Abstract
Annotation of gene expression in prokaryotes often finds itself corrected due to small variations of the annotated gene regions observed between different (sub)-species. It has become apparent that traditional sequence alignment algorithms, used for the curation of genomes, are not able to map the full complexity of the genomic landscape. We present DeepRibo, a novel neural network utilizing features extracted from ribosome profiling information and binding site sequence patterns that shows to be a precise tool for the delineation and annotation of expressed genes in prokaryotes. The neural network combines recurrent memory cells and convolutional layers, adapting the information gained from both the high-throughput ribosome profiling data and ribosome binding translation initiation sequence region into one model. DeepRibo is designed as a single model trained on a variety of ribosome profiling experiments, used for the identification of open reading frames in prokaryotes without a priori knowledge of the translational landscape. Through extensive validation of the model trained on various sets of data, multiple species sequence similarity, mass spectrometry and Edman degradation verified proteins, the effectiveness of DeepRibo is highlighted.
INTRODUCTION
After >20 years of genome sequencing, it has become clear that the genomic diversity in bacteria is much larger than expected, not only between species but also within (1). GenBank for example currently holds over 10 000 genome assemblies for Escherichia coli, one of the prokaryotic model organisms, displaying stunning diversity. The vast number of sequenced prokaryotes across all different phyla makes it impractical to perform genome comparison based on sequence alignments to unravel the genomic complexity (2). Even though sequence alignment is conventionally used, annotation of de novo genes by similarity of properties (i.e. DNA sequence) between previously annotated genes is biased and has shown to propagate errors from anteceding misannotations (3). The novel prediction tool presented in this article is based only on features extracted from the short DNA sequence covering the ribosome binding site and expression data.
The delineation of the open reading frame (ORF) is an essential element in gene annotation and is mostly performed in silico (4,5). ribosome profiling (also called ribo-seq) measures mRNA that is associated with ribosomes by sequencing ribosome-protected fragments (6,7). Ribo-seq experimentally enables the ORF delineation, and the technique has already been successfully adopted for prokaryotes (8,9). An important aspect of the ORF delineation is the determination of the Translation Initiation Site (TIS). Here also, specific prediction tools are in place to perform this task (10–12), but these TISs can also be detected by applying a specific antibiotic treatment (e.g. chloramphenicol or tetracycline) preceding the ribo-seq protocol enriching for initiating ribosomes (13). Recently, prediction methods based on machine learning algorithms have been devised to either delineate the ORF (14) or predict the TIS (15) based on a combination of ribosome profiling and sequence features for prokaryotic genomes. A multitude of tools are available for eukaryotic organisms (16–21).
Alternative proteoform usage can also be investigated by specific mass spectrometry protocols measuring N-terminal peptides (22,23). Although the technology is recognized, it suffers from drawbacks (e.g. peptide physical properties and modifications, mass spectrometry measurement range…), limiting the number of detectable N-termini. In order to attain a more comprehensive map of proteoform usage, proteogenomics studies have combined the aforementioned high-throughput sequencing and mass spectrometry information, resulting in more precise ORF and TIS validation and thus genome annotation. (24,25).
In this article, we present DeepRibo, a novel neural network implementation applying ribosome profiling data and binding site patterns for the precise annotation of TISs in prokaryotes. The use of artificial neural networks, which have proven to be highly effective in solving complex problems given the availability of sufficient data, is still confined to few applications in the field of bioinformatics. Examples are the use of convolutional neural networks for the prediction of DNA- or RNA-binding with a target protein (26) or precise variant calling on next-generation sequencing (BioRxiv: https://doi.org/10.1101/092890). DeepRibo is an artificial neural network that applies both convolutional neural network (CNN) and recurrent neural network (RNN) architectures in order to process information from the DNA sequence and ribosome profiling signal. Only a short DNA sequence of 30 nucleotides covering the ribosome binding region is processed by the neural network. Predictions are based on features extracted from this region, selected through prior knowledge, and enhanced with features extracted from the ribosome profiling signal.
DeepRibo is trained on a combination of available experiments for different bacteria and has been tested to work equally well on de novo ribo-seq data of bacterial genomes. We managed to successfully train a highly precise model that is able to process ribo-seq data without loss of resolution. We further validated our results with multiple species sequence similarity comparison (27), available mass spectrometry data and translation initiation site annotations (28).
MATERIALS AND METHODS
DeepRibo is trained on data collected from ribosome profiling data. Ribo-seq data has the advantage that it does not map the untranslated regions of the transcribed mRNA. It upholds a high resolution and low background noise, making precise gene annotation possible. In prokaryotes, no splicing of the mRNA occurs, giving rise to more straightforward patterns of the signal along the coding regions as compared to eukaryotes. Conversely, bacterial genes are tightly packed and are frequently overlapping, which impedes a straightforward annotation. In order to detect genomic features, the model is designed to evaluate a set of possible ORFs containing ribo-seq signal, from which the top k ranked probability scores are selected to be expressed genes. The model is furthermore trained on a short DNA sequence covering a 30nt region overlapping with both the the Shine-Dalgarno (SD) motif in SD-led genes (up to 20nt upstream of the TIS) and ribosome binding region in leaderless genes (up to 10nt downstream the TIS). The ribosome binding site has proven to be of major importance in predicting the presence of a TIS (10,29). Sequences no longer than 30nt are considered to prevent DeepRibo from training on intragenic DNA patterns.
Sample selection using the four parameter S-curve
The input (candidate ORF) samples, labeled using the latest genome assemblies of the species, is the collection of all possible ORFs meeting a minimum signal strength. As ribosome profiling changes according to the expression profile of the organism at the time of the experiment, no signal is present along several parts of the genome. Practically, it is not possible to make any predictions about these regions based on the expression data. Before selection of the positive and negative data, all candidate ORFs containing low ribosome profiling signal are therefore not considered when training/evaluating the model. The remaining data is afterwards labeled using the annotations retrieved from the NCBI Reference Sequence (RefSeq) database. The selection of data is based upon two properties of the samples, the coverage and signal read count. The coverage indicates the nucleotide fraction of the candidate ORF at which signal is present. The signal read count, expressed as Reads Per Kilobase Million (RPKM), expresses the amount of reads within the sample as compared to the dataset read count. Since the biggest partition of the considered dataset has zero to low coverage, a more balanced distribution of read count, coverage, and label values is obtained from the filtered input samples. Moreover, the final dataset contains about one-fifth of the input samples as compared to all candidate ORFs present in the collected data.
To determine the minimum cut-off values for coverage and RPKM, a method introduced by Ndah et al. (14) has been applied. The method is based upon threshold dose-response estimation done by Lutz et al. (30). For this, a four parameter S-curve is fitted on the coverage in function of the RPKM. Only the positive samples are considered when fitting the S-curve. By predicting the lower bend of the fitted S-curve, minimum cut-off values of the signal coverage and RPKM for each dataset are obtained. This point is of importance as it separates the positive samples that can be distinguished from the background noise. This point is defined as the point from which an increase in RPKM within the positively labeled candidate ORFs is correlated to the coverage of the ribo-seq signal in said dataset. Using this technique, it is possible to pool the data from several individual experiments, as the S-curve is fitted on each dataset individually.
To label the samples, the public genome annotations of the referred species are used. Indeed, the assumption is made that DeepRibo, trained on data labeled via sequence alignment, can offer precise predictions by learning from the ribo-seq signal instead of using the full DNA sequences as an input. Although it is expected that the annotated genomes contain errors because of the shortcomings of prevalent but more conservative DNA sequence alignment methods, this behaviour is not mimicked as the model does not learn the DNA sequences of the coding sequences.
Neural network architecture
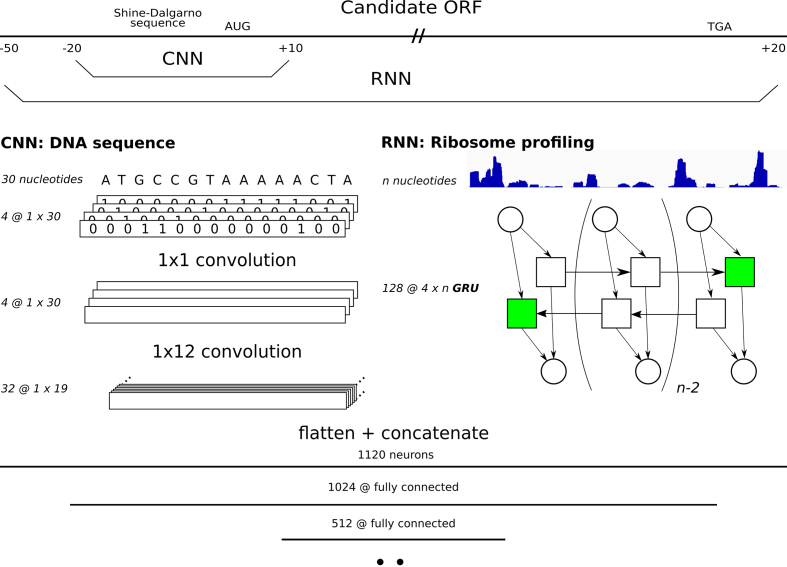
DeepRibo is a neural network built in PyTorch (31), of which the architecture is presented in Figure 1. It is specifically designed to process two types of data: strings (i.e. DNA sequences) and floats (i.e. ribo-seq signal). The model first processes each type of data in parallel before combining the features created from both inputs into a set of fully-connected layers. The DNA sequence is transformed into a binary image with four channels, a method proposed by Alipanahi (26). This image is consecutively processed by two convolutional layers. The first layer transforms the sparse matrix into a dense matrix using four 1 × 1 convolutional kernels. Afterwards, 32 kernels of 1 × 12 convolutions process the data in the second and last convolutional layer. The ribosome profiling data is fed into a double-layered, bidirectional Gated Recurrent Unit (GRU). The gated recurrent unit was selected instead of the long short-term memory cell as it showed to train better models and it was overall faster to train. Only the final hidden states of the memory cell are retrieved for further processing, making the use of varied length inputs (i.e. candidate ORFs) possible. After each type of data is processed, the output nodes of both networks are concatenated and fed into a fully-connected layer. The final layers of the network consist of three fully connected layers that combine the features of both the Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN) to obtain a final prediction. The rectified linear unit is applied as the activation function for each layer but the last. The binary cross entropy is used as the loss function during training.
Figure 1.
The architecture of the neural network DeepRibo. For each candidate ORF two types of data are processed and fed into their respective parts of the neural network. The convolutional layers train on a 30 nucleotide DNA sequence ranging from 20 nucleotides upstream to 10 nucleotides downstream of the TIS. The recurrent neural network covers the complete ORF from 50 nucleotides upstream of the start codon, including the SD region, and extending 20 nucleotides downstream of the stop codon. The DNA sequence is first translated in a binary image before being processed by four 1 × 1 and 32 1 × 12 convolutional kernels, respectively. The ribosome profiling data is processed by a double layered bidirectional GRU of 128 hidden nodes. The outputs of both neural networks are flattened and concatenated and fed into three consecutive fully-connected layers of length 1024, 512 and 2.
Dataset construction
Several databases have been included for training, consisting of experiments performed on prokaryotes grown under standard conditions. The experiments cover both Gram-negative (Salmonella typhimurium (14), Escherichia coli (32), Caulobacter crescentus (33)) and Gram-positive bacteria (Bacillus Subtilis (34), Mycobacterium smegmatis (35), Staphylococcus aureus (36), Streptomyces coelicolor (37)). The model is trained with the ribosome profiling coverage signal. The S-curve is fitted on each dataset to obtain the minimum required coverage and RPKM signal of the ribosome profiling signal of the samples within each dataset. Table 1 gives an overview of the used datasets, and the amount of samples each contributes to the the positive/negative dataset.
Table 1.
The ribosome profiling datasets used to train and validate DeepRibo
| Original data | S-curve selection | |||
|---|---|---|---|---|
| Dataset | Negative set | Positive set | Negative set | Positive set |
| S. typhimurium (14) | 432 983 | 4938 | 117 301 | 3586 |
| E. coli (32) | 439 895 | 4144 | 148 921 | 3544 |
| C. crescentus (33) | 274 390 | 3855 | 52 637 | 2179 |
| M. smegmatis (35) | 576 574 | 6716 | 148 909 | 4607 |
| B. subtilis (34) | 417 850 | 4154 | 91 010 | 2798 |
| S. coelicolor (37) | 547 814 | 7 766 | 27 421 | 1342 |
| S. aureus (36) | 311 296 | 2767 | 21 601 | 852 |
| Total | 3 000 802 | 34 340 | 607 800 | 18 908 |
To obtain a more balanced distribution of the labels and RPKM, each dataset has been filtered by applying a minimum threshold on coverage and RPKM. Cut-off values have been determined by estimating the lower bend point of the fitted S-curve.
To make sure no bias is introduced during the creation of the input data, the first step selects all candidate ORFs of the genome for each of the included ribo-seq datasets. It has been shown that ATG, GTG and TTG are the three nucleotide combinations that almost exclusively make up all start codons in a wide variety of bacteria (38). Therefore, all DNA sequences within the genomes starting with either ATG, GTG and TTG up until a stop codon (TAA, TGA or TAG) are considered candidate ORFs in this study. Since a large number of ORFs exists with lengths too short to be translated into a functional protein, a pseudo-arbitrary cutoff of 30 nucleotides is chosen to be the minimum length of the samples.
The study is built up as follows: the training data is created from six out of the seven available datasets, using the remaining dataset as the test set. A total of seven models have been trained and evaluated for this study, using each of the available datasets as a test set. Furthermore, the performance of two models has been highlighted in this study. In the first set-up we exclude data of S. aureus from the training set. In the second set-up, data from E. coli is excluded from the training set. Both set-ups cover the datasets with both the lowest and highest correlation between RPKM and coverage of the annotated genes. All experiments evaluate the performance of DeepRibo on de novo data (i.e. transfer learning), in accordance to the design goals discussed. The training data, constructed out of six datasets, is split up in a training set (95%) and validation set (5%). The loss of the validation set is used to determine the point at which training is stopped. Supplementary Figures S3 and S4 visualize the loss of the model on the training, validation and test sets for all evaluated models.
Evaluation and post-processing
To evaluate the model, the Area Under the Precision–Recall Curve (PR AUC) performance measure is used. The labels of the input samples are highly imbalanced due to the experimental set-up. Therefore, a large change in false positives leads to only a small change in the false positive rate. As the eventual use of the model is focused on the prediction of the top k ranked genes, PR AUC is known to be a more informative measure (39). Indeed, measured Area Under Receiver operating characteristic Curve (ROC AUC) values can be high even in cases in which the absolute amount of false positives (heavily) outweighs the absolute amount of true positives.
An important post-processing step of the annotations given by the model is the decision whether or not only one TISs can be annotated for each stop codon. The sequencing depth, reflected by the translation rates of the RNA, varies strongly between different gene regions. Differences in the distribution of probability scores exist between gene regions due to varying RPKM rates. Hence, it occurs that multiple start sites are annotated within one region while not obtaining TISs in another region. To compare the model with the annotations retrieved from NCBI (that do not support multiple start sites), focus is given to only the highest prediction probability between two stop codons (single start site setting). In order to obtain a set of postive predictions, a threshold on the probability scores has to be set, determining the annotation of the top k ranked predicted ORFs. In this study, the threshold for each organism is set in order to obtain an equal amount of positive predictions as positively labeled ORFs.
Multiple sequence comparison based on local alignment
Given the performance measures for each of the models, a more in depth exploration of the results is made. Assuming the existence of incompletions and mistakes in the annotation files, discrepancies between the annotations made by DeepRibo and the assembly have been investigated using the Basic Local Alignment Search Tool (BLAST) (27). The false positive predictions of the model are compared to a database containing a collection of proteins that have been previously discussed in literature, forming a good criterion to evaluate the existence of the predicted ORF. A query of the false positive predictions on ‘the non-redundant protein sequences’ (containing non-redundant sequences from GenBank translations together with sequences from Refseq, PDB, SwissProt, PIR and PRF (40)) has been performed using protein-protein BLAST (pBLAST). A maximum cut-off value of 0.1 for the expect (E) value is taken. The E value gives the expected amount of hits covering a similar alignment given the size of the database. For the sake of clarity, false postive predictions are considered as possible proteoforms or novel proteins, and are thus labeled as such. Specifically, proteoforms constitute false positive predictions with a varying start site compared to the positively labeled ORFs. Novel proteins cover any predicted ORF for which no previous annotation was present.
RESULTS
S-curve estimation for cut-off values filters high-quality from low-quality data
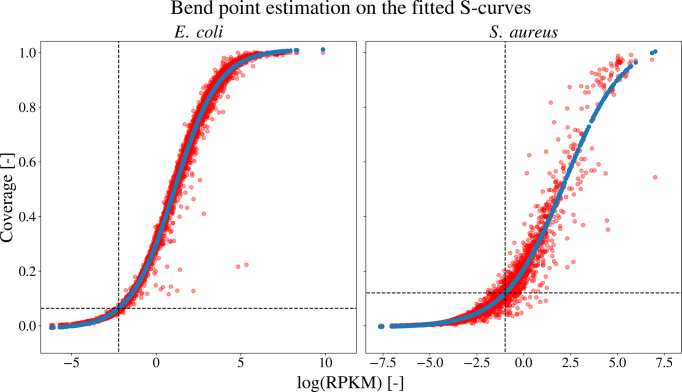
To normalize the total signal counts between multiple datasets, the expression rates of the different experiments are assumed to be equal. Since we are not working with repetitions of the same experiment, no normalization is performed before merging the datasets. However, differences in overall signal strengths between different experiments can be caused either by differences in expression profiles of the organisms, varying growth conditions, or technical variance introduced when performing the study. To filter candidate ORFs with signal strengths indistinguishable from the background noise, minimum cut-off values are estimated for each dataset using the S-curve methodology (30) (Supplementary Figure S1). Interestingly, datasets containing a high amount of low expression values give rise to more stringent cut-off values (e.g. S. aureus). In case of a clear distinction between expressed and non-expressed genes, a relatively low cut-off value is obtained (e.g. E. coli). Therefore, depending on the quality of the data, the amount of samples selected from each dataset can vary greatly. The positive samples and the fitted S-curves for the E. coli and S. aureus datasets are plotted in Figure 2. In the case of an incorrectly annotated dataset, a decreased correlation between the coverage and RPKM of the positive samples is expected, with a shift of the data points towards lower RPKM and coverage values. As these elements create a more gradual fit of the lower bend point of the S-curve on the data, these estimated cut-off values will be higher.
Figure 2.
Bend point estimation on the fitted S-curves of the coverage in function of the log RPKM for both the E. coli (left) and S. aureus (right) dataset. The positive samples for each dataset (red) are plotted with the predicted (blue) ones for the fitted S-curve. For each dataset, the lower bend point of the fitted curve is estimated using the bent-cable function to obtain the minimum cut-off values.
High performance values for predictions in the context of both single and multiple start codons
For the purpose of evaluating the performance, the test set is filtered to exclude any positively labeled data with low expression rates. As these genes are not being expressed, positive samples with non-existent or low ribo-seq data are filtered out (see Table 1). In parallel with the selection of the training set, minimum cut-off values have been determined using the fitted S-curve. Table 2 shows the performances of all the models on the independent dataset. Even though DeepRibo is trained on a dataset for which a maximum of one positively labeled ORF within two stop codons is present, this is not reflected into the predictions of the model. As genome assemblies are annotated using a maximum of one start codon for each stop codon, AUC and PR AUC scores are overall better including only the highest ranked start site for each stop codon. The performance of the model varies only slightly between the different experimental set-ups. A PR AUC as high as 0.965 and 0.943 on the test set is obtained for S. aureus and E. coli, respectively. Although the existence of multiple start sites within prokaryotes has been confirmed (13), it can be expected that the predictions have shifted distributions between different regions due to a varying ribo-seq signal. However, even when considering the predictions which allow multiple ORFs sharing a stop site, PR AUC scores are as high as 0.874.
Table 2.
The ROC AUC and PR AUC performance values for the different experimental set-ups in which the listed dataset is used as the test set
| Gram-negative | Gram-positive | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metric | Model | S. typhimurium | E. coli | C. crescentus | M. smegmatis | B. subtilis | S. coelicolor | S. aureus | |||||||
| MS | SS | MS | SS | MS | SS | MS | SS | MS | SS | MS | SS | MS | SS | ||
| ROC AUC | Full | 0.983 | 0.991 | 0.991 | 0.995 | 0.971 | 0.973 | 0.930 | 0.956 | 0.985 | 0.993 | 0.973 | 0.966 | 0.983 | 0.995 |
| CNN | 0.943 | 0.962 | 0.969 | 0.976 | 0.918 | 0.946 | 0.877 | 0.929 | 0.956 | 0.974 | 0.935 | 0.949 | 0.969 | 0.987 | |
| RNN | 0.939 | 0.980 | 0.934 | 0.980 | 0.923 | 0.958 | 0.809 | 0.854 | 0.942 | 0.982 | 0.907 | 0.913 | 0.933 | 0.965 | |
| PR AUC | Full | 0.804 | 0.910 | 0.860 | 0.943 | 0.710 | 0.842 | 0.522 | 0.717 | 0.796 | 0.922 | 0.777 | 0.863 | 0.874 | 0.965 |
| CNN | 0.574 | 0.706 | 0.640 | 0.763 | 0.562 | 0.730 | 0.419 | 0.627 | 0.639 | 0.779 | 0.622 | 0.760 | 0.812 | 0.910 | |
| RNN | 0.533 | 0.777 | 0.531 | 0.812 | 0.576 | 0.781 | 0.114 | 0.175 | 0.508 | 0.768 | 0.478 | 0.637 | 0.485 | 0.707 | |
| ROC AUC | REP | - | 0.916 | - | 0.916 | - | 0.838 | - | 0.821 | - | 0.933 | - | 0.838 | - | 0.944 |
| PR AUC | REP | - | 0.735 | - | 0.799 | - | 0.344 | - | 0.285 | - | 0.889 | - | 0.272 | - | 0.910 |
The performance metrics for are given in case multiple start sites are considered possible (MS) and in case each stop codon can only have a single predicted start site (SS). Performances of DeepRibo using either the DNA sequences as input (CNN) or ribo-seq data (RNN) highlights the improved performance if both features are combined in one model (Full). The performances on REPARATION (REP) are furthermore given. Note that these models are both trained and evaluated on the listed dataset using cross-validation.
DeepRibo combines sequence information and ribosome profiling data
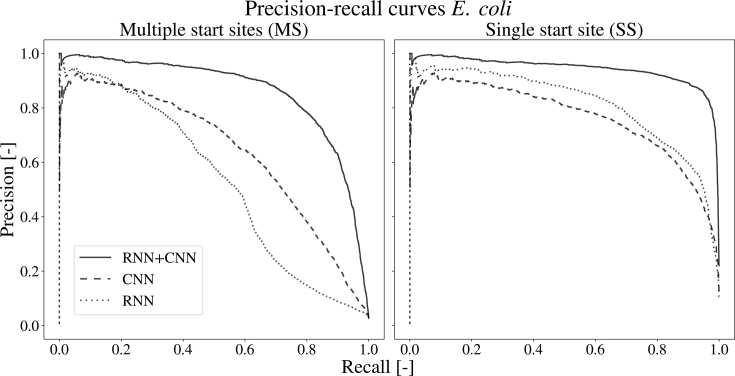
To confirm the ability of the neural network to apply the ribosome profiling signal for its predictions, custom models have been trained on either the sequence of the ribosome binding region (based on the CNN) or the ribosome profiling data (based on the RNN). The architectures of the models are kept similar, except for the loss of the recurrent or convolutional section, in case of the model trained on the DNA sequence and ribo-seq data, respectively. Table 2 lists the performances of both models for each set-up. Figure 3 displays the precision-recall curve for the model using E. coli as a test set. Related plots for each of the other models, each showing similar results, are given by Supplementary Figure S5 through S11. Both approaches prove to be effective at training from their specific data, with AUC values of 0.965 and 0.987 for the RNN and CNN for S. aureus. Overall, the CNN performs better than the RNN, shown by the PR AUC scores between the two architectures. The combination of both neural network partitions brings an improvement to the performances as compared to the individual parts. An increase for the PR AUC score of about seven percent compared to the CNN and 23 percent compared to the RNN shows that the model is able to combine both types of information in a meaningful way.
Figure 3.
The precision-recall curves of the different networks on the E. coli dataset. the precision-recall curves are given in case of the multiple start site and the single start site set-up. The full model (full line), combining the RNN and CNN outperforms both the single CNN (dashed) and RNN (dotted) architecture.
Evaluation of leaderless and SD-led genes
The fraction of genes carrying a Shine-Dalgarno region varies within each phylum. Actinobacteria (M. smegmatis, S. coelicolor) have on average 19.2%, α-Proteobacteria (C. crescentus) 6.3%, γ-proteobacteria (E. coli, S. typhimurium) 4.5% and Firmicutes (B. subtilis, S. aureus) 4.2% leaderless genes (41). Unlike leaderless genes, SD-led genes are defined by the consensus sequence “AGGAGG”, present 0–20 nt upstream of the TIS. Previous studies revealed no pattern downstream of the TIS of leaderless genes (35). The overall lower performances of M. smegmatis and S. coelicolor suggest a correlation with the fraction of leaderless genes in the genome of the evaluated organisms. In contrast, no correlation between the performances of the CNN on Actinobacteria is observed, showing results competitive with those of other set-ups. A high discrepancy of performances is observed however between the performances of the RNNs, with an PR AUC as low as 0.175 for M. smegmatis. Investigation on the ribo-seq data showed a high fraction of duplicated reads (92%), resulting in the lowest count of unique reads per positively labeled ORF (459.9) of all used datasets. This is more than four times lower than S. coelicolor (1952.0) and C. crescentus (2109.7), and well below S. aureus (8114.3), B. Subtilis (8328.6), S. typhimurium (23268.5) and E. coli (26908.0). The high correlation between these counts and the performance of the RNNs underlines the importance a high quality data. As a result, ribosome profiling from M. abscessus (42) has been evaluated to verify the applicability of DeepRibo on organisms with a higher fraction of leaderless genes. A PR AUC of 0.865 and 0.577 for both the complete and RNN model used to evaluate M. smegmatis was obtained, a score in line with the results of the model on the other organisms. The performance using the full model increases slightly when trained on all seven datasets (PR AUC: 0.898). The performance is slightly reduced for the RNN model (0.569), indicating the negative impact of the lower quality ribosome profiling data of M. smegmatis. Shell et al. (35) proposed a re-annotation of 150 genes for M. smegmatis. 30 out of 116 re-annotated ORFs present in the test set are present in the annotations given by DeepRibo (top 4607 predictions).
Comparison of DeepRibo with REPARATION
REPARATION (14) is the only existing tool that performs a similar task for prokaryotes. However, REPARATION follows a different approach on certain key aspects. A positive set is created by comparative genomics using all candidate ORFs -given the start codons ATG, GTG or TTG- in the target genome. The negative set is assembled out of the set of all possible ORFs with the start codon CTG. Specifically, for each set of ORFs, sharing the in frame stop codon, the longest sequence is taken. REPARATION applies Random Forests to distinguish the set of ORFs matched through comparative genomics (ATG, GTG, TTG) from the subset of all ORFs with the start codon CTG (negative set). In comparison, the negative set in our approach is assembled out of all possible ORFs not positively labeled by the assembly file, ignoring ORFs with the start codon CTG for both the positive and negative set. Therefore, DeepRibo handles a higher fraction of negatively labeled data, with no bias (start codon, length) existent between the positive and negative set. It can therefore be stated that DeepRibo handles a more complex problem. DeepRibo outperforms REPARATION on all seven datasets (Table 2), showing more robust performances as compared to REPARATION. However, the comparison should be interpreted with the knowledge that both tools perform a different function. It should furthermore be noted that performances evaluated by REPARATION are also correlated to the quality of the different experiments, with performances returned on M. smegmatis, S. coelicolor and C. crescentus being unexpectedly low. REPARATION indicates to be more sensitive to the quality of the ribo-seq data as compared to DeepRibo. DeepRibo offers several more advantages: (i) no resolution loss of the input experimental data, (ii) no limits in the amount of datasets a single model can be trained on and (iii) applicability of a pre-trained model by the user. Also, (iv) performances have been evaluated on independent test sets (as compared to using cross-validation for each experiment).
Edman degradation assisted validation of predictions
Through sequencing of the N-terminal residues of the matured proteome using Edman degradation, the creation of certain proteins within a cell can be verified. A collection of 922 proteins within E. coli K-12, featuring all the verified proteins discussed in literature, is featured by Ecogene (28). Of the 922 proteins, a total amount of 838 ORFs are expressed within the E. coli dataset, determined using the S-curve methodology. The positive predictions are composed of the top 3544 predictions, using the single start site setting, in accordance to previous methods. 744 (88.8%) of the genes have been predicted correctly by the model. 23 (2.7%) verified proteins have TISs differing from the annotation, resulting in 815 proteins for which the annotation and verified protein set agree. None of the predicted TISs in agreement with the verified proteins were in disagreement with the labeled dataset. 71 out of 815 (8.7%) TISs present in the annotations and Ecogene dataset are not picked up by the model. More importantly, 28 out of the 71 (39.4%) false negatives are actually present in the top 4400 ranked predictions. Due to the annotation of novel ORFs by DeepRibo, some of the positively labeled input samples are bound to be excluded from the pool of 3544 positive predictions. This means only 43 out of 815 (5.27%) of the false negatives have predicted TISs up- or downstream of the labeled gene.
N-terminal proteomics based validation of predictions
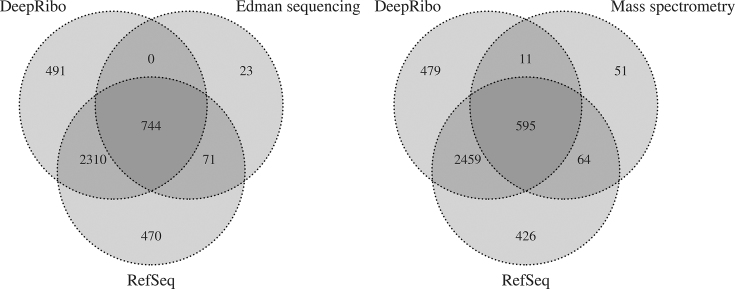
Next to the Edman sequencing (Ecogene dataset), mass spectrometry based proteomics can serve to validate annotations made by DeepRibo. N-terminal proteomics, more specifically, is a technology that enables us to detect N-terminal peptides compliant with the rules of initiator methionine processing. 781 such N-termini were previously determined for E. coli (14). 721 N-terminal peptide sequences that are aligned with coding sequences are expressed and are therefore present in the test set. 659 out of 721 samples (91.4%) are in accordance with the annotation. 64 (9.7%) of these are not predicted by the model, of which 34 have differing TISs and 30 fell out of the top 3544 predictions. Interestingly, of the 62 peptide sequences that indicate a TIS in disagreement with the RefSeq annotation, 11 have been predicted by DeepRibo. Although the presence of a TIS at a site differing from the annotation can be suggested as indicated by the ribosome profiling data, this is tangible proof that the annotation is not waterproof, negatively influencing the performance measure of the model. Figure 4 gives an overview of the overlap between the two validation datasets with the NCBI annotations and predictions.
Figure 4.
Venn diagram displaying the distributions of the proteins verified by Edman sequencing (left) and mass spectrometry (right) within the annotations provided by DeepRibo and the NCBI RefSeq database (labels). Distributions only include expressed ORFs, determined using the S-curve methodology.
Multiple sequence alignment of the false positive predictions
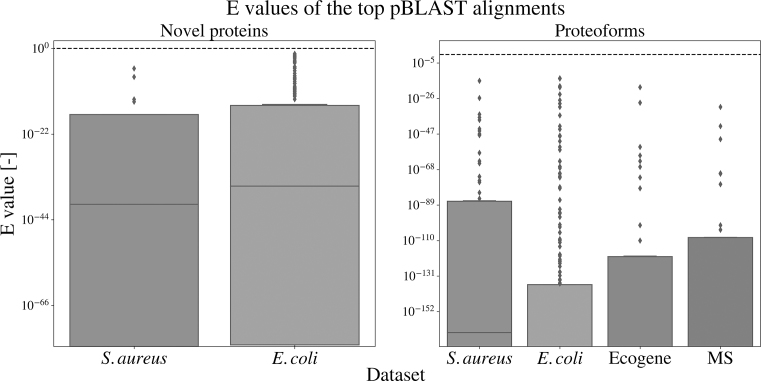
Multiple proteoforms exist for a large amount of the annotated proteins. Yet, only one variety of each protein has been annotated in the genome assembly. Biological variation, growth condition or growth phase are some of the factors influencing protein expression rates. Accordingly, variety in protein expression between different experiments creates variation from the annotated genome. pBLAST searches have been performed to investigate whether false positive predictions could be caused by expressed proteoforms not present in the annotation. A summary is created by simply taking the best aligned protein for each of the false positive predictions. pBLAST searches have been performed on the complete set of false positives for S. aureus and E. coli. Comparing the annotations curated by DeepRibo with the mass spectrometry and Edman sequencing datasets resulted in 34 and 42 ORFs with differing TISs as proposed by the model. These two sets of alternative annotations by the model have furthermore been included for sequence similarity comparison. Table 3 gives an overview of the results. As expected, all proteoforms have been successfully aligned, given they are partly identical to the annotated gene. As much as 73 out of 79 (92.4%) and 198 out of 232 (85.3%) annotated proteoforms for S. aureus and E. coli have been fully aligned with a protein site in the databank, having a shared TIS and stop site. Of all novel proteins annotated by DeepRibo, more than half have a match that is fully aligned, summing up to a total of 15 out of 25 (60%) and 137 out of 258 (53.1%) protein sequences for S. aureus and E. coli. Interestingly, a considerable percentage of the novel proteins are described as ‘hypothetical’. The model predictions that annotated a differing TIS as compared to the MS and Ecogene dataset mostly indicate perfect alignment with proteins present in the non-redundant database, with 28 out of 34 (82.4%) and 36 out of 43 (83.7%) matches, respectively. Figure 5 gives the spread of the E values for each of the aligned proteins. A complete list of the false positive and false negative predictions for E. coli and S. aureus, including the two validation sets and the BLAST results is provided in Supplementary File 2.
Table 3.
Results of the BLAST search on the false positive set of E. coli and S. aureus, and specifically on the false positives in disagreement with the annotation of the Mass Spectrometry (MS) and Edman sequencing (Ecogene) dataset
| Set-up | type | # | aligned total | TIS | TIS + stop | description hypothetical |
|---|---|---|---|---|---|---|
| S. aureus | Proteoform | 79 | 79 | 77 | 73 | 12 |
| Novel protein | 25 | 19 | 17 | 15 | 6 | |
| E. coli | Proteoforms | 232 | 232 | 217 | 198 | 39 |
| Novel protein | 258 | 204 | 157 | 137 | 106 | |
| MS | Proteoforms | 34 | 34 | 22 | 28 | 1 |
| Ecogene | Proteoforms | 43 | 43 | 40 | 36 | 1 |
These predictions can be divided into proteoforms, which have a TIS that is either up- or downstream of the annotated ORF, or novel proteins, constituting ORFs with a non-annotated stop site. A BLAST search of these proteins was performed on the non-redundant protein database. A maximum cut-off value of 0.1 for the E score is taken. The total amount of false positives are given for each type. Taking only the best aligned protein (i.e. highest E score) for each of the false positive results, the total amount of matches that were aligned by start site or both start and stop site are given. Finally, the total amount of proteins described as ‘hypothetical’ are given.
Figure 5.
E value distributions for the pBLAST results on newly predicted proteins (left) and proteoforms (right) for the different datasets. The E values are given for the best hit (if existent) for each of the false positives. The dashed line indicates the E value of 1.
DISCUSSION
The success of deep learning methods on popular topics involving big data is slowly finding its way to the field of bioinformatics involving multi-omics. Although big data created by high-throughput methods has been available since the arrival of second generation sequencing, it has so far mainly been explored using statistical methods, excluding machine learning. Deep learning has proven to be considerably successful, allowing the use of a black box approach when the interpretation of the features is not desirable or feasible. In this study, we present a deep neural network for the precise annotation of expressed proteins on the genome using ribosome profiling data. This tool uses data from in vivo expression profiles to annotate the genome without the use of comparative sequence alignment. DeepRibo learns from information contained in both DNA sequences and ribo-seq, using a novel architecture that combines both convolutional layers and recurrent memory cells. Results obtained from machine learning models, which are trained and evaluated on the same dataset, can be overestimates of their performance on new data due to overfitting. The use of a single model trained on a variety of existing datasets and evaluated on independent test sets makes due with this problem. Moreover, building the model on a combination of datasets trains it to differentiate between useful features present over all the datasets and dataset-dependent variations, making the need for normalization steps redundant. DeepRibo is the first tool for the precise delineation of ORFs in prokaryotes trained and validated on multiple datasets. It furthermore outperforms REPARATION on all datasets tested.
When evaluating the results of DeepRibo, a certain cut-off has to be determined to specify the positive predictions from the negative. To evaluate the model, the amount of positive ORFs has been set equal to the ORFs present in the annotations. However, due to novel predictions being made, a fraction of the annotated samples are bound to have a rank lower than the top k predictions (especially in a multiple start site setting). This is furthermore reflected by the fraction of proteins in the validation sets not picked up by the top k predictions of the model. No cut-off is optimal for every instance and has to be determined in line with the application of the tool, which postulates the desired precision/recall.
The performance of DeepRibo is consistent on all seven test sets, reaching PR AUC scores of >0.90 for four datasets. No difference is observed on the performance between gram-positive and gram-negative bacteria. Even though a relatively low performance was returned for M. smegmatis, evaluation of the dataset and performances returned for M. abscessus, another member of the Mycobacterium family, showed no relation with the fraction of leaderless genes present. Instead, the importance of the quality of the ribosome profiling experiment is highlighted, with unique reads per positively labeled ORFs showing correlation to the performance of the individual RNNs. Although the absolute number of reads mapped to the genome is sufficient, a high level of duplication, and therefore lower number of unique reads, results in a lower resolution of the ribosome profiling. To guarantee the quality of the ribosome profiling experiment several tools are available: FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) for the evaluation of reads and mQC (https://github.com/Biobix/mQC) for the evaluation of mapped reads.
Since the majority of the candidate ORFs share their stop sites with other samples, selecting only the ORF with the highest predicted probabilities within each group gives consistently better performances. Even though an increase in performance is observable when comparing the single start site with the multiple start site setting, the performance of the latter is still noteworthy. Specifically, 105 595 out of the 113 228 (89.5%) candidate ORFs share stop sites with other samples in the E. coli dataset. Some regions have as much as one hundred possible TISs. Although the model has no way of processing this information, making a prediction on every sample individually, remarkable PR AUC scores are achieved on the test sets (MS setting), ranging from 0.710 to 0.874 (excluding M. smegmatis). Part of this error is expected to be caused by differences in RPKM values existent between different genome regions. Yet, the models’ performances indicate this effect to be minimal. Moreover, recent studies have discovered genes with multiple translation initiation sites (13,15,44). As this feature is not supported by the annotations, correct evaluation of the model in a multiple stop site setting is currently unfeasable.
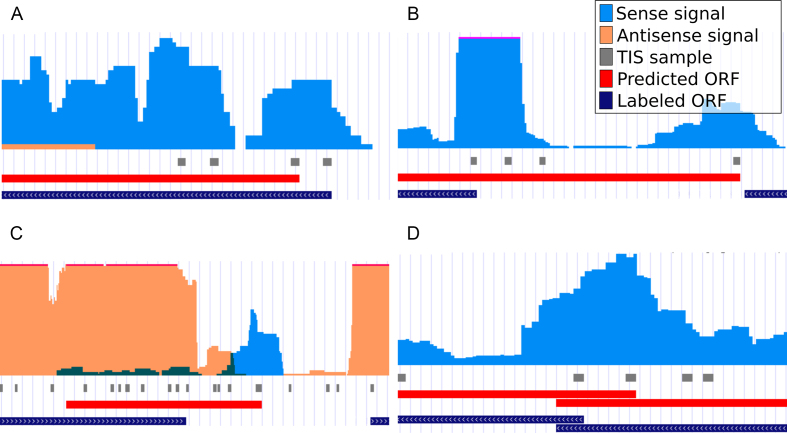
Many prokaryotic systems have closely knit operon structures (45), creating a ribo-seq signal that can overlap different regions of interest. Several false positive annotations made by DeepRibo are situated in these regions (Figure 6D). The inclusion of a padded region around the ribosome profiling signal processed by the RNN has previously increased the resulting performance.
Figure 6.
DeepRibo example annotations displayed alongside the ribo-seq input signal and RefSeq annotations. The data is formatted using the GWIPS-viz browser (43) and is hosted publicly (see Supplementary Data). On every track is displayed (from top to bottom): ribo-seq signal (sense: orange, antisense: blue), TISs of all ORF samples present in the test set, annotations predicted by DeepRibo not in agreement with the RefSeq assembly (Predicted ORF) and the RefSeq genome annotations used to label the data (Labeled ORF). (A) The highest ranking proteoform prediction (gene: PqqL, rank: 231) for E. coli. (B) The highest ranking proteoform prediction (gene: UbiE, rank: 131) for S. aureus. (C) The highest ranking novel protein for E. coli with no pBLAST alignments (rank: 1302). (D) An example of a predicted proteoform in a region with overlapping genes (gene: ybhF, rank: 941).
The applied antibiotic in the ribosome profiling experiment is known to influence the resulting signal. In this study, all experiments apply chloramphenicol treatment, with the exception of S. coelicolor, which applies thiostrepton. As the overall lower score of S. coelicolor might be related to its lower count of unique reads, it is uncertain as to whether the use of thiostrepton influences the performance of DeepRibo. Although this effect seems to be minimal, the effect of different antibiotic treatments on DeepRibo needs to be further investigated. New antibiotic treatments can also offer improvements to the model’s performance. Meydan et al. (46) discuss the use of the antibiotic retapamulin that increases the resolution of the ribo-seq signal. The increased resolution offered by retapamulin might thereby improve the resulting annotations thereof, especially for regions containing overlapping genes.
In case of the E. coli model, many of the novel predictions are situated within a pseudogene. Typically, no candidate ORFs overlapping the complete pseudogene regions were present in the training/testing samples, as these annotated features cover regions with multiple stop codons. Therefore, no positively labeled samples are present. However, ribo-seq signal is often measured at these sites, creating a hot-spot for novel (false positive) predictions.
The identification of a high amount of novel small open reading frames (sORFs) by the model presents another contrast with the annotation. The novel ORF predictions given by the models have a median length of 270 and 63 nucleotides for E. coli and S. aureus. These are well below the median length of the annotated genes within each species (827 and 723). The size of the ORFs influences the power of the statistical methods used for the identification of the sORFs by in silico methods (47) applying sequence alignment. It is hypothesized that a higher amount of sORFs is present in the genome than there are currently present in the annotation. DeepRibo annotates a higher amount of sORFs in comparison to the amount present in the annotation. Specifically, VanOrsdel et al. recently proposed 32 new sORFs for E. coli (48). Of the 21 sORFs present in the dataset, 5 were included in the annotation (top 3544) presented by DeepRibo. In comparison, only one of the proposed sORFs was actually present in the annotations from RefSeq. An example of a novel sORF for E. coli is given in Figure 6C. A distribution of the lengths of the ORFs for both the positive annotations and novel predictions is shown in Supplementary Figure S12.
Corroborated by the results obtained from the pBLAST searches, it is likely that a fraction of the false positives observed when evaluating the predictions of the single start site setting are due to an annotation that does not fully map the translational complexity of the organisms, subsequently negatively influencing the performance of DeepRibo. This is especially expected for prokaryotes which are less known such as S. coelicolor, C. crescentus, M. smegmatis and M. abscessus. In further support, detailed evaluation of the predictions with the ribo-seq signal shows that many false positive results are backed by manual evaluation (Figure 6 A/B/C).
DeepRibo has proven to be a tool with a novel approach and high performance. The model enables the discovery of multiple ORFs sharing a single stop codon, small ORFs or ORFs situated in pseudogenic regions. Training DeepRibo is not bound by any number of datasets, it distinguishes useful features shared between datasets and can be further improved as more data is added. In contrast with sequence alignment methods, the ribosome profiling signal offers actual proof for the annotation of ORFs. The exclusion of DNA sequences from genes ensures the model is not biased towards gene patterns it is trained on. In conclusion, DeepRibo has shown to be a viable tool for the annotation of the genome without the use of gene similarity algorithms, and can be applied to aid the discovery of translated proteins in prokaryotes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the support of Ghent University. Special thanks to Dr Audrey Mannion-Michel for her work on GWIPS-viz and her provided help with the ribosome profiling data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Special Research Fund [BOF24j2016001002 to P.R.] from Ghent University; Research Foundation-Flanders (FWO-Vlaanderen) Postdoctoral Fellowship (to G.M.). Funding for open access charge: Bijzonder Onderzoeksfonds [BOF24j2016001002].
Conflict of interest statement. None declared.
REFERENCES
- 1. Land M., Hauser L., Jun S.-R., Nookaew I., Leuze M.R., Ahn T.-H., Karpinets T., Lund O., Kora G., Wassenaar T. et al.. Insights from 20 years of bacterial genome sequencing. Funct. Integrative Genomics. 2015; 15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richardson E.J., Watson M.. The automatic annotation of bacterial genomes. Brief. Bioinformatics. 2013; 14:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T. et al.. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015; 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delcher A. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999; 27:4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Connor P.B.F., Li G.W., Weissman J.S., Atkins J.F., Baranov P.V.. RRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013; 29:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mohammad F., Woolstenhulme C.J., Green R., Buskirk A.R.. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016; 14:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tech M., Morgenstern B., Meinicke P.. TICO: a tool for postprocessing the predictions of prokaryotic translation initiation sites. Nucleic Acids Res. 2006; 34:W588–W590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ou H.Y., Guo F.B., Zhang C.T.. GS-Finder: a program to find bacterial gene start sites with a self-training method. Int. J. Biochem. Cell Biol. 2004; 36:535–544. [DOI] [PubMed] [Google Scholar]
- 12. Zhu H.-Q., Hu G.-Q., Ouyang Z.-Q., Wang J., She Z.-S.. Accuracy improvement for identifying translation initiation sites in microbial genomes. Bioinformatics. 2004; 20:3308–3317. [DOI] [PubMed] [Google Scholar]
- 13. Nakahigashi K., Takai Y., Kimura M., Abe N., Nakayashiki T., Shiwa Y., Yoshikawa H., Wanner B.L., Ishihama Y., Mori H.. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 2016; 23:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ndah E., Jonckheere V., Giess A., Valen E., Menschaert G., Van Damme P.. REPARATION: ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acids Res. 2017; 45:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giess A., Jonckheere V., Ndah E., Chyzyńska K., Van Damme P., Valen E.. Ribosome signatures aid bacterial translation initiation site identification. BMC Biol. 2017; 15:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S., Liu B., Lee S., Huang S.-X., Shen B., Qian S.-B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crappé J., Ndah E., Koch A., Steyaert S., Gawron D., De Keulenaer S., De Meester E., De Meyer T., Van Criekinge W., Van Damme P. et al.. PROTEOFORMER: Deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 2015; 43:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bazzini A.A., Johnstone T.G., Christiano R., MacKowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C. et al.. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014; 33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chew G.-L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E.. Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Development. 2013; 140:2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao Z., Huang R., Xing X., Chen Y., Deng H., Yang X.. De novo annotation and characterization of the translatome with ribosome profiling data. Nucleic Acids Res. 2018; 46:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erhard F., Halenius A., Zimmermann C., L’Hernault A., Kowalewski D.J., Weekes M.P., Stevanovic S., Zimmer R., D”olken L.. Improved Ribo-seq enables identification of cryptic translation events. Nat. Methods. 2018; 15:363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Staes A., Impens F., Damme P.V., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K.. Selecting protein n-terminal peptides by combined fractional diagonal chromatography. Nat. Protocols. 2011; 6:1130–1141. [DOI] [PubMed] [Google Scholar]
- 23. Berry I.J., Steele J.R., Padula M.P., Djordjevic S.P.. The application of terminomics for the identification of protein start sites and proteoforms in bacteria. PROTEOMICS. 2016; 16:257–272. [DOI] [PubMed] [Google Scholar]
- 24. Hartmann E.M., Armengaud J.. N-terminomics and proteogenomics, getting off to a good start. PROTEOMICS. 2014; 14:2637–2646. [DOI] [PubMed] [Google Scholar]
- 25. Van Damme P., Gawron D., Van Criekinge W., Menschaert G.. N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol. Cell. Proteomics. 2014; 13:1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alipanahi B., Delong A., Weirauch M.T., Frey B.J.. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015; 33:831–838. [DOI] [PubMed] [Google Scholar]
- 27. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 28. Zhou J., Rudd K.E.. EcoGene 3.0. Nucleic Acids Res. 2013; 41:D613–D624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu H., Hu G.-Q., Yang Y.-F., Wang J., She Z.-S.. MED: a new non-supervised gene prediction algorithm for bacterial and archaeal genomes. BMC Bioinformatics. 2007; 8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lutz R.W., Stahel W.A., Lutz W.K.. Statistical procedures to test for linearity and estimate threshold doses for tumor induction with nonlinear dose-response relationships in bioassays for carcinogenicity. Regul. Toxicol. Pharmacol. 2002; 36:331–337. [DOI] [PubMed] [Google Scholar]
- 31. Paszke A., Gross S., Chintala S., Chanan G., Yang E., DeVito Z., Lin Z., Desmaison A., Antiga L., Lerer A.. Automatic differentiation in PyTorch. NIPS-W. 2017; [Google Scholar]
- 32. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schrader J.M., Zhou B., Li G.W., Lasker K., Childers W.S., Williams B., Long T., Crosson S., McAdams H.H., Weissman J.S. et al.. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 2014; 10:e1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., Gawande R., Ahmad R., Sarracino D.A., Ioerger T.R. et al.. Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape. PLOS Genet. 2015; 11:e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davis A.R., Gohara D.W., Yap M.-N.F.. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeong Y., Kim J.N., Kim M.W., Bucca G., Cho S., Yoon Y.J., Kim B.G., Roe J.H., Kim S.C., Smith C.P. et al.. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 2016; 7:11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Panicker I.S., Browning G.F., Markham P.F.. The effect of an alternate start codon on heterologous expression of a PhoA fusion protein in mycoplasma gallisepticum. PLoS ONE. 2015; 10:e0127911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis J., Goadrich M.. The relationship between precision-recall and ROC curves. Proceedings of the 23rd International Conference on Machine Learning. 2006; NY: ACM ICML ’06; 233–240. [Google Scholar]
- 40. Pruitt K.D., Tatusova T., Maglott D.R.. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007; 35(Suppl. 1):D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X., Hu G.-Q., She Z.-S., Zhu H.. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011; 12:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miranda-CasoLuengo A.A., Staunton P.M., Dinan A.M., Lohan A.J., Loftus B.J.. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics. 2016; 17:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michel A.M., Fox G.M., Kiran A., De Bo C., O’Connor P.B., Heaphy S.M., Mullan J.P., Donohue C.A., Higgins D.G., Baranov P.V.. GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res. 2014; 42:D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai Y., Shortreed M.R., Scalf M., Frey B.L., Cesnik A.J., Solntsev S., Schaffer L.V., Smith L.M.. Elucidating Escherichia coli proteoform families using intact-mass proteomics and a global PTM discovery database. J. Proteome Res. 2017; 16:4156–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pallejà A., Harrington E.D., Bork P.. Large gene overlaps in prokaryotic genomes: result of functional constraints or mispredictions?. BMC Genomics. 2008; 9:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meydan S., Vázquez-Laslop N., Mankin A.S.. Genes within genes in bacterial genomes. Microbiol. Spectrum. 2018; 6:doi:10.1128/microbiolspec.RWR-0020-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pauli A., Valen E., Schier A.F.. Identifying (non-)coding RNAs and small peptides: Challenges and opportunities. BioEssays. 2015; 37:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. VanOrsdel C.E., Kelly J.P., Burke B.N., Lein C.D., Oufiero C.E., Sanchez J.F., Wimmers L.E., Hearn D.J., Abuikhdair F.J., Barnhart K.R. et al.. Identifying new small proteins in Escherichia coli. Prpteomics. 2018; 18:1700064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.