Abstract
Assessment of the fidelity of gene expression is crucial to understand cell homeostasis. Here we present a highly sensitive method for the systematic Quantification of Rare Amino acid Substitutions (QRAS) using absolute quantification by targeted mass spectrometry after chromatographic enrichment of peptides with missense amino acid substitutions. By analyzing incorporation of near- and non-cognate amino acids in a model protein EF-Tu, we show that most of missense errors are too rare to detect by conventional methods, such as DDA, and are estimated to be between <10−7–10-5 by QRAS. We also observe error hotspots of up to 10−3 for some types of mismatches, including the G-U mismatch. The error frequency depends on the expression level of EF-Tu and, surprisingly, the amino acid position in the protein. QRAS is not restricted to any particular miscoding event, organism, strain or model protein and is a reliable tool to analyze very rare proteogenomic events.
INTRODUCTION
Fidelity of gene expression is an important determinant of cellular homeostasis. Errors of transcription or translation can lead to formation of non-functional or toxic proteins which disrupt cellular fitness, multiply the energy waste, and increase the costs of quality control in the cell (1). One major type of errors is substitution of a correct amino acid encoded by the mRNA by an incorrect amino acid (missense errors). Missense errors can arise during transcription caused by the mistakes of the RNA polymerase or by mRNA derivatization (2). Alternatively, errors can arise during translation due to either charging a tRNA with an incorrect amino acid or to misreading of an mRNA codon by an incorrect aminoacyl-tRNA on the ribosome. Transcription and translation machineries make very few mistakes due to intricate selection mechanisms that allow their active sites to select the correct, and reject incorrect, substrates. Amino acid substitutions may lead to incorrectly folded proteins (3–5) that are recognized by the cellular quality control systems which remove misfolded proteins (1,6). As a result, the steady-state error frequency in the cell reflects the balance between error-making and error-removing processes.
Despite the eminent importance of translation errors for understanding cellular fitness and evolution, a comprehensive catalogue of error frequencies for different codons, types of substitutions or different protein contexts is not available. So far, the sensitivity of the available analytical methods limits the quantification depth, which sets the lower limit to measured missense error frequencies. Early reports quantified error frequencies in vivo based on the misincorporation of a radioactive amino acid into a protein that normally lacks this amino acid (e.g. Cys into the Cys-free flagellin (7) or L7/12 (8)). Alternatively, error frequencies were estimated using reporter systems that measure how the activity of an enzyme with deleterious mutations is restored by amino acid substitutions reverting it to the wild-type sequence (e.g. in β-lactamase (9), chloramphenicol acetyl transferase (10), β-galactosidase (11), luciferases (12,13) or green fluorescent protein (14)). These estimations of the overall error frequency in the cell, together with the direct quantification of the transcription errors, suggested that mRNA decoding on the ribosome is the most error-prone step in gene expression (with an error frequency of about 10−5–10−3), whereas transcription is more accurate (10−6–10−5) (reviewed in (1)). When used to estimate how many miscoded proteins are produced, an error frequency of 10−3 translates into a prediction that about 30% of cellular proteins will have at least one error (assuming an average protein length of 300 amino acids). This means that cells have to cope with a high error load, which has evolutionary and medical implications (1). However, the reporter assays used so far are restricted in the choice of model proteins, positions and types of amino acid substitutions and limited sensitivity. In addition, these assays rely on the overexpression of the reporter protein, which may activate cellular stress responses leading to altered error rates, and limit the choice of the model organism to strains that allow protein overexpression.
Mass spectrometry is routinely applied to detect low level sequence variants in proteins, such as used in quality control of biopharmaceutical products (15,16). Recently, mass spectrometry was also used to quantify error frequencies in model proteins (17–19) or proteome-wide in cell lysates (BioRxiv: https://doi.org/10.1101/255943) in Escherichia coli and Saccharomyces cerevisiae. These studies identified a set of frequent errors which can be classified into two major classes. One class entails highly abundant substitutions induced by various types of stress, such as the incorporation of non-proteinogenic amino acids (19), protein overexpression in combination with suboptimal mRNA codon usage, aminoglycoside treatment (BioRxiv: https://doi.org/10.1101/255943), or mutations in the protein synthesis machinery (20). In these cases, the error frequency can reach 10%, as observed for norvaline incorporation induced by an error-prone Leu-tRNA synthetase (20). Errors are also abundant in some organisms such as Microsporidia or Ascoidea asiatica which have elevated miscoding levels in comparison to classical model organisms (21,22).
The second class of errors comprises stress-independent amino acid substitutions reflecting the basal level of errors that escaped the cellular correction mechanisms. The most prominent members of this class are substitutions that have a G–U mismatch in the codon–anticodon helix, such as Gly (GGC) → Asp, Val (GUU) → Ile or Arg (CGC) → Gln (17) (BioRxiv: https://doi.org/10.1101/255943). The error frequency for this class is ∼10−4–10−3, consistent with the results of reporter assays (11). The G–U base pair can adopt a Watson-Crick-like geometry due to tautomerization and thus escape shape discrimination used by the ribosome to distinguish between correct and mismatched codon–anticodon base pairs (23). G-U mismatches can pass the initial selection phase during ribosome decoding (24), but are rejected at the proofreading phase (25). Alternatively, these errors can be attributed to erroneous transcription, because the corresponding C→U replacement represents an error hotspot (26) due to preferential cytosine deamination (2).
Notably, even the most complete mass spectrometry datasets comprise only a small subset of all possible amino acid substitutions (17) (BioRxiv: https://doi.org/10.1101/255943). It is unclear whether this reflects technical limitations of common data dependent acquisition approaches or a low cellular error frequency. Thus, new specialized workflows are needed to detect rare misreading events. Here, we introduce a mass spectrometry workflow for deep Quantification of Rare Amino acid Substitutions (QRAS). We combined chromatographic enrichment of peptides that contain amino acid substitutions with targeted mass spectrometric approaches to overcome the dynamic range restrictions. This allowed us to probe the high-fidelity regions on the error frequency landscape and thus to draw a more comprehensive map of misreading errors.
MATERIALS AND METHODS
AQUA peptides
Chemicals were purchased from Merck or Sigma Aldrich if not stated otherwise. Chemicals used for chromatographic separation were of HPLC/MS grade. All protein and peptide handling was performed in low retention reaction cups (Eppendorf). Quantified heavy isotope-labeled AQUA peptides (5 μM) were purchased from Thermo Scientific. AQUA peptides for the quantification of correct peptides were Ultimate grade with a guaranteed concentration error <5%; AQUA peptides for the quantification of missense peptides were QuantPro grade with a guaranteed concentration error <25%.
Bacterial strains and cell growth
EF-Tu (wild type) was prepared from E. coli MRE-600 (ATCC29417) purchased as freeze-dried pellet from UAB School of Medicine, Birmingham, AL, USA. Cells were grown in enriched medium and harvested at mid-logarithmic growth phase. E. coli BL21 (DE3) (Merck Millipore) strain was used for the overexpression of EF-Tu and EF-Tu mutants. For overexpressed EF-Tu, tufA gene from E. coli BL21(DE3) was cloned into the expression vector pet24(+)(kanamycin resistance cassette, C-terminal His-tag; Novagen) using NdeI and XhoI. Cells were grown at 37°C in Terrific broth medium in a Biostat B-plus 5 L fermenter (Sartorius) in the presence of 30 μg ml−1 kanamycin (Serva Electrophoresis). Protein expression was initiated at ∼0.7–0.8 OD600 by addition of 1 mM IPTG (Roth) for 2 h and cells were harvested by centrifugation after 4 h of induction. E. coli W3110 (K12) (Deutsche Sammlung von Mikroorganismen und Zellkulturen) was used to generate the chromosome-encoded C-terminally His-tagged EF-Tu (27). Cells were grown in LB medium at 37°C up to ∼1 OD600 and harvested by centrifugation. For DDA analysis cells were grown to 0.3 OD600 and treated with streptomycin (4 μM) for 2 h.
EF-Tu purification
Cells for the purification of EF-Tu without an affinity tag were resuspended and lysed in buffer A (50 mM HEPES–KOH pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM β-mercaptoethanol, containing Complete Protease inhibitor (1 tablet per 50 ml, Roche Diagnostics) and traces of DNase I (Sigma Aldrich)). Cells were lysed using the EmulsiFlex C3 (Avestin). Cell debris were removed by centrifugation. Lysate was loaded on a HighTrap Q HP anion exchange column (5 ml, GE Healthcare) and eluted using a salt gradient in buffer B (5–400 mM KCl in 25 mM HEPES–KOH pH 7.5, 3 mM MgCl2, 5 mM β-mercaptoethanol, 30 μM GDP). EF-Tu-containing fractions were applied to two sequential purification steps by SEC (HiLoad 26/60 Superdex 75, prep grade, GE Healthcare). EF-Tu-containing fractions were pooled, re-buffered into buffer C (50 mM HEPES–KOH pH 7.5, 50 mM KCl, 10 mM MgCl2), concentrated, and stored at −80°C.
Cells for His-tagged EF-Tu purification under native conditions were solubilized in B-PER reagent (Thermo Scientific) supplemented with 200 mM KCl, 3 mM MgCl2, Complete Protease inhibitor (1 tablet per 50 ml, Roche Diagnostics), 30 μM GDP, 5 mM β-mercaptoethanol and traces of DNase I (Sigma Aldrich). Solubilized cells were sonicated for 1 min and cell debris were precipitated by centrifugation. EF-Tu was purified using Ni-IDA Protino columns (Macherey-Nagel) according to manufacturer's protocol. EF-Tu was stored in buffer D (50 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2) at −80°C. For DDA analysis His-tagged EF-Tu (chromosome-encoded) was purified under denaturating conditions in urea. Cells were opened in buffer E (25 mM HEPES–KOH pH 7.5, 8 M urea, 200 mM KCl, 10 mM MgCl2, 5 mM β-mercaptoethanol) by sonification. Affinity purification was carried out using a Protino Ni-IDA column according to manufacturer's protocol. After elution, the sample complexity was further reduced by running EF-Tu on a 15% SDS-PAGE. The EF-Tu band was excised and proteolysed as described by Shevchenko et al. (28), except that n-methylmaleimide was used for alkylation instead of iodoacteamide.
In-solution proteolysis
For proteolysis of EF-Tu prepared under native conditions, EF-Tu (3000–100 000 pmol) was precipitated overnight with 5 volumes of ice-cold acetone at −20°C. Protein was collected by centrifugation, washed with ice-cold 80% ethanol and the pellet dried. EF-Tu was resuspended in 1% RapiGest (Waters) in 25 mM NH4HCO3 and incubated for 10 min at 37°C. Disulfide bonds were reduced by addition of 20 mM DTT (in 25 mM NH4HCO3) in two incubation steps, at 60°C for 10 min and at 37°C for 20 min. Alkylation of thiols was performed in 30 mM iodoacetamide (in 25 mM NH4HCO3) and incubating the sample at RT for 30 min in the dark. RapiGest in the sample was diluted to 0.1% with 25 mM NH4HCO3. Trypsin (1 μg/μl) (Trypsin Gold, Promega) was added to the sample (final concentration 0.01 μg/μl) and EF-Tu proteolysed overnight at 37°C.
Data dependent aquisition
LC–MS/MS analysis was performed on a nanoflow liquid chromatography system (Ultimate 3000RSLC, Thermo Fisher Scientific) coupled to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Samples were desalted on a self-packed reversed phase C-18 pre-column (20 mm × 0.1 mm inner diameter, Reprosil-Pur C18-AQ 5 μm resin, Dr. Maisch). Peptide separation was carried out on a self-packed RP C18 analytical column (100 mm × 0.05 mm inner diameter, ReproSil-Pur C18-AQ 3 μm resin, Dr Maisch) packed into a Silica Tip emitter (FS360–50-8-N, New Objective). Peptides were separated using a segmented linear gradient of 6.4–72% acetonitrile over 88 min using 0.1% formic acid as ion pair reagent at a flow of 300 nl/min.
Acquisition was performed using two acquisition schemes to maximize identifications while keeping consistent quantification. Quantification runs were performed in positive ion mode and a top 5 method with four micro scans per MS spectrum was applied. MS survey spectra were acquired at a resolution of 120 000 FWHM in the range of 350–1500 m/z. Precursors with charge states z = 2–7 that reached the intensity threshold of 5.0e3 were selected for fragmentation. Ions with unassigned charge states were excluded from fragmentation selection. Masses of fragmented precursor were dynamically excluded for 30 s. Higher energy collisional dissociation (HCD) with a normalized collision energy setting of 35% was applied for peptide fragmentation and fragments were detected in the ion trap. To gain additional identifications that are aligned to the quantification runs, identification runs using the same gradient were performed. Identification runs were acquired at a resolution of 120 000. Precursors with the charge states z = 2–7 that reached the intensity threshold of 5.0e3 were selected for fragmentation in the top speed mode. Masses of fragmented precursor were dynamically excluded for 15 s. HCD with a normalized collision energy setting of 35% was applied for peptide fragmentation and fragments were detected in the ion trap. Proteolysed EF-Tu from Str-treated and untreated samples was analyzed in four technical replicates using the quantification method. The quantification runs were complemented with two identification runs of the Str-treated sample. To avoid carry over of Str-induced missense peptides untreated replicates were analyzed before treated replicates and identification runs.
Acquired raw data were processed using MaxQuant software (version 1.5.5.1) (29). We constructed two sequence databases to allow for systematic detection of sequence variants. In the first search the sequence database contained all proteins that in sum contributed to >99.9% of all iBAQ values and a file with 248 common laboratory contaminants. In the main search the EF-Tu peptide sequences with all possible amino acid substitutions were included in the database. Leucine and isoleucine were considered to be the same amino acid. The main search mass tolerance was set to 20 ppm. N-methylmaleimide was used as fixed modification. Peptide identifications were filtered using a target-decoy approach at a false discovery rate of 0.01. In many cases, several peaks in one run or varying peaks in different chromatographic runs were identified as the same missense peptide. To achieve consistent quantifications the data were further analyzed using the Skyline software (30). Max Quant IDs were imported and the MS signal of the precursor ions (z = 2–5) of correct and missense EF-Tu peptides were extracted at a resolution setting of 60 000. Substitutions were considered to be identified by the globally highest scoring identification and the corresponding peaks were integrated in the same elution window in every chromatographic run. All significantly populated, interference-free charge states were integrated and summed up to one integrated MS signal. Two cognate tryptic peptides and their corresponding missense peptides were excluded from analysis for the following reasons. One peptide was lost during data acquisition due to instability. The other showed poor chromatographic properties and eluted over several minutes. In addition, atryptic peptides, missed-cleaved peptides, and identifications with non-integratable MS signals or inconsistent isotope patterns were excluded from analysis. In ambiguous cases, amino acid substitutions were interpreted as results of near-cognate misincorporations. Because the integration borders in Skyline may differ from the MaxQuant retention times, the average idotproduct (≥0.8) and the mass accuracy (delta mass ≤ 10 ppm) of the Str-induced samples were used as cut-off filters to reduce the number of false positives. In some cases individual features were assigned to different isobaric amino acid substitutions (e.g. N→Q, D→E, V→L/I). Because such regioisomers often co-elute and lead to chimeric spectra, this problem is hard to resolve in DDA approaches. However, a stricter filtering that assigns the features only to the highest-scoring identification would reduce the number of identifications and thus strengthen the notion that DDA datasets are incomplete while all other conclusions would be not effected. The integrated MS signals of treated and untreated samples were compared by a one-tailored t-test. For better comparison all MS signals were normalized to the median MS signal of the correct EF-Tu peptides.
Targeted mass spectrometry
Selected reaction monitoring (SRM)
Samples were analyzed on an Easy nLCII Nano LC or Ultimate 3000RSLC system coupled to a TSQ Vantage or TSQ Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific). Peptides were separated using an in-house packed column (14 cm length, 50 μm inner diameter, packed with Reprosil-Pur 120 C18 3 μm material) at 50°C or Acclaim PepMapRSLC (15 cm length, 75 μm inner diameter, 2 μm RP18 material) and eluted in a 45 min linear gradient from 5% acetonitrile with 0.1% formic acid to 50% acetonitrile with 0.1% formic acid at 0.3 μl/min flow. Q1 was set to unit resolution 0.7 Full Width at Half Maximum (FWHM) except for non-cognate peptides analysis where it was set to 0.5 to reduce interference. A spray voltage of 1800 V (TSQ Vantage) and 2100 V (TSQ Quantiva) was used with a heated ion transfer tube setting of 270°C (TSQ Vantage) and 325°C (TSQ Quantiva), respectively. The declustering voltage was kept at 10 V (TSQ Vantage) and a Chromfilter setting of 4 (TSQ Vantage) or 3 (TSQ Quantiva). Collision energies were optimized as described elsewhere (30). Scheduled transitions were recorded in a 5 min window and a cycle time of 3 s (TSQ Vantage) or 1s (TSQ Quantiva) was applied, typically resulting in dwell times of 100–200 ms per transition.
The open source program Skyline version 3.5 was used for the SRM method set up and results analysis (30). For each peptide of the SRM method, the predominant charge state of the precursor was experimentally assessed and 3–5 transitions per peptide were chosen (31) (Supplementary Table S5). For data analysis, raw files were imported into Skyline that automatically calculates the area under each transition peak to yield the light/heavy ratio for each peptide. To achieve high identification reliability, only peptides with a ratio dot-product (rdotp) close to 1 were considered and the light/heavy ratio of each peptide was ultimately used to calculate the error frequency.
Parallel reaction monitoring (PRM)
The identity of enriched peptides was further verified by targeted selected ion monitoring (tSIM) and parallel reaction monitoring (PRM) on a QExactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were separated by a 58 min linear reversed phase gradients from 5% acetonitrile with 0.1% formic acid to 50% acetonitrile with 0.1% formic acid on in-house packed columns (28 cm length, packed with Reprosil 1.9 μm C18 material) at 60°C. Eluted peptides were sprayed by an ESI-source set at 2400 V and capillary temperature 275°C in a Q-Exactive Plus mass spectrometer and t-SIM method was set at resolution 70 000, AGC target 5e4, maximum injection time 70 ms and scan range 150–2000 m/z and a 3.0 m/z isolation window. PRM method was set at a resolution of 35 000, AGC target 1e6, maximum injection time 300 ms and isolation windows of 0.4 m/z. Raw files were analyzed using Skyline software. MS1 and MS/MS filtering settings were set at a 60000 m/z and 35 000 m/z resolving power, respectively.
Multidimensional chromatography
For absolute quantification of correct peptides the final volume of tryptic digest was determined. 2 μl of the digestion mixture were diluted 1:20 with 5% acetonitrile with 0.5% formic acid and mixed with varying ratio of cognate AQUA Peptides 1–4. The ratios of endogenous: AQUA peptides were determined by SRM on TSQ Vantage or TSQ Quantiva mass spectrometer in triplicate (see below for details). The ratios calculated for each peptide were averaged and used to determine the amount of digested EF-Tu.
The tryptic digest was spiked with substoichiometric amounts of AQUA peptides containing the amino acid substitutions of interest (AQUA: proteolyzed EF-Tu was 1:1000–10 000). Normally 10–15 missense peptides were quantified in a single enrichment. Prior to enrichment, RapiGest was degraded by incubating the sample at acidic pH for 30 min at 37°C and its hydrolytic by-products removed by centrifugation. To remove any particles, the supernatant was filtered using Costar Spin-X Centrifuge Tube Filter 0.45 μm Cellulose Acetate and lyophilized in a SpeedVac vacuum concentrator. Peptides were dissolved in 200 μl 20% acetonitrile with 0.1% formic acid and separated by size-exclusion chromatography (SEC) on a Superdex Peptide 10/300 GL column in an isocratic HPLC run (20% acetonitrile with 0.1% formic acid; 0.8 ml/min flow, fraction size 0.4 ml) as a first chromatographic dimension. For large amounts of EF-Tu (>15000 pmol) 2–3 SEC runs were necessary to fractionate the entire digest. From fractions expected to contain target peptides, an aliquot was taken and diluted 1:5 with 0.1% formic acid to dilute the final concentration of acetonitrile and analyzed by SRM. Depending on the peptides′ distribution, 1–3 fractions were pooled and lyophilized. Peptides were redissolved in 10 mM ammonium acetate in 2% acetonitrile and separated by reversed phase chromatography at neutral pH in the second dimension (33) using a LiChrospher WP300 RP-18 (5 μm) column. Peptides were eluted with a 2–82% acetonitrile gradient in 10 mM ammonium acetate in 45 min run and 0.8 ml/min flow; fraction size 0.8 ml. The elution time for each peptide was established in an independent chromatographic run performed with AQUA peptides alone, by screening each fraction by SRM or MALDI analysis. The respective fractions from the second dimension were selected accordingly, lyophilized and resuspended in 50 μl 5% acetonitrile with 0.1% formic acid. Missense peptides were either quantified or further enriched in a third chromatographic separation (reversed phase chromatography at acidic pH). Peptides were eluted from a LiChrospher WP300 RP-18 (5 μm) column with a 0–65% acetonitrile gradient in 0.1% trifluoroacetic acid in 65 min run and 0.8 ml/min flow; fraction size 0.8 ml.
RESULTS
Amino acid substitutions quantified by data dependent acquisition (DDA)
We first estimated the missense error frequencies by DDA mass spectrometry in a bottom-up approach (Figure 1 and Supplementary Figure S1). To achieve the maximum coverage for rare missense peptides, we selected a model protein, elongation factor Tu (EF-Tu), as a highly conserved and highly abundant E. coli protein (Supplementary Figure S1A). EF-Tu is an essential GTPase that delivers aminoacyl-tRNA to the ribosome. EF-Tu is easy to purify in large quantities without a tag or with a C-terminal His-tag that does not interfere with its function (32), including translation fidelity (25,33). The use of a model protein decreases sample complexity and allows utilizing the full dynamic range of quantifications.
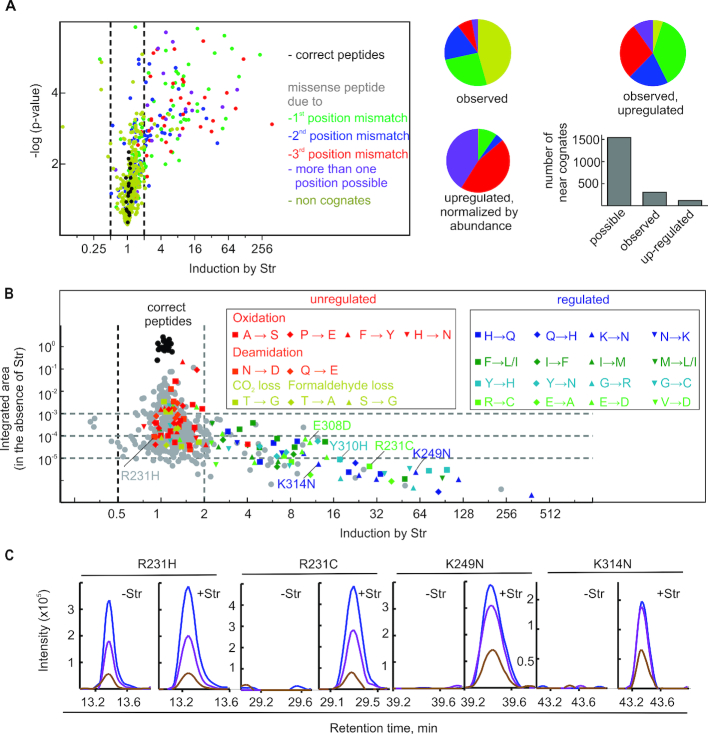
Figure 1.
Estimation of cellular error frequencies by DDA. (A) Str-induced missense peptides. The miscoding events leading to the respective amino acid substitutions are classified by the number of mismatches in the codon–anticodon helix: no mismatch, correct (black); one mismatch, near-cognate (green, blue, red; dependent on the position of the mismatch); if more than one particular mismatch can lead to the substitution (violet), or non-cognate (ocher) with two or three mismatches. Left panel, the volcano plot and statistical analysis are based on the integrated peak areas in four technical replicates (±4 μM Str) and a one tailored t-test. Right panel, pie diagrams: ratio of near- and non-cognates on the basis of the EF-Tu peptides included in the DDA analysis. Bar graph: number of near-cognate substitutions based on the EF-Tu peptides included in the DDA analysis. (B) Estimation of error frequencies in cells in the absence of Str. Integrated areas for amino acid subsitutions were normalized by the median of the integrated areas of the correct tryptic EF-Tu peptides. Identical time windows after retention time alignment between the runs were chosen for integration. For strong Str-induction this can lead to integration over noise in the absence of Str leading to error frequencies beyond the dynamic range of the mass spectrometer (see figure C). Left box: unregulated identifications that are isobaric to common artefacts. Right box: Amino acid substitutions that are consistently upregulated by Str. (C) Extracted molecular ion isotope peaks of selected missense peptides after MS1 filtering: with and without Str as indicated; M (blue), M+1 (violet), M+2 (brown).
To identify missense peptides, purified EF-Tu was digested with trypsin and peptides were analyzed by LC–MS/MS. Data were analyzed with MaxQuant software (29,34) and searched against a database that contained the correct native EF-Tu peptide sequences and all possible amino acid substitutions. Identifications with appropriate delta mass shifts were considered to represent peptides with amino acid substitutions (missense peptides). Extracted ion chromatograms (XICs) of missense peptides were manually integrated by MS1 filtering using the Skyline software (30,35). Error frequencies were estimated as the ratio of XICs of missense peptides to the median of XICs of correct peptides (Supplementary Table S1).
One caveat of this type of analysis is the prevalence of false positives caused by difficulties in distinguishing whether peptides of the expected delta masses originate from degradation or posttranslational modifications, rather than from true missense amino acid substitutions. The comparison with predicted fragmentation spectra and retention times, the use of additional fragmentation techniques, or simply the removal of known false positives can help to curate the dataset (reviewed in (16)). Alternatively, error frequency of gene expression can be altered, e.g. by antibiotics which increase miscoding. This increases the abundance of true missense peptides, but does not affect the frequency of false positives, allowing to distinguish between false positives and true missense peptides in the score-based peptide identification.
We used the latter approach and modulated the error frequency experimentally by adding the misreading-inducing aminoglycoside antibiotic streptomycin (Str) to the growth medium. Str binds to the decoding center of the ribosome and impairs its ability to discriminate between cognate tRNAs that fully match the codon and near-cognate ones with a single mismatch in the first, second, or third position of the codon–anticodon helix. As expected, treatment of E. coli cells with Str did not affect the peak intensities of the correct EF-Tu peptides (Figure 1A), but increased the abundance of missense peptides. Str-induced missense peptides have a higher mass accuracy (<Δppm, Supplementary Figure S1B) and a closer to expected isotope pattern (>ion-dot products, Supplementary Figure S1C) than those that do not respond to Str treatment. Database search and data annotation revealed 558 missense peptides for EF-Tu (Figure 1A, Supplementary Table S1). Amino acid substitutions isobaric to peptide modifications due to common degradation reactions (such as deamidation, oxidation or fragmentation, see Supplementary Table S4) were in most cases not affected by Str (Figure 1B). Therefore, missense peptides with the same mass as false positives were excluded by this approach. About 50% of peptides that were not induced by Str had substitutions that appeared to arise from non-cognate tRNA recognition, i.e. with more than one mismatch in the codon–anticodon complex. As non-cognate tRNAs are normally rejected by the ribosome very efficiently (36), this type of amino acid substitutions more likely represents false positives or amino acid substitutions that were introduced at different steps of protein synthesis such as misacylation by tRNA synthetases (37); as expected, they were not upregulated by Str treatment. In contrast, among the missense peptides that were induced by Str (122) only <5% were non-cognate (Figure 1A). Among the Str-upregulated missense peptides, 46 were due to first-position codon–anticodon mismatches, 24 to the second position mismatches, 34 to third position mismatches, and 12 were of ambiguous origin. When the number of identifications was normalized by the distribution of all possible substitutions based on the sequence of EF-Tu, errors due to third-position mismatches were overrepresented, consistent with recent report (BioRxiv: https://doi.org/10.1101/255943) (Figures 1A). Identifications corresponding to non-native peptides were found at a ratio of 10−5 to 10−1 to the respective correct peptides, which covers the entire dynamic range of the mass spectrometer (38,39), whereas the frequency of true miscoding events was below 10−3. Of note, basal level of F→L/I errors is relatively high, in the range of 10-4, which compares well with error frequencies expected from the invitro experiments, 10-5-10-3 (40). While Str-induced errors are due to mistranslation, the basal error in the absence of antibiotic is more difficult to assess, and can represent the combination of errors at any step of gene expression together with the activity of quality control machinery, or even chemical noise of mass-spectrometry measurements. Thus, quantifications of the basal error frequencies in the DDA setup should be considered as an upper limit for the corresponding amino acid substitution.
Surprisingly, amino acid substitutions introduced by G-U mismatches in the codon–anticodon complex, i.e. with G in the mRNA read by a tRNA that has a U in the anticodon instead of a canonical C, which were reported to be the main source of translation errors (11,17,23,25), did not increase upon Str treatment (Supplementary Figure S1D). Similarily, most U–G mismatches (U in the codon) were not induced by streptomycin. Only Y→H substitutions resulting from a U–G mismatch in the first codon position of a UAU/C codon, were systematically affected by Str treatment. To demonstrate that errors based on G–U mismatches do exist in the cellular proteome of the reference strain MG1655, we quantified the frequency of Arg to His (R→H) substitutions, which result from a G–U mismatch at the second codon position; the G–U base pair tautomerization is particularly well tolerated at this position (41) (Supplementary Figure S1E). The R→H error frequency did not change within the tested range of Str concentrations, whereas frequency of Y→H (U–G mismatch at the first position) and other unrelated missense errors (e.g. E→D and R→C) increased by almost 10-fold. We observed that the R→H error frequency is modulated by mutations in ribosomal proteins that render ribosomes either hyper-accurate or error-prone (Supplemenatry Figure S1F) supporting the notion that G–U misreading occurs during the decoding step. Comparison of translation errors induced by streptomycin or by mutations in ribosomal proteins reveals that G–U mismatches and second position U–G mismatches are relatively unaffected by Str treatment but are induced by ribosomal mutations. In contrast, decoding involving first position U–G mismatches such as Y→H, S→P and C→R can be strongly affected by streptomycin (Supplemental Figure 1G). We then tested whether other aminoglycoside antibiotics that affect fidelity increase G–U misreading. Neamine, ribostamycin, and paromomycin, which also bind to the decoding center and impair the ability of the ribosome to discriminate between cognate and near-cognate tRNAs, strongly induced R→H errors (Supplementary Figure S1H), in agreement with earlier studies ((42) and BioRxiv: https://doi.org/10.1101/255943). In summary, these results show that, in agreement with recent reports (43), the details of decoding involving G–U and U–G mismatches at different positions in the codon–anticodon helix differs in detail. They also underscore differencial error profiles of various aminoglycoside antibiotics, which can be rationalized by their different error-induction mechanisms (44).
Similarly to the most comprehensive datasets ((18) and BioRxiv: https://doi.org/10.1101/255943), coverage of missense peptides in our DDA analysis is rather limited. Out of >1500 possible individual near-cognate amino acid substitutions (calculated for sequences of the peptides analyzed) only 304 were observed, while 48 types of amino acid substitutions, e.g. C→Y, R→G, H→P or M→V, were not observed for any of the possible positions in the EF-Tu sequence. Among the 304 near-cognate substitutions sequence variants that are isobaric to common degradation products such as oxidations or deamidations (e.g. A→S and N→D) were overrepresented and could be detected for almost each position. Together with the fact that only 116 near-cognate substitutions are induced by Str treatment, this indicates that the true missense peptides coverage is even smaller. These qualitative considerations are supported by the integrated peak intensities (XICs) of the Str-responsive amino acid substitutions. Half of the identified peptides detected in the presence of Str could not be quantified in the samples without Str treatment, indicating that their cellular levels either are obscured by noise or are outside of the dynamic range of the instrument (Figures 1B and C). Thus, a systematic analysis of error frequencies by simultaneous quantification of correct and incorrect peptides in the linear dynamic range of current mass spectrometers using DDA does not appear feasible even for purified proteins.
The QRAS workflow
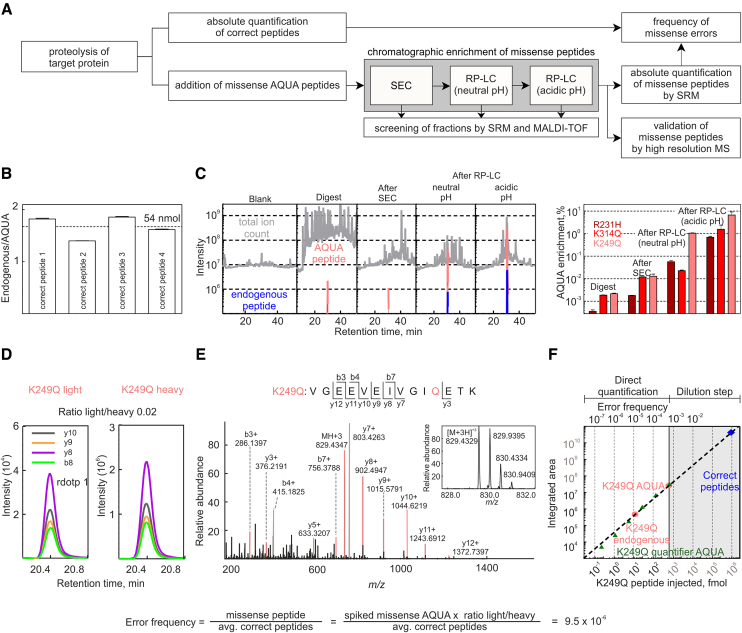
To overcome the limitations in the detection of miscoding events by DDA, we developed a workflow in which missense peptides are chromatographically enriched (Figure 2A). Correct and missense peptides are quantified independently of each other before and after enrichment, respectively. First, the amount of correct peptides is determined by selected reaction monitoring (SRM, reviewed in (31)) using four isotopically labeled Absolute QUAntification (AQUA) peptides of known concentration (45) (Figure 2B). Then, missense AQUA peptides containing amino acid substitutions are spiked into the digest at substoichiometric amounts and missense peptides are enriched in sequential chromatographic steps (Figure 2A and Supplementary Figure S2A). In the first chromatographic dimension, peptides are separated by size-exclusion chromatography (SEC). Fractions are screened by MALDI/SRM analysis targeting the missense AQUA peptides. Target missense peptides are further purified by reversed phase chromatography (RP) at neutral pH. Depending on the sample complexity, target peptides are either quantified or further enriched in a third RP step at acidic pH. Because the subsequent chromatographic steps are at least partially orthogonal (Supplementary Figure S2B), sample complexity is stepwise reduced (Figure 2C and Supplementary Figure S2C). Typically, after the multidimensional enrichment all highly abundant correct peptides were removed and the target missense AQUA peptides were enriched by >1000-fold, which made them very abundant in the sample fraction.
Figure 2.
QRAS workflow. (A) Schematic of QRAS workflow. (B) Quantification of correct peptides. Four tryptic peptides are quantified using highly quantified AQUA peptides (guaranteed concentration error <5%). Their mean concentration and the volume of the digest are used to calculate the amount of proteolysed EF-Tu. (C) Reduction of sample complexity. MS runs of the digest and K249Q missense peptide containing fractions after each enrichment step (left panel). Total ion current (TIC) is shown in grey, extracted ion chromatograms (XIC) of the enriched AQUA are in red and the endogenous peptide is in blue (two most abundant charge states with their 3 most abundant ions, extracted with 10ppm resolution). Bar graph (right panel) shows the contribution of the integrated XICs to the integrated TIC. Error bars represent the standard deviation of three technical replicates. (D) Quantification of missense peptide in the sample relative to the AQUA peptide by SRM analysis. The perfect co-elution and identical fragmentation pattern as the missense AQUA peptide are reflected in the ratio dot product (the vector product of the elution pattern of endogenous and AQUA peptides). (E) High resolution MS/MS spectra of the endogenous missense peptide; inset: corresponding MS spectrum. (F) Linear dynamic range of the K249Q quantification. To determine the dynamic range, a second quantifier AQUA peptide (dark green) sharing same sequence but having an additional isotope-label was titrated. Injected amounts of AQUA peptides as indicated.
After enrichment, missense peptides were quantified by SRM analysis (Figure 2D and Supplementary Figure S3) and error frequencies were calculated as the ratio between of the missense and the correct peptides. The identity of the target peptide was ensured by co-elution and the identical fragmentation pattern of the endogenous and AQUA target peptide and was further validated by high-resolution MS and MS/MS spectra (Figure 2E, Supplementary Figures S2F and S4). In those cases where the MS and MS/MS spectra were ambiguous, we used the more selective parallel reaction monitoring (PRM, reviewed in (46)) (Supplementary Figure S5). The strong reduction of the sample complexity eliminated most interferences and allowed us to apply larger amounts of target peptides leading to higher signal intensities and a lower limit of quantification. The linear dynamic range of this quantification covered 6–7 orders of magnitude (Figure 2F). For additional validation, we controlled all AQUA peptides for contaminations with unlabeled peptides or interfering peptide derivatives (Supplementary Figures S4 and S5). We also estimated the precision and accuracy of quantifications. When error frequencies determined in different digestions of one EF-Tu preparation were compared, the average coefficient of variation was ∼0.05, suggesting high precision of technical replicates (Supplementary Figure S2D). Because error frequencies vary over several orders of magnitude, such small variations are negligible and technical replicates were not further acquired in favor of biological replicates. Furthermore, we evaluated the quantitative accuracy of our results using EF-Tu mutants which contained the target amino acid substitutions. The resulting missense peptides should appeared in a 1:1 stoichiometry to the correct peptides. Overall the average deviation from the accurate stoichiometry was ∼33% without a systematic bias in one direction (Supplementary Figure S2E). Considering the guaranteed accuracy of AQUA peptide concentrations (25% in the QuantPro grade (Thermo Scientific) used for missense peptides) and the correctness of absolute quantification in other studies (47), this accuracy is expected and is well suited to study error frequencies that differ over orders of magnitude.
Error frequencies determined by QRAS
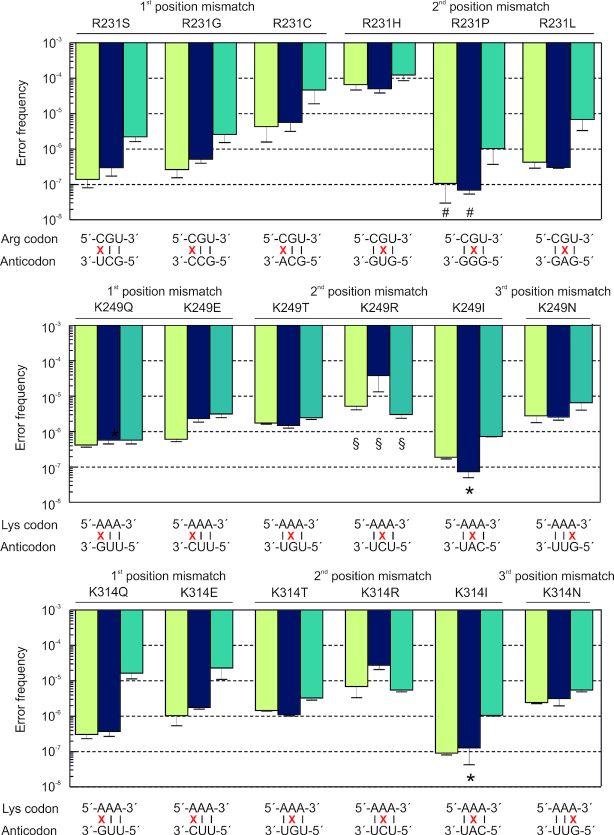
We then applied QRAS to determine error frequencies resulting from various types of codon–anticodon mismatches and at different positions in the protein. First, we selected three positions in EF-Tu (R231, K249 and K314) for which four missense peptides (R231C, R231H, K249N and K314N) were identified by DDA (Figure 1 and Supplementary Table S1); of them, only R231H could be detected in the absence of Str. Based on the genetic code, 6 near-cognate mismatches are possible for each position. Thus, a total of 18 missense substitutions should be detected, 17 of which we were able to enrich and quantify (Figure 3, Supplemenatry Figure S3 and Supplementary Table S2). Error frequencies spanned three orders of magnitude from 10−7 to 10−4. Consistent with the DDA data (Figure 1C, Supplementary Table S1), R231H is the only amino acid substitution that was abundant enough to be directly detected (error frequency of 10−4), supporting the notion that a G–U mismatch in the second position is a common source of codon misreading (11,17,23,25). Error frequencies of all other 16 substitutions were <10−5. We note that the conditions of EF-Tu expression had a significant effect on some error frequencies. In general, EF-Tu overexpression resulted in higher error levels; in an extreme case the difference was two orders of magnitude (K249Q, 3.7 × 10−7 versus 3.4 × 10−5, respectively) (Figure 3). The C-terminal His-tag had very little effect on error frequency at most positions tested in EF-Tu.
Figure 3.
Error frequencies of near-cognate misreading at three individual positions. Green bars, wild type chromosome-encoded EF-Tu from MRE600; blue bars, chromosome-encoded EF-Tu carrying a C-terminal His-tag (K12 strain); teal bars, plasmid-encoded EF-Tu overexpressed in BL21(DE3). Error bars represent the standard deviation of 3–5 biological replicates. For some amino acid substitutions a quantification was not possible and the bars represent an upper limit: * the endogenous peptide was too rare to be detected; # contaminations in the AQUA peptide masked the endogenous peptide; or § there were interferences even after multidimensional enrichment.
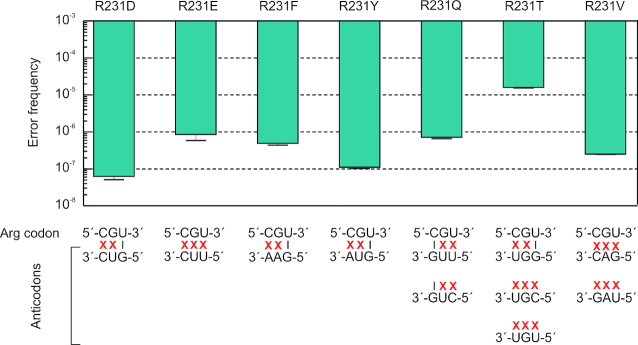
We then expanded our analysis to non-cognate amino acid substitutions, as the frequency of such substitutions is expected to be very low and has never been estimated so far. We systematically studied all possible substitutions at one position in EF-Tu, R231. Out of 13 possible R substitutions to non-cognate amino acids, 11 were tested (A, D, E, F, I, N, Q, T, V, W, Y). R231M and R231K were excluded from the analysis, because methionine is reactive and quantification of R231K would require to use a different protease. We quantified 7 non-cognate amino acid substitutions (Figure 4, Supplementary Figure S5 and Supplementary Table S3), while the remaining four substitutions could not be detected. The range of error frequencies for such substitutions was 10−7–10−5, i.e. not very different from some near-cognate substitutions at the same position. Whether non-cognate amino acids are incorporated during translation or arise from other steps of gene expression, such as incorrect tRNA charging (37), remains unclear.
Figure 4.
Error frequencies of non-cognate decoding at R231 in plasmid-encoded EF-Tu overexpressed in E. coli BL21(DE3). Error bars represent the standard deviation of three biological replicates.
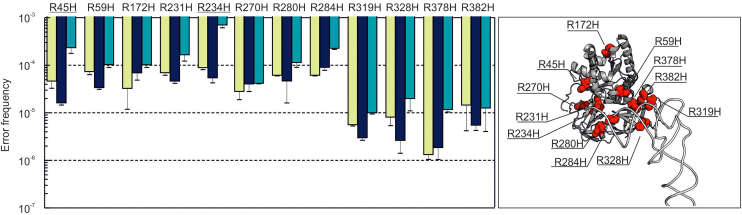
The low error frequencies measured by QRAS for the majority of positions and types of mismatches may explain the low missense peptide coverage of in the DDA data. However, it does not explain why even relatively abundant amino acid substitutions, e.g. R→H, were not detected for each position in EF-Tu by DDA. For example, R172H, R231H, R234H, R263H were detected in DDA, whereas 19 other R → H substitutions were not detected. To test whether the position of Arg in the protein sequence affects the observed error frequency, we enriched peptides with the R→H substitution for 12 out of 23 possible positions in EF-Tu. The error frequencies varied by more than two orders of magnitude between 10−6 and 10−3 depending on the Arg codon and the amino acid position (Figure 5, Supplementary Figure S3 and Supplemenatry Table S2). The R172H, R231H and R234H substitutions were relatively abundant (10−3–10−5), consistent with the DDA estimations, whereas R319H, R328H, R378H and R382H (all located in domain III of EF-Tu) were significantly less abundant (10−6–10−5). Overexpression of EF-Tu resulted in a consistently higher error frequency, in some cases up to 10−3 (R234H). Thus, steady-state levels of missense peptides depend not only on the type of codon–anticodon mismatch, but also on the position of the amino acid in the peptide sequence and the protein expression level.
Figure 5.
Position dependence of R to H substitutions in EF-Tu. Left panel: Error frequencies. Green bars, wild type chromosome-encoded EF-Tu from MRE600; blue bars, chromosome-encoded EF-Tu carrying a C-terminal His-tag (K12 strain); teal bars, plasmid-encoded EF-Tu overexpressed in BL21(DE3). Error bars represent the standard deviation of 3–6 biological replicates. Most of the R→H errors included in the analysis result from reading the 5′-CGT-3′Arg codon except for R45H and R234H substitutions (underlined) that occur on the 5′-CGC-3′ codon. Right panel: The positions of the substitutions are shown in the structure of EF-Tu in the complex with tRNAPhe (PDB file 1OB2).
DISCUSSION
We developed the QRAS workflow to quantify very rare protein sequence variants and to determine steady-state error frequencies for cellular proteins. The enrichment of missense peptides by QRAS is conceptually similar to biomarker approaches (48), which in combination with targeted MS opens a dynamic range encompassing ∼7 orders of magnitude and is also suitable for analysis of rare substitutions in heterogeneous protein mixtures from any strain or organism. We envisage that QRAS approach can be used not only to probe the frequency of amino acid substitutions, but also to study diverse proteogenomic events which are often predicted by bioinformatics analysis, but need validation and quantification by mass spectrometry. For most proteogenomic events such as frameshifting, premature termination, stop-codon readthrough or alternative splicing there are no established affinity enrichment workflows and QRAS might be the only option to detect them. For other events such as alternative initiation (49) or translation of pseudogenes (50) or non coding regions enrichment procedures were reported and QRAS might help for systematic and accurate quantifications. QRAS is also applicable for analysis of post-translational modifications (PTMs), especially for those cases where no specific PTM enrichment strategies are available, or when different PTMs should be analyzed in one sample. QRAS can be also used to better control the quality of biotherapeutic products. Different impurities such as PTMs or amino acid substitutions where proposed to cause immunogenicity (15,51) and tracking such impurities QRAS might help to improve production of biotherapeutics. Another example of a potential application is to detect rare splicing variants or amino acid substitutions in disease variants, e.g. in cancer cells, which are predicted by DNA or RNA sequencing, but the quantification of sequence variants is crucial to better understand how they drive tumor biology (52).
The combination of co-purification, co-elution, matching SRM transition ratio with the corresponding AQUA peptide and high resolution MS and MS/MS spectra allows for high confidence identifications that significantly supersede other analytical methods that capture only relatively abundant errors and are therefore biased towards higher median error frequencies. In contrast to reporter assays (11,12,14) where low-frequency errors are often lost in the background noise, QRAS can reliably detect very low amounts of peptides against the unspecific background signal. The dynamic range can be further expanded by increasing the amount of starting material and applying more chromatographic separation steps. In practice, the dynamic range is limited by the availability of the model protein, the limited choice of truly orthogonal separations methods, the chemical purity of AQUA peptides, ionizability of target peptides and the physicochemical differences between correct and target peptide. In the current work most target peptides differed from the correct parental peptides by their tryptic cleavage pattern, allowing for a separation by size exclusion chromatography. However, even peptides without changes in the tryptic pattern, such as K314R or R328H, could also be efficiently enriched. Because the AQUA peptides as internal standards help to correct for spray instabilities, matrix effects and differences in the ionization efficiencies between correct and missense peptides, QRAS has the potential to be more accurate than direct label-free quantification. Finally, QRAS does not require specialized equipment and can be applied using almost every mass spectrometric setup. A recent review concluded that the diversity of proteoforms cannot be studied currently due to dynamic range constraints and that the quantification awaits new analytical technologies to come (53). QRAS may provide an important step in this direction.
The limitations of QRAS are (i) the relatively large amounts of proteins required for the enrichment (due to high sample loss in chromatographic off-line separations (54)); (ii) the high cost of precisely quantified AQUA peptides; and (iii) the considerable measuring time required for fraction screening, which are the main bottlenecks in applying QRAS for large-scale analysis. To reduce the necessary measuring time, retention times of missense peptides can be first determined in reference runs by MALDI-TOF and then confirmed by SRM analysis in the respective enrichment run. The combination of a semi-preparative HPLC fractionation with a splitter mediated online LC–MS/MS detection would further minimize the measuring time. Furthermore, isobaric tagging (e.g. by tandem mass-tagging, TMT (55)) prior to the multidimensional chromatographic enrichment would allow analysis of a model protein from up to ten biological states in a single chromatographic enrichment. This would strongly reduce the required amount of protein and allow the analysis of comparative experiments, i.e. time courses or titrations, after a single enrichment. Moreover, new data acquisition regimes (56,57) might reduce the number of necessary enrichment steps while the use of bi- or triphasic HPLC columns (58)—which combine different resins in one column—could alleviate sample loss in multidimensional chromatography.
Using QRAS we determined the in-vivo steady-state level of amino acid substitutions in the cell. We found a very broad range of error frequencies (<10−7–10−3) including the lowest ever reported level of substitutions. This is consistent with recent results obtained using specialized reporter assays which, although constrained by the signal-to-noise ratio, provided examples of error frequencies of <10−6 for a number of amino acid substitutions (11). Recent in vitro data also suggest that the error frequency can vary between 10−8–10−3 (25), implying that translation can be surprisingly accurate. In comparison, the fidelity landscape of transcription is rather uniform and independent of growth conditions and sequence context, with individual error frequencies ranging between 1 × 10−6 and 5 × 10−5 (26). These results suggest that translation errors are not systematically more common than transcription errors, in contrast to the current notion that translation is the most error-prone step of gene expression.
Notably, some near-cognate substitutions in the steady-state pool of EF-Tu detected in this work are even less abundant than expected from the error rate of transcription in vivo (26), which suggests that erroneous proteins may be efficiently removed from the cellular pool by the quality control machinery. Amino acid substitutions might destabilize EF-Tu and make it more accessible to proteases. Under stress conditions—which are expected to increase the error frequency (16,20,37,59) (BioRxiv: https://doi.org/10.1101/255943)—EF-Tu is enriched in cellular aggregates (60,61), preferentially carbonylated (62,63) and interacts with chaperones such as GroEL, IbpB or Hsp33 (64–66). Hsp33 was reported to guide EF-Tu to Lon-mediated degradation (64), suggesting that the cell can respond to appearance of aberrant proteins by specifically targeting them to degradation. This also may explain why error frequency is higher for the overexpressed EF-Tu than for the wild-type and the chromosomally encoded protein, as most strains used for overexpression lack the Lon protease and might therefore be deficient in quality control. Alternatively, aberrant EF-Tu located in aggregates might be removed by asymmetrical segregation (67,68).
We also found that the error frequency varies for different positions in EF-Tu. Positions with low error frequencies cluster at the aminoacyl-tRNA binding interface of EF-Tu (69), which leads us to speculate that the removal of aberrant EF-Tu molecules may depend on their impaired functional activity. For instance, the mutation R378A results in a 10-fold larger destabilization of the EF-Tu–aminoacyl-tRNA complex, compared to mutations at positions R59 and R283 (69). Similar qualitative observations of context-dependent errors were reported for other model proteins expressed under stress conditions, and the effect was attributed to differences in protein stability (19) or in the local lower accuracy of translation due to rare codon clusters (70). Thus, in the few cases for which quantitative information is available, the observed lower fidelity correlates with a higher complex stability. Proteins that have lost the ability to bind their interaction partners may be less protected and therefore easier for proteases to degrade.
In summary, our results suggest that the amounts of incorrect proteins in the cell are very small, except for a few hotspots, e.g. misincorporations caused by G–U mismatches (11,17,23) (BioRxiv: https://doi.org/10.1101/255943). This high-fidelity steady-state proteostasis may rapidly change under conditions of cellular stress, e.g. due to protein overexpression or the addition of antibiotics, which results in the accumulation of miscoded proteins that are not removed by the quality control machinery. This may also explain why the reporter assays that monitor activity of heterologously overexpressed model proteins often show higher error frequencies. Those relatively frequent errors can be analyzed by conventional approaches, such as DDA. In contrast, the QRAS approach provides an insight into the high-fidelity areas of the cellular error landscape, which have been not accessible by other methods so far.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wolfgang Wintermeyer for critical reading of the manuscript and Dr Hani Zaher for providing E. coli strains. We thank Olaf Geintzer, Franziska Hummel, Sandra Kappler, Christina Kothe, Theresia Niese, Anna Pfeifer, Uwe Plessmann, Monika Raabe, Annika Reinelt, Tanja Wiles, and Michael Zimmermann for expert technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Science Foundation (Deutsche Forschungsgemeinschaft) via Research Unit FOR [1805]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Drummond D.A., Wilke C.O.. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009; 10:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frederico L.A., Kunkel T.A., Shaw B.R.. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990; 29:2532–2537. [DOI] [PubMed] [Google Scholar]
- 3. Pakula A.A., Sauer R.T.. Genetic analysis of protein stability and function. Annu. Rev. Genet. 1989; 23:289–310. [DOI] [PubMed] [Google Scholar]
- 4. Tokuriki N., Tawfik D.S.. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009; 459:668–673. [DOI] [PubMed] [Google Scholar]
- 5. Stefani M., Dobson C.M.. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol. Med. (Berl.). 2003; 81:678–699. [DOI] [PubMed] [Google Scholar]
- 6. Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003; 426:895–899. [DOI] [PubMed] [Google Scholar]
- 7. Edelmann P., Gallant J.. Mistranslation in E. coli. Cell. 1977; 10:131–137. [DOI] [PubMed] [Google Scholar]
- 8. Bouadloun F., Donner D., Kurland C.G.. Codon-specific missense errors in vivo. EMBO J. 1983; 2:1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toth M.J., Murgola E.J., Schimmel P.. Evidence for a unique first position codon–anticodon mismatch in vivo. J. Mol. Biol. 1988; 201:451–454. [DOI] [PubMed] [Google Scholar]
- 10. Stansfield I., Jones K.M., Herbert P., Lewendon A., Shaw W.V., Tuite M.F.. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 1998; 282:13–24. [DOI] [PubMed] [Google Scholar]
- 11. Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P.J.. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014; 20:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kramer E.B., Farabaugh P.J.. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007; 13:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salas-Marco J., Bedwell D.M.. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 2005; 348:801–815. [DOI] [PubMed] [Google Scholar]
- 14. Meyerovich M., Mamou G., Ben-Yehuda S.. Visualizing high error levels during gene expression in living bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:11543–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris R.P., Kilby P.M.. Amino acid misincorporation in recombinant biopharmaceutical products. Curr. Opin. Biotechnol. 2014; 30:45–50. [DOI] [PubMed] [Google Scholar]
- 16. Wong H.E., Huang C.J., Zhang Z.. Amino acid misincorporation in recombinant proteins. Biotechnol. Adv. 2018; 36:168–181. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Z., Shah B., Bondarenko P.V.. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry. 2013; 52:8165–8176. [DOI] [PubMed] [Google Scholar]
- 18. Mohler K., Aerni H.R., Gassaway B., Ling J., Ibba M., Rinehart J.. MS-READ: Quantitative measurement of amino acid incorporation. Biochim. Biophys. Acta. 2017; 1861:3081–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song Y., Zhou H., Vo M.N., Shi Y., Nawaz M.H., Vargas-Rodriguez O., Diedrich J.K., Yates J.R., Kishi S., Musier-Forsyth K. et al.. Double mimicry evades tRNA synthetase editing by toxic vegetable-sourced non-proteinogenic amino acid. Nat. Commun. 2017; 8:2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cvetesic N., Semanjski M., Soufi B., Krug K., Gruic-Sovulj I., Macek B.. Proteome-wide measurement of non-canonical bacterial mistranslation by quantitative mass spectrometry of protein modifications. Sci. Rep. 2016; 6:28631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melnikov S.V., Rivera K.D., Ostapenko D., Makarenko A., Sanscrainte N.D., Becnel J.J., Solomon M.J., Texier C., Pappin D.J., Söll D.. Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E6245–E6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mühlhausen S., Schmitt H.D., Pan K.T., Plessmann U., Urlaub H., Hurst L.D., Kollmar M.. Endogenous stochastic decoding of the CUG codon by competing Ser- and Leu-tRNAs in Ascoidea asiatica. Curr. Biol. 2018; 28:2046–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. New structural insights into translational miscoding. Trends Biochem. Sci. 2016; 41:798–814. [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Ieong K.W., Johansson M., Ehrenberg M.. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J., Ieong K.W., Mellenius H., Ehrenberg M.. Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs. RNA. 2016; 22:896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Traverse C.C., Ochman H.. Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:3311–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Link A.J., Phillips D., Church G.M.. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1997; 179:6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shevchenko A., Wilm M., Vorm O., Jensen O.N., Podtelejnikov A.V., Neubauer G., Shevchenko A., Mortensen P., Mann M.. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 1996; 24:893–896. [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 30. MacLean B., Tomazela D.M., Abbatiello S.E., Zhang S., Whiteaker J.R., Paulovich A.G., Carr S.A., MacCoss M.J.. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 2010; 82:10116–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lange V., Picotti P., Domon B., Aebersold R.. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008; 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boon K., Vijgenboom E., Madsen L.V., Talens A., Kraal B., Bosch L.. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur. J. Biochem. 1992; 210:177–183. [DOI] [PubMed] [Google Scholar]
- 33. Wohlgemuth I., Pohl C., Rodnina M.V.. Optimization of speed and accuracy of decoding in translation. EMBO J. 2010; 29:3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tyanova S., Temu T., Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016; 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 35. Schilling B., Rardin M.J., MacLean B.X., Zawadzka A.M., Frewen B.E., Cusack M.P., Sorensen D.J., Bereman M.S., Jing E., Wu C.C. et al.. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol. Cell Proteomics. 2012; 11:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A.L., Rodnina M.V.. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R Soc. Lond. B Biol. Sci. 2011; 366:2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reynolds N.M., Lazazzera B.A., Ibba M.. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 2010; 8:849–856. [DOI] [PubMed] [Google Scholar]
- 38. Nagaraj N., Kulak N.A., Cox J., Neuhauser N., Mayr K., Hoerning O., Vorm O., Mann M.. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell Proteomics. 2012; 11:doi:10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makarov A., Denisov E., Lange O., Horning S.. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006; 17:977–982. [DOI] [PubMed] [Google Scholar]
- 40. Gromadski K.B., Daviter T., Rodnina M.V.. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006; 21:369–377. [DOI] [PubMed] [Google Scholar]
- 41. Satpati P., Åqvist J.. Why base tautomerization does not cause errors in mRNA decoding on the ribosome. Nucleic Acids Res. 2014; 42:12876–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matt T., Ng C.L., Lang K., Sha S.H., Akbergenov R., Shcherbakov D., Meyer M., Duscha S., Xie J., Dubbaka S.R. et al.. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:10984–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rozov A., Wolff P., Grosjean H., Yusupov M., Yusupova G., Westhof E.. Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria. Nucleic Acids Res. 2018; 46:7425–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Demirci H., Murphy F 4th., Murphy E., Gregory S.T., Dahlberg A.E., Jogl G.. A structural basis for streptomycin-induced misreading of the genetic code. Nat. Commun. 2013; 4:1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P.. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bourmaud A., Gallien S., Domon B.. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. Proteomics. 2016; 16:2146–2159. [DOI] [PubMed] [Google Scholar]
- 47. Mirzaei H., McBee J.K., Watts J., Aebersold R.. Comparative evaluation of current peptide production platforms used in absolute quantification in proteomics. Mol. Cell Proteomics. 2008; 7:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Immler D., Greven S., Reinemer P.. Targeted proteomics in biomarker validation: Detection and quantification of proteins using a multi-dimensional peptide separation strategy. Proteomics. 2006; 6:2947–2958. [DOI] [PubMed] [Google Scholar]
- 49. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G.R., Vandekerckhove J.. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 2003; 21:566–569. [DOI] [PubMed] [Google Scholar]
- 50. Parle-McDermott A. Methods to study translated pseudogenes: In vitro translation, fusion with a tag/reporter gene, and complementation assay. Methods Mol. Biol. 2014; 1167:243–252. [DOI] [PubMed] [Google Scholar]
- 51. Yin L., Chen X., Vicini P., Rup B., Hickling T.P.. Therapeutic outcomes, assessments, risk factors and mitigation efforts of immunogenicity of therapeutic protein products. Cell Immunol. 2015; 295:118–126. [DOI] [PubMed] [Google Scholar]
- 52. Ruggles K.V., Tang Z., Wang X., Grover H., Askenazi M., Teubl J., Cao S., McLellan M.D., Clauser K.R., Tabb D.L. et al.. An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol. Cell Proteomics. 2016; 15:1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aebersold R., Agar J.N., Amster I.J., Baker M.S., Bertozzi C.R., Boja E.S., Costello C.E., Cravatt B.F., Fenselau C., Garcia B.A. et al.. How many human proteoforms are there. Nat. Chem. Biol. 2018; 14:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Magdeldin S., Moresco J.J., Yamamoto T., Yates J.R. 3rd.. Off-line multidimensional liquid chromatography and auto sampling result in sample loss in LC/LC-MS/MS. J. Proteome Res. 2014; 13:3826–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McAlister G.C., Huttlin E.L., Haas W., Ting L., Jedrychowski M.P., Rogers J.C., Kuhn K., Pike I., Grothe R.A., Blethrow J.D. et al.. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012; 84:7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gallien S., Kim S.Y., Domon B.. Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM). Mol. Cell Proteomics. 2015; 14:1630–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meier F., Geyer P.E., Virreira Winter S., Cox J., Mann M.. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods. 2018; 15:440–448. [DOI] [PubMed] [Google Scholar]
- 58. Link A.J., Eng J., Schieltz D.M., Carmack E., Mize G.J., Morris D.R., Garvik B.M., Yates J.R. 3rd.. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999; 17:676–682. [DOI] [PubMed] [Google Scholar]
- 59. Mohler K., Ibba M.. Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2017; 2:17117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B.. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 2001; 40:397–413. [DOI] [PubMed] [Google Scholar]
- 61. Ling J., Cho C., Guo L.T., Aerni H.R., Rinehart J., Söll D.. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell. 2012; 48:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Luo S., Levine R.L.. Methionine in proteins defends against oxidative stress. FASEB J. 2009; 23:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fredriksson A., Ballesteros M., Dukan S., Nyström T.. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J. Bacteriol. 2005; 187:4207–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bruel N., Castanié-Cornet M.P., Cirinesi A.M., Koningstein G., Georgopoulos C., Luirink J., Genevaux P.. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J. Biol. Chem. 2012; 287:44435–44446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Houry W.A., Frishman D., Eckerskorn C., Lottspeich F., Hartl F.U.. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999; 402:147–154. [DOI] [PubMed] [Google Scholar]
- 66. Fu X., Shi X., Yan L., Zhang H., Chang Z.. In vivo substrate diversity and preference of small heat shock protein IbpB as revealed by using a genetically incorporated photo-cross-linker. J. Biol. Chem. 2013; 288:31646–31654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lindner A.B., Madden R., Demarez A., Stewart E.J., Taddei F.. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3076–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Winkler J., Seybert A., König L., Pruggnaller S., Haselmann U., Sourjik V., Weiss M., Frangakis A.S., Mogk A., Bukau B.. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010; 29:910–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yikilmaz E., Chapman S.J., Schrader J.M., Uhlenbeck O.C.. The interface between Escherichia coli elongation factor Tu and aminoacyl-tRNA. Biochemistry. 2014; 53:5710–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu Y., Sharp J.S., Do D.H., Kahn R.A., Schwalbe H., Buhr F., Prestegard J.H.. Mistakes in translation: Reflections on mechanism. PLoS One. 2017; 12:e0180566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.