Abstract
Magnetic resonance (MR) images of the brain are of immense clinical and research utility. At the atomic and subatomic levels, the sources of MR signals are well understood. However, we lack a comprehensive understanding of the macromolecular correlates of MR signal contrast. To address this gap, we used genome-wide measurements to correlate gene expression with MR signal intensity across the cerebral cortex in the Allen Human Brain Atlas (AHBA). We focused on the ratio of T1-weighted and T2-weighted intensities (T1-w/T2-w ratio image), which is considered to be a useful proxy for myelin content. As expected, we found enrichment of positive correlations between myelin-associated genes and the ratio image, supporting its use as a myelin marker. Genome-wide, there was an association with protein mass, with genes coding for heavier proteins expressed in regions with high T1-w/T2-w values. Oligodendrocyte gene markers were strongly correlated with the T1-w/T2-w ratio, but this was not driven by myelin-associated genes. Mitochondrial genes exhibit the strongest relationship, showing higher expression in regions with low T1-w/T2-w ratio. This may be due to the pH gradient in mitochondria as genes up-regulated by pH in the brain were also highly correlated with the ratio. While we corroborate associations with myelin and synaptic plasticity, differences in the T1-w/T2-w ratio across the cortex are more strongly linked to molecule size, oligodendrocyte markers, mitochondria, and pH. We evaluate correlations between AHBA transcriptomic measurements and a group averaged T1-w/T2-w ratio image, showing agreement with in-sample results. Expanding our analysis to the whole brain results in strong positive T1-w/T2-w correlations for immune system, inflammatory disease, and microglia marker genes. Genes with negative correlations were enriched for neuron markers and synaptic plasticity genes. Lastly, our findings are similar when performed on T1-w or inverted T2-w intensities alone. These results provide a molecular characterization of MR contrast that will aid interpretation of future MR studies of the brain.
Keywords: Molecular neuroanatomy, Gene expression, Transcriptomics, Myelin map, Cortex
Introduction
Magnetic resonance imaging (MRI) is of immense clinical utility and has revolutionized studies of human brain structure and function (Lerch et al., 2017). T1-weighted and T2-weighted MR images are commonly used to study the structure of the brain. Contrast in a T1-weighted (T1-w) MR image is related to the time required for protons (hydrogen ions) to return to equilibrium magnetization after excitation by a radio-frequency pulse (T1 or longitudinal relaxation). A T2-weighted (T2-w) MR signal approximates the time required for the T2 or spin-spin vectors in the magnetic transverse plane to return to equilibrium (loss of phase coherence or alignment). Contrast in these images is associated with molecule size, iron content, diffusion/flow, pH, water content, water binding, and proton density (Koenig, 1995; Kucharczyk et al., 1994; MacKay and Laule, 2016; Stüber et al., 2014; Vymazal et al., 1995). The T1-w and T2-w MRI signals have been associated with histologically assayed myelin content in a study of diseased and normal white matter (Schmierer et al., 2008). These contrasts allow identification of pathologies and segmentation of tissues; for example, white matter lesions that mark demyelination and inflammation are bright in T2-w images and black in T1-w images (Sahraian et al., 2010). While coarse tissue differences are visible across the brain and attributable to general properties, a more detailed understanding of the molecular determinants of MR image contrast in homogeneous brain regions is lacking.
To enhance contrast in the cortex, Glasser and Van Essen proposed and evaluated the ratio of the T1-w and T2-w intensities as a cortical ‘myelin map’ (Glasser and Van Essen, 2011). The T1-w/T2-w ratio image attenuates the receiver-coil bias, which is present in both source images, and because the noise in the two volumes is largely uncorrelated, combining the two measurements increases the accuracy of the myelin-content signal (Glasser and Van Essen, 2011). By taking the T1-w/T2-w ratio across the cortical surface as representative of local relative myelin content, it is possible to use this estimate of the cortical myeloarchitecture to generate finer cortical segmentations (Glasser et al., 2016). The T1-w/T2-w ratio was found to identify more robust markers of schizophrenia than was possible with the individual T1-w or T2-w images (Ganzetti et al., 2015; Iwatani et al., 2015). It has also been used for segmentation of the human habenula and to assay hippocampal myelin in subjects with post-traumatic stress disorder (Chao et al., 2015; Kim et al., 2016). In addition to myelin content, aerobic glycolysis and synaptic plasticity have been associated with the T1-w/T2-w ratio (Glasser et al., 2014).
This paper focuses on the Allen Human Brain Atlas (AHBA) which provides transcriptomic information and MR images for six postmortem brains (Hawrylycz et al., 2012). The AHBA is unprecedented in its size and scope, representing a comprehensive “all genes, all structures” view of the human brain from both an anatomical and transcriptomic perspective (Shen et al., 2012). This data has been related to neuroimaging-based data from positron emission tomography (PET) (Rizzo et al., 2016), fMRI derived networks (Vértes et al., 2016), adolescent cohorts that measure brain maturation (Shin et al., 2017; Whitaker et al., 2016), and connectivity (Hawrylycz et al., 2015; Richiardi et al., 2015; Romme et al., 2017). More recently, the anatomically parcellated T1-w/T2-w ratio information from the Human Connectome Project has been combined with the AHBA to study the anatomical hierarchy of the cortex (Burt et al., 2017). These integrative studies combined imaging data from other studies and subjects with the transcriptomic profiles of the six donor brains [reviewed in (Mahfouz et al., 2017)]. More directly, two studies have also associated expression with connectivity obtained from the diffusion tensor images of two of the Atlas brains (Forest et al., 2017; Li et al., 2016). However, to our knowledge, no studies have used the MR images of the six donor brains to examine relationships between gene expression and MR signal intensity.
The wide clinical and research use of structural neuroimaging motivates our work to characterize the molecular basis of contrast in these images. In the current paper, we investigate whether the expression of genes associated with specific diseases, cell-types, cellular components, processes, or functions have higher or lower spatial correlations with magnetic resonance intensity (Fig. 1). Like the majority of neuroimaging studies, we focus on the cerebral cortex, but also examine these associations throughout the whole-brain. We use T1-w/T2-w ratio as our primary contrast measure, but also compare results obtained from T1-w and T2-w intensities alone.
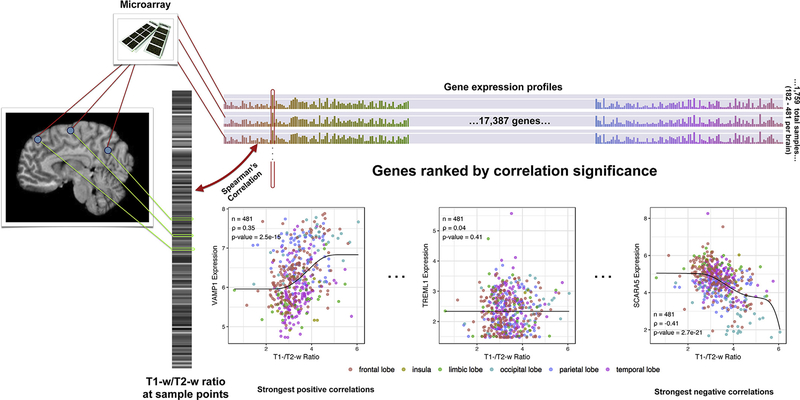
Fig. 1.
Overview of the correlation analysis. MR intensities are extracted from the images on the left, depicted as a single grayscale vector which each bar corresponding to an Allen Atlas sample. The colored bar on top visualizes the expression matrix with samples as rows and genes as columns. Each gene/column forms an expression profile across the cortex (outlined in red) and is correlated with the grayscale intensity vector. Genes are then sorted from strong positive to strong negative correlation for gene set enrichment analysis to test if a particular set is enriched for positive or negative correlations. VAMP1 (high positive correlation), TREML1 (no correlation), and SCARA5 (strong negative correlation) provide examples across the correlation range using data from donor H0351.2001.
Materials and methods
Gene expression and MRI data
The Allen Human Brain Atlas (AHBA) is a multimodal dataset containing comprehensive transcriptomic, neuroimaging and histological information for brains obtained from 6 healthy adult human donors (1 female and 5 male, aged 24–57 years) (Hawrylycz et al., 2012). Custom 64K Agilent microarrays were used to assay genome-wide expression in 3702 spatially-resolved samples. The Allen Institute normalized the data with a seven-step process that adjusted expression for array-specific biases, batch, and dissection method. Full details of the procedure used by Allen Institute researchers are available in the AHBA technical white paper (http://help.brain-map.org/display/humanbrain/Documentation).
To improve quality of the expression data, we filtered out microarray probes that lacked correlation with RNA sequencing data in a subset of samples, using quality control data calculated by Miller et al. (2014). The authors developed a procedure to remove noisy probes with non-specific, off-target, or saturation effects. Specifically, probes that survive the procedure show a significant positive correlation between RNA sequencing and microarray expression values in 231 of the AHBA samples (q < 0.1). Of the 58,692 probes on the array, 37,796 remained after removing the probes that failed the Miller et al. test or that were not mapped to a known gene. Of these 37,796 probes, 6344 lacked a Miller et al. test result since they were mapped to genes not included in the RNA sequencing analysis. We included these probes in our analysis. At the gene level, the mean average was used to summarize expression across multiple probes that map to a single gene (17,387 genes total, average 2.2 probes per gene).
In addition to transcriptomic measurements, structural postmortem MR images were acquired for the same donors by the Allen Institute on 3T Siemens Magnetom Trio scanners (Erlangen, Germany). There was variation in imaging parameters and sites across the subjects, including changes to pulse sequences [page 12 of (Allen Institute for Brain Science, 2013)]. For four of the brains 3D T1-weighted magnetization-prepared rapid gradient echo (MPRAGE) structural MRI images were acquired with 0.98 × 0.98 × 1 mm voxels, three averages, TI = 900 ms, TR = 1900 ms, TE = 2.63 or 3.03 ms and 9° flip angle resulting in an image matrix of 256 × 256 × 176. T1-weighted images of the remaining two brains were acquired with the same parameters except for TE = 2.63 ms and 1 mm isotropic voxels, providing an image matrix of 256 × 192 × 256. For four of the brains, 3D T2-weighted images were acquired with 0.98 × 0.98 × 1 mm voxels, TR = 3200 ms, TE = 449 ms, 120° flip angle resulting in an image matrix of 256 × 256 × 192. T2-weighted images of the remaining two brains were acquired with the same parameters except for 0.9 mm isotropic voxels and TR = 3200–3210 ms, TE = 535–540 ms resulting in an image matrix of 224 × 256 × 160. Two brains were imaged in cranio and four were imaged ex cranio. We obtained the T1-w and T2-w MR images directly from the AHBA. Bias field correction was performed using the SPM12 software package (http://www.fil.ion.ucl.ac.uk/spm/software/spm12/). Default parameters for bias correction were used for all scans (60 mm cutoff and very light regularisation (0.0001)). Upon visual inspection, default parameters did not produce adequate results for one scan (H0351.1012 T2 image), thus they were rerun using a Bias FWHM of 140 mm cutoff and heavy regularisation (0.1).
For each transcriptomic sample, histological information allowed for a determination of the native MRI coordinates and brain region from which the sample was taken. These coordinates and regions were published by the AIBS along with a structural part-of-ontology or reference atlas. For our main analysis, we restricted our analysis to the measurements located in the cerebral cortex (region ID of 4008 in the AHBA reference atlas) so that results would be directly applicable to characterization of the T1-w/T2-w ratio within the cortex. To focus on the neocortex, allocortical regions (hippocampal formation and piriform cortex, region IDs 4249 and 10142) from the AHBA cerebral cortex region were excluded.
In addition to the T1-w/T2-w image, the analysis was repeated for the T1-w and T2-w images, so that comparison of results could be performed between the three imaging measures.
Human Connectome group average T1-w/T2-w image
We obtained a group averaged T1-w/T2-w ratio image from the Human Connectome Project. We used the S1200_Aver-ageT1wDividedByT2w.nii.gz file from the S1200 group average release which was derived from 1113 healthy young adult participants. The HCP 3D T1-weighted MPRAGE images were acquired with the following parameters: TR = 2400 ms, TE = 2.14 ms, TI = 1000 ms, 8° flip angle, 0.7 mm isotropic voxels, and a field of view of 224 × 224 mm. The HCP 3D T2-weighted turbo spin echo sequence (SPACE) images were acquired with the following parameters: TR = 3200 ms, TE = 565 ms, variable flip angle, 0.7 mm isotropic voxels and a field of view of 224 × 244 mm [page 20 of (Human Connectome Project, 2013)].
Correlation analysis
Within each donor, we computed Spearman rank correlations between mean expression for each gene and T1-w/T2-w intensity at the sample points, using the native MRI coordinates. Across the donors, Fisher’s method was used to generate a single combined meta p-value for each gene and direction (Fisher, 1925). This was done by combining six one-sided p-values for positive correlation and another six for negative correlation, resulting in two meta-p values per gene. The lowest was then chosen to provide a single p-value per gene. Finally, multiple test correction was performed using the Bonferroni-Holm family-wise error rate (FWER) method. This procedure extends the simple Bonferroni technique by adding a step-down calculation (Holm, 1979).
To generate correlations between AHBA transcriptomic profiles and the Human Connectome Project (HCP) group average ratio image, the Allen Institute-provided MNI coordinates (resulting from affine and deformable registration methods (Allen Institute for Brain Science, 2013)) for each expression sample were used to link the HCP data. Correlation analysis was performed within each donor, and results were combined at the gene level as described above.
To obtain a genome-wide ranking of genes for enrichment analysis, we ordered genes by the meta p-value and direction (from the most positively correlated gene with the lowest p-value to the most negatively correlated gene with the lowest p-value).
The SCAM R package was used to plot curves representing the rank correlation in the scatter plots (Pya and Wood, 2015).
Linear modeling
The five most positively and negatively correlated genes from the above correlation analysis were used as predictors in an ordinary least squares linear model for each donor. This model was fit separately for each of the donors to determine the percentage of variance explained by the top results.
Gene ontology enrichment analysis
We employed gene set enrichment analysis to summarize the genome-wide results that contain a large number of significantly correlated genes. Through literature curation, the Gene Ontology (GO) classifies genes into sets associated with specific biological processes, molecular functions and cellular locations (Ashburner et al., 2000). This allows us to test if the genes correlated with the T1-w/T2-w ratio are enriched for a common location, function or process using the area under the receiver operator curve (AUROC) statistic. The AUROC for a set of genes is equivalent to the probability that a gene associated with that set will be found first in the genome-wide ranking compared to an unassociated gene. The ranking we use arranges genes from strongest positive correlation to strongest negative correlation. In this context, AUROC > 0.5 for a gene set means that samples with high expression of this gene group will be more likely to have a higher T1-w/T2-w value (genes are enriched for positive correlation). For AUROC < 0.5 the reverse is true, with a bias towards higher expression in samples with a lower or darker T1-w/T2-w ratio (enriched for negative correlation).
A Gene Ontology (GO) enrichment analysis was performed on the genome-wide gene rankings generated from the signed p-values. Annotations for GO gene groups were taken from the GO. db and org. Hs.eg.db packages in R (Carlson, 2017a, 2017b). Annotations were dated March 29, 2017. We calculated the AUROC values for GO groups comprising more than 10 and less than 200 genes, after intersection with genes present in the Allen microarray data (6885 GO groups annotating 13,384 unique genes). AUROC values were generated using the tmod analysis package in R (Weiner and Domaszewska, 2016). The Mann-Whitney U test was used to determine statistical significance. Multiple test correction of the resultant p-values for the many GO groups tested was performed using the Bonferroni-Holm family-wise error rate (FWER) method.
We investigated ten myelin-related GO groups, which were selected by filtering for GO terms containing the strings “myelin” or “ensheathment” and excluding the strings “peripheral” and “sphingomyelin”. Because of the a priori hypothesis of a correlation between myelin-associated genes and the T1-w/T2-w ratio, we performed a separate analysis for these GO groups of interest and did not consider non-myelin-related GO groups while performing multiple test correction. We also used the Benjamini-Hochberg false discovery rate (FDR) procedure for multiple test correction instead of the family-wise error rate method (Benjamini and Hochberg, 1995). The FDR procedure better handles multiple correlated tests, which are expected in this case due to the high overlap of genes between these myelin sets (Benjamini and Yekutieli, 2001).
Myelin fraction marker genes
A transcriptomic study of myelin extracted from whole brain provided a list of genes that were more abundant in myelin from six-month-old mice compared to cortex (Thakurela et al., 2016). This transcriptomic list was obtained from Supplement Table S2 of Thakurela et al. which contains genes with a normalized read count of greater than 100 which are at least twice as abundant in myelin than cerebral cortex. The proteomic list of distinct proteins identified in myelin was obtained from Supplemental Table S7.
Cell type enriched genes
Lists of genes that mark specific transcriptomic cell types in the mouse and human brain were used as a source of cell type markers. Enrichment of these gene lists was determined using the previously described AUROC statistic. All cell type-enriched gene lists taken from mouse studies were first mapped to human homologs using the Homologene database (NCBI Resource Coordinators, 2016) and Ogan Mancarci’s “homologene” R package (https://github.com/oganm/homologene). While these lists have significant overlap for matching cell-types, we note that these sources of markers differ in size of gene lists, number of cells assayed, cortical regions assayed, and experimental design.
Zeisel marker genes
Cell type-specific genes from the mouse brain were obtained from a single cell analysis of 3005 cells from the adult mouse somatosensory cortex (S1) and hippocampus (Zeisel et al., 2015). We used genes from the nine modules provided in Supplemental Table 1 of Zeisel et al. (2015). These modules were identified by clustering of both cells and genes and correspond to 9 major classes (interneurons, SS pyramidal neurons, CA1 pyramidal neurons, oligodendrocytes, microglia, endothelial cells, mural cells, astrocytes and ependymal cells).
NeuroExpresso marker genes
We used cell type marker lists obtained from a cross-laboratory analysis of cell type specific transcriptomes (Mancarci et al., 2017). Mancarci et al. aggregated publicly-available gene expression values from microarray and single-cell RNA-seq studies to identify marker genes for 36 major cell types. We only used the markers obtained from cortex datasets, which included marker lists for astrocytes, endothelial cells, oligodendrocytes, OPCs, pyramidal neurons, GABAergic neurons (further labelled as PV, RelnCalb, and VIPReln), and microglia (further classified as activation state independent, activated, and deactivated). The gene lists were obtained from https://github.com/oganm/neuroExpressoAnalysis.
Darmanis marker genes
From the first single cell transcriptome analysis of healthy human cortex, we obtained marker gene lists corresponding to six main cell types (Darmanis et al., 2015). This study profiled expression in 466 cells from the human cortical tissue of eight adult subjects and four embryos. Clustering was employed to group the cells into six main groups (astrocytes, oligodendrocytes, oligodendrocyte precursor cells (OPCs), neurons, microglia, and endothelial cells) based on their transcriptomic profile, which was obtained using RNA sequencing (Darmanis et al., 2015). We used the top 21 enriched genes in each of the groups from Table S3 of Darmanis et al. and excluded the gene groups associated with fetal cell types. These groups were obtained from unbiased genome-wide results and not the focused clustering that was also applied by the authors.
Perineuronal oligodendrocyte marker genes
Genes expressed at higher levels in perineuronal oligodendrocytes as marked by cell-specific tags were obtained from DataS1 (oligodendrocyte precursor comparison, A2B5+ marker) and DataS2 (committed oligodendrocyte comparison, O4+ marker) in Szuchet et al. (2011). These two lists of rat genes with more than a twofold difference between cell types were filtered for higher expression in perineuronal oligodendrocytes (as marked by A2B5+/OTMP+) and mapped to human homologs. We used the intersection of the two lists to provide a single list of 493 genes that were enriched in A2B5+/OTMP+ cells in comparison to both A2B5+ and O4+ cells.
Laminar marker genes
Cortical layer enriched genes were obtained from a transcriptomic analysis performed by He et al. (2017). In this study, unsupervised sectioning and RNA sequencing were used to assay genome-wide expression profiles of the human prefrontal cortex. Specifically, we used annotations in the ‘Layer marker in human’ column of Supplementary Table S2 (He et al., 2017). This use of laminar marker genes for characterizing the T1-w/T2-w ratio was inspired by an early version of Burt et al. (2017).
pH associated genes
A cross-laboratory analysis of human gene expression profiles provided a source of pH associated genes (Mistry and Pavlidis, 2010). This meta-analysis tested the effects of brain pH on gene expression in eleven datasets that primarily assayed the frontal cortex of the normal human brain. Genes up- and down-regulated with pH were obtained from Supplementary Table 6 at http://hdl.handle.net/11272/10572 (Mistry and Pavlidis, 2010). A subset of core pH up-regulated genes with robust associations was also used.
Disease gene lists
Disease-associated gene sets were downloaded from the Phenocarta database on October 28th, 2016. Phenocarta consolidates data on gene-to-phenotype mappings taken from 17 distinct resources (Portales-Ca-samar et al., 2013). The three sources that provide the majority of the annotations in the database are the Disease and Gene Annotations, The Rat Genome Database and the Comparative Toxicogenomics Database (Davis et al., 2017; Peng et al., 2013; Shimoyama et al., 2015). We investigated all disease-associated gene lists in the size range of 5–200 genes, a total of 1177 diseases.
Availability
Scripts, supplementary tables, and data files for reproducing the analyses are available online at http://www.github.com/jritch/mri_transcriptomics and https://figshare.com/articles/Transcriptomic_characterization_of_MRI_contrast_focused_on_the_T1-w_T2-w_ratio_in_the_cerebral_cortex/5270926.
Results
Allen Human Brain Atlas samples per donor ranged from 182 to 481 cortical regions. We found that intensities of T1-w, T2-w and the ratio image within the single brains were highly correlated across the samples (absolute Spearman’s rho > 0.72). In single brains, correlations between gene expression and T1-w/T2-w ratio have Spearman rank correlation values ranging from −0.47 to 0.41. Combined, median correlation across the six donors ranged from −0.31 to 0.26. After multiple test correction, 1025 positively correlated genes and 2581 negatively correlated were identified at pfwer < 0.05 (complete list in Supplement Table 1). Inconsistency across brains is observed with only 47.6% of the 3606 significant genes showing the same correlation direction across all six brains and 85.6% with consistent direction in at least five.
We built linear models to assay the amount of variance in the T1-w/T2-w ratio explained by the top genes in each brain. The top five negatively and five positively correlated genes together explain 14.6% of the variance in T1-w/T2-w ratio (average adjusted R2 across the six brains). This metric varies from 6.6% (donor 12876/H0351.1009) to 21.6% (donor 15496/H0351.1015). This explanatory power and the thousands of significant genes motivates our characterization of both the top genes and broader genome-wide signals.
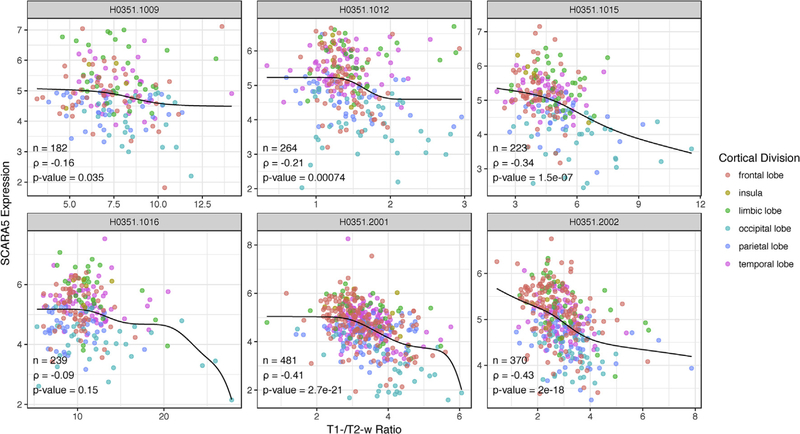
The most significant gene was negatively correlated with T1-w/T2-w ratio (SCARA5, scavenger receptor class A member 5, Fig. 2). Across the six brains, SCARA5 showed a median Spearman correlation of ‒0.27 (ranging from ‒0.093 to ‒0.43). The median across donors, when restricted to data for each of the cortical divisions colored in Fig. 2 ranges from ‒0.13 to ‒0.28, suggesting the relationship is maintained within anatomical divisions.
Fig. 2.
Sample-wise plots of T1-w/T2-w ratio and SCARA5 gene expression. Each donor represented as a single scatter plot of samples. To visualize the rank correlation, monotonically decreasing constrained splines are plotted in black (knots = 10). Samples are colored by their cortical lobe/division.
Gene ontology enrichment analysis
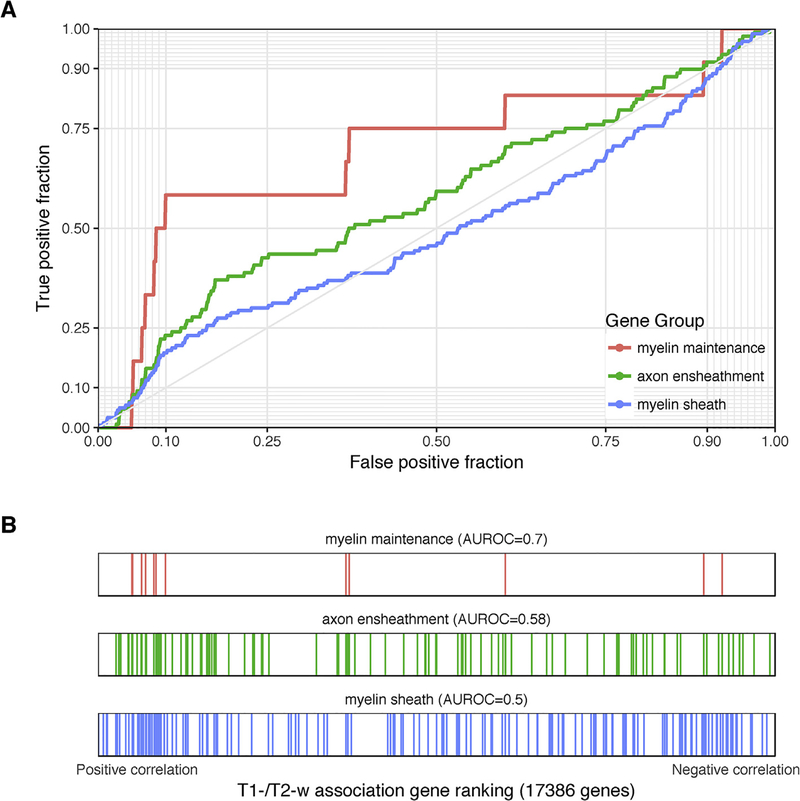
We first performed a targeted analysis of myelin-related gene ontology groups. Two of the ten myelin groups (ensheathment of neurons and myelination) were enriched for genes with positive correlations with the T1-w/T2-w ratio (pfdr < 0.05, Table 1 and Fig. 3). However, we note that these GO groups have high overlap. Broadly, all the myelin groups were enriched for positively correlated genes (AUC > 0.5 and four groups with an uncorrected p < 0.05). Relative to all tested 6230 GO groups, the myelin-related groups are less enriched than 194 other GO groups. While the 12 genes annotated to the “myelin maintenance” group have a high AUROC value of 0.697, we find that 41 of the 893 tested GO groups of equal or smaller size have a more extreme AUROC. Also, as shown in Table 1, they are ranked slightly higher in the enrichment results in separate T1-w or T2-w analyses (AUROC values are similar). In contrast, an independent list of myelin enriched genes obtained from transcriptomic profiling of whole mouse brain myelin fraction is enriched for negatively correlated genes (1493 genes, AUROC = 0.454, p < 10–8) (Thakurela et al., 2016). The same result is found using genes corresponding to proteins extracted from the myelin fraction (2212 genes, AUROC = 0.454, p < 10−11). Within the top ten most positively correlated genes, VAMP1 and STY2 appear in the myelin associated transcriptomic list; and VAMP1 and VAV3 are in the myelin proteome lists. One gene, BAIAP3 appears in the top ten most negatively correlated genes and the myelin associated transcriptomic list and none are in the list of myelin proteins.
Table 1. Enrichment statistics for myelin-related Gene Ontology groups using the T1-w/T2-w ratio ranking.
| Name | Gene Count | p | pFDR | AUROC | Rank | T1 rank | T2 rank |
|---|---|---|---|---|---|---|---|
| ensheathment of neurons | 108 | 0.00283 | 0.0283 | 0.583 | 195 | 148 | 268 |
| myelination | 105 | 0.00822 | 0.0411 | 0.575 | 312 | 255 | 426 |
| myelin maintenance | 12 | 0.0182 | 0.0607 | 0.697 | 491 | 349 | 773 |
| compact myelin | 14 | 0.0347 | 0.0868 | 0.663 | 708 | 267 | 1233 |
| myelin assembly | 18 | 0.0544 | 0.099 | 0.631 | 908 | 868 | 1218 |
| central nervous system myelination | 15 | 0.0594 | 0.099 | 0.641 | 968 | 999 | 874 |
| structural constituent of myelin sheath | 10 | 0.208 | 0.297 | 0.615 | 2086 | 2364 | 2821 |
| positive regulation of myelination | 11 | 0.392 | 0.49 | 0.575 | 3114 | 2955 | 3043 |
| regulation of myelination | 29 | 0.859 | 0.908 | 0.51 | 5535 | 5607 | 4849 |
| myelin sheath | 160 | 0.908 | 0.908 | 0.503 | 5777 | 3125 | 4283 |
Fig. 3.
Associations with T1-w/T2-w ratio intensities for selected myelin-related GO groups. (A) ROC curves for GO groups selected from Table 1. The curves show the proportion of GO group genes that overlap (y-axis, true positive fraction) in varying lengths of the T1-w/T2-w gene ranking (approximated by the x-axis, false positive fraction). Colored lines mark genes in different GO groups. (B) Distributions of the three significant GO groups across the T1-w/T2-w associated gene ranking with each annotated gene representing a single colored line.
The second process of interest is synaptic plasticity, which has been indirectly associated with the T1-w/T2-w ratio based on the inhibitory and stabilizing properties of myelin (Glasser et al., 2014). The regulation of synaptic plasticity GO group is enriched for negative correlations (AUROC = 0.41, uncorrected p < 0.0005, rank = 95th), as is the learning GO group (AUROC = 0.43, uncorrected p < 0.005, rank = 228th). In the context of aerobic glycolysis, we did not see enrichment for groups with titles containing “pyruvate”, “glycolysis”, or “lactate” (all p > 0.05).
Extending to the 6230 GO groups considered in the enrichment analysis, 17 were significantly enriched (pfwer<0.05). Like the gene-wise results, a larger number of negative relations is observed, with 15 GO groups enriched for negative correlations (AUROC < 0.5, Table 2) and two enriched for positive correlations (AUROC > 0.5, Table 3). Full results are provided in Supplement Table 2.
Table 2. Gene Ontology groups that are enriched for significant negative correlations with T1-w/T2-w ratio in cortex (AUROC < 0.5).
CC: cellular component; MF: molecular function; BP: biological process.
| Name | Gene Count | AUROC | p | pfwer | Aspect | T1 AUROC | T2 AUROC |
|---|---|---|---|---|---|---|---|
| regulation of cellular amino acid metabolic process | 55 | 0.265 | 1.78E-09 | 1.11E-05 | BP | 0.286 | 0.705 |
| mitochondrial protein complex | 118 | 0.347 | 1.05E-08 | 6.57E-05 | CC | 0.338 | 0.577 |
| regulation of cellular amine metabolic process | 71 | 0.305 | 1.37E-08 | 8.54E-05 | BP | 0.315 | 0.675 |
| SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 67 | 0.305 | 3.49E-08 | 0.000217 | BP | 0.317 | 0.669 |
| peptidase complex | 79 | 0.323 | 5.39E-08 | 0.000336 | CC | 0.345 | 0.642 |
| cellular amine metabolic process | 112 | 0.353 | 8.92E-08 | 0.000556 | BP | 0.362 | 0.635 |
| endopeptidase complex | 61 | 0.302 | 9.03E-08 | 0.000562 | CC | 0.326 | 0.657 |
| structural constituent of ribosome | 144 | 0.372 | 1.25E-07 | 0.000777 | MF | 0.355 | 0.571 |
| proteasome complex | 60 | 0.303 | 1.48E-07 | 0.000919 | CC | 0.328 | 0.656 |
| inner mitochondrial membrane protein complex | 100 | 0.351 | 2.81E-07 | 0.00175 | CC | 0.33 | 0.571 |
| amine metabolic process | 117 | 0.364 | 3.8E-07 | 0.00236 | BP | 0.375 | 0.626 |
| ribosomal subunit | 168 | 0.391 | 1.19E-06 | 0.00742 | CC | 0.384 | 0.556 |
| mitochondrial membrane part | 168 | 0.394 | 2.45E-06 | 0.0152 | CC | 0.397 | 0.546 |
| negative regulation of cell cycle G2/M phase transition | 75 | 0.349 | 6.62E-06 | 0.0412 | BP | 0.357 | 0.638 |
| cellular response to zinc ion | 17 | 0.184 | 6.66E-06 | 0.0414 | BP | 0.188 | 0.819 |
Table 3. Top 10 Gene Ontology groups for T1-w/T2-w ratio in cortex enriched for positive correlations (AUROC > 0.5).
CC: cellular component; MF: molecular function; BP: biological process.
| Name | Gene Count | AUROC | p | pfwer | Aspect | T1 AUROC | T2 AUROC |
|---|---|---|---|---|---|---|---|
| detection of chemical stimulus involved in sensory perception | 144 | 0.621 | 5.63E-07 | 0.0035 | BP | 0.573 | 0.365 |
| detection of stimulus involved in sensory perception | 189 | 0.601 | 1.77E-06 | 0.011 | BP | 0.557 | 0.389 |
| detection of chemical stimulus | 172 | 0.598 | 9.58E-06 | 0.0595 | BP | 0.559 | 0.391 |
| detection of chemical stimulus involved in sensory perception of smell | 121 | 0.614 | 1.53E-05 | 0.0947 | BP | 0.557 | 0.371 |
| bitter taste receptor activity | 10 | 0.867 | 5.74E-05 | 0.355 | MF | 0.873 | 0.138 |
| MAP kinase kinase kinase activity | 20 | 0.75 | 0.000111 | 0.685 | MF | 0.738 | 0.277 |
| sensory perception of chemical stimulus | 182 | 0.582 | 0.000156 | 0.962 | BP | 0.544 | 0.404 |
| cell-cell junction assembly | 84 | 0.618 | 0.000183 | 1 | BP | 0.597 | 0.396 |
| intermediate filament | 111 | 0.602 | 0.000209 | 1 | CC | 0.581 | 0.387 |
| chromatin DNA binding | 74 | 0.624 | 0.00023 | 1 | MF | 0.604 | 0.38 |
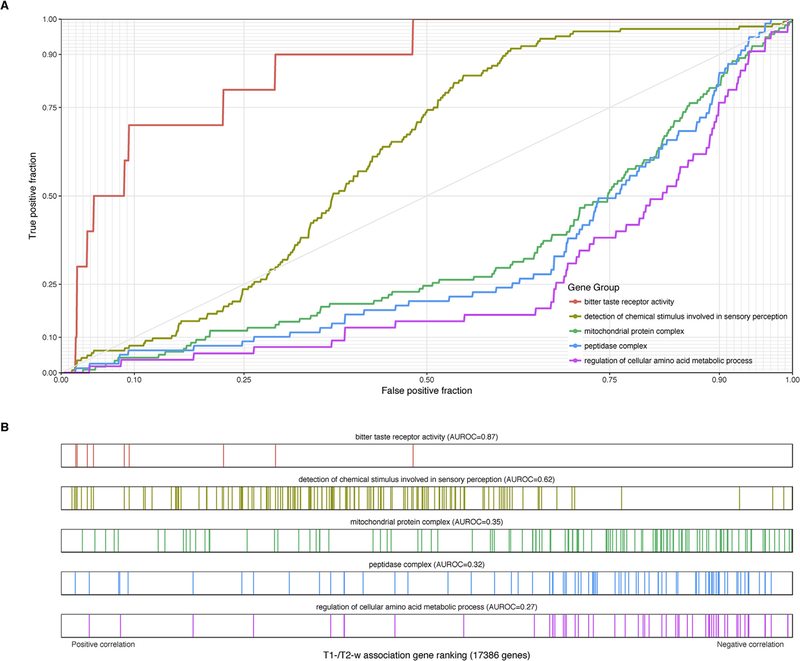
The 15 GO groups with significant enrichment for negative correlations are primarily related to metabolic and catabolic processes (Table 2). ROC curves and raster plots are provided for select groups in Fig. 4. Several overlapping GO groups containing peptidases and proteasome subunits share many genes. “Mitochondrial protein complex” is the second-ranked group (118 genes, AUROC = 0.35, pfwer < 0.0001) and several of the other significant groups contain similar genes. For example, the top-ranked ribosomal subunit genes are primarily specific to mitochondrial ribosomes. Mitochondria maintain a pH gradient by pumping hydrogen protons from the mitochondrial matrix (pH = 7.7) to the inner mitochondrial space (pH = 6.8, more acidic than the cytosol), supporting oxidative phosphorylation (Santo-Domingo and Demaurex, 2012). The “regulation of cellular amine metabolic process” group also suggests a pH association, due to the elevated pH typical of amines. To more directly test pH associated genes, we used genes up- and down-regulated with pH in postmortem human brain samples (Mistry and Pavlidis, 2010). In agreement with the mitochondrial enrichment, we found that pH up-regulated genes are biased for negative correlations with the T1-w/T2-w ratio (1027 genes, AUROC = 0.34, p < 10−62) with a core set of 11 genes showing stronger enrichment (AUROC = 0.18, p < 0.0005) and the pH down-regulated genes showing the opposite enrichment (AUROC = 0.54, 2137 genes, p < 10−8). Other significant and distinct gene sets are cellular response to zinc ion (AUROC = 0.18,17 genes, pfwer < 0.05) and negative regulation of cell cycle G2/M phase transition (AUROC = 0.35, 75 genes, pfwer < 0.05).
Fig. 4.
Selected GO groups with high enrichment of T1-w/T2-w ratio associations in the cortex. (A) ROC curves for GO groups selected from Tables 2 and 3 The curves show the proportion of GO group genes that overlap (y-axis, true positive fraction) in varying lengths of the T1-w/T2-w gene ranking (approximated by the x-axis, false positive fraction). Colored lines mark genes in different GO groups. (B) Distributions of the five significant GO groups across the T1-w/T2-w associated gene ranking with each annotated gene representing a single colored line.
Two GO groups have significant enrichment for positive correlations with other weaker signals that are of interest (Table 3 and Fig. 4). Genes annotated to “Detection of chemical stimulus involved in sensory perception” have the most significant bias for positive correlations with T1-w/T2-w ratio (144 genes, AUROC = 0.621, pfwer < 0.005). This group consists primarily of taste and olfactory receptors. Visualization of the ROC and raster plots in Fig. 3 shows this result is driven by a lack of genes with negative correlations, with most genes having little or no correlation. Several largely overlapping groups are also top ranked, for example, bitter taste receptors (10 genes, AUROC = 0.87, uncorrected p < 0.0005). The intermediate filament group is of note because it marks neurofilament genes (111 genes, AUROC = 0.6, uncorrected p < 0.0005). Its most correlated genes are brain-associated: NEFH, NEFM, INA, and NEFL.
Cell type marker enrichment analysis
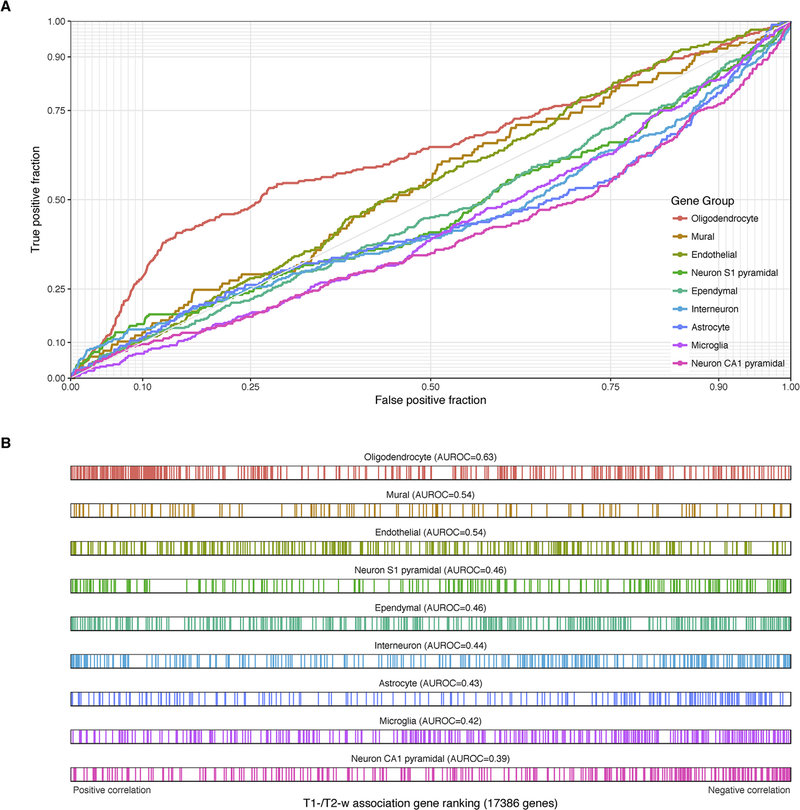
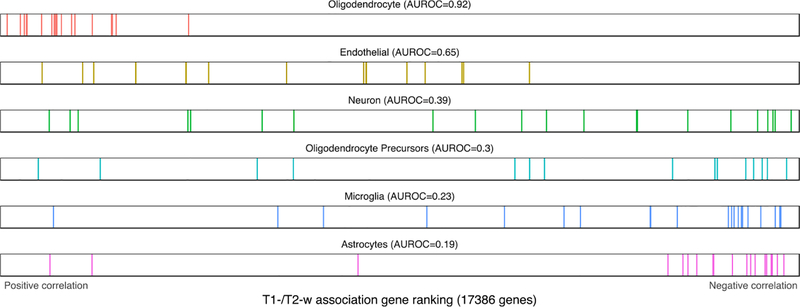
Following the same procedure used for GO gene sets, we tested for enrichment of cell-type specific marker genes. Because the results were mostly consistent across the different sources of marker genes, we grouped the results by cell-type while providing figures and tables for each source. Enrichment results for the Zeisel marker genes that were obtained from a single cell analysis of the adult mouse somatosensory cortex (S1) and hippocampus are in Table 4 and Fig. 5. A second set of mouse markers from the NeuroExpresso cross-laboratory database were used for Supplementary Table 3 (Mancarci et al., 2017). Results for marker gene lists obtained from the Darmanis et al. study of the human temporal cortex are in Supplementary Table 4 and Fig. 6.
Table 4.
Enrichment statistics for Zeisel marker gene sets using the T1-w/T2-w ratio ranking.
| Cell-type or class | Gene Count | AUROC | p | pFWER |
|---|---|---|---|---|
| Oligodendrocyte | 366 | 0.628 | 6.03E-17 | 5.43E-16 |
| Neuron CA1 pyramidal | 314 | 0.39 | 2.29E-11 | 1.83E-10 |
| Microglia | 336 | 0.422 | 8.56E-07 | 5.99E-06 |
| Interneuron | 293 | 0.439 | 0.000311 | 0.00186 |
| Astrocyte | 199 | 0.43 | 0.000714 | 0.00357 |
| Ependymal | 355 | 0.463 | 0.016 | 0.0641 |
| Endothelial | 290 | 0.538 | 0.0264 | 0.0793 |
| Neuron S1 pyramidal | 218 | 0.463 | 0.0625 | 0.125 |
| Mural | 117 | 0.538 | 0.154 | 0.154 |
Fig. 5.
Associations with T1-w/T2-w ratio intensities for Zeisel cell type marker genes. (A) ROC curves showing the proportion of marker genes that overlap (y-axis, true positive fraction) in varying lengths of the T1-w/T2-w gene ranking (approximated by the x-axis, false positive fraction). Colored curves mark genes in different cell type marker lists. (B) Distributions of the cell type markers across the T1-w/T2-w associated gene ranking. Each marker gene is represented by a single colored line.
Fig. 6.
Distributions of the Darmanis cell type markers across the T1-w/T2-w associated gene ranking. Each marker gene is represented by a single colored line. The endothelial and neuron marker list are not significantly enriched after test correction.
Oligodendrocyte genes are the most strongly enriched, showing high positive correlations with T1-w/T2-w intensity. This is consistent across all three marker gene sources (AUROC > 0.62, pfwer < 10–9). We examined the 20 marker genes in the Darmanis oligodendrocyte gene set to investigate whether the enrichment of this group was driven the by the myelin-associated genes in the list (MAG, MOG and OPALIN). None were significantly correlated with T1w/T2w ratio after genome-wide correction (pfwer > 0.09, ranked 7, 12, and 18th of 20 genes; Supplementary Table 5). In contrast, two peptidase-coding genes, CNDP1 and KLK6 were top-ranked (ranked first and third). The TF gene, which codes for transferrin, an iron transport protein, was also top-ranked (4 of 20). For a related cell-type, oligodendrocyte precursors, markers from the Darmanis source were enriched for negative correlations (16 genes, AUROC = 0.30, pfwer < 0.02), but the mouse-brain-derived list from NeuroExpresso was not (135 genes, AUROC = 0.53, pfwer = 1). Astrocyte markers were enriched for negative correlations in all three sources of markers (AUROC < 0.43, pfwer < 0.005). Microglia are enriched for negative correlations in the human list (21 genes, AUROC = 0.23, pfwer < 0.0001) and the Zeisel mouse source (AUROC = 0.42, pfwer < 0.00001) but not the NeuroExpresso markers. Endothelial markers were biased towards positive correlations in all three sources but were significantly enriched in only the NeuroExpresso source (AUROC = 0.59, pfwer < 0.005).
In general, markers of glial cells have stronger signals than neuronal markers. Only a single neuron marker list is available for the human source and is not significantly enriched (21 genes, AUROC = 0.39, pfwer < 0.08). The Zeisel source provides markers for interneurons, and pyramidal neurons from the S1 and CA1 regions. Interneuron markers were enriched for negative correlations (293 genes, AUROC = 0.44, pfwer < 0.002). Pyramidal neuron markers from the hippocampus CA1 are enriched for negative correlations (314 genes, AUROC = 0.39, pfwer < 10−9), while those from S1 are not significantly enriched. The pyramidal neuron markers from NeuroExpresso were also enriched for negative correlations (29 genes, AUROC = 0.32, pfwer < 0.01). None of the three NeuroExpresso GABAergic neuron marker lists were significantly enriched.
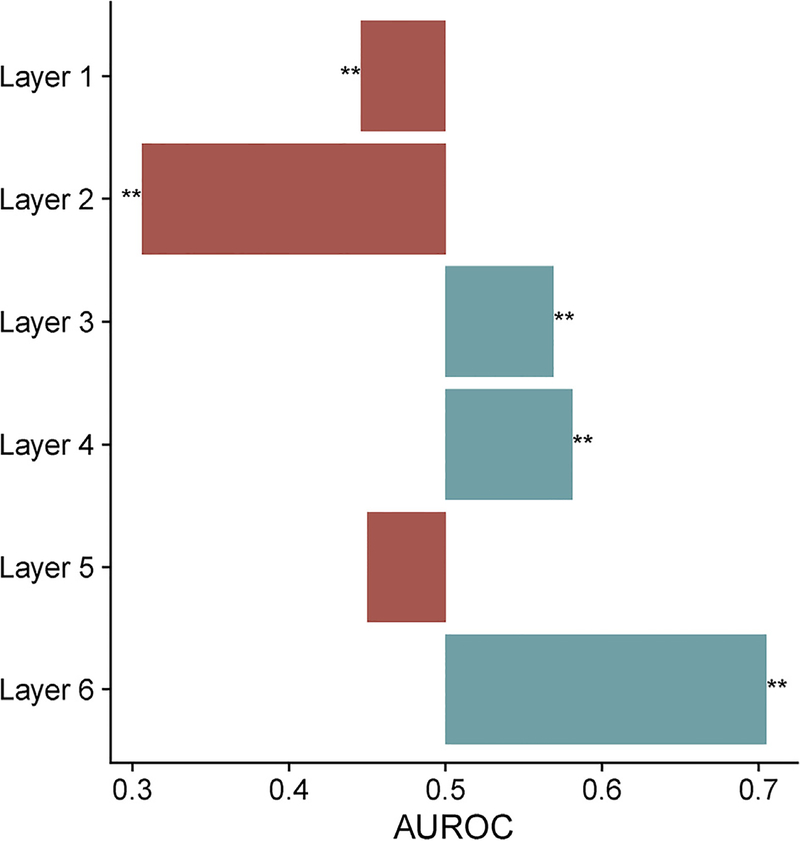
Laminar marker enrichment analysis
We next tested marker genes derived from sectioning of the human prefrontal cortex (He et al., 2017). Genes specifically expressed in layers 1 and 2 were enriched for negative correlations with T1-w/T2-w ratio (AUROC < 0.45, pfwer < 1 0–4, Fig. 7 and Table S6). Genes marking layers 3, 4, and 6 were enriched for positive correlations (AUROC > 0.56, pfwer < 10–3). The strongest enrichment was in layer 6 (1816 genes, AUROC = 0.7, pfwer < 10–170), which is estimated to contain a higher proportion of oligodendrocytes (He et al., 2017).
Fig. 7.
Barplot of enrichment in the T1-w/T2-w associated gene ranking across the He et al. laminar markers. ** indicates pfwer < 0.0005.
Disease-associated gene set analysis
Testing of 1071 disease-associated gene sets revealed significant enrichment of human immunodeficiency virus (HIV) associated genes (163 genes, AUROC = 0.40, pfwer < 0.02). While no clear theme is evident, we note that several proteasome subunit genes are top-ranked in the HIV list. While no other disease-associated gene lists were significant after multiple test correction, we note that cocaine dependence is ranked 6th (41 genes, AUROC = 0.37, uncorrected p < 0.005), temporal lobe epilepsy is ranked eighth (80 genes, AUROC = 0.41, uncorrected p < 0.005) and demyelinating disease is fifteenth (15 genes, AUROC = 0.70, uncorrected p < 0.01).
Human Connectome group average analysis
We repeated the analysis using the HCP group average T1-w/T2-w ratio image in place of the six individual AHBA ratio images. We observe less significantly correlated genes (247 positively correlated and 903 negatively correlated at pfwer < 0.05, Supplement Table 7). Within those genes, agreement across the donors is lower, with 34.6% having consistent correlation direction in all six brains. Within the top ten most positively correlated genes, WNT3 and ADAM10 are in the myelin transcriptomic and proteomic gene lists, respectively (no overlap was observed with the most negative ten).
Using gene set enrichment analysis, we find that 69 GO groups are significantly enriched, compared to 17 when using individual scans (full results in Supplement Table 8). Thirteen of these groups are also significant in the subject-specific analyses above. Four groups are significantly enriched for positive correlations with T1-w/T2-w ratio. The strongest is histone demethylase activity which mostly contains lysine demethylases (26 genes, AUROC = 0.76, pfwer < 0.02). A similar group is ranked 3rd (histone demethylation). Transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding is ranked second (92 genes, AUROC = 0.64, pfwer < 0.03). Ranked fourth is ensheathment of neurons (108 genes, AUROC = 0.627, pfwer < 0.04). Six of the ten myelin GO groups have significant enrichment for positive correlations using the less conservative correction method (n = 10, FDR). As before, enrichment of the myelin transcriptome and proteome obtained from the whole mouse brain is biased for negative correlations (both AUROC < 0.458, p-value < 10–10). The 65 GO groups that are enriched for negative correlations with T1-w/T2-w ratio include 13 of the 15 groups enriched when using the individual scans. The majority of remaining GO groups are related to mitochondria, amine metabolic processes, the proteasome and the ribosome. Other distinct GO groups of note are “antigen processing and presentation of exogenous antigen”, proton transport, and hydrogen transport (all AUROC < 0.39 and pfwer < 0.01).
Cell-type marker enrichment analysis of the average image again highlighted oligodendrocyte marker genes as having higher expression in regions with high T1-w/T2-w ratio. In all three sources of cell-type markers, this AUROC value was higher in the HCP averaged results than with the individual scans. Again, classical myelin associated genes are not key to the oligodendrocyte marker enrichment. Unlike the previous analyses, astrocyte marker genes lack enrichment in any source of markers. Microglia markers from Darmanis and Zeisel sources are enriched for negative correlations (both AUROC < 0.44, pfwer < 0.0005). Neuron markers are also enriched for negative correlations in the same sources (both AUROC < 0.45, pfwer < 0.05). Endothelial markers were again biased towards positive correlations but are now significant in two of the three sources (AUROC > 0.57, pfwer < 0.01). Across cortical layers, a different pattern emerged with all layers enriched for negative correlations except for layer 6 (pfwer any set of laminar markers < 0.002). No disease associated gene lists were significantly enriched.
Overall, the Human Connectome result that used a population-average image had good agreement with the in-sample results. This demonstrates that population averages can be joined with the Allen Human Brain Atlas to further our understanding of molecular neuroanatomy.
Whole brain analysis
While we focus on the neocortex in this paper, the AHBA has comprehensively assayed the brain, allowing a wider analysis. We provide only a summary characterization of whole brain correlation results due to space constraints, and because the T1-w/T2-w ratio was originally proposed for the cerebral cortex (Glasser and Van Essen, 2011).
Across the whole brain, the majority of genes are significantly correlated with T1-w/T2-w ratio intensities after multiple test correction. In a reversal of the neocortical findings, there are less negatively correlated genes (6,593) than positively correlated genes (8,441). Expression patterns of the top genes primarily show contrast between subcortical and cortical regions (including cerebellar cortex). Enrichment of myelin related GO groups are similar to the original analyses with ensheathment of neurons having the strongest enrichment (108 genes, AUROC = 0.61, pfwer < 0.002). Again, we observe many other GO groups with stronger enrichment than this myelin group which was ranked 163rd amongst the other groups. Unlike previous results, genes with high expression in the mouse brain myelin fraction were enriched for positive correlations (1493 genes, AUROC = 0.6, p < 10−33). Genes encoding proteins extracted from myelin were not enriched (AUROC = 0.501). The top GO group that is enriched for positive correlations is type I interferon signaling pathway (65 genes, AUROC = 0.74), followed by many other immune system related groups (Supplementary Table 9). The group with the strongest enrichment of negative correlations is regulation of synaptic plasticity (141 genes, AUROC = 0.30), followed by similar GO groups such as postsynaptic density (194 genes, AUROC = 0.33). We again observe that pH up-regulated genes have negative correlations with T1-w/T2-w ratio. While our cell-type gene markers are from studies of the cortex, we note that endothelial and microglial markers switch from weak negative to strong positive enrichment, in comparison to the neocortical results. This is observed across all three of the marker sources. The strongest cell-type marker enrichments are for oligodendrocytes and neurons that correspond to positive and negative correlations respectively. In contrast to the neocortical results, 35 of the 1178 disease-associated gene lists were significant (Supplementary Table 10). In general, these whole brain results are also found when using the Human Connectome Project average image (data not shown).
Discussion
MR images of the brain provide contrast at the submillimetre scale, allowing segmentation of many structures and identification of abnormalities. At the atomic and subatomic levels, the sources of MR signals are well understood. However, at the macromolecular scale, we lack a comprehensive understanding of what determines MR signal intensity. To address this gap, we have used genome-wide measurements to correlate gene expression with MR signal intensity. By leveraging gene annotations, we found biological processes, cellular components, molecular functions, diseases, and cell-types that are related to MR contrast. This work elucidates the macromolecular correlates of MR signal intensity and will aid interpretation of future MR studies of the brain.
Top-ranked genes
At the level of individual genes, our most significant finding of a negative relationship between T1-w/T2-w ratio and SCARA5 expression is perplexing. This gene is a receptor for the protein ferritin, which provides a transferrin-independent method for cellular delivery of iron (Li et al., 2009). This receptor has been targeted with ferritin loaded with MRI contrast agents to diagnose and target cancer (Geninatti Crich et al., 2015). The negative correlation suggests the ferritin receptor SCARA5 is not a proxy measure of iron content. This is because iron appears dark in T2-w scans and is enriched in white matter, while, in contrast, SCARA5 is positively correlated with T2-w intensity (Schenck, 2003). This counterintuitive direction may be due to binding preference of SCARA5 for the light subunit of ferritin over the heavy chain. The ratio of heavy to light chains is known to vary across cell-types and brain structures (Connor and Menzies, 1996). Transferrin, an oligodendrocyte marker used in iron transport, is the 576th most positively correlated gene. Iron-associated genes as a group, are not significantly enriched for T1-w/T2-w associations.
Retinol Binding Protein 4 (RBP4), which is the second most negatively correlated gene, has been indirectly associated with multiple sclerosis. Specifically, retinol levels in blood were correlated with new lesions in multiple sclerosis patients, as detected with MR imaging (Løken-Amsrud et al., 2013). Within the top ten negative associations, we note two calcium associated genes (PCDH20 and C2CD4C). The top two positively correlated genes encode synaptic vesicle membrane proteins (SYT2 and VAMP1). We also note Estrogen Related Receptor Gamma (ESRRG), which is the 3rd most significant positive correlation. A large genetic study found ESRRG to have a highly suggestive association with brain infarcts (defined by abnormal MRI signal intensity) (Debette et al., 2010). FKBP5 is ranked 9th and has been associated with white matter integrity (Fani et al., 2014). Additionally, GPAT3 which is involved in the production of lysophosphatidic acid ranks 4th. Lysophosphatidic acid is associated with multiple sclerosis (Schmitz et al., 2017) and its receptor regulates myelination in the mouse cerebral cortex (García-Díaz et al., 2015). Given the large number of significant genes, we employed gene set enrichment analysis to examine genes beyond the top ten.
Myelination and related genes
In this paper, we focused on the T1-w/T2-w ratio in the cerebral cortex which has been previously associated with myelin content, aerobic glycolysis and synaptic plasticity (Glasser et al., 2014; Glasser and Van Essen, 2011). Here we use gene expression to provide a new perspective on these past associations revealed by PET imaging and histology. Myelin-associated GO groups are enriched for positive correlations in the T1-w/T2-w gene ranking as expected. In the context of other gene groups, this finding is not strong, as there are over 190 GO groups with stronger enrichment (Table 2). When using the Human Connectome Project average image, myelin GO groups are ranked higher, with 66 groups having stronger enrichment. For this average image, axon ensheathment genes are the fourth most enriched group for positive correlations. This improved ranking may be due to the blurring of cortical boundaries in the average image or the tuning of the scanner used in the project. Specifically, T2w TE was lengthened for improved intracortical myelin contrast (Glasser et al., 2013; Glasser and Van Essen, 2011). We emphasize that all of the samples and cell-type marker lists used were derived from cortical gray matter samples which are known to have low myelin content (MacKay and Laule, 2016). In contrast to the myelin-associated GO groups, markers obtained from the myelin fraction of the mouse brain were enriched for negative correlations. This contradictory enrichment may be due to differences between human cortical myelin and myelin obtained from the whole mouse brain. Previous association of T1-w/T2-w ratio with myelin content may be due to myelin transcripts that were transcribed in white matter and transported to distal sheaths after translation into protein. In this context, genes annotated to the ‘paranode region of axon’, which is where the myelin sheath ends, has the 4th highest AUROC value of all tested GO groups (0.765, 11 genes, uncorrected p < 0.003). However, we note that myelin basic protein is translated locally at the axon contact site (Müller et al., 2013).
Our findings suggest a positive correlation between T1-w/T2-w intensity and axon caliber. The three neurofilament genes are positively correlated with the ratio in order of decreasing weight, with the heavy chain (NEFH) ranked 16th genome-wide. Expression of NEFH plays a major role in the development of high-caliber axons (Elder et al., 1998). A positive correlation has been observed between axon caliber and myelin thickness in neurons, both in developing nerves (Friede, 1972) and in the adult brain (Fraher and Dockery, 1998). NEFH has been linked to an oligodendrocyte associated expression pattern that is expressed at higher levels in ‘relay-like’ regions with few connections (French et al., 2011). This link to connectivity is also demonstrated by PVALB and CALB1 which are ranked the 106th most positive and 64th most negatively correlated genes, respectively. These genes have been previously associated with connectivity in the macaque thalamus (Jones, 1998). This corroborates a similar study of the T1-w/T2-w ratio and transcriptomic patterns that found strong hierarchical relationships in the human cortex (Burt et al., 2017). In addition, we note that PVALB, CALB1, and NEFH are among the 136 genes with robust correlations between expression and functional connectivity networks (Richiardi et al., 2015). PVALB marks fast-spiking GABAergic interneurons that are frequently myelinated and may explain a large proportion of myelin in the cortex (Stedehouder et al., 2017). Therefore, our finding of T1-w/T2-w ratio correlation for neurofilaments, parvalbumin and calbindin provides indirect support for associations with increased myelin content and connectivity.
Synaptic plasticity GO groups were enriched for negative correlations at levels similar to the neuron markers (AUROC < 0.43), supporting previously hypothesized links with plasticity and myelination (Glasser et al., 2014). While not strong in the cortical results, synaptic plasticity genes were the most strongly enriched GO group in the whole brain analyses.
Metabolic activity and pH
We find support for metabolic associations, but not aerobic glycolysis specifically. We observe higher expression of genes localized to mitochondria in regions with low T1-w/T2-w ratio. In contrast, aerobic glycolysis measures glucose that is not used by oxidative phosphorylation in mitochondria and is not strongly linked to oxygen consumption (Vaishnavi et al., 2010). More directly, GO groups related to aerobic glycolysis are not enriched but several genes are top ranked. For example, SLC16A6, MPC1, and SLC16A7 which transport monocarboxylates such as lactate and pyruvate, are the 10th, 149th and 215th most positively correlated genes. High aerobic glycolysis has been linked to the energy use of membrane-bound processes that are commonly used by astrocytes (Vaishnavi et al., 2010). This is supported by our findings of astrocyte marker gene enrichment. Aerobic glycolysis is also associated with biosynthesis or anabolic processes (Glasser et al., 2014; Vaishnavi et al., 2010). In support, we observe enrichment of metabolic processes and ribosomal genes. In the opposite direction, repressors of transcriptional activity are enriched for positive correlations (using the averaged HCP T1-w/T2-w image). While we do not observe a direct association of aerobic glycolysis, we find support for increased biosynthesis and astrocyte proportion in regions with low T1-w/T2-w ratio.
Over 75% of genes in the inner mitochondrial membrane are anti-correlated with T1-w/T2-w ratio, where a pH gradient is maintained. Crossing this membrane changes pH from 6.8 to 7.7 (Santo-Domingo and Demaurex, 2012), which is used to spin the ATP synthase complex through the transfer of hydrogen ions (H+). We note that MR scanners primarily measure signals from hydrogen protons and MR signals white matter lipids were found to be pH dependent (Kucharczyk et al., 1994). To further examine this signal, we tested genes associated with pH in postmortem brain studies (Mistry and Pavlidis, 2010). This provided a more direct comparison of H+ relations. In agreement, we found that genes correlated with pH were also correlated with T1-w/T2-w ratio intensity. This suggests that regions with low T1-w/T2-w ratio intensities have elevated concentrations of H+ while regions with high T1-w/T2-w ratio are acidic. This pH signal may be useful for neuroimaging studies psychiatric disorders, as decreased brain pH has been observed in animal models and postmortem studies of patients with schizophrenia and bipolar disorder (Hagihara et al., 2017).
While a clear mitochondrial-associated signal is present and negatively correlated with T1-w/T2-w ratio, we observe much weaker signals in the positive direction. Fewer genes and GO groups have significant positive correlations with T1-w/T2-w ratio. Also, the genes that are significantly positively correlated have lower average expression (6.34 ± 2.3 [standard deviation]) than the negative direction (7.07 ± 2.1, Mann-Whitney U test p < 10‒9). This may be due to active repression as “transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding” is the 15th most enriched GO group for positive correlations (ranked second in the Human Connectome results). Low expression is also linked to pH, as nonquiescent cells have a higher pH than quiescent (Bright et al., 1987). Only two GO groups are significantly enriched for positive correlations. These groups are largely overlapping and primarily contain taste and olfactory receptors to detect stimuli involved in sensory perception. These genes have little or no expression in the brain, and have been found to be inconsistently expressed across the six AHBA brains in a broad cortical parcellation (French and Paus, 2015). High sequence homology across these receptors and nonspecific binding to the microarray may explain the enrichment of the group. However, bitter taste receptor transcripts are expressed in several regions of the rat brain, and cells containing these transcripts are somewhat functional, being capable of responding to tastants (Singh et al., 2011; Tomás et al., 2016). Also, an olfactory receptor has been shown to function as a hypoxia sensor in glomus cells (Chang et al., 2015). Interestingly, RTP1, which encodes one of the few proteins that facilitate transfer of olfactory receptors to the surface of the cell is the 9th most strongly correlated gene in the negative direction (Saito et al., 2004). Further analysis is needed to determine the function and role of these receptors in the brain.
Molecule size
Genes linked to molecule size show clear enrichment in our analysis. The peptidase complex GO group, which consists primarily of proteasome subunit genes, is enriched for negative correlations (ranked 6th). These genes code for proteins that degrade proteins into smaller peptides or component amino acids by hydrolysis. In the other direction, we find enrichment for the histone demethylase activity GO group in the analysis of the T1-w/T2-w ratio image. This group consists primarily of lysine demethylation genes. A product of the lysine demethylation reaction is endogenous formaldehyde (Shi et al., 2004). Formaldehyde is a fixative that crosslinks proteins, creating larger molecules. Fixation of brain tissue with formalin is known to produce a bright band of T1-w intensities as fixation proceeds (Schmierer et al., 2008; Yong-Hing et al., 2005). Of the 6876 genes in our analysis and the UniProt database (The UniProt Consortium, 2017), protein mass is correlated with T1-w/T2-w association ranking (Spearman’s rho = 0.13, p < 10‒15). For the significantly T1-w/T2-w correlated genes in the positive direction the median weight is 60,692 Da, for the negatively correlated genes it is 45,141 Da (Mann-Whitney U test, p < 10‒11). In agreement, molecule weight and mobility have been previously linked to MRI contrast (Koenig, 1995; Sprawls, 2000). Overall, the association with protein weight and the opposing signals between genes that join and break proteins suggest a significant proportion of MR contrast can be explained by molecule size.
Cell types
Genes specifically expressed in oligodendrocytes are highly expressed in regions with high T1-w/T2-w ratio. This was the strongest and most consistent cell-type in our marker enrichment analysis. In previous research, oligodendrocytes demonstrated significantly lower counts of detected RNA molecules per cell when compared to neurons (Zeisel et al., 2015), in agreement with the above findings associating T1-w/T2-w hyperintensity with transcriptional repression and lowly expressed genes. Though generation of the myelin sheath is a primary function of oligodendrocytes, we observe low enrichment of myelin-associated genes compared to unassociated genes in the human oligodendrocyte marker gene list. This lack of a strong myelin signal suggests that the oligodendrocyte signal we observe may be driven by satellite perineuronal oligodendrocytes that are primarily non-myelinating (Battefeld et al., 2016; Takasaki et al., 2010). To test this hypothesis we used genes from a study of whole rat brain that compared expression between perineuronal oligodendrocytes and other oligodendrocytes (Szuchet et al., 2011). We found enrichment for negative correlations in this set (AUROC = 0.439, gene count = 493, p < 10‒5), but note that the strongest marker (ETV4) has a strong positive correlation in our results (ranked 214th most positively enriched gene, pfwer < 10‒7). Better characterization of glial cell types in the human cortex would facilitate understanding of this result of high oligodendrocyte and low myelin signal.
Beyond the consistent enrichment of oligodendrocyte markers, we observe enrichment of genes that are specifically expressed in astrocyte, neuron, endothelial and microglial cells. Astrocyte, neuron, and microglia marker gene sets were enriched for negative correlations with T1-w/T2-w ratio. Within the neuron markers, there is not a clear difference between interneuron and pyramidal markers. However, markers for CA1 pyramidal neurons are more strongly enriched than those from S1. While hippocampal samples were not used in our analysis of the cortex, the CA1 markers may gauge synaptic plasticity and agree with enrichment directions for plasticity GO groups (Shin et al., 2017).
Disease-associated genes
Our analysis of disease-associated genes found that HIV linked genes were enriched for negative correlations. The top-ranked HIV genes include proteasome subunits, which have been previously associated with HIV (Krishnan and Zeichner, 2004; Schubert et al., 2000; Seeger et al., 1997). In some patients, HIV RNA is detected in cerebrospinal fluid and has been associated with dementia and diffuse white matter signal abnormalities (Kugathasan et al., 2017). Beyond HIV, no other disease gene lists are significant after correction. However, we note that demyelinating disease is ranked 15th of the 1071 disease associated gene lists and is enriched for positive correlations. A few top-ranked genes are indirectly related to multiple sclerosis.
Whole brain analysis
In our analysis of the whole brain, we observe that immune-related genes and marker genes for oligodendrocytes and activated microglia are positively correlated with T1-w/T2-w ratio. In contrast, neuron markers and related GO groups such as ‘postsynaptic density’ were negatively correlated on average. This is also reflected in the disease-associated gene set analysis with significant enrichment of inflammatory and neural associated disease gene lists in opposing directions. Most of the top listed diseases are inflammatory, enriched for positive correlations, and not clearly linked to the brain (‘pustulosis of palm and sole’ for example). However, the Behcet’s disease gene list, which is ranked second, affects the brain (84 genes, AUROC = 0.69, pfwer <0.0001). Behcet’s disease is an autoimmune condition that manifests as vasculitis across many organs of the body, with neurological symptoms in 5–50% of cases (van der Knaap and Valk, 2011). In particular, it can cause lesions of the basal ganglia, midbrain, and pons, as well as white matter abnormalities in the cerebral hemispheres which are similar to the patterns of demyelination that occur in multiple sclerosis (Al-Araji and Kidd, 2009; van der Knaap and Valk, 2011). For negative correlations, genes associated with Angelman’s syndrome had the strongest enrichment (10 genes, AUROC = 0.113, pfwer <0.05). Delayed or reduced myelin formation has been identified in children suffering from Angelman’s syndrome (Harting et al., 2009), as well as in mouse models of the disease (Grier et al., 2015). In comparison to the cortical analysis, the whole brain results show a broad neuron-versus-glia pattern with stronger correlations for endothelial, microglia, and immune-related genes. This suggests that across the whole brain, the variance in MR signal intensity may be partially explained by immune and circulatory activity.
Conclusion
Our results inform the use of the T1-w/T2-w ratio to estimate cortical myelin by characterizing genes that are associated with the ratio. Specifically, molecule size and transcription marking oligodendrocytes, axon calibre, and pH are more strongly associated with the ratio than myelin associated genes. While our transcriptomic perspective supports associations with aerobic glycolysis and synaptic plasticity, future work is needed to understand why the ratio is a good proxy for myelin content in the cortex.
We emphasize that our results vary little when our analysis is performed on T1-w or inverted T2-w intensities alone. While our enrichment values are improved when the ratio is used, it is possible the noise-attenuating properties of the ratio are of limited importance in our study because the MRI scans were acquired post mortem. As such, our findings illustrate the potential utility of detailed transcriptomic characterization of imaging measures other than the T1-w/T2-w ratio, and offer insight into the macromolecular correlates of T1-w and T2-w images.
Supplementary Material
Acknowledgements
We thank the Allen Institute for Brain Science for creating the Allen Human Brain Atlas. We are grateful to the three anonymous reviewers for their helpful comments that improved this analysis.
This study was supported by the CAMH Foundation, a National Science and Engineering Research Council of Canada (NSERC) Discovery Grant to LF and a NIMH K01MH108721 to SPP.
Data were provided in part by the Human Connectome Project, WUMinn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.neuroimage.2018.03.027 and https://figshare.com/articles/Transcriptomic_characterization_of_MRI_contrast_focused_on_the_T1-w_T2-w_ratio_in_the_cerebral_cortex/5270926.
References
- Al-Araji A, Kidd DP, 2009. Neuro-Behçet’s disease: epidemiology, clinical characteristics, and management. Lancet Neurol. 8, 192–204. [DOI] [PubMed] [Google Scholar]
- Allen Institute for Brain Science, 2013. Technical White Paper: Microarray Survey. Available at: http://help.brain-map.org/download/attachments/2818165/WholeBrainMicroarray_WhitePaper.pdf.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battefeld A, Klooster J, Kole MHP, 2016. Myelinating satellite oligodendrocytes are integrated in a glial syncytium constraining neuronal high-frequency activity. Nat. Commun 7, 11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol 57,289–300. [Google Scholar]
- Benjamini Y, Yekutieli D, 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat 29, 1165–1188. [Google Scholar]
- Bright GR, Fisher GW, Rogowska J, Taylor DL, 1987. Fluorescence ratio imaging microscopy: temporal and spatial measurements of cytoplasmic pH. J. Cell Biol 104, 1019–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt JB, Demirtas M, Eckner WJ, Navejar NM, Ji JL, Martin WJ, et al. , 2017. Hierarchy of Transcriptomic Specialization across Human Cortex Captured by Myelin Map Topography. bioRxiv, p. 199703 10.1101/199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, 2017a. GO.db: a set of annotation maps describing the entire Gene Ontology. Available at: http://ctan.uib.no/pub/bioconductor/2.7/data/annotation/html/GO.db.html.
- Carlson M, 2017b. org.Hs.eg.db: Genome Wide Annotation for Human. [Google Scholar]
- Chang AJ, Ortega FE, Riegler J, Madison DV, Krasnow MA, 2015. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 527, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LL, Tosun D, Woodward SH, Kaufer D, Neylan TC, 2015. Preliminary evidence of increased hippocampal myelin content in veterans with posttraumatic stress disorder. Front. Behav. Neurosci 9, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JR, Menzies SL, 1996. Relationship of iron to oligodendrocytes and myelination. Glia 17, 83–93. [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. , 2015. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U. S. A 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. , 2017. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 45,D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debette S, Bis JC, Fornage M, Schmidt H, Ikram MA, Sigurdsson S, et al. , 2010. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke 41, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL Jr., Kang C, Bosco P, Gourov A, Tu PH, et al. , 1998. Requirement of heavy neurofilament subunit in the development of axons with large calibers. J. Cell Biol 143, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani N, King TZ, Reiser E, Binder EB, Jovanovic T, Bradley B, et al. , 2014. FKBP5 genotype and structural integrity of the posterior cingulum. Neuropsychopharmacology 39, 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA, 1925. Statistical Methods for Research Workers. Oliver and Boyd. [Google Scholar]
- Forest M, Iturria-Medina Y, Goldman JS, Kleinman CL, Lovato A, Oros Klein K, et al. , 2017. Gene networks show associations with seed region connectivity. Hum. Brain Mapp 38, 3126–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher J, Dockery P, 1998. A strong myelin thickness-axon size correlation emerges in developing nerves despite independent growth of both parameters. J. Anat 193 (Pt 2), 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Paus T, 2015. A FreeSurfer view of the cortical transcriptome generated from the Allen Human Brain Atlas. Front. Neurosci 9, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Tan PPC, Pavlidis P, 2011. Large-scale analysis of gene expression and connectivity in the rodent brain: insights through data integration. Front. Neuroinf 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friede RL, 1972. Control of myelin formation by axon caliber (with a model of the control mechanism). J. Comp. Neurol 144, 233–252. [DOI] [PubMed] [Google Scholar]
- Ganzetti M, Wenderoth N, Mantini D, 2015. Mapping pathological changes in brain structure by combining T1- and T2-weighted MR imaging data. Neuroradiology 57, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Díaz B, Riquelme R, Varela-Nieto I, Jiménez AJ, de Diego I, Gómez-Conde AI, et al. , 2015. Loss of lysophosphatidic acid receptor LPA1 alters oligodendrocyte differentiation and myelination in the mouse cerebral cortex. Brain Struct. Funct 220, 3701–3720. [DOI] [PubMed] [Google Scholar]
- Geninatti Crich S, Cadenazzi M, Lanzardo S, Conti L, Ruiu R, Alberti D, et al. , 2015. Targeting ferritin receptors for the selective delivery of imaging and therapeutic agents to breast cancer cells. Nanoscale 7, 6527–6533. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, et al. , 2016. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Goyal MS, Preuss TM, Raichle ME, Van Essen DC, 2014. Trends and properties of human cerebral cortex: correlations with cortical myelin content. Neuroimage 93 (Pt 2), 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, et al. , 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC, 2011. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci 31, 11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier MD, Carson RP, Lagrange AH, 2015. Of mothers and myelin: Aberrant myelination phenotypes in mouse model of Angelman syndrome are dependent on maternal and dietary influences. Behav. Brain Res 291, 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Catts VS, Katayama Y, Shoji H, Takagi T, Huang FL, et al. , 2017. Decreased brain pH as a shared endophenotype of psychiatric disorders. Neuropsychopharmacology. 10.1038/npp.2017.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harting I, Seitz A, Rating D, Sartor K, Zschocke J, Janssen B, et al. , 2009. Abnormal myelination in Angelman syndrome. Eur. J. Paediatr. Neurol 13, 271–276. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. , 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, et al. , 2015. Canonical genetic signatures of the adult human brain. Nat. Neurosci 18, 1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Han D, Efimova O, Guijarro P, Yu Q, Oleksiak A, et al. , 2017. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci 20, 886–895. [DOI] [PubMed] [Google Scholar]
- Holm S, 1979. A simple sequentially rejective multiple test procedure. Scand. Stat. Theory Appl 6, 65–70. [Google Scholar]
- Human Connectome Project, 2013. HCP Pilot 1 Data Sharing Fact Sheet. Available at: https://www.humanconnectome.org/storage/app/media/documentation/q1/Q1_Release_Reference_Manual.pdf.
- Iwatani J, Ishida T, Donishi T, Ukai S, Shinosaki K, Terada M, et al. , 2015. Use of T1-weighted/T2-weighted magnetic resonance ratio images to elucidate changes in the schizophrenic brain. Brain Behav. 5, e00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG, 1998. Viewpoint: the core and matrix of thalamic organization. Neuroscience 85, 331–345. [DOI] [PubMed] [Google Scholar]
- Kim J-W, Naidich TP, Ely BA, Yacoub E, De Martino F, Fowkes ME, et al. , 2016. Human habenula segmentation using myelin content. Neuroimage 130, 145–156. [DOI] [PubMed] [Google Scholar]
- Koenig SH, 1995. Classes of hydration sites at protein-water interfaces: the source of contrast in magnetic resonance imaging. Biophys. J 69, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Zeichner SL, 2004. Host cell gene expression during human immunodeficiency virus type 1 latency and reactivation and effects of targeting genes that are differentially expressed in viral latency. J. Virol 78, 9458–9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk W, Macdonald PM, Stanisz GJ, Henkelman RM, 1994. Relaxivity and magnetization transfer of white matter lipids at MR imaging: importance of cerebrosides and pH. Radiology 192, 521–529. [DOI] [PubMed] [Google Scholar]
- Kugathasan R, Collier DA, Haddow LJ, El Bouzidi K, Edwards SG, Cartledge JD, et al. , 2017. Diffuse white matter signal abnormalities on magnetic resonance imaging are associated with human immunodeficiency virus type 1 viral escape in the central nervous system among patients with neurological symptoms. Clin. Infect. Dis 64, 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, van der Kouwe AJW, Raznahan A, Paus T, Johansen-Berg H, Miller KL, et al. , 2017. Studying neuroanatomy using MRI. Nat. Neurosci 20, 314–326. [DOI] [PubMed] [Google Scholar]
- Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, et al. , 2009. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev. Cell 16, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang T, Liu T, Lv J, Hu X, Guo L, et al. , 2016. A data-driven method to study brain structural connectivities via joint analysis of microarray data and dMRI data. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 829–832. [Google Scholar]
- Løken-Amsrud KI, Myhr K-M, Bakke SJ, Beiske AG, Bjerve KS, Bjørnarå BT, et al. , 2013. Retinol levels are associated with magnetic resonance imaging outcomes in multiple sclerosis. Mult. Scler 19, 451–457. [DOI] [PubMed] [Google Scholar]
- MacKay AL, Laule C, 2016. Magnetic resonance of myelin water: an in vivo marker for myelin. BPL 2, 71–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz A, Huisman SMH, Lelieveldt BPF, Reinders MJT, 2017. Brain transcriptome atlases: a computational perspective. Brain Struct. Funct 222, 1557–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancarci BO, Toker L, Tripathy SJ, Li B, Rocco B, Sibille E, et al. , 2017. Crosslaboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro, ENEURO 0212–0217, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Menon V, Goldy J, Kaykas A, Lee C-K, Smith KA, et al. , 2014. Improving reliability and absolute quantification of human brain microarray data by filtering and scaling probes using RNA-Seq. BMC Genom. 15, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry M, Pavlidis P, 2010. A cross-laboratory comparison of expression profiling data from normal human postmortem brain. Neuroscience 167, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Bauer NM, Schafer I, White R, 2013. Making myelin basic protein -from mRNA transport to localized translation. Front. Cell. Neurosci 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators, 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Xu W, Zheng J, Huang K, Wang H, Tong J, et al. , 2013. The Disease and Gene Annotations (DGA): an annotation resource for human disease. Nucleic Acids Res. 41, D553–D560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Ch’ng C, Lui F, St-Georges N, Zoubarev A, Lai AY, et al. , 2013. Neurocarta: aggregating and sharing disease-gene relations for the neurosciences. BMC Genom. 14, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pya N, Wood SN, 2015. Shape constrained additive models. Stat. Comput 25, 543–559. [Google Scholar]
- Richiardi J, Altmann A, Milazzo A-C, Chang C, Chakravarty MM, Banaschewski T, et al. , 2015. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348, 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Veronese M, Expert P, Turkheimer FE, Bertoldo A, 2016. MENGA: a new comprehensive tool for the integration of neuroimaging data and the allen human brain transcriptome atlas. PLoS One 11 e0148744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romme IAC, de Reus MA, Ophoff RA, Kahn RS, van den Heuvel MP, 2017. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol. Psychiatry 81, 495–502. [DOI] [PubMed] [Google Scholar]
- Sahraian MA, Radue E-W, Haller S, Kappos L, 2010. Black holes in multiple sclerosis: definition, evolution, and clinical correlations. Acta Neurol. Scand 122, 1–8. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H, 2004. RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691. [DOI] [PubMed] [Google Scholar]
- Santo-Domingo J, Demaurex N, 2012. Perspectives on: SGP symposium on mitochondrial physiology and medicine: the renaissance of mitochondrial pH. J. Gen. Physiol 139, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck JF, 2003. Magnetic resonance imaging of brain iron. J. Neurol. Sci 207, 99–102. [DOI] [PubMed] [Google Scholar]
- Schmierer K, Wheeler-Kingshott CAM, Tozer DJ, Boulby PA, Parkes HG,Yousry TA, et al. , 2008. Quantitative magnetic resonance of postmortem multiple sclerosis brain before and after fixation. Magn. Reson. Med 59, 268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K, Brunkhorst R, de Bruin N, Mayer CA, Haussler A, Ferreiros N, et al. , 2017. Dysregulation of lysophosphatidic acids in multiple sclerosis and autoimmune encephalomyelitis. Acta Neuropathol. Commun 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, et al. , 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. U. S. A 97, 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Ferrell K, Frank R, Dubiel W, 1997. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem 272, 8145–8148. [DOI] [PubMed] [Google Scholar]
- Shen EH, Overly CC, Jones AR, 2012. The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci. 35, 711–714. [DOI] [PubMed] [Google Scholar]
- Shimoyama M, De Pons J, Hayman GT, Laulederkind SJF, Liu W, Nigam R, et al. , 2015. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 43, D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, French L, Xu T, Leonard G, Perron M, Pike GB, et al. , 2017. Cell-specific gene-expression profiles and cortical thickness in the human brain. Cereb. Cortex 1–11. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. , 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. [DOI] [PubMed] [Google Scholar]
- Singh N, Vrontakis M, Parkinson F, Chelikani P, 2011. Functional bitter taste receptors are expressed in brain cells. Biochem. Biophys. Res. Commun 406, 146–151. [DOI] [PubMed] [Google Scholar]
- Sprawls P, 2000. Magnetic Resonance Imaging: Principles, Methods, and Techniques. Medical Physics Publishing. [Google Scholar]
- Stedehouder J, Couey JJ, Brizee D, Hosseini B, Slotman JA, Dirven CMF, et al. , 2017. Fast-spiking parvalbumin interneurons are frequently myelinated in the cerebral cortex of mice and humans. Cereb. Cortex 27, 5001–5013. [DOI] [PubMed] [Google Scholar]
- Stüber C, Morawski M, Schafer A, Labadie C, Wahnert M, Leuze C, et al. , 2014. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. Neuroimage 93 (Pt 1), 95–106. [DOI] [PubMed] [Google Scholar]
- Szuchet S, Nielsen JA, Lovas G, Domowicz MS, de Velasco JM, Maric D, et al. , 2011. The genetic signature of perineuronal oligodendrocytes reveals their unique phenotype. Eur. J. Neurosci 34, 1906–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki C, Yamasaki M, Uchigashima M, Konno K, Yanagawa Y, Watanabe M, 2010. Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur. J. Neurosci 32, 1326–1336. [DOI] [PubMed] [Google Scholar]
- Thakurela S, Garding A, Jung RB, Müller C, Goebbels S, White R, et al. , 2016. The transcriptome of mouse central nervous system myelin. Sci. Rep 6, 25828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium, 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás J, Santos CRA, Quintela T, Gonçalves I, 2016. “Tasting” the cerebrospinal fluid: another function of the choroid plexus? Neuroscience 320, 160–171. [DOI] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME, 2010. Regional aerobic glycolysis in the human brain. Proc. Natl. Acad. Sci. U. S. A 107, 17757–17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Valk J, 2011. Magnetic Resonance of Myelination and Myelin Disorders. Springer; Berlin Heidelberg. [Google Scholar]
- Vártes PE, Rittman T, Whitaker KJ, Romero-Garcia R, Vasa F, Kitzbichler MG, et al. , 2016. Gene transcription profiles associated with inter-modular hubs and connection distance in human functional magnetic resonance imaging networks. Philos. Trans. R. Soc. Lond. B Biol. Sci 371 10.1098/rstb.2015.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vymazal J, Hajek M, Patronas N, Giedd JN, Bulte JW, Baumgarner C, et al. , 1995. The quantitative relation between T1-weighted and T2-weighted MRI of normal gray matter and iron concentration. J. Magn. Reson. Imaging 5, 554–560. [DOI] [PubMed] [Google Scholar]
- Weiner J 3rd, Domaszewska T, 2016. tmod: an R package for general and multivariate enrichment analysis. PeerJ Prepr. 10.7287/peerj.preprints.2420v1. [DOI] [Google Scholar]
- Whitaker KJ, Vártes PE, Romero-Garcia R, Vása F, Moutoussis M, Prabhu G, et al. , 2016. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc. Natl. Acad. Sci. U. S. A 113, 9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Hing CJ, Obenaus A, Stryker R, Tong K, Sarty GE, 2005. Magnetic resonance imaging and mathematical modeling of progressive formalin fixation of the human brain. Magn. Reson. Med 54, 324–332. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juráus A, et al. , 2015. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.