Summary
Integrins, which bind laminin, a major component of the mammary basement membrane, are strongly expressed in basal stem cell-enriched populations, but their role in controlling mammary stem cell function remains unclear. We found that stem cell activity, as evaluated in transplantation and mammosphere assays, was reduced in mammary basal cells depleted of laminin receptors containing α3- and α6-integrin subunits. This was accompanied by low MDM2 levels, p53 stabilization, and diminished proliferative capacity. Importantly, disruption of p53 function restored the clonogenicity of α3/α6-integrin-depleted mammary basal stem cells, while inhibition of RHO or myosin II, leading to decreased p53 activity, rescued the mammosphere formation. These data suggest that α3/α6-integrin-mediated adhesion plays an essential role in controlling the proliferative potential of mammary basal stem/progenitor cells through myosin II-mediated regulation of p53 and indicate that laminins might be important components of the mammary stem cell niche.
Key words: mammary gland, cre-lox gene deletion, integrin, laminin, stem cells
Highlights
-
•
α3- and α6-integrins are required for mammary basal stem cell function
-
•
p53 is activated in mammary basal cells depleted of α3- and α6-integrins
-
•
RHO and myosin II mediate p53 activation in α3- and α6-integrin-depleted cells
Faraldo and colleagues report that major cellular receptors for laminins, α3-and α6-subunit-containing integrins play an important role in the control of the proliferative potential of mammary basal stem/progenitor cells regulating the p53 activity through an RHOA/myosin II-dependent mechanism. These findings indicate that laminins might be essential components of the mammary stem cell niche.
Introduction
The mammary epithelium is organized as a bilayer, with a luminal layer of steroid hormone receptor-positive and -negative epithelial cell populations and an underlying layer of basal myoepithelial cells (Macias and Hinck, 2012). The extraordinary proliferative potential of the mammary epithelium, replenishing the gland during lobulo-alveolar development in successive pregnancies and repopulating the cleared mammary fat pad following the transplantation of tissue fragments or dispersed cells, strongly suggests that stem and progenitor cells are present in this tissue (Visvader and Smith, 2011). Only basal cells can regenerate bilayer mammary ducts and alveoli following the transplantation of sorted epithelial cell populations from adult mouse mammary gland (Shackleton et al., 2006, Sleeman et al., 2007, Stingl et al., 2006). These observations suggested that the mammary basal cell layer harbors multipotent stem cells able of generating various mammary cell types during lobulo-alveolar development and mature gland homeostasis. However, many lineage-tracing studies have provided evidence refuting this hypothesis and strongly indicating that, in adult glands, the luminal and the basal mammary epithelial compartments are maintained by unipotent stem cells (Davis et al., 2016, Prater et al., 2014, Van Keymeulen et al., 2011, Van Keymeulen et al., 2017). The possible presence of functional multipotent basal stem cells in adult glands therefore remains a matter of debate (Rios et al., 2014, van Amerongen et al., 2012).
Integrins are heterodimeric transmembrane extracellular matrix (ECM) receptors consisting of an α and a β subunit. To date, eight β and 18 α subunits have been described. These subunits can assemble into 24 integrins with different ligand specificities (for review see Barczyk et al., 2010, Hynes, 2002). Associations between the cytoplasmic tail of the integrin and various intracellular adaptor proteins control integrin ligand-binding activity. Conversely, ligand binding triggers intracellular signaling events and extracellular force sensing increases integrin clustering, thereby amplifying biochemical cascades (Barczyk et al., 2010, DuFort et al., 2011).
Integrins have been implicated in the control of stem cell maintenance and progenitor cell differentiation in various embryonic and adult tissues (Brizzi et al., 2012, Chen et al., 2013, Glukhova and Streuli, 2013, Raymond et al., 2012). The α2-, α6-, β1-, and β3-integrin subunits serve as surface markers for the isolation of stem cell-enriched populations from basal and luminal mammary epithelial layers. The ECM may, therefore, be an important component of the mammary stem cell microenvironment, the stem cell niche (Asselin-Labat et al., 2007, Shackleton et al., 2006, Shehata et al., 2012, Stingl et al., 2006).
We have shown that the specific deletion of β1-integrins from the basal layer affects mammary stem cell self-renewal, suggesting that β1-integrins play an essential role in maintaining the mammary stem cell population (Taddei et al., 2008). Other teams reported that β1-integrin deletion from luminal cells reduced epithelial outgrowth development from transplanted mammary tissue (Li et al., 2005) and implicated β1-integrins in the self-renewal of progenitors in mammary organoid culture (Olabi et al., 2018). However, the roles of specific integrin dimers in the maintenance of functional mammary stem cell population(s) remain unclear.
Laminins are major constituents of the basement membrane, a specialized ECM underlying the mammary epithelial bilayer, and the laminin-binding integrin dimers α3β1, α6β1, and α6β4 are expressed by mammary basal epithelial cells (Raymond et al., 2012). We evaluated the contribution of laminin-binding integrins to the control of mammary stem cell activity, using a loss-of-function approach in which the Itga3 and/or Itga6 genes were deleted from the mammary basal cells with the Cre-Lox system. We show here that laminin-binding integrins are essential for mammary stem cell function, although α3β1- and α6-containing integrin dimers may have at least partially redundant functions. Mechanistically, we found that lack of α3β1- and α6-integrins led to increased myosin II activity and induced p53 accumulation leading to growth arrest.
Results
Simultaneous Deletion of the α3- and α6-Integrin Chains Affects Mammary Basal Stem Cell Activity
Mammary epithelial cells express on their surface several integrin receptors, including those for laminins, collagens, and fibronectin (Figure S1). To study the role of laminin-binding integrins in the control of mammary stem/progenitor cell function, we deleted the Itga3 and/or Itga6 genes in vitro by transduction of mammary basal cells freshly isolated from mice carrying the corresponding conditional alleles (Itga3F/F and Itga6 F/F) with an adenovirus expressing Cre recombinase (Adeno-Cre). Basal cells were sorted as described previously (Stingl et al., 2006); a typical profile is shown in Figure S1A. We used the Rosa26LacZ-reporter allele (R26RF/+) to monitor integrin deletion. Following Adeno-Cre transduction, stem and progenitor cell activities were assessed in control (R26RF/+) and integrin-depleted basal cells, in limiting dilution transplantation and mammosphere assays. Basal cells depleted of Itga3, Itga6, or both of these genes, are hereafter referred to as α3KO, α6KO, and α3α6KO, respectively.
Simultaneous deletion of the Itga3 and Itga6 genes greatly decreased the capacity of basal cells to regenerate mammary epithelium following their transplantation into cleared mammary fat pads (Figures 1A and 1B). Deletion of the α3 chain did not affect the regenerative potential of mammary basal cells, and basal cells depleted of α6 presented only a moderate decrease in capacity to repopulate the fat pad (Figures S2A and S2B).
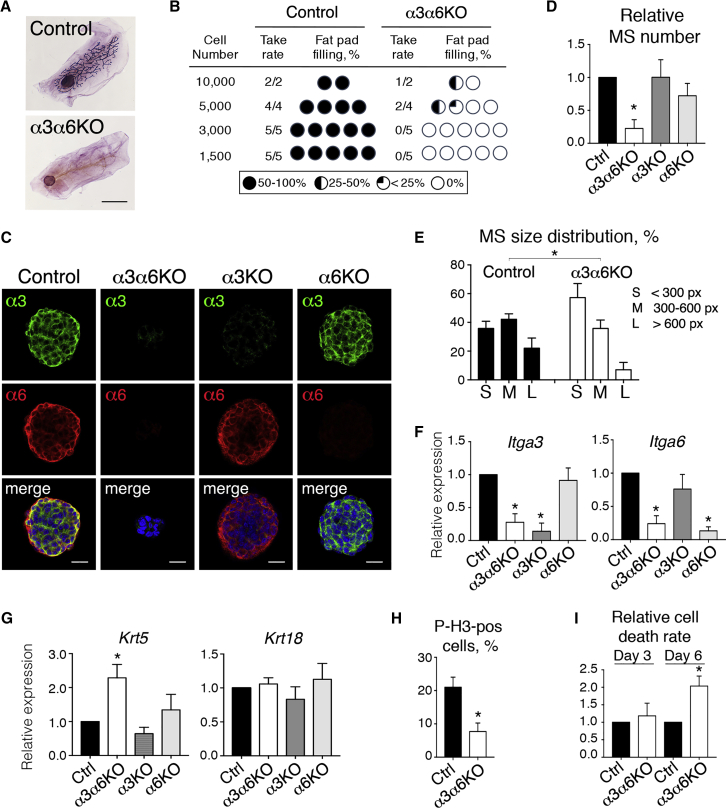
Figure 1.
Deletion of α3- and α6-Integrin Chains from Mammary Basal Cells Affects Stem Cell Activity
Basal (CD24LOW/ITGα6HIGH) mammary cells were isolated from mammary tissue as described previously (Stingl et al., 2006); a typical profile is shown in Figure S1A.
(A) Recipient mouse mammary fat pads grafted with control or α3α6KO mammary basal cells dissected 10 weeks after transplantation and stained with LacZ and Carmine-Alum in whole mounts. Representative images. Scale bar, 5 mm.
(B) Take rate and fat pad filling in the outgrowths developed by control and α3α6KO mammary basal cells in limiting dilution transplantations. Pool of three independent experiments.
(C) Confocal representative images of mammospheres formed by control (Ctrl) and integrin-depleted mammary basal cells after 12 days of culture immunolabeled with anti-integrin antibodies. Nuclei were visualized with DAPI. Scale bars, 20 μm.
(D) Mammospheres formed by integrin-depleted cells counted after 12–14 days of culture. The graph shows means ± SD obtained in 10, 3, and 4 independent experiments for α3α6KO, α3KO, and α6KO cells, respectively; p < 0.0001 for α3α6KO, p = 0.98 for α3KO, and p = 0.06 for α6KO.
(E) Size distribution of mammospheres formed by control and α3α6KO mammary basal cells. The graph shows means ± SD from 4 independent experiments. S, small; M, medium; L, large. p < 0.0001.
(F) qRT-PCR analysis of Itga3 and Itga6 gene expression in mammospheres formed by integrin-depleted cells. The graph shows means ± SD from n independent experiments. For α3α6KO, n = 6, p < 0.0001 for both, Itga3 and Itga6 genes; for α3KO, n = 3, p = 0.007 for Itga3 and p = 0.2 for Itga6; for α6KO, n = 3, p = 0.5 for Itga3, p = 0.002 for Itga6.
(G) qRT-PCR analysis of Krt5 and Krt18 gene expression in cells obtained from mammospheres formed by integrin-depleted and control (Ctrl) mammary basal cells. The graph shows means ± SD from three independent experiments. For α3α6KO, p = 0.048 for Krt5.
(H) The percentage of P-H3-positive cells in mammospheres formed by control or α3α6KO cells evaluated on day 3 of culture. The graph shows means ± SD obtained in three independent experiments, p = 0.005.
(I) Cell death rates in mammosphere cultures of control or α3α6KO cells assessed by CellTox Green assay on days 3 and 6 after seeding. Values shown are means ± SD from three independent experiments. 3 days, p = 0.5, 6 days, p = 0.02.
In (D), (F), (G), and (I) values obtained for control cells were set as 1 in each experiment. See also Figures S1 and S2.
Consistent with the transplantation assay results, α3α6KO cells exhibited poor clonogenic capacity and formed few spheres, most of which were of small size, whereas α3KO and α6KO cells formed spheres of a similar size and number to those observed in control cultures (Figures 1C–1E and S2C). At least 90% of the mammospheres were devoid of integrin surface expression as assessed by immunolabeling (Figure 1C), and qRT-PCR analyses of dissociated mammospheres confirmed integrin chain deletion (Figures 1C and 1F). The addition of collagen 1 or fibronectin to the mammosphere medium did not rescue the capacity of α3α6KO cells to form spheres (Figure S2D).
Mammospheres were characterized by analyzing the expresson of differentiation markers. In line with our previous studies, we found that in the control mammospheres, part of the cells, often basally located, co-expressed keratin 5 (K5) and K8, whereas the inner cells displayed a typical K5−/K8+ luminal phenotype (Chiche et al., 2013) (Figure S2E). The α3α6KO mammospheres where enriched in K5+ cells, particularly the small mammospheres (Figure S2E). Consistently, qRT-PCR analysis showed that Krt5 expression was significantly increased in the mammospheres formed by α3α6KO cells but not in those formed by α3KO and α6KO cells, while Krt18 levels were unchanged (Figure 1G). These data indicate that the absence of laminin-binding integrins does not completely prevent but affects the differentiation of basal cells into the luminal lineage. Interestingly, relative expression of Lgr5, a basal cell marker used to enrich for mammary stem cell activity (Fu et al., 2017) is decreased in α3α6KO mammospheres (Figure S2F).
To understand the cellular mechanisms leading to the weak clonogenic capacity of α3α6KO cells, we analyzed their proliferation rates at early stages of culture. Immunolabeling with anti-phospho (ser10)-histone H3 antibody (P-H3), which specifically labels cells in mitosis (Hendzel et al., 1997), revealed that, on day 3, the percentage of cycling cells had decreased by two-thirds in α3α6KO cultures (Figure 1H). Bromodeoxyuridine (BrdU) incorporation assays revealed that proliferation was decreased in both basal and luminal cells (Figure S2G). Cell death rates in α3α6KO cultures were similar to those in control ones at day 3, not increasing until day 6 (Figures 1I and S2H). Cleaved caspase-3 staining indicated that apoptosis contributed to the observed cell death (Figure S2I). Thus, the deletion of laminin-binding integrins from mammary basal cells prevented these cells from entering the cell cycle and increased cell death rates during subsequent culture, resulting in the production of small spheres by the surviving cells.
p53 Is Activated in α3α6KO Mammary Basal Epithelial Cells
Earlier studies implicated p53 in cellular responses to various stress conditions (Horn and Vousden, 2007), whereas an important role of p53 in stem cell biology has emerged in the last years (Jain and Barton, 2018). In particular, p53 has been found to act as an essential regulator of mammary stem cells by restricting their proliferation and self-renewal (Chiche et al., 2013, Cicalese et al., 2009). We therefore investigated whether the p53 pathway was activated in the mammospheres derived from basal cells depleted of laminin-binding integrins. Immunolabeling showed that most cells, basal and luminal, in small α3α6KO-mammospheres presented nuclear p53 (Figures 2A and S3A). Moreover, qRT-PCR analyses performed with RNA samples obtained from dissociated mammospheres revealed an upregulation of p53 transcriptional targets such as Cdkn1a (coding for the cell cycle regulator p21) and Mdm2 in α3α6KO cells, suggesting an activation of the p53 pathway in these cells (Figure 2B). Expression of Cdkn1a and Mdm2 was not changed in α3KO or α6KO cell (Figure 2B).
Figure 2.
The p53 Pathway Is Activated in Mammary Basal Cells Depleted of α3- and α6-Integrin Chains
(A) Confocal representative images of mammospheres formed by control and α3α6KO mammary basal cells after 12 days of culture immunolabeled with anti-p53 antibody. Phalloidin served to visualize F-actin, nuclei were labeled with DAPI. Scale bar, 20 μm.
(B) qRT-PCR analysis of Cdkn1a and Mdm2 gene expression in cells obtained from mammospheres formed by integrin-depleted and control (Ctrl) mammary basal cells. The graph shows means ± SD from n independent experiments. For α3α6KO, n = 6, p = 0.0003 for Cdkn1a and p = 0.008 for Mdm2; for α3KO, n = 3, p = 0.3 for Cdkn1a and p = 0.4 for Mdm2; for α6KO, n = 3, p = 0.25 for Cdkn1a and p = 0.27 for Mdm2.
(C) Dot plot showing flow cytometry separation of basal and luminal cells from mammary explants from Itga3F/F;Itag6F/F;Rosa26LacZ (control) or K5-CreERT2+/−;Itga3F/F;Itag6F/F;Rosa26LacZ mice, treated with 4-OHT (4-hydroxytamoxifen). B, basal cell population.
(D) qRT-PCR analysis of Itga3 and Itga6 gene expression in α3α6KO basal cells isolated from mammary explants (n = 3). p = 0.0038 for Itga3 and p = 0.0036 for Itga6.
(E) qRT-PCR analysis of p53 target gene expression in α3α6KO basal cells isolated from mammary explants. The graph shows means ± SD from three independent experiments. p = 0.008 for Cdkn1a; p = 0.027 for Mdm2; p = 0.025 for Trp53inp1; p = 0.005 for Rchy1; p = 0.003 for Gadd45.
(F) Western blotting analysis of p53 and MDM2 protein levels in extracts of control and α3α6KO mammary basal cells isolated from 4-OHT-treated explants. Tubulin served as a loading control.
(G) Heatmap based on RNA-seq analysis of freshly sorted control and α3α6KO basal cells isolated from mammary explants.
In (B) and (E), values obtained for control cells were set as 1 in each experiment. See also Figure S3 and Table S1.
We also assessed the p53 pathway activation in cells depleted of the α3- and α6-integrins in explant cultures, an approach with better viable cell yields than Adeno-Cre gene deletion. Fragments of mammary ducts isolated from K5Cre- ERT2;Itga3F/F;Itga6 F/F mice, in which Cre expression is controlled by an estrogen-responsive element, were allowed to attach to plastic, then, treated with tamoxifen to induce Cre expression leading to integrin gene deletion. Mammary epithelial cells migrated from duct fragments onto the plastic. After several days of culture, the cells were collected, and basal populations were sorted by flow cytometry with CD24/ICAM-1 labeling, as described by Di-Cicco et al. (2015) (Figures 2C and S3B). Specific deletion of the Itga3 and Itga6 genes from basal cells was confirmed by qRT-PCR (Figure 2D). Consistently, immunocytofluorescence staining showed that most basal cells in the explants were devoid of α3-integrin and flow cytometry analyses revealed α6-integrin deletion in at least 70% of basal cells (Figures S3C and S3D). Only the α6-negative-sorted basal cell population was studied further in the experiments illustrated in Figures 2E–2G. No significant changes in the surface expression of α1- and α5-integrins were detected; α2 and β4 were moderately downregulated, whereas αv and β3 were slightly upregulated in α3α6KO cells (Figure S3D). Our qRT-PCR data suggested that, as in cells from α3α6KO mammospheres, several p53 target genes were upregulated in freshly sorted α3α6KO basal cells obtained from explant cultures (Figure 2E).
Finally, we analyzed the levels of p53 and MDM2 proteins in control and α3α6KO basal cells by western blotting. The E3 ubiquitin-protein ligase MDM2 plays a central role in the control of p53 stability, and its binding to p53 favors the proteosomal degradation of this protein (Haupt et al., 1997, Kubbutat et al., 1997). We found that p53 protein levels were notably increased in α3α6KO cells in explant culture (Figure 2F). By contrast, MDM2 protein levels were decreased, providing conditions for p53 stabilization and nuclear accumulation (Haupt et al., 1997, Kubbutat et al., 1997). Mdm2 gene is positively regulated by p53 at the transcriptional level; therefore, increased p53 activity is expected to be accompanied by higher Mdm2 transcript levels. However, additionally, MDM2 protein expression is regulated by posttranscriptional mechanisms, including control of protein stability. Previous studies have shown that, p53 activation in stress response occurs due to MDM2 degradation (Carr and Jones, 2016).
RNA sequencing (RNA-seq) analysis on freshly sorted α3α6KO mammary basal cells confirmed that the p53 pathway was activated and revealed the perturbation of several other pathways including those associated with G2/M transition, mitosis, Rho GTP-ases, collagen and keratan sulfate biosynthesis, and cell surface integrin interactions (Figure 2G; Table S1). Notably, we observed altered expression of integrin subunits from dimers interacting with ECM proteins other than laminin (Figure S3E). Consistent with the flow cytometry data, RNA-seq analysis revealed that Itga2 expression was downregulated, whereas, Itga11 (encoding a collagen receptor subunit), Itga9, and Itgb5 genes (encoding subunits of RGD-binding integrins, i.e., receptors for ECM components, such as tenascin-C or osteopontin), were upregulated. In addition, Arhgap11a and Racgap1, coding for two RhoGAPs inactivating, respectively, Rho and Rac, were downregulated in integrin-depleted cells (Figure S3E).
These data strongly indicate that alteration of cell-ECM interactions in mammary basal cells depleted of laminin-binding integrins triggers p53 activation, preventing their entry into the cell cycle and leading to a lack of clonogenic capacity.
Suppression of p53 Function Restores Clonogenic Potential in α3α6KO Mammary Basal Cells
We investigated whether p53 activation was responsible for the low clonogenic potential of α3α6KO cells, by introducing a dominant-negative p53 mutant (p53-DN) into sorted mammary basal cells carrying conditional alleles for Itga3 and Itga6 genes and then treating the cells with Adeno-Cre virus to delete integrin genes. The expression of p53-DN had no significant effect on control cultures, but restored the capacity of α3α6KO basal cells to form mammospheres (Figures 3A and 3B). Consistently, expression of Cdkn1a was diminished in α3α6KO spheres expressing p53-DN (Figure 3C), whereas expression of Krt5 was similar to that found in control spheres, indicating that downregulation of p53 function re-establish normal differentiation of α3α6KO basal cells (Figure S4A).
Figure 3.
Inhibition of p53 Rescues Clonogenic Capacity of Mammary Basal Cells Depleted of α3- and α6-Integrins
(A) Microphotographs of mammospheres formed by control or α3α6KO mammary basal cells expressing p53-dominant-negative (p53-DN) mutant lentivirus. Representative images. Scale bar, 200 μm.
(B) Quantification of mammosphere formation by mammary basal cells expressing p53-DN. Values shown represent means ± SD from three independent experiments, p = 0.03.
(C) qRT-PCR analysis of Cdkn1a gene expression in mammospheres formed by α3α6KO mammary basal cells. The graph shows means ± SD from three independent experiments, p = 0.008.
(D) Confocal representative images of mammospheres formed by α3α6KO and α3α6p53KO cells double immunolabeled with anti-α3 and anti-α6 antibodies. Scale bars, 30 μm.
(E) Quantification of mammosphere formation by α3α6KO and α3α6p53KO mammary basal cells. Values shown represent means ± SD from three independent experiments, p = 0.005.
(F) qRT-PCR analysis of Cdkn1a gene expression in mammospheres formed by α3α6KO and α3α6p53KO mammary basal cells. The graph shows means ± SD from four independent experiments, p = 0.0002.
In (B), (C), (E), and (F) values obtained for control cells were set as 1 in each experiment. See also Figure S4.
We then simultaneously deleted Trp53, Itga3, and Itga6 genes from mammary basal cells isolated from mice carrying the corresponding conditional alleles. Trp53 deletion restored the number and size of the spheres formed by α3α6KO cells and, as expected, downregulated the expression of Cdkn1a and Mdm2 genes (Figures 3D–3F, S4B, and S4C).
Altogether these results indicate that increased p53 activity is responsible for the perturbed clonogenic capacity induced by the loss of laminin-binding integrins in basal mammary cells.
Inhibition of Myosin II Function Restores Clonogenic Capacity in α3α6KO Mammary Basal Cells by Suppressing p53
The activity of the small Rho GTPases RHOA was particularly high in α3α6KO cells (Figure 4A). RHOA activates ROCK, and an inhibitor of ROCK, Y27632, is often used to improve the viability of embryonic stem cells or mammary stem cells in culture (Guo et al., 2012, Watanabe et al., 2007). We therefore tested whether ROCK inhibition by Y27632 could improve the sphere-forming capacity of mammary basal cells depleted of the α3- and α6-integrin chains. Decreased phospho-MLC levels were detected in Y27632-treated cultures, confirming ROCK inhibition (Figure S5A). Strikingly, in the presence of Y27632, the clonogenic capacity of α3α6KO cells was restored (Figures 4B and 4C) and the formed mammospheres were of similar size to that of control spheres (Figure S5B). ROCK controls the phosphorylation/activation status of the regulatory myosin light chain essential for myosin II interaction with actin required for ATPase activation and contraction. Another drug, blebbistatin, inhibits myosin II activity downstream of ROCK. Blebbistatin had similar effects to Y27632 on mammary basal cells: it restored the number and size of mammospheres formed by α3α6KO cells and enhanced sphere formation by control cells (Figures 4B, 4C, and S5B). Interestingly, Lgr5 expression levels were restored by these drugs in the integrin-depleted mammospheres (Figure S5C).
Figure 4.
Inhibition of ROCK or myosin II Leads to MDM2 Activation, Prevention of p53 Accumulation and Restores Mammary Stem/Progenitor Cell Activity
(A) Relative activated RHOA levels in α3α6KO cells. Activated RHOA, evaluated by ELISA assay, was normalized to total RHOA levels measured by western blotting and represent means ± SD from three independent experiments, p = 0.03.
(B) Confocal representative images of mammospheres formed by control or α3α6KO mammary basal cells grown in the presence of Y27632 or blebbistatin double immunolabeled with anti-α3 and anti-α6 antibodies. DAPI served to visualize nuclei. Scale bars, 30 μm.
(C) Quantification of mammosphere formation. The values shown represent means ± SD obtained in 10 independent experiments for untreated and Y27632-treated cells and 4 independent experiments for blebbistatin-treated cells. For α3α6KO mammospheres, p = 0.001 for Y27632-treated cells compared with non-treated cells, p = 0.01 for blebbistatin-treated cells compared with non-treated cells.
(D and E) qRT-PCR analysis of Cdkn1a and Mdm2 gene expression in cells obtained from mammospheres formed by integrin-depleted mammary basal cells in the presence of Y27632 or blebbistatin. The graph shows means ± SD from n independent experiments. For Y27632 treatment, n = 6; for blebbistatin treatment, n = 3. (D) for α3α6KO mammospheres, Y27632-treated versus untreated, p = 0.0004, blebbistatin-treated versus untreated, p = 0.01; (E) Y27632-treated versus untreated, p = 0.001, blebbistatin-treated versus untreated, p = 0.02.
(F) Western blotting analysis of p53, MDM2, and Phospho-MDM2 protein levels in extracts of mammosphere formed by control, α3α6KO and α3α6p53KO cells. Y27632 or blebbistatin were present, where indicated. GAPDH was used as a loading control. The graphs show the means ± SEM from at least three independent experiments. For all indicated comparisons, p < 0.05.
In (A), (C), (D), and (E) values obtained for control cells were set as 1 in each experiment. Through the whole figure: Y, Y27632, B, blebbistatin. See also Figure S5.
We next studied the effects of Y27632 and blebbistatin on p53 activity in control and integrin-depleted cells. Expression of the p53 target genes Cdkn1a and Mdm2 was downregulated in the presence of the drug, suggesting an inhibition of p53 transcriptional activity (Figures 4D and 4E). In line with these data, p53 protein levels were significantly lower in the presence of Y27632 or blebbistatin (Figure 4F). Phosphorylation of MDM2 favors its binding to p53 facilitating its proteosomal degradation (Carr and Jones, 2016). Both phosphorylated and total MDM2 levels were decreased in α3α6KO cells, whereas the inhibition of myosin II by Y27632 or blebbistatin was accompanied by an increase in phosphorylated and total MDM2 protein levels in control and mutant cells (Figure 4F). Taken together, these data strongly indicate that myosin II inhibition favors p53 degradation enabling the cell to enter the cell cycle.
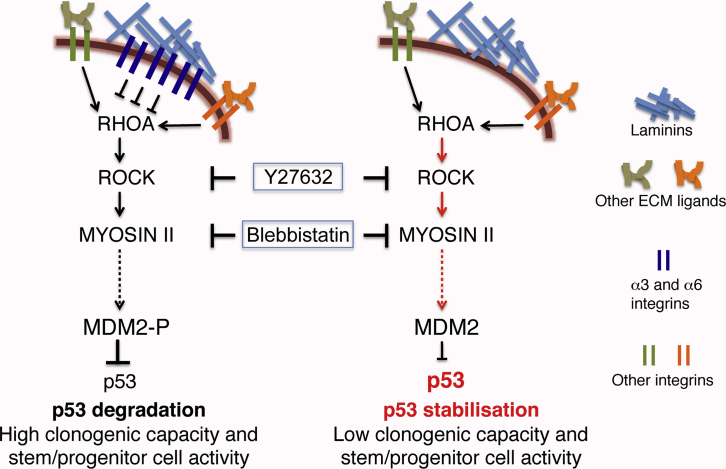
The scheme presented in Figure 5 summarizes the results of this study, implicating laminin receptors, α3- and α6-integrins, in the myosin II-mediated control of p53 activity and, thereby, regulation of mammary basal stem cell function.
Figure 5.
Intracellular Signaling Events Underlying Regulation of Mammary Basal Stem Cell Activity by Laminin-Binding Integrins: Hypothetic Scheme
Left: adhesion of mammary basal cells to laminin via α3- and α6-integrins restricts RHOA activation, thereby limiting myosin II contractility. These events serve for MDM2 phosphorylation/activation thus favoring p53 degradation and providing clonogenic capacity to stem cells. Right: mammary basal cells depleted of laminin-binding integrins display elevated active RHOA triggering activation of myosin II accompanied by low MDM2 activity allowing p53 stabilization and suppression of clonogenicity in mammary basal stem cell. Inhibition of myosin II activity by Y27632 or blebbistatin rescues clonogenic activity of integrin-depleted cells. Red color indicates abnormally activated state of the molecule/pathway in α3/α6-integrin-depleted cells.
Discussion
Together with hormone and soluble growth factor signaling, integrin-mediated adhesion to ECM plays an important role in the maintenance of the mammary stem cell population (Taddei et al., 2008, Visvader and Stingl, 2014). However, knowledge of the mechanisms controlling mammary stem cell functions and the composition of their niche remains incomplete. Laminins are major components of the mammary basement membrane, and the integrin receptor subunits β1 and α6 serve as markers for isolating stem cell-enriched preparations from mammary epithelium (Raymond et al., 2009, Shackleton et al., 2006, Stingl et al., 2006). Laminins have been identified as components of the stem cell microenvironment in other tissues, such as skeletal muscle, the mammalian neural central system, and mouse testis (Chen et al., 2013, Rayagiri et al., 2018, Raymond et al., 2009). We therefore reasoned that signaling via integrins interacting with laminins might contribute to the control of mammary stem function.
The in vivo transplantation model remains the gold standard assay for stem or progenitor cell activity. In addition, high proliferative potential, one of the essential characteristics of stem cells and their progeny can be assessed in vitro, in clonogenicity assay, i.e., mammosphere test in case of mammary stem cells. Here, we show that deletion of α3- and α6-integrins affects stem cell activity. Mammary basal cells depleted of laminin-binding integrins failed to regenerate mammary epithelium upon transplantation into cleared mouse mammary fat pads and displayed poor clonogenic activity in culture, due to the cell-cycle arrest associated with p53 activation.
Strikingly, the clonogenic activity of mammary basal cells depleted of α3- and α6-integrins was rescued by the myosin II inhibitors Y27632 and blebbistatin. Fuchs' laboratory previously showed that, in squamous cell carcinoma, MYH9, a non-muscle myosin IIA, regulates the posttranscriptional stabilization of p53, acting as a tumor suppressor (Schramek et al., 2014). A connection between functional myosin IIA and p53 accumulation associated with tumor suppressor activity has also been reported for head and neck carcinomas (Coaxum et al., 2017). The exact molecular mechanisms underlying p53 activation by myosin II remain to be unraveled. Consistent with our findings, in keratinocytes treated with doxorubicin to induce DNA damage response, the inhibition of myosin II with Y27632 or blebbistatin prevents p53 stabilization (Schramek et al., 2014). By contrast, the inhibition of actin polymerization by latrunculin did not interfere with p53 accumulation, indicating that the effects of myosin II on p53 were not relevant to its role in stimulation of actin polymerization (Schramek et al., 2014). Accordingly, in our model, the treatment of α3α6-integrin-depleted cells with latrunculin did not rescue their clonogenic capacity, which was abolished by p53 accumulation (data not shown).
A pioneering study by the Damsky laboratory showed that cell interactions with fibronectin provide survival signals that suppress a p53-regulated cell death pathway (Ilic et al., 1998). The involvement of p53 in cell death induced by detachment from ECM, anoikis, was subsequently studied mostly in the context of cancer (Grossmann, 2002, Horbinski et al., 2010). By contrast, in this study, rather than focusing on anoikis-associated events, we analyzed the signaling mechanisms activated in the surviving mammary basal cells after the depletion of laminin-binding integrins.
Streuli's laboratory has shown that mammary epithelium from late (embryonic day [E] E16.5–E17.5) embryos with germline inactivation of Itga3, Itga6, or Itgb4 may develop normally following transplantation into cleared mammary fat pads (Klinowska et al., 2001). However, this study provided no quantitative evaluation of stem cell activity in the mutant epithelium studied. By contrast, we report here the results of limiting dilution transplantations and clonogenicity assays performed with sorted α3α6-integrin-depleted cells, allowing a quantitative characterization of stem cell function. We found that only simultaneous deletions of both α3- and α6-integrin subunits resulted in p53 activation leading to a complete loss of stem/progenitor cell activity. Our data indicate that the functions of α3β1- and α6-containing integrins in the control of stem/progenitor cell proliferative potential may overlap at least partially. However, the possibility that α3β1- and α6-integrins are involved in different signaling events that, when combined, lead to p53 activation and stem cell perturbation cannot be excluded.
Previous studies have revealed non-redundant cellular functions of these integrins in different physiological contexts. We recently showed that the α3β1-integrin is essential for the development of mammary tumors with basal-like characteristics in K5ΔNβcat mice presenting a constitutive activation of WNT signaling in the basal cell layer (Cagnet et al., 2014, Moumen et al., 2013, Teuliere et al., 2005). Further, loss of α3-integrin prevented tumor formation in the two-stage skin carcinogenesis mouse model (Sachs et al., 2012). Of note, those studies have analyzed the effects of Itga3 deletion in tumor context, where ECM composition and organization are severely perturbed, involving integrin receptors different from those acting in normal homeostasis. Another recent report provided evidence that Itgb4 gene deletion in vivo in an MMTV-Cre model, leading to embryonic α6β4-integrin depletion, affected early mammary development, including basal cell differentiation (Li et al., 2015). However, adult basal stem cell activity was not examined in these mutant mice.
Our current knowledge of laminin variant diversity in mammary basal membrane remains incomplete, but several laminin chains, including α1, α5, β1-3, γ1, and γ2, have been identified in mammary ducts and alveoli by immunolabeling (Prince et al., 2002). Stem/progenitor cells can alter their proximal ECM composition, thereby regulating their function (Rayagiri et al., 2018). Thus, the diverse ligand specificity of laminin-binding integrins, with a preference of α3β1 and α6β4 for laminins 511/521 and laminin 332, and a broader binding specificity for α6β1, which can also interact with laminin 111 (Yamada and Sekiguchi, 2015), appears to be of great importance, as it can provide highly specific responses to a complex ECM. Thus, it will be interesting to determine which combination of integrins (α3β1 plus α6β1 and/or α6β4) are involved in stem cell regulation.
RNA-seq-based comparative analysis of α3α6-depleted and control mammary basal cells revealed perturbation of several important intracellular pathways including those associated with the control of proliferation and cell-ECM interactions. Notably, altered expression of several integrins interacting with ECM proteins other than laminins, such as collagens, fibronectin, and osteopontin, were observed in α3α6-integrin-depleted cells. Thus, alterations of interactions with the ECM may underlie the stronger RHOA activation observed in α3α6-deficient mammary basal cells. It has been shown previously that laminins preferentially trigger RAC1 and CDC42 activation, while other ECM components, such as fibronectin or collagen, sustained strong RHO activation (Gu et al., 2001, Zhou and Kramer, 2005). Interestingly, the transcripts of two RhoGAPs, ARHGAP11A and RACGAP1, inactivating RHO and RAC, respectively, and recently implicated in the control of proliferation (Lawson et al., 2016), were found downregulated in integrin-depleted cells. The reduced Arhgap11A expression may account for the higher levels of RHO activation in α3α6KO cells. Consistently, laminin-binding integrins in association with tetraspanins have been proposed to generate RHO-suppressive signals (Novitskaya et al., 2014).
Integrins are mechanosensing receptors, and the depletion of laminin-binding integrins alters cellular responses to external mechanical signals induced by cell confinement and ECM stiffness. Of note, the loss of α3 from mammary basal cells in vivo impairs RAC1 activation, resulting in sustained phosphorylation of myosin light chains and a hypercontractile state (Raymond et al., 2011). Integrins are well-known modulators of the activities of Rho GTP-ases, which control epithelial cell polarization, lumen formation during morphogenesis, centrosome positioning, and spindle orientation during mitosis, with an impact on symmetric and asymmetric cell divisions (Akhtar and Streuli, 2013, Rodriguez-Fraticelli and Martin-Belmonte, 2014). Future studies will show whether these alterations of intracellular signaling are relevant to the impaired stem cell properties of integrin-deficient mammary epithelial cells documented here.
In brief, we show here that cell interactions involving α3β1- and α6-containing integrins binding laminins, control the proliferative potential of mammary basal stem cells by inhibiting p53 activation via mechanisms involving myosin II. These findings provide new insights into the molecular mechanisms controlling mammary stem cell activity, and indicate that laminins are highly likely to contribute to mammary stem cell niche. Future studies are required to characterize the role of specific laminin variants in the mammary stem cell niche.
Experimental Procedures
Mice
The generation of Itga6F/F, Trp53F/F, and Itga3F/F mice has been described previously (De Arcangelis et al., 2017, Jonkers et al., 2001, Margadant et al., 2009). K5Cre-ERT2 mice and the Rosa26LacZ reporter strain, were kindly provided by Dr. Metzger (Indra et al., 1999) and by Dr. Soriano (Soriano, 1999), respectively. All mice were bred in a mixed DBA/C57Bl6 background. In Adeno-Cre infection experiments Rosa26LacZ mice were used as controls. The care and use of animals were conducted in accordance with the European and National Regulations for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (facility license C750517/18). All Experimental Procedures were ethically approved (ethical approval 02,265.02).
Mammary Gland Dissociation, Cell Sorting, and Flow Cytometry Analysis
Thoracic and inguinal mammary glands of three to four 15–20-week-old virgin females were pooled, dissociated, and processed for single-cell suspension and flow cytometry as described elsewhere (Stingl et al., 2006, Taddei et al., 2008). For basal cell isolation, cells were incubated at 4°C for 20 min with the following antibodies: anti-CD45-APC (clone 30-F11), anti-CD31-APC (clone MEC13.3), anti-CD24-BViolet421 (clone M1/69), and anti-CD49f-PeCy7 (clone GoH3), or anti-ICAM1-PeCy7 (clone YN1/1.7.4), all from BioLegend. Labeled cells were analyzed and sorted out using either a FACSVantage flow cytometer (BD Biosciences) or a MoFlo Astrios cell sorter (Beckman Coulter). Sorted cell population purity was at least 95%. The antibodies used for the analysis of cell surface integrin expression in basal and luminal cells are presented in Table S2. Data were analyzed using FlowJo software.
AdCre-Mediated Gene Deletion
For conditional gene deletion, freshly sorted basal cells (CD31/45−, CD24low, CD49fhigh population) were incubated with an adenovirus expressing the Cre recombinase under the control of the CMV promotor (AdCre, SignaGen Laboratories) at MOI of 5,000 for 1 h at 37°C. Trypan blue staining showed that at least of 90%–95% were viable after infection.
After washing, cells were resuspended in mammosphere medium and plated in low attachment 24-well plates at 5,000 cells/well in duplicate as described previously (Chiche et al., 2013). For ROCK or myosin II-ATP-ase inhibition, Y27632 (1 μM, Millipore) or Blebbistatin (5 μM, R&D Systems) were added to the mammosphere medium and replenished twice a week. After 12–14 days of culture, wells were photographed and the number and size of mammospheres was analyzed using ImageJ software. When indicated, collagen-I (200 μg/mL) and fibronectin (200 μg/mL) were added to the mammosphere medium containing 2% Matrigel. For BrdU incorporation assays, the mammospheres were incubated during 1 h with 5 μM BrdU, before fixation in Methacarn (methanol:chloroform:acetic acid, 6:3:1) for 15 min. Quantification of RHOA activation was performed using G-LISA RhoA Activation Assay Biochem Kits, following the manufacturer's instructions (Cytoskeleton).
Transplantation
After Adeno-Cre infection, sorted basal cells were resuspended in 10 μL of 50% growth factor-reduced Matrigel (BD Biosciences) and injected into the inguinal fat pads of 3-week-old nude BALB/c females cleared of endogenous epithelium as described elsewhere (Taddei et al., 2008). Ten weeks after transplantation, fat pads were fixed in 2.5% paraformaldehyde for 1 h at 4°C and stained overnight at 30°C with X-gal (Biology of the Mammary Gland, http://mammary.nih.gov).
Mammary Explant Culture
Mammary organoids were obtained from mammary glands dissected from K5-CreERT2+/−;Itga3F/F;Itag6F/F;Rosa26LacZ or K5-CreERT2−/−;Itga3F/F;Itag6F/F;Rosa26LacZ mice according to Ewald's protocol (Ewald et al., 2008). A total of 60,000 organoids were plated on P150 plastic plates in 15 mL of DMEM:F12 medium containing 5% fetal bovine serum, 5 μg/mL insulin, 10 ng/mL epidermal growth factor, and penicillin/streptomycin. After 48 h, 4-hydroxytamoxifen was added at 100 nM (Sigma) and replenished 24 h later at the same concentration. The explants were trypsinized after 6 days of culture and basal cells (CD31/45−, CD24low, ICAM1high) were FACS-sorted.
RNA Extraction and qRT-PCR
RNA was extracted with the RNeasy Micro Kit including DNase treatment (QIAGEN), and reverse-transcribed using MMLV H(−) Point reverse transcriptase (Promega). qPCR was performed using the QuantiNova SYBR Green PCR Kit (QIAGEN) on a LightCycler 480 real-time PCR system (Roche). The values obtained were normalized to Gapdh levels. The primers used for qRT-PCR analysis were purchased from SABiosciences/QIAGEN or designed using Oligo 6.8 software (Molecular Biology Insights) and synthesized by Eurogentec. Primers used in this study are listed in Table S3.
Statistical Analysis
All values are shown as mean ± SD or SEM. The p values were determined using Student's t test with two-tailed distribution and unequal variance (Welch's correction). For mammosphere size comparison, a Pearson's chi-square test was applied.
Author Contributions
M.R., S.C., A.C., L.B., K.R., M.-A.D., and M.M.F. designed and performed the experiments. S.B. and P.d.l.G. performed the RNA-seq analyses. A.D.A., M.K., E.G.-L., and A.S. provided the mouse strains. M.A.G. and M.M.F. designed the experimental plan and wrote the manuscript. All authors contributed to data analysis and interpretation and final approval of the manuscript.
Acknowledgments
We are particularly grateful to A. Di Cicco for expert technical assistance, to Dr. I. Grandjean, S. Jannet, and the personnel of the animal facilities at Institut Curie for taking care of the mice, to Z. Maciorowski, A. Viguier, and S. Grondin for excellent assistance with FACS analyses and to the Cell and Tissue Imaging (PICT-IBiSA), Institut Curie, member of the French National Research Infrastructure France-BioImaging (ANR10-INBS-04). We also thank Drs. D. Metzger, P. Soriano, and J. Jonkers for providing mouse strains. The work was supported by grants from Agence Nationale de la Recherche (ANR-13-BSV2-0001), Canceropôle Ile de France (2014-1-SEIN-01-ICR-1), La Ligue Nationale Contre le Cancer Comité de Paris (RS16/75-70), and Labex Celtisphybio (ANR-10-LABX-0038), part of the Idex PSL. M.R. received funding from Marie Curie Fellowship Program; S.C. from Association pour la Recherche sur le Cancer; A.C. from Agence Nationale de la Recherche, and L.B. from Canceropôle Ile de France. M.A.G. is Directeur de Recherche, M.M.F., M.-A.D., and K.R. are Chargé de Recherche at the Institut National de la Santé et de la Recherche Médicale (INSERM).
Published: March 21, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.02.008.
Accession Numbers
RNA-seq data have been deposited in the GEO under accession number GEO: GSE117120.
Supplemental Information
References
- Akhtar N., Streuli C.H. An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat. Cell Biol. 2013;15:17–27. doi: 10.1038/ncb2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat M.L., Sutherland K.D., Barker H., Thomas R., Shackleton M., Forrest N.C., Hartley L., Robb L., Grosveld F.G., van der Wees J. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Barczyk M., Carracedo S., Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzi M.F., Tarone G., Defilippi P. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr. Opin. Cell Biol. 2012;24:645–651. doi: 10.1016/j.ceb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Cagnet S., Faraldo M.M., Kreft M., Sonnenberg A., Raymond K., Glukhova M.A. Signaling events mediated by alpha3beta1 integrin are essential for mammary tumorigenesis. Oncogene. 2014;33:4286–4295. doi: 10.1038/onc.2013.391. [DOI] [PubMed] [Google Scholar]
- Carr M.I., Jones S.N. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl. Cancer Res. 2016;5:707–724. doi: 10.21037/tcr.2016.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Lewallen M., Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140:255–265. doi: 10.1242/dev.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche A., Moumen M., Petit V., Jonkers J., Medina D., Deugnier M.A., Faraldo M.M., Glukhova M.A. Somatic loss of p53 leads to stem/progenitor cell amplification in both mammary epithelial compartments, basal and luminal. Stem Cells. 2013;31:1857–1867. doi: 10.1002/stem.1429. [DOI] [PubMed] [Google Scholar]
- Cicalese A., Bonizzi G., Pasi C.E., Faretta M., Ronzoni S., Giulini B., Brisken C., Minucci S., Di Fiore P.P., Pelicci P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Coaxum S.D., Tiedeken J., Garrett-Mayer E., Myers J., Rosenzweig S.A., Neskey D.M. The tumor suppressor capability of p53 is dependent on non-muscle myosin IIA function in head and neck cancer. Oncotarget. 2017;8:22991–23007. doi: 10.18632/oncotarget.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F.M., Lloyd-Lewis B., Harris O.B., Kozar S., Winton D.J., Muresan L., Watson C.J. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun. 2016;7:13053. doi: 10.1038/ncomms13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arcangelis A., Hamade H., Alpy F., Normand S., Bruyere E., Lefebvre O., Mechine-Neuville A., Siebert S., Pfister V., Lepage P. Hemidesmosome integrity protects the colon against colitis and colorectal cancer. Gut. 2017;66:1748–1760. doi: 10.1136/gutjnl-2015-310847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Cicco A., Petit V., Chiche A., Bresson L., Romagnoli M., Orian-Rousseau V., Vivanco M., Medina D., Faraldo M.M., Glukhova M.A. Paracrine Met signaling triggers epithelial-mesenchymal transition in mammary luminal progenitors, affecting their fate. Elife. 2015;4 doi: 10.7554/eLife.06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort C.C., Paszek M.J., Weaver V.M. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Brenot A., Duong M., Chan B.S., Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu N.Y., Rios A.C., Pal B., Law C.W., Jamieson P., Liu R., Vaillant F., Jackling F., Liu K.H., Smyth G.K. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 2017;19:164–176. doi: 10.1038/ncb3471. [DOI] [PubMed] [Google Scholar]
- Glukhova M.A., Streuli C.H. How integrins control breast biology. Curr. Opin. Cell Biol. 2013;25:633–641. doi: 10.1016/j.ceb.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann J. Molecular mechanisms of “detachment-induced apoptosis–Anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- Gu J., Sumida Y., Sanzen N., Sekiguchi K. Laminin-10/11 and fibronectin differentially regulate integrin-dependent Rho and Rac activation via p130(Cas)-CrkII-DOCK180 pathway. J. Biol. Chem. 2001;276:27090–27097. doi: 10.1074/jbc.M102284200. [DOI] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zurrer-Hardi U., Bell G. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Horbinski C., Mojesky C., Kyprianou N. Live free or die: tales of homeless (cells) in cancer. Am. J. Pathol. 2010;177:1044–1052. doi: 10.2353/ajpath.2010.091270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Ilic D., Almeida E.A., Schlaepfer D.D., Dazin P., Aizawa S., Damsky C.H. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J. Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.K., Barton M.C. p53: emerging roles in stem cells, development and beyond. Development. 2018;145 doi: 10.1242/dev.158360. [DOI] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Klinowska T.C., Alexander C.M., Georges-Labouesse E., Van der Neut R., Kreidberg J.A., Jones C.J., Sonnenberg A., Streuli C.H. Epithelial development and differentiation in the mammary gland is not dependent on alpha 3 or alpha 6 integrin subunits. Dev. Biol. 2001;233:449–467. doi: 10.1006/dbio.2001.0204. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., Vousden K.H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lawson C.D., Fan C., Mitin N., Baker N.M., George S.D., Graham D.M., Perou C.M., Burridge K., Der C.J., Rossman K.L. Rho GTPase transcriptome analysis reveals oncogenic roles for rho GTPase-activating proteins in basal-like breast cancers. Cancer Res. 2016;76:3826–3837. doi: 10.1158/0008-5472.CAN-15-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sun H., Feltri M.L., Mercurio A.M. Integrin beta4 regulation of PTHrP underlies its contribution to mammary gland development. Dev. Biol. 2015;407:313–320. doi: 10.1016/j.ydbio.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang Y., Naylor M.J., Schatzmann F., Maurer F., Wintermantel T., Schuetz G., Mueller U., Streuli C.H., Hynes N.E. Beta1 integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. EMBO J. 2005;24:1942–1953. doi: 10.1038/sj.emboj.7600674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias H., Hinck L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant C., Raymond K., Kreft M., Sachs N., Janssen H., Sonnenberg A. Integrin alpha3beta1 inhibits directional migration and wound re-epithelialization in the skin. J. Cell Sci. 2009;122:278–288. doi: 10.1242/jcs.029108. [DOI] [PubMed] [Google Scholar]
- Moumen M., Chiche A., Decraene C., Petit V., Gandarillas A., Deugnier M.A., Glukhova M.A., Faraldo M.M. Myc is required for beta-catenin-mediated mammary stem cell amplification and tumorigenesis. Mol. Cancer. 2013;12:132. doi: 10.1186/1476-4598-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskaya V., Romanska H., Kordek R., Potemski P., Kusinska R., Parsons M., Odintsova E., Berditchevski F. Integrin alpha3beta1-CD151 complex regulates dimerization of ErbB2 via RhoA. Oncogene. 2014;33:2779–2789. doi: 10.1038/onc.2013.231. [DOI] [PubMed] [Google Scholar]
- Olabi S., Ucar A., Brennan K., Streuli C.H. Integrin-Rac signalling for mammary epithelial stem cell self-renewal. Breast Cancer Res. 2018;20:128. doi: 10.1186/s13058-018-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prater M.D., Petit V., Alasdair Russell I., Giraddi R.R., Shehata M., Menon S., Schulte R., Kalajzic I., Rath N., Olson M.F. Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol. 2014;16:942–950. doi: 10.1038/ncb3025. 941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince J.M., Klinowska T.C., Marshman E., Lowe E.T., Mayer U., Miner J., Aberdam D., Vestweber D., Gusterson B., Streuli C.H. Cell-matrix interactions during development and apoptosis of the mouse mammary gland in vivo. Dev. Dyn. 2002;223:497–516. doi: 10.1002/dvdy.10070. [DOI] [PubMed] [Google Scholar]
- Rayagiri S.S., Ranaldi D., Raven A., Mohamad Azhar N.I.F., Lefebvre O., Zammit P.S., Borycki A.G. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat. Commun. 2018;9:1075. doi: 10.1038/s41467-018-03425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K., Cagnet S., Kreft M., Janssen H., Sonnenberg A., Glukhova M.A. Control of mammary myoepithelial cell contractile function by alpha3beta1 integrin signalling. EMBO J. 2011;30:1896–1906. doi: 10.1038/emboj.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K., Deugnier M.A., Faraldo M.M., Glukhova M.A. Adhesion within the stem cell niches. Curr. Opin. Cell Biol. 2009;21:623–629. doi: 10.1016/j.ceb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Raymond K., Faraldo M.M., Deugnier M.A., Glukhova M.A. Integrins in mammary development. Semin. Cell Dev. Biol. 2012;23:599–605. doi: 10.1016/j.semcdb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A.E., Martin-Belmonte F. Picking up the threads: extracellular matrix signals in epithelial morphogenesis. Curr. Opin. Cell Biol. 2014;30:83–90. doi: 10.1016/j.ceb.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Sachs N., Secades P., van Hulst L., Kreft M., Song J.Y., Sonnenberg A. Loss of integrin alpha3 prevents skin tumor formation by promoting epidermal turnover and depletion of slow-cycling cells. Proc. Natl. Acad. Sci. U S A. 2012;109:21468–21473. doi: 10.1073/pnas.1204614110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D., Sendoel A., Segal J.P., Beronja S., Heller E., Oristian D., Reva B., Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K.E., Kendrick H., Robertson D., Isacke C.M., Ashworth A., Smalley M.J. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Taddei I., Deugnier M.A., Faraldo M.M., Petit V., Bouvard D., Medina D., Fassler R., Thiery J.P., Glukhova M.A. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 2008;10:716–722. doi: 10.1038/ncb1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuliere J., Faraldo M.M., Deugnier M.A., Shtutman M., Ben-Ze'ev A., Thiery J.P., Glukhova M.A. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132:267–277. doi: 10.1242/dev.01583. [DOI] [PubMed] [Google Scholar]
- van Amerongen R., Bowman A.N., Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Fioramonti M., Centonze A., Bouvencourt G., Achouri Y., Blanpain C. Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep. 2017;20:1525–1532. doi: 10.1016/j.celrep.2017.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Smith G.H. Murine mammary epithelial stem cells: discovery, function, and current status. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Muguruma K., Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Yamada M., Sekiguchi K. Molecular basis of laminin-integrin interactions. Curr. Top. Membr. 2015;76:197–229. doi: 10.1016/bs.ctm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Zhou H., Kramer R.H. Integrin engagement differentially modulates epithelial cell motility by RhoA/ROCK and PAK1. J. Biol. Chem. 2005;280:10624–10635. doi: 10.1074/jbc.M411900200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.