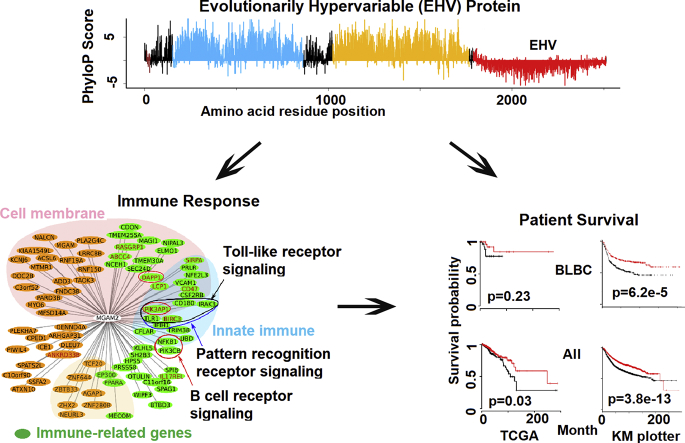
Abstract
Maltase-glucoamylase (MGAM) and MGAM2 both belong to the glycoside hydrolase family 31. MGAM, a therapeutic target for type 2 diabetes, is α-1,4-glucosidase and expressed in the intestine to catalyze starch digestion. MGAM2, however, is largely uncharacterized. By investigating The Cancer Genome Atlas data, we found that among breast cancer subtypes, MGAM2 expression is nearly exclusive to basal-like breast cancers (BLBCs), whereas MGAM tends to express in luminal A breast cancers. Moreover, MGAM2 expression is associated with better patient survival and correlated with immune genes/signatures, unlike MGAM. Both genes have emerged in mammals, but diverged after the placental-marsupial split. In placentals, MGAM2 has likely lost its α-1,4-glucosidase activity due to mutations in key catalytic sites, and has acquired a large domain that is extracellular, threonine-rich and evolutionarily hypervariable (EHV). Guided by MGAM2 findings, our genome-wide search identified >1000 human proteins with EHV regions. These proteins are enriched in immune functions and molecules, including major histocompatibility complex proteins. Their genes are expressed higher in BLBCs and are associated with better patient survival, like MGAM2. Their EHV-coding sequences are rich in simple repeats and harbor more cancer passenger mutations. In conclusion, MGAM2 diverges from MGAM structurally and likely functionally in placentals. MGAM2 is among >1000 human proteins with EHV regions and associated with immune response. We propose that these EHV molecules may have significant implication in cancer immunotherapy and BLBC treatment.
Keywords: MGAM2; MGAM; BLBC; α-1,4-glucosidase; EHV; Immune response; Patient survival; Placental-marsupial split
Graphical Abstract
1. Introduction
Breast cancer is a very heterogeneous disease. Several molecular subtypes, including luminal A, luminal B, HER2-enriched, basal-like and normal-like, have been identified via PAM50 classification [1,2]. With the shortest disease-free survival and overall survival, basal-like breast cancer (BLBC) is regarded as the worst subtype [[3], [4], [5]]. Over 70% of BLBCs are triple negative [[6], [7], [8]], expressing neither estrogen receptor nor progesterone receptor and without HER2 amplification/overexpression. Consequently, therapies targeting hormone receptors (e.g., tamoxifen) or HER2 (e.g., trastuzumab) are often inapplicable. Chemotherapy with drugs such as anthracyclines and taxanes is the only choice of adjuvant systemic therapy currently available for many BLBC patients [9]. Thus, BLBCs present a significant clinical challenge [3,4,10].
Maltase-glucoamylase (MGAM), an α-1,4-glucosidase that belong to the glycoside hydrolase family 31 (GH31) [11] of the carbohydrate-active enzyme (CAZy) database, is well studied. MGAM is an integral membrane protein with its two catalytic domains, maltase and glucoamylase, facing the extracellular environment. MGAM is expressed in the intestine to catalyze the final step of starch digestion to glucose. As such, MGAM has been a major therapeutic target for treating type 2 diabetes [[12], [13], [14]]. FDA approved antidiabetic drugs, such acarbose (Precose) and miglitol (Glyset), are potent MGAM inhibitors [15,16].
MGAM2 is a homolog of MGAM, emerged via a tandem duplication event that likely occurred in primitive mammals [17]. It is also a member of the GH31 family. However, unlike MGAM, MGAM2 is largely uncharacterized with functions unknown.
By studying The Cancer Genome Atlas (TCGA) breast cancer RNA-seq data [6,7], we have found that while MGAM tends to express in luminal A breast cancers (LABCs), MGAM2 expression is highly specific to BLBCs. To understand the significance of this, we performed the study described below.
2. Methods
2.1. Breast Cancer Data Analyses
RNA-seq and clinical data of breast cancer were downloaded from TCGA data portal version 5.0 (portal.gdc.cancer.gov). The subtype information of these breast cancers was obtained from published studies [7,18]. Gene expression data of normal tissues were obtained from the Genotype-Tissue Expression (GTEx) database [19].
Log-rank tests were used for patient survival analyses. Differentially expressed genes were identified with DES eq. [20]. Pearson and Spearman correlation coefficients were both used in correlation analyses. Two tailed Fisher exact tests and Wilcoxon tests were used to determine the difference of gene expression among different breast cancer subtypes. Paired t-tests were performed to determine expression differences between primary tumors and their matching normal tissue samples. In most cases, only genes with a FPKM (fragments per kilobase of transcript per million mapped) value of ≥1 in at least one sample were selected and used in an analysis. All statistical tests were conducted using R package (version 3.0.3). Gene functional enrichment analysis were performed using GSEA [21] and DAVID [22]. ssGSEA (version 9.0.9) were performed with various signature genes.
To identify the intrinsic connection of MGAM2 correlated genes (Fig. 2B), we calculated a similarity weight wij between gene i and gene j, given by , where and , with μ = 1, 2, … , 19,817 and representing a gene encoded in the human genome. We set aij = 1 if r > 0.3 and ρ > 0.3,where r and ρ represent Pearson or Spearman correlation coefficient respectively, and aij = 0 otherwise.
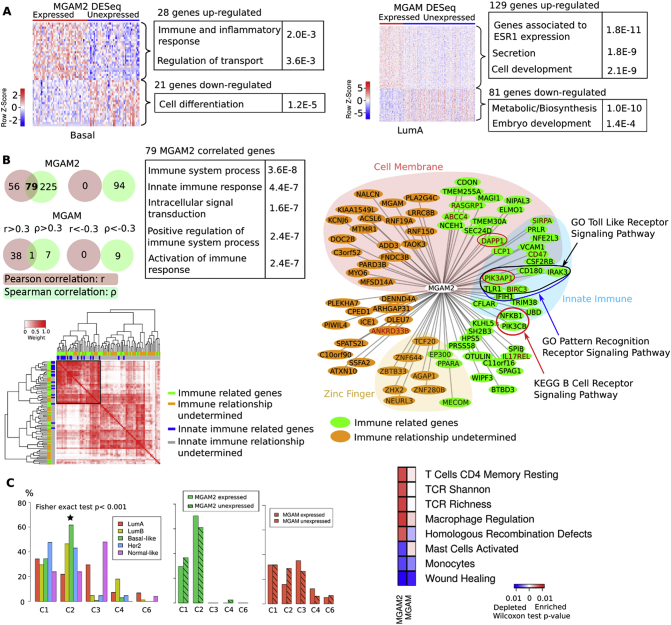
Fig. 2.
MGAM2 expression is correlated with immune response in BLBCs.
A: Heatmap indicates the log2(FPKM) values of differentially expressed genes between MGAM2 (or MGAM) expressing (FPKM >1.0) and not-expressing (FPKM <0.1) BLBCs (or LABCs). Upregulated genes are shown in red and downregulated genes are shown in green, with their enriched functions indicated.
B: Venn Diagrams indicate 79 positively correlated genes with MGAM2 found by both Pearson and Spearman correlations, with their enriched functions shown. The heatmap specifies the connectivity and correlation among the 79 genes. The right image designates the distribution of 79 genes in functions and other properties, where the distance between MGAM2 and each gene is 1 − r, with r being the Pearson correlation coefficient.
C: BLBCs are significantly enriched in the C2 (IFN-γ dominant) immune subtype, indicated by the distribution of the intrinsic subtypes among the immune subtypes (left plot). MGAM2-expression further increases in the C2:C1 ratio, unlike MGAM (middle two plots). The heatmap (right) specifies immune features that differ significantly between MGAM2 (or MGAM) expressed and not expressed BLBCs (or LABCs).
See also Table S2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. EHV Coding Sequence Exon (CDS) and Gene Identification
All analysis is based on the hg38 human genome assembly. PhyloP scores of 100 vertebrates and 20 mammals were downloaded from the UCSC genome database (hgdownload.cse.ucsc.edu). We developed a pipeline (Figs. 3C and S3B) to identify EHV CDSs and genes, based on these phyloP scores of all coding exons or CDSs in the human genome. First, we calculated the average phyloP score for each CDS and selected those CDSs with negative values. Second, we performed z test, , where x is the phyloP score of a CDS selected above, μ is the mean and σ is the standard deviation of x, and l is the CDS length. Based on the test, we identified CDSs with significantly lower score at q < 0.01. Then, we did further selection using cutoffs on minimal CDS length (45 bp) and phyloP score shown in Fig. 3C and S3B.
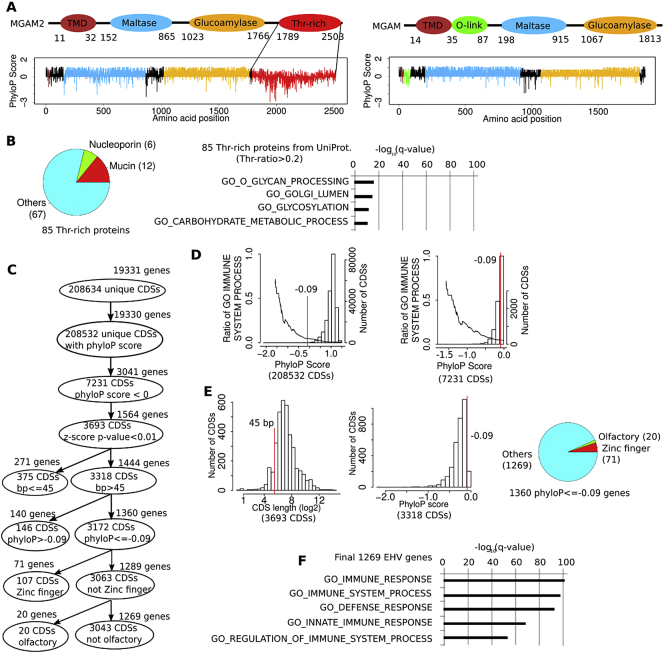
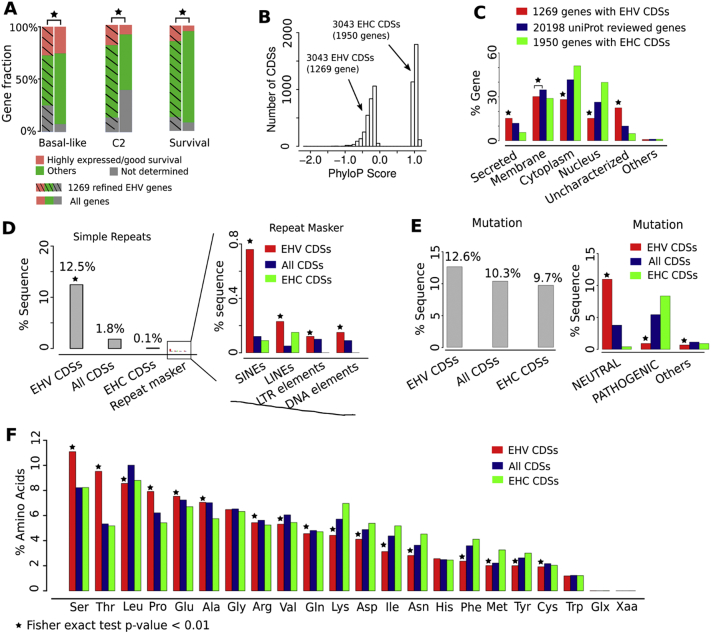
Fig. 3.
Evolutionarily hypervariable (EHV) genes, including MGAM2, are associated with immune response.
A: MGAM2 contains an extra threonine-rich and EHV domain at the C-terminus, compared to MGAM. PhyloP scores are 20 mammals-based, with positive values indicating conservation and negative values indicating variation.
B: A total of 85 proteins with threonine-rich domains, identified from UniProt, are not enriched in immune-related functions.
C: We established a pipeline to identify genes with at least one EHV CDS via multi-step selections, including z-score tests and cutoffs on CDS length and phyloP score (see Methods).
D: The proportion of immune-related genes increases as the phyloP scores of their CDSs decrease. The bars (the right Y-axis) indicate the distribution the average phyloP scores of all CDSs (left image) or CDSs with negative phyloP scores (right image) in the human genome. The line (the left Y-axis) represents the distribution of the proportion of immune-related genes. Red line indicates the average phyloP score of the last CDS of MGAM2.
E: Left two plots indicate the distribution of the CDS length and phyloP score after z-score test selection shown in C, with red lines specifying cutoffs used for further selection. The pie chart indicates the composition of the selected 1360 genes with EHV CDSs.
F: The final EHV genes are significantly enriched in immune response-related functions.
See also Table S3 and Fig. S3 (based on conservation of 100 species). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Sequence and Other Analyses
Genomic synteny and annotation data of MGAM2 from various species were obtained from both the UCSC and Ensembl (www.ensembl.org) databases. Simple repeat content data of human genomic regions were also obtained from the UCSC genome site. Other repeats were identified by using the RepeatMasker program (version 4.0.7). Protein subcellular location and other data were obtained from the UniProt database (released on 2017/07; www.uniprot.org). Cancer mutation data were obtained from the COSMIC database v81 (cancer.sanger.ac.uk/cosmic).
2.4. Protein 3D Structure
The crystal structures of ntMGAM [14] and ctMGAM [23] have been determined. These structures were obtained from the PDB database (2QLY and 3TON respectively), along with those in complex with acarbose (2QMJ and 3TOP respectively). No structures of MGAM2 have been published yet. We hence used I-TASSER [24], a popular tool for protein structure modeling, to predict the 3D structure of ntMGM2 (residues 33–904) and ntMGAM2 (residues 905–1788), with default parameters and no predefined models. MGAM2 sequence were aligned to MGAM with the software Multalin [25] to identify putative active sites.
2.5. Substrate Docking
Substrate docking was performed with AutoDock-Vina (version 1.1.2) [26] with the predicted ntMGAM2 and ctMGM2 structures, to which hydrogen atoms and partial charge were added with AutoDock-ADT (version 1.5.6). The substrates being docked include acarbose, maltose and dextrin. The substrate binding center was set at the midpoint of D577 and E478 for ntMGAM2 and of D1375 and N1480 for ctMGAM2. The binding site covers a radius of 25 Å, as suggested by Atuodock-Vina.
3. Results
3.1. MGAM2 is Expressed in BLBCs while MGAM Tends to Express in LABCs
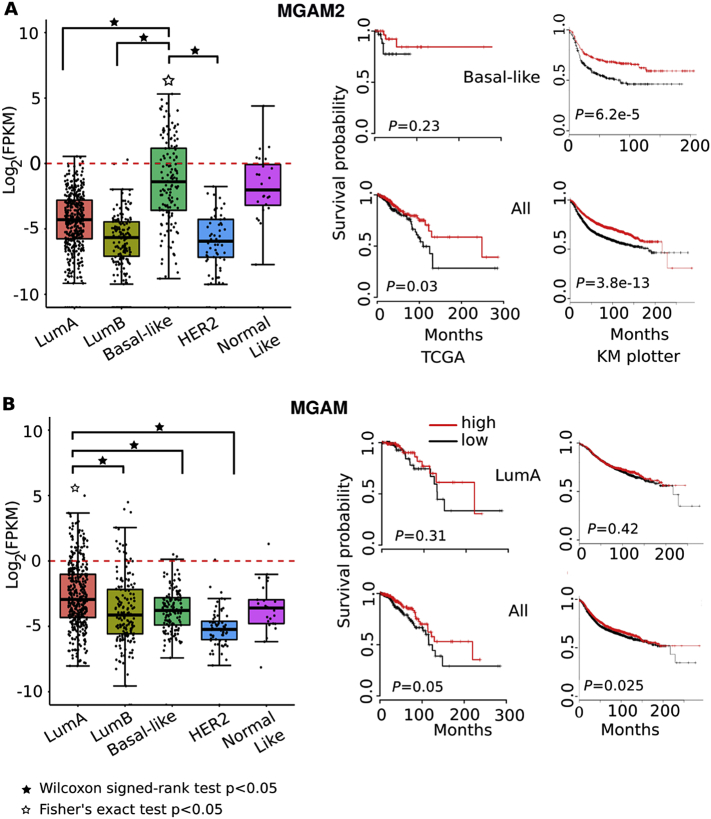
We examined the mRNA expression of MGAM and MGAM2 in TCGA breast cancers [7], which consist of 824 primary tumors (420 luminal A, 174 luminal B, 140 BLBC, 65 Her2-enriched and 25 normal-like) from 814 cases (Table S1A). If considering a FPKM (fragments per kilobase of transcript per million mapped) value of ≥1 as expressed, MGAM and MGAM2 expression distributes differently among the five subtypes (Fig. 1A and B). MGAM is expressed in about 14% LABCs but in <10% tumors of other subtypes (p = .05) (Fig. 1B). MGAM2 expression, however, is highly BLBC-specific (p < 1e-5). MGAM2 is expressed in ~33% BLBCs, in 24% normal-like breast cancers (which share numerous molecular features with BLBCs), but in <1% in each of other subtypes (Fig. 1A; Table S1A). We examined the 2000 breast cancers of the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) [27] and also concluded that MGAM2 is expressed higher in BLBCs (Fig. S1).
Fig. 1.
MGAM2 expression is significantly enriched in BLBCs and is associated with better patient survival, unlike MGAM.
A: MGAM2 expression is highly specific to BLBCs among the five subtypes of breast cancers from TCGA (the left plot), and is associated with better patient survival for both TCGA samples (middle plots) and samples in the KM plotter database, which includes 3951 total breast cancer samples and 618 BLBCs (right plots). Note that in the KM plotter database, MGAM2 is called LOC93432 (Affymetrix Id 216666_at), and relapse free survival (RFS) is used as it yields the largest sample size.
B: MGAM data are presented as A. The KM plotter database contains a total of 1933 luminal A breast cancers.
See also Figs. S1 and S2, and Table S1.
MGAM and MGAM2 are not expressed or expressed lowly in normal breast tissues. In TCGA normal breast samples (113 total), FPKM values range from 0 to 4 with a median of 0.08 for MGAM and range 0 to 10 with a median of 1.3 for MGAM2 (Fig. S1). Likewise, normal breast samples (290 total) from the GTEx database [19] have a TPM (transcripts per million) range of 0–10.4 for MGAM and of 0–14.2 for MGAM2, with a median of 0.25 for both genes (Fig. S1).
MGAM and MGAM2 both belong to the GH31 family. To determine if the observed MGAM and MGAM2 expression distribution among the breast cancer subtypes (Fig. 1A and B) is related to GH31, we investigated other GH31 members. These include SI (sucrase isomaltase), GAA (lysosomal α-glucosidase) and others. We found none of them resembling MGAM or MGAM2 (Fig. S2A). We also studied GH13, another family that contains α-glucosidases as well, including amylases and others. We did not find any MGAM or MGAM2-like pattern either (Fig. S2B). In summary, neither BLBC-expression of MGAM2 (Fig. 1A) nor LABC-expression of MGAM (Fig. 1B) is a common feature of either the GH31 or GH13 family.
3.2. MGAM2 Expression is Associated with Better Patient Survival
By investigating TCGA clinical data, we found that MGAM2 expression is associated with better patient survival, in both BLBCs and all breast cancers (Fig. 1A; Table S1B). For MGAM, however, the association appears to be unclear (Fig. 1B; Table S1B). These conclusions are supported by breast cancer cases (3951 total, of which 618 are BLBCs) presented at the Kaplan-Meier Plotter site (kmplot.com/) [28], investigated with all datasets combined (Fig. 1; Table S1C) and individual dataset separately (Fig. S2C).
3.3. MGAM2 Expression is Associated with Immune Response
To better understand the significance of MGAM2 expression, we first identified genes that are differentially expressed between MGAM2-expressing and not-expressing BLBCs. The analysis revealed 28 upregulated genes, which are significantly enriched in functions related to immune and inflammatory response, in MGAM2-expressing BLBCs (Fig. 2A; Table S2A). As a comparison, we also identified 129 upregulated genes in MGAM-expressing LABCs, which however are not enriched in immune functions (Fig. 2A; Table S2A).
We then investigated genes that correlate with MGAM2 in expression in BLBCs. A total of 79 positively-correlated genes, but no negatively-correlated genes, were identified by both Pearson and Spearman correlations (Fig. 2B; Table S2B). Importantly, the 79 genes are significantly enriched in immune functions (Fig. 2B; Table S2B). Innate immune is especially notable, with at least 17 relevant gene found. These include TLR1, BIRC3 and other members of the Toll-like receptor signaling pathway and pattern recognition receptor pathway (Fig. 2B). Meanwhile, for MGAM, only one positively correlated gene was found (Fig. 2B; Table S2B).
To determine if MGAM2 is expressed by tumor cells and/or by tumor-infiltrating immune cells, we investigated tissue-specific alternative splicing (AS) of MGAM2, using RNA-seq data from GTEx. We identified blood-specific and epithelial-specific AS forms, which differ in the first exon (and hence the promoter) (Fig. S2D). We found that TCGA breast cancers only express the epithelial-specific AS form of MGAM2. Hence, MGAM2 is expressed by tumor cells, which have an epithelial cell origin.
Lastly, we investigated the data from a recent paper on pan cancer immune landscape [18]. Among the six cancer immune subtypes identified, BLBCs are enriched in the C2 (IFN-γ dominant), 64%, and C1 (wound healing) subtypes, ~35% (Fig. 2C; Table S3C). MGAM2 expression further increases the C2 to C1 ratio (69% versus 30%) (Fig. 2C). Consistent with this, MGAM2-expressing BLBCs have significantly increased enrichment scores on macrophage regulation and T cell receptor (TCR) diversity (Fig. 2C; Table S2C). None of these was observed for MGAM, however.
3.4. MGAM2 Has an Extra threonine-Rich Domain that is Evolutionarily Hypervariable (EHV)
The largest difference in composition between MGAM and MGAM2 is that MGAM2 harbors an extra C-terminal domain (Fig. 3A). This domain consists of 715 amino acid residues, 32% of which are threonine, hence threonine-rich. The entire domain is encoded by a single large exon, with a conservation score lower than a typical exon, intron or intergenic region (Fig. 3A; Fig. S3A and Table S3A). Thus, this exon represents a fast-evolving and EHV region in the human genome.
3.5. Proteins with Threonine-Rich Domain Appear Not Enriched in Immune Functions
MGAM2 is associated with immune response (Fig. 2) and harbor an extra domain that is threonine-rich(Fig. 3A). To determine if the immune response is linked to being rich in threonine, we identified 85 proteins with threonine-rich domains (with the threonine ratio of >0.2, based on the UniProt annotation) among 20,198 reviewed human proteins from the UniProt database (Table S3B). With 12 mucins and 6 nucleoporins, these 85 proteins are enriched in glycosylation, but not immune-related functions (Fig. 3B).
3.6. Proteins with EHV Regions Are Enriched in Immune Functions
The threonine-rich domain of MGAM2 is also EHV (Fig. 3A), encoded by a fast-evolving coding sequence exon (CDS) in the human genome. To determine if MGAM2-associated immune response (Fig. 2) is linked to this EHV feature, we identified all human proteins encoded by at least one CDS that is EHV as described below.
We developed a pipeline (see Methods) that examines the phyloP scores, which measure evolutionary conservation based on sequence alignment of 20 mammals (Fig. 3C; see Fig. S3B for 100 species), of all CDSs (>208,000 from 19,331 genes) in the human genome. We first identified CDSs with negative phyloP scores, which as expected only account for <4% of all CDSs (Fig. 3C). Furthermore, as the phyloP score decreases, the proportion of immune-related genes increases (Fig. 3D; Table S3D). We then performed z-score tests and identified 3693 CDSs with significantly low phyloP scores (q < 0.01) (Fig. 3C). We used two more cutoffs, CDS length > 45 bp (chosen based on its distribution; see Fig. 3E) and phyloP score ≤ −0.09 (the value of the CDS encoding the EHV domain of MGAM2; see Fig. 3A). This reduces EHV CDSs to 3172 in total, which involve 1360 genes (Fig. 3C). A substantial portion of these genes are zinc fingers (71 total) and olfactory receptors (20 total) (Fig. 3E; Table S3E), two gene families known to be fast-evolving. This supports the accuracy of our pipeline. After removing zinc fingers and olfactory receptors (many not known to be immune molecules), 1269 genes remain and are significantly enriched in immune response (Fig. 3F; Table S3F). The analysis supports that immune response of MGAM2 is linked to its EHV region.
We repeated the same analysis with the phyloP scores of 100 vertebrates sequenced (Fig. S3), ranging from fish to primates. The analysis identified 2747 EHV CDSs and 1489 genes (after excluding zinc fingers and olfactory receptors), of which about 53% CDS and 73% genes overlap with those found with 20 mammals (Fig. 3). Hence, the same conclusions were reached.
3.7. EHV Genes and CDSs Harbor Unique Features
Like MGAM2, significantly more of the EHV genes identified in Fig. 3C are expressed higher in the BLBC and C2 subtypes, and are associated with better patient survival, compared to the entire human gene set (Fig. 4A; Table S4A). To better understand them, we identified equal number of evolutionarily hyperconserved (EHC) CDSs (3043 total), along with their genes (1950 total) in the human genome (Fig. 4B; Table S4B). We then compared EHV and EHC CDSs/genes, along with the entire CDS/gene set of the human genome.
Fig. 4.
EHV genes and CDSs harbor unique features.
A: Significantly more EHV genes (identified by Fig. 3C) are expressed higher (p ≤ .05) in the BLBC and C2 subtypes, and are associated with better (p ≤ .05) patient survival, compared to the entire human gene set. TCGA data are used.
B: The phyloP score distribution of EHV and evolutionarily hyperconserved (EHC) CDSs and their genes used for comparison in C-F.
C: EHV genes encode proteins that are more likely to be secreted or not characterized, but less likely to be in either cytoplasm or nucleus, compared to EHC genes and the entire gene set. UniProt data are used.
D-F: EHV CDSs consist of more simple repeats (obtained from the UCSC genome database) and other repeats (D) (by RepeatMasker), harbor more passenger mutations (E) (COSMIC data are used), and encode more threonine and serine residues (F).
In cellular location, EHV genes are more likely to encode secreted or uncharacterized proteins, but less likely to encode cytoplasm or nucleus proteins (Fig. 4C; Table S4C). The opposite was observed for EHC genes. In sequence composition, EHV CDSs are made of 12.5% simple repeats, 100 times higher compared to EHC CDSs and 10 times higher compared to the entire CDS set (Fig. 4D; Table S4D). Notably, our analysis reveals that 1/3 of the EHV CDS of MGAM2, 730 bp in total, has arisen from several tandem duplications of a 60 bp sequence (Fig. S4–1). EHV CDSs also harbor significantly more SINEs and LINEs. In total cancer mutations, EHV CDSs are largely the same as EHC CDSs and the entire CDS set. However, >5 time more of their mutations are neutral, not pathogenic (Fig. 4E; Table S4E). Lastly, EHV CDSs encode significantly more threonine (nearly twice) and serine, compared to the other two types of CDS(Fig. 4 F; Table S4F).
We also performed the same analysis with the 2747 EHV CDSs and 1489 EHV genes identified with 100 vertebrates (Fig. S3). We reached the same conclusions (Fig. S4 and Table S4).
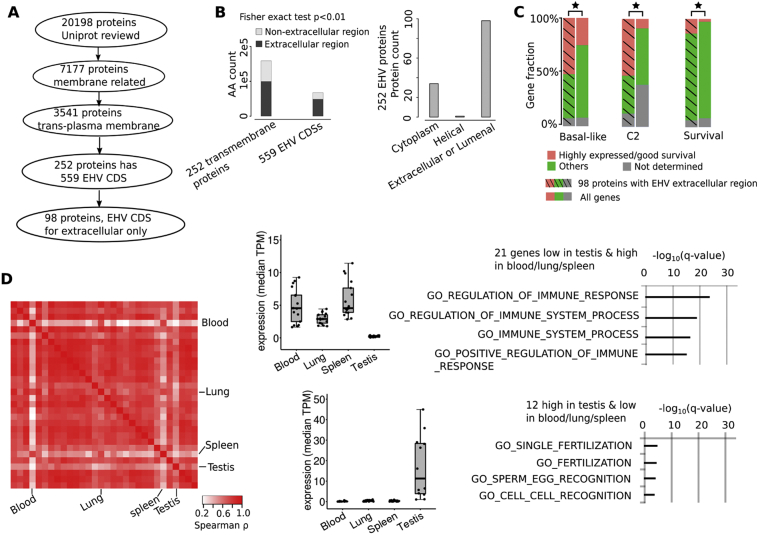
3.8. Transmembrane Proteins with EHV Extracellular Regions and Expressed in Blood are Associated with Immune Response
MGAM2 is a transmembrane protein with an EHV region that is extracellular (Fig. 3A). To better understand this, we developed a pipeline (Fig. 5A) to identify other proteins with the same feature. Using protein topology information from UniProt, we identified 252 transmembrane proteins with regions encoded by at least one EHV CDS (Fig. 5A; Table S5A). We noted that these EHV regions are more likely to be extracellular (Fig. 5B; Table S5B). Indeed, among the 252 proteins, 98 have their EHV regions being solely extracellular, like MGAM2 (Fig. 3A), compared to 34 and one proteins with their EHV regions being solely cytoplasmic or transmembrane respectively (Fig. 5B; Table S5B). In breast cancers, we found significantly more of the 98 proteins (Fig. 5A) to be expressed higher in the BLBC and C2 subtypes, and to be associated with better patient survival, compared to the entire human gene set (Fig. 5C; Table S5C). These properties resemble MGAM2. As a comparison, we did not find the 34 proteins with EHV cytoplasmic regions (Fig. 5B; Table S5B) to be associated with better patient survival.
Fig. 5.
Transmembrane proteins with EHV extracellular regions and expressed in blood are associated with immune response.
A: Our pipeline identified 98 transmembrane proteins with EHV extracellular regions, like MGAM2 (Fig. 3A).
B: EHV CDSs of the 252 transmembrane proteins (A) encode significantly more amino acids that are extracellular (left) and regions that are entirely extracellular (right).
C: Significantly more of the 98 EHV transmembrane proteins are expressed higher (p ≤ .05) in the BLBC and C2 subtypes, and are associated with better (p ≤ .05) patient survival, compared to the entire human gene set. TCGA data are used.
D: The heatmap indicates the Spearman correlation coefficient, ρ, between any pairs of tissues using median expression of genes encoding the 98 EHV transmembrane proteins (A). Blood shows the largest divergence from most other tissues, especially testis. Box plots indicate genes that are expressed highly in blood but lowly in testis, and vice versa, with each dot representing the median expression of a gene and significantly enriched functions of each gene group shown. TPM: transcripts per million. GTEx data are used.
With the data from the Genotype-Tissue Expression (GTEx) database [19], we investigated the genes encoding the 98 MGAM2-like proteins in 30 human tissues. Interestingly, most tissues are highly correlated by the mRNA expression of these genes, with Spearman correlation coefficient ρ > 0.8 (Fig. 5D). Blood, however, is an exception, with high corrections with spleen and lung (ρ ≈ 0.8), but low corrections with other tissues especially testis (ρ ≈ 0.2) (Fig. 5D; Table S5D). We identified 21 genes that are expressed highly in blood and lowly in testis, and found that they are significantly enriched in immune functions (Fig. 5D; Table S5D), including HLA-A and HLA-B that encode major histocompatibility complex (MHC) molecules. To the contrary, the 12 genes that are expressed highly in testis but lowly in blood are enriched in fertilization-related functions (Fig. 5D; Table S5D).
We performed the same analysis with EHV CDSs obtained with100 vertebrate conservation (Fig. S3), and reached the same conclusions (Fig. S5 and Table S5). Note that MGAM2 is also expressed highly in blood.
3.9. MGAM2 Likely Has Lost its α-1,4-Glucosidase Activity
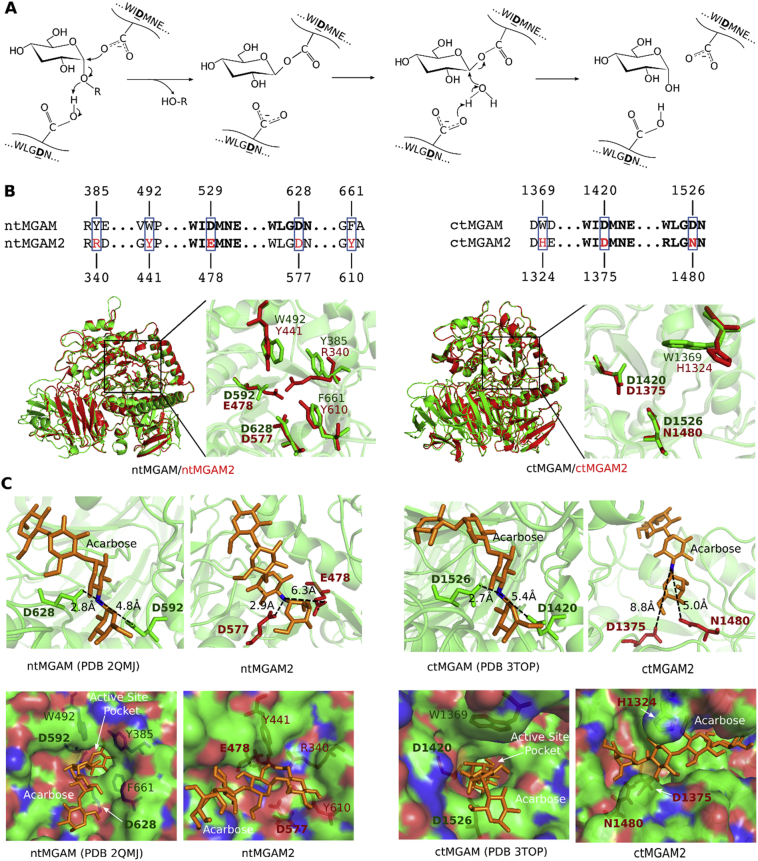
Active glucosidases of the GH31 family harbor two catalytic aspartic acid (D) residues, one acting as a nucleophile and another acting as a proton donor (Fig. 6A) [29]. The two residues locate in highly conserved GH31 signature peptides, WIDMNE and WLGDN (Fig. S6A). In MGAM, they are D529 and D628 for N-terminal MGAM (ntMGAM), a maltase, and D1420 and D1526 for C-terminal MGAM (ctMGAM), a glucoamylase (Fig. 6B). In MGAM2, however, D529 is mutated to glutamic acid (E) and D1526 is mutated to asparagine (N) (Fig. 6B; Fig. S6B), indicating that MGAM2 is likely no longer active.
Fig. 6.
MGAM2 likely lacks α-1,4-glucosidase activity due to mutations of key amino acids.
Additional mutations were identified at the active sites (Fig. 6B). To better understand their impact, we performed molecular modeling of MGAM2, as its structure has not been experimentally determined, unlike MGAM. Our predicted structures of ntMGAM2 and ctMGAM2 match well with the crystal structures of their MGAM counterparts [14,23] overall, with root-mean-square deviation (RMSD) of <1 Å (Fig. 6B; Table S6A). However, larger RMSD values were observed for some mutated residues at catalytic sites (Fig. 6B; Table S6A), indicating that substrate-binding may be affected. To test this, we performed substrate docking. In crystal structures [14,23], the distances between the two catalytic D residues to the oxygen (replaced by nitrogen in acarbose) of α-1,4-glycosidic bond to be cleaved are 2.8 Å and 4.8 Å for ntMGAM, and 2.9 Å and 5.4 Å for ntMGAM (Fig. 6C). However, the corresponding numbers were predicted to be 2.9 Å and 6.3 Å for ntMGAM2, and 8.8 Å and 5.0 Å for ctMGAM2 (Fig. 6C; Tables S6B and S6C). Modeling analysis indicates that substrate-binding pockets in MGAM2 are altered, and substrates are not binding in the correct orientation for catalysis (Fig. 6C; Fig. S6C). These analyses support that MGAM2 is likely no longer an active α-glucosidase, unlike MGAM.
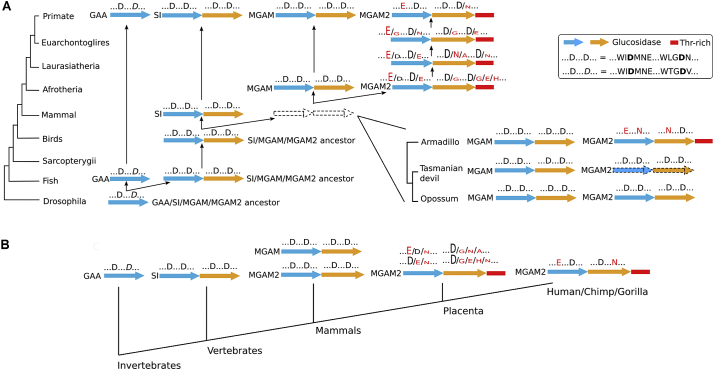
3.10. MGAM2 Lost Key Catalytic Residues and Acquired an EHV Domain during Placental-Marsupial Split
A previous study [17] reports that MGAM, MGAM2 and SI, three GH31 members with two catalytic sites, emerged via tandem duplications. To better understand this, we investigated >100 species whose genomes are sequenced, ranging from drosophila to the human. In drosophila, we did not find homologues of human MGAM, MGAM2 or SI. We however identified an α-glucosidase that appears to be a homolog of human GAA, a GH31 member with only one catalytic site (Fig. 7A; Table S7A). In fishes and birds, we found homologues of both human GAA and SI/MGAM/MGAM2 (Fig. 7A; Fig. S7 and Table S7A). Thus, in the GH31 family, one catalytic site members (e.g., GAA) are more ancient (Fig. 7B). Then, a tandem duplication occurred in vertebrates, yielding two catalytic site members (e.g., SI) (Fig. 7B).
Fig. 7.
GH31 α-glucosidases evolve through duplications and mutations.
A: Two catalytic domain members (SI, MGAM and MGAM2) emerge via duplication of an ancestor of one catalytic domain member (e.g., GAA). MGAM2 has lost key catalytic residues and acquired an EHV domain during placental-marsupial divergence.
B: The summary of GH31 α-glucosidase evolution.
See also Table S7 and Fig. S7.
Additional tandem duplications took place in mammals, leading to the split of SI,MGAM and MGAM2 (Fig. 7A and B). Interestingly, in marsupial species examined (opossum, Tasmanian devil, wallaby), MGAM2 retains the GH31 α-glucosidase signature peptides WIDMNE and WLGDN, and lacks the EHV domain (Fig. 7A; Tables S7B-7D). In placentals examined (from armadillo to primates), however, MGAM2 consistently harbors mutations of the catalytic D residues (Table S7B), and meanwhile acquires the EHV domain (Fig. 7A). The data indicate the likelihood that MGAM2 has lost its α-glucosidase activity and acquired new functions in placentals (see Discussion).
4. Discussion
4.1. MGAM2 may Function in Immune Response in Placentals and could be a Biomarker in BLBC Immunotherapy
MGAM and MGAM2 are both GH31 members. MGAM, a known α-glucosidase, is expressed abundantly in the small intestine to catalyze the final step of starch digestion. MGAM is an anti-diabetics target, and numerous inhibitors have been designed to block its α-1,4 glucosidase activity [12,14,[30], [31], [32], [33]]. To the contrary, MGAM2 is much less understood. Like MGAM, it also has maltase and glucoamylase domains, both of which have however lost their key catalytic residues in humans, revealed by our study. Hence, human MGAM2 is likely no longer an α-glucosidase, which of course needs future experimental validation.
Our analysis reveals a remarkably clean evolution of α-glucosidases of GH31. Found in both invertebrates and vertebrates, α-glucosidases with a single catalytic site, including GAA, are more ancient. Then, tandem duplications led to the emergence of α-glucosidases with two catalytic sites in vertebrates, as well as MGAM and MGAM2 in mammals. Finally, MGAM2 diverged from MGAM during the placental-marsupial split. In monotremes and marsupials, MGAM2 closely resembles MGAM and likely also functions as an α-glucosidase. In placentals however, MGAM2 has lost its key catalytic residues, hence likely no longer an α-glucosidase, and meanwhile has acquired a large threonine-rich extracellular domain that is EHV. Because proteins with EHV domains are significantly associated with immune response and for reasons discussed below, we propose that MGAM2 function in the immune system in placentals instead.
Marsupials and placentals differ in the tolerance of the fetal tissue by the maternal immune system, trophoblast development and invasion, and ultimately the gestation time [34]. It would be interesting to determine if the MGAM2-MGAM split plays a role in these marsupial-placental differences. Interestingly, among normal human tissues investigated by GTEx [19], MGAM2 is expressed in blood, gastrointestinal tract, breast and nerve. Importantly, except nerve, MGAM2 expression is significantly correlated with gene signatures of immune cells, especially neutrophils in blood. Future studies are required to determine if MGAM2 indeed plays a role in the immune system in placentals.
Among cancers investigated by TCGA, MGAM2 is expressed in GI cancers and breast cancer. Notably, in breast cancers, MGAM2 expression is highly specific to the BLBC subtype and, more importantly, is associated with better patient survival. Based on our differentially expressed gene and correlation studies, we propose that this better patient survival is due to a stronger host immune response elicited by the expression of MGAM2. Interestingly, MGAM2 expression is associated with the C2 (IFN-γ dominant) subtype, identified from pan cancer immune landscape analysis [18], and with macrophage regulation and TCR diversity. More studies are clearly required to explain these observations, including experiments to validate if MGAM2 expression indeed elicits immune response.
In BLBCs, MGAM2 is positively correlated in expression with CD47 and SIRPA, which constitutes an immune checkpoint that generates “don't eat me” signal for cancer cells [35] and is currently being targeted for cancer treatment [36]. CD47 is a ubiquitously expressed transmembrane protein marking the cell as “self” [35], while SIRPα is a cell surface receptor on phagocytic immune cells (e.g., macrophages). The binding of CD47 to SIRPA prevents phagocytosis [35], and blocking the CD47/SIRPA interaction results in tumor reduction in preclinical models [37,38]. Further studies are clearly required to understand the relationship between MGAM2 and the CD47/SIRPA checkpoint in BLBC, and to evaluate if MGAM2 expression provides any diagnostic value in the CD47/SIRPA blockade therapy. BLBC represents the worst subtype of breast cancer, lacking targeted therapies and with the lowest patient survival rate. Hence, these future studies are important.
4.2. Proteins with EHV Regions may Represent a New Class of Molecules Useful in Cancer Immunotherapy
In the human genome, CDSs are typically more conserved than introns or intergenic regions. Our pipeline reveals that about 1.4% of human CDSs, approximately 3000 in total and including the last CDS of MGAM2, are however opposite and EHV. This is likely due to their high content of simple repeats and SINEs [[39], [40], [41], [42]]. We have identified >1000 human proteins that are partially or entirely (very rare) encoded by these EHV CDSs, including MGAM2. Among them, 20% are zinc fingers or olfactory receptors, two families known to be fast-evolving. The EHV protein regions are more likely to be extracellular and threonine/serine/proline-rich, and harbor more cancer passenger mutations. Importantly, many of these EHV proteins function in immune and host defense response. Most studies have focused on conserved regions; as a result, EHV CDSs and proteins are poorly characterized [43]. More efforts [44] should be spent on understanding this unique class of genomic sites and proteins, particularly their roles in host immune response to pathogen infection and carcinogenesis.
Our preliminary TCGA pan cancer analysis (Fig. S8) indicates that EHV genes are associated with better patient survival in skin cancer, mesothelioma and colon cancer, but with worse survival in chromophobe renal cell carcinoma, acute myeloid leukemia, lower grade glioma and uveal melanoma. Interestingly, EHV genes are associated with better survival in the C4 (lymphocyte depleted) immune subtype, but with worse survival in the C3 (inflammatory) immune subtype. Furthermore, EHV genes are positively correlated with macrophage regulation, lymphocyte infiltration, IFN-γ response and TGF-β response across different cancer types. They are negatively correlated with cancer testis antigen score and Th2 cells. Future research is required to understand the significance of these observations.
Our study sheds some light on a subset of these EHV proteins that resemble MGAM2, being transmembrane and with EHV regions extracellular. These proteins, about 100 in total, include classical immune molecules such as HLA-A, HLA-B, HLA-C. Among 30 tissues investigated, their genes present a unique expression pattern in blood, with >90% of those that are highly expressed in blood functioning in immune system. We propose that EHV regions may be necessary for blood cells to quickly adapt to the dynamic pathogens that enter the host. Interestingly, these EHV genes tend to express higher in BLBC and associate with better patient survival, like MGAM2. Examples include HLA-DQB2 and HLA-DRB5, which encode MHC class II proteins. Both have EHV extracellular region, and are expressed in BLBC and associated with good patient survival. As immunotherapy becomes increasingly important in cancer treatment, understanding this unique class of molecules would clearly be important.
Author Contributions
SX performed all data analyses. SZ designed the study and wrote the original manuscript. All authors read and approved the final manuscript.
Disclosure of Conflicts of Interest
The authors declare no conflict of interest.
See also Table S4 and Fig. S4 (based on conservation of 100 species).
See also Table S5 and Fig. S5 (based on conservation of 100 species).
A: The catalytic mechanism of α-1,4-glycosidase of ntMGAM and ctMGAM requires one aspartic acid (D) as nucleophile and another D as proton donor. The two signature peptides are shown. The figure is modified from a published work [29].
B: Top panel indicates mutated residues at the two catalytic sites between MGAM and MGAM2. Bottom images indicate the overall agreement between nt/ct-MGAM (cystal structures) and nt/ct-MGAM2 (predicted structures) is excellent. Each catalytic site is enlarged to indicate overlap between mutant residues.
C: Substrate docking indicates ntMGAM2 and ctMGAM2 are inactive. The numbers in each image in top panel indicate the distances, in crystal structures for MGAM and through docking for MGAM2, between the nitrogen atom (blue) of α-1,4-glycosidic bond of acarbose (substrate) and the two catalytic D residues. Surface view of acarbose binding to MGAM (crystal structures) or MGAM2 (predicted structures), in bottom panel, indicates that the binding pockets of MGAM2 are altered.
See also Table S6 and Fig. S6.
Acknowledgements
We thank Drs. Kelley Moremen and Christopher Mark West for insightful discussion on this work. This work is funded by NCI R01 CA182093 to S. Zhao.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.03.008.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Supplementary material 6
Supplementary material 7
Supplementary material 8
Supplementary material 8
References
- 1.Prat A., Perou C.M. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Marotta L.L., Polyak K. Unraveling the complexity of basal-like breast cancer. Oncotarget. 2011;2(8):588–589. doi: 10.18632/oncotarget.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith O.L., Gray J.W. Omic approaches to preventing or managing metastatic breast cancer. Breast Cancer Res. 2011;13(6):230. doi: 10.1186/bcr2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soung Y.H., Pruitt K., Chung J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci Rep. 2014;4 doi: 10.1038/srep03846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A. Comprehensive molecular portraits of invasive lobular breast Cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F., Ren C., Zhao H., Yang L., Su F., Zhou M.M. Identification of novel prognostic indicators for triple-negative breast cancer patients through integrative analysis of cancer genomics data and protein interactome data. Oncotarget. 2016;7(44):71620–71634. doi: 10.18632/oncotarget.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badve S., Dabbs D.J., Schnitt S.J., Baehner F.L., Decker T., Eusebi V. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24(2):157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 10.Hudis C.A., Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16 doi: 10.1634/theoncologist.2011-S1-01. Suppl 1:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Nichols B.L., Eldering J., Avery S., Hahn D., Quaroni A., Sterchi E. Human small intestinal maltase-glucoamylase cDNA cloning. Homology to sucrase-isomaltase. J Biol Chem. 1998;273(5):3076–3081. doi: 10.1074/jbc.273.5.3076. [DOI] [PubMed] [Google Scholar]
- 12.Ren L., Cao X., Geng P., Bai F., Bai G. Study of the inhibition of two human maltase-glucoamylases catalytic domains by different alpha-glucosidase inhibitors. Carbohydr Res. 2011;346(17):2688–2692. doi: 10.1016/j.carres.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Rossi E.J., Sim L., Kuntz D.A., Hahn D., Johnston B.D., Ghavami A. Inhibition of recombinant human maltase glucoamylase by salacinol and derivatives. FEBS J. 2006;273(12):2673–2683. doi: 10.1111/j.1742-4658.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- 14.Sim L., Quezada-Calvillo R., Sterchi E.E., Nichols B.L., Rose D.R. Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J Mol Biol. 2008;375(3):782–792. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- 15.van de Laar F.A., Lucassen P.L., Akkermans R.P., van de Lisdonk E.H., Rutten G.E., van Weel C. Alpha-glucosidase inhibitors for patients with type 2 diabetes: results from a Cochrane systematic review and meta-analysis. Diabetes Care. 2005;28(1):154–163. doi: 10.2337/diacare.28.1.154. [DOI] [PubMed] [Google Scholar]
- 16.Scheen A.J. Is there a role for alpha-glucosidase inhibitors in the prevention of type 2 diabetes mellitus? Drugs. 2003;63(10):933–951. doi: 10.2165/00003495-200363100-00002. [DOI] [PubMed] [Google Scholar]
- 17.Naumov D.G. Structure and evolution of mammalian maltase-glucoamylase and sucrase-isomaltase genes. Mol Biol (Mosk) 2007;41(6):1056–1068. [PubMed] [Google Scholar]
- 18.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H. The immune landscape of Cancer. Immunity. 2018;48(4) doi: 10.1016/j.immuni.2018.03.023. 812–30 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium G.T. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Ren L.M., Qin X.H., Cao X.F., Wang L.L., Bai F., Bai G. Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell. 2011;2(10):827–836. doi: 10.1007/s13238-011-1105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q.Y. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 29.Chiba S. Molecular mechanism in alpha-glucosidase and glucoamylase. Biosci Biotechnol Biochem. 1997;61(8):1233–1239. doi: 10.1271/bbb.61.1233. [DOI] [PubMed] [Google Scholar]
- 30.Cao X.F., Zhang C., Dong Y.Y., Geng P., Bai F., Bai G. Modeling of cooked starch digestion process using recombinant human pancreatic alpha-amylase and maltase-glucoamylase for in vitro evaluation of alpha-glucosidase inhibitors. Carbohydr Res. 2015;414:15–21. doi: 10.1016/j.carres.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Ren L.M., Cao X.F., Geng P., Bai F., Bai G. Study of the inhibition of two human maltase-glucoamylases catalytic domains by different alpha-glucosidase inhibitors. Carbohydr Res. 2011;346(17):2688–2692. doi: 10.1016/j.carres.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Sim L., Jayakanthan K., Mohan S., Nasi R., Johnston B.D., Pinto B.M. New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata. Biochemistry. 2010;49(3):443–451. doi: 10.1021/bi9016457. [DOI] [PubMed] [Google Scholar]
- 33.Simsek M., Quezada-Calvillo R., Ferruzzi M.G., Nichols B.L., Hamaker B.R. Dietary phenolic compounds selectively inhibit the individual subunits of maltase-glucoamylase and sucrase-isomaltase with the potential of modulating glucose release. J Agric Food Chem. 2015;63(15):3873–3879. doi: 10.1021/jf505425d. [DOI] [PubMed] [Google Scholar]
- 34.Renfree M.B. Review: marsupials: placental mammals with a difference. Placenta. 2010;(31 Suppl):S21–S26. doi: 10.1016/j.placenta.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 35.McCracken M.N., Cha A.C., Weissman I.L. Molecular pathways: activating T cells after Cancer cell phagocytosis from blockade of CD47 "Don't eat me" signals. Clin Cancer Res. 2015;21(16):3597–3601. doi: 10.1158/1078-0432.CCR-14-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veillette A., Chen J. SIRPalpha-CD47 immune checkpoint blockade in anticancer therapy. Trends Immunol. 2018;39(3):173–184. doi: 10.1016/j.it.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Tseng D., Volkmer J.P., Willingham S.B., Contreras-Trujillo H., Fathman J.W., Fernhoff N.B. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc Natl Acad Sci U S A. 2013;110(27):11103–11108. doi: 10.1073/pnas.1305569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willingham S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109(17):6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12(12):236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 41.Liu G.E., Alkan C., Jiang L., Zhao S., Eichler E.E. Comparative analysis of Alu repeats in primate genomes. Genome Res. 2009;19(5):876–885. doi: 10.1101/gr.083972.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerdes P., Richardson S.R., Mager D.L., Faulkner G.J. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0965-5. [doi:ARTN 100 10.1186/s13059-016-0965-5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huddleston J., Eichler E.E. An incomplete understanding of human genetic variation. Genetics. 2016;202(4):1251–1254. doi: 10.1534/genetics.115.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantsilieris S., Nelson B.J., Huddleston J., Baker C., Harshman L., Penewit K. Recurrent structural variation, clustered sites of selection, and disease risk for the complement factor H (CFH) gene family. Proc Natl Acad Sci U S A. 2018;115(19):E4433–E4442. doi: 10.1073/pnas.1717600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Supplementary material 6
Supplementary material 7
Supplementary material 8