Abstract
Mutations in the Leucine-rich repeat kinase 2 (LRRK2) gene have been implicated in the pathogenesis of Parkinson’s disease (PD). Identification of PD-associated LRRK2 mutations has led to the development of novel animal models, primarily in mice. However, the characteristics of human LRRK2 and mouse Lrrk2 protein have not previously been directly compared. Here we show that proteins from different species have different biochemical properties, with the mouse protein being more stable but having significantly lower kinase activity compared to the human orthologue. In examining the effects of PD-associated mutations and risk factors on protein function, we found that conserved substitutions such as G2019S affect human and mouse LRRK2 proteins similarly, but variation around position 2385, which is not fully conserved between humans and mice, induces divergent in vitro behavior. Overall our results indicate that structural differences between human and mouse LRRK2 are likely responsible for the different properties we have observed for these two species of LRRK2 protein. These results have implications for disease modelling of LRRK2 mutations in mice and on the testing of pharmacological therapies in animals.
Keywords: Parkinson’s disease, kinase activity, protein stability, level of expression, mouse model
Introduction
Genetics has been shown to contribute substantially to the lifetime risk of Parkinson’s disease (PD), with several different genes identified [1]. Of these, mutations in the Leucine-rich repeat kinase 2 (LRRK2) gene are a relatively common cause of inherited PD [2–4]. Additionally, variation around the LRRK2 locus is associated with lifetime risk of sporadic PD [5]. The LRRK2 gene encodes a large protein with several protein-protein interaction domains surrounding regions that have GTPase and kinase activities. Familial mutations alter one or more of these activities and, therefore, may contribute to disease pathogenesis [6].
Supporting this concept, kinase dead versions of LRRK2 are less toxic in culture models than their kinase active counterparts [7, 8] and inhibitors of kinase activity can ameliorate toxicity in animal models containing LRRK2 mutations [9–11]. Similarly, chemical entities that bind the GTPase region of LRRK2 are reported to be neuroprotective [12]. These observations have led to the concept that LRRK2 might be able to be targeted for therapeutic intervention by inhibition [13]. However, some mutations in LRRK2, particularly the G2385R variant that increases lifetime risk of PD by ~2-fold, may cause disease by mechanisms other than gain of function as these apparently have lower kinase activity than the wild-type protein [14]. Therefore, an important extant question for therapeutic development is whether approaches such as kinase inhibition will be effective for all LRRK2 mutations and, by extension, for sporadic disease [15].
The identification of LRRK2 mutations has also led to the development of novel animal models for PD. Although several species have been used to understand LRRK2 function and dysfunction, the greatest variety of models have been generated in mice, in part because the LRRK2 gene is evolutionarily conserved between man and rodents [16]. Because key residues of the protein are identical in these species, knock-in alleles for G2019S [17] and R1441C [18] have been made. Although these animals do not develop Parkinson’s disease, they do have convergent phenotypes related to synaptic vesicle exocytosis that suggest common effects [19]. Some aspects of the effects of mutations are conserved between species, as G2019S increases kinase activity of murine Lrrk2 by ~2-fold [17] as it does for the human protein [20]. However, the properties of human and mouse LRRK2 have not previously been directly compared.
In the present study, we examine how mutations and risk factors affect protein properties of human and mouse LRRK2. We report that conserved substitutions, such as G2019S, have consistent effects on protein function but that this is not the case for variation around position 2385, which is not fully conserved between humans and mice. More interestingly, we find that proteins from different species have different properties with human LRRK2 prominently having more kinase activity than its murine counterpart. These results have implications for the modelling of LRRK2 mutations in mice and on the testing of kinase inhibitors in animals.
Materials and Methods
Plasmids
3xFLAG-HD-LRRK2 constructs were described previously [14, 21, 22]. The QuikChange II XL Site-Directed Mutageneis Kit (Agilent) was used to introduce point mutations. Plasmids were fully sequenced.
Measurement of mRNA and protein expression
HEK293FT cells in a 6-well plate were transiently transfected with either reagent only (Mock) or 3xFlag-HD-tagged Gus, Human LRRK2 WT, Human LRRK2 G2385R, Human LRRK2 G2385E, Mouse LRRK2 WT, Mouse LRRK2 E2385R, or Mouse LRRK2 E2385G plasmid using Lipofectamine 2000 (Invitrogen). Cells were collected 40–48 hours after transfection, with half of the cell samples intended for western blotting and the other half for quantitative real-time PCR. RNA was extracted from the latter set of samples using Trizol (Invitrogen). Following the spectrophotometric determination of RNA concentration on a Nanodrop instrument (Thermo Scientific), the TURBO DNA-free Kit (Ambion) was used to remove any contaminating plasmid DNA from 2 μg of each RNA sample. Then SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) was used to transform 1 μg of the treated RNA into cDNA via reverse transcription. qRT-PCR was performed using power SYBR-green dye mix (Applied Biosystems) and primer pairs that amplified both the human and mouse LRRK2 genes as well as the housekeeper gene human cyclophilin B (PPIB). Samples were run in quadruplicate on a 7900HT Fast Real-time PCR system (Applied Biosystems). As previously described, LRRK2 mRNA levels were calculated by normalization to those of cyclophilin B [23].
Primer sequences used were the following: 3xFLAG-HD forward 5’- GTTTTCCCAGTCACGACGTT-3’, 3xFLAG-HD reverse 5’- ATGGCGGTCATATTGGACAT-3’, human PPIB forward 5’-GCACAGGAGGAAAGAGCATC-3’, and human PPIB reverse 5’-AGCCAGGCTGTCTTGACTGT-3’.
Cell pellets collected for western blotting were lysed in 1x Cell Lysis Buffer (Cell Signaling) with 1x Halt phosphatase inhibitor cocktail (Thermo Scientific) and protease inhibitor cocktail (Roche) by rotation for 30 minutes at 4°C. Lysates were centrifuged for 10 minutes at 13,000 RPM. The cleared lysates were separated by SDS/PAGE (4–20% Criterion TGX gels) and analyzed by immunoblotting for Flag-LRRK2 (Sigma-Aldrich mouse anti-Flag M2, 1:2000) and Cyclophilin B (rabbit anti-Cyclophilin B, Abcam, 1:5000) as a loading control.
Generation of E2385R knock-in mice and measurement of endogenous Lrrk2 protein expression and activity
An arginine residue was inserted at position 2385 of mouse Lrrk2, equivalent to human G2385R, using Crispr-Cas9 by Applied Stem Cell (Menlo Park). A CAAmixture of active guide RNA molecules (gRNAs), one single stranded oligo donor nucleotide (ssODN) and qualified Cas-9 mRNA were injected into the cytoplasm of C57BL/6 embryos. Embryos were implanted into pseudopregnant CD1 mice and live mice born from these litters were genotyped by amplification using primers flanking exon 48 of mouse Lrrk2 (TCAAGTACAGCCCAAACCAGTTA and ATTAGCTTCAATGCTCTCTGTG) followed by sequencing.
Genotype was confirmed by PCR. The mice were housed in a facility with 12-hour light/dark cycles and given access to food and water ad libitum. Homozygous E2385R and WT littermate animals were generated from E2385R heterozygous breeding pairs. Mice were sacrificed at 1 month of age, and brain (bisected along the longitudinal fissure), lung, and kidney tissues were removed and immediately frozen. Samples were thawed and homogenized in 1x Cell Lysis Buffer (Cell Signaling) with 1x Halt phosphatase inhibitor cocktail (Thermo Scientific) and protease inhibitor cocktail (Roche) over a period of 30 minutes on ice. Lysates were centrifuged for 10 minutes at 13,000 RPM, separated by SDS/PAGE and analyzed by immunoblotting for Lrrk2 (Abcam MJFF2 (c41–2) antibody) and cyclophilin B. Separately, lysates were used to assess the kinase activity of endogenous Lrrk2 by immunoblotting for phospho-serine 935 (rabbit anti-pS935, Abcam), phospho-serine 1292 (rabbit anti-pS1292, Abcam), phospho-T72 Rab8A (rabbit anti-Rab8A T72, Abcam), and phospho-T73 Rab10 (rabbit anti-Rab10 T73, Abcam). Measurements from these blots were normalized to total Lrrk2 detected by Abcam MJFF2 c41–2 anti-Lrrk2 antibody or total Rab (rabbit anti-Rab8A or mouse anti-Rab10, Abcam) as appropriate.
LRRK2 pulse-chase protein degradation assay
HEK293FT cells in a Matrigel-coated 6-well plate were transiently transfected with either 3xFlag-HD Human LRRK2 or 3xFlag-HD Mouse Lrrk2 plasmid using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were incubated with cysteine- and methionine-free media for 1 hour. Next, cells were incubated with EasyTag™ EXPRESS35S Protein Labeling Mix (Perkin Elmer) containing both 35S-L-methionine and 35S-L-cysteine for 3 hours, 150 mCi per well. The radioactive media was then aspirated, and cells were washed 2 times with normal media (DMEM, 10% FBS, 1% L-glutamine) supplemented with “cold” amino acids (2mM Methionine and 2mM Cysteine). A set of cells (3 wells each of human and mouse LRRK2-transfected cells) was collected in 1mL of cold PBS immediately to establish 0 hour protein levels. Cells were collected at 4, 8, 18, and 24 hours. Additionally, cells were collected at either 2 hours or 32 hours. Cells were lysed as described above, and lysates were centrifuged for 5 minutes at 13,000 RPM. Immunoprecipitation was performed by 1 hour rotation at 4°C with EZview Red anti-Flag M2 affinity gel (Sigma Aldrich). Samples were washed gently 4 times with wash buffer: 25mM Tris-HCl, 400mM NaCl, and 1% triton. Then Flag-tagged protein was eluted as described below and subjected to SDS/PAGE. Membranes were dried and exposed to a storage phosphor screen for 48–72 hours. Detected 35S-LRRK2 was normalized to total amount of Flag-LRRK2 which was measured via immunoblotting for Flag-LRRK2 (Sigma-Aldrich mouse anti-Flag M2, 1:10000).
Co-immunoprecipitation
HEK293FT cells in a 6-well plate were transiently transfected with 3xFlag-HD LRRK2 plasmid using Lipofectamine 2000 (Invitrogen). After 36–40 hours, cells were collected and lysed in buffer containing 50% Glycerol, 1.5% Triton, 0.1 M Tris-HCl pH 7.5, 0.75 M NaCl and 5 mM EDTA supplemented with 1x Halt phosphatase inhibitor cocktail (Thermo Scientific) and protease inhibitor cocktail (Roche) by rotation for 30 minutes at 4°C. Lysates were centrifuged and further cleared by a 30-minute incubation with EZview Red Protein G affinity gel (Sigma Aldrich) to minimize non-specific binding. Immunoprecipitation was accomplished by overnight rotation at 4°C with EZview Red anti-Flag M2 affinity gel (Sigma Aldrich). Samples were washed gently 4 times with wash buffer: 10% Glycerol, 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 0.1% Triton. Then Flag-tagged protein was eluted in 1x kinase buffer (Cell Signaling) containing 400 mM NaCl, 0.02% Triton and 150 ng/μL of 3xFlag peptide (Sigma Aldrich) by shaking for 20 minutes at 1000 RPM. Standard western blotting procedure was followed and blots were probed for 14-3-3 (mouse anti-14-3-3, Santa Cruz), Hsp90 (rabbit anti-Hsp90, Cell Signaling), Hsc70 (rat anti-Hsc70, Abcam), CHIP (rabbit anti-CHIP, Cell Signaling), phospho-serine 935 (rabbit anti-pS935, Abcam), all at a 1:1000 concentration, and for Flag-LRRK2 (mouse anti-Flag M2, Sigma-Aldrich, 1:2000). Each co-immunoprecipitation was repeated in 7–8 independent experiments. Quantifications were made by calculating the ratio of immunoprecipitated binding partner to amount of LRRK2. Residual calculations were also performed to regress out the effect of individual experiment by making day-to-day variation a factor in a linear model. Graphs show the range of values collected over n=8 independent experiments, normalized across experiments to maintain variance as indicated.
Size-exclusion chromatography
HEK293FT cells in 10-cm dishes were transiently transfected with 3xFlag-HD-LRRK2 plasmid using poly(ethylenimine). Samples were collected 48 hours after transfection and fast protein liquid chromatography (FPLC) was performed as previously described [14].
In vitro assays of LRRK2 enzymatic activity
HEK293FT cells in 15-cm dishes were transiently transfected with 3xFlag-HD-LRRK2 using lipofectamine 2000 (Invitrogen). Cells were lysed 36–40 hours after transfection in buffer containing 10% Glycerol, 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton with 1x Halt phosphatase inhibitor cocktail (Thermo Scientific) and protease inhibitor cocktail (Roche). LRRK2 was purified according to the same protocol used for co-immunoprecipitation with slight modification. Lysates were rotated with anti-Flag M2 affinity gel for just 1 hour, and then the affinity gel was washed 6 times with kinase assay wash buffer (25 mM Tris-HCl pH 7.5, 400 mM NaCl, and 1% Triton) and once with the kinase assay reaction buffer which was also used for elution (1x Cell Signaling Kinase Buffer with 150 mM NaCl and 0.02% Triton). Protein was eluted with 150 ng/μL of 3xFlag peptide by shaking at 1000 RPM for 30 minutes.
Autophosphorylation experiments and GTP binding assays were performed as described previously [14, 21]. Phosphorylation of physiological substrate Rab8A was measured similarly using a reaction mixture containing LRRK2 and Rab8A in a 1:4 ratio.
Immunopurified LRRK2 was incubated at 30°C for 30 minutes with 10 μM non-radioactive ATP, approximately 3 μCi of radioactive ATP [γ−33P] (Perkin Elmer), Rab8A protein that had been immmunopurified alongside LRRK2, and 5x kinase assay reaction buffer diluted with water to reach a total volume of 30 μL with continuous shaking at 300 RPM. The kinase reaction was terminated by addition of SDS sample buffer and heated at 95°C for 5 minutes. Samples were subjected to SDS-PAGE as described above. Membranes were incubated with Coomassie stain, rinsed in water, then dried and exposed overnight to a storage phosphor screen. Measured phosphorylated Rab8A was normalized to the total amount of Rab8A apparent on Coomassie stain.
Cellomics assay for LRRK2 relocalization to the TGN
HEK293FT cells were seeded at 2.5 × 104 cells per well in a Matrigel-coated 96-well plate and transfected with 3xFlag-HD-LRRK2 and either 2xmyc-Gus, 2xmyc-Rab29 or mutant 2xmyc-Rab29 Q67L plasmid. 24 hours after transfection cells were fixed and stained, then imaged and analyzed on a high throughput Cellomics VTI arrayScanner as previously described [21].
Hybrid LRRK2 constructs
Hybrid LRRK2 constructs composed of various human LRRK2 and mouse Lrrk2 sequences were made using the Infusion HD cloning kit (Takara Bio USA). Primers for PCR amplification of the desired fragments were designed using full-length human LRRK2 and mouse Lrrk2 plasmid DNA as a template. This resulted in a large N-terminal fragment (amino acids 1–1883), a kinase domain fragment (amino acids 1884–2135), and a C-terminal region fragment (amino acids 2136–2527) of both human and mouse LRRK2 with 20 base pair overhangs to promote linkage of the pieces in the desired arrangement. The fragments were spliced together in the pCR8 entry vector, then transferred to a 3x-Flag destination vector using the Gateway LR Clonase II Enzyme Mix (Invitrogen). Proper fusion of the plasmid DNA was confirmed by full sequencing of the resulting cDNA constructs. Protein expression of these 3x-Flag N-terminal-tagged hybrid constructs was measured as described above.
Phosphorylation of model substrate Nictide by hybrid LRRK2
3x-Flag hybrid LRRK2 proteins were immunopurified from lysates of transiently transfected HEK293FT cells. The kinase reaction mixtures were prepared on ice as described for measurement of autophosphorylation activity, with the addition of 200 μM of peptide substrate Nictide [24]. 10 μL of each sample were collected at the zero-time point before the kinase reaction was initiated at 30°C and re-suspended with 10 μL of stop solution (0.5 M EDTA in bromophenol blue). After 30 minutes at 30°C, another 10 μL of each sample were re-suspended with 10 μL of stop solution. All collected time points were transferred to P81 paper and left to air-dry overnight. The following day the P81 paper was washed 4 times with 75 mM phosphoric acid for 40 minutes total, then radioactivity of each sample was measured via liquid scintillation counting.
Results
Mouse Lrrk2 is more stable than human LRRK2 and resistant to variation at residue 2385
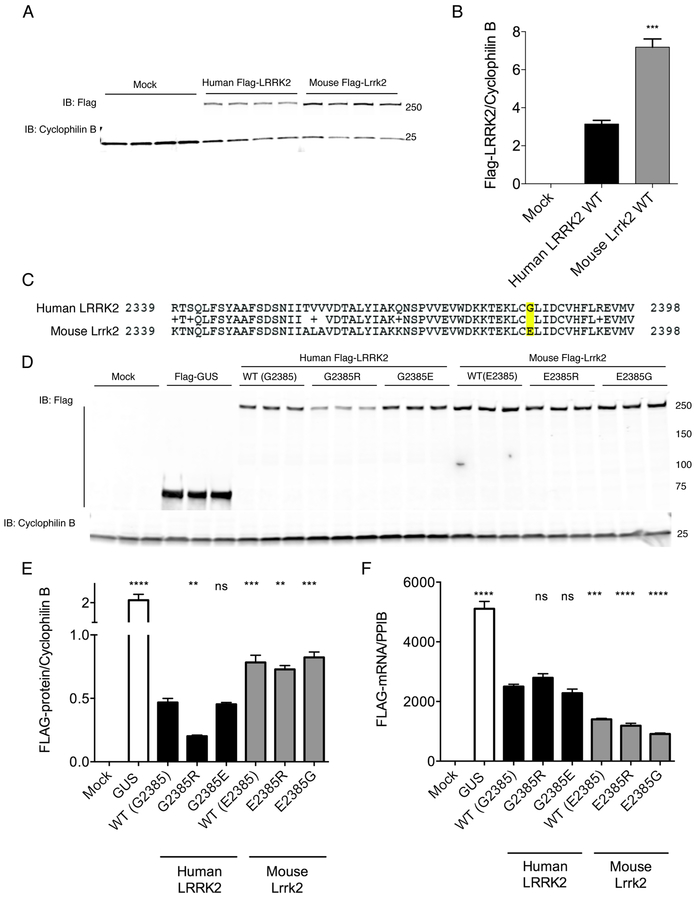
As an initial comparison of human and mouse LRRK2 homologues, we measured the steady-state protein expression levels of 3xFlagHD-LRRK2 cDNA constructs after transient transfection in HEK293FT cells. Using the same antibody against the tagged proteins, we found that mouse Lrrk2 was expressed at higher steady state levels compared to the human version (Fig. 1A, B; n=4 samples per group, F2,9=177, p<0.001, one-way ANOVA).
Figure 1. Human and mouse LRRK2 protein levels and variation at residue 2385.
A. HEK293FT cells were transfected with Flag-tagged human or mouse LRRK2, or mock transfected and protein levels measured by western blot using a flag antibody. Each lane is a separate transfection reaction and is representative of two independent experiments. Cyclophilin B is used as a loading control for each lane.
B. Quantification of protein expression in (A) related to cyclophilin B. Error bars indicate SEM. One way ANOVA indicated significant differences between groups (F2.9=176.9, p<0.0001, n=4 samples per construct); ***, p<0.001 by Tukey’s post-hoc test comparing human and mouse proteins.
C. Region around position 2385 of human (upper sequence) and mouse (lower) LRRK2 proteins.
D. Cells as in A were transfected with the negative control protein GUS or with variants of human and mouse LRRK2. Each lane is a separate transfection reaction and is representative of two independent experiments.
E. Quantification of protein levels in D relative to cyclophilin B. Error bars indicate SEM. **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA with a significant overall effect (F6.14=226.2, p<0.0001, n=3 samples per construct).
F. mRNA expression using primers within the Flag region of the construct relative to cyclophilin B (gene PPIB). Error bars indicate SEM; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA with a significant overall effect (F6.14=125.6, p<0.0001, n=3 samples per construct).
We have shown previously that the G2385R mutation in human LRRK2 is associated with lower steady state protein levels [25]. Upon inspection of the sequence around G2385 in human LRRK2 and its mouse homologue, we noted that this residue was not conserved between species, having a glutamic acid in mouse (Fig. 1C). We therefore looked at G/E/R substitutions at position 2385 in mouse and human LRRK2. As previously described human G2385R was expressed at lower levels than wild-type protein, but substitution of the mouse residue to make G2385E had no effect (Fig. 1D, E). In contrast, mouse Lrrk2 was tolerant of substitution of the variant (E2385R) or wild-type (E2385G) human amino acids.
The differences between human and mouse LRRK2 steady state levels and effects of variation at position 2385 could be related to differences in expression at the RNA level or protein turnover. To exclude altered mRNA levels as a potential mechanism to explain these results, we performed real-time quantitative PCR. In contrast to the protein effects, mRNA expression was lower for the mouse LRRK2 constructs, but variation at the 2385 position did not alter mRNA levels for either species of LRRK2 (Fig. 1F). Therefore, differing protein levels for human and mouse LRRK2 do not appear to be related to mRNA expression. We performed a radioactive 35S protein turnover assay to compare half-lives of human and mouse LRRK2, which yielded calculated half-lives of ~3.5 hours for both species of protein (Supplementary Fig. S1). Thus, differing rates of protein degradation also fails to explain the observed differences in protein levels for human and mouse LRRK2.
We also considered that, by choosing to use a human cell line for consistent transfection, mouse proteins might generally be less efficiently metabolized than their human counterparts. However, mouse Lrrk2 was also more highly expressed than human LRRK2 in other cell types, including primary mouse astrocytes, mouse neuroblastoma N2a cells, and mouse embryonic fibroblast NIH 3T3 cells (Supplementary Fig. S2), indicating that the difference in expression level was not due to our choice of cell line. Overall, these results show that mouse LRRK2 is expressed at higher steady state levels than the human protein and that this is not explained by differences in mRNA expression, protein half-life, or choice of cell line.
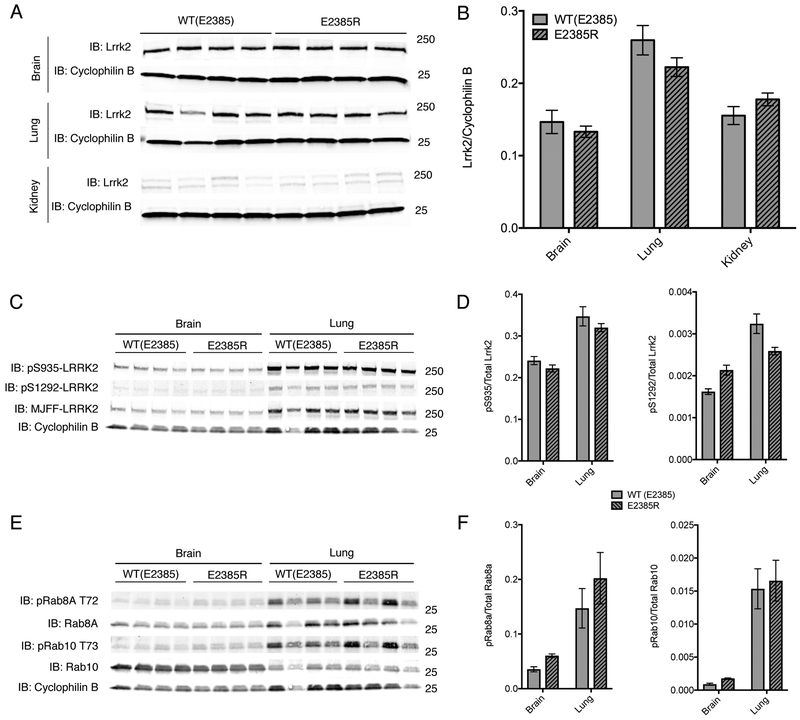
Endogenous expression and activity of Lrrk2 E2385R is similar to wild-type in knock-in mice
A prediction of the above results is that mouse Lrrk2 will be tolerant of substitution of amino acid at position 2385. We therefore knocked-in the human risk variant into the mouse genome, resulting in an E2385R mouse. In vivo expression of Lrrk2 E2385R was measured in half-brain, lung, and kidney of homozygous E2385R knock-in mice and compared to Lrrk2 expression in wild-type littermates. There was no difference observed in Lrrk2 expression between these animals (Fig. 2 A, B) consistent with our in vitro results. Additionally, there were no differences observed in Lrrk2-dependent phosphorylation events based on immunoblotting for LRRK2 autophosphorylation sites (phospho-serines 935 and 1292) and established LRRK2 phosphorylation sites on Rab proteins (phospho-Rab8A T72 and phospho-Rab10 T73) [26].
Figure 2. Knockin of E2385R in mouse Lrrk2 does not affect protein levels in vivo.
A. Protein lysates from the brain (upper panels), kidney (middle panels) or lung (lower panels) of WT or E2385R knockin mice. Each lane is a separate animal. In each tissue, cyclophilin B was used as a loading control.
B. Quantification of steady state of mouse Lrrk2 levels relative to cyclophilin B in each genotype and tissue. In kidney tissue, the double bands detected as Lrrk2 were each quantified as one band. Error bars indicate SEM, n=4 animals per genotype.
C. Protein lysates from the brain (left panel) or lung (right panel) of WT or E2385R knockin mice. Each lane is a separate animal. Cyclophilin B was used as a loading control.
D. Quantification of phospho-serine 935 and phospho-serine 1292 relative to total Lrrk2 protein in each genotype and tissue. Two-way ANOVA indicated that there was a statistically significant difference in pS935 and pS1292 phosphorylation between tissues but not between genotypes (pS935: F tissue (1, 12) = 52.51, p<0.0001; F genotype (1, 12) = 2.645, p=0.1298. pS1292: F tissue (1, 12) = 53.29, p<0.0001; F genotype (1, 12) = 0.2364, p=0.6356; n = 4 animals per genotype).
E. Protein lysates from the brain (left panel) or lung (right panel) of WT or E2385R knockin mice. Each lane is a separate animal. Cyclophilin B was used as a loading control.
F. Quantification of phospho-T72 Rab8A and phospho-T73 Rab10 relative to total Rab protein in each genotype and tissue. Two-way ANOVA indicated that there was a statistically significant difference in Rab8a phosphorylation between tissues but not between genotypes (F tissue (1, 12) = 18.07, p=0.001; F genotype (1, 12) = 0.2564, p=0.6218; n=4 animals per genotype). Similarly, twoway ANOVA indicated that there was a statistically significant difference in Rab10 phosphorylation between tissues but not between genotypes (F tissue (1, 12) = 45.55, p<0.0001; F genotype (1, 12) = 0.239, p=0.6337; n = 4 animals per genotype).
Mouse Lrrk2 has altered binding to chaperones and co-chaperones compared to human LRRK2
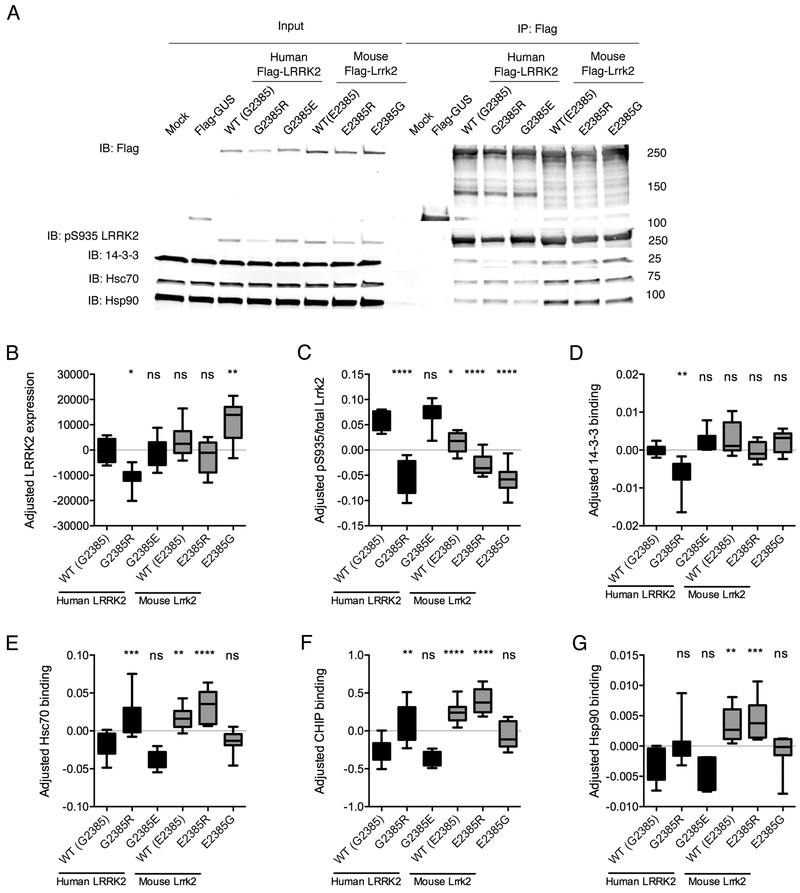
Although there may be multiple mechanisms by which the stability of mouse and human LRRK2 might differ, one possibility is that the two species have different affinities for interacting proteins that regulate stability. We therefore compared interactions with heat shock protein 90 (Hsp90) and heat shock cognate protein 70 (Hsc70), the E3 ubiquitin ligase CHIP (C-terminus of Hsp70-Interacting Protein) [27], and 14-3-3 [28] (Fig. 3). In these experiments we also measured phosphorylation at Ser935, which is one of two phosphosites known to mediate 14-3-3 interaction with LRRK2 [28].
Figure 3. Mouse Lrrk2 binds protein chaperones more than human LRRK2.
A. HEK293FT cells were transfected with the indicated flag-tagged mouse or human LRRK2 constructs, with mock transfected or flag-GUS transfected cells as negative controls, and lysates subjected to immunoprecipitation (IP) using Flag antibodies then blotted for (from top to bottom) flag, pS935-LRRK2, 14-3-3, Hsc70 and Hsp90. Representative data from n=8 independent experiments.
B-G. Quantification of blots for LRRK2 (B) and pS935-LRRK2 (C) and relative IP of 14-3-3 (D), Hsc70 (E), CHIP (F) or Hsp90 (G) binding. In each case, protein levels were normalized across experiments and expressed as standard deviations from the mean, centered at zero for each mean. Lower and upper bounds of the boxes indicate interquartile range, central line indicates the mean while range bars indicate full range of values from n=7–8 independent experiments. **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test from one-way ANOVA compared to WT human LRRK2. In each case, the underlying ANOVA showed significant differences between groups; total LRRK2, F (5, 42) = 11.86, p<0.001; pS935, F (5, 42) = 41.01, p<0.001; 14-3-3, F (5, 36) = 8.221, p<0.001; Hsc70, F (5, 42) = 18.89, p<0.001; CHIP, F (5, 42) = 23.09, p<0.001; Hsp90, F (5, 42) = 10.41, p<0.001.
Confirming previously published results [14], human LRRK2 G2385R showed significantly decreased Ser935 phosphorylation along with a corresponding disruption of 14-3-3 binding. The G2385R mutant also had a higher binding to Hsp90, Hsc70 and CHIP. However, human LRRK2 G2385E, designed to be like mouse protein, did not behave differently from wild type protein
Mouse LRRK2 had increased binding of Hsp90, Hsc70, and CHIP compared to human LRRK2 WT. Furthermore, the human-like E2385G substitution, but not E2385R returned binding to levels similar to the human protein. These results show that mouse and human LRRK2 have differential protein interactions and, given that Hsp90 is a candidate for controlling turnover of LRRK2 [29], one possible interpretation is that the enhanced binding stabilizes mouse over human LRRK2.
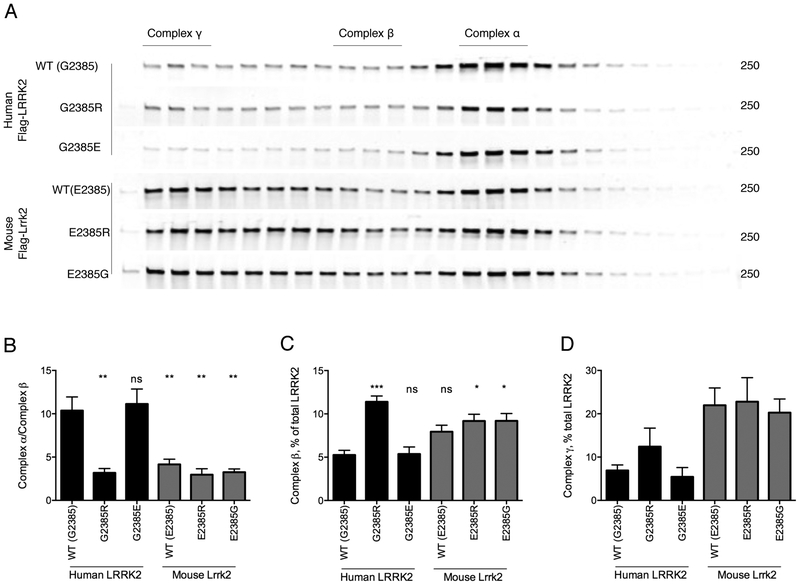
We next used fast protein liquid chromatography (FPLC) to characterize variation in complex formation and possible conformational differences between our set of LRRK2 proteins. The majority of human LRRK2 WT elutes with an apparent molecular weight of around 600 kDa, consistent with prior reports [14, 30–32]. The elution profile of human LRRK2 G2385E was very similar to wild-type, while human LRRK2 G2385R and the mouse LRRK2 proteins showed a significantly larger amount of protein eluting in the higher molecular weight fractions (Fig. 4). However, capacity for self-interaction was not different between species (Supplementary Fig. S3), and therefore the differences in FPLC mobility are not likely due to formation of dimers but rather due to differences in binding chaperone proteins.
Figure 4. Differential mobility of human and mouse LRRK2 in FPLC gradients.
A. HEK293FT cells were transfected with the indicated flag-tagged human and mouse LRRK2 constructs and protein extracts separated by FPLC. Regions above the separated fragments indicate assignment of LRRK2 to each of three major complexes, α,β,γ.
B-D. Quantification relative amounts of LRRK2 in complex a relative to complex b (B), complex b relative to all LRRK2 (B) and complex g relative to all LRRK2 (C) from n=3 independent experiments. **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test from one-way ANOVA compared to WT human LRRK2. In each case, the underlying ANOVA showed significant differences between groups; complex a/complex b, F (5, 18) = 13.2, p<0.001; complex b/all LRRK2, F (5, 18) = 10.5, p<0.001; complex g/all LRRK2, F (5, 18) = 4.44, p=0.0082.
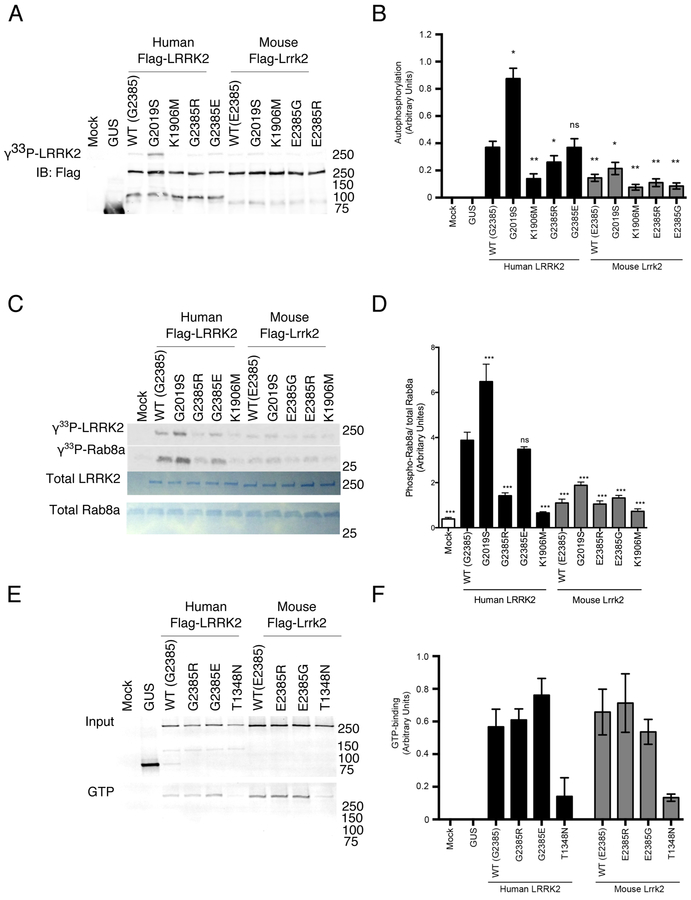
G2385R-LRRK2 and mouse Lrrk2 have low kinase activity in vitro
We therefore examined the enzymatic activities of LRRK2, as these have previously been shown to correlate with protein stability [33]. To address whether human and mouse LRRK2/Lrrk2 have different activities, we used three different in vitro kinase assays, with hyperactive G2019S and kinase dead LRRK2 K1906M [7, 20] used as controls. Mouse Lrrk2 variants had significantly lower autophosphorylation activity than human LRRK2, while human G2385R had lower activity as expected (Fig. 5A, B). Similar results were seen with the model peptide Nictide [24] (Supplementary Fig. S4). Finally, we compared activity against the recently described substrate Rab8A [33] and also found the same pattern of activity (Fig. 5C, D). No significant differences in GTP-binding capacity were observed between any of the variants (Fig. 5E, F). GTP binding-deficient mutants LRRK2 K1347A and T1348N [34, 35] were used as controls. Overall, these results highlight that mouse Lrrk2 has lower kinase activity compared to its human homologue.
Figure 5. Enzymatic activity of human and mouse LRRK2.
A. HEK293FT cells were transfected with human or mouse LRRK2 as indicated, with mock transfected cells and GUS transfected cells as controls. LRRK2 was purified by immunoprecipitation and subjected to autophosphorylation reaction in vitro. Lower blot shows Flag-protein levels in the same reactions. K1906M LRRK2 are kinase dead to evaluate background non-specific reactions.
B. Quantification of n=3 independent experiments as in A. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F9,50=29.24, p<0.001, n=6 independent experiments).
C. Purified LRRK2 variants as in A were incubated with recombinant Rab8a and phosphorylation visualized by autoradiography (upper two panels). Equivalent loading of proteins was confirmed by Coomassie blue staining (lower two panels).
D. Quantification of n=3 independent experiments as in C. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F10,55=44.25, p<0.001, n=3 independent experiments).
E. HEK293FT cells were transfected with human or mouse LRRK2 as indicated, with T1384N as a negative control for GTP binding. Protein lysates were blotted for total LRRK2 (upper panel) or bound to GTP-agarose, eluted and blotted for LRRK2 (lower panel).
F. Quantification of n=3 independent experiments as in E. Mean values per construct were significantly different by one-way ANOVA (F7,16=4.87, p=0.0042, n=3) only the T1348N (‘TN’) constructs were different from the others by post-hoc Tukey’s tests.
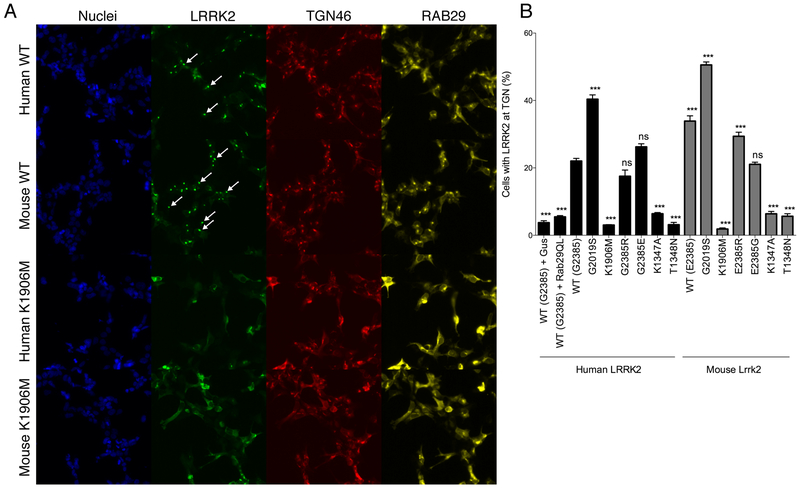
Mouse Lrrk2 relocalizes to trans-Golgi network in the presence of Rab29/Rab7L1
We have reported that LRRK2 will relocalize to the trans-Golgi network (TGN46) in the presence of Rab29/Rab7L1 [22]. As this is a kinase-dependent phenomenon, given that K1906M LRRK2 does not relocalize to the TGN, we used this assay as a functional measure of the overall activity of human and mouse LRRK2 in cells. Both mouse and human LRRK2 were able to support TGN localization (Fig. 6A, B), suggesting that the net effect of higher protein levels (Fig 1) but lower kinase activity (Fig 5) of mouse vs human LRRK2 results in a similar net recruitment to the TGN.
Figure 6. Mouse Lrrk2 can be recruited to the trans-Golgi network.
A. HEK293FT cells were transfected with wild type (WT, upper panels) or kinase dead (K1906M) human or mouse LRRK2 along with human RAB29, then stained for nuclei (blue), LRRK2 (green), the trans-Golgi marker TGN46 (red) and RAB29 (yellow). Arrows on the LRRK2 channel show examples of cells where LRRK2 is recruited to the TGN.
B. Quantification of the percentage of cells with relocalization of LRRK2 to the TGN relative to the overall percentage of transfected cells. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F15,80=275.8, p<0.001, n=6 replicate cultures per construct).
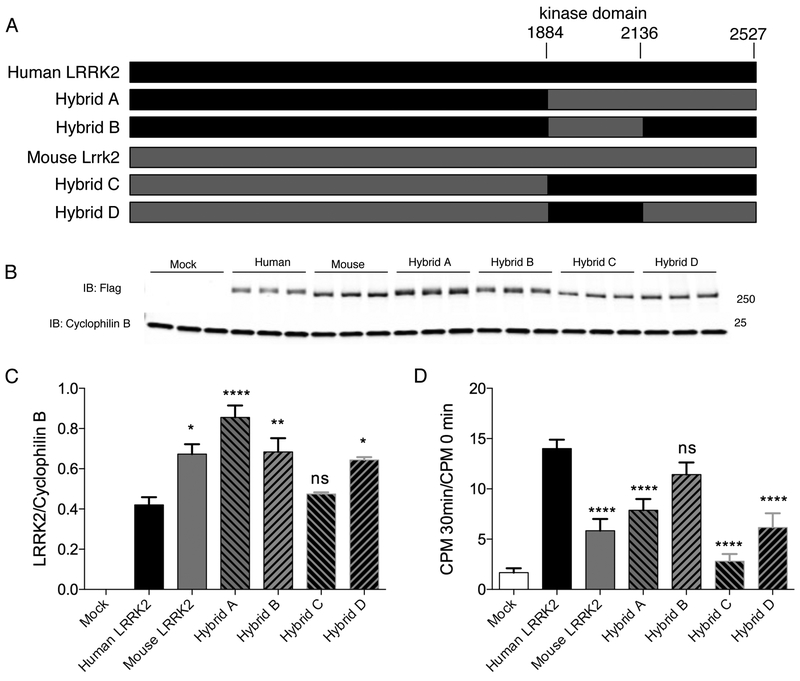
Human-mouse LRRK2 hybrids implicate underlying structural differences in control of steady state protein levels
We made a series of constructs where the kinase and WD40 domains of LRRK2 were exchanged between human and mouse LRRK2 (Fig. 7A). We then measured steady state protein levels of tagged proteins in HEK293FT cells (Fig. 7B, C). Substitution of the C-terminal region, including both kinase and WD40 domains, of mouse LRRK2 into the human protein results in higher protein levels, while substitution of the C-terminal region of human LRRK2 into the mouse protein resulted in lower steady state levels (Fig. 7C).
Figure 7. Human and mouse hybrid constructs.
A. Schematic representation of constructs generated to include human and mouse sequences.
B. HEK293FT cells were transfected in triplicate with flag-tagged versions of the indicated hybrid constructs and blotted for LRRK2 (upper panel). Cyclophilin B is used as a loading control for each lane.
C. Quantification of protein expression in (B) related to cyclophilin B. Error bars indicate SEM, n=3. *, p<0.05; **, p<0.01; ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F6,14=43.8, p<0.0001, n=3 experiments).
D. Quantification of n=3 independent experiments using hybrid constructs of the phosphorylation of the Nictide peptide. ****, p<0.0001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F6,14=54.61, p<0.0001, n=3 experiments).
We also measured kinase activity of the hybrid constructs (Fig. 7D) We found that adding the C-terminal region of mouse LRRK2 to human protein diminished kinase activity (Fig. 7D). However, addition of human C-terminal region did not increase kinase activity of mouse LRRK2.
Discussion
Here, we have compared the biochemical properties of murine Lrrk2 and human LRRK2, and the functional effects of PD-associated mutations and risk factors on proteins. As the amino acid sequence of the LRRK2 protein is over 88% conserved between humans and rodents [36], we expected that human and mouse LRRK2 would behave similarly in general. However, we have found that mouse and human LRRK2 have different properties in terms of steady state levels and kinase activity.
We found that mouse LRRK2 was expressed at higher steady state levels than its human counterpart. As we did not see differences in mRNA expression between species, we considered that protein stability could explain the differences in steady state levels. Measurement of protein turnover did not demonstrate a significant difference in the half-life of human vs. mouse LRRK2, excluding major differences in turnover between the two species. We speculate that the increased steady state levels of mouse Lrrk2 protein may be due to more efficient translation of mouse Lrrk2 rather than a difference in protein stability, although this will need to be confirmed or refuted in future studies. Co-immunoprecipitation experiments demonstrated increased binding by mouse Lrrk2 of chaperone proteins Hsp90, Hsc70, and CHIP, supported by differentiation of elution patterns using FPLC. These chaperones are important mediators of stability and their enhanced interaction with another protein may be indicative of altered protein folding, as with the human G2385R mutant that shows a decreased level of protein expression compared to human wild-type LRRK2 [25].
Another major difference between human LRRK2 and mouse LRRK2 was in enzymatic activity. Although GTP-binding was similar between the two, using several different assays human LRRK2 had significantly greater kinase activity than mouse protein. Prior results suggest that both the kinase activity and the C-terminal region of LRRK2 have been found to affect protein stability [7, 30]. We therefore suggest that the species-dependent effects on steady state levels and kinase activity are inter-related. Because both steady state levels and kinase activity contribute to recruitment of LRRK2 to the TGN, the net effect of the differences between species on this specific cellular assay counteract each other.
Altogether these data suggest that human LRRK2 and mouse Lrrk2 proteins differ structurally from each other and therefore have different biochemical properties. These results could have important implications for modeling Lrrk2 disease in rodent species.
Conclusions
Human and mouse LRRK2 have different steady state levels when expressed in cells from the same plasmid backbone. The non-conserved E2385R mutation in mouse Lrrk2 does not behave like its human LRRK2 G2385R counterpart. Observed differences in protein-protein interactions and enzymatic activities of human and mouse LRRK2 are likely due to differences in protein structure and/or conformation. Implications of these differences for LRRK2-related disease modeling and drug testing in animals remain to be delineated.
Supplementary Material
Supplementary figure S1. Higher mouse LRRK2 protein expression is not explained by increased protein stability.
A. HEK293FT cells were transfected with Flag-tagged human or mouse LRRK2 and subjected to a 35S-cysteine/35S-methionine “pulse” followed by a “chase” with media enriched with with media enriched with “cold” cysteine and methionine. Cell lysates were subjected to immunoprecipitation (IP) for Flag and exposed to a storage phosphor screen (upper panel), then blotted for Flag (lower panel). The representative blots shown are taken from an experiment in which cells were collected at 32 hours. In a separate experiment cells were collected at 2 hours, and not at 32 hours.
B. Quantification of 35S-LRRK2 relative to Flag-LRRK2 for human and mouse LRRK2 from n=3 independent experiments, with technical n=3 for each construct at each time point. Error bars indicate SEM. The best-fit lines shown are semilog lines, where X is linear, and Y is log. A one phase decay non-linear regression equation was used to calculate half-life of each protein (Human LRRK2: half-life = 3.83h, 95%CI=2.71–5.68, R2=0.84. Mouse Lrrk2: half-life = 3.36h, 95%CI=2.60–4.38, R2=0.904).
Supplementary figure S2. Mouse LRRK2 is expressed at higher levels than human protein in mouse cells.
A-C. Primary mouse glial cells (A), N2a cells (B) or NIH 3t3 cells (C) were transfected with Flag-tagged human or mouse LRRK2, or mock transfected and protein levels measured by western blot using a flag antibody. Cyclophilin B is used as a loading control for each lane.
Supplementary figure S3. Mouse and human LRRK2 self-interact at similar levels.
A. HEK293FT cells were co-transfected with GFP-tagged LRRK2 and indicated Flag-tagged human or mouse LRRK2 constructs, with GUS as a negative control. Cell lysates were subjected to immunoprecipitation (IP) for GFP and blotted for GFP (upper panel) and Flag (lower panels). Inputs for the IP are shown on the left.
B. Quantification of IP for Flag-tagged proteins relative to inputs for indicated human and mouse constructs from n=3 independent experiments. Error bars indicate SEM, **, p<0.01; ns, non-significant by Tukey’s post-hoc test from one way ANOVA compared to WT human LRRK2.
Supplementary figure S4. Enzyme activity of mouse and human LRRK2 estimated using model peptide, Nictide.
Quantification of n=3 independent experiments using indicated constructs of the phosphorylation of the Nictide peptide. *, p<0.05; ***, p<0.001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F10,55=20.87, p<0.001, n=6 samples per construct).
Acknowledgements:
We are grateful to Dr. Matthew S. Goldberg (University of Texas Southwestern Medical Center, USA) for sharing a mouse Lrrk2 cDNA construct. This work was supported by the Intramural Research Program of the National Institute on Aging, NIH, by The Michael J. Fox Foundation for Parkinson’s Research, and by a Parkinson’s Foundation-American Parkinson Disease Association Summer Student Fellowship, PF-APDA-SFW-1742.
References
- 1.Kumaran R, Cookson MR (2015) Pathways to Parkinsonism Redux: convergent pathobiological mechanisms in genetics of Parkinson’s disease. Hum Mol Genet 24:R32–44. doi: 10.1093/hmg/ddv236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimprich A, Biskup S, Leitner P, et al. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44:601–607. doi: 10.1016/j.neuron.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 3.Paisán-Ruíz C, Jain S, Evans EW, et al. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44:595–600. doi: 10.1016/j.neuron.2004.10.023 [DOI] [PubMed] [Google Scholar]
- 4.Funayama M, Hasegawa K, Ohta E, et al. (2005) An LRRK2 mutation as a cause for the parkinsonism in the original PARK8 family. Ann Neurol 57:918–921. doi: 10.1002/ana.20484 [DOI] [PubMed] [Google Scholar]
- 5.Hernandez DG, Reed X, Singleton AB (2016) Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J Neurochem 139 Suppl 1:59–74. doi: 10.1111/jnc.13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cookson MR (2010) The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci 11:791–797. doi: 10.1038/nrn2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greggio E, Jain S, Kingsbury A, et al. (2006) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23:329–341. doi: 10.1016/j.nbd.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 8.Skibinski G, Nakamura K, Cookson MR, Finkbeiner S (2014) Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci Off J Soc Neurosci 34:418–433. doi: 10.1523/JNEUROSCI.2712-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee BD, Shin J-H, VanKampen J, et al. (2010) Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med 16:998–1000. doi: 10.1038/nm.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao C, Johnson WM, Gao Y, et al. (2013) Kinase inhibitors arrest neurodegeneration in cell and C. elegans models of LRRK2 toxicity. Hum Mol Genet 22:328–344. doi: 10.1093/hmg/dds431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Hamamichi S, Lee BD, et al. (2011) Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum Mol Genet 20:3933–3942. doi: 10.1093/hmg/ddr312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, He X, Thomas JM, et al. (2015) A novel GTP-binding inhibitor, FX2149, attenuates LRRK2 toxicity in Parkinson’s disease models. PloS One 10:e0122461, doi: 10.1371/journal.pone.0122461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AB (2015) Ten years and counting: moving leucine-rich repeat kinase 2 inhibitors to the clinic. Mov Disord Off J Mov Disord Soc 30:180–189. doi: 10.1002/mds.26075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudenko IN, Kaganovich A, Hauser DN, et al. (2012) The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem J 446:99–111. doi: 10.1042/BJ20120637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudenko IN, Chia R, Cookson MR (2012) Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson’s disease? BMC Med 10:20. doi: 10.1186/1741-7015-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langston RG, Rudenko IN, Cookson MR (2016) The function of orthologues of the human Parkinson’s disease gene LRRK2 across species: implications for disease modelling in preclinical research. Biochem J 473:221–232. doi: 10.1042/BJ20150985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue M, Hinkle KM, Davies P, et al. (2015) Progressive dopaminergic alterations and mitochondrial abnormalities in LRRK2 G2019S knock-in mice. Neurobiol Dis 78:172–195. doi: 10.1016/j.nbd.2015.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Y, Pisani A, Martella G, et al. (2009) R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc Natl Acad Sci U S A 106:14622–14627. doi: 10.1073/pnas.0906334106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greggio E, Cookson MR (2009) Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro 1:. doi: 10.1042/AN20090007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West AB, Moore DJ, Biskup S, et al. (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A 102:16842–16847. doi: 10.1073/pnas.0507360102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chia R, Haddock S, Beilina A, et al. (2014) Phosphorylation of LRRK2 by casein kinase 1α regulates trans-Golgi clustering via differential interaction with ARHGEF7. Nat Commun 5:5827. doi: 10.1038/ncomms6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beilina A, Rudenko IN, Kaganovich A, et al. (2014) Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc Natl Acad Sci U S A 111:2626–2631. doi: 10.1073/pnas.1318306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RTPCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols RJ, Dzamko N, Hutti JE, et al. (2009) Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. Biochem J 424:47–60. doi: 10.1042/BJ20091035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudenko IN, Kaganovich A, Langston RG, et al. (2017) The G2385R risk factor for Parkinson’s disease enhances CHIP-dependent intracellular degradation of LRRK2. Biochem J 474:1547–1558. doi: 10.1042/BCJ20160909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steger M, Tonelli F, Ito G, et al. (2016) Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5:. doi: 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding X, Goldberg MS (2009) Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PloS One 4:e5949. doi: 10.1371/journal.pone.0005949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols RJ, Dzamko N, Morrice NA, et al. (2010) 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J 430:393–404. doi: 10.1042/BJ20100483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Xie C, Greggio E, et al. (2008) The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci Off J Soc Neurosci 28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen ND, Peng Y, Ho CC-Y, et al. (2009) The WD40 domain is required for LRRK2 neurotoxicity. PloS One 4:e8463. doi: 10.1371/journal.pone.0008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greggio E, Zambrano I, Kaganovich A, et al. (2008) The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem 283:16906–16914. doi: 10.1074/jbc.M708718200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein CL, Rovelli G, Springer W, et al. (2009) Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J Neurochem 111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x [DOI] [PubMed] [Google Scholar]
- 33.Zhao J, Molitor TP, Langston JW, Nichols RJ (2015) LRRK2 dephosphorylation increases its ubiquitination. Biochem J 469:107–120. doi: 10.1042/BJ20141305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito G, Okai T, Fujino G, et al. (2007) GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry (Mosc) 46:1380–1388. doi: 10.1021/bi061960m [DOI] [PubMed] [Google Scholar]
- 35.West AB, Moore DJ, Choi C, et al. (2007) Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet 16:223–232. doi: 10.1093/hmg/ddl471 [DOI] [PubMed] [Google Scholar]
- 36.West AB, Cowell RM, Daher JPL, et al. (2014) Differential LRRK2 expression in the cortex, striatum, and substantia nigra in transgenic and nontransgenic rodents. J Comp Neurol 522:2465–2480. doi: 10.1002/cne.23583 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1. Higher mouse LRRK2 protein expression is not explained by increased protein stability.
A. HEK293FT cells were transfected with Flag-tagged human or mouse LRRK2 and subjected to a 35S-cysteine/35S-methionine “pulse” followed by a “chase” with media enriched with with media enriched with “cold” cysteine and methionine. Cell lysates were subjected to immunoprecipitation (IP) for Flag and exposed to a storage phosphor screen (upper panel), then blotted for Flag (lower panel). The representative blots shown are taken from an experiment in which cells were collected at 32 hours. In a separate experiment cells were collected at 2 hours, and not at 32 hours.
B. Quantification of 35S-LRRK2 relative to Flag-LRRK2 for human and mouse LRRK2 from n=3 independent experiments, with technical n=3 for each construct at each time point. Error bars indicate SEM. The best-fit lines shown are semilog lines, where X is linear, and Y is log. A one phase decay non-linear regression equation was used to calculate half-life of each protein (Human LRRK2: half-life = 3.83h, 95%CI=2.71–5.68, R2=0.84. Mouse Lrrk2: half-life = 3.36h, 95%CI=2.60–4.38, R2=0.904).
Supplementary figure S2. Mouse LRRK2 is expressed at higher levels than human protein in mouse cells.
A-C. Primary mouse glial cells (A), N2a cells (B) or NIH 3t3 cells (C) were transfected with Flag-tagged human or mouse LRRK2, or mock transfected and protein levels measured by western blot using a flag antibody. Cyclophilin B is used as a loading control for each lane.
Supplementary figure S3. Mouse and human LRRK2 self-interact at similar levels.
A. HEK293FT cells were co-transfected with GFP-tagged LRRK2 and indicated Flag-tagged human or mouse LRRK2 constructs, with GUS as a negative control. Cell lysates were subjected to immunoprecipitation (IP) for GFP and blotted for GFP (upper panel) and Flag (lower panels). Inputs for the IP are shown on the left.
B. Quantification of IP for Flag-tagged proteins relative to inputs for indicated human and mouse constructs from n=3 independent experiments. Error bars indicate SEM, **, p<0.01; ns, non-significant by Tukey’s post-hoc test from one way ANOVA compared to WT human LRRK2.
Supplementary figure S4. Enzyme activity of mouse and human LRRK2 estimated using model peptide, Nictide.
Quantification of n=3 independent experiments using indicated constructs of the phosphorylation of the Nictide peptide. *, p<0.05; ***, p<0.001; ns, non-significant by Tukey’s post-hoc test compared to WT human LRRK2 from one way ANOVA (F10,55=20.87, p<0.001, n=6 samples per construct).