Abstract
G-quadruplex ligands exert their antiproliferative effects through telomere-dependent and telomere-independent mechanisms, but the inter-relationships among autophagy, cell growth arrest and cell death induced by these ligands remain largely unexplored. Here, we demonstrate that the G-quadruplex ligand 20A causes growth arrest of cancer cells in culture and in a HeLa cell xenografted mouse model. This response is associated with the induction of senescence and apoptosis. Transcriptomic analysis of 20A treated cells reveals a significant functional enrichment of biological pathways related to growth arrest, DNA damage response and the lysosomal pathway. 20A elicits global DNA damage but not telomeric damage and activates the ATM and autophagy pathways. Loss of ATM following 20A treatment inhibits both autophagy and senescence and sensitizes cells to death. Moreover, disruption of autophagy by deletion of two essential autophagy genes ATG5 and ATG7 leads to failure of CHK1 activation by 20A and subsequently increased cell death. Our results, therefore, identify the activation of ATM by 20A as a critical player in the balance between senescence and apoptosis and autophagy as one of the key mediators of such regulation. Thus, targeting the ATM/autophagy pathway might be a promising strategy to achieve the maximal anticancer effect of this compound.
INTRODUCTION
G-quadruplexes (G4) are non-canonical DNA or RNA structures found in guanine-rich regions of the genome (1). G4 structures are formed by stacking of two or more G-quartets and are further stabilized by monovalent cations. G4-prone motifs are enriched in telomeres, promoters and the first introns of genes (2). Evidence for G4 formation in cells comes from studies using DNA structure-specific antibodies, in vivo nuclear magnetic resonance, and small compounds capable of selective binding to G4 and from analysis of genomic instability (for a review: (3)). Compounds that bind to G4 are called G-quadruplex ligands (G4L), and the most promising compounds exhibit exquisite selectivity for this unusual structure (for a review: (4)). G4L were initially developed as telomerase inhibitors, and some G4Ls have antiproliferative effects that are associated with stabilization of the telomeric G4 structures and telomere erosion (5,6).
Evidence suggests that antiproliferative effects of certain G4Ls result from telomere-independent mechanisms. For example, the majority of G4-antibody foci are actually not found at telomeres (7), and a number of G4Ls alter the expression of genes, such as the KRAS and c-MYC oncogenes, that contain G4 motifs in their promoters (for a review on G4 in promoters: (8)). In addition, some G4Ls may act by targeting RNA G4 (for recent reviews on G4 RNA: (9,10)). As a general mechanism, G4Ls promote the DNA damage response (DDR) (11), which ultimately leads to senescence (a permanent growth arrest) or, when the damage is left unrepaired, cell death (12). These properties make G4Ls attractive for cancer therapy. In addition, some G4Ls are able to activate the p53/p21 pathway, which is implicated in the regulation of DDR, senescence and cell death (13,14). It is not clear, however, what determines whether cells undergo senescence or apoptosis in response to a G4L.
A few G4Ls such as RHPS4 (14,15), napthalene diimides (16), acridine derivatives (6) and EMICORON (17) exhibit antitumor activity in animal models either alone or in combination with other anticancer agents (for a review: (18)). Despite a flurry of G4Ls described in the literature recently (for a recent review: (19)), only a few G4-related compounds have been tested in clinical trials, and none have progressed through the drug-development pipeline. There is, therefore, an urgent need to identify G4Ls with better drug-like properties. The 2,4,6-triarylpyridines bind to G4-DNA with fair to excellent selectivity (20). Among these derivatives, compound 20A (compound #3 in reference (20)) has a good affinity and selectivity for G4, and the structure of the G4-ligand complex was recently solved (21). Its ability to inhibit the proliferation of HeLa cells (20) prompted us to study its anticancer mechanism of action in vitro and in vivo.
Macroautophagy (often simply referred to autophagy) is a vesicular mechanism by which cellular components are transported to the lysosome for degradation and recycling (22). This process is orchestrated by autophagy-related proteins and is regulated through complex signaling pathways that converge mostly on the AMPK/MTORC1 axis (22–24). Autophagy serves as an adaptive mechanism allowing cell viability in response to stress including those to which cancer cells are exposed (e.g. hypoxia, environmental changes and DNA damage) (25). Autophagy is also important in cell homeostasis as it induces selective degradation of unwanted mitochondria, aggregated proteins and specific signaling proteins (26). Autophagy is now considered indispensable for development, cell survival, differentiation and immune responses. Over the last two decades, it has been established that autophagy and cancer are interconnected (27–29). Notably, autophagy is activated in response to a variety of anticancer agents (30). In most cases, the activation of autophagy by cancer therapies confers tumor protection that results in therapy resistance. However, in response to some anticancer therapies, autophagy induces immunogenic and non-immunogenic cell death or growth arrest (30–33). Upon DNA damage, autophagy modulates the DDR through regulation of cell-cycle checkpoints, induction of apoptotic cell death and the activation of the DNA repair machinery (34,35).
Few studies have examined the regulation and the role of autophagy in response to G4 ligands (13,36,37). The inter-relationships between autophagy, cell growth arrest and cell death induced by these ligands are largely unexplored, however. Using a variety of approaches, we investigated the molecular connections between DDR and autophagy and explored how these regulatory mechanisms influence cell fate choice between senescence and apoptosis in response to the G4 ligand 20A.
MATERIALS AND METHODS
Reagents
ATM inhibitor KU-55933 (#SML1109), Hoechst 33258 (#14530), E64d (#516485), bafilomycin A1 (#B1793), 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide, MTT (#M2128), blasticidine (#3513-03-9), Fluoromount (#F4680), puromycin (#58-58-2), Quinoline-Val-Asp-Difluorophenoxymethyl Ketone, QVD-OPH (# SML0063) and doxycycline (Dox) (#D9891) were purchased from Sigma-Aldrich. Pepstatin A methyl ester (#516485) and epoxomycin (# 134381-21-8) were obtained from Calbiochem. Tetramethyl rhodamine methyl ester perchlorate dye (#T-668) and carboxyfluorescein succinimidyl ester (CFSE) (#C34554) were purchased from Molecular Probes. LY2603618 (#S2626) was purchased from Selleckchem. 20A was synthesized as previously described (compound #3 in (20)). Sentragor (#AR8850020) for antibody-enhanced detection of senescence cells was provided by Arriani Pharmaceuticals.
Antibodies
Antibodies against the following proteins were used: phospho-ATM (Ser 1981) (#5883), ataxiatelangiectasia mutated, ATM (#2873), phospho-AMPK (Thr172) (#2535), AMPKα1 (#2795), phospho-AMPK substrates (#5759), ATG7 (#8558), cleaved caspase 3 (#9664), phospho-CHK1 (Ser 345, 133D3) (#2348), CHK1 (2G1D5) (#2360), phospho-CHK2 (Thr 68) (#2197), CHK2 (#2662), p21 Waf1/CIP1 (12D1) (#2947), p27 Kip 1 (D69C12) XP (#3686), phospho 4EBP1 (Thr 37/46) (#9459), 4EBP1 (#9452), phospho-p70S6 kinase (Thr389) (#9205), p70S6 kinase (#9202), γH2AX (Ser139) (#9718), H2A.X (D17A3) XP (#7631) and phospho-MTOR (Ser2448) (#2971) all from Cell Signaling Technology); Ki67 (30-9) (#790-4286) from Roche; PARP1 (C-2-10) (#BML-SA249-0050) from Enzo Life Sciences; p62 (#610832) from BD Biosciences; ATG5 (#0262-100/ATG5-7C6) from Nanotools; ACTIN β (#NB600-501) from Novus Biologicals; LC3 (#M152-3) from MBL; phospho-p62 (Ser403) (#MABC186) from Merck Millipore; p16 (#805-4713) from Ventana; horseradish peroxidase-conjugated anti-rabbit (#111-035-003) and horseradish peroxidase (HRP)-conjugated anti-mouse (#115-035-174) from Jackson ImmunoResearch; and anti-mouse (#A11001) and anti-rabbit Alexa Fluor 488 (#A11008) from Invitrogen.
Cell culture
The human cervical cancer cell line HeLa and the human lung carcinoma A549 cell line were purchased from the American Type Culture Collection. HeLa and A549 cells were grown in RPMI 1640 and Dulbecco's modified Eagle's medium (DMEM) culture media, respectively, supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine (Gibco-Life Technologies). Saos-2 cells were grown in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco-Life Technologies). p53 expression was induced in Saos-2 cells following addition of 0.5 μg/ml Dox to the cell medium. After 6 hours, the medium was replaced with medium without Dox and then supplemented with the appropriate treatment. All cell lines used in this study were cultivated at 37°C in a humidified atmosphere with 5% CO2.
Cell viability assay
Cell viability was evaluated using the MTT assay. Cells were seeded in a 96-well plate (4 × 103 cells/well). After 24 h of drug treatment, MTT was added to each well to a final concentration of 0.5 mg/ml, followed by incubation at 37°C for 3 h. The medium was then removed, and 100 μl of dimethylsufoxide (DMSO) was added per well. The absorbance in each well was measured at 570 nm and 630 nm using a Flexstation 3 microplate reader (Molecular Devices). To determine the IC50 of 20A, a non-linear regression curve was fit using GraphPad Prism software.
Cell division assay
HeLa cells were labeled with 1 μM CFSE on day zero and then subjected to flow cytometry analysis to monitor CFSE dilution, which occurs with cell division on indicated days. The median fluorescence intensity (MFI) was scored every day, and the results are expressed as the mean percentage of MFI normalized to that obtained from day zero.
Apoptosis analysis
Apoptosis was determined by measuring the mitochondrial transmembrane potential (ΔΨm) using tetramethyl rhodamine methyl ester perchlorate dye as previously described (38). Apoptosis was also evaluated by western blotting analyses of cleaved forms of either PARP1 or caspase 3.
Senescence assay
Cells were stained for senescence-associated β-galactosidase activity using a kit from Cell Signaling Technology (#9860) as per the manufacturer’s instructions. Ten images were acquired on a CKX41 Olympus microscope 20× objective for each experimental condition. The percentages of β-galactosidase-positive cells were scored using Image J software. Results are given as means ± S.D. of four independent experiments. The in vivo senescence assay was performed in tumor sections using SenTraGor™, a Sudan Black B analog conjugated with biotin, which reacts with lipofuscin granules that have been shown to accumulate during the senescence process (39).
Meta-TIF assay
The meta-TIF assay for detection of telomere-induced foci (TIF) in metaphase spreads was performed as described previously (40). See also the experimental procedure in the Supplementary Data (part I).
Protein expression analysis
Cell extracts were prepared in 10 mM Tris, pH 7.4, 1% sodium dodecyl sulphate, 1 mM sodium vanadate, 2 mM phenylmethylsulfonylfluoride (Sigma-Aldrich), 1% Protease Inhibitor Cocktail (Sigma-Aldrich) and 1% Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Extracts were treated with benzonase endonuclease (Merck Millipore) and then heated for 5 min at 95°C. For western blotting, aliquots of cellular extracts (20–50 μg) were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis using a Tris/glycine buffer system based on the method of Laemmli as previously described (41). After electrophoresis, proteins were transferred to a nitrocellulose membrane (GE Healthcare Life Sciences). The blots were then probed with primary antibodies using the manufacturer’s protocol and then incubated with the appropriate HRP-conjugated secondary antibody. Staining for ACTIN β and staining with Ponceau Red were scored to evaluate the protein loading levels of the samples. Immunostained proteins were visualized on a chemiluminescence detector equipped with a camera (FUSION FX7, Fisher Bioblock Scientific) using the enhanced chemiluminescence detection system. The densitometry quantification was performed using the ImageJ software. For all of the immunoblots, representative images of at least two experiments are shown.
Label-free quantitative proteomics
The proteomic experiments and analyses were performed by the Proteomics Core Facility at the University of Bordeaux (https://proteome.cgfb.u-bordeaux.fr/en). The steps of sample preparation, protein digestion and nano-liquid chromatography-tandem mass spectrometry analysis were performed as previously described (42). For protein identification, Sequest HT and Mascot 2.4 algorithms through Proteome Discoverer 1.4 Software (Thermo Fisher Scientific) were used for protein identification in batch mode by searching against a Homo sapiens database (68 978 entries, Reference Proteome Set, release 2013_12 from UniProt website). Two missed enzyme cleavages were allowed. Mass tolerances in MS and MS/MS were set to 10 ppm and 0.02 Da. Oxidation of methionine, acetylation of lysine and deamidation of asparagine and glutamine were searched as dynamic modifications. Carbamidomethylation on cysteine was searched as static modification. Peptide validation was performed using Percolator algorithm (43), and only ‘high confidence’ peptides were retained corresponding to a 1% false discovery rate (FDR) at the peptide level. Feature detection, alignment and quantification were performed with Progenesis QI with parameters as previously described (44). Only non-conflicting features and unique peptides (FDR <1%) were considered for calculation of quantification at the protein level. A minimum of two peptides matched to a protein was used as the criteria for identification as a differentially expressed protein.
RNA extraction and microarray experiments
Total RNAs were extracted from cells with Trizol reagent (Life Technologies) and purified with the RNeasy Min Elute Cleanup Kit (Qiagen) according to the manufacturer’s procedures. RNA was quantified using a Nanodrop 1000 spectrophotometer (Thermo Scientific), and RNA quality was checked on an Agilent 2100 bioanalyzer (Agilent Technologies). Microarray experiments were performed using Agilent-014 850 Whole Human Genome Microarray 4 × 44K G4112F array according to the manufacturer’s recommended protocol.
Transcriptome and proteome analyses
The experimental designs and data filtering results are summarized in Supplementary Figure S4A. Briefly, the variability between replicates for each gene was computed as the standard deviation divided by the mean. Thresholds of 0.5 and 0.1 were set according to distributions in proteomic and transcriptomic analyses, respectively. Using a principal component analysis (Supplementary Figure S4B and C), we identified samples not clustered properly with their respective replicates and discarded them from subsequent analysis. We first monitored changes in gene expression induced by 20A by comparison of transcriptomes of untreated and 20A-treated HeLa cells using Agilent microarrays. We found a good correlation between log fold changes at 6 h (x-axis) and 16 h (y-axis) in transcriptomic (R = 0.68) and proteomic data (R = 0.81) (Supplementary Figure S5A and B). The number of up-regulated genes was higher than the number of down-regulated genes (595 versus 95 after 6 h of treatment; 317 versus 184 after 16 h) as evidenced by the volcano plots (Supplementary Figure S5C).
Differential expression
Statistical analyses were performed in R version 3.3.1. We processed the proteomic data using DESeq2 package version 1.14.1 with default parameters (45). Proteomic data have the same biases as gene expression data in that the high abundance of some proteins and lengths of the proteins are known to distort the signal. We used the normalized abundance per gene as counts. We processed the transcriptomic data using limma package version 3.30.13 following usual preprocessing steps (46): agilent files were loaded and processed, background correction was applied, control probes were removed, quantile normalization was applied, average signal per gene were computed and results were annotated using R package biomaRt version 2.30.0 (47) with Ensembl release 87. We used the data from 6-h control samples for comparisons to data from both 6 and 16 h 20A-treated samples and modeled gene expression with linear model functions from the limma package. The full transcriptome/proteome analysis is provided as a separate xls file.
Functional enrichment and KEGG pathways
Functional enrichment was computed using command line version of GSEA software and the Hallmark datasets (48–50). We then deepened the analyses using the KEGG datasets (51). We put a maximum threshold of 0.01 on the FDR. We next used the pathview R package version 1.14.0 to visualize the behavior of genes and proteins at 6 and 16 h in relevant KEGG pathways (52).
G4 enrichment
G4 regions were predicted by analysis of the human genome version hg38 using G4Hunter with default parameters (threshold = 1.52; window = 25) (2). A threshold of 1.2 on the score was used. Statistical enrichment of G4 complexes around the transcription start sites and transcription terminal sites of differentially expressed transcripts (FDR <0.01 and the absolute log fold change >1) was computed by a binomial test. The expected frequencies were estimated from the observed frequency in all genes present in the transcriptomic analysis. We used a sliding window of 100 bp with 50-bp step from the transcription start site or the termination site to 1 kb upstream (promoter, 3′UTR) and downstream (5′UTR, downstream of the gene).
Immunofluorescence analyses
Cells were fixed at room temperature with 4% (v/v) paraformaldehyde for 10 min and then permeabilized with 50 μg/ml digitonin before incubation with the LC3 antibody. Cells were then stained with appropriate Alexa Fluor-conjugated secondary antibody. Nuclei were counterstained for 10 min with Hoechst 33258. The fluorescence of cells was examined using a Zeiss LSM510 META confocal microscope (Zeiss) with an ApoPLAN ×63 objective. Identical exposures were used for each channel throughout individual experiments.
Lentiviral vector production and cell transduction
The CRISPR-Cas9 lentivirus system was used to generate autophagy-deficient HeLa cell lines. The pLenti CRISPR (pXPR) vector expressing Cas9 and the single guide (sg)RNAs that target ATG5 or ATG7 or non-specific sgRNA control were constructed and validated in the laboratory of Prof. Kevin M Ryan (Beatson Institute of Health, University of Glasgow). The lentiviral tdTomato-expressing vector (a generous gift from Prof. Richard Iggo, University of Bordeaux) was used to monitor tumor growth in the in vivo experiments. For cell transduction experiments, cells were plated in a 6-well plate (105 cells/well) 24h prior to infection with either the tdTomato vector or the CRISPR expressing vectors (sgControl, sgATG5 and sgATG7) for 24 h. The titer of each lentiviral batch was determined on HeLa cells. At 24h post infection, cells expressing the vector tdTomato or the CRISPR-Cas9 vectors were selected in culture media supplemented with either 10 μg/ml blasticidine for 5 days or 2 μg/ml puromycin for 72 h, respectively.
Transient transfection with small interfering RNA
A non-specific control small interfering RNA (siRNA) (Sense: 5′-CUUACGCUGAGUACUUCGAtt-3′) and a specific siRNA that targets ATM (sense 5′-AACATACTACTCAAAGACAtt-3′) were previously validated (53) and were synthesized by Eurogentec. Cells were plated in a 12-well plate (4 × 104 cells/well) and were transiently transfected with 100 nM siRNA using Oligofectamine (ThermoFisher) following the manufacturer’s instructions. After 24 h, cells were treated with drugs at the indicated concentrations.
Autophagy assay
Autophagy was evaluated by (i) western blotting analysis of the abundance of the autophagosome marker LC3 in the presence and absence of the lysosomal inhibitors, (ii) a fluorescence microscopy counting of cells with LC3 puncta and (iii) cellular ultrastructure analysis of the presence of autophagosome by transmission electron microscopy (TEM). Briefly, cell pellets were fixed in a 1.6% glutaraldehyde solution in 0.1 M cacodylate buffer at room temperature and stored overnight at 4°C. After three rinses in 0.1 M cacodylate buffer (15 min each), pellets were post-fixed in a 1% osmium tetroxide and 1% potassium ferrocyanide solution in 0.1 M cacodylate buffer for 1 h at room temperature to enhance the staining of cytoplasmic membranes. The cells were subsequently dehydrated in a series of acetone baths (90%, 100% three times, 15 min each) and progressively embedded in Epon 812 (Fluka) resin (acetone/resin 1:1, 100% resin two times, 2 h for each bath). Resin blocks were hardened in a 60°C oven for 2 days. Ultrathin sections (70 nm) were obtained with a Reichert Ultracut S ultramicrotome equipped with a Drukker International diamond knife and collected on 200-mesh copper grids. The sections were stained with lead citrate and uranyl acetate. TEM observations were performed with a JEOL JEM-1400 transmission electron microscope equipped with a Morada camera at a 100 kV acceleration voltage.
Xenograft tumor formation and immunohistochemistry
The study was performed in accordance with European Community Standards of Care (animal house authorization number B33063916). Approval for the animal experiments was obtained by the Comité d’Ethique pour l’Experimentation Animale (CEEA50), Bordeaux (project number APAFIS≠ 9321-2017031417171591). For xenograft tumor formation, 2 × 106 tdTomato-expressing HeLa cells were suspended in 100 μl of RPMI medium and subcutaneously injected in the right flank of 8-week-old male NSG mice (NOD-scid IL2RG−/−, Jackson Laboratory) (54). When the tdTomato fluorescence average within the tumors reached 6 × 109, the tumor-bearing mice were randomly assigned to three groups (10 mice/group) and received peritumoral injections of 100 μl of vehicle (15% DMSO) or 20A at either 10 or 20 mg/kg per mouse 5 days a week for a total of 12 days. Weights were scored weekly. The tdTomato fluorescence within the tumor was measured twice a week using a PhotonIMAGER (Biospace) from day 35 after cell injection. Results are expressed in photon intensity (photon/second/steradian) and normalized for each group to the values of fluorescence obtained at day 35 after cell injection. At the day of the sacrifice, tumor blocks were fixed in 4% formol and paraffin embedded for hematoxylin-eosin-saffron staining and immunohistochemical labeling of cell proliferation marker Ki67, apoptosis marker cleaved-caspase 3 and Sudan Black B for detection of lipofuscin-containing senescent cells following the respective manufacturer’s instruction. High-quality images were made with slide scanner Nanozoomer (Hamamatsu) in the Bordeaux Imaging Center. Tumor morphology and immunohistochemical staining were analyzed by a pathologist.
Statistical analysis
Data are presented as means and error bars as standard deviations (SD) of at least three independent experiments unless otherwise stated. Comparisons of the different groups were made with either Mann–Whitney unpaired test or unpaired t test. For the in vivo experiments, the results are expressed as means, error bars as standard errors of the mean (SEM) and comparisons of the different groups were with Mann–Whitney unpaired test. Values of *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 were considered statistically significant. The software used was GraphPad Prism.
RESULTS
Antiproliferative effects of 20A result from induction of senescence and apoptosis
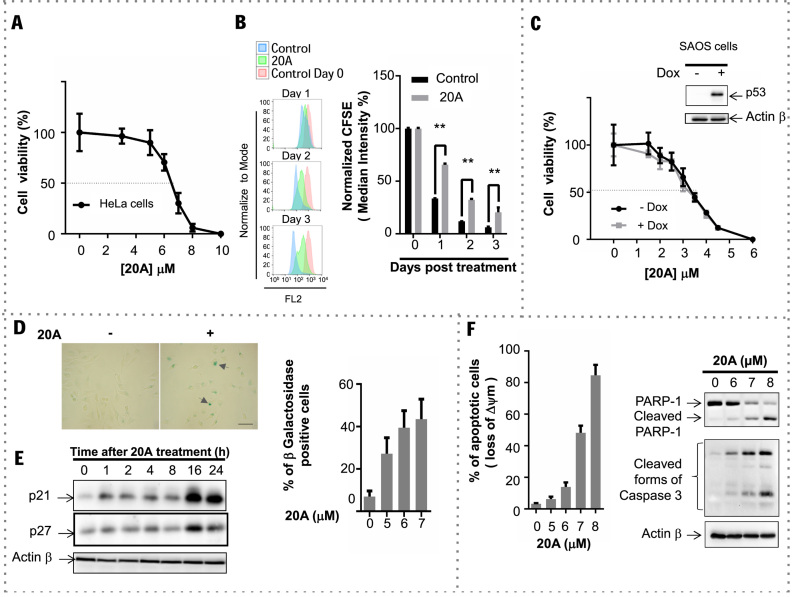
20A inhibits the viability of HeLa cells in a dose-dependent manner with a half maximal inhibitory concentration (IC50) of 6.5 μM after 24 h of treatment (Figure 1A). 20A treatment compromises cell division as demonstrated by the delay of CFSE dye dilution, which occurs due to cell division (55), as compared to untreated cells (Figure 1B).
Figure 1.
20A treatment results in cancer cell growth arrest through the induction of senescence and apoptosis. (A) Viability of HeLa cells after a 24-h exposure to 20A at concentrations ranging from 1 to 10 μM. Plotted are means ± S.D. of three independent experiments, each performed in quadruplicate. (B) HeLa cells were labeled with 1 μM CFSE (day 0) and then treated with 6 μM 20A. Flow cytometry analysis of the CFSE fluorescence intensity was monitored on indicated days. Left, representative flow cytometry traces in which the red histogram corresponds to the day of cell staining (day 0). Control cells were mock treated. Right, results expressed as MFI normalized to that obtained from day zero. Plotted are means ± S.D. of six values obtained from two independent experiments, each performed in triplicate. **P < 0.01 using Mann–Whitney test. (C) Saos-2 cells were grown in either the presence or absence of 0.5 μg/ml Dox for 6 h and then treated for 24 h with the indicated concentration of 20A. Plotted are means ± S.D. of three independent experiments, each performed in quadruplicate. Inset represents western blot analysis of p53 expression levels in Saos-2 cells grown in the presence or absence of 0.5 μg/ml Dox for 6 h; ACTINβ was detected as a loading control. (D) HeLa cells were treated with 6 μM 20A for the indicated time and then stained for β-galactosidase activity to detect senescence. Left, representative images of cells stained for β-galactosidase (arrowheads indicate senescent cells; scale bar = 50 μm). Total cell number and total number of β-galactosidase-positive cells were scored to determine the fraction of senescent cells. Data are means ± S.D. of four independent experiments; at least 100 cells from 10 randomly chosen fields were counted in each experiment. *P < 0.05 using Mann–Whitney test. (E) Representative immunoblot analysis of p21, p27 and ACTINβ levels in HeLa cells treated with 6 μM 20A for the indicated time. (F) Apoptotic cell death evaluated after treatment of HeLa cells with the indicated concentration of 20A for 24 h. Left, percentage of cells that have lost mitochondrial transmembrane potential (Δψm) plotted versus 20A concentration. Results are means ± S.D. of triplicate data obtained from one experiment. Right, immunoblot analysis of cleaved forms of caspase 3 and PARP1 in cells treated with the indicated concentrations of 20A; ACTINβ was detected as a loading control.
To determine if p53 status influences the loss in cell viability caused by 20A treatment, we used the Saos-2 osteosarcoma cell line. These cells display no endogenous p53 expression but contain a Dox-inducible p53 transgene (56). As illustrated in Figure 1C, both Saos-2 cells that do not express p53 (-Dox) and Saos-2 cells expressing the p53 transgene (+Dox) were equally sensitive to 20A as shown by nearly superimposable survival curves (IC50 values are 3.3 and 3.2 μM, respectively). This experiment demonstrated that the antiproliferative effect of 20A is independent of the presence or absence of p53 in cells.
To explore the mechanisms responsible for the antiproliferative action of 20A, we further examined senescence and apoptotic cell death, two well-defined processes involved in cell growth arrest and programmed cell death, respectively. Senescence was evaluated by using Senescence Associated β-Galactosidase assay. 20A treatment resulted in a dose-dependent increase in the percentage of cells stained for β-galactosidase (Figure 1D), suggesting that 20A causes senescence. We also evaluated the levels of two senescence markers, p21 and p27. As shown in Figure 1E, 20A treatment elicited a significant time-dependent up-regulation of both p21 and p27 proteins in HeLa cells.
Treatment of cells with 20A also induced a dose-dependent activation of apoptosis as evidenced by loss of the mitochondrial transmembrane potential as well as the appearance of cleaved forms of caspase 3 and PARP1 (Figure 1F). Accordingly, in cells treated with QVD-OPh, an inhibitor of caspases, cell death induced by 20A was reduced (Supplementary Figure S1A). Moreover, senescence and apoptosis were observed after 48 and 72 h (Supplementary Figure S1B and C) suggesting that the cells do not escape senescence over time. 20A treatment also led to senescence and apoptosis in other cancer cells such as lung carcinoma A549 cells (which express p53) (Supplementary Figure S2A) and Saos-2 cells expressing or not p53 (Supplementary Figure S2B). Together, these results demonstrate that 20A treatment causes a loss of cell viability by inducing senescence and caspase-driven apoptotic cell death in a p53-independent manner.
20A has antitumor effects in vivo
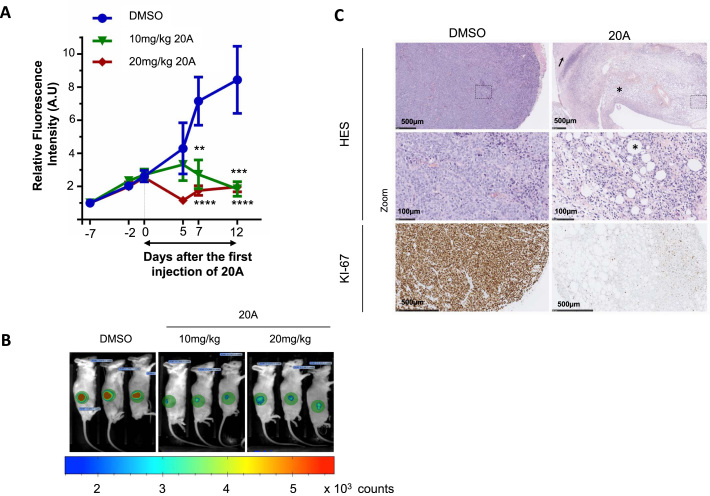
The significant antiproliferative effect of 20A in HeLa cells prompted us to determine whether this compound had antitumor properties in a mouse model of cancer. To this end, HeLa cells that stably express tdTomato, which encodes a red fluorescent protein, were subcutaneously injected into the dorsal flank of NOD scid γ mice. Tumor growth was monitored by live imaging of the red fluorescence within the xenografts. When the tumors reached a sufficient volume, peritumoral daily injections of either DMSO (vehicle control) or 10 or 20 mg/kg 20A were given over a period of 12 days. Tumor growth was significantly reduced after 12-day treatment with both doses of 20A as evidenced in live animal images (Figure 2A, 3 representative animals out of 10 mice are presented for each group in Figure 2B). During the period of the treatment, the average weight of the mice from the two 20A-treated groups did not differ significantly from that of the control group, suggesting that 20A had low toxicity in this in vivo model (Supplementary Figure S3A). At the end of the treatment, the tumor characteristics were analyzed. Hematoxylin, eosin and saffron staining of DMSO-treated xenografts revealed the presence of marked atypia and mitotic activity with scattered inflammatory cells in HeLa tumor xenografts (Figure 2C, left panel). In contrast, 20A-treated tumor samples had a low density of malignant cells and areas of necrosis, fibrosis and adiponecrosis were observed in HeLa xenografts from mice treated with 20 mg/kg 20A (Figure 2C, right panel) and 10 mg/kg 20A (Supplementary Figure S3B, right panel). Immunohistochemistry labeling for the DNA replication marker Ki67 revealed that the percentage of proliferating cells in 20A-treated tumor xenografts was reduced compared to DMSO-treated tumors (Figure 2C). Tumor xenografts treated with 10 mg/kg 20A manifested also some features of apoptosis or senescence as evidenced by the cleaved forms of caspase 3 and Sudan Black B labeling, respectively (Supplementary Figure S3B). Taken together, these in vivo data confirm those obtained in vitro and indicate that 20A treatment promotes an arrest of tumor growth along with induction of apoptosis and senescence.
Figure 2.
20A treatment compromises tumor growth in mice bearing HeLa xenografts. (A) The tdTomato fluorescence within tumors was recorded in HeLa xenografts in mice during the treatment period with DMSO (control) or 10 or 20 mg/kg 20A per injection. Tumor growth is plotted as means ± S.E.M. of the relative fluorescence intensity within the xenografts scored on indicated days following tumor implantation from 10 mice per group. P **<0.01; ***<0.001; ****<0.0001 with Mann–Whitney test compared to DMSO-treated group. (B) For each treatment group, images of three representative mice are shown at day 12 after treatment initiation. The fluorescence intensity is presented as a rainbow scale with increasing intensity from blue to red. The green circles represent areas used to measure total fluorescence intensity. (C) Representative images of hematoxylin-eosin-saffron stained DMSO-treated and 20 mg/kg 20A-treated tumor sections performed at the end of the treatment period. Arrow indicates tumor cells with necrosis and asterisk shows fibrosis and adiponecrosis. Immunohistochemistry (IHC) analyses for Ki67 expression were also performed on the paraffin-embedded sections of tumor tissues.
Molecular pathways altered by 20A treatment
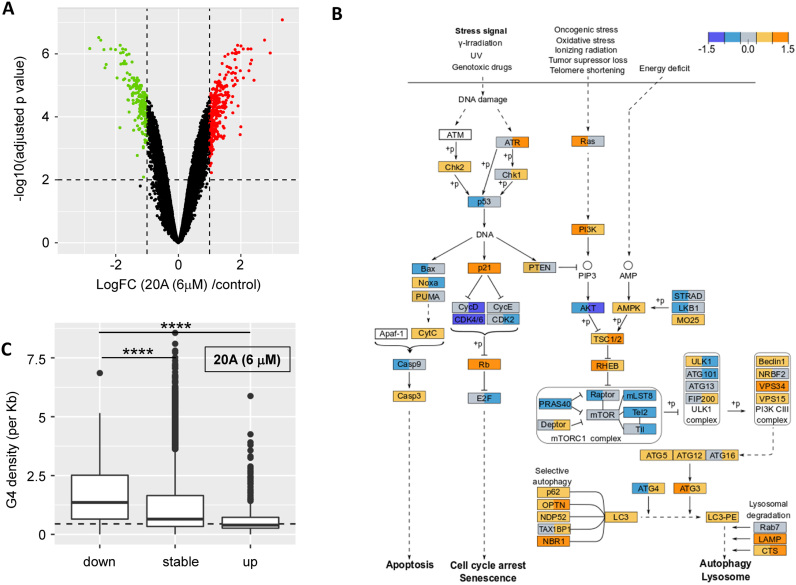
To identify the biological pathways and signaling networks affected by 20A, we investigated its global effects on gene transcription and protein expression levels in HeLa cells (Supplementary Figures S4 and S5). 20A induces both up- and down-regulation of genes (595 versus 95 after 6 h of treatment, Supplementary Figure S5C; 317 versus 184 after 16 h, as shown in Figure 3A). An enrichment analysis performed using the Broad Gene Set Enrichment Analysis (GSEA) and the Molecular Signatures Database (MSigDB) (50,52) revealed that 20A induces a significant enrichment in a number of biological pathways including pathways involved in DNA damage (p53_PATHWAY), apoptosis (APOPTOSIS) and lysosomal function (MTORC1_SIGNALING, PROTEIN_ SECRETION) (Table 1; a complete list of genes up- or down-regulated is provided as a Supplementary File 1 in excel format). Using the KEGG database, we found a significant enrichment of biological pathways related to the DNA damage (MISMATCH_REPAIR, up-regulated) and cell growth (CELL_CYCLE, down-regulated) as well as pathways related to autophagy (REGULATION_OF_AUTOPHAGY and LYSOSOME) (Supplementary Table S1). The diagram in Figure 3B recapitulates the transcriptome analysis of three interconnected KEGG signaling pathways (p53, MTORC1 and AUTOPHAGY) that are significantly enriched in response to 20A treatment (Supplementary Table S2 and Supplementary Figure S6). This figure reveals that 20A elicits (i) activation of DDR as evidenced by up-regulation of Chk1 and Chk2, which encode downstream effectors of ATR and ATM, respectively; (ii) up-regulation of two senescence effectors, p21 and pRb, and the down-regulation of the Rb target, E2F; (iii) the modulation of apoptosis as evidenced by up-regulation of Cytc and Casp3 and down-regulation of both Bax and Casp9; and (iv) a significant down-regulation of expression of genes encoding elements of the MTORC1 complex associated with increased expression of a large panel of autophagic and lysosomal genes (e.g. Beclin1, VPS34, ATG5, ATG12, ATG3, LC3, p62, OPTN, NBR1, Lamp and CTS) (23,57,58).
Figure 3.
Transcriptome analysis of 20A treated cells. (A) Global changes in gene expression in 20A treated cells. limma differential expression results are plotted with the x-axis showing the log fold changes between 6 μM 20A treatment versus control cells (after 16 h, Figure 3A) or 6 h, Figure S2C) and the y-axis showing the significance of the statistical tests (p corrected for multi-testing minus log transformed). Green and red dots highlight significantly down- and up-regulated genes (absolute log fold change above 1 and significance above 2; P < 0.01). Results related to the proteomics analysis are presented in Figure S2A and S2B and Table S1. (B) Results obtained on three interconnected KEGG signaling pathways that are significantly enriched in response to 20A treatment (p53, MTORC1 and autophagy; each pathway is individually shown in Figure S6). For each gene product, the value of log fold change at 6 h (in the left part of the box) and log fold change at 16 h (in the right part of the box) are presented and denoted with the color code presented on the top of the figure. Log fold change values between −0.3 and 0.3 are colored in gray, those between 0.3 and 0.9 are in light orange, above 0.9 are in orange, between -0.3 and -0.9 are in light blue and those below -0.9 are in dark blue. Solid lines represent direct regulation, dotted lines represent indirect regulation and ‘+p’ indicates regulation by phosphorylation. (C) G4 density in gene bodies for genes down-regulated by 20A, not significantly affected (‘stable’) by 6 μM 20A, or up-regulated after 16 h of treatment with 20A. Results obtained after 6 h are presented in Figure S7B. ****p< 0.0001 versus genes down-regulated.
Table 1.
Hallmark gene sets significantly enriched (False discovery rate (FDR) q-value <0.05) after either 6 or 16 h of treatment with 6 μM 20A in transcriptomic or the proteomic data
| 20A treatment (6 h) | 20A treatment (16 h) | ||||
|---|---|---|---|---|---|
| HALLMARK | NES | FDR q-val | HALLMARK | NES | FDR q-val |
| Transcriptomics | |||||
| MYC_TARGETS_V1 | 2.6544 | 0 | KRAS_SIGNALING_DN | −2.1145 | 0 |
| MTORC1_SIGNALING | 2.6135 | 0 | CHOLESTEROL_HOMEOSTASIS | 2.6057 | 0 |
| PROTEIN_SECRETION | 2.4228 | 0 | MTORC1_SIGNALING | 2.4009 | 0 |
| CHOLESTEROL_HOMEOSTASIS | 2.4154 | 0 | PROTEIN_SECRETION | 2.2930 | 0 |
| ANDROGEN_RESPONSE | 2.2860 | 0 | FATTY_ACID_METABOLISM | 2.0078 | 0.00077 |
| TNFA_SIGNALING_VIA_NFKB | 2.1413 | 0 | ANDROGEN_RESPONSE | 1.9120 | 0.00081 |
| COMPLEMENT | 1.9466 | 0.00032 | OXIDATIVE_PHOSPHORYLATION | 1.7578 | 0.00396 |
| HYPOXIA | 1.8538 | 0.00125 | BILE_ACID_METABOLISM | 1.7664 | 0.00422 |
| GLYCOLYSIS | 1.7893 | 0.00273 | COMPLEMENT | 1.6984 | 0.00605 |
| TGF_BETA_SIGNALING | 1.8056 | 0.00281 | EPITHELIAL_MESENCHYMAL_TRANSITION | −1.7906 | 0.00639 |
| UNFOLDED_PROTEIN_RESPONSE | 1.7613 | 0.00297 | GLYCOLYSIS | 1.6756 | 0.00713 |
| OXIDATIVE_PHOSPHORYLATION | 1.7665 | 0.00305 | KRAS_SIGNALING_UP | 1.6542 | 0.00778 |
| KRAS_SIGNALING_DN | −1.9267 | 0.00306 | HEDGEHOG_SIGNALING | −1.6975 | 0.00938 |
| FATTY_ACID_METABOLISM | 1.7571 | 0.00309 | NOTCH_SIGNALING | −1.7226 | 0.00940 |
| E2F_TARGETS | 1.7529 | 0.00321 | PEROXISOME | 1.6287 | 0.01003 |
| HEME_METABOLISM | 1.6345 | 0.01004 | APICAL_JUNCTION | −1.6543 | 0.01156 |
| G2M_CHECKPOINT | 1.6470 | 0.01020 | HYPOXIA | −1.5587 | 0.01568 |
| IL2_STAT5_SIGNALING | 1.6357 | 0.01067 | MYC_TARGETS_V2 | −1.5682 | 0.01711 |
| UV_RESPONSE_DN | 1.5911 | 0.01544 | HEME_METABOLISM | 1.5718 | 0.01773 |
| P53_PATHWAY | 1.5564 | 0.01965 | TNFA_SIGNALING_VIA_NFKB | −1.4714 | 0.03031 |
| APOPTOSIS | 1.5230 | 0.02433 | MYOGENESIS | −1.4066 | 0.04704 |
| KRAS_SIGNALING_UP | 1.4755 | 0.04037 | XENOBIOTIC_METABOLISM | 1.4561 | 0.04803 |
| ESTROGEN_RESPONSE_LATE | 1.4584 | 0.04467 | UNFOLDED_PROTEIN_RESPONSE | 1.4620 | 0.04819 |
| E2F_TARGETS | 1.4496 | 0.04832 | |||
| Proteomics | |||||
| OXIDATIVE_PHOSPHORYLATION | 2.7854 | 0 | OXIDATIVE_PHOSPHORYLATION | 2.4181 | 0 |
| INFLAMMATORY_RESPONSE | 2.0673 | 0.00315 | ESTROGEN_RESPONSE_EARLY | 1.8408 | 0.00976 |
| ADIPOGENESIS | 1.9909 | 0.00724 | ADIPOGENESIS | 1.8952 | 0.00976 |
| COMPLEMENT | 1.8457 | 0.01836 | PROTEIN_SECRETION | 1.8448 | 0.01083 |
| APOPTOSIS | 1.8347 | 0.01894 | APOPTOSIS | 1.8111 | 0.01252 |
| FATTY_ACID_METABOLISM | 1.8702 | 0.01937 | HEME_METABOLISM | 1.9069 | 0.01302 |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 1.8049 | 0.02273 | IL2_STAT5_SIGNALING | 1.6552 | 0.04166 |
| P53_PATHWAY | 1.6676 | 0.04325 | |||
| COAGULATION | 1.6358 | 0.04325 | |||
| ESTROGEN_RESPONSE_LATE | 1.5982 | 0.04647 | |||
| EPITHELIAL_MESENCHYMAL_TRANSITION | 1.5855 | 0.04767 | |||
NES is the Normalized Enrichment Score given by GSEA, positive values mean an up-regulation of the majority of the genes or proteins involved in the corresponding Hallmark set, whereas negative values indicate down-regulation of the majority of genes involved in the Hallmark set. The FDR q-value represents the adjusted P-value.
The proteome data were also analyzed with the GSEA, MSigDB and KEGG databases (50,52), which revealed that several Hallmark sets were affected by 20A treatment (Table 1 and Supplementary Table S1). These sets included those found in the transcriptome experiments (i.e. apoptosis, the p53 pathway and protein secretion).
To determine whether a causal link existed between changes in transcript or protein levels and the presence of G4 sequences within key regulatory elements of the corresponding genes, we performed a bioinformatics analysis using G4-Hunter (2). We analyzed for the enrichment of sequences with G4-forming potential around transcription start and end sites in up- and down-regulated transcripts. The analysis showed an enrichment of sequences with G4-forming potential directly upstream (50–150 bp) of transcription termination sites of genes down-regulated after both 6 and 16 h of treatment with 20A (Supplementary Figure S7A). This observation is in line with previous results showing that G4 ligands may not only regulate transcription via the regulation of promoter activity but may also influence later transcriptional events as well as the maturation of specific messenger RNAs (10).
We then performed an analysis of G4 density in gene bodies of up-regulated, and down-regulated transcripts and transcripts unchanged by 20A treatment. Interestingly, and in agreement with its proposed mode of action, down-regulated gene bodies carry higher G4 densities (expressed as number of G4 sites per kb) than unchanged genes and up-regulated genes (Figure 3C for analysis after 16 h; 6-h data shown in Supplementary Figure S7B). This observation, which is significant both after 6 and 16 h, suggests that a high density of G4 structures bound to 20A block transcription, whereas the quasi-absence of G4-forming motifs in the gene bodies of up-regulated genes protects these genes from down-regulation by 20A.
20A promotes a global DNA damage response but not telomeric damage
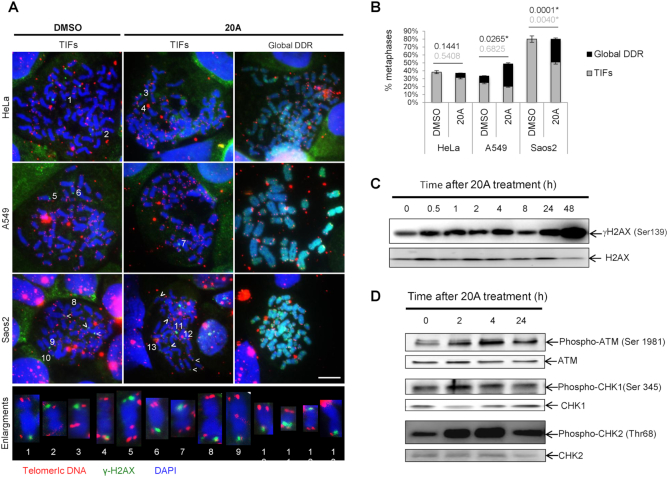
As several G4Ls have been shown to induce telomeric DNA damage, we investigated whether this is the case for 20A using the meta-TIF assay (40). We scored the abundance of TIF, which correspond to sites of DNA damage (as evidenced by the DNA damage marker γH2AX) that co-localize with telomeres revealed by a telomere-specific peptide nucleic acid probe. As shown in Figure 4A, 20A did not augment the frequency of TIF in any of the cell lines tested. This observation, together with the quantitative polymerase chain reaction (qPCR) assay results (Supplementary Figure S8), reinforces our conclusion that no important telomere shortening occurred after 20A treatment.
Figure 4.
20A causes global DNA damage but not telomeric damage. (A) Representative metaphases for HeLa, A549 and Saos2 cells displaying TIFs or global DDR under two experimental conditions, 24-h treatment with 20A or DMSO. Telomeric DNA (TelC, red), (γH2AX, green) and (4′,6-Diamidino-2-phenylindoledihydrochloride (DAPI), blue). Enlargements of chromosomes with TIFs are shown; scale bar = 10 μM. (B) Quantification of metaphases with at least one TIF or global DDR. Two biological replicates, n = 35–45 metaphases per condition. P-values, statistics of proportions for Global DDR (black print) and TIFs (gray print), * p value < 0.05 versus untreated cells. (C) HeLa cells were treated with 6 μM 20A at time indicated and then subjected to western blotting analysis for γH2AX and total H2AX. (D) Western blotting analysis of the levels of phospho-ATM (Ser 1981), ATM, phospho-CHK1 (Ser 345), CHK1, phospho-CHK2 (Thr68) and CHK2 in HeLa cells following treatment with 6 μM 20A for the indicated times.
However, we observed that 20A treated cells elicit increased in the frequencies of metaphase chromosomes entirely covered by γH2AX (which we call here global DDR) as compared to untreated cells, which was statistically significant for A549 and Saos-2 cells (Figure 4B). These results indicate that 20A induces DNA damage without enrichment at chromosomal ends. A time-course experiment showed the γH2AX levels increased as early as 30 min following 20A treatment and remained elevated after 48 h of treatment. (Figure 4C; the quantification of four independent experiments over a 4-h time course is presented in Supplementary Figure S9).
This global DDR prompted us to examine whether the effects of classical DNA damaging agents correlate with the transcription profile of 20A. To test this hypothesis, we located publicly available expression data from HeLa cells treated with X-rays or chemicals. After abstract curation and dataset exploration, we selected three studies: GSE19526 (X-ray irradiation); GSE95227 (UV-C irradiation) and GSE72905 (ruthenium complex treatment). We computed expression changes between treated (X-ray, neutron, RuT7) and control HeLa cells and observed no correlation between the effects of these treatments and 20A effects. These results are presented in detail in Supplementary Information Part II. These analyses clearly demonstrate that the observed transcriptional alterations induced by 20A do not correspond to a classical general response to DNA damage.
To gain further insight into the regulation of the DDR pathway by 20A, we explored the activation of the ATM and ATR, two key DDR sensors (59), after 20A treatment. As shown in Figure 4D, cells subjected to 20A treatment displayed a rapid and robust stimulation of ATM and its downstream target CHK2 (as indicated by increased phosphorylation on Ser 1981 and Thr 68, respectively) along with a rapid activation of the downstream target of ATR, CHK1 (as indicated by its increased phosphorylation Ser 345). These results suggest that both ATM and ATR pathways are activated in response to 20A (Figure 4D).
ATM activation drives cells to senescence to prevent cell death induced by 20A
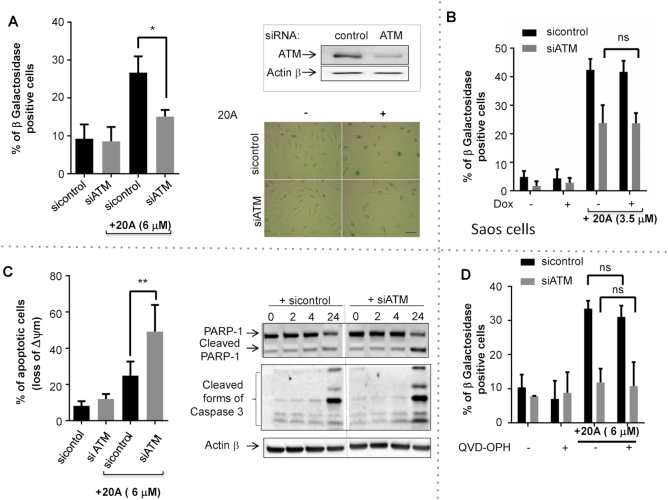
To evaluate the role of ATM in senescence and apoptosis induced by 20A, we used siRNAs to silence expression of ATM. As shown in Figure 5A, loss of ATM significantly reduced senescence induced by 20A, irrespective of the presence or absence of p53 protein (Figure 5B). Moreover, ATM silencing markedly enhanced cell killing induced by 20A as evidenced by loss of mitochondrial transmembrane potential and the appearance of the cleaved forms of PARP1 and caspase 3 (Figure 5C), and this response also occurred in a p53-independent manner (Supplementary Figure S10C). Similar results were found with KU55933, an ATM kinase inhibitor (Supplementary Figure S10A and B). Of note, inhibition of ATM activation compromises 20A-induced senescence in the presence or absence of the broad caspase inhibitor QVD-OPh, suggesting that reduced senescence in absence of ATM is not due to increased apoptosis (Figure 5D). We also investigated the consequences of ATM inhibition on the expression of CHK1 and the cyclin dependent kinase inhibitors, p16, p21 and p27. As shown in Supplementary Figure S10D-E, depletion of ATM following 20A treatment reduced CHK1 expression and led to p27 up-regulation, but no significant effect was observed on the levels of p21 or p16. This suggests that ATM regulates the stability of both CHK1 and p27 through an as yet unidentified mechanism. Altogether, these results suggest that ATM is involved in the establishment of senescence onset to protect cells against apoptotic cell death induced by 20A.
Figure 5.
ATM is activated by 20A to direct cells to senescence instead of apoptosis. In panels A, C and D HeLa cells were transiently transfected with either ATM siRNA or control siRNA. 20A (6 μM) was added 24 h later for a period of 24 h. (A) Left: the percent of senescent cells determined as described in Figure 1D. Data are means ± S.D. of four independent experiments. *P < 0.05 using Mann–Whitney test. Upper right, immunoblot analysis of the levels of ATM protein expression in either the presence or absence of siRNA ATM. Lower right, representative images of cells treated as indicated and stained for β-galactosidase; scale bar = 50 μm. (B) Saos-2 cells were grown in either the presence or absence of 0.5 μg/ml Dox for 6 h and then treated for 24 h with 3.5 μM 20A. The percentage of senescent cells was determined as described in Figure 1D. Data correspond to the mean ± S.D. of three independent experiments. (C) Left, apoptotic cell death was scored by measurement of loss of Δψm. The data are presented as means ± S.D. of eight values obtained from four independent experiments each performed in duplicate. **P < 0.01 using Mann–Whitney test. Right, immunoblot images of the appearance of the cleaved forms of both caspase 3 and PARP1 are shown. (D) Cells were incubated for 2 h with QVD-OPH (20 μM) prior the addition of 6 μM 20A for 24 h. The percent of senescent cells determined as described in Figure 1D. Data correspond to the mean ± S.D. of three independent experiments. *ns: not significant by t-test.
20A treatment promotes ATM-dependent autophagy activation and p62 up-regulation
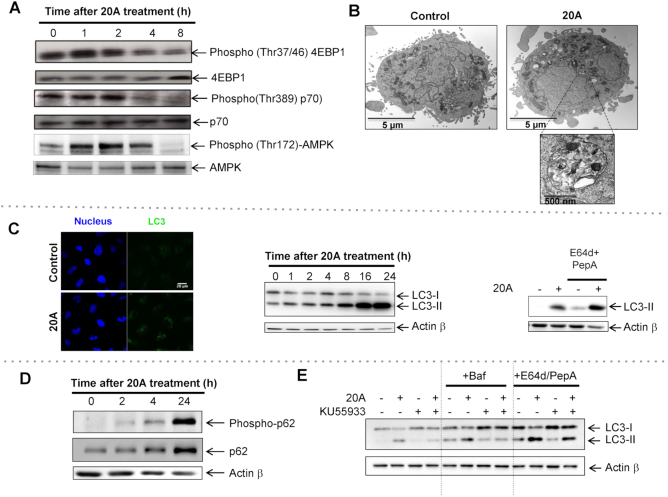
Based on the transcriptome and proteome results, we reasoned that the autophagy pathway is triggered by 20A. We thus explored the activation of the MTOR and AMPK axes, two pivotal pathways that control autophagy negatively and positively, respectively (22). As illustrated in Figure 6A and Supplementary Figure S11A, treatment of HeLa cells with 20A causes MTOR inhibition as evidenced by a decrease in the phosphorylation of mTOR (on Ser 2488) and of its two substrates, p70 (on Thr 389) and 4-EBP1 (on Thr 37/46), as early as 4 h after treatment with 20A was begun. We also observed a rapid and transient activation of AMPK (as indicated by an increase of its phosphorylation on Thr 172 and activation of its substrates; Figure 6A and Supplementary Figure S11B).
Figure 6.
20A promotes autophagy induction through a mechanism that involves ATM activation. (A) HeLa cells were treated for the indicated times with 6 μM 20A. The activities of the MTORC1 and AMPK pathways were determined by immunoblot analysis using antibodies directed against phospho-4EBP1 (Thr37/46), 4EBP1, phospho-p70 (Thr 389), p70, phospho-AMPK (Thr 172) and AMPK. (B) Representative electron micrographs of HeLa cells untreated or treated for 16 h with 6 μM 20A. (C) Left, immunofluorescence analysis for the abundance of LC3 puncta (green dots) in HeLa cells after 3 h in either the absence or presence of 6 μM 20A; Hoechst was used to stain nuclei (blue). Representative confocal images are shown; scale bar = 10 μm. Center, immunoblot images of LC3-II levels in HeLa cells treated with 6 μM 20A for the indicated times. Right, immunoblot of cells treated with or without E64d (10 μg/ml) plus pepstatin A (2 μg/ml) 2 h prior 20A treatment (6 μM, 8 h). (D) Immunoblot analysis of phospho-p62 (Ser 403) and p62 levels in HeLa cells treated with 6 μM 20A for the indicated times. (E) Immunoblot analysis of LC3-II levels in HeLa cells incubated with or without 10 μM KU55933 for 2 h and then treated with 6 μM 20A for 8 h. Where indicated, cells were incubated with 10 μg/ml E64d plus 2 μg/ml pepstatin A for 2 h prior 20A treatment. Where noted, bafilomycin A1 (50 nM) was added 3 h before cell lysis.
To further evaluate the autophagy activity in response to 20A, we determined the abundance of autophagosomes by performing cellular ultrastructure analysis and by studying changes in the subcellular localization of the autophagosome marker LC3. Electron micrographs of HeLa cells revealed that 20A treatment caused accumulation of multi-membrane vesicles containing electron-dense materials with features of autophagosomes (Figure 6B). The increase in abundance of autophagosomes induced by 20A was confirmed by immunofluorescence, which showed a significant increase in LC3 fluorescent dots in 20A-treated cells as compared to untreated cells (Figure 6C, left). Western blotting analysis also showed a time-dependent accumulation of LC3-II, the form of LC3 protein conjugated to phosphatidylethanolamine (Figure 6C, center), a marker of the autophagosomal membrane that is eventually degraded by lysosomal proteases (22). Moreover, 20A-induced accumulation of LC3-II was enhanced in the presence of lysosomal protease inhibitors E64d and pepstatin A, indicating that the material sequestered within the autophagosomes was successfully addressed to lysosomes and degraded after 8 h of treatment (Figure 6C, right). We also examined p62, a protein involved in the selective transport of specific cellular cargos to autophagosomes (60). As shown in Figure 6D, 20A promoted up-regulation of p62 levels concomitantly with an increase in the phosphorylation of p62 on serine 403, known to regulate the selective autophagy-mediated degradation of ubiquitinated proteins. Of note, the expression levels of p62 were also regulated by the proteasome system in these cells since inhibition of proteasome by Epoxomicin markedly enhanced 20A-induced up-regulation of p62 protein levels (Supplementary Figure S11C).
As ATM activation has been previously reported to stimulate autophagy following DNA damage (34), we further explored the influence of this pathway on autophagy induction by 20A. The effect of the ATM inhibitor KU55933 on the accumulation of the autophagy marker LC3-II in the presence and absence of two autophagy flux inhibitors was evaluated. As presented in Figure 6E, the inhibition of ATM activation by KU55933 prevented 20A-induced accumulation of LC3-II. This effect was not attributable to the activation of the autophagy flux, as under this condition the levels of LC3-II were not enhanced compared to that of 20A-treated cells when autophagy flux was inhibited by bafilomycin A1 or E64d and pepstatin A. Similar results were obtained when ATM expression was silenced by siRNA treatment (Supplementary Figure S11D). Altogether, these data indicate that inhibition of ATM activation counteracts 20A-induced autophagy, supporting the idea that ATM activation is responsible for the activation of autophagy.
Loss of autophagy impairs 20A-induced senescence and drives cells to apoptosis
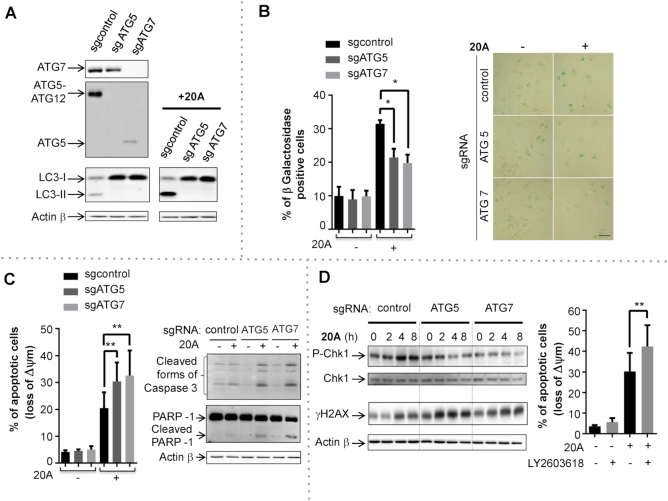
In order to evaluate the role of autophagy on global DDR and cell growth arrest induced by 20A, we used the CRISPR-Cas9 system to generate two HeLa cell lines with complete autophagy disruption resulting from deletion of key autophagy genes ATG5 or ATG7. ATG5 or ATG7 levels were undetectable in the respective CRISPR-targeted cells as shown by western blotting (Figure 7A). It is worth noting that disruption of autophagy did not have an effect on cell viability under these growth conditions (Supplementary Figure S11E). As expected, loss of ATG5 or ATG7 abrogated the induction of basal levels of autophagy as well as of 20A-induced autophagy as evidenced by immunoblot analysis of LC3-II expression levels (Figure 7A). Moreover, the disruption of autophagy led to accumulation of the autophagy substrate p62 both in control and 20A-treated cells (Supplementary Figure S11F).
Figure 7.
Autophagy disruption causes a deficiency in CHK1 activation and sensitizes cells to apoptosis induced by 20A. (A) Autophagy disruption in HeLa cells was validated by immunoblot analysis of ATG5, ATG7, LC3-II in autophagy-proficient (sgControl) and autophagy-deficient (sgATG5 and sgATG7) cells following treatment with 6 μM 20A for 16 h. (B) Left, the percentages of senescent cells were scored by β-galactosidase staining as described in Figure 1D in sgControl, sgATG5 and sgATG7 cells after 24h treatment with 6 μM 20A. The data are presented as means ± S.D. of four independent experiments. *P < 0.05 using Mann–Whitney test. Right, representative cell images; scale bar = 50 μm. (C) Autophagy-proficient (sgControl) and autophagy-deficient (sgATG5 and sgATG7) cells were treated with 6 μM 20A for 24 h. Left, loss of mitochondrial transmembrane potential was evaluated by flow cytometry. Data are means ± S.D. of 12 values obtained from four independent experiments each performed in triplicate. **P < 0.01 using Mann–Whitney test. Right, immunoblot analysis of cleaved forms of caspase 3 and PARP1. (D) Left, immunoblot analysis of phospho-CHK1 (Ser 345) and γH2AX (Ser 139) levels in sgControl, sgATG5 and sgATG7 cells treated with 6 μM 20A for the indicated time. Right, loss of mitochondrial transmembrane potential was evaluated by flow cytometry assay in HeLa cells pretreated for 2 h with the CHK1 inhibitor LY2603618 (0.5 μM) prior to treatment with 6 μM 20A for an additional 24 h. The data are presented as means ± S.D. of nine values obtained from three independent experiments each performed in triplicate. **P < 0.01 using Mann–Whitney test.
To determine the functional role of autophagy in the antiproliferative effects of 20A, we next examined the effect of autophagy disruption on senescence and apoptosis triggered by 20A treatment. As shown in Figure 7B, the 20A-induced senescence phenotype was significantly reduced in both ATG5- and ATG7-deficient cells as compared to control cells. Concomitantly, autophagy-deficient cells exhibited higher levels of apoptosis relative to control cells as evidenced by loss of mitochondrial transmembrane potential and cleavage of both Caspase 3 and PARP1 (Figure 7C). These results corroborate with those observed when ATM was inhibited (Figure 5) underscoring the contribution of autophagy in modulating the balance between senescence and apoptosis upon 20A treatment.
Because loss of autophagy following DNA damage has been reported to cause dephosphorylation of CHK1 (at Ser 345) (61), we explored whether this effect occurs in response to 20A. As shown in Figure 7D (left, panel), autophagy-deficient cells exhibited significantly lower levels of CHK1 phosphorylation in response to 20A as compared to autophagy-proficient cells. Similarly, loss of ATM led to a significant decrease of 20A-induced both CHK1 activation and expression arguing for the implication of ATM in the control of CHK1 stability (Supplementary Figure S10D). Furthermore, loss of autophagy resulted in an enhanced accumulation of γH2AX at earlier time points after the addition of 20A, in a time window where apoptotic cell death was not observed, supporting the idea that the accumulation of γH2AX is, at least in part, independent of apoptotic DNA fragmentation. To determine whether inhibition of CHK1 activation can phenocopy the loss of autophagy, we examined the effect of LY2603618, a specific inhibitor of CHK1 in HeLa cells. Pretreatment of wild-type HeLa cells with LY2603618 enhanced 20A-induced apoptotic cell death to the same extent than what was observed in HeLa cells in which autophagy was disrupted Figure 7D (right, panel). We thus speculate that CHK1 is one of the regulators of autophagy-mediated DDR and cell survival following 20A treatment.
DISCUSSION
Given that the outcome of cancer therapy relies on the cancer cell fate—survival, senescence or death—the identification of regulatory mechanisms that impact this fate is required for optimization of the efficacy of cancer treatment (33,62). The triarylpyridine family of G4 ligands represents a novel class of DNA ligands that are potential anticancer agents (21). In the present study, we provide evidence that 20A, a representative compound of this family of G4 ligands, inhibits cancer cell growth both in vitro and in vivo. We show that this compound induces senescence and apoptosis in a p53-independent manner. Notably, we demonstrated that the activation of the ATM/autophagy pathway by 20A operates as a linchpin between senescence and apoptosis. To our knowledge, this is the first demonstration that the ATM/autophagy axis is involved in the cell fate choice between senescence and apoptosis in cancer cells treated with a G4 ligand.
The ability of G4 ligands to promote senescence was previously attributed to stabilization of telomeric G4 structures, resulting in telomerase inhibition, slowed telomere attrition and eventually telomere uncapping (5,12). Replicative senescence is triggered by telomere shortening and appears after several weeks of treatment with G4 ligands at sub-lethal doses (5,14). However, increasing evidence indicates that non-telomeric sites also play essential roles in the biological effects of G4 ligands (7–10). We observed no increase in telomeric damage upon treatment of HeLa cells with 20A (Figure 4A and B), arguing against preferential action of this compound at chromosomal ends. We therefore assume that 20A does not promote replicative senescence but rather a senescence phenotype that resembles stress-induced (premature) senescence, which appears within a period as short as a day after cells are exposed to stress of chemical or physical nature that induces DNA damage (63–65).
In support of this mechanism, our transcriptomic and proteomic analyses revealed a strong stimulation of the senescence pathway (including up-regulation of p21) with the concurrent activation of the DDR in response to 20A treatment. We found that 20A promoted a rapid activation of two key pathways implicated in the DDR: ATM/CHK2 and ATR/CHK1. Inhibition of ATM following 20A treatment reduced the senescence onset and exacerbated apoptotic cell death, indicating that ATM functions as a linchpin between senescence and apoptosis. Our findings extend previous evidence that ATM is required for survival of cancer cells that undergo premature senescence in response to DNA damage agents and oxidative stress conditions (i.e. doxorubicin, camptothecin and hydrogen peroxide) (66). ATM has been shown to regulate metabolic reprogramming in cancer cells (by controlling the levels of glucose and glutamine) to prevent bypass of senescence (67). Here, we showed that in response to 20A, senescence is accompanied by up-regulation of p21 and both processes occur in a p53-independent manner. A growing body of evidence suggests that in addition to p53, alternative pathways downstream of ATM (e.g. cyclin-dependent kinase inhibitors, CHK2 and C-MYC) control senescence mediated by ATM (59,66–69). A number of p53-independent pathways are also involved in the transcriptional induction of p21 and its stability (e.g. transcription factors including SP1, SP2, AP2 and STAT1 as well as signaling pathways governed by CHK2, JNK, p38 and AKT) and this might contribute to p21-mediated senescence (68,70,71). Of note, we found that ATM regulates senescence through both autophagy-dependent and autophagy-independent mechanisms as disruption of autophagy following 20A treatment only partially inhibited senescence. Further studies are required to shed light on the precise network that underlies senescence induced by G4 ligands such as 20A.
Interestingly, it was reported that the cell sensitivity to G4 ligands is correlated with the basal level of DNA damage and telomere deprotection (11). Moreover, some G4 ligands preferentially target cancer cells deficient for DNA repair, as shown specifically for cells harboring BRCA1 and BRCA2 mutations (15,72). In the latter study, inhibition of DNA repair using a PARP inhibitor enhanced the antitumor effect of G4 ligand RHPS4 in vitro and in tumor xenografts (51). In the same vein, evidence indicates that a dual G4/PARP ligand has a significant antiproliferative potential in cancer cells (52). Collectively, these data support the idea that the modulation of DDR can influence the susceptibility of cells to the cytotoxic effect of a G4 ligand.
In a transcriptome analysis, we found that 20A caused an enrichment of the lysosomal regulatory pathways (i.e. the MTOR and Protein Secretion Hallmarks as well as the KEGG pathway REGULATION_OF_AUTOPHAGY). In particular, this analysis provides evidence that ATM activation is responsible for the induction of autophagy by 20A. Such regulation of autophagy has been previously reported in response to other DNA damage agents and may constitute a mechanism through which ATM enables cells to escape death by entering senescence (74,75). In keeping with this scenario, we found that disruption of autophagy by deletion of either of two key autophagy genes, ATG5 or ATG7, directed cells toward apoptosis instead of senescence. The relationship between autophagy and senescence is complex as in some settings autophagy accelerates the establishment of the senescence phenotype, whereas in other circumstances autophagy impedes this process (32,76). The activation of autophagic flux and the up-regulation of the autophagy receptor p62 by 20A support the idea that autophagy participates in changes in metabolism and extensive cellular remodeling that occur during cellular senescence, a mechanism that may confer cell adaptation to DNA damage (34). In addition, we found that 20A promoted activation of AMPK with a concurrent inhibition of MTOR activity, two pathways that link autophagy to metabolism and DDR (34). Further investigation is required to determine the exact mechanism through which autophagy operates to maintain the integrity of senescent cells treated with 20A.
Although the induction of senescence by chemo- or radiotherapies was initially considered as a tumor-suppressive mechanism, emerging evidence supports the notion that this response can also promote tumorigenesis (76). In contrast to apoptotic cells, senescent cells are viable and have the ability to influence tumor growth as well as tumor response to treatment by secretion of pro-inflammatory and mitogenic factors (76). Whether or not 20A promotes the secretion of such factors warrant further investigation.
We found that the activation of autophagy by 20A regulates DDR through the stabilization of CHK1 phosphorylated at Ser 345; this phosphorylation is known to coordinate the DDR and the cell-cycle checkpoint response (77). Deficiency in autophagy coupled with 20A treatment led to failure in CHK1 activation and was associated with increased levels of γH2AX. Similar results were observed upon loss of ATM suggesting that CHK1 stability is also regulated by ATM. Such interplay between ATM and ATR/CHK1 has been also reported in response to some DNA damage agents (78). Our data are in line with a recent study showing that autophagy-deficient cells have impaired homologous recombination through a mechanism that involves proteasome degradation of phospho-CHK1 (61). Interestingly, the dephosphorylation of CHK1 has been also reported in A549 cells treated with the G4 ligand 12459 (79). The phosphatase PPM1D (also known as WIP1) was shown to be responsible for CHK1 inactivation in cell treated with G4L 12459.
Autophagy induction by G4 ligands Ant1,5, SYUIQ-5 and CX-5461, three compounds unrelated to 20A, was previously reported (13,36,37). Zaffaroni et al. showed that Ant1,5 causes cell growth arrest, increased p21 expression and induction of cytoprotective autophagy in cells (13). It is noteworthy that these responses occurred without induction of senescence, which is different from the mechanism identified here for 20A. The G4 ligand SYUIQ-5 causes cell death through a mechanism that depends on autophagy (37). This suggests that the activation of autophagy in response to a G4 ligand can have different cellular outcomes that may be related to the chemical properties of the compound, the concentration used, and the specific signaling pathways activated by the ligand. Further investigations are required to decipher the context-dependent role of autophagy in response to different classes of G4 ligands. Given that the mechanism underlying the regulation of autophagy by G4 ligands is yet not understood, it will be interesting to determine if this regulation relies on binding of the ligand to particular G4 structures.
Collectively, our data provide evidence for the antiproliferative effects of 20A in cancer models in vivo and in vitro. Our study also revealed that targeting of the ATM and autophagy pathways is a promising strategy to achieve the optimal efficiency of this G4 ligand in cancer cells. This property and its p53-independent effects on senescence and caspase-driven apoptotic cell death underline the therapeutic potential of 20A in a large panel of tumors.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (80) partner repository with the dataset identifier PXD009430. We have submitted the list of genes ordered by the log fold changes (returned by DESeq2 and limma) to microarray data GEO accession number GSE113424.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all reviewers for helpful suggestions. The authors gratefully acknowledge the BIC platform in Bordeaux and L. Lacroix for G4Hunter bed files. The authors also acknowledge the Cell and Tissue Imaging (PICT-IBiSA) and Nikon Imaging Centre, Institut Curie, member of the French National Research Infrastructure France-BioImaging (ANR10-INBS-04).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ligue Régionale d’Aquitaine, comité de Dordogne (to J.L.M., M.D.M.); INCa-DGOS-Inserm 6046 (to M.D.M.); FR Transbiomed (to M.D.M.); SYMBIT project (reg. no. CZ.02.1.01/0.0/0.0/15_003/0000477) financed by the ERDF (to J.L.M.). Funding for open access charge: INSERM funding. CCMA electron microscopy equipments have been funded the Région Sud-Provence-Alpes-Côte d'Azur, the Conseil Général des Alpes Maritimes, and the GIS-IBiSA.
Conflict of interest statement. None declared.
REFERENCES
- 1. Huppert J.L., Balasubramanian S.. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bedrat A., Lacroix L., Mergny J.-L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 4. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 5. Riou J.F., Guittat L., Mailliet P., Laoui A., Renou E., Petitgenet O., Mégnin-Chanet F., Hélène C., Mergny J.L.. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S.. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005; 65:1489–1496. [DOI] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rigo R., Palumbo M., Sissi C.. G-quadruplexes in human promoters: a challenge for therapeutic applications. Biochim. Biophys. Acta. 2017; 1861:1399–1413. [DOI] [PubMed] [Google Scholar]
- 9. Cammas A., Millevoi S.. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017; 45:1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fay M.M., Lyons S.M., Ivanov P.. RNA G-Quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017; 429:2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salvati E., Rizzo A., Iachettini S., Zizza P., Cingolani C., D’Angelo C., Porru M., Mondello C., Aiello A., Farsetti A. et al.. A basal level of DNA damage and telomere deprotection increases the sensitivity of cancer cells to G-quadruplex interactive compounds. Nucleic Acids Res. 2015; 43:1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang F.-C., Chang C.-C., Wang J.-M., Chang T.-C., Lin J.-J.. Induction of senescence in cancer cells by the G-quadruplex stabilizer, BMVC4, is independent of its telomerase inhibitory activity. Br. J. Pharmacol. 2012; 167:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orlotti N.I., Cimino-Reale G., Borghini E., Pennati M., Sissi C., Perrone F., Palumbo M., Daidone M.G., Folini M., Zaffaroni N.. Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy. 2012; 8:1185–1196. [DOI] [PubMed] [Google Scholar]
- 14. Salvati E., Leonetti C., Rizzo A., Scarsella M., Mottolese M., Galati R., Sperduti I., Stevens M.F.G., D’Incalci M., Blasco M. et al.. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Invest. 2007; 117:3236–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmer J., Tacconi E.M.C., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A. et al.. Targeting BRCA1 and BRCA2 deficiencies with G-Quadruplex-Interacting compounds. Mol. Cell. 2016; 61:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohnmacht S.A., Marchetti C., Gunaratnam M., Besser R.J., Haider S.M., Di Vita G., Lowe H.L., Mellinas-Gomez M., Diocou S., Robson M. et al.. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015; 5:11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Porru M., Zizza P., Franceschin M., Leonetti C., Biroccio A.. EMICORON: a multi-targeting G4 ligand with a promising preclinical profile. Biochim. Biophys. Acta. 2017; 1861:1362–1370. [DOI] [PubMed] [Google Scholar]
- 18. Ohnmacht S.A., Neidle S.. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014; 24:2602–2612. [DOI] [PubMed] [Google Scholar]
- 19. Duarte A.R., Cadoni E., Ressurreição A.S., Moreira R., Paulo A.. Design of modular G-quadruplex ligands. ChemMedChem. 2018; 13:869–893. [DOI] [PubMed] [Google Scholar]
- 20. Smith N.M., Labrunie G., Corry B., Tran P.L.T., Norret M., Djavaheri-Mergny M., Raston C.L., Mergny J.-L.. Unraveling the relationship between structure and stabilization of triarylpyridines as G-quadruplex binding ligands. Org. Biomol. Chem. 2011; 9:6154–6162. [DOI] [PubMed] [Google Scholar]
- 21. Kerkour A., Mergny J.-L., Salgado G.F.. NMR based model of human telomeric repeat G-quadruplex in complex with 2,4,6-triarylpyridine family ligand. Biochim. Biophys. Acta. 2017; 1861:1293–1302. [DOI] [PubMed] [Google Scholar]
- 22. Klionsky D.J., Codogno P.. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013; 5:427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I. et al.. Molecular definitions of autophagy and related processes. EMBO J. 2017; 36:1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamb C.A., Yoshimori T., Tooze S.A.. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013; 14:759–774. [DOI] [PubMed] [Google Scholar]
- 25. Kroemer G., Mariño G., Levine B.. Autophagy and the integrated stress response. Mol. Cell. 2010; 40:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johansen T., Lamark T.. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011; 7:279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosenfeldt M.T., Ryan K.M.. The multiple roles of autophagy in cancer. Carcinogenesis. 2011; 32:955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White E. The role for autophagy in cancer. J. Clin. Invest. 2015; 125:42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galluzzi L., Pietrocola F., Bravo-San Pedro J.M., Amaravadi R.K., Baehrecke E.H., Cecconi F., Codogno P., Debnath J., Gewirtz D.A., Karantza V. et al.. Autophagy in malignant transformation and cancer progression. EMBO J. 2015; 34:856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levy J.M.M., Towers C.G., Thorburn A.. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017; 17:528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang C., Elledge S.J.. How autophagy both activates and inhibits cellular senescence. Autophagy. 2016; 12:898–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gewirtz D.A. Autophagy and senescence in cancer therapy. J. Cell. Physiol. 2014; 229:6–9. [DOI] [PubMed] [Google Scholar]
- 33. Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W. et al.. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018; 25:486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eliopoulos A.G., Havaki S., Gorgoulis V.G.. DNA damage response and autophagy: a meaningful partnership. Front. Genet. 2016; 7:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hewitt G., Korolchuk V.I.. Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol. 2017; 27:340–351. [DOI] [PubMed] [Google Scholar]
- 36. Zhou W.-J., Deng R., Feng G.-K., Zhu X.-F.. [A G-quadruplex ligand SYUIQ-5 induces autophagy by inhibiting the Akt-FOXO3a pathway in nasopharyngeal cancer cells]. Ai Zheng Aizheng Chin. J. Cancer. 2009; 28:1049–1053. [DOI] [PubMed] [Google Scholar]
- 37. Zhou W.-J., Deng R., Zhang X.-Y., Feng G.-K., Gu L.-Q., Zhu X.-F.. G-quadruplex ligand SYUIQ-5 induces autophagy by telomere damage and TRF2 delocalization in cancer cells. Mol. Cancer Ther. 2009; 8:3203–3213. [DOI] [PubMed] [Google Scholar]
- 38. Trocoli A., Mathieu J., Priault M., Reiffers J., Souquere S., Pierron G., Besançon F., Djavaheri-Mergny M.. ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy. 2011; 7:1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Georgakopoulou E.A., Tsimaratou K., Evangelou K., Fernandez Marcos P.J., Zoumpourlis V., Trougakos I.P., Kletsas D., Bartek J., Serrano M., Gorgoulis V.G.. Specific lipofuscin staining as a novel biomarker to detect replicative and stress-induced senescence. A method applicable in cryo-preserved and archival tissues. Aging. 2013; 5:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cesare A.J., Kaul Z., Cohen S.B., Napier C.E., Pickett H.A., Neumann A.A., Reddel R.R.. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 2009; 16:1244–1251. [DOI] [PubMed] [Google Scholar]
- 41. Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Souquère S., Pierron G., Codogno P.. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J. Biol. Chem. 2006; 281:30373–30382. [DOI] [PubMed] [Google Scholar]
- 42. Henriet E., Abou Hammoud A., Dupuy J.-W., Dartigues B., Ezzoukry Z., Dugot-Senant N., Leste-Lasserre T., Pallares-Lupon N., Nikolski M., Le Bail B. et al.. Argininosuccinate synthase 1 (ASS1): a marker of unclassified hepatocellular adenoma and high bleeding risk. Hepatol. Baltim. Md. 2017; 66:2016–2028. [DOI] [PubMed] [Google Scholar]
- 43. Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J.. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007; 4:923–925. [DOI] [PubMed] [Google Scholar]
- 44. Pineda E., Thonnus M., Mazet M., Mourier A., Cahoreau E., Kulyk H., Dupuy J.-W., Biran M., Masante C., Allmann S. et al.. Glycerol supports growth of the Trypanosoma brucei bloodstream forms in the absence of glucose: analysis of metabolic adaptations on glycerol-rich conditions. PLoS Pathog. 2018; 14:e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. et al.. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 49. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P.. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo W., Brouwer C.. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinforma. Oxf. Engl. 2013; 29:1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li J., Stern D.F.. Regulation of CHK2 by DNA-dependent protein kinase. J. Biol. Chem. 2005; 280:12041–12050. [DOI] [PubMed] [Google Scholar]
- 54. Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J. et al.. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. Baltim. Md 1950. 2005; 174:6477–6489. [DOI] [PubMed] [Google Scholar]
- 55. Fulcher D., Wong S.. Carboxyfluorescein succinimidyl ester-based proliferative assays for assessment of T cell function in the diagnostic laboratory. Immunol. Cell Biol. 1999; 77:559–564. [DOI] [PubMed] [Google Scholar]
- 56. Crighton D., Wilkinson S., O’Prey J., Syed N., Smith P., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M.. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006; 126:121–134. [DOI] [PubMed] [Google Scholar]
- 57. Mizushima N., Yoshimori T., Ohsumi Y.. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011; 27:107–132. [DOI] [PubMed] [Google Scholar]
- 58. Boya P., Reggiori F., Codogno P.. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013; 15:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith J., Tho L.M., Xu N., Gillespie D.A.. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010; 108:73–112. [DOI] [PubMed] [Google Scholar]
- 60. Lamark T., Svenning S., Johansen T.. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017; 61:609–624. [DOI] [PubMed] [Google Scholar]
- 61. Liu E.Y., Xu N., O’Prey J., Lao L.Y., Joshi S., Long J.S., O’Prey M., Croft D.R., Beaumatin F., Baudot A.D. et al.. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmitt C.A., Fridman J.S., Yang M., Lee S., Baranov E., Hoffman R.M., Lowe S.W.. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002; 109:335–346. [DOI] [PubMed] [Google Scholar]
- 63. Chen J.-H., Hales C.N., Ozanne S.E.. DNA damage, cellular senescence and organismal ageing: causal or correlative?. Nucleic Acids Res. 2007; 35:7417–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013; 75:685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakamura A.J., Chiang Y.J., Hathcock K.S., Horikawa I., Sedelnikova O.A., Hodes R.J., Bonner W.M.. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin. 2008; 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maréchal A., Zou L.. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aird K.M., Worth A.J., Snyder N.W., Lee J.V., Sivanand S., Liu Q., Blair I.A., Wellen K.E., Zhang R.. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep. 2015; 11:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aliouat-Denis C.-M., Dendouga N., Van den Wyngaert I., Goehlmann H., Steller U., van de Weyer I., Van Slycken N., Andries L., Kass S., Luyten W. et al.. p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol. Cancer Res. MCR. 2005; 3:627–634. [DOI] [PubMed] [Google Scholar]
- 69. Zhang X., Li J., Sejas D.P., Pang Q.. The ATM/p53/p21 pathway influences cell fate decision between apoptosis and senescence in reoxygenated hematopoietic progenitor cells. J. Biol. Chem. 2005; 280:19635–19640. [DOI] [PubMed] [Google Scholar]
- 70. Gartel A.L., Tyner A.L.. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp. Cell Res. 1999; 246:280–289. [DOI] [PubMed] [Google Scholar]
- 71. Jung Y.-S., Qian Y., Chen X.. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell. Signal. 2010; 22:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J. et al.. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Salvati E., Botta L., Amato J., Di Leva F.S., Zizza P., Gioiello A., Pagano B., Graziani G., Tarsounas M., Randazzo A. et al.. Lead discovery of dual G-Quadruplex stabilizers and Poly(ADP-ribose) polymerases (PARPs) inhibitors: a new avenue in anticancer treatment. J. Med. Chem. 2017; 60:3626–3635. [DOI] [PubMed] [Google Scholar]
- 74. Singh K., Matsuyama S., Drazba J.A., Almasan A.. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy. 2012; 8:236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kang H.T., Park J.T., Choi K., Kim Y., Choi H.J.C., Jung C.W., Lee Y.-S., Chul Park S.. Chemical screening identifies ATM as a target for alleviating senescence. Nat. Chem. Biol. 2017; 13:616–623. [DOI] [PubMed] [Google Scholar]
- 76. Campisi J., d’Adda di Fagagna F.. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007; 8:729–740. [DOI] [PubMed] [Google Scholar]
- 77. Sørensen C.S., Hansen L.T., Dziegielewski J., Syljuåsen R.G., Lundin C., Bartek J., Helleday T.. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005; 7:195–201. [DOI] [PubMed] [Google Scholar]
- 78. Yan S., Sorrell M., Berman Z.. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell. Mol. Life Sci. CMLS. 2014; 71:3951–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Douarre C., Mergui X., Sidibe A., Gomez D., Alberti P., Mailliet P., Trentesaux C., Riou J.-F.. DNA damage signaling induced by the G-quadruplex ligand 12459 is modulated by PPM1D/WIP1 phosphatase. Nucleic Acids Res. 2013; 41:3588–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al.. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (80) partner repository with the dataset identifier PXD009430. We have submitted the list of genes ordered by the log fold changes (returned by DESeq2 and limma) to microarray data GEO accession number GSE113424.