to ATR inhibition, we performed CRISPR/Cas9-based whole-genome screens in 3 independent cell lines treated with a highly selective ATR inhibitor, AZD6738. These screens uncovered a comprehensive genome-wide profile of ATR inhibitor sensitivity. From the candidate genes, we demonstrated that RNASEH2 deficiency is synthetic lethal with ATR inhibition both in vitro and in vivo. RNASEH2-deficient cells exhibited elevated levels of DNA damage and, when treated with AZD6738, underwent apoptosis (short-time treated) or senescence (long-time treated). Notably, RNASEH2 deficiency is frequently found in prostate adenocarcinoma; we found decreased RNASEH2B protein levels in prostate adenocarcinoma patient derived xenograft (PDX) samples. Our findings suggest that ATR inhibition may be beneficial for cancer patients with reduced levels of RNASEH2 and that RNASEH2 merits further exploration as a potential biomarker for ATR inhibitor-based therapy.
Introduction
The maintenance of genomic integrity is critically important for cell survival. Safeguarding the genome is a challenge because cells face enormous stresses all the time, which include extrinsic stressors, such as ultraviolet light, ionizing radiation, environmental mutagens, and intrinsic stressors, such as reactive metabolic byproducts, RNA-DNA hybrids, and secondary DNA structures1–3. The serine/threonine protein kinase ATR functions in the cellular response to a broad range of DNA-damaging stressors and DNA lesions. ATR is also the major orchestrator of protecting cells from replication-related stresses4,5. Moreover, ATR plays important roles in cell cycle checkpoint regulation, telomere maintenance, meiosis, and the cellular response to mechanical and osmotic stress6–8. These multi-functional nature of ATR, especially its essential functions in maintaining the viability of replicating cells, make ATR a promising target for cancer therapy9–11.
Although ATR has been studied extensively, targeting ATR for cancer therapy has been challenging because ATR is an essential protein in mouse and human cells5–8. Several studies found that ATR inhibition induces synthetic lethality in cancer cells with defects in DNA repair pathways, such as loss of ERCC1 or XRCC111,12. ATR inhibition may also have a synergistic antitumor effect on cell lines with specific genetic alterations, including ATM, TP53, or ARID1A deficiency13–15. In xenograft models, ATR inhibition sensitized cancer cells to chemotherapy and radiotherapy16,17. However, there remains a pervasive view that ATR inhibitors could be toxic in patients, which has hampered advances in targeting ATR for cancer therapy. A comprehensive understanding of the genes with a synthetic lethal relationship with ATR inhibition will not only provide new markers for use in future clinical studies, but also reduce the misconception that ATRi are highly toxic.
Genome-wide CRISPR screening is a powerful approach for the identification of genetic interactions in human cells. For example, several studies using pooled CRISPR/Cas9 libraries demonstrated that CRISPR technology offers vast improvement in mammalian genetic screens18–21. Genome-wide CRISPR knockout screens in human cell lines uncovered far more essential genes than those performed using RNA interference screens22. CRISPR technology has several advantages over RNA interference-based screens: (1) CRISPR technology generates depletion of genes in a more consistent manner at the genomic level; (2) CRISPR screens have a much higher sensitivity for detecting genes with low mRNA expression levels or short mRNA half-lives; and (3) CRISPR technology could reduce the heterogeneity of RNA interference among different cell lines18,22. Moreover, better standards and analytic tools for assessing genome-wide screens have been established and can now be applied to functional genomic screens22–24.
To get a comprehensive understanding of the genes with a synthetic lethal relationship with ATR inhibition and identify possible targets for ATRi therapy, we performed genome-wide CRISPR-based functional screens to detect genes that, when depleted, showed synthetic lethality with ATR inhibition. AZD6738, a highly selective, orally active, and bioavailable ATR kinase inhibitor, shows a good margin of selectivity against other kinases in broad in vitro assay screens (0/442 kinases showed > 50% inhibition at 1 μM) and no significant inhibition of other PI3K-like kinases, such as DNA-PK, ATM, or mTOR25,26. Using AZD6738, we conducted single guide RNA (sgRNA) screens in 293A, HCT116, and MCF10A cells and obtained a comprehensive list of genes associated with ATR inhibition.
The RNASEH2 complex plays important roles in nucleic acid metabolism to preserve genome stability27. RNASEH2 contains 3 subunits: a catalytic subunit, RNASEH2A, and 2 noncatalytic subunits, RNASEH2B and RNASEH2C. Dysfunction mutations in any of the 3 subunits of the human RNASEH2 complex can result in Aicardi-Goutières syndrome (AGS), a severe autoimmune disease28,29. RNASEH2 mutations could invoke cGAS-STING pathway which is believed to be a possible cause of AGS30,31. RNASEH2 is also essential for removing ribonucleotides incorporated in genomic DNA during replication as well as R-loop resolving32–35. Depletion of RNASEH2 in human cells could impair cell cycle progression and induce genome stability30,33. Moreover, it was reported recently that ribonucleotide incorporation caused by deletion of RNASEH2 could result in PARP-trapping lesions, which makes it a promising clinical target36.
In this study, through genome-wide CRISPR screens, we found that RNASEH2 depletion led to dramatic synthetic lethality with ATR inhibition. We validated that RNASEH2 deficiency induced ATRi sensitivity both in vitro and in vivo. We found that RNASEH2 deficiency not only induces DNA damage but also promotes apoptosis and senescence when ATR is inhibited. Notably, RNASEH2 reduction or depletion is frequently observed in prostate adenocarcinoma. Collectively, these data provide a rationale for future exploration of RNASEH2 deficiency as a potential biomarker for ATRi-based cancer therapy.
Results
Pooled genome-wide CRISPR/Cas9-based screens in 3 cell lines with ATR inhibition
To identify genes whose depletion endows cells with sensitivity or resistance to ATR inhibition, we carried out pooled CRISPR/Cas9-based screens in 293A, HCT116, and MCF10A cells treated with a highly selective ATR inhibitor, AZD6738. The guide RNA (gRNA) library we chose was the Toronto Knock Out Library v3 (TKOv3), which contains 70,948 gRNAs targeting 18,053 protein-coding genes37. Cells were infected with viruses encompassing the TKOv3 library at a low multiplicity of infection (MOI) (< 0.3), and infected cells were selected with puromycin. Cells were treated with either dimethyl sulfoxide (DMSO) as a control or with AZD6738 and passaged for about 20 doubling cycles. Cells were collected at each passage point, and each group had duplicate repeats. The initial cell populations were marked as _T0, and the final cell populations were marked as _DMSO or _ATRi depending on the treatment. Genomic DNA was extracted and labeled with barcodes using a polymerase chain reaction (PCR). The final PCR amplicons were deep-sequenced and analyzed (Figure 1A). For these screens, we chose IC20 (the concentration that inhibits cell proliferation by 20%) as the treatment concentration. This concentration was used to reduce the random loss of sgRNA information caused by high chemical concentrations and at the same time ensure that some cells were killed by drug treatment with every passage (Supplementary Figure 1A). The proportion of surviving cells were confirmed at each time point (Supplementary Figure 1B).
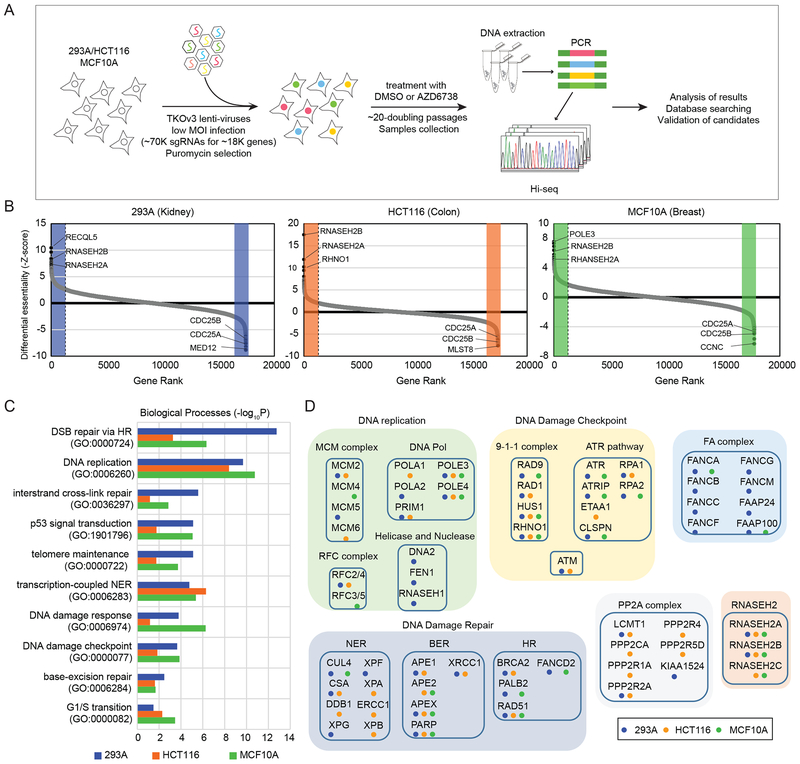
Figure 1. Pooled CRISPR/Cas9-based genome-wide screens were performed in 3 cell lines.
A. Schematic representation of the workflow for CRISPR screens performed in 293A, HCT116, and MCF10A cells.
B. Ranking of ATRi co-essential genes based on drugZ analysis of the results of CRISPR/Cas9-based screening in 3 cell lines. The z-score was used to define a possible synthetic lethal interaction with ATR inhibition. All genes targeted by the Toronto Knock Out Library v3 were scored according to the fold change of levels of their sgRNAs (ATRi treatment vs. DMSO treatment). High-confidence candidate genes, those with a false discovery rate (FDR) of less than 0.05, are shaded in color. Genes whose loss of function led to ATRi sensitivity appear on the left side, and genes whose loss of function led to ATRi resistance appear on the right side. Some high-confidence genes are marked.
C. The top10 significantly enriched Gene ontology (GO) terms (P< 0.01). High-confidence candidate genes from B in each cell lines are categorized based on the biological process. Different color represent different cell line. Blue: 293A, Orange: HCT116, Green: MCF10A.
D. Candidate ATRi co-essential genes grouped according to their roles in specific pathways. The colored dots indicate the cell lines in which the candidate genes were identified. Blue: 293A, Orange: HCT116, Green: MCF10A.
Abbreviations: TKOv3, Toronto Knock Out Library v3; MOI, multiplicity of infection; sgRNA, single guide RNA; DMSO, dimethyl sulfoxide; PCR, polymerase chain reaction; T0, time point 0 (baseline); ATRi, ATR inhibitor
For these screens, we ensured that the depth of sequencing (> 200-fold of gRNA numbers) and distribution of sequencing reads were more than adequate for subsequent bioinformatic analysis (Supplementary Figure 1C, 1D). To determine whether the CRISPR-based screens were successful, we analyzed changes in essential genes and nonessential genes from the starting time point (T0) to the ending time point (T21) as previously described38. We found that sgRNAs targeting essential genes were reduced while sgRNAs targeting nonessential genes remained the same between these time points, indicating that the gRNAs worked well in these screens (Supplementary Figure 1E). We then used the Bayesian Analysis of Gene Essentiality (BAGEL) algorithm to calculate a log Bayes factor for each gene24. The Pearson correlation coefficients of the Bayes factor distributions for all pairs of screens are shown in Supplementary Figure 1F. Notably, the high correlation of 293A_T0, HCT116_T0, and MCF10A_T0 (correlation index> 0.9) indicated the consistency of the screen results at the starting time point. The high correlations among different groups between 293A, HCT116, and MCF10A screens (> 0.7) indicated that the results of these 3 screens were comparable. Precision-recall curves were also used to evaluate the screens’ performance (Supplementary Figure 1G) as described previously22. All 3 screens showed high performance, with more than 800 essential genes identified in each screen at a false discovery rate (FDR) of 5%. Among them, 540 essential genes were identified in all 3 screens (Supplementary Figure 1H). Taken together, these analyses demonstrated that the screens were successful and that the information derived from them is likely to be reliable.
DrugZ analysis revealing genes whose depletion affects ATRi sensitivity
Next, we focused on looking for genes whose depletion specifically affected cell viability with ATRi. Drug Z analysis was used to compare the AZD6738-treated group and the DMSO-treated group in each screened cell line. The genes were ranked by their drugZ scores and comprehensive information was included in Supplementary Table 1. The colored regions of Figure 1B show genes with an FDR of less than 0.05 and several top-ranked genes are highlighted.
To provide an unbiased view of the functional categories of genes identified in our screens, we performed gene set enrichment analysis for these genes. We first looked for co-essential genes—genes whose depletion made cells more sensitive to ATRi. The top 10 gene sets identified by gene ontology analysis are listed in Figure 1C. Seven of the top 10 sets of ATRi co-essential genes were linked with the cellular response to DNA damage, including nucleotide excise repair, base excision repair, interstrand cross-link repair, and homologous recombination (HR) repair. Moreover, the top 2 enriched gene sets were double-strand break (DSB) repair via HR and DNA replication, suggesting that cells with defective DNA replication and/or HR are more sensitive to ATR inhibition than are cells in which these mechanisms are intact. The identified ATRi co-essential genes were also categorized by their functions and protein complexes (Figure 1D). Genes involved in the ATR pathway and its upstream factors (RPA1, RPA2, ATRIP, ATR, ETAA1, CLSPN, RAD9-HUS1-RAD1, and RHNO1) were significantly enriched; each of these genes was identified in at least 2 screens. These results suggest that a defective ATR-dependent signaling pathway makes cells more sensitive to modest ATR inhibition. DNA replication-related complexes (MCM complex, DNA polymerase, helicases, and RFC complex), the Fanconi anemia complex, and genes participating in various DNA damage-repair pathways were also enriched in our dataset. Because defects in many of these genes, such as BRCA2, PALB2, POLE3, and POLE4, are linked with cancer development9,39,40, these results suggest that ATR inhibition could be an effective treatment strategy for cancers harboring defects in these DNA repair and/or replication-associated genes. The other 2 gene sets identified with high confidence in our screens were the PP2A complex and the RNASEH2 complex. To further validate our screen results and uncover new ATRi co-essential genes, we combined the high-confidence gene lists for the 3 screens and focused on the ones that overlapped (Figure 2A).
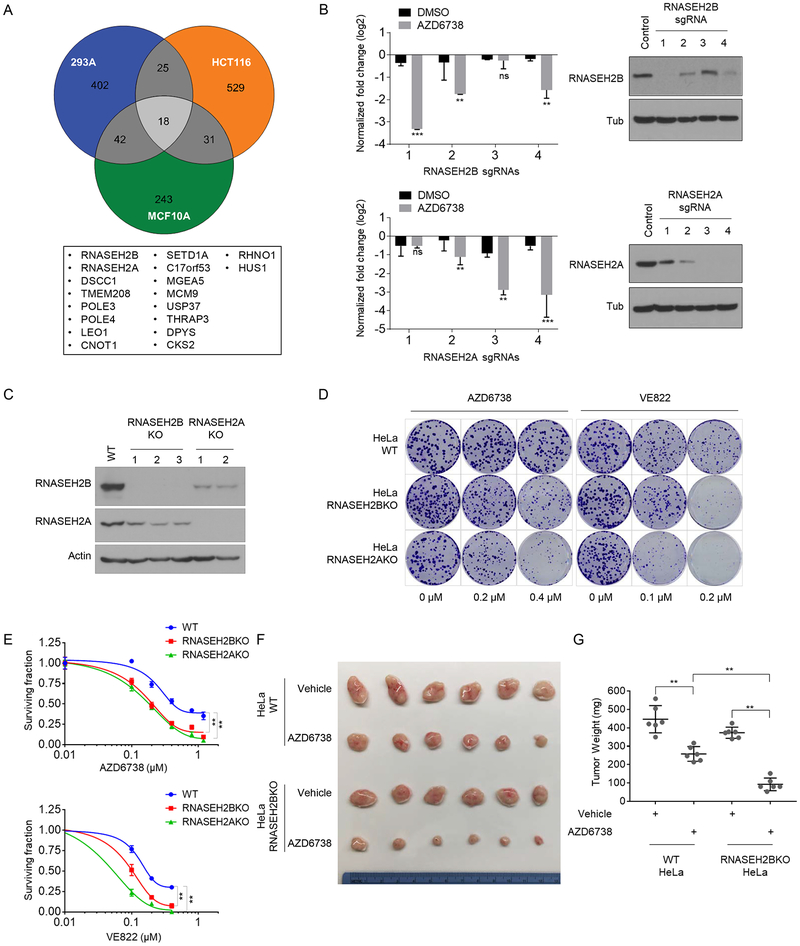
Figure 2. ATRi co-essential genes identified in CRISPR-based sgRNA screens.
A. Venn diagram showing overlapping identified ATRi co-essential genes. The genes that overlapped in all 3 cell lines are listed below the diagram.
B. Normalized sgRNA fold changes in DMSO treated group and AZD6738 treated group from the screen conducted in 293A cells. The counts of each sgRNA were divided by the counts of sgRNA in T0 (start point). Bar charts illustrating the log2 fold change of the indicated sgRNAs from DMSO-treated cells and ATRi-treated cells at Day21 (T21). Student T tests were performed to evaluate differences between the groups. ns, not significant; **P<0.01. Western blot were performed to determine the efficiency of sgRNAs. 293A cells were infected with lentiviruses expressing the indicated sgRNAs at low MOI and selected with puromycin. Cell lysates were blotted with the indicated antibodies.
C. Validation of RNASEH2B and RNASEH2A knockout (KO) in HeLa cells by Western blotting using indicated antibodies. Note the marked decrease in the RNASEH2B protein level when RNASEH2A was knocked out. In contrast, RNASEH2A protein levels were only slightly decreased when RNASEH2B was knocked out.
D. Loss of RNASEH2B/A sensitize cells to ATRi treatment in clonogenic assay. Images of colonies in colony formation assay were presented. HeLa wild-type (WT), RNASEH2B KO, and RNASEH2A KO cells were exposed to increasing concentrations of different ATR inhibitors and grew for 12 days. Results are representative of duplicate biological experiments.
E. Dose-response survival curves of HeLa-WT, HeLa-RNASEH2BKO, HeLa-RNASEH2AKO cells exposed to increasing concentration of AZD6738 (upper) or VE822 (lower). Error bar represent s.d. (n=3), ANOVA p-value<0.01, results are representative of duplicate biological experiments.
F. Loss of RNASEH2B sensitizes HeLa derived xenograft tumors to ATRi treatment. Two million of WT or RNASEH2BKO HeLa cells were subjected to xenograft assay and treated with AZD6738 (60mg/kg, 5 × weekly by oral gavage) or drug vehicle. Mice were treated for 3 weeks and then sacrificed. Xenograft tumors were shown. n=6.
G. The weights of the tumors from F were quantified. n=6 mice, mean±s.d. Paired student t-test were used to analyze the result, *P<0.05, **P<0.01.
We also looked for genes whose depletion would make cells resistant to ATR inhibition. The enriched gene sets included genes involved in transcription regulation, mitochondrial regulation, and G2/M transition regulation (Supplementary Figure 2A). Among them, loss of genes in the cyclin C complex (CCNC, MED12, MED13, MED24, CDK8), CDC25A/B, CDK2, and KEAP1 was associated with resistance to ATRi. The genes that overlapped in 2 or more screens are presented in Supplementary Figure 2B. Among the overlapping genes, loss of MED12 is known to be highly correlated with multiple-drug resistance41. Similarly, SLFN11 loss also correlates with resistance to many chemotherapeutic agents42,43. Moreover, another CRISPR screen using a different ATRi also reported that CDC25A/B loss can lead to ATRi resistance21. All these results further confirmed the reliability of our screens.
RNASEH2-deficient cells are sensitive to ATR inhibition
Studies of ATRi co-essential genes should allow us not only to uncover potential biomarkers for ATRi-based therapy, but also to identify new components that may function in or together with the ATR-dependent replication stress pathway. In the follow-up experiments, we focused on genes identified with high confidence in all 3 screens. Among them, RNASEH2 was of particular interest because 2 RNASEH2 subunits—RNASEH2A, RNASEH2B, were identified in all 3 screens and the other subunit RNASEH2C was identified in 2 screens. (Figure 1D, Figure 2A).
Deep-sequencing results showed that levels of sgRNAs targeting RNASEH2B or RNASEH2A were significantly decreased in ATRi-treated cells than in control DMSO-treated cells (Figure 2B and Supplementary Table 2). To validate that these sgRNAs targeted the intended genes, we infected 293A cells with a low-MOI lentivirus containing the sgRNAs. Western blotting demonstrated that these sgRNAs indeed efficiently decreased RNASEH2B or RNASEH2A protein levels (Figure 2B). It is of note that the sgRNA efficiency correlated with ATRi sensitivity, which further confirmed the robustness of the screens. We also used shRNAs targeting RNASEH2B and RNASH2A to confirm the screen results. As shown in Supplementary Figure 3A and B, both knocking-down of RNASEH2B and RNASEH2A in 293A cells could lead to sensitivity to ATRi treatment in cell viability assay. This further suggests that the screen results were unlikely to be an off-target sgRNA effect. Moreover, the sgRNAs targeting RNASEH2C were also shown dramatic decreased in ATRi-treated group in HCT116 cells and MCF10A cells (Supplementary Figure 3C and Supplementary table 2). We speculated the less changes of sgRNAs targeting RNASEH2C in 293A cells was due to the variation between screens.
To further demonstrate that RNASEH2 deficiency-induced ATRi sensitivity is a general phenomenon, we generated 2 or 3 independent clones of HeLa-derived KO cell lines for each gene (Figure 2C). Notably, as reported previously30,33,36, KO of RNASEH2A dramatically decreased RNASEH2B protein levels; RNASEH2A protein levels were also decreased in RNASEH2B KO cells (Figure 2C). With these cells, we performed clonogenic survival assays using 2 different ATRi, AZD6738 and VE822. Knockout of RNASEH2B or RNASEH2A led to notable growth inhibition following ATRi treatment (Figure 2D, 2E), suggesting that the synthetic lethality was not specific to AZD6738. To further determine whether this sensitivity was caused by ATR pathway inhibition, we conducted clonogenic assays using a Chk1 inhibitor (LY2606368), an ATM inhibitor (AZD0156), and a DNA-PK inhibitor (NU7441). As shown in Supplementary Figure 4, RNASEH2B KO and RNASEH2A KO cells showed increased sensitivity to Chk1 inhibition, but not to ATM or DNA-PK inhibition. These data suggested that the increased ATRi sensitivity caused by RNASEH2B or RNASEH2A depletion is specific to the ATR pathway.
We next assessed whether the clinical ATRi, AZD6738, could inhibit RNASEH2 deficient tumors in vivo. Nude mice with established xenograft tumors derived from either HeLa-WT or HeLa-RNASEH2BKO cells were treated with either AZD6738 or drug vehicle for 21 days. We found that, WT-HeLa-derived tumor growth could be suppressed by AZD6738 while loss of RNASEH2 significantly enhanced this growth inhibition (Figure 2F, G). We did not perform similar experiments using HeLa-RNASEH2AKO cells, since xenograft tumors of HeLa-RNASEH2AKO grew slowly (Supplementary Figure 5), which is similar with the phenotype as previously reported33. Taken together, these in vitro and in vivo results suggest that RNASEH2 deficiency is able to sensitize tumors to ATR inhibitor.
ATR inhibition enhanced DNA damage induced by RNASEH2 deficiency
To uncover the reasons why loss of RNASEH2A or RNASEH2B led cells to be sensitive to ATRi, we determined the levels of endogenous DNA damage in control and RNASEH2-depleted cells. As expected from previous reports27,44, we observed more intense of γ-H2A.X staining or level in RNASEH2B KO and RNASEH2A KO cells than in wild-type cells (Figure 3A, B, C), indicating that the KO cells bore more spontaneous DNA damage. Consistent with the staining results, we found that ATR phosphorylation (p-ATR-T1989) and Chk1 phosphorylation (p-Chk1-S317) were increased in both RNASEH2B KO and RNASEH2A KO cells (Supplementary Figure 6A). These results suggested that RNASEH2-deficient cells harbor more spontaneous DNA damage than do cells with intact RNASEH2 and the ATR-Chk1 pathway is activated in RNASEH2-deficient cells. We did not find significant difference in the number of 53BP1 foci between control and RNASEH2-depleted cells (Supplementary Figure 6B, C) or significant changes of pChk2 or pATM (Supplementary Figure 6D), suggesting that spontaneous DNA damage observed in RNASEH2-depleted cells is likely attributable to lesions that induce replication stress, but not DSBs.
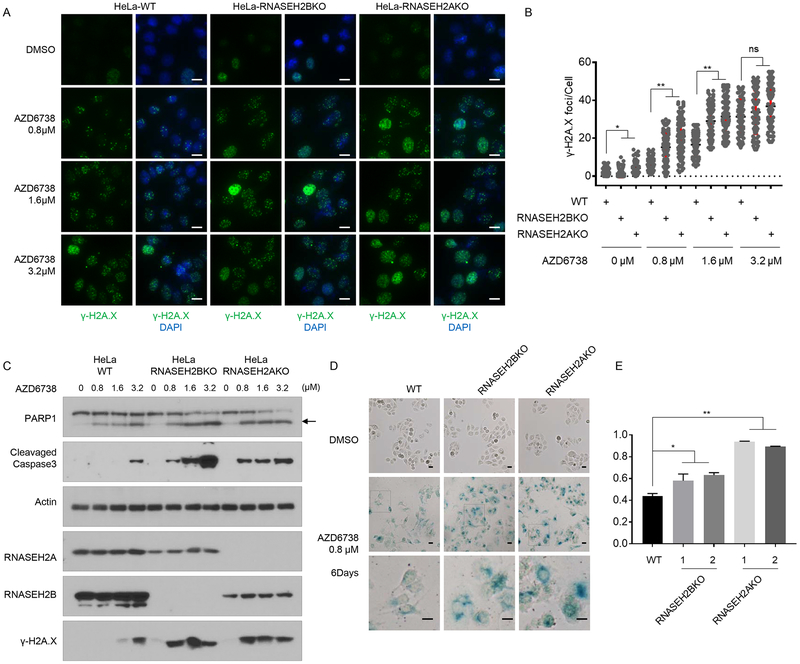
Figure 3. ATRi treatment induced more DNA damage, apoptosis and senescence in RNASEH2 deficient cells.
A. Wild-type (WT) HeLa cells, RNASEH2B KO cells, or RNASEH2A KO cells were treated with DMSO or increasing concentrations of AZD6738 for 48 hrs. Representative immunofluorescence images of γ-H2A.X foci in each cell line were shown. Scale bar=10 μm.
B. Quantification of γ-H2A.X foci in different groups of cells in A. n=100 cells, *P<0.05, **P<0.01, student t-test.
C. ATRi treatment induced apoptosis in RNASEH2 deficient cells. HeLa WT cells, RNASEH2B KO cells, or RNASEH2A KO cells were treated with DMSO or increasing concentrations of AZD6738 for 48 hrs. Cell lysates were blotted with the indicated antibodies. Arrow marked the cleaved PARP1.
D. Prolonged ATRi treatment induced senescence in RNASEH2 deficient cells. HeLa WT cells, RNASEH2B KO cells, or RNASEH2A KO cells were treated with DMSO or 0.8 μM AZD6738 for 6 Days. β-Galactosidase (β-Gal) staining was used to identify senescent cells. The boxed region is enlarged 3 times on the lower panel. Scale bar=10 μm. Results are representative of duplicate biological experiments.
E. Quantification of β-Gal positive staining cells in D (mean±s.d.). ns=not significant, *P<0.05, **P<0.01. Student t-test.
Treatment with ATRi decreased levels of both pATR and pChk1 (Supplementary Figure 4A), suggesting that ATRi treatment was effective. Compared with WT cells, RNASEH2BKO or RNASEH2AKO cells harbored dramatic elevated γ-H2A.X foci and γ-H2A.X levels even at low concentration ATRi-treatment (Figure 3A, B, C). There was no significant differences in the number of γ-H2A.X foci or γ-H2A.X levels between control and RNASEH2-depleted cells after high concentration ATRi treatment (Figure 3A, B, C), which suggest high concentration ATRi-treatment may saturate the γ-H2A.X changes.
RNASEH2 is important for removal of ribonucleotides incorporated into DNA as well as longer RNA-DNA hybrid (R-loop)32,33,35. We checked the DNA-RNA hybrid contents in HeLa-WT and HeLa-RNASEH2 deficient cells treated with DMSO or AZD6738. In our experiment, we did not detect any significant increase of S9.6 signals in RNASEH2 deficient cells in either control or under ATRi treatment (Supplementary Figure 7). We speculated that the ATRi sensitivity of RNASEH2 deficient cells may not be caused by accumulation of R-loop. A recent paper reported that it is likely that the role of RNASEH2 in ribonucleotide excise repair (RER) may be responsible of PAPRi sensitivity in RNASEH2 deficient cells36. Given the similarity between our results and those reported for PAPRi sensitivity, it is likely that the role of RNASEH2 in RER may also be responsible for ATRi sensitivity in these KO cells.
ATRi treatment accelerates apoptosis and senescence in RNASEH2-depleted cells
We checked cleaved PARP1 protein levels and cleavage Caspase-3 protein levels in cells with and without ATRi treatment. We found that compared with WT cells, RNASEH2BKO and RNASEH2AKO cells exhibited significant increase in these apoptotic markers and these increases were associated with ATRi dosasge (Figure 3C). This result suggests that apoptosis may be one of the reasons for ATRi sensitivity in RNASEH2 deficient cells.
We also determined cellular senescence in control and RNASEH2 KO cells with and without ATRi treatment. Senescence-associated β-galactosidase staining was conducted 6 days after cells were exposed to ATRi. Compared to control cells, both RNASEH2B KO and RNASEH2A KO cells exhibited increased β-galactosidase staining (Figure 3D, E), indicating that ATRi treatment accelerates cellular senescence in RNASEH2-depleted cells.
It was reported that RNASEH2 deficiency could invoke cGAS-STING pathway and cGAS-STING pathway is considered as essential for cellular senescence30,31,45. We also checked expression levels of cGAS-STING pathway target genes in HeLa-WT cells and RNASEH2-deficient cells. As shown in Supplementary Figure 8, expression levels of cGAS-STING target genes (e.g. IFN-β, ISG54 and CCL5) were higher in RNASEH2 deficient cells and could be further enhanced by ATRi treatment, which suggest the activation of cGAS-STING pathway may be another reason for increased cellular senescence caused by ATR inhibition.
RNASEH2 genes are frequently deleted in prostate adenocarcinoma
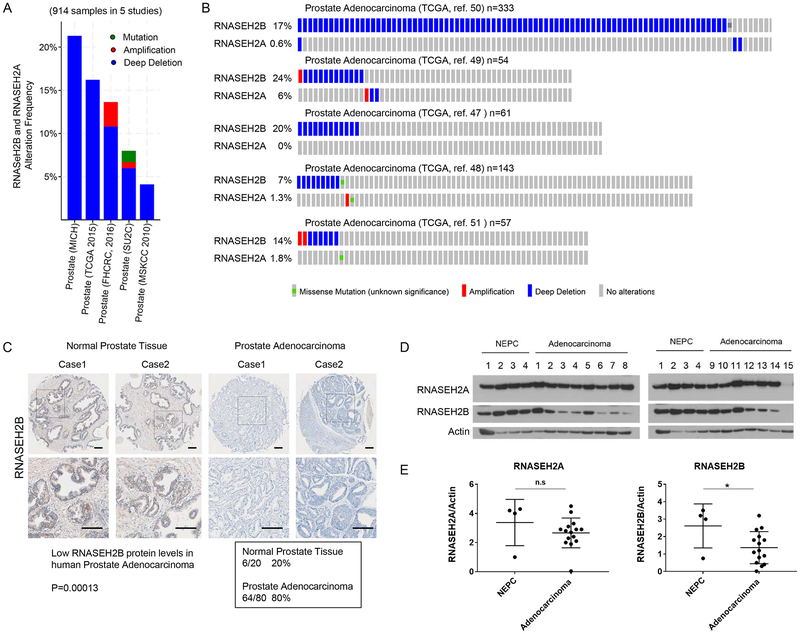
Although the role of RNASEH2 in Aicardi-Goutières syndrome has been well documented28,29,46, the relationship between RNASEH2 and cancer is unclear. By searching public databases and tools available in The Cancer Genome Atlas, cBioPortal, and the Human Protein Atlas, we found that RNASEH2B is frequently deleted in prostate adenocarcinoma (Figure 4A, B)47–53. RNASEH2A depletion was also reported in prostate adenocarcinoma, but less often (Figure 4A, B). RNASEH2C depletion was rarely reported (Supplementary Figure 9A). Using human prostate adenocarcinoma cancer tissue arrays, we confirmed that the protein levels of RNASEH2B were decreased in prostate adenocarcinoma: 80% in prostate adenocarcinoma compared to 30% in normal prostate tissues (P=0.00013) (Figure 4C). The data from the Human Protein Atlas also indicated that RNASEH2B protein levels are lower in prostate cancers (Supplementary Figure 9B).
Figure 4. Frequent RNASEH2B depletion in prostate adenocarcinoma.
A. Genomic alternation of RNASEH2B and RNASEH2A in TCGA prostate cancer databases (914 samples from 5 independent studies). Blue: deep deletion; Red: amplification; Green: mutation.
B. OncoPrint showing detailed genetic alterations from the prostate cancer samples in A.
C. Low protein levels of RNASEH2B were frequently found in prostate adenocarcinoma cancer. Immunohistochemical staining of RNASEH2B was performed in normal prostate and prostate adenocarcinoma cancer samples. Gray staining indicates positive immunoreactivity. Representative cases of RNASEH2 staining were shown. The low RNASEH2B stained samples in prostate cancer tissues were summarized in the box. Statistical significance of low RNASEH2B protein levels and prostate adenocarcinoma was determined by Peason χ2 analysis. Scale bar=100 μm.
D. Western blots showing decreased RNASEH2B protein levels in patient-derived prostate adenocarcinoma xenografts.
E. Plots comparing RNASEH2A or RNASEH2B protein levels normalized to Actin in neuroendocrine prostate cancer and prostate adenocarcinoma samples. T tests were used to evaluate differences between the groups. n.s., not significant; *P < 0.05.
*NEPC= neuroendocrine prostate cancer
To further confirm that RNASEH2B level is decreased in prostate adenocarcinoma, we collected patient-derived xenograft samples (4 neuroendocrine prostate cancer samples and 15 prostate adenocarcinoma samples) (Supplementary Table 3) and examined RNASEH2B and RNASEH2A protein levels by Western blotting. While there was no significant difference in RNASEH2A protein levels between these 2 types of prostate tumors, RNASEH2B protein levels were lower in the prostate adenocarcinoma samples than those in the neuroendocrine prostate tumor samples (Figure 4E, F). These data indicate that lower protein level of RNASEH2B is frequently observed in prostate adenocarcinoma.
Discussion
ATR is a potential target for cancer therapy, and several ATR inhibitors currently in clinical trials have shown promising results. In this study, to determine the genetic contexts that have synthetic lethality with ATR inhibition, we performed genome-wide functional screens in 3 different cell lines using CRISPR/Cas9 technology. We identified RNASEH2 deficiency as a genetic determinant of ATRi sensitivity. Moreover, we found that lower protein level of RNASEH2B is frequently observed in prostate adenocarcinoma. We speculate that patients with lower RNASEH2B expression may benefit from ATRi-based therapy, which warrants further investigation.
In order to advance the use of ATRi in cancer therapy, we need to achieve a comprehensive understanding of the genetic alterations that endow tumors with hypersensitivity to ATRi. Functional screens using CRISPR/Cas9 technology represent a major advance in the search for such co-essential genetic interactions. Besides RNASEH2, our screens also identified many other synthetic lethal interactions with ATRi, including several previously reported candidates such as ATM, TP53, and ERCC112,15. Additionally, we noticed that several groups of genes with similar functions had synthetic lethality with ATRi, including a group of genes directly involved in the ATR pathway (ATR, ATRIP, CHEK1, CLSPN, HUS1, RHNO1)5,12. This observation agrees with those made in a previous report indicating that the ATR pathway itself is essential for cell survival12. Another group of genes identified in our screens includes HR-related genes (e.g., BRCA2, PALB2, BRIP1, MAD2L2), suggesting that ATRi-based therapy may also benefit BRCA-mutant cancers. Our screens also uncovered several other genes, such as LCMT1 and LEO1; how they may act in an ATR-dependent pathway or in a parallel pathway remains to be determined. We anticipate that further investigation of these synthetic lethality interactions will lead to a better understanding of how ATR functions to regulate complex replication processes. In short, the genome-wide sgRNA screen is a powerful tool to reveal co-essential genetic interactions and uncover novel components and/or redundant pathways involved in a particular cellular process.
The roles of the RNASEH2 complex in nucleic acid metabolism and genome maintenance are well documented27. Here, we generated RNASEH2BKO and RNASEH2AKO HeLa cells and demonstrated that RNASEH2-depleted cells are hypersensitive to ATRi both in vivo and in vitro. It is of note that, in our experiments, RNASEH2AKO were more sensitive to ATRi treatment in multiple assays. There are at least two possible reasons for these results. First, RNASEH2A is the catalytic subunit, which is essential for the function of RNASEH2. Second, loss of RNASEH2A leads to dramatic decrease of RNASEH2B protein level, while KO of RNASEH2B only leads to slight decrease in RNASEH2A level, implying that RNASE2A may have some residual activity in the absence of RNASEH2B. Our in vivo xenograft experiments also showed that RNASEH2A loss could significantly reduce tumor growth, which is similar to an early report33.
Our experiments showed RNASEH2-deficient cells bore more spontaneous DNA damage (indicated by γ-H2A.X) which is consistent to the previous reports30,33. It is of note that we could not detect any obvious 53bp1 foci in non-treated RNASEH2-deficient cells. We speculate that spontaneous DNA damage observed in our RNASEH2-depleted cells is likely attributable to lesions that induce replication stress, but not DSBs, which is slightly different from the previous reports. RNASEH2 is responsible for RER and the removal of longer RNA-DNA hybrid (R-loop). Here, we checked R-loop level with S9.6 antibody but did not observe any significant increase in RNASEH2 deficient cells with or with ATRi-treatment, which may not be surprising since other enzymes such as RNASEH1 also play a role in R-loop removal54,55. A recent paper reported that it is likely that the role of RNASEH2 in RER may be responsible of PAPRi sensitivity in RNASEH2 deficient cells36. Given the similarity between our results that those reported for PAPRi sensitivity, it is possible that the role of RNASEH2 in RER may also be responsible for ATRi sensitivity in these KO cells.
Our results also showed that inhibition of ATR induced significant apoptosis in RNASEH2-depleted cells, which may be one of the reasons for the ATRi hypersensitivity observed in these cells. Furthermore, prolonged treatment with ATRi induced more senescence in RNASEH2 deficient cells which may be another reason for ATRi hypersensitivity. We also observed increased expressions of cGAS-STING pathway target genes in RNASEH2 deficient cells and ATRi-treatment further enhanced this increase. As it is reported that RNASEH2 deficiency could invoke cGAS-STING activation and because of the essential role of cGAS-STING pathway in cellular senescence30,31,56,57, we assume that the activation of cGAS-STING pathway may also contribute to senescence phenotype in our study.
A recent reports showed that RNASEH2B depletion is frequently observed in chronic lymphocytic leukaemia (CLL) cases and metastatic prostate cancer36. Here, we found frequent reduction of RNASEH2B protein levels in prostate adenocarcinoma using tissue microarray and PDX samples, suggesting that RNASEH2B could be a promising biomarker to guide ATRi-based cancer therapy, an application that should be explored in future clinical studies.
Methods
Cell lines and cell culture
293A, HCT116, MCF10A, and HeLa cells were obtained from ATCC. Dulbecco’s modified Eagle medium (DMEM) with 10% fetal calf serum was used to culture 293A, HCT116, and HeLa cells. MCF10A cells were cultured in DMEM/F12 Ham’s mixture supplemented with 5% equine serum (Gemini Bio), 20 ng/mL epidermal growth factor (Sigma), 10 μg/mL insulin (Sigma), 0.5 mg/mL hydrocortisone (Sigma), 100 ng/mL cholera toxin (Sigma), 100 units/mL penicillin, and 100 μg/mL streptomycin. All the cells were passed the test of mycoplasma.
Chemicals
AZD6738, VE822, NU7441, AZD0156, and LY2606368 were all purchased from Selleck Chemicals.
sgRNA screening
The TKOv3 library was a gift from Traver Hart’s laboratory. This library contains 70,948 gRNAs targeting 18,053 protein coding genes (4 gRNAs/gene) with 142 control gRNAs targeting EGFP, LacZ, and luciferase (71,090 total). Library virus production was conducted as described in a previous paper38. Briefly, 9 million 293T cells were seeded per 15-cm plate, and the cells were transfected 24 h later with a mix of 8 μg lentiviral pLCKO vector containing the library, 8 μg packaging vector psPAX2, 4 μg envelope vector pMD2.G, 60 μL X-treme Gene transfection reagent (Roche), and 2.0 mL Opti-MEM medium (Life Technologies). After 24 h, the medium was changed to a serum-free, high-bovine serum albumin (BSA) growth medium (DMEM, 1.1 g/100 mL BSA, 1% penicillin/streptomycin). The virus-containing medium was harvested 48 and 72 h after transfection, centrifuged at 1500 rpm for 5 min, and frozen.
For sgRNA screening, 120 million 293A, HCT116, or MCF10A cells were infected with the TKOv3 library lentiviruses at a low MOI (< 0.3). Twenty-four hours after infection, the medium was replaced with fresh medium containing puromycin (2 μg/mL for 293A and HCT116, 1.5 μg/mL for MCF10A). After selection, cells were split into 3 replicates containing ~20 million cells each, passaged every 3 days, and maintained at 200-fold coverage. At day 0 and every 3 days from day 6 to day 21, 25 million cells (> 300-fold coverage) were collected for genomic DNA extraction. Genomic DNA was extracted from cell pellets using the QIAamp Blood Maxi Kit (Qiagen), precipitated using ethanol and sodium chloride, and resuspended in Buffer EB (10 mM Tris-HCl, pH 7.5). gRNA inserts were amplified via PCR using primers harboring Illumina TruSeq adapters with i5 and i7 barcodes as previously reported58, and the resulting libraries were sequenced on an Illumina HiSeq 2500 system. The sequencing results were used for further analysis. The BAGEL (Bayesian Analysis of Gene Essentiality) algorithm was used to calculate essentiality scores. Drug-Z analysis was used to calculate the difference between the DMSO- and ATRi-treated groups.
Functional genomic analysis of co-essential genes
We chose an FDR of 0.05 as the cutoff. Co-essential genes were tested for enrichment in various biological processes using the DAVID server at http://david.abcc.ncifcrf.gov/ and also tested for enrichment using the GO_BP_ALL category. The top 10 GO_BP terms were listed.
Generation of KO cells
pLentiCRISPRv2 was used to generate KO cells. Cells were transiently transfected with the indicated plasmids and selected using puromycin (2 mg/mL). Single cells were then plated into 96-well plates. After 10 days, clones were picked and checked by Western blotting. The sgRNA sequences used were:
RNASEH2A_sgRNA_1_Forward: GACCGTGTGCCCGCGGTGTGCCGCA; RNASEH2A_sgRNA_1_Reverse: AAACTGCGGCACACCGCGGGCACAC; RNASEH2A_sgRNA_2_Forward: GACCGATAACAGATGGCGTAGACCA
RNASEH2A_sgRNA_2_Reverse: AAACTGGTCTACGCCATCTGTTATC; RNASEH2B_sgRNA_1_Forward: GACCGAAATAGAGGATCCACAGGTG; RNASEH2B_sgRNA_1_Reverse: AAACCACCTGTGGATCCTCTATTTC
RNASEH2B_sgRNA_2_Forward: GACCGTATCCACCACAACTTGATCA; RNASEH2B_sgRNA_2_Reverse: AAACTGATCAAGTTGTGGTGGATAC.
β-Galactosidase staining
Cells were plated in 6-well plates and treated with DMSO or AZD6738 (0.8μM) for 6 days. β-Galactosidase staining was performed according to the manufacturer’s instructions (Cat# 9860, Cell Signaling Technology).
Western blotting
Cells were lysed in NET-N buffer (20 mM Tris [pH 7.6], 1 mM ethylenediaminetetraacetic acid, 1% NP40, 150 mM NaCl) supplemented with protease inhibitor cocktail tablets (Roche). The following primary antibodies were used in this study: γ-H2A.X antibody (Cat# 05–636, Millipore); 53BP1 (Cat# MAB3802, Millipore); pATR (Cat# GTX128145, GeneTex); pChk1 (Cat# 2344s, Cell Signaling Technology); α-tubulin (Cat# 2148, Cell Signaling Technology); RNASEH2B (Cat# HPA040084, Sigma); and RNASEH2A (Cat# NBP1–76981, Novus).
Cell survival assay
For cell growth assays, cells were seeded in 6-well plates (104 per well) and passaged every 3 days until cell numbers were determined. For clonogenic assays, cells were seeded in 6-well plates (400 cells per well) and continuously exposed to ATRi treatment for 14 days, beginning 24 h after seeding. Cells were fixed with 10% trichloroacetic acid and stained with sulphorhodamine B (Sigma-Aldrich). Colonies were counted manually. All cell survival assays were performed at least in triplicate.
Immunofluorescence staining
Cells were grown on coverslips for 24 h before treatment. After the indicated treatment, cells were fixed in 4% paraformaldehyde and permeablized with phosphate-buffered saline (PBS) with 0.5% Triton X-100. Then, cells were incubated with primary antibodies diluted in PBS with 0.05% Triton X-100 and 1% BSA (PBST-BSA) for 1h at room temperature. After 3 washes with PBS, fluorescently labeled secondary antibodies in PBST-BSA were added for 1 h. Cells were then washed in PBS with Hoechst stain (1:10,000). Slides were imaged at 40× on a Leica microscope.
RNA extraction, reverse transcription and real-time PCR
RNA samples were extracted with TRIZOL reagent (Invitrogen). Reverse transcription assay was performed by using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA) according to the manufacturer’s instructions. Real-time PCR was performed by using Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA). For quantification of gene expression, the 2 − ΔΔCt method was used. Actin expression was used for normalization.
Human tissue IHC analysis
The human prostate adenocarcinoma tissue microarray (PR806) were purchased from US Biomax. It contains 80 prostate adenocarcinoma samples and 20 normal prostate tissue samples. Samples were deparaffinized and rehydrated. Antigen retrieval was done using 0.01 M sodium citrate buffer (pH 6.0) in a microwave oven. To block endogenous peroxidase activity, the sections were treated with 1% hydrogen peroxide in methanol for 30 min. After 1 h of preincubation in 10% normal goat serum to prevent nonspecific staining, the samples were incubated with anti-RNASEH2B (1:50; Cat# HPA040084, Sigma), overnight at 4°C. The sections were then incubated with a biotinylated secondary antibody (1:200; Vector Laboratories, PK-6101) and incubated with avidin–biotin peroxidase complex solution (1:100) for 30 min at room temperature. Color was developed with the 3-amino-9-ethylcarbazole (AEC) solution. Counterstaining was carried out using Mayer’s hematoxylin. All immunostained slides were scanned on the Automated Cellular Image System III (ACIS III, Dako) for quantification by digital image analysis. A total score of protein expression was calculated from both the percentage of immune-positive cells and the immunostaining intensity. High and low protein expressions were defined using the mean score of all samples as a cutoff point.
Xenograft assays
All the xenograft tumor experiments were strictly followed institutional guidelines that are approved by the MD Anderson Cancer Center Animal Care and Use Committee and performed under veterinary supervision. The 4-week old male nude mice were obtained from Jackson Laboratory and kept in a pathogen-free environment. HeLa (wild-type, RNASEH2BKO and RNASEH2AKO DKO) cells (2×106) were injected subcutaneously. When tumors were approximately 30 mm3 in size, 12 mice for each cell line were randomly assigned into two groups (6 mice per group) and subjected to the indicated compound treatment (vehicle control or 60 mg/kg/day AZD6738) through oral gavage. After 21 days, tumors were collected and analyzed.
Statistics
Statistical analyses were performed using GraphPad Prism software version 7.0. All of the statistical methods used are described in the main text. Each experiment was repeated twice or more, unless otherwise noted. No samples or animals were excluded from the analysis. For the mouse experiment, no statistical method was used to predetermine sample size. The samples or animals were randomly assigned to different groups. A laboratory technician who provided animal care was blinded to the group allocation during all animal experiments and outcome assessment. Differences between groups were analyzed by the Student t-test and Pearson χ 2 analysis. A P-value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank the members of Dr. Junjie Chen’s lab for their kind help and Dr. Lei Li for his suggestions regarding the experimental design. We also thank Amy Ninetto from the Department of Scientific Publications at MD Anderson for editing the manuscript. This work was supported in part by CPRIT (RP160667) and NIH grants (CA157448, CA193124, CA210929, CA216911, and CA216437) to J.C. and MD Anderson’s NIH Cancer Center Support Grant (CA016672).
Footnotes
Conflict of interest statement: The authors declare no competing financial interests.
Data availability
All relevant data not presented in the main figures or Supplementary Data are available from the authors.
References
- 1.Saldivar JC, Cortez D & Cimprich KA The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 18, 622–636, doi: 10.1038/nrm.2017.67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keszthelyi A, Minchell NE & Baxter J The Causes and Consequences of Topological Stress during DNA Replication. Genes (Basel) 7, doi: 10.3390/genes7120134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeman MK & Cimprich KA Causes and consequences of replication stress. Nat Cell Biol 16, 2–9, doi: 10.1038/ncb2897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Techer H, Koundrioukoff S, Nicolas A & Debatisse M The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet 18, 535–550, doi: 10.1038/nrg.2017.46 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Saldivar JC, Cortez D & Cimprich KA The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nature reviews. Molecular cell biology, doi: 10.1038/nrm.2017.67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Klein A et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Current Biology 10, 479–482, doi:Doi 10.1016/S0960-9822(00)00447-4 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Brown EJ & Baltimore D ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & development 14, 397–402 (2000). [PMC free article] [PubMed] [Google Scholar]
- 8.Wright JA et al. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. P Natl Acad Sci USA 95, 7445–7450, doi:DOI 10.1073/pnas.95.13.7445 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rundle S, Bradbury A, Drew Y & Curtin NJ Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers 9, 10.3390/cancers9050041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charrier JD Discovery of potent and selective inhibitors of Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. Abstr Pap Am Chem S 242 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Hall AB et al. Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget 5, 5674–5685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohni KN, Kavanaugh GM & Cortez D ATR Pathway Inhibition Is Synthetically Lethal in Cancer Cells with ERCC1 Deficiency. Cancer Research 74, 2835–2845, doi: 10.1158/0008-5472.Can-13-3229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson CT et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun 7, 10.1038/ncomms13837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok M, Davies N & Agathanggelou A ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells (vol 127, pg 582, 2016). Blood 127, 2647–2647, doi: 10.1182/blood-2016-03-708008 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Reaper PM et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 7, 428–430, doi: 10.1038/Nchembio.573 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Prevo R et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther 13, 1072–1081, doi: 10.4161/cbt.21093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fokas E et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis 3, 10.1038/cddis.2012.181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, Zhou Y & Wei W Genome-Wide CRISPR/Cas9 Screening for High-Throughput Functional Genomics in Human Cells. Methods in molecular biology 1656, 175–181, doi: 10.1007/978-1-4939-7237-1_11 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Steinhart Z et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med 23, 60–68, doi: 10.1038/nm.4219 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Sidik SM et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 166, 1423–1435 e1412, doi: 10.1016/j.cell.2016.08.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz S et al. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Molecular cell 62, 307–313, doi: 10.1016/j.molcel.2016.03.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart T et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, doi: 10.1016/j.cell.2015.11.015 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hart T, Brown KR, Sircoulomb F, Rottapel R & Moffat J Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol 10, 10.15252/msb.20145216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart T & Moffat J BAGEL: a computational framework for identifying essential genes from pooled library screens. Bmc Bioinformatics 17, 10.1186/s12859-016-1015-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Checkley S et al. Bridging the gap between in vitro and in vivo: Dose and schedule predictions for the ATR inhibitor AZD6738. Scientific reports 5, 10.1038/Srep13545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guichard SM et al. The pre-clinical in vitro and in vivo activity of AZD6738: A potent and selective inhibitor of ATR kinase. Cancer Research 73, doi: 10.1158/1538-7445.Am2013-3343 (2013). [DOI] [Google Scholar]
- 27.Hiller B et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. The Journal of experimental medicine 209, 1419–1426, doi: 10.1084/jem.20120876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrino FW, Harvey S, Shaban NM & Hollis T RNaseH2 mutants that cause Aicardi-Goutieres syndrome are active nucleases. Journal of molecular medicine 87, 25–30, doi: 10.1007/s00109-008-0422-3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pendergraft WF 3rd & Means TK AGS, SLE, and RNASEH2 mutations: translating insights into therapeutic advances. The Journal of clinical investigation 125, 102–104, doi: 10.1172/JCI78533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie KJ et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35, 831–844, doi: 10.15252/embj.201593339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokatayev V et al. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med 213, 329–336, doi: 10.1084/jem.20151464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams JS, Gehle DB & Kunkel TA The role of RNase H2 in processing ribonucleotides incorporated during DNA replication. DNA Repair 53, 52–58, doi: 10.1016/j.dnarep.2017.02.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizzi S et al. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet 24, 649–658, doi: 10.1093/hmg/ddu485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chon H et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res 41, 3130–3143, doi: 10.1093/nar/gkt027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiller B et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med 209, 1419–1426, doi: 10.1084/jem.20120876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann M et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289, doi: 10.1038/s41586-018-0291-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart T et al. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3-Genes Genom Genet 7, 2719–2727, doi: 10.1534/g3.117.041277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart T et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526, doi: 10.1016/j.cell.2015.11.015 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Kim H et al. Targeting the ATR/CHK1 axis in BRCA1/2 mutant ovarian cancer using an orthotopic patient-derived xenograft (PDX) model. Clin Cancer Res 22, doi:Doi 10.1158/1557-3265.Ovca15-A08 (2016). [DOI] [Google Scholar]
- 40.Kim H et al. Targeting the ATR/CHK1 Axis with PARP Inhibition Results in Tumor Regression in BRCA-Mutant Ovarian Cancer Models. Clinical Cancer Research 23, 3097–3108, doi: 10.1158/1078-0432.CCR-16-2273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell 151, 937–950, doi: 10.1016/j.cell.2012.10.035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He T et al. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 9, 849–862, doi: 10.2217/epi-2017-0019 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Mu Y et al. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO reports 17, 94–109, doi: 10.15252/embr.201540964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams JS, Gehle DB & Kunkel TA The role of RNase H2 in processing ribonucleotides incorporated during DNA replication. DNA Repair (Amst) 53, 52–58, doi: 10.1016/j.dnarep.2017.02.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartsch K et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26, 3960–3972, doi: 10.1093/hmg/ddx283 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Rice GI et al. Synonymous mutations in RNASEH2A create cryptic splice sites impairing RNase H2 enzyme function in Aicardi-Goutieres syndrome. Human mutation 34, 1066–1070, doi: 10.1002/humu.22336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243, doi: 10.1038/nature11125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson D et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 162, 454, doi: 10.1016/j.cell.2015.06.053 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Kumar A et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22, 369–+, doi: 10.1038/nm.4053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abeshouse A et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025, doi: 10.1016/j.cell.2015.10.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor BS et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 18, 11–22, doi: 10.1016/j.ccr.2010.05.026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419, doi: 10.1126/science.1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skourti-Stathaki K & Proudfoot NJ A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28, 1384–1396, doi: 10.1101/gad.242990.114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akman G et al. Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc Natl Acad Sci U S A 113, E4276–4285, doi: 10.1073/pnas.1600537113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li T & Chen ZJ The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215, 1287–1299, doi: 10.1084/jem.20180139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang H, Wang H, Ren J, Chen Q & Chen ZJ cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A 114, E4612–E4620, doi: 10.1073/pnas.1705499114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hart T et al. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 (Bethesda) 7, 2719–2727, doi: 10.1534/g3.117.041277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.