Abstract
The DNA methylation epigenetic signature is a key determinant during development. Rules governing its establishment and maintenance remain elusive especially at repetitive sequences, which account for the majority of methylated CGs. DNA methylation is altered in a number of diseases including those linked to mutations in factors that modify chromatin. Among them, SMCHD1 (Structural Maintenance of Chromosomes Hinge Domain Containing 1) has been of major interest following identification of germline mutations in Facio-Scapulo-Humeral Dystrophy (FSHD) and in an unrelated developmental disorder, Bosma Arhinia Microphthalmia Syndrome (BAMS). By investigating why germline SMCHD1 mutations lead to these two different diseases, we uncovered a role for this factor in de novo methylation at the pluripotent stage. SMCHD1 is required for the dynamic methylation of the D4Z4 macrosatellite upon reprogramming but seems dispensable for methylation maintenance. We find that FSHD and BAMS patient's cells carrying SMCHD1 mutations are both permissive for DUX4 expression, a transcription factor whose regulation has been proposed as the main trigger for FSHD. These findings open new questions as to what is the true aetiology for FSHD, the epigenetic events associated with the disease thus calling the current model into question and opening new perspectives for understanding repetitive DNA sequences regulation.
INTRODUCTION
SMCHD1 is a 230 kDa protein grouped in the SMC family of chromosomal proteins based on the presence of an SMC hinge domain (1). However, SMCHD1 is a non-canonical family member owing to its distinct domain architecture, including the presence of an N-terminal GHKL rather than bipartite ABC-type ATPase domain (2). Additionally, SMCHD1 exclusively homodimerises via its hinge domain (2,3), and as a result does not heterodimerise like other SMC proteins, nor participate in the tripartite ring complex formed by other cohesins (2). In the mouse, Smchd1 loss of function results in early lethality in female embryos, attributed to derepression of genes on the inactive X chromosome (1,4,5). SMCHD1 is also involved in the silencing of repetitive DNA sequences, regulation of clustered imprinted genes, the monoallelically expressed protocadherin genes (5–7) and Hox genes (8). SMCHD1 is preferentially loaded onto H3K9me3-enriched chromatin in association with LRIF1 and HP1 (9,10). In addition, SMCHD1 has been found at telomeres with a direct correlation between telomere length and SMCHD1 enrichment (11,12) but its role in the regulation of telomeric chromatin is unknown.
Recently, heterozygous germline mutations in the SMCHD1 gene have been identified in type 2 Facio-Scapulo-Humeral muscular dystrophy (FSHD2) (13–15). FSHD is one of the most fascinating syndrome involving methylation changes. This autosomal dominant muscular dystrophy is ranked as one of the most common myopathies. FSHD is linked to a complex chromosomal abnormality at the 4q35 subtelomeric locus (16–18). In the majority of patients, a heterozygous deletion of an integral number of GC-rich repetitive macrosatellite elements, D4Z4, in the distal region of the 4q arm is found. This deletion segregates with a ‘permissive’ qA subtelomeric haplotype downstream of this repetitive array (19,20). In 5% of FSHD cases (FSHD2), there is no D4Z4 array shortening but a large fraction of these patients carry a heterozygous mutation in the SMCHD1 gene. D4Z4 is extremely GC-rich (70%) (21) and contains an open reading frame encoding the DUX4 transcription factor (22). In FSHD1 and 2, D4Z4 is hypomethylated (13,23–26) and D4Z4 chromatin relaxation has been associated with expression of the DUX4 retrogene encoded by the most distal D4Z4 repeat and adjacent qA haplotype leading to activation of a cascade of genes which perturbs skeletal muscle homeostasis (20,27).
More recently, germline SMCHD1 mutations have been found in patients affected with Bosma Arhinia and Microphthalmia Syndrome (BAMS), an extremely rare condition characterized by absence of the nose with or without ocular defects. Intriguingly, BAMS patients show no sign of muscular dystrophy. With <50 patients reported to date (28,29), arhinia is presumed to result from a specific defect of the nasal placodes or surrounding neural crest-derived tissues during embryonic development. In FSHD, SMCHD1 missense or splice and truncating mutations are likely loss of function and have been described across the whole coding sequence while in BAMS, mutations are likely gain of function and mainly clustered within exons 3 to 13, spanning a GHKL-type ATPase domain and the associated region immediately C terminal to it (2,9), (28,29). Although there is some controversy surrounding whether BAMS missense mutations are loss- or gain of function, Arhinia has been associated with an increased ATPase activity (28–30). Intriguingly, D4Z4 hypomethylation is observed in both diseases indicating that loss or gain of function mutations are all associated with epigenetic changes at this macrosatellite but with completely different phenotypical outcomes (28,29).
In order to investigate the impact of SMCHD1 mutations in FSHD2 and BAMS, we created a collection of human induced pluripotent stem cells (hiPSCs) from patients with either diseases. By first analyzing methylation of the D4Z4 macrosatellite involved in FSHD, we showed that D4Z4 methylation is dynamically methylated upon reprogramming. In pluripotent cells, D4Z4 methylation is regulated by SMCHD1. We further show that BAMS and FSHD2 cells are permissive for DUX4 expression suggesting that besides SMCHD1’s pleiotropic role in chromatin regulation, SMCHD1 BAMS and FSHD mutations also have tissue specific effects, which remain to be identified.
MATERIALS AND METHODS
Study samples
DNA was extracted from the different types of samples using the Qiagen DNA prep kit. All individuals have provided written informed consent for the collection of samples and subsequent analysis for medical research. The study was done in accordance with the Declaration of Helsinki. Controls are randomly selected individuals or patient's relatives, selected in the same age range and sex representation as patients. Controls are neither carrier of any genetic mutation nor affected by any constitutive pathology. Samples are listed in Supplementary Tables S1 and S3.
Cell culture
The human embryonic kidney 293 cells (CRL-1573) and the male colon carcinoma cell line (CCL-247) were obtained from ATCC. The HCT116 DKO cells were obtained from Dr. B. Vogelstein Laboratory (1). In HCT116 and 293 cells, SMCHD1 expression was invalidated by transfection of Zn finger nucleases using CompoZr knockout Zinc Finger Nucleases (CKOZFN19780, Sigma-Aldrich) targeting exon 1. After transfection, individual clones were selected and characterized for homozygous gene deletion. Primary fibroblasts and HEK293 cells were grown in DMEM with l-alanyl-l-glutamine (GlutaMAX™I), d-glucose and sodium pyruvate (Life technologies). The human colon carcinoma HCT116 cells were maintained in McCoy's medium. Media were supplemented in with 10% fetal bovine serum (FBS, Gibco), 1% penicillin/streptomycin (P/S). Cells were grown at 37°C, 5% CO2, in a humidified atmosphere.
Human iPSCs clones
All iPSCs clones were derived from primary fibroblasts (Supplementary Table S1) generated after transfection of different vectors by electroporation (pCXCLE-hOCT3/4-shp53-F (Addgene ref 27077), pCXLE-hSK (Addgene, ref 27078, encoding SOX2 and KLF4), pCXLE-hUL (Addgene,ref 27080, encoding L-Myc and LIN28). Human iPS (hiPSCs) colonies were picked 4–6 weeks after transfection based on their ES cell-like morphology. Colonies were grown and expanded in mTeSR™1 medium (Stem cells) on BD Matrigel™ (BD Biosciences, cat. no. 354277) coated dishes. For each cell line two to three clones were fully characterized using classical procedures (2) such as expression of pluripotency markers, absence of expression of the reprogramming transgenes, phosphatase alkaline staining and karyotype for checking the absence of chromosomal abnormalities. Expression of the Keratin Sulfate antigens Tra1-60 and Tra1-81 and the glycolipid antigens SSEA3 and SSEA4 was verified by flow cytometry.
Sodium bisulfite sequencing
One μg of genomic DNA was denatured for 30 min at 37°C in NaOH 0.4 N and incubated overnight in a solution of 3 M sodium bisulfite pH 5 and 10 mM hydroquinone using previously described protocol (4). Converted DNA was purified using the Wizard DNA CleanUp kit (Promega) following manufacturer's recommendation and precipitated by ethanol precipitation for 5 h at −20°C. After centrifugation, DNA pellet was resuspended in 40μL of water and stored at −20°C until use. In order to amplify both methylated and unmethylated DNA with the same efficiency, converted DNA was amplified using primer sets designed with the MethPrimer software (5) avoiding the presence of CpGs in the primer sequence (Supplementary Table S6). For Ion Torrent sequencing, primers were modified by 5′ addition of barcode/adaptors sequences and 3′ addition of BSP sequences. Amplification was carried out using the High Fidelity Taq polymerase (Roche) according to manufacturer's instructions. After initial denaturation at 94°C for 2 min, amplification was done after 10 cycles at 94°C for 20 s, 56°C for 30 s, 72°C for 1 min and 25 cycles at 94°C for 20 s, 54°C for 30 s, and an amplification step of 4 min and 30 s for the first cycle and an implement of 10 s at each subsequent cycle followed by a final extension step at 72°C for 7 min.
PCR products were purified using the Ampure XP kit (Beckman Coulters) and quantified using the Qubit® dsDNA High Sensitivity Assay Kit (Thermofisher). An equimolar mix of all regions and samples/barcodes is prepared. DNA high-throughput sequencing was performed using Ion PGM Sequencing 400 kit (Life Technologies) reagents. Ion Sphere Particles (ISP) were loaded onto an Ion 316R sequencing chip (Life Technologies) and DNA sequencing was performed with the Ion PGM instrument at 650 run flows. Raw data were processed using the Ion Torrent platform-specific pipeline software Torrent Suite v5.0.4. Sequences were identified by the presence of a 17-base pair index at the beginning or at the end of the sequence. After barcode trimming, data are analyzed using the Bamtools software (BamTools 2.4.0, https://github.com/pezmaster31/bamtools, (6)). After sequencing, sense and antisense sequences were assembled in a single sequence and bam file converted to fastq file. A trimming of each BSP primers is performed using the fastx_barcode_splitter.pl software and the fastq_to_fasta software (Barcode Splitter & FASTX_ToolKit, 0.0.13, https://github.com/agordon/fastx_toolkit) is used to prepare data for the BiQ Analyser HiMod software (http://biq-analyzer.bioinf.mpi-inf.mpg.de) (7). Processed data were exported into the R software (version 3.4.2). Aligned sequences with a conversion rate >90% and a recovery rate >50% were kept for further analysis. BiQ Analyzer HiMod converts sequencing data by using ‘1’ for a methylated CG, ‘0’ for unmethylated and ‘x’ in case of misalignment. The percentage of coverage is calculated as the ratio between the numbers of ‘x’ divided by the total number of letters per sequence. Based on the number of ‘1’ or ‘0’, three methylation score are calculated, (i) the CpG methylation score corresponds to the percentage of methylation for each CpG in the reference sequence calculated as the ratio of methylated CpGs on the number of CpGs aligned to the reference sequence, (ii) the sequence methylation score, corresponds to the average methylation level of each sequence calculated as the ratio of methylated CpG of all aligned CpG for a given sequence and (iii) the global methylation score corresponds to the global level of methylation for each biological sample in a given region calculated as the ratio of methylated CpG with the number of aligned CpG for all sequences and CpG positions for a given biological sample. Graphics representations were performed using barplot, boxplot and hist functions.
RNA extraction and quality control
Total RNA was extracted using the RNAeasy kit (Qiagen) following manufacturer's instructions. Quality, quantification and sizing of total RNA was evaluated using the RNA 6000 Pico assay (Agilent Technologies Ref. 5067-1513) on an Agilent 2100 Bioanalyzer system.
Quantitative RT-PCR
Reverse transcription of 1 μg of total RNA was performed using the Superscript II kit and oligo dT following manufacturer's instructions at 42°C for 50 min followed by inactivation at 70°C for 15 min (Life Technologies). Primers were designed using Primer Blast and Primer3 (Supplementary Table S7). Real-time PCR amplification was performed on a LightCycler 480 (Roche) using the SYBR green master mix. All PCR were performed using a standardized protocol and data were analyzed with the Lightcycler 480 software version 1.5.0.39 (Roche). Primer efficiency was determined by absolute quantification using a standard curve. For each sample, fold-change was obtained by comparative quantification and normalization to expression of the HPRT, GAPDH and PPIA (DUX4 and DUX4 target genes) or 36B4 (characterization of hiPSCs) housekeeping genes used as standard. Data are expressed as means ± SEM.
RT-PCR for DUX4
One μg of total RNA was used for first strand cDNA synthesis using SuperScript IV reverse transcriptase and a mixture of oligo dT and hexanucleotide primers. Primary PCR reactions were performed with Taq DNA polymerase (Euromedex) using 7% of the first strand reaction as template in a total reaction volume of 30 μl. Nested PCR reactions were performed on 1 μl of the primary reaction. In order to discard any risk of contamination by genomic DNA, each RNA sample was treated with DNAseI and PCR amplification was performed on RNA samples incubated in the absence of reverse transcriptase (RT–) or without addition of cDNA. In all cases, amplification products were only observed when RNA samples were reverse-transcribed. For DUX4-fl detection the following primers were used as described (8) (Supplementary Table S7). PCR products were examined on 3% Molecular Biology Grade Agarose gels stained with ethidium bromide. All PCR products were sequenced and compared to Genbank accession number KX467569 and KX467570.
3′ UTR RACE
For the 3′ UTR RACE, reverse transcription was realized with an oligo-dT adapter primer: (GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTT) as described (9). PCRs were performed in a final volume of 15 μl with 1 μl of RT product, 3 μl of reverse primer at 20 μM and 1 μl of primer at 20 μM, 7.5 μl 2× PCR MasterMix (ThermoScientific). The PCRcycling conditions were 94°C for 5 min, followed by 5 cycles at 94°C for 20 s and 72°C for 50 s, then 5 cycles at 94°C for 20 s and 70°C for 30 s and 72°C for 20 s and 25 cycles at 94°C for 20 s and 68°C for 20 s and 72°C for 30 s, followed by a final extension at 72°C for 7 min. The nested PCRs were realized in a final volume of 15 μl with 1 μl of primary PCR product, 1 μl of reverse and/or forward primers 20 μM, 7.5 μl 2× PCR MasterMix (Thermo Scientific).The PCR cycling conditions were 94°C for 5 min, followed by 35 cycles at 94°C for 20 s and 60°C for 20 s and 72°C for 30 s, followed by a final extension at 72°C for 7 min. Nested PCR products were purified using a NucleoSpin Gel and PCR Clean-Up column (Macherey-Nagel).Fragments were cloned into TOPO-TA vector with a TOPO-TA Cloning kit (LifeTechnologies).Cleavage sites were determined by Sanger sequencing of at least 10 Individual clones.
Chromatin immunoprecipitation (ChIP)
Between 10–20 × 106 cells were cross-linked with 1% formaldehyde for 10 min at room temperature then for 40 min at 4°C. Cross linking reaction was stopped by addition of glycine (final concentration 0.125M). Cells were washed twice in ice-cold PBS, harvested and incubated for 10 min in 1 ml ice-cold lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP40, protease inhibitors). Nuclei were extracted with using a dounce homogeneizer and collected by centrifugation at 5000 rpm for 5 min at 4°C. Nuclei were then resuspended in a 50 mM Tris–HCl pH 8, 10 mM EDTA, 1% EDTA solution. Chromatin was sonicated using a Bioruptor instrument (Diagenode) to an average length of 400–600 bp. After centrifugation for 10 min at 14 000 rpm, supernatants were collected and diluted 10 times in a dilution buffer (16.7 mM Tris–HCl pH 8, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA). Pre-clearing was performed using a mixture of Protein A and G sepharose (50:50, Sigma) for 1 h at 4°C. ChIP-ready chromatin was pulled down with either anti-H3 (Milipore, 04–928), anti-H3K9me3 (Millipore, 07-442) or anti-SMCHD1 (Abcam, ab31865) overnight under rotation at 4°C. Chromatin was then incubated with magnetic beads coupled with protein A or G agarose (Milipore, 16-661; 16-662) for 1 h at 4°C under rotation. Beads were washed under rotation for 5 min in different buffers: low salt wash buffer (TE 2×, 150 mM NaCl, 1% Triton X-100, 0.1% SDS), high salt wash buffer (TE 2×, 500 mM NaCl, 1% Triton X-100, 0.1% SDS), LiCl wash buffer (TE 1×, 0,25 mM LiCl, 1% NP40, 1% sodium deoxycholate), TE 1×. Immune complexes were eluted by addition of 500 μl elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed by addition of 20 μl of NaCl 5 M for 4 h at 65°C. DNA samples were then recovered by phenol/chloroform extraction, ethanol precipitation and resuspended in water. Enrichment of the immunoprecipitated fraction/input was quantified by real time qPCR. Primers were designed using Primer Blast and Primer3 (Supplementary Table S8).
Digital droplet PCR
For ChIP quantification, chromatin immunoprecipitated using anti-histone H3 antibodies (positive control) and 1% input were used as controls for ddPCR analysis. Briefly, we used 1 μl of ChIP samples in 20 μl of 0.1 mM primers and 10 μl of either 2× ddPCR Supermix (no dUTP) for probes or 2× ddPCR Evagreen supermix. After droplets generation, samples were transferred into a 96-well plate for PCR amplification. The plate was processed with a ddPCR reader, using appropriate settings depending on the reaction mixture (Evagreen or Mix for probes). Only wells with >13 000 droplets counted were kept for analysis. Results are normalized to input and a unique intergenic chromosome 5 locus. PCR primers were tested on genomic DNA to verify specificity and efficiency.
Detection of telomerase activity in hiPSCs was performed as described (10). Approximately 10,000 hiPSCs cells were lyzed in 40 μl of NP-40 buffer (10 mM Tris–HCl, pH 8.0, 1 mM MgCl2, 1 mM EDTA, 1% (vol/vol) NP-40, 0.25 mM sodium deoxycholate, 10% (vol/vol) glycerol, 150 mM NaCl, 5 mM β-mercaptoethanol; 0.1 mM AEBSF (4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride). One microliter of this lysate was added to a 50 μl extension reaction containing 1× TRAP reaction buffer (10× concentration: 200 mM Tris–HCl, pH 8.3, 15 mM MgCl2), 0.4 mg/ml BSA, TS telomerase extension substrate (HPLC purified, 200 nM; 5′-AATCCGTCGAGCAGAGTT), dNTPs (2.5 mM each) and incubated for 40 min at 25°C followed by telomerase inactivation at 95°C for 5 min, then held at 4°C. The ddPCR reaction contained 1× EvaGreen ddPCR Supermix v2.0 (Bio-Rad, Hercules, CA, USA), 50 nM TS primer, 50 nM ACX primer, 50 cell equivalents or less of extension product and dH2O to 20 μl per sample. Droplets were produced in the droplet generator according to the manufacturer's instructions (Bio-Rad, Hercules, CA, USA), the emulsions transferred to a 96-well PCR plate for the PCR reaction. PCR reaction was performed as described (10) with a ramp rate of 2.5°C/s between all steps. On average 17 000 droplets were analyzed per 20 μl PCR.
Statistical analysis
For DNA methylation, statistical analyses were done with R (version 3.4.2) and RStudio softwares, dplyr library and ComplexHeatmap library from Bioconductor (version 3.6). DNA methylation levels were compared using a Wilcoxon non-parametric test (wilcox.test function). The significance threshold (α = 0.05) was corrected for multiple comparisons using the Bonferroni method for False Discovery Rate (FDR) determination. Only values showing a significant correlation with a P-value <0.05 are presented. Heatmaps and Manhattan distance calculation were used to simultaneously visualize clusters of samples and global methylation of the different sequences. Clusters of CpG by CpG differentially methylated were determined using an Euclidean distance calculation. For each sample, we used the Mixtools version 1.1.0 package in R that estimates the sampling distribution using a random sampling method and confidence intervals calculation. Bootstrap was used to determine a confidence interval for the ratio of methylated CG divided by the number of CG analyzed for each sample by constructing an interval centered at a point estimate with a margin of error equal to twice the standard error.
RESULTS
D4Z4 is remethylated upon reprogramming in cells from FSHD1 patients
Using sodium bisulfite modification (31) coupled with deep-sequencing (Roche et al. Submitted), we investigated the methylation profile of the D4Z4 macrosatellite in primary fibroblasts and derived hiPSCs clones from FSHD1 patients carrying a shortened D4Z4 array and healthy donors (Supplementary Table S1). For each sample, two to three hiPSC clones were isolated and amplified for at least 20 passages to erase donor cell memory, which is usually maintained at early passages (32). We selected pluripotent clones by checking OCT4, NANOG and SOX2 expression (Supplementary Figure S1), verifying extinction of the episomal genes and presence of a normal karyotype (not shown) and finally ensuring that these clones had a differentiation potential (Supplementary Figure S2).
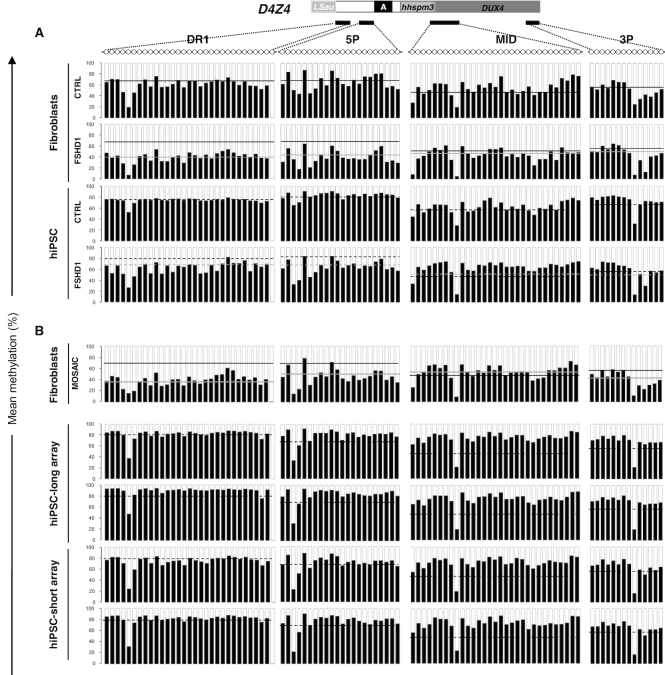
In primary fibroblasts from FSHD1 patients, D4Z4 is hypomethylated with most methylation changes occurring at the DR1 and 5P regions, located upstream of the DUX4 coding sequence while methylation of the MID (DUX4 promoter) and 3P (end of DUX4 exon 1 coding sequence) are not modulated (Figure 1A), as observed in blood (15,33). In control hiPSC clones, D4Z4 methylation is slightly increased compared to the corresponding fibroblasts with an average increase of methylation of 8.4% for DR1 and 11% for 5P between fibroblasts and pluripotent cells (Figure 1A, Supplementary Table S2), a trend of hypermethylation described for other subtelomeric regions (34,35). This increased level of methylation is significantly more pronounced in FSHD1 hiPSCs with an average increase of 27.9% for DR1 (P value < 0.04) and 29.2% for 5P (P value < 0.036) between fibroblasts and corresponding pluripotent cells (Figure 1A, Supplementary Table S2). When we compared the global methylation level and distribution at the individual CpGs level between clones from the same donor, we did not observe any striking difference. Altogether, this indicates that upon reprogramming, D4Z4 methylation level is systematically increased in cells derived from FSHD1 patients carrying a shortened D4Z4 array.
Figure 1.
Methylation of the D4Z4 macrosatellite increases upon reprogramming in FSHD1 cells. (A) DNA methylation was determined after sodium bisulfite modification for four subdomains across D4Z4 by PCR amplification and high throughput DNA sequencing. DR1 and 5P are located upstream of the DUX4 coding sequence, MID corresponds to the DUX4 promoter and 3P, to the end of DUX4 exon 1 encoding the protein. For each sequence, dots represent individual CpG. For the different types of samples (control or FSHD1 fibroblasts; controls or FSHD1 hiPSCs), we plotted the mean methylation level of each CpG for the four D4Z4 subdomains. Histogram bars represent the average of methylated (black) or unmethylated (white) CpG for each position in DNA from primary fibroblasts from 8 controls (row 1, mean methylation level for DR1: 71.4% ± 8.8, mean methylation level for 5P, 73.1% ± 13.1) and four FSHD1 (row 2, mean methylation level for DR1: 39.9% ± 4.4, mean methylation level for 5P, 40.5% ± 3). For hiPSCs derived from control (row 3, DR1: 79.8% ± 16.6 and 5P: 84.1% ± 7.4) and FSHD1 fibroblasts (row 4, DR1: 67.8% ± 23.4 and 5P: 69.7% ± 11.1), the average mean methylation level of each CG is represented as above. For each histogram, the mean methylation level in control fibroblasts is indicated by a black line, or a grey line in FSHD1 fibroblasts. In hiPSCs, the mean methylation level is indicated by a dashed black line for conrols or a dashed grey line for FSHD1 cells. Methylation level is significantly increased in FSHD1 hiPSCs for DR1 (P value <0.04) and for 5P (P value <0.036) compared to fibroblasts. (B) We analyzed the D4Z4 methylation pattern in fibroblasts from a mosaic patient clinically affected with FSHD and carrying a short 2 D4Z4 units repeat in 25% of cells and a long 11 D4Z4 units repeat in 75% of cells. DNA methylation was also analyzed in two hiPSCs clones carrying the 11 D4Z4 units repeat and two hiPSCs clones carrying the 2 D4Z4 units repeat derived from this patient's fibroblasts. Mean methylation level was plotted as detailed above. Values are given in the Supplementary Table S2. In fibroblasts from this mosaic patient, the mean methylation level is indicated by a gray line. The black lines correspond to the mean methylation levels in control fibroblasts. In hiPSCs, the dashed black line corresponds to the mean methylation level in control hiPSCs. The methylation level in hiPSCs is not significantly different between the different clones or compared to the controls.
This observation was corroborated by analyzing fibroblasts and hiPSC clones from a patient carrying a mosaic of short and long D4Z4 alleles (25% of cells carrying a short D4Z4 array of two units in fibroblasts) (Figure 1B, Supplementary Table S2). As observed in FSHD1 patients, D4Z4 methylation level is higher in hiPSC clones compared to fibroblasts (DR1: 38.6% and 5P: 47.7%). Methylation level, in this mosaic patient is similar between clones but more importantly between the two hiPSC clones carrying the short D4Z4 array (two repeats; mean methylation level at DR1: 76.8% ± 4.75%; 5P: 76% ± 4.14%) and the two clones carrying the long array (11 repeats; mean methylation level at DR1: 84.1% ± 4.89%, 5P: 82.8% ± 2.52%) (Figure 1B, Supplementary Table S2). Importantly, by analyzing the distribution of methylated DNA molecules in this mosaic patient (Supplementary Figure S3A; B), we observed similar distributions for hypo- (number of molecules with a methylation level <25%) or highly methylated (number of molecules with a methylation level >60%) molecules between both types of clones (short D4Z4 array corresponding to an FSHD allele or long array, healthy allele).
Taken together, these results reveal a higher level of methylation in pluripotent stem cells compared to the fibroblasts-of-origin. This indicates that D4Z4 remethylation occurs after cell reprogramming and suggests a dynamic methylation of the repeat. Furthermore, as evidenced by comparing somatic and stem cells from patients with a shortened D4Z4 array (FSHD1) or from a mosaic patient, D4Z4 remethylation shows that the level of methylation of the macrosatellite does not depend on the residual number repeats. The analysis of different clones per patient and different patients excludes clonal variability as an explanation of our findings.
High D4Z4 methylation is a feature of pluripotency
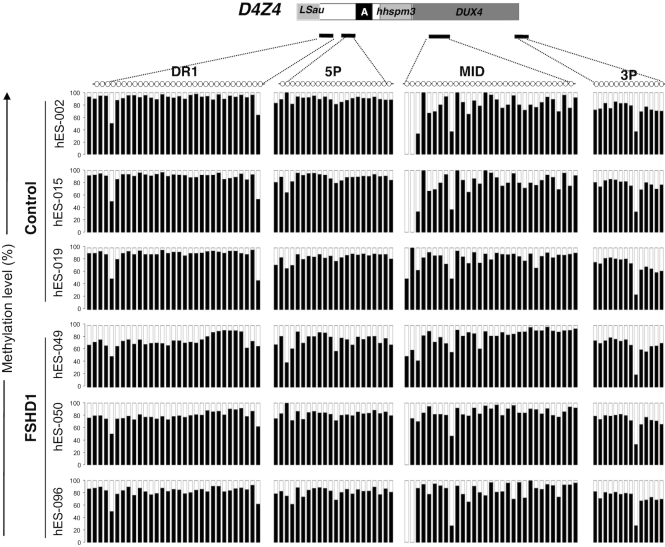
To further investigate whether remethylation in hiPSCs is dependent on reprogramming or a feature of pluripotency, we analyzed D4Z4 methylation in human Embryonic Stem Cells (hESCs) from three healthy donors or three donors carrying a short D4Z4 array (FSHD1) (Figure 2, Supplementary Table S1). As observed in hiPSCs, D4Z4 methylation is not significantly different between healthy donors and FSHD1 hESCs. In the two groups of samples, the D4Z4 repeat is highly methylated for the different regions analyzed without any significant difference between hESCs and hiPSCs (Figure 2, Supplementary Figure S4A, Supplementary Table S2). Altogether this indicates that D4Z4 is methylated to a level >75% of methylated CGs in pluripotent cells (either induced or embryonic), independently of the number of copies of the macrosatellite. Overall these results showing no difference between FSHD1 and control hiPSCs and hESCs indicate that D4Z4 methylation depends on stemness rather than on the number of residual copies.
Figure 2.
D4Z4 methylation profiles in control and FSHD1 embryonic stem cells. We compared the methylation profile of the four different D4Z4 subdomains (DR1, 5P, MID and 3P) in three different human embryonic stem cells (hESCs) from healthy donors and three different donors carrying a short D4Z4 array (FSHD1). Each sample is represented individually. Histogram bars represent the percentage of methylated (black) or unmethylated (white) CpG for each position and each individual sample.
D4Z4 remethylation is reduced in cells carrying mutations in SMCHD1
The majority of type 2 FSHD patients displaying a marked D4Z4 hypomethylation carry a mutation in the SMCHD1 gene. D4Z4 hypomethylation and SMCHD1 mutations have also been found in BAMS (28,29). In FSHD2, SMCHD1 mutations are associated with a loss of function of the protein while a gain of ATPase activity is suggested in most BAMS cases. These results strongly suggest that D4Z4 hypomethylation is not specific to FSHD but likely controlled by dosage of the functional homodimeric complex formed by this chromatin-associated factor (30). To test this hypothesis and evaluate the influence of SMCHD1 gene dosage on D4Z4 methylation, we first analyzed its methylation profile in blood DNA from patients carrying a deletion of the 18p11.32 region encompassing the SMCHD1 gene and phenocopying the FSHD2 loss-of-function situation (Supplementary Figure S5A; Supplementary Table S3). As in BAMS (29), we observed a marked decrease in D4Z4 methylation in the four 18p deletion samples analyzed (DR1: 22.7% of methylated CG ± 9.6; 5P: 51.7% ± 5.4) compared to controls (DR1: 71.4% of methylated CG; 5P: 75%, Roche et al. submitted). Interestingly, D4Z4 methylation is not increased in individuals carrying an 18p11.32 duplication and three SMCHD1 copies (Supplementary Figure S5B; Supplementary Table S2). Thus, D4Z4 methylation is sensitive to the decrease in SMCHD1 gene dosage but not increased by additional SMCHD1 gene copies.
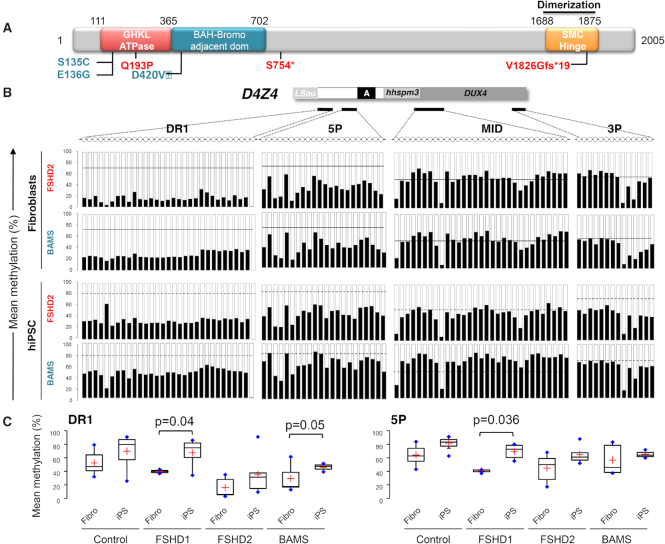
To investigate the role of SMCHD1 in the regulation of D4Z4 methylation in stem cells, we assessed D4Z4 methylation in fibroblasts from FSHD2 and BAMS patients and their corresponding hiPSCs clones by sodium bisulfite sequencing (Figure 3, Supplementary Figure S6). For DR1, D4Z4 methylation only slightly increases in FSHD2 hiPSC clones (n = 4; DR1: 28% of methylated CGs ± 12.7 compared to 15.7% of methylated CGs ± 14.1 in fibroblasts) compared to BAMS patients (n = 3; DR1: 46.2% ± 6.3 in hiPSCs compared to 21.3% of methylated CGs ± 11.7 in fibroblasts, P value = 0.05) (Figure 3, Supplementary Table S2). We concluded from these results that SMCHD1 might be implicated in the deposition of methyl marks at the pluripotent stage. Differences between FSHD2 and BAMS cells in the dynamics of D4Z4 methylation also suggest that, as observed for the ATPase activity, the type of mutations impacts the regulation of D4Z4 methylation differently.
Figure 3.
D4Z4 remethylation is impaired by mutation in SMCHD1. (A) Schematic representation of the SMCHD1 protein and position of mutations in BAMS (cyan) or FSHD2 patients (red). BAMS-1 (E136G) and BAMS-2 (S135C) carry a missense mutation in the ATPase domain reported as a gain of function. BAMS-9 carries a missense mutation in the C-terminal region of the extended GHKL-like ATPase domain (D420V). FSHD2 patient #14586 also carries a mutation in the ATPase domain (Q193P) while FSHD2-11440 and FSHD2-11491 carry a truncating mutation (p.S754* and p.V1826Gfs*19 respectively). (B) DNA methylation was determined after sodium bisulfite modification for 4 regions across D4Z4 (DR1, 5P, MID, 3P) by PCR amplification and high throughput DNA sequencing. For each sequence, dots represent individual CpGs. The mean methylation level of each CpG was plotted for the 4 D4Z4 subdomains. Histogram bars represent the average of methylated (black) or unmethylated (white) CpG for each position in DNA from fibroblasts (rows 1; 2) from FSHD2 patients (n = 3) and patients with BAMS (n = 3) and corresponding hiPSCs clones (rows 3; 4). For each histogram, the mean methylation level in control fibroblasts is indicated by the black line. In hiPSCs, the mean methylation level is indicated by a dashed black line for controls. (C) Boxplot representation of the mean methylation level for the DR1 and 5P differentially methylated sequences in primary fibroblasts and hiPSCs from control donors, FSHD1, FSHD2 and BAMS patients. Significant differences are indicated by brackets with the corresponding pvalues determined using a Kruskal–Wallis non parametric test.
Methylation changes linked to mutations in SMCHD1 only occur for a specific subset of repetitive sequences
To test whether methylation differences observed in the different types of samples are specific to D4Z4 or concern other types of repeats, we applied the same methodology to analyze the methylation level of other types of repetitive elements. We selected primers for dispersed repeats such as interspersed AluI or LINEs considered as surrogate markers of the global epigenetic status, the RS447 macrosatellite or TAR1 subtelomeric sequences. In hiPSCs or hESCs from controls or FSHD1 patients, we did not see any difference in the level of methylation for the AluI, LINE-1, and RS447 repetitive elements (Supplementary Figures S4; 7, Supplementary Table S4). Regarding the TAR1 sequence, methylation is decreased in fibroblasts from FSHD1 patients but highly variable between individuals (46.7% ± 27.2 compared to 67.11% ± 11.2 in controls). We made the same observations in hiPSCs from FSHD2 patients. Thus, TAR1 methylation is not sensitive to heterozygous SMCHD1 mutations but more variable between samples, as observed for other subtelomeric sequences (34,35).
We concluded that upon reprogramming, remethylation of the D4Z4 repeat is not linked to its subtelomeric position. Specific differences in D4Z4 methylation between FSHD1 and SMCHD1-mutated cells strengthen the hypothesis of a role for this protein in D4Z4 regulation.
Imprinting and X inactivation are not altered in FSHD2 and BAMS cells
Based on inactivation studies in the mouse, Smchd1 has been involved in X inactivation (1) and more recently, in the regulation of imprinted genes and monoallelically expressed loci (6,7,36). To test whether methylation of some of these loci is affected by SMCHD1 mutations found in FSHD2 and BAMS patients, we first analyzed the methylation status of the IGF2/H19 differentially methylated region (DMR) corresponding to the imprinting control region of this locus in the different cell types (Figure 4A, Supplementary Table S5). By plotting the mean methylation level for the different categories of samples we did not observe any difference between the frequency of methylated versus unmethylated alleles in the different conditions. This indicates that heterozygous SMCHD1 mutations in BAMS and FSHD2 patients do not modify the regulation of imprinting of this locus in primary somatic cells or after reprogramming in pluripotent cells.
Figure 4.
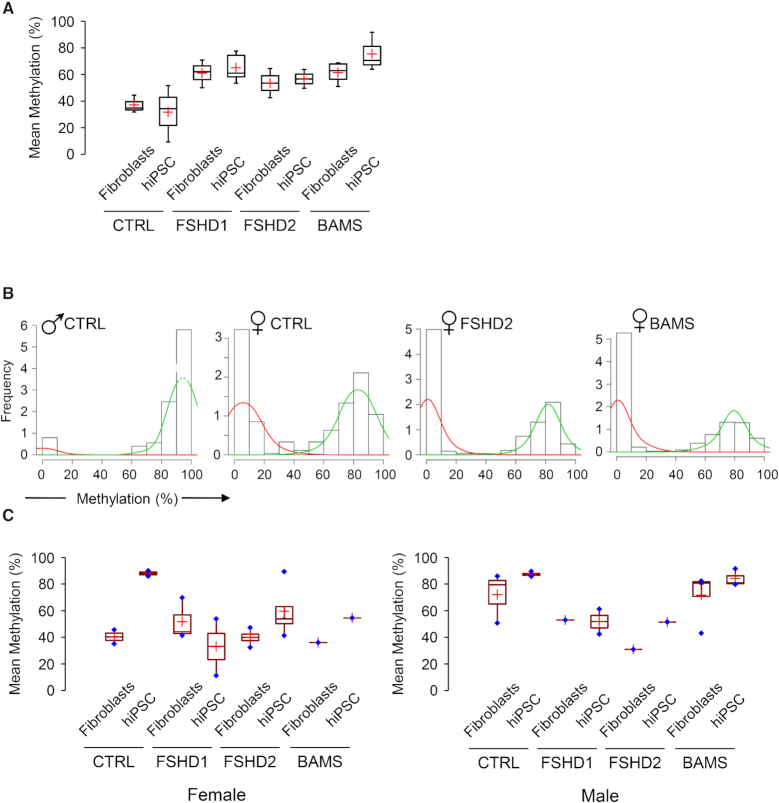
FSHD2- or BAMS-linked SMCHD1 mutations do not alter H19 imprinting or X chromosome conformation and methylation. (A) We plotted the mean methylation level for the H19 imprinted locus differentially methylated region (DMR) after bisulfite sequencing. Values are not significantly different between samples with approximately the same amount of methylated or unmethylated molecules in the different samples. This indicates that mutations in SMCHD1 do not modify imprinting of this locus in primary cells or after reprogramming. (B) Representative distribution of the frequency of sequences relative to their level of methylation for the DXZ4 macrosatellite element in the different contexts (control male, control female, female FSHD2, female BAMS). Histogram bars represent the frequency of methylated molecules from low (0–10%) to high (90–100%) methylation. The mean methylation level for each group is determined as the area under the curve. Green curves, high methylation level; red curves, low methylation level. (C) We plotted the mean methylation level for the DXZ4 macrosatellite element in female (left graph) or male (right graph) fibroblasts and hiPSCs.
Neither FSHD2 nor BAMS have been linked to changes or skewed X inactivation. Nonetheless, we tested for X inactivation changes in female cells with SMCHD1 mutations. To this aim, we first used 3D DNA FISH and hybridization with a whole X painting probe to determine X chromosome volume in fibroblasts and hiPSCs. As expected, the majority of female cells had two signals corresponding to the X probe while only one signal was detectable in male cells attesting to the specificity of the probe (Supplementary Figure S8). In the different contexts (controls, FSHD2, BAMS), we calculated the volume of X signals in male fibroblasts (6.035 μm3) and hiPSCs (3.49 μm3) and compared these values used as control of active X to the volumes measured in the other samples (Supplementary Figure S8). In female cells from controls, FSHD2 or BAMS, the two X chromosomes were analyzed separately (red and black dots). Volumes are equivalent to controls in FSHD2 hiPSCs and BAMS cells. We observed a broad distribution of volumes in female control hiPSCs and FSHD2 primary fibroblasts. This indicates that chromosome X volume is not modified in female cells with mutations in SMCHD1.
We next sought to determine the methylation profile of the X-linked DXZ4 macrosatellite repeat in the different contexts (Figure 4B, C; Supplementary Table S5). This element is a GC-rich tandem repeat of between 12 and 120 uninterrupted 3 kb units located on Xq23. Males are hemizygous for DXZ4 while females carry several copies of this macrosatellite on the active (Xa) and inactive (Xi) chromosomes (37). In males and on the female Xa from somatic cells or male hESCs, DXZ4 is heavily methylated but hypomethylated on the Xi in female cells (38).
To test whether SMCHD1 mutations influence the methylation profile of this macrosatellite, we analyzed DXZ4 methylation in the different contexts after bisulfite modification and deep sequencing (Figure 4B). As done earlier, we analyzed the distribution of hypo (mean methylation <25%) or hypermethylated sequences in male cells. We observed a main peak of methylation with a mean methylation level of 90–100% methylated CG per DNA molecule (green curves). This indicates that in male cells, the large majority of DXZ4 molecules are highly methylated in all samples tested, regardless of SMCHD1 status. As expected from the difference of methylation reported for the Xa and Xi, female cells display two peaks of methylation, with molecules harboring a low methylation level (0–10% of methylated CGs, red curves) likely corresponding to the hypomethylated Xi and molecules with a high methylation level (90–100%, green curves) corresponding to the Xa (Figure 4C). We observed the same distribution in fibroblasts from female FSHD2 or BAMS patients indicating that SMCHD1 mutations do not affect the methylation of this macrosatellite. By comparing the mean methylation level in the different samples, we observed a high methylation level in male control and BAMS fibroblasts. Surprisingly the level of methylation is decreased in fibroblasts from male FSHD1 and FSHD2 patients, regardless of the SMCHD1 status (Figure 6C). Despite this decreased methylation in male FSHD cells, we observed no notable difference between fibroblasts and hiPSCs indicating that DXZ4 methylation is maintained after reprogramming in the different subsets of samples.
Figure 6.
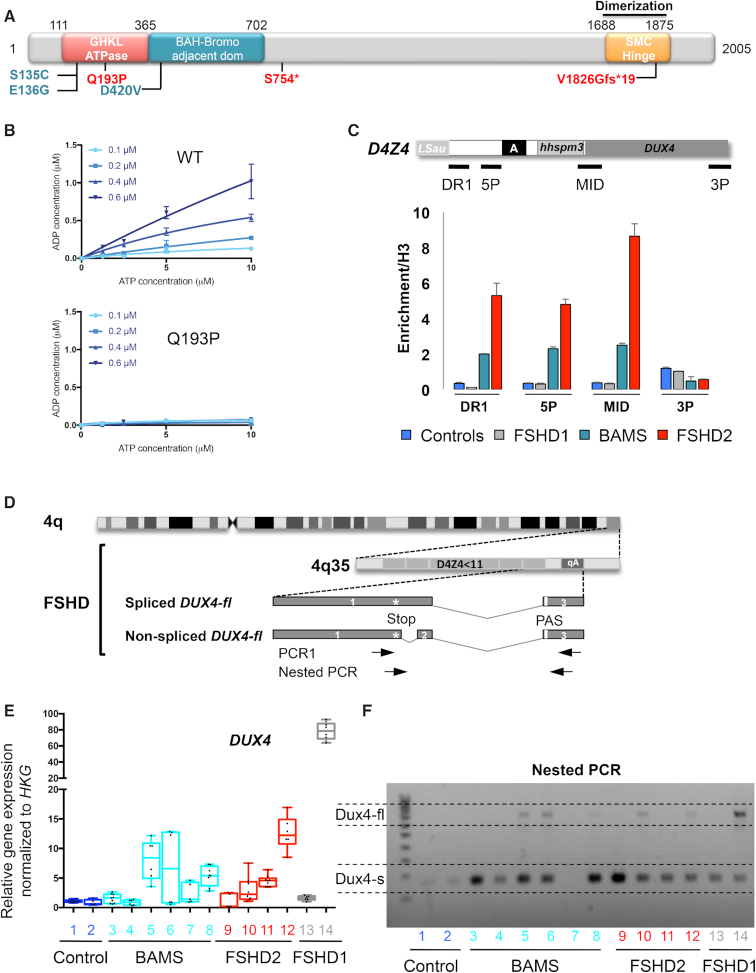
BAMS patients express the pathogenic DUX4-fl transcript and DUX4 targets. (A) Schematic representation of the SMCHD1 protein and position of mutations in BAMS (cyan) or FSHD2 patients (red). BAMS-1 (E136G) and BAMS-2 (S135C) carry missense mutations in the ATPase domain reported as gain of function. BAMS-9 carries a missense mutation in the C-terminal region of the extended GHKL-like ATPase domain (D420V). FSHD2 patient #14586 also carries a mutation in the ATPase domain (Q193P) while FSHD2-11440 and FSHD2-11491 carry a truncating mutation (p.S754* and p.V1826Gfs* respectively). (B) In vitro analysis of ATPase activity using a recombinant wild-type extended SMCHD1 ATPase domain and a recombinant extended SMCHD1 ATPase domain containing the FSHD2 Q193P mutation. Each graph shows ATP concentration (x axis, μM) versus concentration of ADP produced (y axis, μM) for different concentrations of recombinant protein. The protein concentration is indicated in the inset legend adjacent to the y axis. Means ± S.D. are shown for triplicate measurements. (C) Chromatin Immunoprecipitation assay for SMCHD1 in hiPSCs. Histogram bars correspond to quantification of SMCHD1 binding to D4Z4 by digital droplet PCR after SMCHD1 immunoprecipitation. Enrichment was determined by comparison to histone H3 immunoprecipitation with error bars corresponding to S.D. (D) Schematic representation of chromosome 4 and 4q35 locus with position of the DUX4 coding sequence. Each D4Z4 monomer contains the first exon of the DUX4 ORF. The second and third exons correspond to the 3′ untranslated region (3′ UTR). The third exon contains a non-canonical polyadenylation signal (PAS; TAATTT). Alternative splicing involving exon 1 (with or without exon 2) and downstream exons 3 to 7 give rise to different transcripts produced in the germline and stabilized by a polyadenylation site downstream of exon 7. Transcripts consisting of exons 1–3 encode the DUX4-fl long isoform produced in FSHD cells and stabilized by the presence of the PAS (20,40). This transcript is also detectable in healthy individuals (41,57,58). The DUX4-fl transcript is detectable using a nested PCR designed to amplify transcripts containing the 3′ end of exon 1 and exon 3 containing the polyadenylation site. Position of primers is indicated by arrows. (E) We evaluated expression of the DUX4-fl transcript in different hiPSCs clones derived from 2 clones from healthy donors (AG08; AG09; blue), three patients with BAMS (BAMS-1; BAMS-2; BAMS-9, cyan); three patients with FSHD2 (11440; 11491; 14586, red), and two clones from FSHD1 patients (TalF carrying two D4Z4 units; 12759 carrying 7 D4Z4 units, gray). Expression level was normalized to three different housekeeping genes (HKG). (F) DUX4 transcripts were analyzed by RT-PCR using primers designed to amplify the short isoform (DUX4-s), detectable in all cells and the DUX4-fl pathogenic transcript. We detected the DUX4-fl transcript in samples 5, 6 (two clones derived from BAMS 9), 8 (BAMS 1), 10 (FSHD2, 11440), 12 (FSHD2, 14586) and 14 (TalF; FSHD1 patient with two RU).
In female cells, where only the Xa is methylated, we observed no difference in patients’ cells, with an average 50% of methylated molecules as expected for the Xa/Xi ratio (Figure 4C). Altogether, these results suggest that in FSHD2 and BAMS patients, SMCHD1 mutations do not affect DXZ4 methylation in somatic cells or in hiPSCs after reprogramming.
D4Z4 remethylation coincides with acquisition of pluripotency upon reprogramming
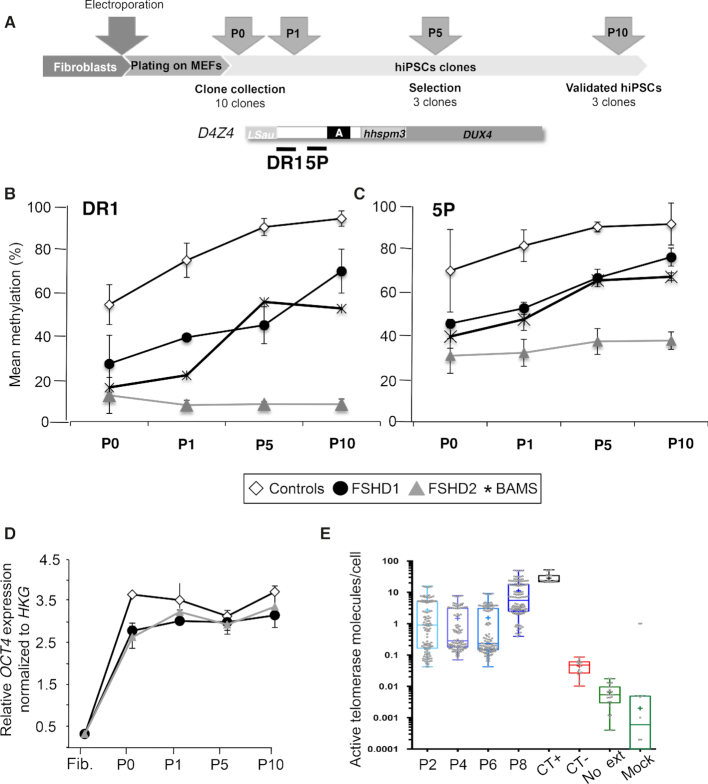
To evaluate the role of SMCHD1 in D4Z4 remethylation upon reprogramming, we analyzed the kinetics of D4Z4 methylation at different time points after transfection of the reprogramming cocktail in controls (n = 2), FSHD1 (n = 2), FSHD2 (n = 2; 11491: p.V1826Gfs*, 14586: p.Q193P) and BAMS (n = 2; BAMS1: p. E136G, BAMS9: p. D420V) primary fibroblasts. Cells were collected at different time points and processed as detailed (Figure 5A). Given the limited amount of DNA available at each step, we focused our analyses on the DR1 and 5P sequences where most remethylation occurs. Methylation of the DR1 and 5P D4Z4 regions is decreased in FSHD1, FSHD2 and BAMS fibroblasts compared to controls (Figure 5B). Upon reprogramming, we observed a progressive increase in the level of methylation in hiPSCs clones from FSHD1 patients (Figure 5B, C) with methylation reaching a level significantly different from the donor cells (P value < 0.05) and almost identical to the controls (Figure 5B, C). This increased methylation appears after reactivation of the endogenous OCT4 pluripotency marker (Figure 5D) and of the telomerase (Figure 5E) suggesting that it is associated with cell stemness. D4Z4 methylation remains low over time in FSHD2 cells (Figure 3B). Interestingly, methylation raises after the first passage in BAMS cells and remains higher in these cells compared to FSHD2 cells mutated for SMCHD1 but never reaches the level of controls or FSHD1 cells.
Figure 5.
D4Z4 remethylation correlates with expression of pluripotency markers and telomerase reactivation. (A) Timeline of the analysis of D4Z4 methylation dynamics upon reprogramming. (B) DNA methylation was analyzed in primary fibroblasts at low passage prior to reprogramming. Reprogramming was performed by electroporation of vectors encoding the different reprogramming factors. Six days after, cells are collected and plated on MEFs. A fraction of them were kept for methylation analysis (P0). Clones emerge between 2 and 3 weeks on the layer of feeders. For each sample, 10 clones were randomly chosen and isolated. For each clone, a fraction was kept for amplification. DNA and RNA extraction was done on the remaining cells pellet. (C, D) DNA methylation was analyzed after sodium bisulfite modification and deep-sequencing for the DR1 (C) and 5P (D) regions at different time points (after plating on MEFs (P0), first (P1), fifth (P5) or tenth (P10) passage) in ten clones/sample from two controls (white bars), two FSHD1 (black bars), two FSHD2 (dark gray) and two BAMS (light grey) patients. For each time point, the average methylation level ± S.D. is reported. Data sets were compared with the Wilcoxon non-parametric test and brackets identify significantly different groups based on post hoc Dunn comparison and Bonferroni correction (** P< 0.001) between P0 and P10. (D) To monitor reprogramming, expression of the OCT4 pluripotency marker was determined by RT-qPCR at the different time points. (E) Telomerase reactivation was monitored by ddTRAP at different time points using previously described conditions (56). The threshold between positive and negative droplets was determined by including controls such as a no-template lysis buffer control (CT−), a control where no primers were present but lysate was added to the ddPCR (Mock) and a control in the absence of enzyme for primer extension (Ext). The U87 cell line was used as positive control (CT+). Once the threshold (i.e. separation of positive droplets and negative droplets) was determined, the output was given in molecules of extension products per microliter of ddPCR reaction. We defined telomerase activity as the number of extended TS molecules counted by ddPCR and expressed in molecules per μl. The measured telomerase activity was then converted to total product generated. The telomerase activity was normalized to a per-cell equivalent basis by dividing total product generated by the number of cell equivalents input into the assay. Telomerase activity is detectable at passage 2, stable between passages two and six and increased between passages 8 and 10.
Altogether, our results strongly support a dynamic D4Z4 remethylation upon reprogramming that follows acquisition of pluripotency. In FSHD1 cells, D4Z4 remethylation to a level similar to control cells and human ESCs indicate that D4Z4 methylation level does not reflect the number of residual copies of the macrosatellite. Weak remethylation in FSHD2 cells also reveals a role for SMCHD1 in the distribution of methylated CGs at the pluripotency stage. Unlike mutations in FSHD2, BAMS mutations do not inhibit this remethylation suggesting that the increased ATPase activity linked to the SMCHD1 gain of function mutations in BAMS cells does not favor D4Z4 remethylation or that the ATPase activity is moderately required for D4Z4 methylation.
SMCHD1 binds to the proximal part of the D4Z4 repeat
Based on the impact of SMCHD1 mutations on the deposition of methyl marks in stem cells, we tested the role of SMCHD1 on D4Z4 regulation in pluripotent cells from two BAMS and two FSHD2 patients (Figure 6A). One BAMS patient carries a mutation in the ATPase domain (E136G, core ATPase adjacent motif I) that causes an increased enzymatic activity (29,30) suggesting that the mutation acts as a gain of function. The second BAMS patient carries a mutation in the C-terminal portion of the extended GHKL ATPase domain (D420V) where using the same ATPase assay in recombinant protein we have not found an effect on the enzymatic ATPase activity (29). One of the FSHD2 patients carries a stop mutation in the hinge domain (11491, p.V1826Gfs*19), likely associated with haploinsufficiency. The second patient carries a missense mutation in the ATPase domain (14856, Q193P, core ATPase adjacent motif II), not reported in BAMS. To assess the impact of this mutation in the catalytic activity of the SMCHD1 protein, we first tested in vitro the SMCHD1 ATPase activity using recombinant proteins in previously described conditions (30). By measuring ADP production after addition of different concentrations of ATP and different quantities of recombinant protein (WT or mutated), we observed the absence of ATP hydrolysis and ADP production for the Q193P mutation compared to the control. Proline confers protein rigidity likely affecting protein folding and in turn protein function and replacement of a Glutamine by a Proline residue abrogates SMCHD1 catalytic activity (Figure 6B).
We next performed chromatin binding analyses by chromatin immunoprecipitation assay and digital droplet PCR quantification for the DR1, 5P, MID and 3P regions (Figure 6C). We observed a significantly stronger association of SMCHD1 to D4Z4 in FSHD2 cells from the Q193P patient compared to FSHD1 and control cells. Interestingly, binding to D4Z4 is also increased in BAMS cells. Thus, despite a low level of D4Z4 methylation in these samples (Supplementary Figure S6) and mutations in the ATPase domain, SMCHD1 is able to bind to the repeat indicating that SMCHD1 binding is not dependent on D4Z4 methylation level and that SMCHD1 can be recruited to hypomethylated repeats (Figure 6C) with a higher affinity for mutations occurring in the ATPase domain. The increase binding in FSHD2 and BAMS cells further suggests that the ATPase domain might be required for loading/release of the protein to/from DNA (Figure 6C).
Both FSHD and BAMS pluripotent cells are permissive for DUX4 expression and express the DUX4-fl pathogenic transcript
SMCHD1 loss-of-function, D4Z4 hypomethylation and chromatin relaxation followed by activation of the DUX4 retrogene encoded by the last D4Z4 repeat and adjacent pLAM sequence is thought to be the driver mechanism in FSHD2 (Figure 6D). The DUX4 open reading frame is fully contained within the first exon encoded by D4Z4 whereas exons 2 and 3 corresponding to the DUX4 3′ untranslated region (3′ UTR) are encoded by the pLAM adjacent region (39). The main polyadenylation site (PAS) required for transcript stabilization and protein production is contained within exon 3 (Figure 6D). Additional alternative polyadenylation sites have also been identified distal to the last repeat suggesting alternative site usage (39). Only the long DUX4 transcript (DUX4-fl) has been associated with FSHD.
Given the methylation changes in FSHD and BAMS, we tested the presence of the pathogenic DUX4-fl transcript in different hiPSC clones (controls, FSHD1, FSHD2 and BAMS patients) using previously described conditions and quantitative PCR (40). We observed the absence of DUX4-fl transcript in hiPSCs from healthy donors (Figure 6E, F). DUX4-fl is expressed in FSHD1 cells despite the high methylation level (Figure 6E, F) including in clones derived from a mosaic patient in which we detected the DUX4-fl transcript in both short- and long-allele cells (Supplementary Figure S9A,B). We also detected an increased expression of DUX4 target genes in these samples (Supplementary Figure S9C). Additionally, the pathogenic DUX4-fl transcript is produced in different hiPSC clones derived from FSHD2 patients but also from BAMS cells with disparities in the level of expression between samples, regardless of the type of disease (Figure 6E, F). Sequences of the different transcripts are identical and match the DUX4-fl sequence containing exons 1–3 (not shown).
The PAS sequence, a degenerated polyadenylation site (AUUAAA) described as the main poly(A) site in FSHD muscle cells and required for stabilization of the DUX4 pathogenic transcript is located 766 bp downstream of the DUX4 stop codon (Figure 6D). We characterized the DUX4-fl transcript produced in the different cell types to test whether different transcripts are produced in BAMS or FSHD cells. To this aim, we performed a 3′ RACE assay and sequenced the DUX4-fl 3′ UTR (Supplementary Figure S10). We confirmed production of the DUX4-fl transcript in the three FSHD2 patients and one out the three BAMS cases tested and showed that the sequence of the 3′ UTR is identical in the two pathologies (Supplementary Figure S10). We inferred from these results that the DUX4-fl pathogenic transcript is produced both in FSHD and BAMS cells and that BAMS cells are permissive for production of the DUX4-fl transcript implicated in FSHD.
Since both BAMS and FSHD cells express DUX4 (Figure 6E, F), we further assessed expression of different DUX4 target genes (ZSCAN4, MBD3L2 and TRIM43) as well as expression of the TRIM28 gene required for DUX4 transcriptional activation (Supplementary Figure S11). TRIM28 expression is not modulated in the different sets of samples (Supplementary Figure S11E) but we noticed increased expression of ZSCAN4, MBD3L2 and TRIM43 with no notable difference between BAMS, FSHD2 and FSHD1 hiPSCs (Supplementary Figure S11A). Furthermore, DUX4-fl and DUX4 target genes are also detectable in embryoid bodies at day 14 of differentiation for the three groups of patients suggesting that DUX4 remains expressed at early differentiation stages in the two pathologies (Supplementary Figure S11 B–D).
Altogether, these results show that DUX4 expression is not only activated in FSHD1 and 2 cells but also in BAMS. In both diseases, the DUX4-fl transcript is identical to the pathogenic transcript described in FSHD muscle including in its 3′ UTR indicating that BAMS cells are also permissive for DUX4 expression. Furthermore, in FSHD1 hiPSCs, production of this transcript is not correlated with a decreased D4Z4 methylation.
SMCHD1 is required for de novo methylation of the D4Z4 repeat
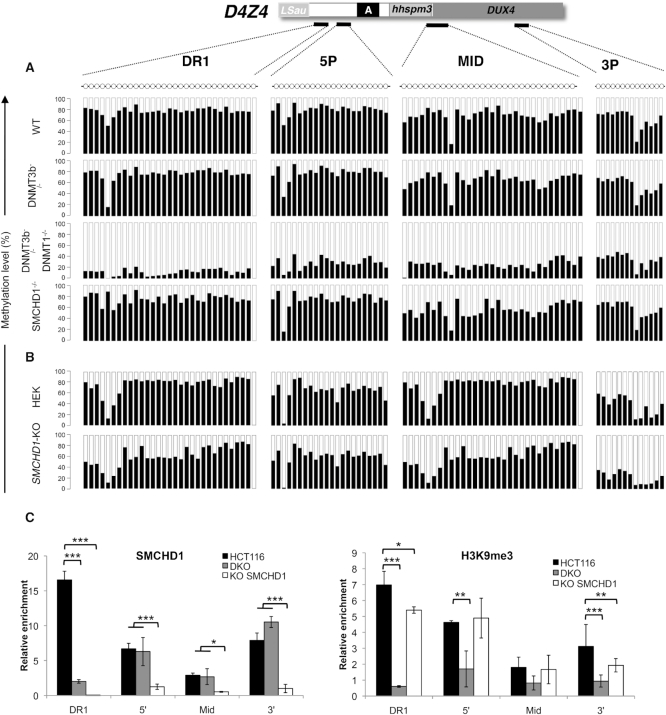
To further investigate the role of SMCHD1 in DNA methylation, we analyzed the distribution of methylated CGs in somatic cancer cells in which SMCHD1 has been deleted using a zinc finger nuclease (HCT116-SMCHD1 KO) in comparison with cells where the de novo DNA methyltransferase (DNMT) 3B (3B KO) is deleted or in cells were both the maintenance enzyme DNMT1 and DNMT3B (dKO cells) are absent (Figure 7A, Supplementary Figure S12, Supplementary Table S2). D4Z4 methylation is strongly decreased in dKO cells but not affected in DNMT3B-KO cells as previously observed (41). More strikingly, D4Z4 methylation is not decreased in SMCHD1-KO HCT116 cells compared to the parental HCT116 cell line (Figure 7A, Supplementary Table S2). This absence of effect is not cell-type dependent because similar results were observed in HEK293 cells in which SMCHD1 has been invalidated the same way (Figure 7B, Supplementary Figure S12B, Supplementary Table S2). These data strongly suggest that when the D4Z4 methylation pattern is established, SMCHD1 is not required for DNA methylation maintenance. In agreement with a role for SMCHD1 in the epigenetic regulation of D4Z4, this factor binds the proximal part of the macrosatellite where most methylation change occurs and highly enriched in the H3K9me3 heterochromatin mark (Figure 7C). Moreover, as observed for DNA methylation, SMCHD1 invalidation does not lead to a massive loss in this repressive H3K9me3 histone mark (Figure 7C). We further showed that in the HCT116 cells carrying a 4qA-type allele and thus permissive for DUX4 activation (supplementary Figure S12C,D), invalidation of both DNMTs or SMCHD1 leads to transcription of the DUX4 retrogene encoded by the macrosatellite and production of the DUX4 long transcript (DUX4-fl) (Supplementary Figure S12E,F).
Figure 7.
Invalidation of SMCHD1 in somatic cells does not lead to D4Z4 hypomethylation. (A) We determined D4Z4 methylation in the HCT116 cell line (HCT116), and in clones in which DNMT3B (3BKO), both DNMT1 and DNMT3B (dKO) or SMCHD1 (SMCHD1-KO) have been invalidated using Zinc finger nucleases. DNA methylation was analyzed after sodium bisulfite modification and deep-sequencing for four regions across D4Z4. The percentage of methylation was calculated for each individual CpG, black bars correspond to methylated CGs; white bars to unmethylated ones. Methylation percentages are provided in the Supplementary Table S2 as the average values ± S.D from three independent experiments. (B) D4Z4 methylation in the HEK 293 cell line and in HEK cells invalidated for SMCHD1 for four regions across D4Z4. (C) Schematic representation of D4Z4 from position 1 to 3303 relative to the two flanking KpnI sites and position of primers used for ChIP-qPCR. We analyzed SMCHD1 binding and distribution of H3K9 trimethylation across D4Z4 after chromatin immunoprecipitation in HCT116, dKO or SMCHD1-KO cells. Enrichment over input was determined by qPCR. Histograms display the average enrichment after normalization over a single copy intergenic region. Values are average from at least three independent biological replicates and a technical duplicate. Error bar represents standard error. Statistical significance was determined using a paired two-tailed Student's t test (*P< 0.01; **P< 0.001; ***P< 0.0001).
As observed in hiPSCs from FSHD1 patients, SMCHD1 inactivation in somatic cells induces expression of the DUX4-fl retrogene without a massive hypomethylation. Altogether, this suggest a role for SMCHD1 in the regulation of DUX4 expression independent of D4Z4 chromatin status as observed for other loci upon acute Smchd1 deletion in mice (8).
DISCUSSION
To investigate the epigenetic mechanisms linked to SMCHD1 mutations in two rare diseases, FSHD2 and BAMS (28,29), we reprogrammed primary fibroblasts from patients affected with these diseases. We also included hiPSCs clones from classical type 1 FSHD using cells from patients with differing numbers of D4Z4 units. In the different contexts, we analyzed DNA methylation of the D4Z4 macrosatellite linked to FSHD and of other repetitive elements in patients’ primary fibroblasts, hiPSCs after reprogramming or human stem cells (FSHD1 and healthy donors) after sodium bisulfite treatment of genomic DNA and deep-sequencing.
We observed that upon reprogramming, D4Z4 methylation increases in FSHD1 to a level similar to control cells. Interestingly, the FSHD1-hiPSCs methylation profile is highly similar to that of hESCs (FSHD1 and controls). Given this high methylation in pluripotent cells, we concluded that D4Z4 methylation level does not necessarily correlate with the number of residual repeats but is a feature of stemness. These observations further suggest that D4Z4 is dynamically remethylated upon reprogramming. We confirmed this hypothesis in individual clones derived from a mosaic patient and a time course analysis showing that D4Z4 remethylation follows acquisition of pluripotency. In FSHD2 patients carrying a mutation in SMCHD1, D4Z4 methylation is low in primary fibroblasts and slightly increases upon reprogramming while methylation increases more in BAMS hiPSCs. These results together with the absence of methylation changes in SMCHD1-deficient somatic cells emphasize a role for this protein in the de novo methylation of the macrosatellite. Hence, different mutations in this gene have different consequences on the epigenetic regulation of D4Z4 but not on its transcription as evidenced by the production of the DUX4-fl pathogenic transcript in both FSHD and BAMS patient's cells.
FSHD is associated with production of the DUX4-fl transcript transcribed from the last D4Z4 unit and adjacent 4qA and pLAM sequences containing a polyadenylation site (PAS) required for stabilization of the transcript and production of the DUX4 protein (20). In FSHD patients, DUX4 accumulation in 1 out of 1000 cells has been proposed as sufficient to generate toxicity in the muscle, by activating a number of target genes of unknown function in muscle biology (27). We detected expression of the long pathogenic DUX4 transcript (DUX4-fl) able to encode the DUX4 protein thanks to the presence of a polyadenylation site in FSHD and BAMS cells as well as some of the DUX4 target genes such as ZSCAN4, TRIM43 or MBD3L2 to a level comparable between samples. Thus, BAMS patients’ cells are permissive for DUX4 transcription. However, BAMS patients do not develop muscular dystrophy while SMCHD1 mutations in FSHD2 are not associated with craniofacial defects as in BAMS (29). The same holds true for patients with deletion of the 18p11.32 region encompassing the SMCHD1 gene showing a marked D4Z4 hypomethylation. Based on the estimation of qA-type haplotype frequency in the Caucasian population (42) required for DUX4-fl production, 12.6% of the 18p deletion patients would be permissive for DUX4 transcription and at risk of developing FSHD (42) but do not present this muscular dystrophy (43).
Our results thus raise important questions on the impact of D4Z4 hypomethylation, chromatin relaxation, the specific role of DUX4 in the FSHD pathogenesis and the relevance of targeting of SMCHD1 or DUX4 for the development of therapeutics. Along the same line, the DUX4 transcription factor activated at the two-cell stage in the mouse and four-cell stage in human embryos is required the Zygotic Genome Activation stage (ZGA) (44–46). This suggests that production of this transcription factor in embryonic stem cells does not depend on the presence of the pLAM sequence and that both qA and qB alleles which are equally common in the population (19), are permissive for DUX4 transcription and production of the DUX4 protein. If the qB allele is unable to support the production of a stable DUX4 protein, individuals carrying two qB alleles should not proceed past the 8-cell stage.
Several DUX4 transcripts have been described, with a germline-specific isoform containing additional exons (exons 4–7) encoding the 3′ UTR. These different isoforms are translated into the same protein encoded by the DUX4 ORF contained within exon 1 of the last D4Z4 repeat (20,40). This raise novel questions on the regulation of splicing of this key transcription factor in healthy versus pathological contexts.
During reprogramming, DNA methylation patterns are not fully erased as in early embryogenesis, but involve profound epigenetic changes and acquisition of an epigenetic profile similar to embryonic stem cells. Hence, reprogrammed cells often keep memory of the epigenetic pattern of the parental somatic cells used for reprogramming but also acquire iPSC-specific DNA methylation profiles (47,48). The consistent increase in D4Z4 methylation in control and FSHD1 hiPSCs and the comparison with other repetitive sequences, including the TAR1 subtelomeric repeats shows that D4Z4 does not retain the methylation pattern inherited from the donor cells but acquires a methylation profile identical to ESCs and thus likely specific to cell stemness. Our results strongly suggest a role for SMCHD1 in this remethylation. D4Z4 methylation is high in human ES cells, which are derived from the inner blastocyst cell mass (ICM), a later developmental stage compared to ZGA. Interestingly, SMCHD1 expression peaks at the 8-cell stage in human embryos (49), a stage corresponding to the silencing of DUX4 (50–52), suggesting a role for this factor in the remethylation of the repeat at early embryonic stages after post-fertilization waves of zygotic genome demethylation.
The high D4Z4 methylation level in FSHD1 hESCs or hiPSCs and decreased methylation in somatic cells further suggests that acquisition of this epigenetic mark in pluripotent cells is followed by demethylation in somatic cells (24,31,33) opening new questions on the molecular mechanisms enabling this process. Methylation of repetitive elements, in particular at early developmental stages, is regulated by specific pathways. This methylation is required to maintain a high level of chromatin compaction and limit genomic instability especially at regions proximal to centromeres or telomeres enriched in the H3K9me3 heterochromatin marks, which are more resistant to the erasure of inherited marks. In normal cells, D4Z4 is highly enriched in H3K9me3 and it remains to be determined whether persistence of this mark acts as a signal that facilitates remethylation of the underlying DNA. Interestingly, the absence of remethylation in FSHD2 is reminiscent of what is observed in cells from ICF1 patients (Immunodeficiency, centromeric instability and facial abnormalities, OMIM #242860) carrying mutations in the de novo DNA methyltransferase 3B (DNMT3B) gene (35) suggesting common pathways. Loss of Smchd1 causes defective X chromosome silencing (1,53) and Xi topological changes (8,54). FSHD2 or BAMS mutations do not affect X inactivation and biochemical analyses revealed that depending on the type of mutation, the impact on the function of the protein is different with a gain of ATPase activity in most BAMS patients and a loss of function in FSHD2 (29,30). By comparing patients with different mutations in the ATPase domain, we show that this domain might be involved in the loading or release from DNA while gain-, loss-of-function are all associated with D4Z4 hypomethylation.
Overall, our results unravel a role for SMCHD1 in the regulation of D4Z4 methylation in pluripotent cells but suggest that SMCHD1 might be dispensable for maintaining DNA methylation in somatic cells. As observed for the ATPase activity (29,30) and consistent with previous results (8), our results also indicate that depending on the type of mutation, the binding of SMCHD1 to chromatin might be different. This opens new perspectives to monitor the impact of SMCHD1 on chromatin regulation but also for understanding the respective involvement of SMCHD1 mutations in FSHD2 or BAMS. Pluripotent cells thus appear as a potent model for studying the dynamic regulation of D4Z4 methylation, the role of SMCHD1, FSHD pathogenesis and associated epigenetic mechanisms. Given the high level of methylation in the FSHD1-hESCs and the decreased methylation in somatic tissues, our results raise interesting questions on the regulation and dynamics of DNA methylation at repetitive sequences, corresponding to the largest fraction of the human genome (55). Our findings also force the revisiting of a number of previously proposed concepts regarding D4Z4 methylation in FSHD and open new avenues for investigating the molecular defects leading to muscle alterations, including in the regulation of DUX4 especially at early differentiation stages.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted and thank all patients for participating in this study. Induced pluripotent stem cells were produced and characterized by Marseille Stem Cells core facility (MaSC; MMG, U1251). We thank Dr Leslie Caron, Ms. Biljana Dumevsaka and Dr Uli Schmidt from Genea Biocells (Sydney, Australia) for their generous gift of hESC line DNA and helpful comments. All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Association Française contre les Myopathies (AFM) [MNHDecrypt and TRIM-RD]; Agence Nationale de la Recherche [ANR-13-BSV1-0001]; Fondation Aix-Marseille Université, Santé, Sport et développement Durable (to F.M.); C.D. was the recipient of a fellowship from the French Ministry of Education and FSH Society; C.L. is the recipient of a fellowship from the French Ministry of Education; A.G. was supported by a research training program scholarship from the Australian government; M.E.B. was supported by a Bellberry-Viertel senior medical research fellowship; J.M.M. was supported by Australian National Health and Medical Research Council (NHMRC) fellowship [1105754]; M.E.B. and J.M.M. acknowledge NHMRC grant [1098290]; Independent Research Institute Infrastructure Scheme (IRIISS) [9000433] support, and Victorian State Government Operational Infrastructure Support. Funding for open access charge: Aix Marseille Univ.
Conflict of interest statement. None declared.
REFERENCES
- 1. Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M., Apedaile A., Hilton D.J., Dunwoodie S.L., Brockdorff N. et al.. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008; 40:663–669. [DOI] [PubMed] [Google Scholar]
- 2. Chen K., Czabotar P.E., Blewitt M.E., Murphy J.M.. The hinge domain of the epigenetic repressor Smchd1 adopts an unconventional homodimeric configuration. Biochem. J. 2016; 473:733–742. [DOI] [PubMed] [Google Scholar]
- 3. Jansz N., Chen K., Murphy J.M., Blewitt M.E.. The Epigenetic regulator SMCHD1 in development and disease. Trends Genet. 2017; 33:233–243. [DOI] [PubMed] [Google Scholar]
- 4. Gendrel A.V., Apedaile A., Coker H., Termanis A., Zvetkova I., Godwin J., Tang Y.A., Huntley D., Montana G., Taylor S. et al.. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell. 2012; 23:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M., Heard E., Brockdorff N.. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell Biol. 2013; 33:3150–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mould A.W., Pang Z., Pakusch M., Tonks I.D., Stark M., Carrie D., Mukhopadhyay P., Seidel A., Ellis J.J., Deakin J. et al.. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenet. Chromatin. 2013; 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen K., Hu J., Moore D.L., Liu R., Kessans S.A., Breslin K., Lucet I.S., Keniry A., Leong H.S., Parish C.L. et al.. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3535–E3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jansz N., Keniry A., Trussart M., Bildsoe H., Beck T., Tonks I.D., Mould A.W., Hickey P., Breslin K., Iminitoff M. et al.. Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 2018; 25:766–777. [DOI] [PubMed] [Google Scholar]
- 9. Brideau N.J., Coker H., Gendrel A.V., Siebert C.A., Bezstarosti K., Demmers J., Poot R.A., Nesterova T.B., Brockdorff N.. Independent mechanisms target SMCHD1 to trimethylated histone H3 lysine 9-modified chromatin and the inactive X chromosome. Mol. Cell Biol. 2015; 35:4053–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nozawa R.S., Nagao K., Igami K.T., Shibata S., Shirai N., Nozaki N., Sado T., Kimura H., Obuse C.. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol. 2013; 20:566–573. [DOI] [PubMed] [Google Scholar]
- 11. Dejardin J., Kingston R.E.. Purification of proteins associated with specific genomic Loci. Cell. 2009; 136:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grolimund L., Aeby E., Hamelin R., Armand F., Chiappe D., Moniatte M., Lingner J.. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 2013; 4:2848. [DOI] [PubMed] [Google Scholar]
- 13. Sacconi S., Camano P., de Greef J.C., Lemmers R.J., Salviati L., Boileau P., Lopez de Munain Arregui A., van der Maarel S.M., Desnuelle C.. Patients with a phenotype consistent with facioscapulohumeral muscular dystrophy display genetic and epigenetic heterogeneity. J. Med. Genet. 2012; 49:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R. et al.. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012; 44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaillard M.C., Puppo F., Roche S., Dion C., Campana E.S., Mariot V., Chaix C., Vovan C., Mazaleyrat K., Tasmadjian A. et al.. Segregation between SMCHD1 mutation, D4Z4 hypomethylation and Facio-Scapulo-Humeral Dystrophy: a case report. BMC Med. Genet. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R. et al.. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992; 2:26–30. [DOI] [PubMed] [Google Scholar]
- 17. Upadhyaya M., Lunt P., Sarfarazi M., Broadhead W., Farnham J., Harper P.S.. The mapping of chromosome 4q markers in relation to facioscapulohumeral muscular dystrophy (FSHD). Am. J. Hum. Genet. 1992; 51:404–410. [PMC free article] [PubMed] [Google Scholar]
- 18. Sarfarazi M., Wijmenga C., Upadhyaya M., Weiffenbach B., Hyser C., Mathews K., Murray J., Gilbert J., Pericak-Vance M., Lunt P. et al.. Regional mapping of facioscapulohumeral muscular dystrophy gene on 4q35: combined analysis of an international consortium. Am. J. Hum. Genet. 1992; 51:396–403. [PMC free article] [PubMed] [Google Scholar]
- 19. Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M.. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002; 32:235–236. [DOI] [PubMed] [Google Scholar]
- 20. Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. et al.. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010; 329:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyle R., Wright T.J., Clark L.N., Hewitt J.E.. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995; 28:389–397. [DOI] [PubMed] [Google Scholar]
- 22. Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D. et al.. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999; 236:25–32. [DOI] [PubMed] [Google Scholar]
- 23. van Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W., van der Maarel S.M.. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2005; 58:569–576. [DOI] [PubMed] [Google Scholar]
- 24. van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M.. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003; 35:315–317. [DOI] [PubMed] [Google Scholar]
- 25. de Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L. et al.. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010; 75:1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M.. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009; 30:1449–1459. [DOI] [PubMed] [Google Scholar]
- 27. Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R. et al.. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012; 22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A. et al.. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017; 49:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gordon C.T., Xue S., Yigit G., Filali H., Chen K., Rosin N., Yoshiura K.I., Oufadem M., Beck T.J., McGowan R. et al.. De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet. 2017; 49:249–255. [DOI] [PubMed] [Google Scholar]
- 30. Gurzau A.D., Chen K., Xue S., Dai W., Lucet I.S., Ly T.T.N., Reversade B., Blewitt M.E., Murphy J.M.. FSHD2- and BAMS-associated mutations confer opposing effects on SMCHD1 function. J. Biol. Chem. 2018; 293:9841–9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gaillard M.C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J. et al.. Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology. 2014; 33:733–742. [DOI] [PubMed] [Google Scholar]
- 32. Polo J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y., Apostolou E., Stadtfeld M., Li Y., Shioda T. et al.. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010; 28:848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartweck L.M., Anderson L.J., Lemmers R.J., Dandapat A., Toso E.A., Dalton J.C., Tawil R., Day J.W., van der Maarel S.M., Kyba M.. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology. 2013; 80:392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sagie S., Ellran E., Katzir H., Shaked R., Yehezkel S., Laevsky I., Ghanayim A., Geiger D., Tzukerman M., Selig S.. Induced pluripotent stem cells as a model for telomeric abnormalities in ICF type I syndrome. Hum. Mol. Genet. 2014; 23:3629–3640. [DOI] [PubMed] [Google Scholar]
- 35. Yehezkel S., Rebibo-Sabbah A., Segev Y., Tzukerman M., Shaked R., Huber I., Gepstein L., Skorecki K., Selig S.. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011; 6:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mason A.G., Slieker R.C., Balog J., Lemmers R., Wong C.J., Yao Z., Lim J.W., Filippova G.N., Ne E., Tawil R. et al.. SMCHD1 regulates a limited set of gene clusters on autosomal chromosomes. Skelet. Muscle. 2017; 7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Giacalone J., Friedes J., Francke U.. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1992; 1:137–143. [DOI] [PubMed] [Google Scholar]
- 38. Chadwick B.P. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X specific role involving CTCF and anti-sense transcripts. Genome Res. 2008; 18:1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marsollier A.C., Joubert R., Mariot V., Dumonceaux J.. Targeting the polyadenylation signal of Pre-mRNA: A new gene silencing approach for facioscapulohumeral dystrophy. Int. J. Mol. Sci. 2018; 19:E1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J. et al.. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLos Genet. 2010; 6:e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das S., Chadwick B.P.. Influence of repressive histone and DNA methylation upon D4Z4 transcription in non-myogenic cells. PLoS One. 2016; 11:e0160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lemmers R.J., van den Boogaard M.L., van der Vliet P.J., Donlin-Smith C.M. Nations, S.P. . Nations, S.P. Ruivenkamp C.A., Heard P., Bakker B., Tapscott S., Cody J.D. et al.. Hemizygosity for SMCHD1 in facioscapulohumeral muscular dystrophy type 2: consequences for 18p deletion syndrome. Hum. Mutat. 2015; 36:679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hasi-Zogaj M., Sebold C., Heard P., Carter E., Soileau B., Hill A., Rupert D., Perry B., Atkinson S., O’Donnell L. et al.. A review of 18p deletions. Am. J. Med. Genet. C Semin. Med. Genet. 2015; 169:251–264. [DOI] [PubMed] [Google Scholar]
- 44. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D.. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017; 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hendrickson P.G., Dorais J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L. et al.. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017; 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J.. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017; 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Urbach A., Bar-Nur O., Daley G.Q., Benvenisty N.. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010; 6:407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Avitzour M., Mor-Shaked H., Yanovsky-Dagan S., Aharoni S., Altarescu G., Renbaum P., Eldar-Geva T., Schonberger O., Levy-Lahad E., Epsztejn-Litman S. et al.. FMR1 epigenetic silencing commonly occurs in undifferentiated fragile X-affected embryonic stem cells. Stem Cell Rep. 2014; 3:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al.. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- 50. Monk M., Boubelik M., Lehnert S.. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987; 99:371–382. [DOI] [PubMed] [Google Scholar]
- 51. Rougier N., Bourc’his D., Gomes D.M., Niveleau A., Plachot M., Paldi A., Viegas-Pequignot E.. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998; 12:2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Howlett S.K., Reik W.. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991; 113:119–127. [DOI] [PubMed] [Google Scholar]
- 53. Blewitt M.E., Vickaryous N.K., Hemley S.J., Ashe A., Bruxner T.J., Preis J.I., Arkell R., Whitelaw E.. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:7629–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang C.Y., Jegu T., Chu H.P., Oh H.J., Lee J.T.. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell. 2018; 174:406–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D.. Repetitive elements may comprise over two-thirds of the human genome. PLos Genet. 2011; 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ludlow A.T., Robin J.D., Sayed M., Litterst C.M., Shelton D.N., Shay J.W., Wright W.E.. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res. 2014; 42:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P. Jr et al.. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012; 21:4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Broucqsault N., Morere J., Gaillard M.C., Dumonceaux J., Torrents J., Salort-Campana E., Maues De Paula A., Bartoli M., Fernandez C., Chesnais A.L. et al.. Dysregulation of 4q35- and muscle-specific genes in fetuses with a short D4Z4 array linked to facio-scapulo-humeral dystrophy. Hum. Mol. Genet. 2013; 22:4206–4214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.