Abstract
Nitric oxide (NO) is an essential signaling molecule in the regulation of multiple cellular processes. It is endogenously synthesized by NO synthase (NOS) as the product of L-arginine oxidation to L-citrulline, requiring NADPH, molecular oxygen, and a pterin cofactor. Two NOS isoforms are constitutively present in cells, nNOS and eNOS, and a third is inducible (iNOS). Despite their biological relevance, the details of their complex structural features and reactivity mechanisms are still unclear. In this review, we summarized the contribution of computational biochemistry to research on NOS molecular mechanisms. We described in detail its use in studying aspects of structure, dynamics and reactivity. We also focus on the numerous outstanding questions in the field that could benefit from more extensive computational investigations.
Keywords: Nitric oxide synthase, computational methods, molecular mechanisms, redox regulation
Graphical Abstract
1. Introduction
Nitric oxide (NO) is a gaseous radical identified as a signaling molecule by Furchgott, Ignarro, and Murad, who were awarded the Nobel prize in Physiology and Medicine in 1998 for their pioneering findings on this small, highly diffusible molecule [1,2]. Before NO was identified and characterized, the molecule responsible for vasodilation was known as “endothelium-derived relaxing factor” [3,4] and many years passed until it became clear that this was NO [1,2,5]. The finding of its involvement in the cardiovascular system [6] was followed by the realization of its relevance in the context of immune response [7,8], neurotransmission [9], respiratory function [10], and a number of different other biological processes, establishing NO as a crucial physiologic regulator of cellular signaling.
Because of its highly diffusible nature and reactivity, NO is a prototypic cellular messenger, as it can signal in a dose-, time-, and organ-dependent manner by directly targeting cellular biomolecules such as proteins and nucleic acids. NO is generated by a class of nicotinamide adenine dinucleotide phosphate (NADPH)-dependent NO synthases (NOSs), from L-arginine and molecular oxygen (Fig. 1). Several cofactors are involved in the reaction, which allow the electron flow from NADPH to the heme group and molecular oxygen, through a molecule of flavin adenine dinucleotide (FAD), and then flavin mononucleotide (FMN). During catalysis, the tetrahydrobiopterin (BH4) cofactor provides an additional electron, which is replaced during the catalytic turnover [11].
Fig. 1.
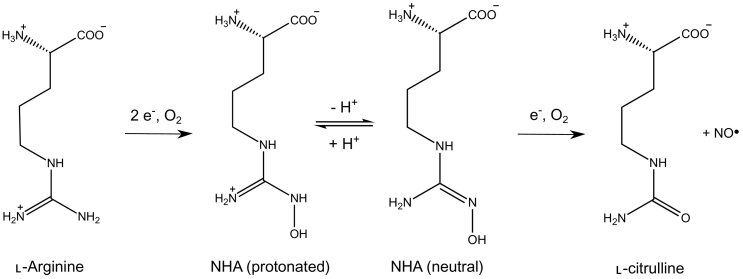
Two-step oxidation of L-arginine catalyzed by NOS. The NHA intermediate is depicted in its protonated (left) and neutral (right) states.
Three isoforms of NOS that require binding to calmodulin to be active have been identified so far in mammals [12,13]. Two are constitutively expressed (cNOSs), while the third is inducible (iNOS or NOS2). Constitutive NOSs comprise the neuronal NOS (nNOS or NOS1) and the endothelial NOS (eNOS or NOS3), both of which have calcium-dependent activity, whereas iNOS can produce NO efficiently and massively without calcium [14]. nNOS plays a pivotal role in the nervous system, as it is involved in synaptic plasticity for regulation of nerves tone [15]. It is also likely to be involved in long term potentiation, because of the neurotransmitter properties of the NO produced in the central nervous system [6,16]. nNOS also has many crucial functions also in skeletal muscle cells [17], where it localizes below the sarcolemma by interacting with dystrophin [18,19], and it regulates blood flow in muscle cells during exercise. Moreover, nNOS is associated with the cardiac sarcoplasmic reticulum, where it regulates myocardial contraction by exerting highly specific, localized NO production that acts on ion channels or transporters involved in calcium cycling [[20], [21], [22]]. eNOS is expressed mainly in vascular endothelium to guarantee vasorelaxation [23,24], cellular proliferation, white blood cells adhesion, and platelet aggregation [24,25]. cNOSs exert their functions functions mainly by producing low, controlled fluxes of NO, whereas iNOS acts mainly as a cytotoxic, antimicrobial enzyme, which is induced by stress and inflammatory conditions [26]. It has been reported that the activation of iNOS by pro-inflammatory cytokines (such as interleukin-1, tumor necrosis factor α, and interferon γ) results in massive production of NO and sustains host immunity as part of the oxidative burst of macrophages. Other immune cell types also respond to NO [7], resulting in an even wider role of iNOS in immunity. For example, it is well documented that iNOS-derived NO can activate T-cells [8] and interfere with lymphocyte development [27] and death [8,28]. iNOS has also been linked to cancer progression and development, but its role in cancer biology has not been fully elucidated, as both tumor promoting and inhibiting activities have been described [29,30].
Multiple molecular effects are induced by NO, ranging from signaling to irreversible modification or damage (Fig. 2), strongly affecting several physiological processes, indicating that NOS defects underlie many human disease conditions. Understanding the structure and mechanisms of action of NOSs is, therefore, fundamental to developing useful clinical interventions. In this review, we summarize the state-of-the-art information on the structure and reactivity of NOSs, with an emphasis on the contributions of computational biochemistry, molecular modeling, and simulations. Current knowledge and pending questions are discussed in the following sections, and perspectives for molecular modeling studies are proposed in the final section.
Fig. 2.
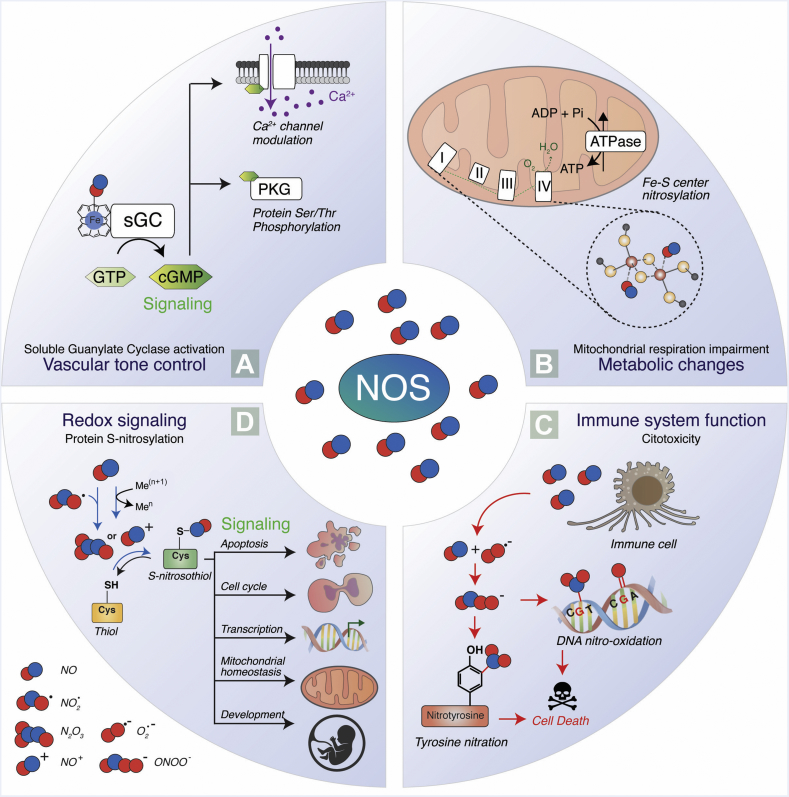
Schematic representation of NO-mediated signaling pathways. (A) NO induces the soluble guanylate cyclase (sGC) by binding its heme-group and stimulates the production of cyclic GMP (cGMP) [182]. cGMP production modulates calcium channels and activates the protein kinase G (PKG), leading to a downstream phosphorylation cascade that is important in muscle tone control. (B) Other central sensors of NO fluxes are mitochondria, which adjust the oxygen consumption rate and energy production according to NO levels. NO can affect the mitochondrial respiration rate by direct attachment to Fe-S centers or by the covalent binding to specific tyrosines (C) and cysteines (D) [[183], [184], [185]]. (C) Large amounts of NO produced by immune cells (e.g. macrophages) react with superoxide (O2·), generating the highly reactive peroxynitrite (ONOO-), which leads to protein tyrosine nitration, DNA nitro-oxidation, cell damage, and death. (D) The reaction between NO and nitrogen dioxide (NO2) or redox metals (e.g., Fe3+, Cu2+) generates dinitrogen trioxide (N2O3) or nitrosonium ion (NO+), respectively. Both these species can bind directly to cysteine residues (-SH) of proteins, forming S-nitrosothiols (-SNO). The reaction, termed S-nitrosylation, acts as a posttranslational modification that affects protein function, stability, localization and signaling. The processes in which protein S-nitrosylation plays a role include apoptosis, cell cycle, cell proliferation, gene transcription, mitochondrial homeostasis, and development [82,186,187].
2. NOS Structure
The NOS structure has two main domains. The N-terminal oxygenase domain (NOSox) harbors the heme porphyrin center (Figs. 3 and 4), that catalyzes oxidation of L-arginine to L-citrulline, requiring BH4 and resulting in the release of NO (Fig. 1), while the C-terminal reductase domain consists of three binding domains for FMN, FAD, and NADPH cofactors (Figs. 5 and 6). The latter domain provides electrons for the reaction, which takes place in the NOSox domain. These two domains are connected through a linker, which includes a calmodulin binding site that is essential for NOS activity. cNOSs binding to calmodulin is responsive to calcium levels, whereas this is not the case for iNOS, which binds calmodulin independently of calcium concentrations.
Fig. 3.
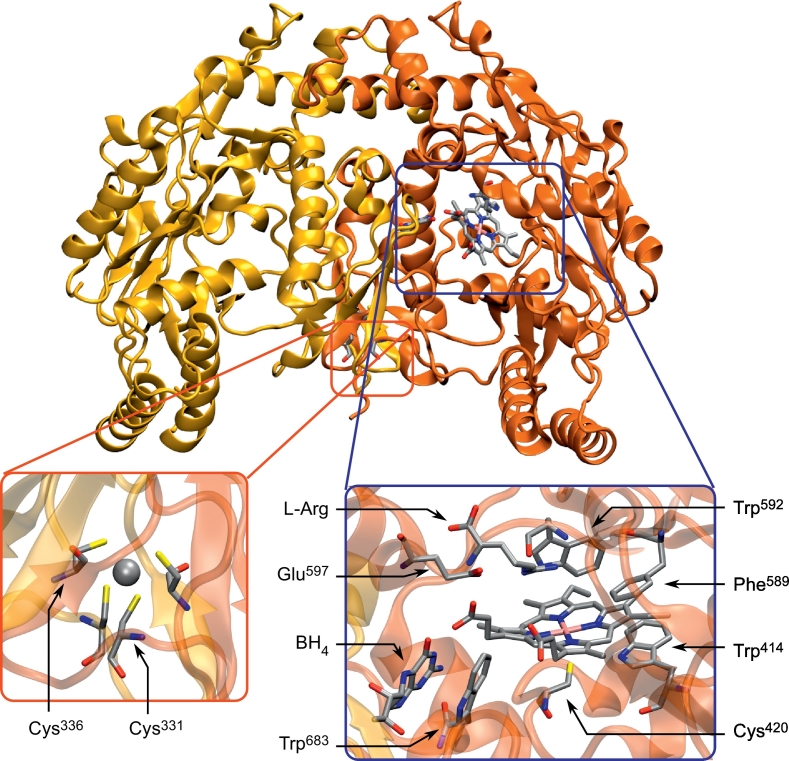
Structure of the human nNOSox dimer (PDB: 4D1N). Monomers are colored differently for sake of clarity. The left magnified section (red) shows the ZnS4 motif, with arrows pointing to the cysteines of the first monomer and the central zinc cation depicted in Van der Waals representation. The right magnified section (blue) provides a view of the active site, with the heme moiety in the center, coordinated to Cys420 on one side and dioxygen on the other. Interacting with the heme carboxylates, the pterin BH4 cofactor forms π-stacking interactions with Trp683. Glu597 stabilizes the substrate above the heme moiety, and the aromatic residues Phe589 and Trp414 sandwich the latter to maintain the proper organization of the active site. Trp592, presumably involved in the electron transfer from NOSred to NOSox, is located at the bottom of the heme-binding pocket. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Sequence alignment of the three human NOS isoforms generated with ClustalW [188] for the oxygenase domain, harboring heme-interacting residues (red squares), Zn-coordinated thiolates (green circles), BH4-interacting tryptophan and L-arginine-binding glutamate (blue circles), post-translationally modified cysteines (pink pentagons), and the tryptophan hypothesized to be involved in the electron transfer from FMN to the heme (black star). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
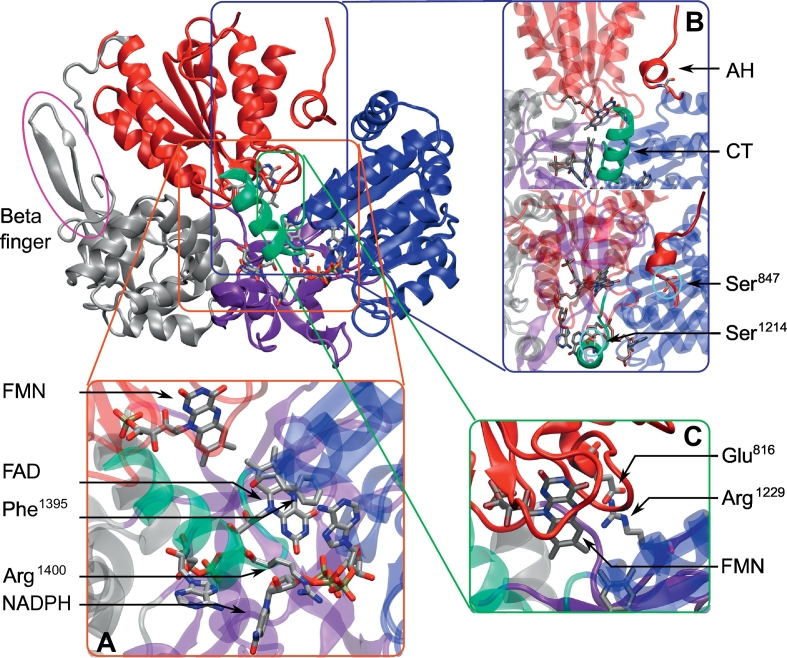
Structure of the rat nNOS reductase domain (PDB: 1TLL, chain A), with the NADPH domain in blue, the FAD domain in violet, the FMN domain in red, the α-helix hinge section in gray, the C-terminal tail in green, and the beta-finger in the pink circle on the left. The magnified section A (bottom left) is the active site of the reductase, revealing the placement of the cofactors within the structure, with Phe1395 stacking with the FAD flavin, and Arg1400 interacting with NADPH phosphates. Magnified section B (right) shows two regulatory elements: CT in green and the autoinhibitory segment of the FMN domain (AH) in red, front view (top) and top view (bottom). The two phosphorylated serine residues are also displayed. Magnified section C (bottom right) shows the conserved salt bridge linking the FMN and FAD domains. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Sequence alignment of the three human NOS isoforms generated with ClustalW [188] for the reductase domain, with calmodulin-binding in orange, FMN-binding in red, connecting regions in gray, FAD-binding in purple, NADPH-binding in blue, and the C-terminal tail in green. Phosphorylation sites are indicated by green squares, the phenylalanine shield and the NADPH-interacting arginine by red circles, aromatic residues that may be involved in electron transfer from FMN to heme by black stars, and the salt bridge between the FMN and FAD domains by light blue circles. The auto-inhibitory and beta-finger regions appear as dashed lines within the FMN and connecting domains, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
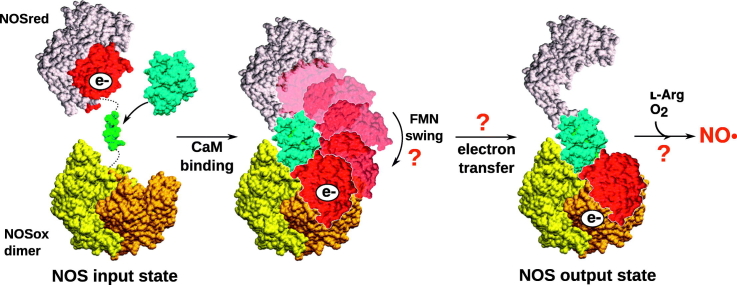
The electrons from the reductase domain are shuttled toward the oxygenase reactive site through long-range displacement of the FMN domain [31]. Although this ‘swing’ has been the subject of many studies, aspects of its mechanisms of regulation are not yet understood [[32], [33], [34], [35], [36], [37], [38], [39]]. The NOS isoforms are functional upon dimerization, and electron transfer is likely to occur in trans from one monomer to the other [40,41]; however, the details of this mechanism, such as the contact surface and the elements that stabilize the dimer interface, are still being investigated (Section 2.3). The three-dimensional (3D) structures of the full-length NOS variants are unknown, although numerous X-ray structures of the individual domains (Table 1) and models built from cryogenic electron microscopy (cryo-EM) densities and other experimental data are available [[32], [33], [34], [35], [36], [37],[42], [43], [44], [45]], so that the architecture of the full-length NOS can be hypothesized.
Table 1.
Available X-ray and NMR structures of the domains of each NOS isoform, along with the PMID associated to the corresponding publication in Pubmed and the corresponding entry in the Protein Data Bank (PDB). We did not report the structures of the heme domain in complex with inhibitors.
| Isoform | Organism | Domain structure | PDB ID | PMID |
|---|---|---|---|---|
| nNOS | Human | Heme | 4D1N | 25286850 |
| Rat | Heme | 4FVW, 3HSN, 2G6H, 1ZVI, 1LZX | 23586781, 19791770, 16804678, 16033258, 12437343 | |
| Reductase | 1TLL | 15208315 | ||
| FNR-like (FAD/NADPH) | 1F20 | 11473123 | ||
| iNOS | Human | Heme | 1NSI | 10409685 |
| calmodulin binding + calmodulin | 5TP6, 2LL6 | 28121131, 22486744 | ||
| FMN/calmodulin binding + calmodulin | 3HR4 | 19737939 | ||
| Mouse | Heme | 3NQS, 1DWV, 1DF1 | 20659888, 10769116, 10562539 | |
| Heme mutant | 3DWJ, 1JWJ | 18815130, 11669619 | ||
| eNOS | Human | Heme | 3NOS, 4D1O | 10074942, 25286850 |
| calmodulin binding + calmodulin | 2MG5, 2LL7, 2N8J, 1NIW | 24495081, 22486744, 27696828, 12574113 | ||
| Bovin | Heme | 1NSE, 2G6O, 1ED6 | 9875848, 16804678, 11331003 |
Computational studies would address several outstanding questions about the structural mechanisms of NOSs; however, only a few molecular dynamics simulations of NOSs have been reported, on very short timescales, no extensive structural analysis, and most as part of more comprehensive experimental studies. Other computational approaches, such as homology modeling and docking, have been used together with experiments to determine the quaternary structure of NOS [[32], [33], [34], [35], [36], [37],[43], [44], [45], [46]]. However, they suffer of major limitations in terms of accuracy and the capability to account for highly flexible and conformationally heterogeneous proteins. The interaction of NOSs with selective inhibitors has been reported [47,48] but there has been no extensive computational investigation of the free enzyme structure.
In this section, we summarized the current knowledge on NOS structural features from studies with computational methods and raise questions that would benefit from more extensive, accurate studies with molecular modeling and simulations.
2.1. The Oxygenase Domain
The NOSox is composed of a unique heme domain, harboring a heme-porphyrin catalytic center, a structural zinc tetrathiolate (ZnS4) motif, and the BH4 pterin cofactor. Numerous high-resolution structures of the three human isoforms of NOSox are available (Table 1), which provide considerable source of information for unraveling the structural features of this catalytic domain.
The geometry of the active site is highly conserved among NOS isoforms and mammalian species [[49], [50], [51]]. The heme iron is axially coordinated to a cysteine thiolate on one side (Cys420 in nNOS), and the other side of the porphyrin is the binding site for the dioxygen molecule required for L-arginine oxidation (Fig. 3). The heme insertion within the NOSox domain might be favored by the interaction of NOS with heat shock protein 90 (hsp90), by a structural deformation that allows access of heme to its binding cleft in the protein [52,53], concomitantly promoting NOS dimerization [54]. A conserved glutamate residue (Glu597 in nNOS) is likely to play an important role in substrate binding [51], as shown by experimental mutagenesis [55,56]. The pterin redox cofactor binds in the vicinity of the active site through interaction with the heme propionate groups, and this process is thought to promote the binding of L-arginine [57]. Conserved aromatic residues near the active site form stacking interactions with the porphyrin moiety (Trp414 and Phe589 in nNOS) and the pterin cofactor (Trp683 in nNOS), which is involved in an extensive hydrogen-bond network. This network is likely to promote the stabilization of the NOSox dimer interface and the binding of L-arginine to the enzyme [49]. An important tryptophan residue located at the back of the heme pocket (Trp592 in nNOS) has been proposed to shuttle the electron from the FMN cofactor to heme (Section 2.3).
Another element that favors dimerization is the zinc tetrathiolate (ZnS4) motif. Indeed, the zinc ion is tetra-coordinated with two thiolates (Cys331 and Cys336 in nNOS) from each monomer, contributing to the maintainance of the architecture of NOS by bridging the two NOSox domains (Fig. 3). The absence of this cation or the modification of one of the coordinated cysteines drastically destabilizes the dimer and thus reduces the NOS catalytic activity [[58], [59], [60]].
Surprisingly, no computational studies have been carried out so far on the dynamic properties of NOSox and the catalytic site with NOS in the closed state. To the best of our knowledge, the only study with MD simulation of isolated NOSox involved a very short trajectory below the nanosecond timescale. This 300-ps MD simulation was performed within a reactivity study of iNOS active site with fixed heme, dioxygen, coordinated cysteine, and surrounding water molecules [61]. The simulations were performed with Turbomole/Jaguar, coupled to DL-POLY. The QM and MM simulations were treated at the B3LYP/LACV3P*+ level of theory and with CHARMM potentials, respectively. Electronic embedding was applied. Given the very short timescale, this study did not allow a proper assessment of the dynamic behavior of the NOSox domain, which would have required longer, unconstrained MD simulations. In another study, the interaction of NOSox with caveolin-1 was studied by docking and short MD simulations (10ns) with a CHARMM force field, and accompanied by experimental investigations [62]. Caveolin specifically decreases eNOS activity by hindering calmodulin binding and further activation [63,64]. The MD simulations were performed to assess the stability of the docking poses and were too short to elucidate the dynamics of the complex between NOSox and caveolin-1. Furthermore, few details were given about the simulation, so that the data would be difficult to reproduce. Their results suggest that caveolin-1 prevents binding of the BH4 cofactor by interacting with the eNOS Trp447 (equivalent to Trp683 in nNOS). However, given the lack of technical details and the short timescale, this hypothesis must be confirmed in studies of the competitive binding of caveolin-1 to eNOS against BH4.
Designing effective isoform-selective inhibitors is important for targeted therapies, and modeling and simulations could be valuable in this context. Docking, Quantitative-Structure-Activity Relations (QSAR), and GRID/Consensus Principal Component Analysis (CPCA) approaches have been used to probe the chemical environment within the heme binding site and the ligand-host interactions [48,65], sometimes coupled with homology modeling, MM-Poisson-Boltzmann (Implicit Solvent Model) Surface Area calculations, thermodynamic integration, and short MD simulations [[66], [67], [68], [69], [70], [71], [72]]. These computational studies shed light on the subtle electrostatic differences among the active sites of the NOS isoforms, opening new avenues for enhanced selective inhibitor design. Many questions remain, however, about NOSox structure and regulation. Numerous experimental data are available on the structure of the NOS heme domain in the closed state, but several aspects remain to be elucidated. For comparison with a homolog, extensive in silico studies (e.g., microsecond all-atom and coarse-grain MD, docking) have been reported on P450 enzymes [[73], [74], [75], [76], [77], [78]], including the investigation of the structure of the active site, substrate tunneling and binding modes, mutational effects, and interaction with membranes. These show the usefulness of molecular modeling in a context similar to NOSox. Use of computational methods with regard to NOSox has essentially focused on the design of selective inhibitors. However, as for P450 enzymes, longer timescale MD investigations with unbiased simulations or enhanced sampling approaches could provide important insight into NOSox domain structural behavior, its interaction with other proteins (including Hsp90 and caveolin [79]), the structural effect of post-translational modifications (PTMs), such as S-nitrosylation/S-sulfhydration of Cys441 [[80], [81], [82]] and S-glutathionylation of Cys382 in eNOS [81], and into the role of BH4 and ZnS4 in the stabilization of the dimer interface.
2.2. The Reductase Domain
The NOS reductase domain (NOSred) has three different sub-domains, namely NADPH, FAD, and FMN, each of which binds a specific cofactor for electron transfer (Fig. 5). This organization is similar to that of the cytochrome P450 reductase protein (CPR), which shares 60% sequence homology with NOSred and catalyzes analog electron transfer from NADPH to P450 reactive site [83].
The NADPH and FAD domains assemble to form the ‘FNR-like’ unit, while the individual FMN part is thought to serve as an electron shuttle toward the heme center, as observed in CPR [84,85]. An α-helix hinge section connects the FMN and the FAD sub-domains, ensuring proper alignment of the two flavins in a position to promote electron transfer. The electron goes from NADPH to FAD then to FMN. Upon activation, the FMN domain undergoes large-scale movements to dock on the NOSox domain and terminate electron transfer toward the heme center. This phenomenon occurs in trans, from the reductase domain of one monomer to the NOSox of the other monomer, and is triggered by binding of calmodulin [40,41,86,87].
Several NOSred elements have been suggested to respond to calmodulin binding and regulate FMN domain, unlocking from NADPH/FAD domains in an isoform-dependent manner. First, a conserved auto-regulatory segment is present in cNOSs FMN-binding domain, whose N-terminal α-helix shares sequence similarity with the calmodulin-binding motif [88]. This segment harbors a phosphorylation site at Ser847, suggesting an additional layer of post-translational regulation. Second, the C-terminal tail, the length of which differs among the three NOS isoforms, has been suggested to modulate FAD-FMN interactions in synergy with the auto-regulatory sequence and calmodulin binding [89,90]. The C-terminal tail of cNOS includes the Ser1412 phosphorylation site, which is also likely to play a role in regulating NOS activity. Third, the beta-finger (a small insertion encompassing the so-called CD2A loop) present in the flexible hinge region may play an important role in the closed-to-open state switch, presumably by modulating interactions with the FMN domain, especially in eNOS [[91], [92], [93]]. The subtleties of such regulatory mechanisms at the atomic level are still poorly understood.
Only one structure of the isolated rat nNOS FAD/NADPH-binding domain and another of the human iNOS FMN- and calmodulin-binding domains bound to calmodulin are available [43,94], as shown in Table 1. Nevertheless, the crystal structure of the rat nNOS reductase resolved by Garcin et al. [32] provided unprecedented insight into the structural features and organization of this domain. This was the only successful attempt to crystallize the three subdomains of a NOS reductase domain with the NADPH and flavins cofactors in their respective binding pockets. The structure revealed the extensive network of hydrogen bonds between the three cofactors and the protonation state of the FMN, which appeared as a semiquinone (Fig. 7). In addition, inspection of the FMN/FAD domains interface revealed important hydrophobic contacts and salt bridges, especially between the Glu816 and Arg1229 residues, both conserved in cNOSs. Arg1400, present in both nNOS and eNOS (Arg1165), was suggested to play a role in the selective binding of NADPH, and in locking of FMN in its electron-acceptor state in the absence of calmodulin. The FAD-shielding residue Phe1395 is thought to be involved in the repression of the electron transfer (Fig. 5) in the calmodulin-free state by acting as an aromatic shield between NADPH and FAD [95], as observed in the crystal structure [32].
Fig. 7.
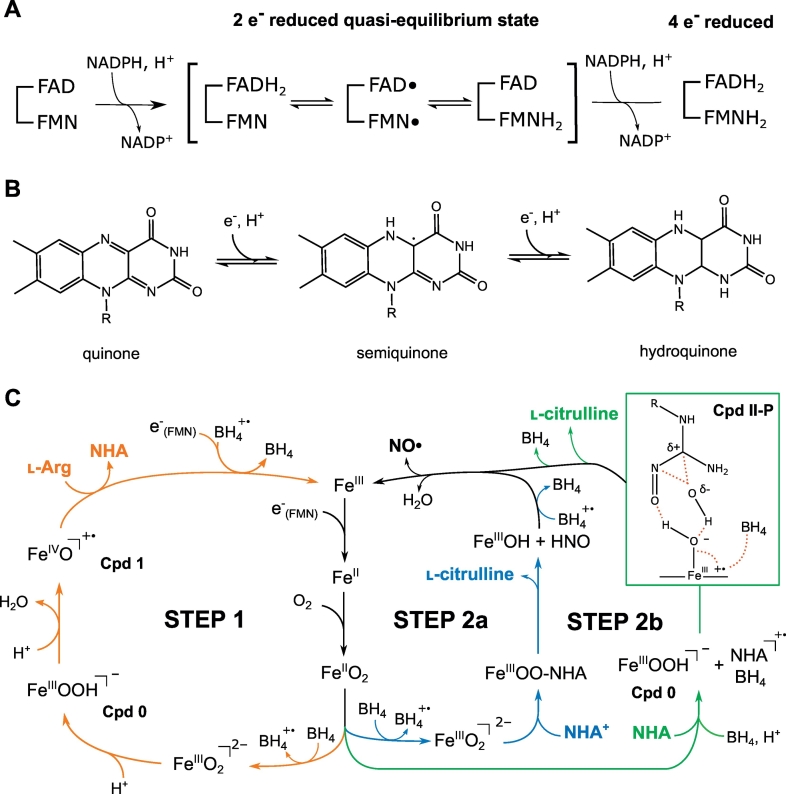
Schematic representation of NOS reactivity. a) Electron and proton transfers in the reductase domain leading to the four-electrons reduced state through two electrons reduced to the quasi-equilibrium state. b) Three different oxidation states of the flavin moiety (present in FAD and FMN). c) Mechanism of L-arginine oxidation: first half-reaction on the left (step1, orange path), second half-reaction on the right, with NHA protonated as reviewed by Santolini et al. [118] (step2a, blue path), and NHA neutral as proposed by Shamovsky et al. [117] (step2b, green path) involving formation of the Cpd II-P intermediate with extensive spin delocalization depicted by red dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The functionality of the dimeric form of nNOSred observed in this crystal is, however, a matter of controversy, as the dimer interface is presumably between the NOSox domains rather than the NOSred in the three isoforms, as suggested by recent models guided by cryo-EM data [33,36,37,46]. Likewise, experimental studies show that iNOS dimer is still functional even in the absence of the reductase domain of one monomer [40], indicating that the essential dimeric interface is localized between the two oxygenase domains. Modeling of the nNOS open-state structure from other EM data suggests that interactions between reductase domains might have a stabilizing effect [35]. This remains compatible with the assumptions mentioned above, as the main dimer interface is still found between the oxygenase domains. Dimerization of the reductase domain of the homologous CYP102A1 system has also been suggested [96].
The chronology of the NADPH and calmodulin binding events is still under discussion. Volkmann et al. proposed that binding of NADPH on the eNOS-calmodulin complex triggers electron transfer [37], whereas it is more commonly considered that NADPH binds calmodulin-free nNOS to lock the FMN domain in its electron-donor state until activation upon subsequent calmodulin binding [97,98].
Few studies involving computational biochemistry to investigate NOSred structure have been reported. Homology modeling and protein-protein docking methods have been used to build models from cryo-EM data (Section 2.3), and only one study reported MD simulations on eNOSred, providing a rationale for the effects of phosphorylation on NOS structure on the basis of the analyses of short trajectories [99]. In this study, Devika et al. performed 40-ns MD simulations using the GROMOS96 43a1 force field on the human structure of eNOSred built by sequence homology. The structure was rebuilt with Modeller and structures of homologs as templates (i.e., the rat nNOSred with PDB entry 1TLL, the rat FMN-like unit with PDB entry 1F20, and the human FMN and calmodulin-binding domains in complex with calmodulin with PDB entry 3HR4). Although the results provide insight into mutational effects on the structure of eNOS reductase, MD simulations with homology model as a starting structure should be considered with extreme caution, as they can result in large deviations from the native structure [100].
Molecular modeling has been of particular interest for studying structural features of the homologous CPR system, and a number of computational studies have been reported [[101], [102], [103]]. Thus, extensive unbiased and enhanced sampling MD simulations could provide additional insight, at the atomic level, into the numerous questions about the inner structural organization of NOSred, such as: i) What are the mechanisms of NOSred activity modulation by the regulatory elements? ii) Which structural changes are induced by PTMs in this domain (including phosphorylation of several serines in cNOSs, phosphorylation of Thr495 and S-glutathionylation of Cys689 and Cys908 in eNOS [81,[104], [105], [106]])? iii) Are the regulatory mechanisms of the NOS isoforms different? iv) Which amino acids are involved in the regulatory events, ligand binding, and interaction among the sub-domains? v) How does Arg1400 discriminate between NADPH and NADH? vi) How does NADPH and calmodulin binding affect the structure and dynamics of the reductase domain? Computational studies could provide new insight into the conformational changes of NOSred triggered by its multiple regulatory elements, which would be of utmost importance for understanding the complex NOSred regulation.
2.3. Inter-Domain Interactions
The full-length structure of NOS remains elusive in X-ray crystallography because of the flexibility of the NOSred domain. The structural and dynamic processes that drive inter-domain electron transfer and the regulatory effects of calmodulin binding are key to understand the molecular mechanisms underlying NOS activity, and have been investigated with structural biology methods.
As high-resolution structural data are limited to the isolated domains, homology modeling, protein-protein docking, and MD simulations have been used to characterize the sites of interaction between the FMN domain, calmodulin, and NOSox/NOSred. Cryo-EM techniques, combined with homology modeling and/or protein-protein docking, have been used to identify the architecture of holo-NOS and the changes of FMN from the closed to the open state [[35], [36], [37],46]. Other full-length NOS models have been built with modeling approaches, directly from the X-ray structures [32,34,43], and by integration of experimental data from hydrogen-deuterium exchange mass spectrometry [33] or electron paramagnetic resonance [44,45].
Overall, modeling of the multi-domain NOS structure indicates that binding of calmodulin might regulate the swing of the FMN domain from the reductase to the oxygenase domain, thus modulating electron shuttling between the two redox partners. The strong mobility of the NOSred domain with respect to the NOSox dimer has been highlighted with the heme domain acting as an anchor within the full-length architecture. The three NOS isoforms can adopt diverse intermediate conformations, ranging from the closed to the open state. The binding of calmodulin is likely to constrain the movement of the FMN-binding domain toward the output conformation [36].
The existence of a rotational pivot has been proposed, which would guide the FMN domain toward the appropriate binding area on the heme domain by restricting the conformational space accessible along the swing from NOSred to NOSox [37,107,108]. The work of Ilagan et al. [45], who reported a full-length model of rat nNOS (constructed manually), suggested that the FMN subdomain has higher affinity for the FNR-like unit than for the heme domain. In their study, calmodulin binding induced the conformational changes required to destabilize this balance and favored the open state, without increasing the binding affinity of FMN to the heme domain. Persechini et al. [46] suggested that activation of eNOS by calmodulin binding is a two-step process. First, unhooking of the FMN domain from the FNR-like unit would be triggered by interactions of calmodulin with the numerous NOSred regulatory elements. Second, calmodulin would dock onto NOSox and facilitate the docking of the FMN domain itself. Unfortunately, there is no consensus on the exact activation mechanisms after calmodulin binding, and the hypothesis that it might be different from different isoforms cannot be ruled out.
The nNOSred crystal structure published by Garcin et al. [32,34,43] brought unprecedented insight into the contact area of the FMN sub-domain with the FNR-like unit (Section 2.2). Despite the many investigations on the subject, however, the FMN/NOSox domains interface is not well described. Full-length NOS models designed from cryo-EM densities provide information about the possible docking area of the FMN domain onto NOSox, although detailed mapping of the interface contacts is not available.
Recently, Hollingsworth et al. [109] published a MD study of the iNOSox-FMN-calmodulin complex, and confirmed the role of intermolecular salt bridges. Their model system, encompassing the iNOSox dimer and the FMN and calmodulin-binding domains in the iNOS output state (FMN domain in its electron-donor state), was built by manual docking, guided by HDX-MS data [33]. 100-ns MD simulations with restraints on the iron atom of the heme were conducted in the model system with and without calmodulin, using the CHARMM22 force field with additional parameters generated by the same group. The results suggest that the FMN/heme interface is stabilized by salt bridges. The stability of this complex appeared to be enhanced by electrostaticinteractions of the two domains with the bound calmodulin. Hence, calmodulin binding stabilizes the inter-domain interactions, and its absence leads to disruption of the interface contacts after 20-ns MD simulation.
Sheng et al. [108] investigated the interactions of the iNOSox/FMN domains with 60-ns MD and steered MD simulations and found specific residues that are important for the efficient binding of the FMN domain to the NOSox docking surface. Their model was built with the same X-ray structures as that of Hollingsworth et al., but they docked the two domains (FMN and NOSox) with ZDOCK [110]. The generated structures were then filtered according to criteria obtained experimentally. The linker region was constructed with the Scigress Explorer Ultra platform, and 60-ns MD simulations for different oxidation states of the system (i.e., before and after electron transfer of FMN to heme) were performed using the CHARMM27 force field and in-house parameters for the heme, FMN, and BH4 cofactors. The results suggest that conformation changes leading to the swing of the FMN domain after the electron transfer are redox-dependent. The inter-domain interaction network they report differs from that observed by Hollingsworth et al. [109], with many more interactions between the FMN and the two NOSox monomers. The distance between the FMN cofactor and the heme was found at about 18 Å by Hollingworth et al. [109], bridged by a conserved tryptophan residue that has often been proposed to shuttle the electron between the two moieties [33,34,43]. Sheng et al. reported that the Trp372 (equivalent to Trp592 in nNOS) center of mass remained around 11.7 Å from FMN and 9.4 Å of the iron atom. This result differs from that found for the cytochrome P450 structure, in which the FMN-heme distance is only 6 Å, suggesting direct transfer from the cofactor to the heme center [85]. The simulations by Sheng et al. [108] suggest that, in some conformations induced by the dynamics of the NOS system, electron transfer involves not only the conserved tryptophan, but also shuttling through Tyr631 or Phe593, both located near the FMN cofactor in the FMN sub-domain. This hypothesis is supported by experimental studies that show a drastic decrease in the rate constant of intermolecular electron transfer after mutation of Tyr631 to Phe, which would disrupt important interactions [111]. The differences between the two MD studies above might be due to the use of different model structures of the iNOSox-FMN-calmodulin complex in the simulations. Unfortunately, lack of experimental data obviate a conclusion, highlighting the importance of finding highly accurate models by multiple cross-validation with experimental data and a more extensive sampling of the conformational space in the simulations.
Enhanced sampling MD approaches would make it possible to observe structural rearrangements over a longer time, which is necessary because the NOS system is prone to large conformational changes promoted by different allosteric effects. Such simulations would also be important to confirm the long-term stability of the key interactions identified in the short MD trajectories and other structural studies published so far. It could provide insight into the role of calmodulin binding in the activation of the different NOS isoforms and the effects of PTMs, which might be isoform-dependent.
The complexity of the NOS architecture, a homodimer of up to 330 kDa with several co-factors, and regulatory events (PTMs, allosteric effects) makes it a challenging system to investigate with molecular modeling and simulations. Nevertheless, force field parameters are available for most of the cofactors and ligands, and numerous structures of the isolated domains have been deposited in the Protein Data Bank (Table 1). Several in silico investigations of the structure of the homologous P450/CPR system have been published, providing a framework for investigation of the full-length NOS structure [101,112]. The scientific community would gain much from further use of computational methods, which would allow visualization of the NOS full-length structure and details on its dynamics at the atom level, which is essential for answering numerous outstanding questions.
3. NOS Reactivity
NOS catalyzes the oxidation of L-arginine to L-citrulline and NO in a two-step mechanism involving dioxygen, the BH4 cofactor, and electron transfers from NOSred (Fig. 1). The production of electrons in the reductase domain is similar to that observed in P450 reductase, and the mechanisms are well described [97,113,114]. First, the binding of NADPH to NOS allows transfer of a first hydride to the FAD cofactor. Then, electron transfer from FAD to FMN and proton addition lead to a quasi-equilibrium state between the two-electron-reduced species [FADH2/FMN ↔ FADH·/FMNH· ↔ FAD/FMNH2] until transfer of a second hydride from NADPH, which results in the four-electron reduced FADH2/FMNH2 state [115], as illustrated in Fig. 7. Complete flavin reduction, which is unlikely to take place physiologically, is gated by the release of NADP+ needed for de integro binding of NADPH and hydride transfer toward the final stable two-electron reduced state of FMN, required for the transfer toward the heme [41,116]. Upon calmodulin binding, the FMN subdomain can swing and dock onto NOSox, where it delivers the electron to the iron center, triggering two-steps L-arginine oxidation. The first mono-oxygenation reaction (first half-reaction) results in the formation of the stable Nω-hydroxy-L-arginine (NHA) intermediate, whose subsequent oxidation (second half-reaction) leads to the production of L-citrulline and NO (Fig. 7). Although NOS has many similarities to the CPR/P450 system [83], the reaction mechanisms in NOS are more complicated, as oxidation implies participation of the BH4 cofactor in electron transfer. Although the mechanisms of NOS reactivity are still mainly hypothetical, recent evidence of P450-like reactivity suggests that it involves distinct heme-oxy species [117]. Considerable research has been conducted on the role of the pterin cofactor, the protonation state of the NHA intermediate, and the heme-oxy species formed during the two steps, as they are essential for understanding the mechanisms of NOS reactivity [11,97,[118], [119], [120], [121]]. Although several computational studies (QM and QM/MM calculations) have been reported, many aspects of NOS reaction mechanisms remain hypothetical, especially with regard to the second half-reaction. Below, we summarized the latest findings on L-arginine oxidation mechanisms, in which computational studies have been of major importance.
3.1. First Half-Reaction
O2 activation in NOS is catalyzed by the heme moiety (Fig. 7) in a manner similar to that for cytochromes P450. Electron transfer from FMN switches the iron from the ferric FeIII to the ferrous FeII state, stimulating coordination of the dioxygen molecule to the heme center [118]. After binding of O2, a second electron transfer induces formation of the ferric-peroxide FeIIIO22- complex, which is required for the launching L-arginine oxidation [122]. Several hypotheses have been proposed for the mechanisms that lead to formation of the so-called compound I (Cpd I) cation-radicaloid porphyrin ferryl-oxo FeIVO+· species, which oxidizes the substrate [61,118]. The commonly preferred hypothesis is a mechanism involving two proton transfers (one from the solvent and the other from the L-arginine substrate) and formation of FeIIIOOH species [118,119], as supported by theoretical QM and QM/MM studies [[123], [124], [125]]. The first proton transfer would lead to the formation of the so-called compound 0 (Cpd 0) anionic ferric-hydroperoxo FeIIIOOH, and the second H+ addition would result in the generation of Cpd I and a water molecule by heterolytic cleavage of the peroxide bond. The latter species then oxidizes the deprotonated substrate, leading to formation of the stable NHA intermediate and FeIII. Density functional theory (DFT) calculations performed on a model of bovine eNOSox active site published by de Visser et al. [124] suggest that the latter process involves formation of the unstable ferryl FeIVO- state (Cpd II). The initial geometry, extracted from the PDB file 4NSE, involved the truncated heme and coordinated cysteine, L-arginine, and surrounding amino acids (Glu363, Trp358, Trp359). Geometry optimizations were performed with Jaguar at the B3LYP/LACVP and 6-311+G* levels of theory for the iron and the other atoms, respectively. It is well accepted that the second electron transfer required for O2 activation is provided by the BH4 cofactor. The pterin moiety would allow fast electron transfer toward the FeIIO2 intermediate, stimulating the L-arginine oxidation and preventing uncoupling from the NADPH oxidation cycle [126,127]. The latter process is involved in numerous diseases, including vascular diseases and cancer, and is therefore of major therapeutic interest [25,82,[128], [129], [130]]. It depends on availability of L-arginine and/or BH4, disruption of the NOSox dimer, and PTMs and results in FeIIO2 decay to the release of superoxide anion-radical O2·- rather than NO [[131], [132], [133], [134], [135], [136]]. Consequently, BH4 is a key player in NOS reactivity, as it ensures the bio-availability of NO and prevents oxidative stress [57,127,130]. Many studies, including DFT investigations, suggest that the pterin cofactor stays bound at the active site in its radical cationic state H4B+·, to be re-used during the second half-reaction [120,[137], [138], [139]].
3.2. Second Half-Reaction
Continuation of NO· catalysis requires prior reduction of H4B+· back to BH4 to prevent its further time-dependent oxidation to BH2 and NOS uncoupling [140,141]. Without it, the NOSox active site does not have the appropriate chemistry for reduction of the iron center from FeIII to FeII, which is necessary for the O2 binding that initiates oxidation of NHA [142,143]. Experimental evidence suggests that the electron is provided by NOSred in a calmodulin-dependent manner, similarly to the iron center reduction, and is transferred from FMN through the heme moiety [120,144]. Whether the electron is transferred to the metallic center or shuttled towards the pterin radical may depend on the redox state of BH4, and hence on the NOS catalytic cycle stage. Theoretical investigations could provide insight to probe this hypothesis. Once BH4 is reduced back to its neutral form, the second half-reaction can take place.
Unlike the first step, the second half-reaction differs from other known enzymatic mechanisms and is poorly understood. Nevertheless, numerous experimental and theoretical investigations suggest possible reaction intermediates for the formation of NO· [118]. As in the first step, FeIII is reduced by electron transfers from NOSred and BH4 to allow O2 activation. Various subsequent mechanisms have been proposed. In most, NHA is considered to be in its protonated form [119,125,145,146]. Oxidation to L-citrulline would then involve a proton transfer from NHA to the ferric-peroxide FeIIIO22- species after formation of a tetrahedral complex that binds the guanidinium moiety to the peroxide. This would induce the release of HNO, L-citrulline, and FeIIIOH. Other possibilities, reviewed by Santolini et al. [118], would involve the transfer of an electron back to H4B+·, terminating the BH4 cycle concomitantly with the binding of the newly formed NO· to the iron center and release of water (Fig. 7). DFT and QM/MM investigations of this hypothesis did not allow theoretical validation but suggested an alternative mechanism, involving double protonation of the active site to formation of an FeIIIHOOH intermediate [146,147], which might eventually lead to the ferryl-oxo FeIVO+· compound (Cpd I) [145]. Hence, no consensus has yet been found on the NOS second half-reaction mechanism.
Recent computational investigations, however, represent new means for understanding the complex chemisty of NOS. Although, it has been accepted that NHA reacts in its protonated state [57,[148], [149], [150]], Shamovsky et al. [117] proposed a new mechanism featuring neutral NHA (Fig. 7). They performed extensive DFT investigations on the active site of the murine iNOSox structure (PDB 1NOD) at the M06-L/6-31+G* level of theory and extracted the coordinates of the heme, NHA, BH4, and 11 surrounding amino acids from the crystal structure to build the initial geometry. Constraints were applied to the heme center during geometry optimization to maintain its location in the crystal structure. Overall, their calculations suggest the participation of BH4 in a sequence of electron transfers coupled with a proton transfer from the heme propionate and involvement of a protonated Cpd II intermediate (Cpd II-P) instead of Cpd I (Fig. 7). They propose that this mechanism would explain several experimental observations and especially the kinetics of the reaction and the elusive character of some intermediates. More details can be found in their elegant publication [117]. More data are now required to validate or refute this promising hypothesis.
Many aspects of NOS reactivity remain to be elucidated, but the work done so far gives encouraging results for further investigations. QM and QM/MM methods have proved their value in this context, as for their application to the P450/CPR system [[151], [152], [153], [154]]. More extensive use of computational approaches could shed light on NOS chemistry aspects, including: (i) the residues involved in electron transfer from NOSred to NOSox in the different NOS isoforms; (ii) how the electrostatic properties of the active site drive the dual chemistry of NOSox; (iii) rationalization of the different reaction yields found for the different isoforms; and (iv) the chemical features of the NHA intermediate and its stability at the active site.
4. Summary and Outlook
NOS is a highly complex chemical system with several layers of regulation, involving numerous players that mediate the interactions between the reductase and the oxygenase domains, both of which are involved in the enzymatic reactivity. The two-step oxidation of L-arginine necessitates a finely tuned organization of the active site, without which uncoupling from NADPH and production of ROS rather than NO· can occur.
Many studies have been conducted to unravel NOS molecular mechanisms, including the usage of computational methods, especially docking and QSAR to study ligand binding, and, to a lesser extent, QM calculations to probe oxygenase reactivity. Nevertheless, the full potential of molecular modeling is underexploited. Computational approaches could provide important insight on NOS structure and reactivity at the atom level. Homology modeling and docking have already provided the models of individual domains and of the holo-enzyme. Coarse-grained force fields could be considered for initial exploration of the dynamic behavior of the holo-enzyme and to identify possible allosteric effects to guide more accurate all-atom approaches. Computational tools are now available to treat systems, including intrinsically disordered regions, by coarse graining, which would be necessary to model the flexible linkers between NOS domains [155]. Furthermore, extensive MD simulations could provide information on the dynamic behavior of isolated domains, long range reorganization induced by PTMs, and allosteric effects [[156], [157], [158], [159], [160], [161]]. The integration of MD-derived ensembles with the protein structure network paradigm could also help in predicting structural pathways of communication as the basis of allosteric mechanisms [[162], [163], [164], [165], [166]]. Methods of enhanced sampling MD would be adequate for studying the allosteric effects triggered from distal sites to the inter-domain interface [159,[167], [168], [169], [170]]. In view of the growing amounts of EM data, the application of integrative structural modeling and EM density maps could also provide important information about NOS structural and dynamic features [[171], [172], [173]]. Conceptual DFT calculations and topological analysis of electron density, which have been applied to P450 and other enzymes [174,175], could provide unprecedented insight into the chemical properties of NOSox active site during oxidation [[176], [177], [178]]. Use of QM/MM and DFTB-QM/MM metadynamics might also be considered for studying electron transfer from the reductase to the oxygenase domain and identifying the key players involved in this process [[179], [180], [181]]. Overall, there is a wide range of possibilities for the study of NOS molecular mechanisms with molecular modeling and simulations, opening perspectives that could lead to important breakthroughs on this topic.
Declarations of interest
None
Acknowledgements
This work has been supported by the Alfred Benzon Principal Investigator Fellowship to EP group, along with grants from the Danish Cancer Society (R146-A9414); the Italian Association for Cancer Research, (AIRC IG2017-20719) to GF group. Both CBL and ROS groups are part of the Center of Excellence in Autophagy, Recycling and Disease (CARD), funded by the Danish National Research Foundation (DNRF125).
Contributor Information
Emmanuelle Bignon, Email: embi@cancer.dk.
Elena Papaleo, Email: elenap@cancer.dk.
References
- 1.Ignarro L.J., Buga G.M., Wood K.S., Byrns R.E., Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc Natl Acad Sci U S A. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer R.M., Ferrige A.G., Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 3.Griffith TM, Edwards DH, Lewis MJ, Newby AC, Henderson AH. The nature of endothelium-derived vascular relaxant factor. Nature n.d.;308:645–7. [DOI] [PubMed]
- 4.Rapoport RM, Draznin MB, Murad F. Endothelium-dependent relaxation in rat aorta may be mediated through cyclic GMP-dependent protein phosphorylation. Nature n.d.;306:174–6. [DOI] [PubMed]
- 5.Knowles R.G., Moncada S. Nitric oxide as a signal in blood vessels. Trends Biochem Sci. 1992;17:399–402. doi: 10.1016/0968-0004(92)90008-w. [DOI] [PubMed] [Google Scholar]
- 6.Hubalek M., Ramoni A., Mueller-Holzner E., Marth C. Malignant mixed mesodermal tumor after tamoxifen therapy for breast cancer. Gynecol Oncol. 2004;95:264–266. doi: 10.1016/j.ygyno.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 7.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 8.García-Ortiz A., Serrador J.M. Nitric oxide signaling in T cell-mediated immunity. Trends Mol Med. 2018;24:412–427. doi: 10.1016/j.molmed.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Steinert J.R., Robinson S.W., Tong H., Haustein M.D., Kopp-Scheinpflug C., Forsythe I.D. Nitric oxide is an activity-dependent regulator of target neuron intrinsic excitability. Neuron. 2011;71:291–305. doi: 10.1016/j.neuron.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antosova M., Mokra D., Pepucha L., Plevkova J., Buday T., Sterusky M. Physiology of nitric oxide in the respiratory system. Physiol Res. 2017;66:S159–S172. doi: 10.33549/physiolres.933673. [DOI] [PubMed] [Google Scholar]
- 11.Stuehr D.J., Haque M.M. Nitric oxide synthase enzymology in the 20 years after the Nobel Prize. Br J Pharmacol. 2019;176:177–188. doi: 10.1111/bph.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marletta M.A. Nitric oxide synthase: aspects concerning structure and catalysis. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 13.Knowles R.G., Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298(Pt 2):249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith O.W., Stuehr D.J. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 15.Hardingham N., Dachtler J., Fox K. The role of nitric oxide in pre-synaptic plasticity and homeostasis. Front Cell Neurosci. 2013;7:190. doi: 10.3389/fncel.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuriyama K., Ohkuma S. Role of nitric oxide in central synaptic transmission: effects on neurotransmitter release. Jpn J Pharmacol. 1995;69:1–8. doi: 10.1254/jjp.69.1. [DOI] [PubMed] [Google Scholar]
- 17.Shenkman B.S., Nemirovskaya T.L., Lomonosova Y.N. No-dependent signaling pathways in unloaded skeletal muscle. Front Physiol. 2015;6:298. doi: 10.3389/fphys.2015.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang W.J., Iannaccone S.T., Lau K.S., Masters B.S., McCabe T.J., McMillan K. Neuronal nitric oxide synthase and dystrophin-deficient muscular dystrophy. Proc Natl Acad Sci U S A. 1996;93:9142–9147. doi: 10.1073/pnas.93.17.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giudice E., Molza A.-E., Laurin Y., Nicolas A., Le Rumeur E., Delalande O. Molecular clues about the dystrophin-neuronal nitric oxide synthase interaction: a theoretical approach. Biochemistry. 2013;52:7777–7784. doi: 10.1021/bi400794p. [DOI] [PubMed] [Google Scholar]
- 20.Wade R.C., Davis M.E., Luty B.A., Madura J.D., McCammon J.A. Gating of the active site of triose phosphate isomerase: Brownian dynamics simulations of flexible peptide loops in the enzyme. Biophys J. 1993;64:9–15. doi: 10.1016/S0006-3495(93)81335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strasen J., Ritter O. Role of nNOS in cardiac ischemia-reperfusion injury. Trends Cardiovasc Med. 2011;21:58–63. doi: 10.1016/j.tcm.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Seddon M., Shah A.M., Casadei B. Cardiomyocytes as effectors of nitric oxide signalling. Cardiovasc Res. 2007;75:315–326. doi: 10.1016/j.cardiores.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Marsden P.A., Schappert K.T., Chen H.S., Flowers M., Sundell C.L., Wilcox J.N. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–293. doi: 10.1016/0014-5793(92)80697-f. [DOI] [PubMed] [Google Scholar]
- 24.Garcia V., Sessa W.C. Endothelial NOS: perspective and recent developments. Br J Pharmacol. 2019;176:189–196. doi: 10.1111/bph.14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Förstermann U., Münzel T. Endothelial nitric oxide synthase in vascular disease: from marvel to menace. Circulation. 2006;113:1708–1714. doi: 10.1161/CIRCULATIONAHA.105.602532. [DOI] [PubMed] [Google Scholar]
- 26.Green S.J., Scheller L.F., Marletta M.A., Seguin M.C., Klotz F.W., Slayter M. Nitric oxide: cytokine-regulation of nitric oxide in host resistance to intracellular pathogens. Immunol Lett. 1994;43:87–94. doi: 10.1016/0165-2478(94)00158-8. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z., Wang Z.-E., Doulias P.-T., Wei W., Ischiropoulos H., Locksley R.M. Lymphocyte development requires S-nitrosoglutathione reductase. J Immunol. 2010;185:6664–6669. doi: 10.4049/jimmunol.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vig M., Srivastava S., Kandpal U., Sade H., Lewis V., Sarin A. Inducible nitric oxide synthase in T cells regulates T cell death and immune memory. J Clin Invest. 2004;113:1734–1742. doi: 10.1172/JCI20225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vannini F., Kashfi K., Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334–343. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukumura D., Kashiwagi S., Jain R.K. The role of nitric oxide in tumour progression. Nat Rev Cancer. 2006;6:521–534. doi: 10.1038/nrc1910. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh D.K., Salerno J.C. Nitric oxide synthases: domain structure and alignment in enzyme function and control. Front Biosci. 2003;8:d193–d209. doi: 10.2741/959. [DOI] [PubMed] [Google Scholar]
- 32.Garcin E.D., Bruns C.M., Lloyd S.J., Hosfield D.J., Tiso M., Gachhui R. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 33.Smith B.C., Underbakke E.S., Kulp D.W., Schief W.R., Marletta M.A. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci U S A. 2013;110:E3577–E3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tejero J., Hannibal L., Mustovich A., Stuehr D.J. Surface charges and regulation of FMN to heme electron transfer in nitric-oxide synthase. J Biol Chem. 2010;285:27232–27240. doi: 10.1074/jbc.M110.138842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokom A.L., Morishima Y., Lau M., Su M., Glukhova A., Osawa Y. Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J Biol Chem. 2014;289:16855–16865. doi: 10.1074/jbc.M114.564005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell M.G., Smith B.C., Potter C.S., Carragher B., Marletta M.A. Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci U S A. 2014;111:E3614–E3623. doi: 10.1073/pnas.1413763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkmann N., Martásek P., Roman L.J., Xu X.-P., Page C., Swift M. Holoenzyme structures of endothelial nitric oxide synthase - an allosteric role for calmodulin in pivoting the FMN domain for electron transfer. J Struct Biol. 2014;188:46–54. doi: 10.1016/j.jsb.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panda K., Haque M.M., Garcin-Hosfield E.D., Durra D., Getzoff E.D., Stuehr D.J. Surface charge interactions of the FMN module govern catalysis by nitric-oxide synthase. J Biol Chem. 2006;281:36819–36827. doi: 10.1074/jbc.M606129200. [DOI] [PubMed] [Google Scholar]
- 39.Haque M.M., Fadlalla M., Wang Z.-Q., Ray S.S., Panda K., Stuehr D.J. Neutralizing a surface charge on the FMN subdomain increases the activity of neuronal nitric-oxide synthase by enhancing the oxygen reactivity of the enzyme heme-nitric oxide complex. J Biol Chem. 2009;284:19237–19247. doi: 10.1074/jbc.M109.013144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddhanta U., Wu C., Abu-Soud H.M., Zhang J., Ghosh D.K., Stuehr D.J. Heme iron reduction and catalysis by a nitric oxide synthase heterodimer containing one reductase and two oxygenase domains. J Biol Chem. 1996;271:7309–7312. doi: 10.1074/jbc.271.13.7309. [DOI] [PubMed] [Google Scholar]
- 41.Miller R.T., Martásek P., Omura T., Siler Masters B.S. Rapid kinetic studies of electron transfer in the three isoforms of nitric oxide synthase. Biochem Biophys Res Commun. 1999;265:184–188. doi: 10.1006/bbrc.1999.1643. [DOI] [PubMed] [Google Scholar]
- 42.Wang E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013;340:261–269. doi: 10.1016/j.canlet.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 43.Xia C., Misra I., Iyanagi T., Kim J.-J.P. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J Biol Chem. 2009;284:30708–30717. doi: 10.1074/jbc.M109.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Astashkin A.V., Elmore B.O., Fan W., Guillemette J.G., Feng C. Pulsed EPR determination of the distance between heme iron and FMN centers in a human inducible nitric oxide synthase. J Am Chem Soc. 2010;132:12059–12067. doi: 10.1021/ja104461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ilagan R.P., Tejero J., Aulak K.S., Ray S.S., Hemann C., Wang Z.-Q. Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry. 2009;48:3864–3876. doi: 10.1021/bi8021087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persechini A., Tran Q.-K., Black D.J., Gogol E.P. Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett. 2013;587:297–301. doi: 10.1016/j.febslet.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulos T.L., Li H. Nitric oxide synthase and structure-based inhibitor design. Nitric Oxide Biol Chem. 2017;63:68–77. doi: 10.1016/j.niox.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tafi A., Angeli L., Venturini G., Travagli M., Corelli F., Botta M. Computational studies of competitive inhibitors of nitric oxide synthase (NOS) enzymes: towards the development of powerful and isoform-selective inhibitors. Curr Med Chem. 2006;13:1929–1946. doi: 10.2174/092986706777585031. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Jamal J., Plaza C., Pineda S.H., Chreifi G., Jing Q. Structures of human constitutive nitric oxide synthases. Acta Crystallogr D Biol Crystallogr. 2014;70:2667–2674. doi: 10.1107/S1399004714017064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Poulos T.L. Structure-function studies on nitric oxide synthases. J Inorg Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Fischmann T.O., Hruza A., Niu X.D., Fossetta J.D., Lunn C.A., Dolphin E. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol. 1999;6:233–242. doi: 10.1038/6675. [DOI] [PubMed] [Google Scholar]
- 52.Ghosh A., Chawla-Sarkar M., Stuehr D.J. Hsp90 interacts with inducible NO synthase client protein in its heme-free state and then drives heme insertion by an ATP-dependent process. FASEB J. 2011;25:2049–2060. doi: 10.1096/fj.10-180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bender A.T., Silverstein A.M., Demady D.R., Kanelakis K.C., Noguchi S., Pratt W.B. Neuronal nitric-oxide synthase is regulated by the Hsp90-based chaperone system in vivo. J Biol Chem. 1999;274:1472–1478. doi: 10.1074/jbc.274.3.1472. [DOI] [PubMed] [Google Scholar]
- 54.List B.M., Klösch B., Völker C., Gorren A.C., Sessa W.C., Werner E.R. Characterization of bovine endothelial nitric oxide synthase as a homodimer with down-regulated uncoupled NADPH oxidase activity: tetrahydrobiopterin binding kinetics and role of haem in dimerization. Biochem J. 1997;323(Pt 1):159–165. doi: 10.1042/bj3230159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tejero J., Hannibal L., Mustovich A., Stuehr D.J. Surface charges and regulation of FMN to heme electron transfer in nitric-oxide synthase. J Biol Chem. 2010;285:27232–27240. doi: 10.1074/jbc.M110.138842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gachhui R., Ghosh D.K., Wu C., Parkinson J., Crane B.R., Stuehr D.J. Mutagenesis of acidic residues in the oxygenase domain of inducible nitric-oxide synthase identifies a glutamate involved in arginine binding. Biochemistry. 1997;36:5097–5103. doi: 10.1021/bi970331x. [DOI] [PubMed] [Google Scholar]
- 57.Tejero J., Stuehr D. Tetrahydrobiopterin in nitric oxide synthase. IUBMB Life. 2013;65:358–365. doi: 10.1002/iub.1136. [DOI] [PubMed] [Google Scholar]
- 58.Li H., Raman C.S., Glaser C.B., Blasko E., Young T.A., Parkinson J.F. Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase. J Biol Chem. 1999;274:21276–21284. doi: 10.1074/jbc.274.30.21276. [DOI] [PubMed] [Google Scholar]
- 59.Hemmens B., Goessler W., Schmidt K., Mayer B. Role of bound zinc in dimer stabilization but not enzyme activity of neuronal nitric-oxide synthase. J Biol Chem. 2000;275:35786–35791. doi: 10.1074/jbc.M005976200. [DOI] [PubMed] [Google Scholar]
- 60.Suvorava T., Pick S., Kojda G. Selective impairment of blood pressure reduction by endothelial nitric oxide synthase dimer destabilization in mice. J Hypertens. 2017;35:76–88. doi: 10.1097/HJH.0000000000001127. [DOI] [PubMed] [Google Scholar]
- 61.Cho K.-B., Derat E., Shaik S. Compound I of nitric oxide synthase: the active site protonation state. J Am Chem Soc. 2007;129:3182–3188. doi: 10.1021/ja066662r. [DOI] [PubMed] [Google Scholar]
- 62.Trane A.E., Pavlov D., Sharma A., Saqib U., Lau K., van Petegem F. Deciphering the binding of caveolin-1 to client protein endothelial nitric-oxide synthase (eNOS): scaffolding subdomain identification, interaction modeling, and biological significance. J Biol Chem. 2014;289:13273–13283. doi: 10.1074/jbc.M113.528695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ju H., Zou R., Venema V.J., Venema R.C. Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. J Biol Chem. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- 64.Michel J.B., Feron O., Sacks D., Michel T. Reciprocal regulation of endothelial nitric-oxide synthase by Ca2+-calmodulin and caveolin. J Biol Chem. 1997;272:15583–15586. doi: 10.1074/jbc.272.25.15583. [DOI] [PubMed] [Google Scholar]
- 65.Ji H., Li H., Flinspach M., Poulos T.L., Silverman R.B. Computer modeling of selective regions in the active site of nitric oxide synthases: implication for the design of isoform-selective inhibitors. J Med Chem. 2003;46:5700–5711. doi: 10.1021/jm030301u. [DOI] [PubMed] [Google Scholar]
- 66.Li H., Evenson R.J., Chreifi G., Silverman R.B., Poulos T.L. Structural basis for isoform selective nitric oxide synthase inhibition by thiophene-2-carboximidamides. Biochemistry. 2018;57:6319–6325. doi: 10.1021/acs.biochem.8b00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suaifan G., Shehadeh M., Al-Ijel H., Al-Jamal K.T., Taha M. Pharmacophore and QSAR Modeling of Neuronal Nitric Oxide Synthase Ligands and Subsequent Validation and In Silico Search for New Scaffolds. Med Chem. 2016;12:371–393. doi: 10.2174/1573406411666151002130609. [DOI] [PubMed] [Google Scholar]
- 68.Chayah M., Carrión M.D., Gallo M.A., Jiménez R., Duarte J., Camacho M.E. Development of urea and thiourea kynurenamine derivatives: synthesis, molecular modeling, and biological evaluation as nitric oxide synthase inhibitors. Chem Med Chem. 2015;10:874–882. doi: 10.1002/cmdc.201500007. [DOI] [PubMed] [Google Scholar]
- 69.Wang X., Ren Z., He Y., Xiang Y., Zhang Y., Qiao Y. A combination of pharmacophore modeling, molecular docking and virtual screening for iNOS inhibitors from Chinese herbs. Biomed Mater Eng. 2014;24:1315–1322. doi: 10.3233/BME-130934. [DOI] [PubMed] [Google Scholar]
- 70.Delker S.L., Ji H., Li H., Jamal J., Fang J., Xue F. Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model. J Am Chem Soc. 2010;132:5437–5442. doi: 10.1021/ja910228a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matter H., Kumar H.S.A., Fedorov R., Frey A., Kotsonis P., Hartmann E. Structural analysis of isoform-specific inhibitors targeting the tetrahydrobiopterin binding site of human nitric oxide synthases. J Med Chem. 2005;48:4783–4792. doi: 10.1021/jm050007x. [DOI] [PubMed] [Google Scholar]
- 72.Rosenfeld R.J., Garcin E.D., Panda K., Andersson G., Aberg A., Wallace A.V. Conformational changes in nitric oxide synthases induced by chlorzoxazone and nitroindazoles: crystallographic and computational analyses of inhibitor potency. Biochemistry. 2002;41:13915–13925. doi: 10.1021/bi026313j. [DOI] [PubMed] [Google Scholar]
- 73.Martiny V.Y., Miteva M.A. Advances in molecular modeling of human cytochrome P450 polymorphism. J Mol Biol. 2013;425:3978–3992. doi: 10.1016/j.jmb.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 74.Leth R., Ercig B., Olsen L., Jørgensen F.S. Both reactivity and accessibility are important in cytochrome P450 metabolism: a combined DFT and MD study of fenamic acids in BM3 mutants. J Chem Inf Model. 2019;59:743–753. doi: 10.1021/acs.jcim.8b00750. [DOI] [PubMed] [Google Scholar]
- 75.Caddell Haatveit K., Garcia-Borràs M., Houk K.N. Computational protocol to understand P450 mechanisms and design of efficient and selective biocatalysts. Front Chem. 2018;6:663. doi: 10.3389/fchem.2018.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mustafa G., Nandekar P.P., Camp T.J., Bruce N.J., Gregory M.C., Sligar S.G. Influence of transmembrane helix mutations on cytochrome P450-membrane interactions and function. Biophys J. 2019;116:419–432. doi: 10.1016/j.bpj.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Don C.G., Smieško M. Microsecond MD simulations of human CYP2D6 wild-type and five allelic variants reveal mechanistic insights on the function. PLoS One. 2018;13 doi: 10.1371/journal.pone.0202534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kontoyianni M., Lacy B. Toward computational understanding of molecular recognition in the human metabolizing cytochrome P450s. Curr Med Chem. 2018;25:3353–3373. doi: 10.2174/0929867325666180226104126. [DOI] [PubMed] [Google Scholar]
- 79.Kone B.C., Kuncewicz T., Zhang W., Yu Z.-Y. Protein interactions with nitric oxide synthases: controlling the right time, the right place, and the right amount of nitric oxide. Am J Physiol Renal Physiol. 2003;285:F178–F190. doi: 10.1152/ajprenal.00048.2003. [DOI] [PubMed] [Google Scholar]
- 80.Altaany Z., Ju Y., Yang G., Wang R. The coordination of S-sulfhydration, S-nitrosylation, and phosphorylation of endothelial nitric oxide synthase by hydrogen sulfide. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005478. ra87. [DOI] [PubMed] [Google Scholar]
- 81.Chen C.-A., De Pascali F., Basye A., Hemann C., Zweier J.L. Redox modulation of endothelial nitric oxide synthase by glutaredoxin-1 through reversible oxidative post-translational modification. Biochemistry. 2013;52:6712–6723. doi: 10.1021/bi400404s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bignon E., Allega M.F., Lucchetta M., Tiberti M., Papaleo E. Computational structural biology of S-nitrosylation of cancer targets. Front Oncol. 2018;8:272. doi: 10.3389/fonc.2018.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorren A.C.F., Mayer B. Nitric-oxide synthase: a cytochrome P450 family foster child. Biochim Biophys Acta. 1770;2007:432–445. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 84.Huang W.-C., Ellis J., Moody P.C.E., Raven E.L., Roberts G.C.K. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugishima M., Sato H., Higashimoto Y., Harada J., Wada K., Fukuyama K. Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase. Proc Natl Acad Sci U S A. 2014;111:2524–2529. doi: 10.1073/pnas.1322034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sagami I., Daff S., Shimizu T. Intra-subunit and inter-subunit electron transfer in neuronal nitric-oxide synthase: effect of calmodulin on heterodimer catalysis. J Biol Chem. 2001;276:30036–30042. doi: 10.1074/jbc.M104123200. [DOI] [PubMed] [Google Scholar]
- 87.Siddhanta U., Presta A., Fan B., Wolan D., Rousseau D.L., Stuehr D.J. Domain swapping in inducible nitric-oxide synthase. Electron transfer occurs between flavin and heme groups located on adjacent subunits in the dimer. J Biol Chem. 1998;273:18950–18958. doi: 10.1074/jbc.273.30.18950. [DOI] [PubMed] [Google Scholar]
- 88.Rhoads A.R., Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 89.Roman L.J., Martásek P., Miller R.T., Harris D.E., de La Garza M.A., Shea T.M. The C termini of constitutive nitric-oxide synthases control electron flow through the flavin and heme domains and affect modulation by calmodulin. J Biol Chem. 2000;275:29225–29232. doi: 10.1074/jbc.M004766200. [DOI] [PubMed] [Google Scholar]
- 90.Roman L.J., Masters B.S.S. Electron transfer by neuronal nitric-oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J Biol Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 91.Jones R.J., Smith S.M.E., Gao Y.T., DeMay B.S., Mann K.J., Salerno K.M. The function of the small insertion in the hinge subdomain in the control of constitutive mammalian nitric-oxide synthases. J Biol Chem. 2004;279:36876–36883. doi: 10.1074/jbc.M402808200. [DOI] [PubMed] [Google Scholar]
- 92.Knudsen G.M., Nishida C.R., Mooney S.D., Ortiz de Montellano P.R. Nitric-oxide synthase (NOS) reductase domain models suggest a new control element in endothelial NOS that attenuates calmodulin-dependent activity. J Biol Chem. 2003;278:31814–31824. doi: 10.1074/jbc.M303267200. [DOI] [PubMed] [Google Scholar]
- 93.Haque M.M., Panda K., Tejero J., Aulak K.S., Fadlalla M.A., Mustovich A.T. A connecting hinge represses the activity of endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:9254–9259. doi: 10.1073/pnas.0700332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J., Martàsek P., Paschke R., Shea T., Siler Masters B.S., Kim J.J. Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2001;276:37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 95.Adak S., Sharma M., Meade A.L., Stuehr D.J. A conserved flavin-shielding residue regulates NO synthase electron transfer and nicotinamide coenzyme specificity. Proc Natl Acad Sci U S A. 2002;99:13516–13521. doi: 10.1073/pnas.192283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang H., Yokom A.L., Cheng S., Su M., Hollenberg P.F., Southworth D.R. The full-length cytochrome P450 enzyme CYP102A1 dimerizes at its reductase domains and has flexible heme domains for efficient catalysis. J Biol Chem. 2018;293:7727–7736. doi: 10.1074/jbc.RA117.000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hedison T.M., Hay S., Scrutton N.S. Trapping methods for probing functional intermediates in nitric oxide synthases and related enzymes. Front Biosci. 2018;23 doi: 10.2741/4678. Landmark Ed. (1874–88) [DOI] [PubMed] [Google Scholar]
- 98.Craig D.H., Chapman S.K., Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J Biol Chem. 2002;277:33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 99.Devika N.T., Amresh P., Hassan M.I., Ali B.M.J. Molecular modeling and simulation of the human eNOS reductase domain, an enzyme involved in the release of vascular nitric oxide. J Mol Model. 2014;20:2470. doi: 10.1007/s00894-014-2470-7. [DOI] [PubMed] [Google Scholar]
- 100.Raval A., Piana S., Eastwood M.P., Dror R.O., Shaw D.E. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 101.Sündermann A., Oostenbrink C. Molecular dynamics simulations give insight into the conformational change, complex formation, and electron transfer pathway for cytochrome P450 reductase. Protein Sci. 2013;22:1183–1195. doi: 10.1002/pro.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sarapusit S., Lertkiatmongkol P., Duangkaew P., Rongnoparut P. Modeling of Anopheles minimus Mosquito NADPH-cytochrome P450 oxidoreductase (CYPOR) and mutagenesis analysis. Int J Mol Sci. 2013;14:1788–1801. doi: 10.3390/ijms14011788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pandey A.V., Flück C.E. NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol Ther. 2013;138:229–254. doi: 10.1016/j.pharmthera.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 104.Sharma N.M., Patel K.P. Post-translational regulation of neuronal nitric oxide synthase: implications for sympathoexcitatory states. Expert Opin Ther Targets. 2017;21:11–22. doi: 10.1080/14728222.2017.1265505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tillery L.C., Epperson T.A., Eguchi S., Motley E.D. Featured Article: Differential regulation of endothelial nitric oxide synthase phosphorylation by protease-activated receptors in adult human endothelial cells. Exp Biol Med (Maywood) 2016;241:569–580. doi: 10.1177/1535370215622584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fleming I. Molecular mechanisms underlying the activation of eNOS. Pflugers Arch. 2010;459:793–806. doi: 10.1007/s00424-009-0767-7. [DOI] [PubMed] [Google Scholar]
- 107.Aaltoma SH, Lipponen PK, Kosma VM. Inducible nitric oxide synthase (iNOS) expression and its prognostic value in prostate cancer. Anticancer Res n.d.;21:3101–6. [PubMed]
- 108.Sheng Y., Zhong L., Guo D., Lau G., Feng C. Insight into structural rearrangements and interdomain interactions related to electron transfer between flavin mononucleotide and heme in nitric oxide synthase: A molecular dynamics study. J Inorg Biochem. 2015;153:186–196. doi: 10.1016/j.jinorgbio.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hollingsworth S.A., Holden J.K., Li H., Poulos T.L. Elucidating nitric oxide synthase domain interactions by molecular dynamics. Protein Sci. 2016;25:374–382. doi: 10.1002/pro.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pierce B.G., Wiehe K., Hwang H., Kim B.-H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen L., Zheng H., Li W., Li W., Miao Y., Feng C. Role of a conserved tyrosine residue in the FMN-heme interdomain electron transfer in inducible nitric oxide synthase. J Phys Chem A. 2016;120:7610–7616. doi: 10.1021/acs.jpca.6b08207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Batabyal D., Richards L.S., Poulos T.L. Effect of redox partner binding on cytochrome P450 conformational dynamics. J Am Chem Soc. 2017;139:13193–13199. doi: 10.1021/jacs.7b07656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pudney C.R., Heyes D.J., Khara B., Hay S., Rigby S.E.J., Scrutton N.S. Kinetic and spectroscopic probes of motions and catalysis in the cytochrome P450 reductase family of enzymes. FEBS J. 2012;279:1534–1544. doi: 10.1111/j.1742-4658.2011.08442.x. [DOI] [PubMed] [Google Scholar]
- 114.Leferink N.G.H., Hay S., Rigby S.E.J., Scrutton N.S. Towards the free energy landscape for catalysis in mammalian nitric oxide synthases. FEBS J. 2015;282:3016–3029. doi: 10.1111/febs.13171. [DOI] [PubMed] [Google Scholar]
- 115.Brenner S., Hay S., Munro A.W., Scrutton N.S. Inter-flavin electron transfer in cytochrome P450 reductase - effects of solvent and pH identify hidden complexity in mechanism. FEBS J. 2008;275:4540–4557. doi: 10.1111/j.1742-4658.2008.06597.x. [DOI] [PubMed] [Google Scholar]
- 116.Knight K., Scrutton N.S. Stopped-flow kinetic studies of electron transfer in the reductase domain of neuronal nitric oxide synthase: re-evaluation of the kinetic mechanism reveals new enzyme intermediates and variation with cytochrome P450 reductase. Biochem J. 2002;367:19–30. doi: 10.1042/BJ20020667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shamovsky I., Belfield G., Lewis R., Narjes F., Ripa L., Tyrchan C. Theoretical studies of the second step of the nitric oxide synthase reaction: Electron tunneling prevents uncoupling. J Inorg Biochem. 2018;181:28–40. doi: 10.1016/j.jinorgbio.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 118.Santolini J. The molecular mechanism of mammalian NO-synthases: a story of electrons and protons. J Inorg Biochem. 2011;105:127–141. doi: 10.1016/j.jinorgbio.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 119.Brunel A., Lang J., Couture M., Boucher J.-L., Dorlet P., Santolini J. Oxygen activation in NO synthases: evidence for a direct role of the substrate. FEBS Open Bio. 2016;6:386–397. doi: 10.1002/2211-5463.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramasamy S., Haque M.M., Gangoda M., Stuehr D.J. Tetrahydrobiopterin redox cycling in nitric oxide synthase: evidence supports a through-heme electron delivery. FEBS J. 2016;283:4491–4501. doi: 10.1111/febs.13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Davydov R., Labby K.J., Chobot S.E., Lukoyanov D.A., Crane B.R., Silverman R.B. Enzymatic and cryoreduction EPR studies of the hydroxylation of methylated N(ω)-hydroxy-L-arginine analogues by nitric oxide synthase from Geobacillus stearothermophilus. Biochemistry. 2014;53:6511–6519. doi: 10.1021/bi500485z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davydov R., Ledbetter-Rogers A., Martásek P., Larukhin M., Sono M., Dawson J.H. EPR and ENDOR characterization of intermediates in the cryoreduced oxy-nitric oxide synthase heme domain with bound L-arginine or N(G)-hydroxyarginine. Biochemistry. 2002;41:10375–10381. doi: 10.1021/bi0260637. [DOI] [PubMed] [Google Scholar]
- 123.Cho K.-B., Carvajal M.A., Shaik S. First half-reaction mechanism of nitric oxide synthase: the role of proton and oxygen coupled electron transfer in the reaction by quantum mechanics/molecular mechanics. J Phys Chem B. 2009;113:336–346. doi: 10.1021/jp8073199. [DOI] [PubMed] [Google Scholar]
- 124.de Visser S.P., Tan L.S. Is the bound substrate in nitric oxide synthase protonated or neutral and what is the active oxidant that performs substrate hydroxylation? J Am Chem Soc. 2008;130:12961–12974. doi: 10.1021/ja8010995. [DOI] [PubMed] [Google Scholar]
- 125.de Visser S.P. Density functional theory (DFT) and combined quantum mechanical/molecular mechanics (QM/MM) studies on the oxygen activation step in nitric oxide synthase enzymes. Biochem Soc Trans. 2009;37:373–377. doi: 10.1042/BST0370373. [DOI] [PubMed] [Google Scholar]
- 126.Abu-Soud H.M., Gachhui R., Raushel F.M., Stuehr D.J. The ferrous-dioxy complex of neuronal nitric oxide synthase. Divergent effects of L-arginine and tetrahydrobiopterin on its stability. J Biol Chem. 1997;272:17349–17353. doi: 10.1074/jbc.272.28.17349. [DOI] [PubMed] [Google Scholar]
- 127.Roe N.D., Ren J. Nitric oxide synthase uncoupling: a therapeutic target in cardiovascular diseases. Vascul Pharmacol. 2012;57:168–172. doi: 10.1016/j.vph.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 128.Rabender C.S., Alam A., Sundaresan G., Cardnell R.J., Yakovlev V.A., Mukhopadhyay N.D. The role of nitric oxide synthase uncoupling in tumor progression. Mol Cancer Res. 2015;13:1034–1043. doi: 10.1158/1541-7786.MCR-15-0057-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xia N., Horke S., Habermeier A., Closs E.I., Reifenberg G., Gericke A. Uncoupling of endothelial nitric oxide synthase in perivascular adipose tissue of diet-induced obese mice. Arterioscler Thromb Vasc Biol. 2016;36:78–85. doi: 10.1161/ATVBAHA.115.306263. [DOI] [PubMed] [Google Scholar]
- 130.Li H., Forstermann U. Pharmacological prevention of eNOS uncoupling. Curr Pharm Des. 2014;20:3595–3606. doi: 10.2174/13816128113196660749. [DOI] [PubMed] [Google Scholar]
- 131.Ramachandran J., Peluffo R.D. Threshold levels of extracellular l-arginine that trigger NOS-mediated ROS/RNS production in cardiac ventricular myocytes. Am J Physiol Cell Physiol. 2017;312:C144–C154. doi: 10.1152/ajpcell.00150.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li H., Förstermann U. Uncoupling of endothelial NO synthase in atherosclerosis and vascular disease. Curr Opin Pharmacol. 2013;13:161–167. doi: 10.1016/j.coph.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 133.Lin M.I., Fulton D., Babbitt R., Fleming I., Busse R., Pritchard K.A. Phosphorylation of threonine 497 in endothelial nitric-oxide synthase coordinates the coupling of L-arginine metabolism to efficient nitric oxide production. J Biol Chem. 2003;278:44719–44726. doi: 10.1074/jbc.M302836200. [DOI] [PubMed] [Google Scholar]
- 134.Chen C.-A., Druhan L.J., Varadharaj S., Chen Y.-R., Zweier J.L. Phosphorylation of endothelial nitric-oxide synthase regulates superoxide generation from the enzyme. J Biol Chem. 2008;283:27038–27047. doi: 10.1074/jbc.M802269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wei C.C., Wang Z.Q., Wang Q., Meade A.L., Hemann C., Hille R. Rapid kinetic studies link tetrahydrobiopterin radical formation to heme-dioxy reduction and arginine hydroxylation in inducible nitric-oxide synthase. J Biol Chem. 2001;276:315–319. doi: 10.1074/jbc.M008441200. [DOI] [PubMed] [Google Scholar]
- 136.Xia Y., Tsai A.L., Berka V., Zweier J.L. Superoxide generation from endothelial nitric-oxide synthase. A Ca2+/calmodulin-dependent and tetrahydrobiopterin regulatory process. J Biol Chem. 1998;273:25804–25808. doi: 10.1074/jbc.273.40.25804. [DOI] [PubMed] [Google Scholar]
- 137.Stoll S., NejatyJahromy Y., Woodward J.J., Ozarowski A., Marletta M.A., Britt R.D. Nitric oxide synthase stabilizes the tetrahydrobiopterin cofactor radical by controlling its protonation state. J Am Chem Soc. 2010;132:11812–11823. doi: 10.1021/ja105372s. [DOI] [PubMed] [Google Scholar]
- 138.Wei C.-C., Wang Z.-Q., Hemann C., Hille R., Stuehr D.J. A tetrahydrobiopterin radical forms and then becomes reduced during Nomega-hydroxyarginine oxidation by nitric-oxide synthase. J Biol Chem. 2003;278:46668–46673. doi: 10.1074/jbc.M307682200. [DOI] [PubMed] [Google Scholar]
- 139.Wei C.-C., Wang Z.-Q., Arvai A.S., Hemann C., Hille R., Getzoff E.D. Structure of tetrahydrobiopterin tunes its electron transfer to the heme-dioxy intermediate in nitric oxide synthase. Biochemistry. 2003;42:1969–1977. doi: 10.1021/bi026898h. [DOI] [PubMed] [Google Scholar]
- 140.Luo S., Lei H., Qin H., Xia Y. Molecular mechanisms of endothelial NO synthase uncoupling. Curr Pharm Des. 2014;20:3548–3553. doi: 10.2174/13816128113196660746. [DOI] [PubMed] [Google Scholar]
- 141.Daff S. NO synthase: structures and mechanisms. Nitric Oxide Biol Chem. 2010;23:1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 142.Krzyaniak M.D., Cruce A.A., Vennam P., Lockart M., Berka V., Tsai A.-L. The tetrahydrobiopterin radical interacting with high- and low-spin heme in neuronal nitric oxide synthase - A new indicator of the extent of NOS coupling. Free Radic Biol Med. 2016;101:367–377. doi: 10.1016/j.freeradbiomed.2016.10.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wei C.-C., Wang Z.-Q., Durra D., Hemann C., Hille R., Garcin E.D. The three nitric-oxide synthases differ in their kinetics of tetrahydrobiopterin radical formation, heme-dioxy reduction, and arginine hydroxylation. J Biol Chem. 2005;280:8929–8935. doi: 10.1074/jbc.M409737200. [DOI] [PubMed] [Google Scholar]
- 144.Wei C.-C., Wang Z.-Q., Tejero J., Yang Y.-P., Hemann C., Hille R. Catalytic reduction of a tetrahydrobiopterin radical within nitric-oxide synthase. J Biol Chem. 2008;283:11734–11742. doi: 10.1074/jbc.M709250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Robinet J.J., Cho K.-B., Gauld J.W. A density functional theory investigation on the mechanism of the second half-reaction of nitric oxide synthase. J Am Chem Soc. 2008;130:3328–3334. doi: 10.1021/ja072650+. [DOI] [PubMed] [Google Scholar]
- 146.Cho K.-B., Gauld J.W. Second half-reaction of nitric oxide synthase: computational insights into the initial step and key proposed intermediate. J Phys Chem B. 2005;109:23706–23714. doi: 10.1021/jp054864o. [DOI] [PubMed] [Google Scholar]
- 147.Cho K.-B., Gauld J.W. Quantum chemical calculations of the NHA bound nitric oxide synthase active site: O2 binding and implications for the catalytic mechanism. J Am Chem Soc. 2004;126:10267–10270. doi: 10.1021/ja049186i. [DOI] [PubMed] [Google Scholar]
- 148.Jansen Labby K., Li H., Roman L.J., Martásek P., Poulos T.L., Silverman R.B. Methylated N(ω)-hydroxy-L-arginine analogues as mechanistic probes for the second step of the nitric oxide synthase-catalyzed reaction. Biochemistry. 2013;52:3062–3073. doi: 10.1021/bi301571v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Giroud C., Moreau M., Mattioli T.A., Balland V., Boucher J.-L., Xu-Li Y. Role of arginine guanidinium moiety in nitric-oxide synthase mechanism of oxygen activation. J Biol Chem. 2010;285:7233–7245. doi: 10.1074/jbc.M109.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tantillo Dean J., Fukuto Jon M., Hoffman Brian M., Silverman Richard B., Houk K.N. 2000. Theoretical studies on NG-hydroxy-l-arginine and derived radicals: implications for the mechanism of nitric oxide synthase. [Google Scholar]
- 151.Shaik S., Cohen S., Wang Y., Chen H., Kumar D., Thiel W. P450 enzymes: their structure, reactivity, and selectivity-modeled by QM/MM calculations. Chem Rev. 2010;110:949–1017. doi: 10.1021/cr900121s. [DOI] [PubMed] [Google Scholar]
- 152.Dubey K.D., Wang B., Vajpai M., Shaik S. MD simulations and QM/MM calculations show that single-site mutations of cytochrome P450BM3 alter the active site’s complexity and the chemoselectivity of oxidation without changing the active species. Chem Sci. 2017;8:5335–5344. doi: 10.1039/c7sc01932g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dubey K.D., Wang B., Shaik S. Molecular dynamics and QM/MM calculations predict the substrate-induced gating of cytochrome P450 BM3 and the regio- and stereoselectivity of fatty acid hydroxylation. J Am Chem Soc. 2016;138:837–845. doi: 10.1021/jacs.5b08737. [DOI] [PubMed] [Google Scholar]
- 154.Postils V., Saint-André M., Timmins A., Li X.-X., Wang Y., Luis J.M. Quantum mechanics/molecular mechanics studies on the relative reactivities of compound I and II in cytochrome P450 enzymes. Int J Mol Sci. 2018;19:1974. doi: 10.3390/ijms19071974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu H., Wolynes P.G., Papoian G.A. AWSEM-IDP: a coarse-grained force field for intrinsically disordered proteins. J Phys Chem B. 2018;122:11115–11125. doi: 10.1021/acs.jpcb.8b05791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klepeis J.L., Lindorff-Larsen K., Dror R.O., Shaw D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 157.Piana S., Klepeis J.L., Shaw D.E. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr Opin Struct Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 158.Dror R.O., Dirks R.M., Grossman J.P., Xu H., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 159.Papaleo E., Saladino G., Lambrughi M., Lindorff-Larsen K., Gervasio F.L., Nussinov R. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev. 2016;116:6391–6423. doi: 10.1021/acs.chemrev.5b00623. [DOI] [PubMed] [Google Scholar]
- 160.Tiberti M., Papaleo E., Bengtsen T., Boomsma W., Lindorff-Larsen K. ENCORE: software for quantitative ensemble comparison. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Valimberti I., Tiberti M., Lambrughi M., Sarcevic B., Papaleo E. E2 superfamily of ubiquitin-conjugating enzymes: constitutively active or activated through phosphorylation in the catalytic cleft. Sci Rep. 2015;5:14849. doi: 10.1038/srep14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Papaleo E. Integrating atomistic molecular dynamics simulations, experiments, and network analysis to study protein dynamics: strength in unity. Front Mol Biosci. 2015;2:28. doi: 10.3389/fmolb.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Karami Y., Laine E., Carbone A. Dissecting protein architecture with communication blocks and communicating segment pairs. BMC Bioinformatics. 2016;17:13. doi: 10.1186/s12859-015-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pandini A., Fornili A., Fraternali F., Kleinjung J. Detection of allosteric signal transmission by information-theoretic analysis of protein dynamics. FASEB J. 2012;26:868–881. doi: 10.1096/fj.11-190868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nygaard M., Terkelsen T., Olsen A.V., Sora V., Salamanca J., Rizza F. The mutational landscape of the oncogenic MZF1 SCAN domain in cancer. Front Mol Biosci. 2016 doi: 10.3389/fmolb.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tiberti M., Invernizzi G., Lambrughi M., Inbar Y., Schreiber G., Papaleo E. PyInteraph : a framework for the analysis of interaction networks in structural ensembles of proteins. J Chem Inf Model. 2014;54:1537–1551. doi: 10.1021/ci400639r. [DOI] [PubMed] [Google Scholar]
- 167.Lambrughi M., De Gioia L., Gervasio F.L., Lindorff-Larsen K., Nussinov R., Urani C. DNA-binding protects p53 from interactions with cofactors involved in transcription-independent functions. Nucleic Acids Res. 2016;44:9096–9109. doi: 10.1093/nar/gkw770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Spiwok V., Sucur Z., Hosek P. Enhanced sampling techniques in biomolecular simulations. Biotechnol Adv. 2015;33:1130–1140. doi: 10.1016/j.biotechadv.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 169.Pietrucci F. Strategies for the exploration of free energy landscapes: unity in diversity and challenges ahead. Rev Phys. 2017;2:32–45. [Google Scholar]
- 170.Elber R. Simulations of allosteric transitions. Curr Opin Struct Biol. 2011;21:167–172. doi: 10.1016/j.sbi.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 171.Bonomi M., Hanot S., Greenberg C.H., Sali A., Nilges M., Vendruscolo M. Bayesian weighing of electron cryo-microscopy data for integrative structural modeling. Structure. 2018 doi: 10.1016/j.str.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Habeck M. Bayesian modeling of biomolecular assemblies with Cryo-EM maps. Front Mol Biosci. 2017;4:15. doi: 10.3389/fmolb.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu X., Yan C., Wohlhueter R., Ivanov I. Integrative modeling of macromolecular assemblies from low to near-atomic resolution. Comput Struct Biotechnol J. 2015;13:492–503. doi: 10.1016/j.csbj.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Valdez C.E., Morgenstern A., Eberhart M.E., Alexandrova A.N. Predictive methods for computational metalloenzyme redesign - a test case with carboxypeptidase A. Phys Chem Chem Phys. 2016;18:31744–31756. doi: 10.1039/c6cp02247b. [DOI] [PubMed] [Google Scholar]
- 175.Grishina M.A., Potemkin V.A. Topological analysis of electron density in large biomolecular systems. Curr Drug Discov Technol. 2018 doi: 10.2174/1570163815666180821165330. [DOI] [PubMed] [Google Scholar]
- 176.Tognetti V., Morell C., Joubert L. Quantifying electro/nucleophilicity by partitioning the dual descriptor. J Comput Chem. 2015;36:649–659. doi: 10.1002/jcc.23840. [DOI] [PubMed] [Google Scholar]
- 177.Morell C., Ayers P.W., Grand A., Chermette H. Application of the electron density force to chemical reactivity. Phys Chem Chem Phys. 2011;13:9601–9608. doi: 10.1039/c0cp02083d. [DOI] [PubMed] [Google Scholar]
- 178.Tognetti V., Joubert L. Density functional theory and Bader’s atoms-in-molecules theory: towards a vivid dialogue. Phys Chem Chem Phys. 2014;16:14539–14550. doi: 10.1039/c3cp55526g. [DOI] [PubMed] [Google Scholar]
- 179.Gillet N., Elstner M., Kubař T. Coupled-perturbed DFTB-QM/MM metadynamics: Application to proton-coupled electron transfer. J Chem Phys. 2018;149 doi: 10.1063/1.5027100. [DOI] [PubMed] [Google Scholar]
- 180.Gillet N., Ruiz-Pernía J.J., de la Lande A., Lévy B., Lederer F., Demachy I. QM/MM study of l-lactate oxidation by flavocytochrome b2. Phys Chem Chem Phys. 2016;18:15609–15618. doi: 10.1039/c6cp00395h. [DOI] [PubMed] [Google Scholar]
- 181.de la Lande A., Gillet N., Chen S., Salahub D.R. Progress and challenges in simulating and understanding electron transfer in proteins. Arch Biochem Biophys. 2015;582:28–41. doi: 10.1016/j.abb.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 182.Stone J.R., Marletta M.A. Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry. 1994;33:5636–5640. doi: 10.1021/bi00184a036. [DOI] [PubMed] [Google Scholar]
- 183.Giulivi C., Kato K., Cooper C.E. Nitric oxide regulation of mitochondrial oxygen consumption I: cellular physiology. Am J Physiol Cell Physiol. 2006;291:C1225–C1231. doi: 10.1152/ajpcell.00307.2006. [DOI] [PubMed] [Google Scholar]
- 184.Dahm C.C., Moore K., Murphy M.P. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J Biol Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- 185.Brown G.C., Borutaite V. Nitric oxide inhibition of mitochondrial respiration and its role in cell death. Free Radic Biol Med. 2002;33:1440–1450. doi: 10.1016/s0891-5849(02)01112-7. [DOI] [PubMed] [Google Scholar]
- 186.Furuta S., Basal S. Nitrosylation is the guardian of tissue homeostasis. Trends Cancer. 2017;3:744–748. doi: 10.1016/j.trecan.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 187.Hess D.T., Matsumoto A., Kim S.-O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 188.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]