Abstract
It has been well established that an accumulation of mutations in DNA, whether caused by external sources (e.g. ultraviolet light, radioactivity) or internal sources (e.g. metabolic byproducts, such as reactive oxygen species), has the potential to cause a cell to undergo carcinogenesis and increase the risk for the development of cancer. Therefore, it is critically important for a cell to have the capacity to properly respond to and repair DNA damage as it occurs. The DNA damage response (DDR) describes a collection of DNA repair pathways that aid in the protection of genomic integrity by detecting myriad types of DNA damage and initiating the correct DNA repair pathway. In many instances, a deficiency in the DDR, whether inherited or spontaneously assumed, can increase the risk of carcinogenesis and ultimately tumorigenesis through the accumulation of mutations that fail to be properly repaired. Interestingly, although disruption of the DDR can lead to the initial genomic instability that can ultimately cause carcinogenesis, the DDR has also proven to be an invaluable target for anticancer drugs and therapies. Making matters more complicated, the DDR is also involved in the resistance to first-line cancer therapy. In this review, we will consider therapies already in use in the clinic and ongoing research into other avenues of treatment that target DNA repair pathways in cancer.
Keywords: DNA damage response, DNA repair, radiation, radiobiology
Conundrum: a Double-edged Sword of DNA Repair Perturbation
It is generally believed that the progression of a cell from a normal to a tumorigenic state occurs through a series of gene-altering steps that ultimately evade genomic stability-maintaining mechanisms and instead lead to genomic instability. Indeed, genomic instability is one of the hallmarks of cancer, reflecting the path from which the cancerous cells arose [1]. In normal cells, genomic integrity is protected by the DNA damage response (DDR), which describes the collection of pathways capable of detecting and repairing different types of DNA damage. Deficiencies in the DDR and its repair pathways, whether acquired or inherited, can accelerate the accumulation of mutations and help augment the loss of stability that ultimately gives rise to malignancy. Paradoxically, however, although deficiencies in DNA repair pathways can first lead to a loss of genomic integrity through the accumulation of mutations, the proteins involved have proven to be viable and efficacious targets for cancer therapies, while at the same time also contributing to resistance to first-line cancer treatments. To reconcile these differences, it is important to recognise that cancerous cells arise from normal, non-malignant cells, and thus share many of the same characteristics as their unaffected kin. A major goal in creating more effective cancer therapies is to find differences between cancerous and non-malignant cells that allow treatments to more specifically target the unhealthy cells, therefore minimising the toxicity cancer patients experience while undergoing therapy. An important clinical term regarding this notion of targeted toxicity is the therapeutic index or ratio, which is defined as the amount of tumour control that can be achieved for a given amount of healthy tissue toxicity [2]. The higher the ratio, the better the efficiency of the treatment in targeting cancerous as opposed to non-cancerous cells, minimising side-effects to healthy tissues. One way to increase the therapeutic ratio is to use synthetic lethality. Synthetic lethality is achieved when the simultaneous deficiency of two or more genes causes cell death, whereas a deficiency in only one is otherwise non-lethal [3]. Cancerous cells generally have higher incidences of DNA repair pathway deficiencies, resulting in a higher dependence on any remaining intact DNA repair pathways to maintain genomic stability [4]. This dependency renders the cancerous cells vulnerable to agents that target the remaining intact pathways in a synthetic lethal way that would otherwise be harmless to normal, healthy cells, and is a rapidly expanding field of research in cancer treatments [5,6]. A closely related concept to synthetic lethality that also offers a promising new strategy for increasing the therapeutic ratio through the targeting of the DDR is ‘acquired vulnerability’, otherwise known as ‘collateral sensitivity’. This term describes the phenomenon in which acquired resistance to one drug confers hypersensitivity towards a different reagent, thereby offering the possibility of clinical exploitation should the vulnerability be identified [7]. Research has already started to find that some of these acquired vulnerabilities are tied to the DDR, further underscoring the notion that the DDR is an invaluable target in the quest to find novel therapeutic approaches in the treatment of cancer [8].
Types of DNA Repair
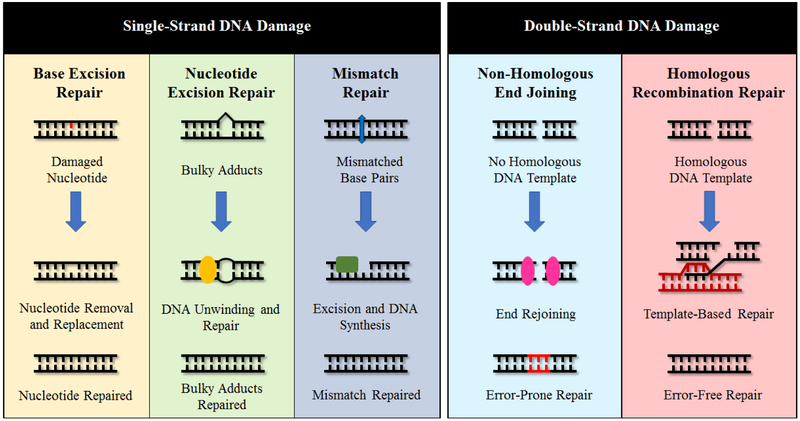
The cell has a complex system of interconnected DNA repair pathways that allows it to respond to a variety of different types of damage and regulate the outcomes of the intended repair (Figure 1). Single-strand DNA (ssDNA) damage can include a wide variety of insults, such as single-strand breaks (SSBs), deamination of bases, errors in base matching incurred during DNA replication and the creation of bulky adducts. There are, in the broadest of terms, three main pathways that are responsible for the repair of ssDNA damage: base excision repair (BER), nucleotide excision repair (NER) and mismatch repair (MMR) [9]. BER removes damaged bases that do not significantly alter the overall structure of DNA, as opposed to NER, which can recognise and repair the bulky, structurally altering ssDNA lesions [10]. Finally, MMR functions as a method of proofreading after DNA replication to catch any mismatched base pairs that were overlooked during DNA synthesis by DNA polymerases [10]. Mutations that impair any of these pathways can lead to disorders that substantially increase the lifetime risk of developing cancer, such as MUTYH-associated polyposis (BER), xeroderma pigmentosa (NER) and Lynch syndrome (MMR) [11]. DNA can also be affected by interstrand crosslinks (ICLs), which form when two base pairs are covalently bound together [12]. ICLs are especially cytotoxic to cells, as the covalent bond can block replication and/or transcription, and when left unrepaired, can lead to mutation and chromosomal breakage [12]. As with ssDNA repair pathways, defects in the DDR pathways responsible for repairing ICLs can lead to diseases that dramatically increase the chances of tumorigenesis: Fanconi anaemia is a rare disease that is caused by mutation of the Fanconi anaemia subtype (FANC) proteins. The FANC proteins function in the Fanconi anaemia pathway, which is essential for the maintenance of genomic integrity through the repair of ICLs [12].
Fig 1.
DNA repair pathways of single-strand DNA (ssDNA) damage and DNA double-strand breaks (DSBs). From left to right for ssDNA repair: base excision repair (BER) is used to repair damage to nucleotides that do not significantly alter the DNA structure. Nucleotide excision repair (NER) mends structurally altering DNA damage, such as bulky adducts. Mismatch repair (MMR) fixes pairs of nucleotides that were mismatched during DNA synthesis. Non-homologous end joining (NHEJ) repairs DNA DSBs without the use of a homologous DNA template by rejoining the broken DNA ends together and is prone to introducing errors. Homologous recombination repair (HRR) uses a homologous DNA template to repair DNA DSBs without error.
In addition to ssDNA damage, cells can also encounter double-strand breaks (DSBs), the most deleterious type of DNA damage. DSBs are repaired by two main pathways: non-homologous end joining (NHEJ) and homologous recombination repair (HRR) [13]. NHEJ is a form of repair template-independent repair and thus is an error-prone mechanism of repair active from G1 through G2. In some cases after a DSB has occurred, the ends of the broken strands will undergo resection, where degradation of overhangs will occur via exo- or endonuclease activity to stick the two broken DNA ends back together [14]. HRR, on the other hand, is considered to be an error-free method of DSB repair and occurs mainly in the S and G2 phases of the cell cycle when there is a sister chromatid available for use as a repair template [15]. More extensive end resection is carried out to the broken ends of the DSB as opposed to in NHEJ, where more minimal resection is needed. Of the two pathways, NHEJ is favoured over HRR, even during the S and G2 phases where HRR is more active, with some studies showing a 4:1 ratio of NHEJ to HRR in mammalian somatic cells [14]. Similar to ssDNA repair pathways, the impairment of NHEJ or HRR through the mutation of regulatory or involved proteins can lead to an increased risk of carcinogenesis and tumorigenesis due to accumulation of unresolved DSBs in the genome. For example, certain mutations in breast cancer 1/2 (BRCA1/2), proteins essential to proper HRR, are linked to an increased risk of developing a wide range of different cancers, including breast, prostate, colon, ovarian and pancreatic, with increased lifetime risks for cancer development as high as 55% [16]. In addition, patients with LIG4 syndrome have a predisposition to developing lymphoid malignancies due to a deleterious mutation in the gene encoding protein DNA Ligase IV, resulting in impaired NHEJ [17].
Poly (ADP-ribose) Polymerase Inhibitors
One of the most developed cancer therapies targeting DNA repair pathways that has risen to prominence within the last few years is the use of poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi). PARPs are a class of 17 nuclear enzymes involved in multiple cellular functions [18]. These proteins, which can transfer either one or multiple (ADP-ribose) units from NAD+ onto substrates to make poly (ADP-ribose) (PAR) chains, are found in all eukaryotes except yeast [19]. PARP1 has been found to play a crucial role in the DDR, including in ssDNA repair, NHEJ and HRR [20–22]. PARP1 is recruited to sites of damage for various types of damage, aided by its DNA-binding domain, and has a number of functions, such as binding SSBs, recruiting downstream DNA repair proteins and promoting HRR at stalled and/or collapsed replication forks [22]. Although PARPi were first found over 30 years ago, only in 2005 was it demonstrated through the work of two independent research groups that the use of PARPi is synthetically lethal in cells deficient in HRR, such as when there are mutations in the BRCA1/2 proteins [21,23]. Indeed, PARPi have had enough success that as of October 2018, four PARPi have been approved by the US Food and Drug Administration (FDA): olaparib (Lynparza), rucaparib (Rubraca), niraparib (Zejula) and the newest one, talazoparib (Talzenna) [24]. These drugs have been approved for use in patients with BRCA1/2 deficiencies in ovarian cancer and by the European Medical Agency in patients who have responded to platinum-based chemotherapy with relapsed BRCA1/2 mutant ovarian, fallopian tube or primary peritoneal cancers [25]. Olaparib, the first PARPi to be approved by the FDA in 2014, has also been approved for clinical use in patients with BRCA1/2 mutations and HER2-negative breast cancer [24,26,27]. These drugs have also shown promise in treating other types of HRR-deficient breast and prostate cancer. However, the exact mechanism describing this synthetic lethal relationship has not yet been fully elucidated [28]. Originally, it was hypothesised that the synthetic lethality between PARP inhibition and BRCA1/2 mutation relied on the induction of persistent SSBs after PARPi inhibition. During replication, the replication fork would collapse when encountering the SSBs, and thus potentially create a DSB that was unable to be properly repaired by HRR [29]. In the absence of HRR, other DNA repair processes more prone to introducing deletions, mutations and potentially genomic rearrangements would take over, often leading to cell death [29]. This model has changed with new evidence suggesting some of the PARPi ‘trap’ PARP1 onto DNA, preventing its release and thus stalling repair [29]. However, as with many other types of cancer treatment, tumour resistance to PARPi is frequently seen and represents a major hurdle in longterm treatments [30]. The mechanism for acquired resistance has been suggested to fall into two broad main categories: secondary mutations restore necessary minimal HRR function, rendering the previously synthetic lethal phenotype ineffective [29]; resistance can occur in an HRR-independent manner, such as through PARP protein expression loss, rendering PARPi ineffective [29,31]. Research is already underway to establish what therapies can be used to prevent and/or counter PARPi resistance, taking advantage of the idea of acquired vulnerability, but more work needs to be done to make this goal a reality [8].
Kinase Inhibitors
Another route of targets that has seen moderate success in the cancer therapeutic field includes the class of DDR kinase inhibitors. As of January 2019, the FDA has approved over 30 kinase inhibitors targeted at the treatment of cancers [32]. Phosphorylation plays a critical role in the regulation of many DDR pathways. Ataxia telangiectasia mutated (ATM), which is a key player in the repair of DSBs through the HRR pathway and a serine/threonine kinase in the phosphatidylinositol 3-kinase (PI3K)-related kinase (PIKK) family, acts as an early signalling protein in the DDR and is responsible for the phosphorylation of hundreds of downstream targets [33,34]. The protein is named after a rare autosomal recessive disorder, ataxia telangiectasia, which results from mutations in the ATM gene. Patients who suffer from this disorder have symptoms such as radiosensitivity, immunodeficiencies and an increased risk of cancer [35]. Studies have shown that ATM is synthetic lethal with PARP deficiencies and that ATM inhibitors can sensitise cells to DSB-inducing reagents and [AQ1]IR [36,37]. ATM inhibitors are currently being explored in a clinical setting: for example, the ATM inhibitor AZD0156 in conjunction with olaparib (a PARPi) or irinotecan (a topoisomerase inhibitor) is currently under review in an early phase clinical [AQ2]phase I trial (clinical trial NCT02588105) [35]. Ataxia telangiectasia and Rad3-related protein (ATR) shares many of the same characteristics as ATM. Another PIKK family member and serine/threonine kinase, ATR also functions as an early signalling kinase in the DDR response, primarily following replication stress. Preclinical studies have found that ATR has a synthetic lethal relationship with several DDR players, including XRCC1 and ATM, and is currently under clinical investigation for its potential as a target in cancer therapies [38,39]. VX-970, or M6620, a potent ATR inhibitor, is currently involved in phase II trials, used either as a single agent or in tumours with DNA repair deficiencies. A phase I trial in which VX-970 was used alone or in combination with carboplatin showed early evidence of potential efficacy [40]. Checkpoint kinase 1 (CHK1), a major downstream effector of ATR, and WEE1, are additional kinases of interest in the clinical setting. CHK1 is involved in a multitude of different functions in the cell and prevents cells with damaged or incompletely replicated DNA after exposure to IR or chemotherapeutic drugs moving from G2 onto mitosis. CHK1 inhibitors have already been used in National Institutes of Health (NIH) phase I clinical trials of monotherapy, such as LY2606368 (prexasertib) (clinical trial NCT02203513) [41]. WEE1, which is involved in triggering the DDR after DNA damage has occurred, is also under clinical investigation as a target for inhibition in the treatment of certain cancers [42]. AZD1775, a WEE1 inhibitor, is currently being tested in phase I and II trials, both as monotherapy and in combination therapies (e.g. clinical trial NCT02593019 and those listed in Table 1, respectively).
Table 1.
A list of clinical trials that have taken place or are currently ongoing in the US and Europe that combine radiation therapy with the listed DNA damage response (DDR) inhibitors. The phase of the trials, cancer types being investigated, current status as of submission, names of the drugs and the respective DDR targets, and trial identifier numbers have been listed
| Phase | Cancer | Status | Drug | Drug target | Trial identifier |
|---|---|---|---|---|---|
| I | Head and neck | Completed | Olaparib | PARP | NCT01758731 |
| I | Triple negative breast cancer | Recruiting | Olaparib | PARP | NCT03109080 |
| II | Inflammatory breast cancer | Recruiting | Olaparib | PARP | NCT03598257 |
| I | Inoperable breast cancer | Recruiting | Olaparib | PARP | NCT02227082 |
| I | Head and neck | Recruiting | Olaparib | PARP | NCT02229656 |
| I/II | Metastatic castration-resistant prostate cancer in bone | Recruiting | Olaparib | PARP | NCT03317392 |
| I | Soft-tissue sarcoma | Recruiting | Olaparib | PARP | NCT02787642 |
| I | Small cell lung cancer | Recruiting | Olaparib | PARP | NCT03532880 |
| I | Head and neck | Recruiting | Olaparib | PARP | NCT02308072 |
| I/II | Unresectable high-grade glioma | Recruiting | Olaparib | PARP | NCT03212742 |
| II | Glioblastoma | Ongoing | Olaparib | PARP | 2014-001216-19 |
| I/II | Diffuse pontine glioma | Completed | Veliparib | PARP | NCT01514201 |
| II | Non-small cell lung cancer with brain metastases | Completed | Veliparib | PARP | NCT01657799 |
| I | Peritoneal carcinomatosis, epithelial ovarian, fallopian, and primary peritoneal | Completed | Veliparib | PARP | NCT01264432 |
| I | Brain metastases | Completed | Veliparib | PARP | NCT00649207 |
| I | Rectal cancer | Completed | Veliparib | PARP | NCT01589419 |
| I | Recurrent breast cancer | Completed | Veliparib | PARP | NCT01477489 |
| II | Malignant glioma without H3 K27M or BRAFV600E mutations | Recruiting | Veliparib | PARP | NCT03581292 |
| I | Pancreatic | Active | Veliparib | PARP | NCT01908478 |
| I | Triple negative breast cancer | Recruiting | Rucaparib | PARP | NCT03542175 |
| I | Castrate-resistant prostate cancer | Recruiting | Niraparib | PARP | NCT03076203 |
| I | Non- and small cell lung cancer, and neuroendocrine | Recruiting | VX-970 (M6620) | ATR | NCT02589522 |
| I | Oesophageal and other | Not yet recruiting | VX-970 (M6620) | ATR | NCT03641547 |
| I | HPV-negative head and neck squamous cell carcinoma | Recruiting | VX-970 (M6620) | ATR | NCT02567422 |
| I | Brain cancer | Recruiting | AZD1390 | ATM | NCT03423628 |
| I | Advanced solid tumours | Recruiting | M3814 | DNA-PK | NCT03724890 |
| I | Advanced solid tumours | Recruiting | MSC2490484A | DNA-PK | NCT02516813 |
| I/II | Rectal cancer | Not yet recruiting | M3814 | DNA-PK | NCT03770689 |
| I | Head and neck | Recruiting | LY2606368 (Prexasertib) | CHK1/CHK2 | NCT02555644 |
| I | Head and neck | Recruiting | AZD1775 (Adavosertib) | WEE1 | NCT03028766 |
| I | Cervical, vaginal, and uterine | Recruiting | AZD1775 (Adavosertib) | WEE1 | NCT03345784 |
| I | Intermediate-/high-risk squamous head and neck | Recruiting | AZD1775 (Adavosertib) | WEE1 | NCT02585973 |
| I/II | Unresectable adenocarcinoma of the pancreas | Active | AZD1775 (Adavosertib) | WEE1 | NCT02037230 |
| I | Diffuse intrinsic pontine glioma | Recruiting | AZD1775 (Adavosertib) | WEE1 | NCT01922076 |
| I | Glioblastoma | Recruiting | AZD1775 (Adavosertib) | WEE1 | NCT01849146 |
| I | Soft-tissue sarcoma | Recruiting | AMG-232 | MDM2 | NCT03217266 |
| I/II | Non-small cell lung cancer | Completed | NFV | AKT | 2006-001031-22 |
PARP, poly (ADP-ribose) polymerase; ATR, ataxia telangiectasia and Rad3-related protein; ATM, ataxia-telangiectasia mutated; DNA-PK, DNA protein kinase; CHK, checkpoint kinase.
MDM2
AKT
Attempts have also been made to inhibit the protein DNA-dependent protein kinase (DNA-PKcs), another member of the PIKK group that serves a crucial role as a main regulator of the NHEJ pathway. DNA-PK, a complex made of the catalytic subunit DNA-PKcs and the Ku heterodimer, is itself a target of ATM and ATR and is dependent on the binding of DNA for activation [43,44]. Once bound to Ku and DNA, DNA-PKcs helps in the recruitment of other NHEJ proteins to begin DNA resection at the broken ends of the DSBs and later DNA ligating complex proteins to join the DNA ends. Interestingly, it has been found that in cell lines with defective DNA-PK function, whether through a lacking of Ku or DNA-PKcs, there exists a hypersensitivity to IR and chemical agents that cause DSBs [44]. On the flip side, upregulation of DNA-PK activity has been correlated with increased resistance to DNA damage in some cancers [44]. Therefore, DNA-PK is a promising target for anticancer therapies. One of the first DNA-PKcs inhibitors, wortmannin, was isolated from the fungi Penicillium funiculosum in 1957 [45]. Wortmannin is a non-specific PI3K family inhibitor and has been found to be an effective radiosensitiser [46]. However, wortmannin has proven to have limited clinical application, given that the substance is poorly soluble in aqueous solutions, non-specific and toxic [47]. Subsequently, other general PI3K inhibitors have been developed with the aim of increasing clinical applicability while maintaining its sensitising properties, such as LY294002 and its prodrug SF1126. Although LY294002 did not reach clinical trials due to similar issues as seen in wortmannin, together with a quick metabolic clearance rate of 1 h, SF1126 has so far successfully completed a phase I clinical trial that ended in 2011, which found the drug to be well-tolerated with promising results, and is undergoing a second phase I trial in patients with advanced hepatocellular carcinoma (NCT00907205 and NCT03059147, respectively) [44,48,49]. NU7441, a DNA-PKcs-specific inhibitor, is also being studied and has shown promising preclinical effects on certain types of cancer, such as non-small cell lung cancer [50].
Radiation Therapy: Combinatorial Approach
Radiation therapy, or the use of IR in the treatment of cancer, has been used for over a century and functions to exploit the genomic instability phenotype of cancerous cells, as a deficiency in the ability to repair DSBs frequently results in an increased sensitivity to IR [51]. Since its inception, radiation therapy has seen significant improvements that reduce toxicity to normal tissue, although this toxicity still remains a limiting factor [4]. One area of improvement that has been of great interest is the induction of ‘artificial synthetic lethality,’ where radiation therapy is combined with specific targets to DNA repair pathways redundant in non-malignant cells, but crucial for cancerous cell survival. This combination of radiation therapy and inhibition of DNA repair pathways comes with the goal to increase the therapeutic index by conferring increased IR sensitivity only to cancerous cells [52]. Current radiotherapy techniques are used such that the dose is targeted to the tumour to spare the surrounding healthy tissue from treatment, which also functions to limit the amount of damage normal tissues would receive after application of a systematic inhibitor of a DNA repair protein [53]. Together, the targeted radiation therapy and inhibited DNA repair in cancer cells would potentially lead to an increase in the therapeutic index, allowing for a reduction in the effective radiation dose while limiting toxic side-effects.
There have been several agents targeting DNA repair pathway proteins that have been found in preclinical studies to be effective radiosensitisers, many of which are under current clinical investigation in phase I and II trials (Table 1). One protein that has been shown in preclinical studies to increase radiosensitivity is Artemis, an endo/exonuclease in the NHEJ pathway that is recruited and activated by DNA-PKcs to process broken DNA ends at DSBs [54]. Patients who have null mutations for Artemis show extreme radiosensitivity, making Artemis a target of interest for its projected efficiency in working synergistically with radiation therapy, as well as etoposide treatment (which, similar to IR, causes DSBs) [55]. However, to date, no inhibitor of Artemis has been found, although compounds are currently being screened to identify potential inhibitors (NIH 5F31GM116569-03). NU7441, mentioned previously as a DNA-PKcs-specific inhibitor, has also been shown to increase cell death after IR and etoposide treatment in different types of colon cancer cells in a DNA-PKcs-dependent manner [56]. However, NU7441 has proven to be clinically unusable due to problems with bioavailability and solubility, and other DNA-PKcs inhibitors are currently being investigated in both preclinical and clinical phases (Table 1) [57]. PARPi, including the four FDA-approved inhibitors mentioned previously, are being heavily investigated in combination therapies with radiation in numerous types of cancer (Table 1). LY2606368, a CHK1 inhibitor, is also currently undergoing a phase I clinical trial in head and neck cancer in conjunction with radiation, with an estimated completion date set for 2019 (Table 1, clinical trial NCT02555644). However, new CHK1 inhibitors are still being developed and tested under preclinical settings in combination with radiation, such as CCT244747, which when used on p53-deficient head and neck squamous cell carcinoma cells increases radiosensitivity to paclitaxel-based chemoradiotherapy [58]. CHIR-124, SAR-020106 and SB-218078, three more CHK1 inhibitors, have also been shown in preclinical studies to increase radiosensitivity in cells withp53 deficiencies or mutations [59–61]. WEE1 inhibitors, such as AZD11775 (adavosertib), have also made it to the clinic in ongoing phase I and II trials in combination with radiation (Table 1). Alongside Artemis, DNA-PK, ATM, CHK1 and WEE1, other DDR factors not described in this review are also being investigated clinically in conjunction with radiation (Table 1, last two trials). Although all of these proteins are promising future clinical targets, success in clinical trials will depend heavily on understanding the conditions under which these treatments will have a therapeutic gain.
Future Targets and Conclusion
In the past few years there has been an explosion of new information about the DDR and its involvement in cancer, of which we have only just begun to understand how to use in a clinically relevant setting. Although there have been exciting steps in clinical advancements in the treatment of cancer, and there is a plethora of extensive preclinical data to support clinical application for many of these new therapies, researchers and clinicians alike have much still to uncover and learn, especially in the area of how the DDR contributes to carcinogenesis and tumorigenesis, as well as how DDR pathways can be exploited for better future treatments. As an example, although most current cancer therapeutic targets are DDR kinases, about only 4% of all DDR proteins are kinases, leaving a large field of novel, non-kinase targets that could be targeted. Indeed, new players in DDR are continually being discovered, many of which are not kinases. In our own laboratory we have found two new non-kinase players involved in the DDR: SIRT2 and SAMHD1. SIRT2, a class III histone deacetylase (HDAC), was found to be directly involved in the replication stress response acting as a DDR regulator that leads to ATR activation [62–64]. SAMHD1 is a dNTP triphosphohydrolase and HIV-1 restrictase known to have an association with cancer when mutated. Our laboratory discovered that SAMHD1 functions independently of its dNTPase activity in the HR pathway and can be targeted for radiation and PARPi sensitisation using virus-like particles containing Vpx, a lentiviral accessory protein, that targets SAMHD1 for proteasomal degradation [65]. Continuing research into DDR and its components, as well as the drugs that affect DNA repair function, will lead to further clinical advancements in the prevention of cancer and effective therapies.
Acknowledgements
Research in the authors’ laboratory is supported by the National Institutes of Health [F31CA225124 to E.V.M.; R01CA178999 to D.S.Y.], Department of Defense [OC160540 to D.S.Y.; CA160771 to D.S.Y.], Lung Cancer Research Foundation (60208 to D.S.Y.], Basser Center for BRCA [32356 to D.S.Y.], and Winship Cancer Institute/Brenda Nease Breast Cancer Research Fund [53237 to D.S.Y.]. We would also like to thank the journal for the invitation to submit a review article, as well as all researchers who have contributed their time and effort to improving cancer therapies and our understanding of cancer pathology. Finally, we would like to thank Dr PamelaSara Head for minor edits.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yao Y, Dai W. Genomic instability and cancer. J Carcinog Mutagen 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zindler JD, Thomas CR Jr, Hahn SM, Hoffmann AL, Troost EG, Lambin P. Increasing the therapeutic ratio of stereotactic ablative radiotherapy by individualized isotoxic dose prescription. J Natl Cancer Inst 2016; 108. [DOI] [PubMed] [Google Scholar]
- [3].Nijman SM. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett 2011;585:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015;60:547–60. [DOI] [PubMed] [Google Scholar]
- [5].Brown JS, O'Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov 2017;7:20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood 2011;117:6074–82. [DOI] [PubMed] [Google Scholar]
- [7].Hall MD, Handley MD, Gottesman MM. Is resistance useless? Multidrug resistance and collateral sensitivity. Trends Pharmacol Sci 2009;30:546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barazas M, Gasparini A, Huang Y, Kucukosmanoglu A, Annunziato S, Bouwman P et al. Radiosensitivity is an acquired vulnerability of PARPi-resistant BRCA1-deficient tumors. Cancer Res 2019;79:452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Y, Rohde LH, Wu H. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr Genom 2009;10:250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dexheimer TS. DNA repair pathways and mechanisms. In: DNA repair of cancer stem cells. 2013. p. 19–32. [Google Scholar]
- [11].Poulsen ML, Bisgaard ML. MUTYH associated polyposis (MAP). Curr Genom 2008;9:420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lopez-Martinez D, Liang CC, Cohn MA. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell Mol Life Sci 2016;73:3097–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008;7:2902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017;18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao X, Wei C, Li J, Xing P, Li J, Zheng S et al. Cell cycle-dependent control of homologous recombination. Acta Biochim Biophys Sin 2017;49:655–68. [DOI] [PubMed] [Google Scholar]
- [16].Lee MV, Katabathina VS, Bowerson ML, Mityul MI, Shetty AS, Elsayes KM et al. BRCA-associated cancers: role of imaging in screening, diagnosis, and management. Radiographics 2017;37:1005–23. [DOI] [PubMed] [Google Scholar]
- [17].Altmann T, Gennery AR. DNA ligase IV syndrome; a review. Orphanet J Rare Dis 2016;11:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer 2011;105:1114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jubin T, Kadam A, Jariwala M, Bhatt S, Sutariya S, Gani AR et al. The PARP family: insights into functional aspects of poly (ADP-ribose) polymerase-1 in cell growth and survival. Cell Prolif 2016;49:421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].del Rivero J, Kohn EC. PARP inhibitors: the cornerstone of DNA repair-targeted therapies. Oncology 2017;31:265–73. [PubMed] [Google Scholar]
- [21].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- [22].Eustermann S, Videler H, Yang JC, Cole PT, Gruszka D, Veprintsev D et al. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol 2011. ;407:149–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434(7035):913–7. [DOI] [PubMed] [Google Scholar]
- [24].Honey K FDA approves a new PARP inhibitor for BRCA-mutant breast cancer. In Cancer Research Catalyst. AACR; 2018. [Google Scholar]
- [25].Nikolaev A, Yang ES. The impact of DNA repair pathways in cancer biology and therapy. Cancers 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lim JSJ, Tan DSP. Understanding resistance mechanisms and expanding the therapeutic utility of PARP inhibitors. Cancers 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Konstantinopoulos PA, Matulonis UA. PARP inhibitors in ovarian cancer: a trailblazing and transformative journey. Clin Cancer Res 2018;24:4062–5. [DOI] [PubMed] [Google Scholar]
- [28].Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015;5:1137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017;355(6330):1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim Y, Kim A, Sharip A, Sharip A, Jiang J, Yang Q et al. Reverse the resistance to PARP inhibitors. Int J Biol Sci 2017;13:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cruz C, Castroviejo-Bermejo M, Gutierrez-Enriquez S, Llop-Guevara A, Ibrahim YH, Gris- Oliver A et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol 2018;29:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bhullar KS, Lagaron NO, McGowan EM, Parmar I, Jha A, Hubbard BP et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer 2018;17:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Minchom A, Aversa C, Lopez J. Dancing with the DNA damage response: next-generation anti-cancer therapeutic strategies. Ther Adv Med Oncol 2018;10:1758835918786658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Durant ST, Zheng L, Wang Y, Chen K, Zhang L, Zhang T et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv 2018;4(6):eaat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pike KG, Barlaam B, Cadogan E, Campbell A, Chen Y, Colclough N et al. The identification of potent, selective, and orally available inhibitors of ataxia telangiectasia mutated (ATM) kinase: the discovery of AZD0156 (8-{6-[3- (Dimethylamino)propoxy]pyridin-3-yl}-3-methyl-1-(tetrahydro-2 H-pyran-4-yl)-1,3-dihydro-2 H-imidazo[4,5- c]quinolin-2-one). J Med Chem 2018;61:3823–41. [DOI] [PubMed] [Google Scholar]
- [36].Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta A, Valenzuela MT, Matinez-Romero R et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol 2007;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004;64:9152–9. [DOI] [PubMed] [Google Scholar]
- [38].Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7:428–30. [DOI] [PubMed] [Google Scholar]
- [39].Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PLoS One 2013;8:e57098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].O'Carrigan B, Luken MJdM, Papadatos-Pastos D, Brown J, Tunariu N, Lopez RP et al. Phase I trial of a first-in-class ATR inhibitor VX-970 as monotherapy (mono) or in combination (combo) with carboplatin (CP) incorporating pharmacodynamics (PD) studies. J Clin Oncol 2016;34(15_suppl):2504. [Google Scholar]
- [41].Qiu Z, Oleinick NL, Zhang J. ATR/CHK1 inhibitors and cancer therapy. Radiother Oncol 2018;126:450–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol 2011;194:567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell 2009;34:285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pospisilova M, Seifrtova M, Rezacova M. Small molecule inhibitors of DNA-PK for tumor sensitization to anticancer therapy. J Physiol Pharmacol 2017;68:337–44. [PubMed] [Google Scholar]
- [45].Brian PW, Curtis PJ, Hemming HG, Norris GLF. Wortmannin, an antibiotic produced by Penicillium wortmanni. Trans Br Mycol Soc 1957;40:365–8. [Google Scholar]
- [46].Davidson D, Amrein L, Panasci L, Aloyz R. Small molecules, inhibitors of DNA-PK, targeting DNA repair, and beyond. Front Pharmacol 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene 2005;24:949–61. [DOI] [PubMed] [Google Scholar]
- [48].Zhao W, Qiu Y, Kong D. Class I phosphatidylinositol 3-kinase inhibitors for cancer therapy. Acta Pharm Sin B 2017;7:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mahadevan D, Chiorean EG, Harris WB, Von Hoff DD, Stejskal-Barnett A, Qi W et al. Phase I pharmacokinetic and pharmacodynamic study of the pan-PI3K/mTORC vascular targeted pro-drug SF1126 in patients with advanced solid tumours and B-cell malignancies. Eur J Cancer 2012;48:3319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yanai M, Makino H, Ping B, Takeda K, Tanaka N, Sakamoto T et al. DNA-PK inhibition by NU7441 enhances chemosensitivity to topoisomerase inhibitor in non-small cell lung carcinoma cells by blocking DNA damage repair. Yonago Acta Med 2017;60:9–15. [PMC free article] [PubMed] [Google Scholar]
- [51].Gianfaldoni S, Gianfaldoni R, Wollina U, Lotti J, Tchernev G, Lotti T. An overview on radiotherapy: from its history to its current applications in dermatology. Open Access Maced J Med Sci 2017;5:521–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nickoloff JA, Boss M-K, Allen CP, LaRue SM. Translational research in radiation-induced DNA damage signaling and repair. Transl Cancer Res 2017:S875–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sishc BJ, Davis AJ. The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Franco S, Murphy MM, Li G, Borjeson T, Boboila C, Alt FW. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J Exp Med 2008;205:557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Woodbine L, Grigoriadou S, Goodarzi AA, Riballo E, Tape C, Oliver AW et al. An Artemis polymorphic variant reduces Artemis activity and confers cellular radiosensitivity. DNA Repair 2010;9:1003–10. [DOI] [PubMed] [Google Scholar]
- [56].Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res 2006;66:5354–62. [DOI] [PubMed] [Google Scholar]
- [57].Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol Ther 2016;160:65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Barker HE, Patel R, McLaughlin M, Schick U, Zaidi S, Nutting CM et al. CHK1 inhibition radiosensitizes head and neck cancers to paclitaxel-based chemoradiotherapy. Mol Cancer Ther 2016;15:2042–54. [DOI] [PubMed] [Google Scholar]
- [59].Tao Y, Leteur C, Yang C, Zhang P, Castedo M, Pierre A et al. Radiosensitization by Chir-124, a selective CHK1 inhibitor: effects of p53 and cell cycle checkpoints. Cell Cycle 2009;8:1196–205. [DOI] [PubMed] [Google Scholar]
- [60].Borst GR, McLaughlin M, Kyula JN, Neijenhuis S, Khan A, Good J et al. Targeted radiosensitization by the Chk1 inhibitor SAR-020106. Int J Radiat Oncol Biol Phys 2013;85:1110–8. [DOI] [PubMed] [Google Scholar]
- [61].Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res 2000;60:566–72. [PubMed] [Google Scholar]
- [62].Zhang H, Park SH, Pantazides BG, Karpiuk O, Warren MD, Hardy CW et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci U S A 2013;110:13546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Head PE, Zhang H, Bastien AJ, Koyen AE, Withers AE, Daddacha WB et al. Sirtuin 2 mutations in human cancers impair its function in genome maintenance. J Biol Chem 2017;292:9919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang H, Head PE, Daddacha W, Park SH, Li X, Pan Y et al. ATRIP deacetylation by SIRT2 drives ATR checkpoint activation by promoting binding to RPA-ssDNA. Cell Rep 2016;14:1435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN et al. SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep 2017;20:1921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]