Abstract
Mpp10 forms a complex with Imp3 and Imp4 that serves as a core component of the ribosomal small subunit (SSU) processome. Mpp10 also interacts with the nucleolar protein Sas10/Utp3. However, it remains unknown how the Mpp10–Imp3–Imp4 complex is delivered to the nucleolus and what biological function the Mpp10–Sas10 complex plays. Here, we report that the zebrafish Mpp10 and Sas10 are conserved nucleolar proteins essential for the development of the digestive organs. Mpp10, but not Sas10/Utp3, is a target of the nucleolus-localized Def-Capn3 protein degradation pathway. Sas10 protects Mpp10 from Capn3-mediated cleavage by masking the Capn3-recognition site on Mpp10. Def interacts with Sas10 to form the Def–Sas10–Mpp10 complex to facilitate the Capn3-mediated cleavage of Mpp10. Importantly, we found that Sas10 determines the nucleolar localization of the Mpp10–Imp3–Imp4 complex. In conclusion, Sas10 is essential not only for delivering the Mpp10–Imp3–Imp4 complex to the nucleolus for assembling the SSU processome but also for fine-tuning Mpp10 turnover in the nucleolus during organogenesis.
INTRODUCTION
In eukaryotes, ribosome biogenesis consumes more than 60% of the total energy of a cell, and this process includes transcription of the pre-ribosomal RNA (rRNA); translation of ribosomal proteins and non-ribosomal proteins for the maturation of rRNAs; maturation of 18S, 5.8S and 28S rRNAs and assembly of the small and large ribosomal subunits (1). The ribosomal small subunit (SSU) contains an 18S rRNA and more than 30 ribosomal proteins. The biogenesis of ribosomal SSU starts from the processing and maturation of 18S rRNA from the 35S (in yeast) pre-rRNA transcript and is a precisely controlled stepwise process. This process involves the participation of ∼70 non-ribosomal factors and various small nucleolar RNAs (snoRNAs), including the U3 snoRNA (2–4). Upon transcription of the 5′-external transcribed spacer (5′-ETS) of the 35S pre-rRNA, 5′-ETS recruits the U Three Protein-A (UTP-A) and UTP-B complexes, followed by the formation of a complex containing mitotic phosphorylated protein 10 (Mpp10), Mpp10-interacting protein 3 (Imp3) and Mpp10-interacting protein 4 (Imp4) (namely, the Mpp10–Imp3–Imp4 complex) as well as the U3 small nucleolar ribonucleoprotein particle (snoRNP). These complexes assemble into a huge complex termed the 90S pre-ribosome or SSU processome (4–7). The SSU processome mediates 18S rRNA maturation by cleavage at A0, A1 and A2 sites (5,8–11).
Mpp10 was first identified in an expression screening for phosphoproteins using the MPM2 antibody, which recognizes a set of phosphorylated proteins (12). Mpp10 is phosphorylated by an unidentified kinase and is co-localized with Fibrillarin (Fib) in the nucleoli during interphase (12). In one study, a yeast two-hybrid experiment revealed that Imp3 and Imp4 interact with Mpp10 (13). In humans, the 327–565-amino acid (aa) region of hMpp10 is required for the interaction with hImp3 and hImp4 (14). The Mpp10–Imp3–Imp4 protein complex is stably associated with the U3 snoRNA (14,15). Imp3 is believed to mediate the association of the heterotrimeric complex with the U3 snoRNA (7). Therefore, the Mpp10–Imp3–Imp4 complex plays an important role in stabilizing the U3 snoRNA/pre-18S rRNA hybrid that guides the site-specific cleavage of the 35S pre-rRNA (7,16). Interestingly, Imp4, Imp3 and Mpp10 proteins are interdependent for both nucleolar localization and protein level maintenance (14,17). However, it remains unclear how the Mpp10–Imp3–Imp4 complex is delivered to the nucleolus to participate in SSU processome assembly.
Something about silencing 10 (Sas10)/Utp3 was first identified as a factor involved in the de-repression of the silenced mating-type genes when overexpressed in yeast (18). Sas10 contains an ∼80-aa-long domain termed as the Sas10/C1D domain, which is found in a small group of proteins (19). The Sas10/C1D domain appears to serve as a binding surface for protein interaction (19). The Sas10/C1D family proteins play diverse biological functions, including RNA processing (19,20), translational control (19,21) and DNA repair (19,22,23). In yeast, Sas10/Utp3 is an essential protein as the loss-of-function mutation of the sas10 gene results in inviable spores. After conditional sas10 knockout, the cells are arrested in the late S or G2/M phase of the cell cycle. A protein interaction study showed that Sas10/Utp3 interacts with the N-terminus of Mpp10 (24). Although Sas10/Utp3 was found to be co-immunoprecipitated with the U3 snoRNA and Mpp10 (5), recent studies have failed to identify the Mpp10–Sas10/Utp3 complex in the 90S pre-ribosome particle (6,7), raising a question regarding the specific role of the Mpp10–Sas10 complex in SSU processome assembly.
Digestive organ expansion factor (Def) was first characterized as a factor essential for digestive organ development in zebrafish (25). Def and its yeast counterpart Utp25 are nucleolar proteins (26–29). Subsequent studies have found that both human and zebrafish Def/Utp25 recruit the cysteine proteinase Calpain 3 (Capn3) to the nucleolus to degrade target proteins, such as the tumour suppressor factor p53 (29,30). Interestingly, protein interaction studies in yeast have revealed the presence of a strong interaction between Utp25 and Sas10 but a weak association between Utp25 and Mpp10 (26,27). It is proposed that this complex serves as a bridge to link different SSU subcomplexes (26); however, the Upt25-Sas10/Utp3-Mpp10 complex is not found in the purified 90S pre-ribosome (7).
Although studies have shown that both Sas10/Utp3 and Mpp10 are essential proteins in yeast and that both play important roles in the biogenesis of 18S rRNA, the biological functions of Sas10 and Mpp10 in higher eukaryotes have not yet been investigated. Furthermore, although Sas10/Utp3 and Mpp10 form a robust complex (24), only the Mpp10–Imp3–Imp4 complex, but not the Mpp10–Sas10/Utp3 complex, has been identified in the 90S pre-ribosome particle (6,7). The fact that Sas10/Utp3 also strongly interacts with Utp25 raises an interesting question regarding the biological significance of the binding of Sas10/Utp3 to Mpp10 and Utp25. In this study, we showed that the zebrafish sas10 and mpp10 genes exhibit similar dynamic expression patterns during early embryogenesis and that both genes are essential for digestive organ development. Sas10/Utp3 strongly interacts with Def/Utp25. Importantly, we showed that Sas10/Utp3 binds to the N-terminus of Mpp10 to protect it from Capn3-mediated protein degradation in the nucleolus. Furthermore, we showed that the nucleolar localization of the Mpp10–Imp3–Imp4 complex depends on Sas10/Utp3. Thus, our work illustrates the role of Sas10/Utp3 in SSU processome assembly.
MATERIALS AND METHODS
Zebrafish lines and maintenance
Zebrafish wild-type (WT) AB line was used in this study, and all mutant lines were derived from this line. Zebrafish were raised and maintained according to the guidelines available at http://zfin.org/. To generate the sas10 mutant, we synthesized gRNA against exon 4 (5′-GAAGAGGATGAAGATAAAGCGG-3′) of the zebrafish sas10 gene. The s△2and s△8 mutant lines were identified by digesting the polymerase chain reaction (PCR) product using the restriction enzyme AciI. To generate the mpp10 mutant, we synthesized gRNA against exon 2 (5′-GGTGGTTGAAAACTTCGACGAGG-3′) of the zebrafish mpp10 gene. The m△10 mutant line was identified by digesting the PCR product using the restriction enzyme PvuII, and the m+2 mutant line was identified by sequencing the PCR products. The zebrafish def−/− (defhi429) mutant line was identified as described previously (25). All animal procedures were performed in accordance with the requirements of the Regulation for the Use of Experimental Animals in Zhejiang Province. This work was approved by the Animal Ethics Committee of the School of Medicine, Zhejiang University (ETHICS CODE Permit NO. ZJU2011-1-11-009Y, issued by the Animal Ethics Committee of the School of Medicine, Zhejiang University).
Cell culture and plasmid transfection
Human 293T/7 (HEK 293T/7) cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, high glucose; Biological Industries) supplemented with 10% foetal bovine serum (GIBCO). Transfection of the cultured cells with plasmids was performed using PolyJet transfection reagent (#100688; SignaGen Laboratories, Rockville, MD, USA) according to the manufacturer’s instructions (Supplementary Table S1). Briefly, 1-μg plasmids were mixed with 3-μl PolyJet in 100-μl DMEM and cultured in six-well plates. The sas10 and mpp10 complementary DNAs and their derivatives were cloned into the pcs2+ vector. Proteins were harvested 48 h post-transfection.
Co-immunoprecipitation (Co-IP) and western blot analysis
Human 293T cells were transfected with various plasmids as described in the main text and total proteins were extracted with IP lysis buffer (containing ‘cell lysis buffer for Western and IP’ and 1 × cOmplete) 48 h post-transfection. Zebrafish livers were dissected under a microscope and sonicated (Branson Sonifier, SLPe) in IP lysis buffer for protein extraction. Zebrafish embryos were collected at 32 h post-fertilization (hpf) and sonicated in IP lysis buffer for protein extraction. Antibodies against the HA tag, the Myc tag, Sas10, Mpp10 and Def were used in Co-immunoprecipitation (Co-IP) (Supplementary Table S1). The input and immunoprecipitated protein samples were subjected to western blot analysis with the corresponding antibodies. Western blot and Co-IP were performed as described previously (31,32).
Northern blot analysis
Total RNA from different samples was extracted using TRIpure Reagent (Aidlab) according to the manufacturer’s instructions. 5′-ETS and internal transcribed spacer 1 (ITS1) probes (Supplementary Table S1) for northern blot analysis were digoxigenin (DIG) labelled according to the manufacturer’s instructions (Roche Diagnostics), and hybridization using the probes was performed as described previously (29,32).
Whole-mount RNA in situ hybridization (WISH)
For WISH, DIG (Roche Diagnostics) was used to label the fabp10a, fabp2, trypsin, gata6, foxa3, hhex, prox1, sas10 and mpp10 probes (Supplementary Table S1). The preparation of the fabp10a, fabp2, trypsin, gata6, foxa3, hhex and prox1 probes and WISH analysis were performed as described previously (25,33).
Enzymatic activity assay
For the experiments shown in Figure 5A and B, the cultured 293T cells expressing Sas10 or Mpp10 (48 h post-transfection) or Capn3 or Capn3C129S (20 h post-transfection) were rinsed twice with phosphate-buffered saline (PBS), followed by centrifugation at 2000 × g and 4°C. The cells were then resuspended in the extraction buffer (40 mM Tris–HCl, 1 mM Dithiothreitol (DTT), pH 7.4) and extracted by subjecting to liquid nitrogen multigelation three times. The cells were then centrifuged at 12 000 × g and 4°C for 10 min, and the supernatant was collected. For enzymatic assay, 20 μl of the supernatant containing Sas10 or Mpp10, 20 μl of the supernatant containing Capn3, and 0.4 μl of 200 mM CaCl2 (CaCl2 final concentration: 2 mM) were mixed to obtain the reaction mixture. For the negative controls, 2 μl of 200 mM ethylenediaminetetraacetic acid (EDTA) (EDTA final concentration: 10 mM) was added to the reaction mixtures prior to incubation. The reaction mixtures were then incubated at 37°C for 15 or 30 min. Reactions (excluding negative controls) were stopped by adding 2 μl of 200 mM EDTA (EDTA final concentration: 10 mM). The reaction mixtures were then denaturized at 100°C for 10 min and subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) for western blot analysis.
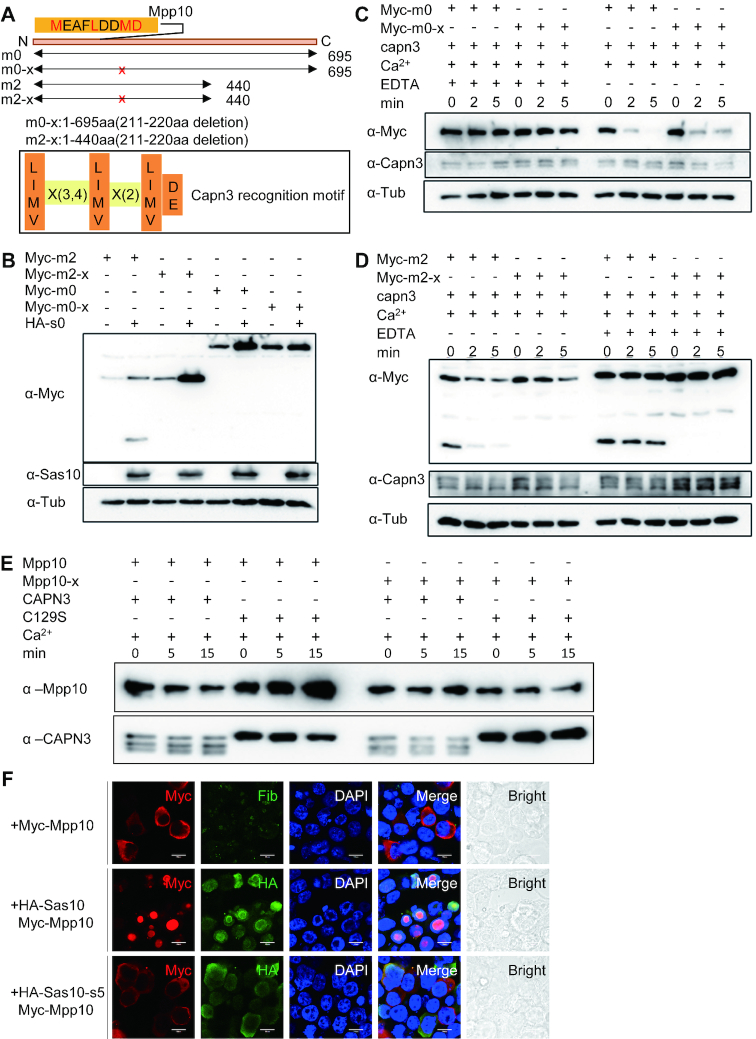
Figure 5.
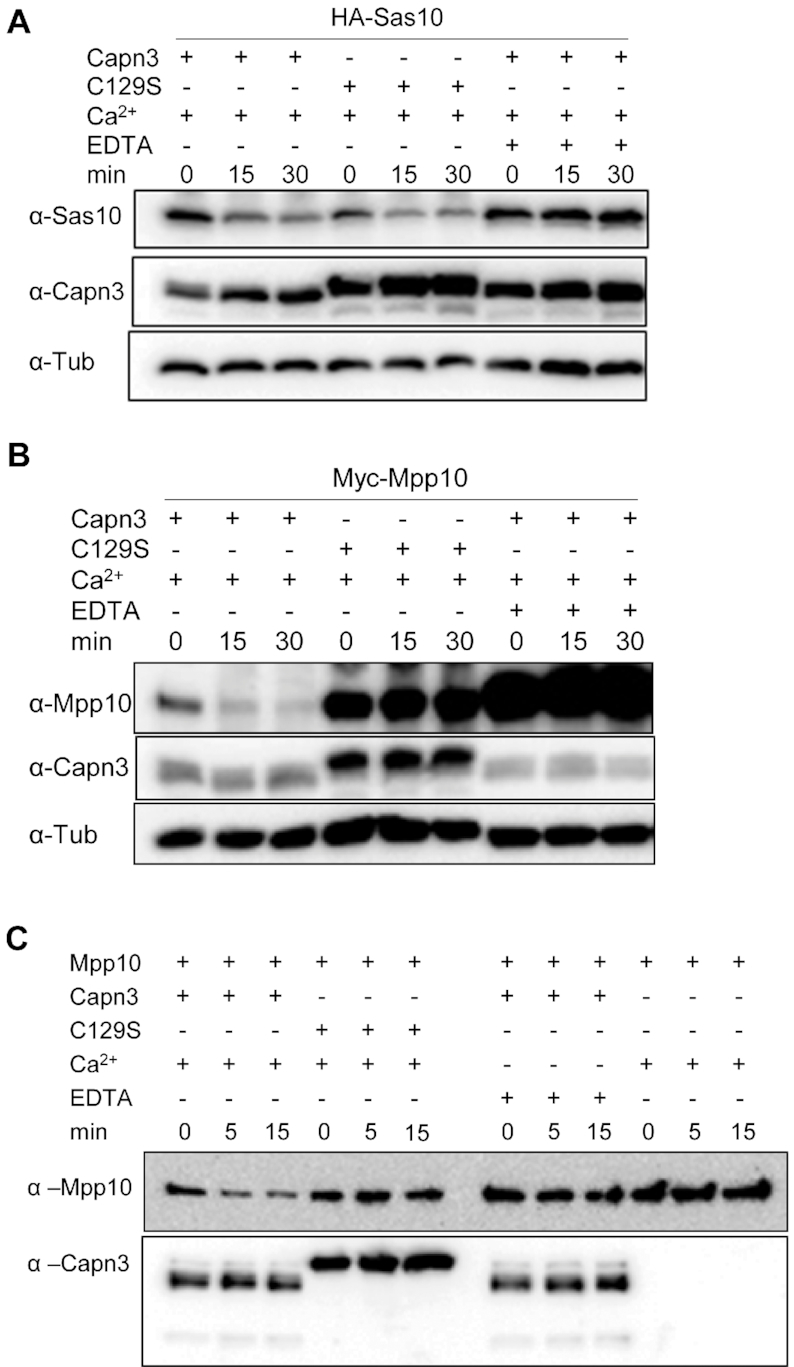
Mpp10 but not Sas10 is a Capn3 target. (A and B) Cell lysates containing Capn3 or its inactive mutant Capn3C129S (C129S) were mixed with cell lysates containing HA-Sas10 (A) or Myc-Mpp10 (B). Reaction mixture (containing 2 mM Ca2+) was incubated at 37°C for 15 and 30 min with or without 10 mM EDTA as indicated. (C) Enzymatic activity assays using purified His-Capn3 or HisCapn3C129S (C129S) as the enzyme and purified His-Mpp10 as the substrate. Reaction mixture containing 10 mM Ca2+ or 20 mM EDTA was incubated at 37°C for 5 and 15 min as indicated.
While assaying the effect of Capn3 on Mpp10, we noticed that Mpp10 was extremely unstable due to the endogenous Ca+2-dependent protease activity (Figure 5B) and underwent quick degradation even when the reaction mixture was placed on ice (Supplementary Figure S8B). To solve this problem, we modified the protein extraction condition by adding EDTA in the extraction buffer to a final concentration of 2 mM EDTA. The final Ca2+ concentration in the reaction mixture then increased to 3 mM. Reactions (excluding negative controls) were stopped by adding 2 μl of 300 mM EDTA (EDTA final concentration: 17 mM). The reaction mixtures were then denaturized at 100°C for 7 min and subjected to SDS-PAGE for western blot analysis (Figures 6C and D and 7B–D).
Figure 6.
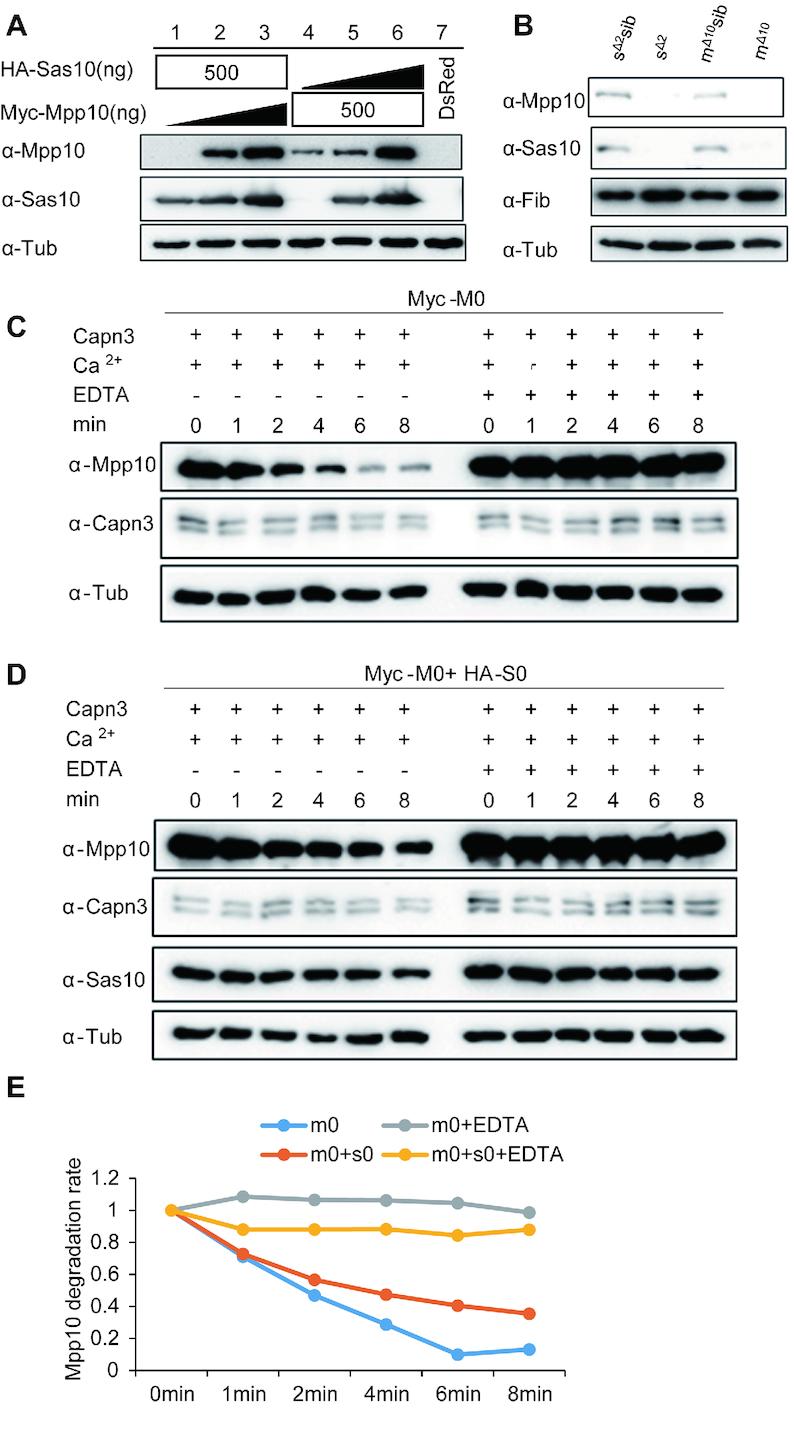
Stabilities of Sas10 and Mpp10 are interdependent. (A) The stabilities of Sas10 and Mpp10 mutually depend on each other in a dosage-dependent manner. Lanes 1–3: HA-Sas10 plasmid (500 ng) was co-transfected with different amount of Myc-Mpp10 plasmid. Lanes 4–6: Myc-Mpp10 plasmid (500 ng) was co-transfected with different amount of HA-Sas10 plasmid.  : 0 ng, 250 ng, 500 ng. The pCS2+ plasmid was used to make up the total amount of 500 ng plasmid DNA for transfection. Lane 7: control cells transfected with the DsRed plasmid. Mpp10 and Sas10 were detected using their specific antibodies, respectively. Total proteins were extracted from 293T cells 48 h post-transfection. (B) Western blot of Mpp10, Sas10, Fib and α-Tub in sas10Δ2 and mpp10Δ10 homozygous mutants and their siblings (WT and heterozygous). Total proteins were extracted from 5dpf-old embryos. (C–E) In vitro assay of Myc-Mpp10 degradation by Capn3. Capn3, Myc-Mpp10 or Myc-Mpp10 together with HA-Sas10 was respectively expressed in 293T cells. Protein extracts were mixed as indicated (C and D) and the mixture was incubated in the reaction buffer containing 3 mM CaCl2 at 37°C for different time intervals (min) as shown. The ratio of Myc-Mpp10 at different time point against Myc-Mpp100 min in Capn3+Myc-Mpp10 or in Capn3+Mpp10+ HA-Sas10 with or without EDTA reaction mixture was plotted against the time interval (E). α-Tub: loading control.
: 0 ng, 250 ng, 500 ng. The pCS2+ plasmid was used to make up the total amount of 500 ng plasmid DNA for transfection. Lane 7: control cells transfected with the DsRed plasmid. Mpp10 and Sas10 were detected using their specific antibodies, respectively. Total proteins were extracted from 293T cells 48 h post-transfection. (B) Western blot of Mpp10, Sas10, Fib and α-Tub in sas10Δ2 and mpp10Δ10 homozygous mutants and their siblings (WT and heterozygous). Total proteins were extracted from 5dpf-old embryos. (C–E) In vitro assay of Myc-Mpp10 degradation by Capn3. Capn3, Myc-Mpp10 or Myc-Mpp10 together with HA-Sas10 was respectively expressed in 293T cells. Protein extracts were mixed as indicated (C and D) and the mixture was incubated in the reaction buffer containing 3 mM CaCl2 at 37°C for different time intervals (min) as shown. The ratio of Myc-Mpp10 at different time point against Myc-Mpp100 min in Capn3+Myc-Mpp10 or in Capn3+Mpp10+ HA-Sas10 with or without EDTA reaction mixture was plotted against the time interval (E). α-Tub: loading control.
Figure 7.
Sas10 protects Mpp10 from Capn3-mediated degradation and determines the nucleolar-localization of Mpp10. (A) Prediction of a Capn3-recognition motif (211MEAFLDDMD) at the N-terminal of Mpp10 based on the Capn3 recognition consensus sequence. The MEAFLDDMD motif was deleted in Myc-tagged m0 (Myc-m0-x) and m2 (Myc-m2-x). (B) Myc-m0, Myc-m0-x, Myc-m2 or Myc-m2-x was respectively transfected into 293T cells together with or without HA-Sas10 (HA-s0) plasmid. Western blot was performed to compare the stability of corresponding proteins. (C and D) Protein samples were extracted from 293T cells expressing, respectively, Capn3, Myc-m0, Myc-m0-x, Myc-m2 or Myc-m2-x. Capn3 protein extract was mixed with Myc-m0 or Myc-m0-x (C) and with Myc-m2 or Myc-m2-x (D) with or without EDTA for 2 and 5 min. Western blot was performed to compare the protein stability in each reaction. (E) Enzymatic activity assays using purified His-Capn3 or His-Capn3C129S (C129S) as the enzyme and purified His-Mpp10 or His-Mpp10-X as the substrate. Reaction mixture containing 10 mM Ca2+ or 20 mM EDTA was incubated at 37°C for 5 and 15 min as indicated. (F) Myc-mpp10 was transfected into 293T cells alone or together with HA-sas10 or HA-sas10-s5 (lacking the Mpp10-interacting domain) plasmid. The cells were subjected to co-immunostaining of Myc-Mpp10 and Fib, Myc-Mpp10 and HA-Sas10, or Myc-Mpp10 and HA-Sas10-s5 48 h post-transfection; bar, 10 μm.
Expression and purification of Capn3 and Capn3C129S in SF9 cells
Capn3 and Capn3C129S were expressed in Spodoptera Frugiperda (SF9) cells as previously described (30). SF-9 cells were collected 72 h post-infection and washed on ice with washing buffer (20 mM Tris–HCl, 0.1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA). Cells were lysed in lysis buffer (20 mM Tris–HCl, 5 mM NaHCO3, 0.1 mM EGTA, 1 mM phenylmethane sulfonyl fluoride (PMSF), 5 mM 2-mercaptoethanol, pH 7.6) by cell scraper and were then gently sonicated. The supernatant after centrifugation were incubated with Ni-NTA agarose beads (YEASEN, #20502ES10) for 2 h. Beads were then washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, 1 mM DTT, 2 mM EDTA, pH 8.0). The purified Capn3 and Capn3C129S were used for enzymatic assay immediately because Capn3 losses its enzymatic activity rapidly. Since the elution buffer contains imidazole that may influence the enzymatic assay, the volume of the purified Capn3 added should be <1/10 of the total reaction volume. The purified Capn3C129S is more stable and can be stored at 4°C for 1 week.
Expression and purification of Mpp10 and Mpp10-X in Escherichiacoli
mpp10 and mpp10-x were cloned into the expression vector pET30a and transfected into Escherichia coli BL21 (DE3), respectively. The expression of His-Mpp10 and His-Mpp10-X was induced by 0.005 mM (for Mpp10) or 0.2 mM (for Mpp10-X) IPTG. Cells were treated with lysozyme (1 mg/ml) in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) for 30 min on ice and were then sonicated. The supernatant after centrifugation was incubated with Ni-NTA agarose beads (YEASEN, #20502ES10) overnight at 4°C. Beads were washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) three times and then eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). The eluent was desalted by ultrafiltration with reaction buffer (40 mM Tris–HCl, 1 mM DTT, 2 mM EDTA, pH 7.6) using protein concentrators (Thermo, #88529) before enzymatic assay. 1.5 μg Mpp10 or Mpp10-X were used as the Capn3 substrate in a 40 ul total reaction volume. The purified Mpp10 and Mpp10-X proteins can be stored in 4°C for 1 week.
Immunofluorescence staining on cryosections
Zebrafish cryosections were prepared as described previously (29,30). The cryosections were permeabilized with PBS-Triton (PBS plus 0.2% Triton X-100) for 30 min. After a brief wash with PBS-Triton containing 0.5% bovine serum albumin (PBB), the sections were blocked with 20% goat serum in PBB. After another brief wash with PBB, the sections were incubated with primary antibody (the desired concentration achieved by dilution with PBB) overnight at 4°C. After three 10-min washes with PBB, the sections were incubated with secondary antibodies (1:400) and 4′,6-diamidino-2-phenylindole (DAPI) (1:500) in PBB for 1 h. After another three washes with PBB, the sections were finally mounted in 80% glycerol and covered with coverslips for image acquisition. Antibodies against Sas10 (polyclonal antibody, 1:200), Sas10 (monoclonal antibody, 1:200), Mpp10 (polyclonal antibody, 1:200), Fib (1:500), phosphorylated Histone 3 (p-H3) (1:600) and Betaine-homocysteine methyltransferase (Bhmt) (monoclonal antibody, 1:300) were used for immunofluorescence staining (Supplementary Table S1).
To quantify the signal intensities of the immunostained Sas10 and Mpp10 in WT and def−/−, images in different samples in an independent experiment were taken under the same voltage for each laser channel by a confocal microscope (Olympus BX61WI). Positive signal intensities for Sas10, Mpp10 and DAPI were acquired using the ImageJ program (Wayne Rasband, National Institutes of Health, USA). The signal intensity of the stained DAPI was used to normalize the Sas10 and Mpp10 signal intensity in each corresponding cryosection. For statistic analysis, the relative signal intensity of Sas10 and Mpp10 in def−/− against WT (taken as 1) was obtained from at least 10 sections from three embryos in each case.
Immunofluorescence staining of cultured cells
The cultured cells were plated onto coverslips and placed in six-well plates. Forty-eight hours post-transfection, the cells on the coverslips were rinsed twice with ice-cold PBS and then fixed with 4% paraformaldehyde (Sigma) on ice for 30 min. The cells were then washed twice with PBS and permeabilized with PBS-Triton (0.2% Triton X-100 in PBS) at room temperature (RT) for 30 min. After blocking in 1 x PBS containing 0.2% Triton X-100, 2% donkey serum, 3% bovine serum albumin (FDB) for 30 min at RT, the coverslips were incubated with primary antibody for 2 h at RT, followed by three 5-min washes with PBS-Triton. Secondary antibodies (1:400 diluted in FDB) and DAPI (1:400 diluted in FDB) were added, and the coverslips were incubated for a further 1 h at RT. After three quick washes with PBS-Triton, the coverslips were finally mounted in 80% glycerol for image acquisition. Anti-Sas10, -Mpp10, -HA and -Myc polyclonal antibodies and anti-Sas10, -HA, -Myc, and -Fib monoclonal antibodies were used for immunostaining (Supplementary Table S1).
Isolation of the nucleoli
The zebrafish liver nucleoli were isolated as described previously with minor modifications (30). Zebrafish livers were collected and homogenized in PBS plus 1 × cOmplete. After centrifugation at 2000 × g and 4°C for 5 min, the precipitates were briefly washed by PBS plus 1 × cOmplete and were resuspended in buffer A (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.9, 10 mM KCl, 1.5 mM MgCl2 and 0.5 mM DTT) at 4°C for 30 min. Nonidet P 40 (NP40, 0.1%) was then added to rupture the cell membranes. After centrifugation at 1500 × g and 4°C for 5 min, the supernatant was retained as the cytoplasmic fraction, whereas the pellet (containing the nuclei) was resuspended in the zfS1 buffer (0.5 M sucrose, 3 mM MgCl2) and sonicated for six 5-s bursts (with 15-s intervals between bursts) at 40% amplitude. The sonicated sample was layered onto the zfS2 buffer (1 M sucrose, 3 mM MgCl2) and centrifuged at 1800 × g for 10 min. The pellet was collected as the nucleolar fraction and the supernatant as the nucleoplasmic fraction.
TUNEL assay
TUNEL assay was performed using the in situ cell death detection kit TMR red (Roche Diagnostics) according to the manufacturer’s instructions. Sample were prepared as described previously (29,34,35). Sections of WT embryos pre-treated with DNase I were used as a positive control.
Yeast two-hybrid assay
pGAD-Sas10, pGAD-Mpp10, pGBK-Sas10 and pGBK-Mpp10 were generated using the pGADT7 AD Cloning Vector and pGBKT7 DNA-BD Cloning Vector plasmids provided in the Matchmaker® Gold Yeast Two-Hybrid System kit (Supplementary Table S1). The yeast two-hybrid assay was performed according to the instructions in the kit’s user manual.
Statistical analysis
Statistical analyses were performed using the Student’s t-test. Significant differences were considered as follows: *P < 0.05; **P < 0.01; ***P < 0.001; n.s., no significant difference.
RESULTS
Zebrafish Sas10 and Mpp10 are nucleolar proteins
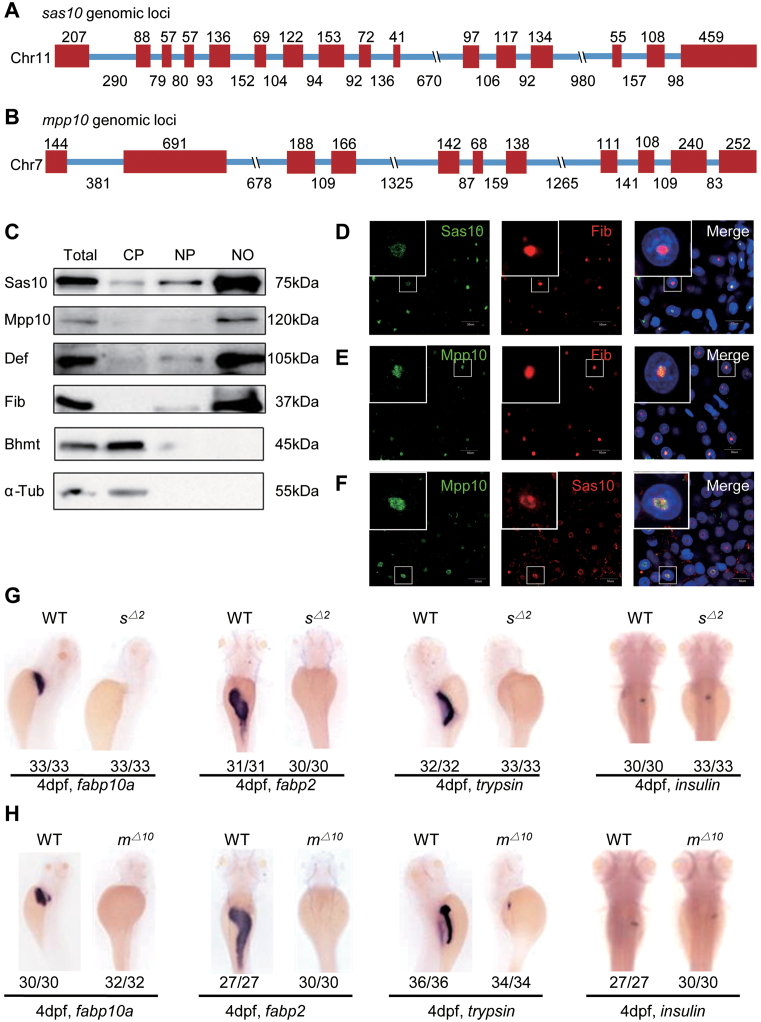
The zebrafish sas10 gene spans an ∼6.4-kb genomic DNA fragment on chromosome 11 and contains 16 exons and 15 introns (Figure 1A). The zebrafish mpp10 gene spans an ∼7.8-kb genomic DNA fragment on chromosome 7 and contains 11 exons and 10 introns (Figure 1B). Protein sequence alignments showed that both Sas10 and Mpp10 are highly conserved proteins across different species and share 47% and 54% identity with their human counterparts, respectively (Supplementary Figures S1A and S2A). We generated a polyclonal antibody and a monoclonal antibody against zebrafish Sas10 (Supplementary Figure S1B and C) and a ployclonal antibody Mpp10 (Supplementary Figure S2B) for determining the subcellular localizations of Sas10 and Mpp10. First, the adult liver from the WT zebrafish was harvested for extracting cytoplasmic protein, nucleoplasmic protein and nucleolar protein. Western blot analysis showed that Sas10 and Mpp10 were enriched in the nucleolar fraction similar to the other two nucleolar proteins Def (25,29) and Fib (Figure 1C), whereas Bhmt (36) and α-Tubulin (α-Tub) were mainly detected in the cytoplasmic fraction (Figure 1C). Next, we co-stained Fib with Sas10 or Mpp10 and found that both Sas10 (Figure 1D) and Mpp10 (Figure 1E) were co-localized with Fib in the nucleoli of the WT adult liver. Sas10 and Mpp10 co-staining also showed that they were co-localized in the nucleoli (Figure 1F). These results indicate that both Sas10 and Mpp10 are conserved nucleolar proteins in zebrafish.
Figure 1.
Zebrafish Sas10 and Mpp10 are nucleolar proteins and display dynamic expression patterns during early embryogenesis. (A and B) Diagram showing the genomic structures of the zebrafish sas10 gene on chromosome 11 (A) and mpp10 gene on chromosome 7 (B). Red bar, exons; blue line, introns. Numbers above each exon and under each intron indicate their length (bp). (C) Western blot of Sas10, Mpp10, Def, Fib, Bhmt and α-Tub in total protein (Total), cytoplasmic fraction (CP), nucleoplasmic fraction (NP) and nucleolar fraction (NO) from adult livers. (D–F) Co-immunostaining of Sas10 and Fib (D), Mpp10 and Fib (E), or Mpp10 and Sas10 (F) in the adult liver. Big box shows the high magnification view of the nucleus in the small box. Nuclei were stained with DAPI; bar, 50 μm. (G and H) WISH using the fabp10a, fabp2, trypsin; insulin probes on 4dpf-old WT, sas10Δ2 (sΔ2) (G) and mpp10Δ10 (mΔ10) (H) embryos. The number of total embryos genotyped (as denominator) and the number of embryos exhibiting the displayed phenotype (as numerator) are shown at the bottom.
sas10 and mpp10 display similar dynamic expression patterns during early organogenesis
The WISH experiments showed that the transcripts of both sas10 and mpp10 were present in unfertilized eggs, suggesting that they are maternal genes, and were maintained in the embryos from 6 to 12 hpf (sas10: Supplementary Figure S3A; mpp10: Supplementary Figure S4A). At one day post-fertilization (dpf), both sas10 and mpp10 transcripts were highly enriched in the brain region and were also detectable in the endoderm region which gives rise to the liver, pancreas and intestine (sas10: Supplementary Figure S3A; mpp10: Supplementary Figure S4A). From 2 to 3 dpf, the levels of sas10 and mpp10 transcripts decreased sharply in the brain region and were low but still distinguishable in the digestive organs (sas10: Supplementary Figure S3A; mpp10: Supplementary Figure S4A). Interestingly, the expression of both sas10 and mpp10 was enriched in the digestive organs from 4 to 5 dpf (sas10: Supplementary Figure S3A; mpp10: Supplementary Figure S4A). The dynamic expression patterns of sas10 and mpp10 suggest that their expression is regulated by the demand of cellular activities during early organogenesis.
Loss-of-function mutation of sas10 or mpp10 results in hypoplastic digestive organs
In recent years, increasing evidence has demonstrated that in addition to their roles in ribosome biogenesis, many nucleolar factors also function as the regulators of organogenesis (25,32,37). To investigate the biological function of Sas10 and Mpp10, we generated sas10 and mpp10 mutants, respectively, using the CRISPR-Cas9 technique. Two sas10 mutant alleles were obtained, one carrying an 8-bp deletion (sas10Δ8) and another a 4-bp deletion together with a 2-bp insertion (sas10Δ2) in exon 4 (Supplementary Figure S3B). We also obtained two mpp10 mutant alleles, one carrying a 13-bp deletion together with a 3-bp insertion (consequently 10-bp deletion, mpp10Δ10) and another carrying a 2-bp deletion together with a 4-bp insertion (mpp10+2) in exon 2 (Supplementary Figure S4B). At 5 dpf, the sas10Δ2 and mpp10Δ10 mutants displayed similar phenotypes, including smaller eyes, cardiac oedema, no swim bladder and abnormal yolk absorption (sas10Δ2: Supplementary Figure S3C; mpp10Δ10: Supplementary Figure S4C). The sas10Δ2 and mpp10Δ10 mutant embryos died at ∼7 dpf. Considering that the expression of sas10 and mpp10 was enriched in the digestive organs during early embryogenesis (sas10: Supplementary Figure S3A; mpp10: Supplementary Figure S4A), we assessed digestive organ development in sas10Δ2 and mpp10Δ10 mutant embryos using liver fatty acid-binding protein 10a (fabp10a, a liver-specific marker), trypsin (an exocrine pancreas-specific marker), intestine fatty acid-binding protein 2 (fabp2, an intestine-specific marker) and insulin (an endocrine pancreas-specific marker) probes. The WISH results showed that, except for the endocrine pancreas, Sas10 or Mpp10 depletion caused severe defects in digestive organ development and the liver, intestine and exocrine pancreas were hardly detectable at 4 dpf (Figure 1G and H). To determine the timepoint when the phenotype is discernible, we performed WISH using the early endoderm markers gata6 and foxa3 and the hepatic markers hhex and prox1. No observable difference was found between the WT and sas10Δ2 or mpp10Δ10 mutant embryos at 30 hpf, suggesting that the initiation of the digestive organ development was not obviously affected (sas10Δ2: Supplementary Figure S3D–G; mpp10Δ10: Supplementary Figure S4D–G). At 50 hpf, the growth of the liver and exocrine pancreatic buds in the sas10Δ2 and mpp10Δ10 mutant embryos appeared to be severely retarded, as revealed by the four markers (sas10Δ2: Supplementary Figure S3D–G; mpp10Δ10: Supplementary Figure S4D–G). To determine whether the defects were caused by cell proliferation arrest or cell apoptosis, we performed the p-H3 (a G2/M-phase transition marker) staining assay and the TUNEL assay (for apoptosis). The p-H3 staining assay revealed that the ratios of p-H3-positive cells in the liver were significantly lower in the sas10Δ2 and mpp10Δ10 mutant embryos than in the WT embryos (Supplementary Figure S5), whereas the TUNEL assay revealed no significant changes in the number of apoptotic cells between the WT and sas10Δ2 or mpp10Δ10 mutant embryos (Supplementary Figure S6A). These results suggest that the cell cycle arrest was the main cause of hypoplastic digestive organs in these two mutants. Therefore, both sas10 and mpp10 genes are essential for digestive organ development.
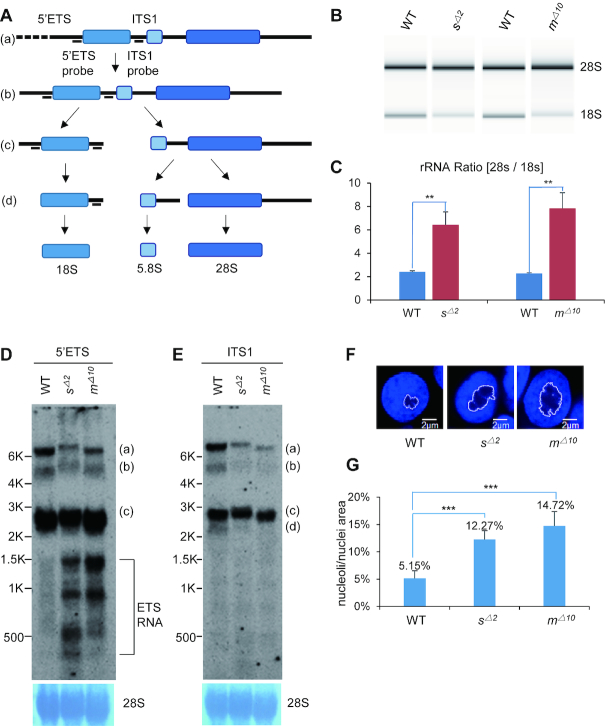
Sas10 and Mpp10 depletion results in pre-rRNA processing defects
The Mpp10–Imp3–Imp4 complex serves as a core component of the SSU processome (6,7,17). Although structural analysis failed to identify Sas10 in the SSU processome, the fact that Sas10 interacts with Mpp10 suggests that Sas10 is likely involved in pre-rRNA processing. Similar to other species, the mature 5.8S, 18S and 28S rRNAs in zebrafish are processed and assembled from an initial pre-rRNA transcript (Figure 2A). We analysed the levels of 18S and 28S rRNAs on an Agilent 2100 Bioanalyzer and found that the mature 18S rRNA levels in the sas10Δ2 or mpp10Δ10 mutant embryos were lower than those in the WT embryos at 5 dpf, whereas the 28S rRNA levels were not evidently different (Figure 2B). This low 18S rRNA level led to a significant increase in the 28S/18S rRNAs ratios in the sas10Δ2 or mpp10Δ10 mutants (Figure 2C). Northern blot analysis using DIG-labelled oligonucleotide probes complementary to the 5′ETS detected unknown intermediate products of pre-rRNA processing in the sas10Δ2 or mpp10Δ10 mutants (Figure 2D). These unknown intermediate products appeared not to harbour the 18S rRNA sequence (Supplementary Figure S6B), suggesting that they were likely products of aberrantly cleaved 5′ETS. In contrast, the ITS1 showed no obvious change in the band pattern but revealed decreased pre-rRNA levels (bands a and b) in the sas10Δ2 or mpp10Δ10 mutants (Figure 2E). These results suggest that Sas10 and Mpp10 play important roles in pre-rRNA processing.
Figure 2.
Depletion of Sas10 and Mpp10 leads to pre-rRNA processing defects. (A) Diagram showing the stepwise processing of the pre-rRNA transcript (a) to the intermediates (b and c) to the mature 18S rRNA. 5′ETS, 5′-external transcribed sequence; ITS1, internal transcribed sequence. (B and C) Detection of 28S and 18S rRNA on an Agilent 2100 Bioanalyzer (B) for analysing the ratio of 28S rRNA versus 18S rRNA (C) in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) embryos.**, P< 0.01 (D and E) Northern blot analysis of rRNA precursors using the 5′ETS (D) and ITS1 (E) probes in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) embryos. 28S: loading control. (F and G) DAPI was used to stain the nuclei. The area of nucleolus (regions lacking or showing faint DAPI signal) and of nucleus in an individual cell was measured (F), respectively, in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) liver. The average area ratios of nucleolus/nucleus from at least 90 hepatocytes from three embryos for each genotype were used in statistic analysis (G); bar, 2 μm. ***, P < 0.001.
Defective ribosome biogenesis often causes a change in the nucleolar morphology (38–40). We observed that the size of the nucleoli increased by 12% and 15% on average in the livers of 5-dpf-old sas10Δ2 and mpp10Δ10 mutants, respectively (Figure 2F and G).
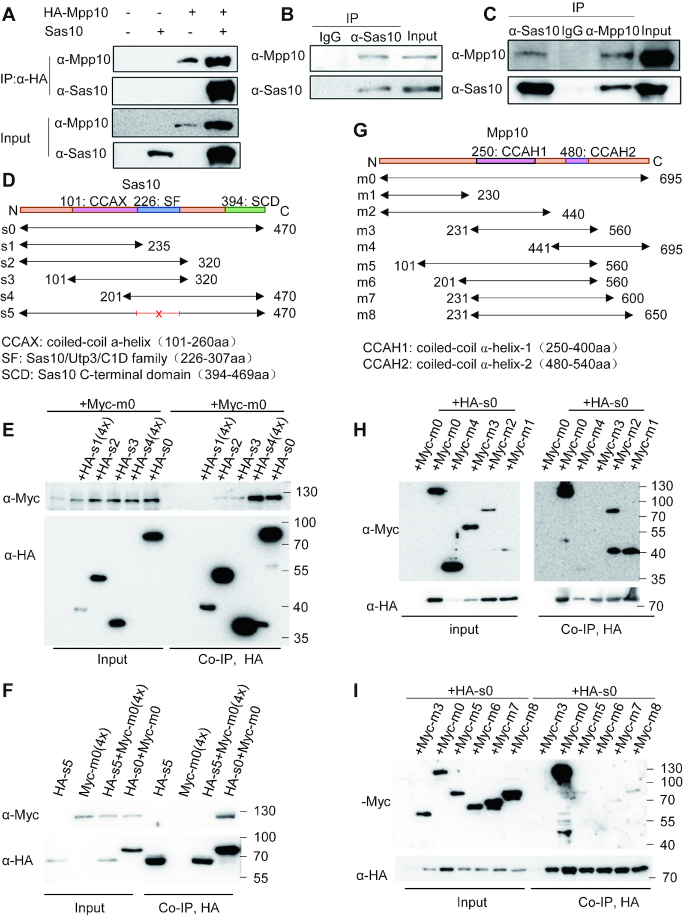
Zebrafish Sas10 and Mpp10 interact with each other
In yeast, it has been shown that Sas10 and Mpp10 interact with each other (26) and that Sas10 binds to the N-terminus of Mpp10 (24). To determine whether the interaction between Sas10 and Mpp10 is conserved in the vertebrates, we co-expressed zebrafish Sas10 and HA-tagged zebrafish Mpp10 (HA-Mpp10) in human 293T cells. Co-IP analysis using the proteins extracted from these cells showed that Sas10 was pulled down by HA-Mpp10 (Figure 3A). To determine whether the endogenous Sas10 and Mpp10 interact with each other, we extracted the total protein from 32-hpf-old embryos (Figure 3B) and adult liver nuclei (Figure 3C) and subjected these protein samples to Co-IP using anti-Sas10 or anti-Mpp10 antibodies, respectively. In both cases, Sas10 and Mpp10 were detected in the Co-IP products (Figure 3B and C). Finally, we cloned sas10 and mpp10 into the expression vectors for yeast two-hybrid assays. The result showed that Sas10-GalBD interacted with Mpp10-GalAD (Supplementary Figure S7A) and that Mpp10-GalBD exhibited auto-activation activity (Supplementary Figure S7A). These results suggest that Sas10 interacts with Mpp10 directly.
Figure 3.
The Sas10/Utp3/C1D domain interacts with the N-terminus of Mpp10. (A) HA-tagged Mpp10 (HA-Mpp10) and Sas10 (without a tag) were co-expressed in 293T cells for Co-IP analysis using an anti-HA antibody. HA-Mpp10 and Sas10 were detected with their specific antibodies, respectively. (B and C) Co-IP of the endogenous Mpp10 and Sas10 extracted from the 32hpf-old embryos (B, using the anti-Sas10 polyclonal antibody for Co-IP) and adult liver (C, using the anti-Sas10 and anti-Mpp10 polyclonal antibodies for Co-IP, respectively). Co-IP with IgG serves as a negative control. (D–F) Co-IP analysis of the interaction between Myc-tagged Mpp10 (Myc-m0) with HA-tagged Sas10 (s0) and its different derivatives (s1 to s5) constructed as shown in (D). s5 is deleted of 226–307 amino acids in Sas10. Myc-Mpp10 interacted with Sas10 (s0) and its derivatives containing the Sas10/Utp3/C1D domain s2 to s4 (E, Co-IP with an anti-Myc antibody) but not with s5 without the domain (F, Co-IP with an anti-HA antibody). 4×: 4-fold amount of plasmids for transfection. (G–I) Co-IP analysis of the interaction between HA-tagged Sas10 (HA-s0) with Myc-Mpp10 (m0) and its different derivatives (m1 to m8) constructed as shown in (G). HA-Sas10 interacted strongly with Myc-Mpp10 (m0) and its N-terminal derivatives m1 and m2 (H) but with negligible interaction with m3-m8 (H and I). In (D–G), numbers in each construct define the corresponding positions of amino acids in Sas10 (D) and Mpp10 (G). Predicted α-helix domains in Mpp10 and conserved domains in Sas10 are also outlined.
The Sas10/Utp3/C1D domain mediates Sas10 and Mpp10 interaction
To determine the domain of Sas10 that interacts with Mpp10, we generated plasmids expressing different HA-tagged Sas10 derivatives, namely full-length WT Sas101-470 (s0), Sas101-235 (s1), Sas101-320 (s2), Sas10101-320 (s3) and Sas10201-470 (s4) (Figure 3D). These HA-sas10-derived plasmids were co-transfected with Myc-tagged full-length mpp10 (Myc-m0) into human 293T cells. Co-IP analysis showed that, except for s1, the full-length WT Sas10 and all other truncated Sas10 successfully pulled down the Myc-Mpp10 (Figure 3E). Interestingly, s4 appeared to pull down the Myc-Mpp10 more efficiently than s2 and s3 (Figure 3E), suggesting that the C-terminus of Mpp10 facilitates the strong interaction between Sas10 and Mpp10. Because s2 (Sas101-320), s3 (Sas10101-320) and s4 (Sas10201-470) shared the 201–320-aa region of Sas10 (Figure 3D), the Co-IP result suggested that this Sas10201-320 region serves as the domain for interaction with Mpp10. Protein sequence analysis revealed that the zebrafish Sas10226-307 region corresponds to the conserved domain in the Sas10/Utp3/C1D family (Supplementary Figure S1A). The Sas10/Utp3/C1D family domain is supposed to serve as a platform for protein interactions (39). We then constructed a plasmid by deleting the sequence encoding the Sas10 226–307-aa region (Sas10Δ226-307, s5). Co-IP analysis showed that s5 (Sas10Δ226-307) lost the ability to interact with Mpp10 (Figure 3F).
The N-terminus of Mpp10 is required for robust interaction with Sas10
To identify the region of Mpp10 that interacts with Sas10, we generated plasmids expressing different Myc-tagged Mpp10 derivatives, namely full-length WT Mpp101-695 (m0), Mpp101-230 (m1), Mpp101-440 (m2), Mpp10231-560 (m3) and Mpp10441-695 (m4) (Figure 3G). These plasmids were co-transfected with HA-tagged sas10 plasmid (HA-s0) into human 293T cells. Co-IP analysis showed that both Mpp101-230 and Mpp101-440 strongly interacted with Sas10 (Figure 3H). Interestingly, two Co-IP products (75 and 38 kDa) were detected when Mpp101-440-expressing cells were used. The 75-kDa fragment matched the size predicted for Mpp101-440 (m2), whereas the 38-kDa fragment was very similar to Mpp101-230 (m1) in size (Figure 3H). This prompted us to investigate the nature of the 38-kDa fragment, which is explained later in the text. Mpp10441-695, but not Mpp10231-560, showed weak interaction with Sas10 (Figure 3H). We generated four additional plasmids expressing Mpp10101-560 (m5), Mpp10201-560 (m6), Mpp10231-600 (m7) and Mpp10231-650 (m8) (Figure 3G). Co-IP analysis showed that compared with the WT Mpp10 (m0), Mpp10101-560 (m5), Mpp10231-600 (m7) and Mpp10231-650 (m8) displayed a negligible weak interaction with Sas10, whereas Mpp10201-560 (m6) showed no interaction (Figure 3I). Therefore, the first 230 aa of Mpp10 are necessary for its interaction with Sas10. This result is consistent with the structural analysis result of the ctSas10–ctMpp10 protein complex obtained from the thermophilic fungus Chaetomium thermophilum, which revealed that the N-terminal 59-90-aa and 125-157-aa regions of ctMpp10 interact with Sas10 (24).
Def interacts with Sas10 to form the Def–Sas10–Mpp10 complex
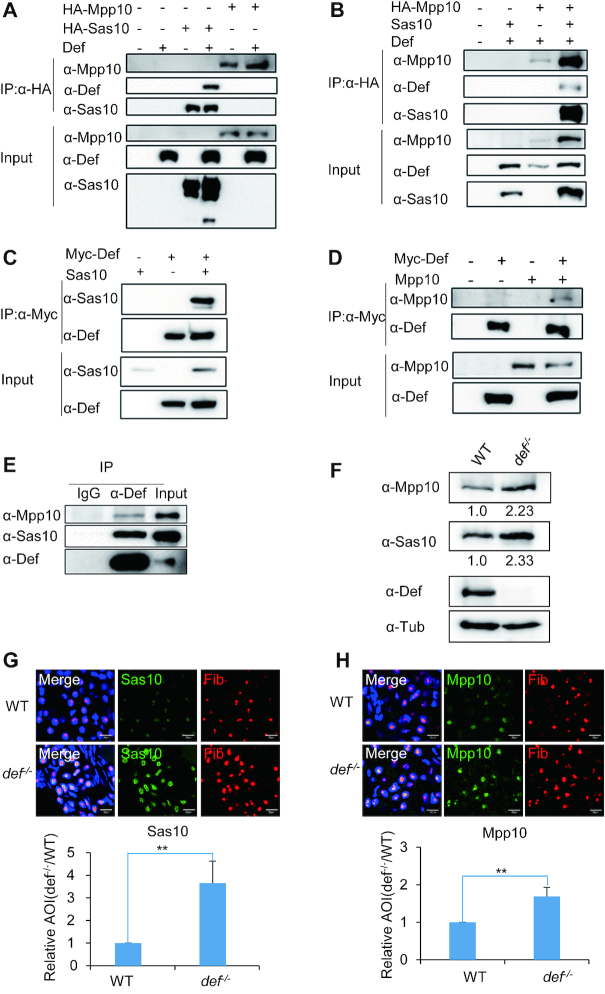
A previous study showed that Sas10 and Mpp10 interact with Utp25, another nucleolar protein, in yeast (26). The counterpart of Utp25 in zebrafish is Def, and Def/Utp25 is conserved in human, mouse and Arabidopsis (25). Therefore, to determine whether zebrafish Def interacts with Sas10 and Mpp10, we co-expressed Def with HA-tagged Mpp10 or Sas10 in human 293T cells. Co-IP using an anti-HA tag antibody showed that Def was successfully pulled down by HA-Sas10 but not by HA-Mpp10 (Figure 4A). Next, we co-expressed Def with Sas10 (untagged) and HA-Mpp10 to determine whether Def, Sas10 and Mpp10 form a complex. Co-IP using the anti-HA-tag antibody showed that Def was detected in the Co-IP product, although at a much lower level, only when Def, Sas10 and HA-Mpp10 were co-expressed in human 293T cells (Figure 4B). This result suggests that Sas10 bridges Mpp10 and Def to form the Mpp10–Sas10–Def complex. Based on this result, we surmised that immunoprecipitating Def would pull down Sas10 together with Mpp10. Indeed, both Sas10 (Figure 4C) and Mpp10 (Figure 4D) were detected in the Co-IP product when the anti-Myc antibody was used to pull down Def. Finally, we extracted the nuclei from the WT adult zebrafish liver and performed Co-IP using the anti-Def antibody. The result showed that Def successfully pulled down Sas10 and Mpp10 (Figure 4E).
Figure 4.
Def interacts with and stabilizes the Sas10–Mpp10 complex. (A and B) Def (without a tag) was co-expressed with HA-Mpp10 or HA-Sas10 (A) or together with HA-Mpp10 and Sas10 (without a tag) (B) in 293T cells for Co-IP analysis using an anti-HA antibody. Def, Mpp10 and Sas10 were detected with their respective specific antibodies as indicated. (C and D) Myc-tagged Def (Myc-Def) was co-expressed with Sas10 (without a tag, C) or Mpp10 (without a tag, D) in 293T cells for Co-IP analysis using an anti-Myc antibody. (E) Proteins samples were extracted from the adult liver for analysis of the interaction between the endogenous Def and the Sas10-Mpp10 using an anti-Def antibody for Co-IP. Def, Sas10 and Mpp10 were detected with their specific antibodies. (F) Western blot of Mpp10, Sas10, Def and α-Tub in the WT and def−/− mutant. Total proteins were extracted from 5dpf-old embryos. Value below the figures indicated the intensity of Mpp10/Tub or Sas10/Tub where the ratio for WT was taken as 1.0. (G and H) Immunostaining of Sas10 and Fib (G) or Mpp10 and Fib (H) in the liver of 5dpf-old WT and def−/− mutant embryos. DAPI was used to stain the nuclei. Statistic analysis of the relative signal intensity of Sas10 and Mpp10 in WT (taken as 1) and def−/− was obtained from at least 10 sections from three embryos in each case. AOI: area of intensity; bar, 50 μm. **, P < 0.01.
Sas10 and Mpp10 protein levels are elevated in the def−/− null mutant
The loss-of-function mutation of def in the defhi429 null mutant (def−/−) results in hypoplastic digestive organs, a phenotype similar to that displayed by the sas10Δ2 and mpp10Δ10 mutants (25). To study the effect of Def depletion on the Sas10 and Mpp10 proteins, we performed a western blot analysis and the result showed that, compared with WT, both Sas10 and Mpp10 protein levels were obviously elevated in the def−/− mutant at 5dpf (Figure 4F). We then performed co-immunostaining of Sas10 and Mpp10 with Fib, respectively, in the WT and def−/− mutant at 5dpf. We found that, compared with WT, both Sas10 (Figure 4G) and Mpp10 (Figure 4H) accumulated to a much higher level in the nucleoli of liver cells in the def−/− mutant, suggesting that the depletion of Def stabilized both Sas10 and Mpp10.
Mpp10 is sensitive to Capn3-mediated protein degradation
Our recent studies have shown that Def recruits Capn3 to the nucleolus to mediate the degradation of the tumour suppressor p53 in the nucleolus (30). Considering that the three nucleolar factors Def, Sas10 and Mpp10 can form a complex and that the levels of Sas10 and Mpp10 were elevated in the def−/− mutant, we hypothesized that Def serves as a scaffold protein for Capn3 to target Sas10 and/or Mpp10. To test the effect of Capn3 on Sas10 and Mpp10, we extracted proteins from cells expressing Capn3 or Capn3C129S (a bio-inactive Capn3 due to the substitution of C129 by S129) (29,41) and mixed them with the protein extracts from cells expressing Sas10 or Mpp10. The reaction mixtures were supplemented with EDTA or no EDTA (an inhibitor of the Capn3 enzymatic activity) and were incubated at 37°C for 15 or 30 min before being subjected to western blot analysis. The result showed that when incubated with Capn3 or Capn3C129S, the Sas10 protein levels declined slowly in a similar pattern (Figure 5A). This decrease in the Sas10 levels was completely inhibited by EDTA, suggesting that this reduction was not due to Capn3 but likely due to an unknown EDTA-sensitive protease in the protein extract. In contrast, the Mpp10 level reduced drastically when the reaction mixture contained Capn3 but showed no reduction when the reaction mixture contained Capn3C129S or was supplemented with EDTA (Figure 5B). We noticed that, in such reaction mixture, Mpp10 appeared also to be targeted by other EDTA-sensitive protease (Figure 5B, compare three lanes for Capn3C129S with three lanes for EDTA; also see Supplementary Figure S8A). Mpp10 underwent degradation even when the reaction mixture was placed on ice (Supplementary Figure S8B).
To exclude the effect of other EDTA-sensitive protease and to prove Mpp10 to be a Capn3 substrate unequivocally, we expressed His-tagged Capn3 and Capn3C129S in the SF9 cells (Supplementary Figure S7B and C) and His-tagged Mpp10 in E. coli (Supplementary Figure S7D) and purified these proteins using the Ni-NTA agarose beads, respectively. Enzymatic activity assays showed Capn3, but not Capn3C129S, decreased the level of Mpp10 at 5 and 15 min (Figure 5C). As expected, the effect of Capn3 on Mpp10 was abolished by EDTA.
Sas10 and Mpp10 stability is interdependent
While studying the interaction between Sas10 and Mpp10 in human 293T cells, we surprisingly found that the co-transfection of HA-sas10 and Myc-mpp10 plasmids greatly enhanced the stability of both Sas10 and Mpp10 proteins (Figures 3A and 4B). We found that this stability was interdependent and dosage dependent (Figure 6A). We reckoned that if Sas10 and Mpp10 stability is really interdependent, it is reasonable to expect a down-regulation of Mpp10 level in the sas10Δ2 mutant and of Sas10 levels in the mpp10Δ10mutant. To explore this possibility, we examined Sas10 and Mpp10 levels in sas10Δ2 and mpp10Δ10 mutants and found that both Sas10 and Mpp10 were almost abolished in both mutants (Figure 6B).
The 211-220-aa region in Mpp10 is required for Capn3-mediated Mpp10 degradation
The interdependency of Sas10 and Mpp10 stability suggests that the interacting domains of these two proteins contain target sites of a certain protease and that the target site is masked once Sas10 and Mpp10 form a complex. Considering that Mpp10 but not Sas10 is the Capn3 target, we hypothesized that the binding of Sas10 to Mpp10 masks the Capn3-recognition site on Mpp10. This hypothesis is supported by the fact that both Mpp10 and Sas10 accumulated in the nucleoli of the def−/− mutant cells (Figure 4G and H), which lacked Capn3 in the nucleoli (30). To further test this hypothesis, we extracted proteins from cells expressing Capn3 and mixed them with proteins extracted from cells expressing Mpp10 alone (Figure 6C) or co-expressing Mpp10 and Sas10 (Figure 6D). The reaction mixtures were incubated with or without EDTA at 37°C for different time intervals before subjecting to western blot analysis (Figure 6C and D). Plotting the signal of Mpp10 at different reaction times against the signal of the starting Mpp10 (Mpp100 min) ratio showed that the Mpp10 level in the protein samples of cells expressing Mpp10 alone decreased much faster than that in the protein samples of cells expressing both Mpp10 and Sas10 (Figure 6E). This finding suggested that Sas10 protected Mpp10 from Capn3-mediated protein degradation.
Next, we assessed whether the Sas10 and Mpp10 interacting domains contained a Capn3-recognition motif (30,42). Indeed, one motif (the 211–220-aa region) in the Mpp10 interacting domain was found to contain a putative Capn3-recognition site (Figure 7A), but no Capn3-recognition motif was found in the Sas10 interacting domain. As shown in Figure 3H, we detected two bands in the Co-IP product: one fragment (75 kDa) that matched the size of m2 (Mpp101-440) and a smaller fragment (38 kDa) similar to the size of m1 (Mpp101-230). Interestingly, both m1 and m2 contain the 211–220-aa motif, but this motif is near the end of m1. This prompted us to investigate whether the 211–220-aa motif is necessary for Capn3-mediated Mpp10 degradation. We deleted the 211–220-aa motif in m0 (m0-x) and m2 (m2-x) (Figure 7A) and compared the stability between m0 and m0-x and between m2 and m2-x in the presence and absence of Sas10 in human 293T cells. The result showed that the deletion of the 211–220-aa motif in either m0 or m2 greatly stabilized the protein (Figure 7B). More importantly, the smaller band (38 kDa) in m2 was abolished in the m2-x sample (Figure 7B). The vast increase in the level of the Myc-m2-x protein in the presence of Sas10 suggests that the binding of Sas10 to Mpp10 not only protects Mpp10 from Capn3-mediated degradation but also from the degradation possibly mediated by other proteinase in the protein extract.
To determine whether the deletion of the 211–220-aa motif would reduce the sensitivity of m0-x and m2-x to Capn3, we mixed the protein extracts containing m0-x or m2-x with Capn3 from human 293T cells and incubated the reaction mixture at 37°C for 2 or 5 min, respectively. Western blot analysis showed that compared with their corresponding controls, both m0-x (Figure 7C) and m2-x (Figure 7D) showed reduced sensitivity to Capn3. As expected, EDTA inhibited the effect of Capn3 on all Mpp10 derivatives (Figure 7C and D), including the 38-kDa fragment in the protein samples from the m2 plasmid-transfected cells (Figure 7D). These results indicate that the 211–220-aa motif is necessary for the cleavage of m2 by Capn3 to produce the smaller 38-kDa fragment.
To exclude the effect of other EDTA-sensitive protease in the protein extracts on Mpp10 degradation, we mixed the Ni-NAT agarose purified Capn3 or Capn3C129S (Supplementary Figure S7B and C) with the purified Mpp10 or Mpp10-X (Supplementary Figure S7D and E) for enzymatic activity assay. The result clearly showed that the level of Mpp10, but not of Mpp10-X, was reduced by Capn3 (Figure 7E). As expected, both Mpp10 and Mpp10-X were insensitive to Capn3C129S (Figure 7E).
Finally, we generated a zebrafish capn3b mutant allele capn3b−/−, which harbours a 14 bp deletion in the exon 1 and a 19 bp deletion in the exon 3 using the CRISPR-Cas9 approach. Western blot analysis showed that the capn3b−/− mutant is absent from Capn3b at 5 dpf (Supplementary Figure S8C), suggesting it is a null allele. Further western blot analysis showed that, compared with WT, the level of Mpp10 in capn3b−/− was obviously elevated (Supplementary Figure S8C).
Nucleolar localization of the Mpp10–Imp3–Imp4 complex depends on Sas10
Next, we investigated whether the interaction between Sas10 and Mpp10 is required for nucleolar localization of these two proteins. For this purpose, Sas10- or Mpp10-expressing plasmids were transfected into human 293T cells alone or in combination. The cells were then subjected to Sas10 and Mpp10 immunostaining at 48 h post-transfection. The staining specificities of anti-Sas10 and anti-Mpp10 antibodies were confirmed by co-staining with the anti-HA-tag and anti-Myc-tag antibodies, respectively (Supplementary Figure S9A). The result showed that when overexpressed alone in the transfected cells, Sas10 was localized both in the nucleoplasm and nucleolus (Supplementary Figure S9A–C), whereas Mpp10 was localized in the cytoplasm (Figure 7F; Supplementary Figure S9A and B). When Sas10 and Mpp10 were co-expressed, both Sas10 and Mpp10 were clearly localized in the nucleolus (Figure 7F and Supplementary Figure S9B). Furthermore, Sas10 and Mpp10 co-expression appeared to enlarge the nucleoli (Figure 7F and Supplementary Figure S9B). In contrast, co-expressing HA-Sas10-s5 (corresponding to s5 in Figure 3D, lacking the Mpp10 interacting domain) with Myc-Mpp10 failed to transfer Mpp10 into the nucleolus while HA-Sas10-s5 was localized in the nucleus (Figure 7F and Supplementary Figure S9C).
As shown in Supplementary Figure S3A, sas10 is a maternal gene. Western blot analysis revealed that, compared with WT, the level of Sas10 in sas10Δ2 at 28 hpf was greatly reduced (Supplementary Figure S10A). As expected, the level of Mpp10 was also reduced (Supplementary Figure S10A). We injected HA-sas10 and HA-sas10-s5 messenger RNA (mRNA), respectively, into the sas10Δ2 embryo at the one-cell stage with the purpose to learn whether the restoration of Sas10 expression would increase the Mpp10 level and restore the nucleolus-localization of Mpp10 in sas10Δ2. We found that HA-sas10 mRNA injection not only partially restored the level (Supplementary Figure S10A) but also the nucleolus-localization (Supplementary Figure S10B) of the endogenous Mpp10 in sas10Δ2. In contrast, HA-sas10-s5 mRNA injection neither restored the level nor nucleolus-localization of the endogenous Mpp10 (Supplementary Figure S10A and B). All of these data suggest that the nucleolar localization of Mpp10 depends on its interaction with Sas10.
In yeast, Imp3 and Imp4 are Mpp10-interacting proteins (13,43,44). We cloned the zebrafish counterparts of Imp3 and Imp4 and co-expressed Imp3 or Imp4 with Mpp10 in human 293T cells. We found that consistent with the results in yeast, zebrafish Mpp10 interacted with Imp3 and Imp4 (Supplementary Figure S10C). Meanwhile, Imp3 or Imp4 co-expression with Mpp10 stabilized Mpp10 (Supplementary Figure S10C), consistent with the findings in yeast (17). Immunostaining showed that Imp3, when overexpressed alone, was localized in the nucleolus. Surprisingly, when co-expressed with Mpp10, both Mpp10 and Imp3 were co-localized in the nucleus (Figure 8A), suggesting that Imp3 can recruit Mpp10 into the nucleus but not into the nucleolus. Imp4, when overexpressed alone, was localized in the nucleus (Figure 8A). Imp4 also recruited most of Mpp10 into the nucleus and formed a clear ring surrounding the nucleolus (Figure 8A). Surprisingly, when Sas10 was co-expressed with Imp3 and Mpp10 or with Imp4 and Mpp10 in human 293T cells, Mpp10 was co-localized with Imp3 or Imp4 in the nucleolus (Figure 8A). These results indicate that Sas10 determines the nucleolar localization of the Mpp10–Imp3–Imp4 complex.
Figure 8.
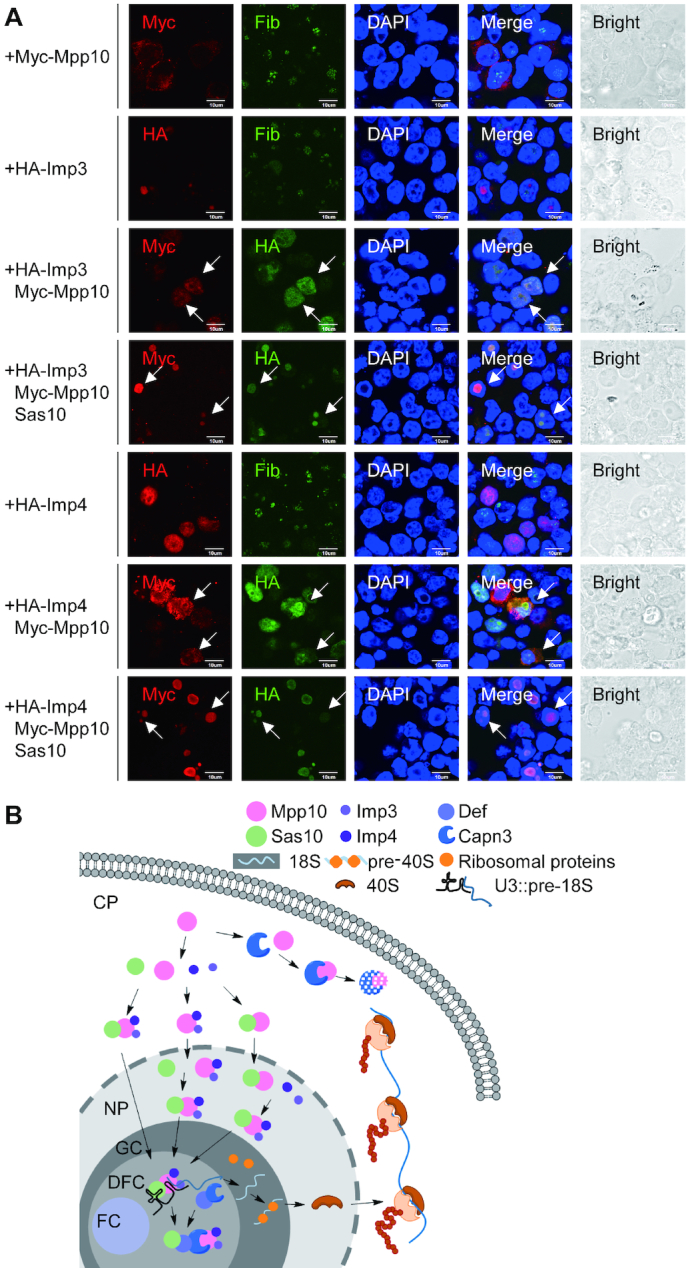
Sas10 determines the nucleolar localization of the Mpp10–Imp3–Imp4 complex. (A) HA-Imp3 and HA-Imp4 were transfected into 293T cells alone or co-transfected with Myc-mpp10 or with Myc-mpp10 and sas10. The cells were subjected to co-immunostaining of HA-tag and Myc-tag, Myc-tag and Fib or HA-tag and Fib 48 h post-transfection. Arrows pointed to co-transfected cells; bar, 10 μm. (B) A model depicts the route of delivery of the Mpp10–Imp3–Imp4 complex into the nucleolus. Once Mpp10 is synthesized it will form the complex with Imp3 and Imp4. Sas10 binds to Mpp10 to protect it from being recognized by cytoplasmic Capn3 and meanwhile to facilitate the translocation of Mpp10 (or the Mpp10–Imp3–Imp4 complex) to the nucleus. Sa10 also binds to the nuclear-localized Mpp10–Imp3–Imp4 complex. After that, Sas10 delivers the Mpp10–Imp3–Imp4 complex to nucleolus for assembling the SSU processome/90S pre-ribosome.
DISCUSSION
The SSU processome is a highly organized complex responsible for 18S rRNA maturation in the nucleolus, but the mechanism by which the SSU processome is assembled has not been sufficiently explored. Here, we report that the Mpp10–Imp3–Imp4 complex, a key component of the SSU processome, is likely formed in the cytoplasm and can enter the nucleoplasm but not the nucleolus. Sas10 binds to Mpp10 that not only facilitates the translocation of the Mpp10–Imp3–Imp4 complex to the nucleus but also delivers the complex to the nucleolus. In addition, we found that the stability of Sas10 and Mpp10 is interdependent and that Sas10 protects Mpp10 from Capn3-mediated degradation by binding to Mpp10 and masking its Capn3-recognition site. We also demonstrated that zebrafish Sas10 and Mpp10 are conserved nucleolar proteins and are essential for digestive organ development.
As observed in the thermophilic fungus C. thermophilum (24), the Sas10/Utp3/C1D domain of the zebrafish Sas10 interacts with the N-terminus of Mpp10. Previous structural studies have revealed that the Mpp10–Imp3–Imp4 complex is a core component of the 90S pre-ribosome/SSU processome in lower eukaryotes (4,6,7,43). Surprisingly, the structural analysis failed to identify the Mpp10–Sas10 complex in the 90S pre-ribosome/SSU processome, raising an interesting question about the role of the Mpp10–Sas10 complex. By examining the Sas10 protein level in the mpp10 null mutant and the Mpp10 protein level in the sas10 null mutant, we found that the stability of Sas10 and Mpp10 are interdependent. This observation was confirmed in the cultured cells where Mpp10 and Sas10 stabilized only when they were co-expressed. A previous yeast two-hybrid study showed that yeast Sas10 and Mpp10 interact with Utp25/Def (26). Here, we showed that a proportion of zebrafish Sas10 interacts with Def and that Sas10 probably serves as a link to form the Def–Sas10–Mpp10 complex. This finding prompted us to speculate that Mpp10 and/or Sas10 is the substrate of Capn3 because we have previously shown that Def recruits Capn3 to the nucleolus and forms the Def-Capn3 protein degradation pathway to cleave its target (e.g. p53) in the nucleolus (29,30). This speculation was supported by the fact that Mpp10 and Sas10 accumulated in the def−/− null mutant. Because Mpp10 and Sas10 were stabilized when they formed a complex, we reckoned that the interaction domains of Mpp10 and/or Sas10 are targeted by Capn3. Bioinformatics analysis revealed that there is indeed a Capn3-recognition motif in the N-terminus of Mpp10 but not in the Sas10/Utp3/C1D domain of Sas10. Detailed biochemical analysis showed that Mpp10 is sensitive to Capn3 and that the deletion of the Capn3-recognition motif in Mpp10 attenuates the effect of Capn3 on Mpp10.
Our data showed that Mpp10 is a cytoplasmic protein, whereas Sas10, Imp3 and Imp4 are localized in the nucleus. Both Imp3 and Imp4 facilitate the translocation of Mpp10 to the nucleus but not the nucleolus, whereas Sas10 determines the nucleolar localization of Mpp10. These data suggest that (i) the interaction between Mpp10 and Imp3, Imp4 or Sas10 in the cytoplasm is a prerequisite for the nuclear translocation of Mpp10 and that (ii) Sas10 interacts with the Mpp10–Imp3–Imp4 complex in the cytoplasm or the nucleus to deliver the Mpp10–Imp3–Imp4 complex to the nucleolus for assembling the SSU processome.
Taken together, our results suggest the following stages in cells undergoing active protein biosynthesis: (i) Sas10 determines the nucleolar localization of the Mpp10–Imp3–Imp4 complex. In this way, they not only stabilize each other but also maintain their molar ratio as 1:1:1:1 to meet the demands of ribosome biogenesis. (ii) Once the Mpp10–Sas10–Imp3–Imp4 complex enters the nucleolus, Sas10 dissociates and only the Mpp10–Imp3–Imp4 complex is recruited to assemble the 90S pre-ribosome/SSU processome. (iii) Once the Mpp10–Imp3–Imp4 complex accomplishes its function and the SSU processome is disassembled, Sas10 interacts with Mpp10 to protect it from Capn3-mediated protein destruction. On the other hand, in cells with low protein biosynthesis activity, Def interacts with Sas10, probably leading to the disassociation of the Sas10-Mpp10 complex to facilitate the Capn3-mediated Mpp10 cleavage (Figure 8B). However, many questions remain to be answered; for example, how is Sas10 degraded in the absence of Mpp10? Does the Mpp10–Sas10 complex play biological functions other than ribosome biogenesis? In summary, the nucleolus, as the ‘nucleus’ of the nucleus, contains over a thousand types of proteins that are responsible not only for ribosome biogenesis but also as a centre for stress sensing and response. Considering the limited volume of the nucleolus, how these various proteins are produced in the most economical way and how they are assembled and work in coordination to accomplish different functions are challenging questions that remain to be investigated.
DATA AVAILABILITY
All data are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Ting Tao, Huabing Wang, Yong Wang, Ce Gao, Yihong Guan, Qinfang Zhu, Hongjian Gong and Caiqiao Zhang and Mr Weifan Chen and all members in JRP, JC and HBW labs for their valuable suggestions. The authors are grateful to Yixin Ye, Xiaocai Du, Zhengxin Xu and Xiangfeng Shen for their technical support with animal studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (http://www.nsfc.gov.cn/) and the ‘973 Program’ of the Ministry of Science and Technology of China in the order of [31330050, 2015CB942802, 2017YFA0504501, 31771596]. Funding for open access charge: National Natural Science Foundation of China [31330050].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I.. The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 2007; 8:574–585. [DOI] [PubMed] [Google Scholar]
- 2. McCann K.L., Baserga S.J.. Genetics. Mysterious ribosomopathies. Science. 2013; 341:849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaker-Margot M., Hunziker M., Barandun J., Dill B.D., Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015; 22:920–923. [DOI] [PubMed] [Google Scholar]
- 4. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R. et al.. Architecture of the 90S Pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 5. Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y. et al.. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002; 417:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 7. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 8. Venema J., Tollervey D.. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999; 33:261–311. [DOI] [PubMed] [Google Scholar]
- 9. Bernstein K.A., Gallagher J.E., Mitchell B.M., Granneman S., Baserga S.J.. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell. 2004; 3:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grandi P., Rybin V., Baßler J., Petfalski E., Strauß D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D. et al.. 90S Pre-Ribosomes include the 35S Pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002; 10:105–115. [DOI] [PubMed] [Google Scholar]
- 12. Matsumoto-Taniura N., Pirollet F., Monroe R., Gerace L., Westendorf J.M.. Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell. 1996; 7:1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S.J., Baserga S.J.. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 1999; 19:5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Granneman S. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res. 2003; 31:1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baserga S.J., Agentis T.M., Wormsley S., Dunbar D.A., Lee S.. Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors. Nucleic Acids Symp. Ser. 1997; 64–67. [PubMed] [Google Scholar]
- 16. Gérczei T., Shah B.N., Manzo A.J., Walter N.G., Correll C.C.. RNA chaperones stimulate formation and yield of the U3 snoRNA–Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J. Mol. Biol. 2009; 390:991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehner K.A., Gallagher J.E., Baserga S.J.. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 2002; 22:7258–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamakaka R.T., Rine J.. Sir- and silencer-independent disruption of silencing in Saccharomyces by Sas10p. Genetics. 1998; 149:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitchell P. Rrp47 and the function of the Sas10/C1D domain. Biochem. Soc. T. 2010; 38:1088–1092. [DOI] [PubMed] [Google Scholar]
- 20. Wiederkehr T., Pretot R.F., Minvielle-Sebastia L.. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998; 4:1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung M.Y., Lorenz L., Richter J.D.. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol. Cell. Biol. 2006; 26:4277–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erdemir T., Bilican B., Cagatay T., Goding C.R., Yavuzer U.. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 2002; 46:947–957. [DOI] [PubMed] [Google Scholar]
- 23. Hieronymus H. Genome-wide mRNA surveillance is coupled to mRNA export. Gene Dev. 2004; 18:2652–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sa-Moura B., Kornprobst M., Kharde S., Ahmed Y.L., Stier G., Kunze R., Sinning I., Hurt E.. Mpp10 represents a platform for the interaction of multiple factors within the 90S pre-ribosome. PLoS One. 2017; 12:e183272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J., Ruan H., Ng S.M., Gao C., Soo H.M., Wu W., Zhang Z., Wen Z., Lane D.P., Peng J.. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005; 19:2900–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charette J.M., Baserga S.J.. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010; 16:2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldfeder M.B., Oliveira C.C.. Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. FEBS J. 2010; 277:2838–2852. [DOI] [PubMed] [Google Scholar]
- 28. Harscoet E., Dubreucq B., Palauqui J.C., Lepiniec L.. NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS One. 2010; 5:e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tao T., Shi H., Guan Y., Huang D., Chen Y., Lane D.P., Chen J., Peng J.. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013; 23:620–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan Y., Huang D., Chen F., Gao C., Tao T., Shi H., Zhao S., Liao Z., Lo L.J., Wang Y. et al.. Phosphorylation of Def regulates nucleolar p53 turnover and cell cycle progression through def recruitment of calpain3. PloS Biol. 2016; 14:e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gong L., Gong H., Pan X., Chang C., Ou Z., Ye S., Yin L., Yang L., Tao T., Zhang Z. et al.. p53 isoform Δ113p53/Δ133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell. Res. 2015; 25:351–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y., Luo Y., Hong Y., Peng J., Lo L.. Ribosome biogenesis factor Bms1-like is essential for liver development in zebrafish. J. Genet. Genomics. 2012; 39:451–462. [DOI] [PubMed] [Google Scholar]
- 33. Huang H., Ruan H., Aw M.Y., Hussain A., Guo L., Gao C., Qian F., Leung T., Song H., Kimelman D. et al.. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008; 135:3209–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu M., Bai Y., Zhang C., Liu F., Cui Z., Chen J., Peng J.. Liver-Enriched gene 1, a glycosylated secretory protein, binds to FGFR and mediates an anti-stress pathway to protect liver development in zebrafish. PloS Genet. 2016; 12:e1005881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niu X., Hong J., Zheng X., Melville D.B., Knapik E.W., Meng A., Peng J.. The nuclear pore complex function of Sec13 protein is required for cell survival during retinal development. J. Biol. Chem. 2014; 289:11971–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang S.L., Aw S.S., Chang C., Korzh S., Korzh V., Peng J.. Depletion of Bhmt elevates sonic hedgehog transcript level and increases beta-cell number in zebrafish. Endocrinology. 2011; 152:4706–4717. [DOI] [PubMed] [Google Scholar]
- 37. Lerch-Gaggl A., Haque J., Li J., Ning G., Traktman P., Duncan S.A.. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 2002; 277:45347–45355. [DOI] [PubMed] [Google Scholar]
- 38. Lahmy S., Guilleminot J., Cheng C.M., Bechtold N., Albert S., Pelletier G., Delseny M., Devic M.. DOMINO1, a member of a small plant-specific gene family, encodes a protein essential for nuclear and nucleolar functions. Plant J. 2004; 39:809–820. [DOI] [PubMed] [Google Scholar]
- 39. Chen Y.C., Wang H.J., Jauh G.Y.. Dual role of a SAS10/C1D family protein in ribosomal RNA gene expression and processing is essential for reproduction in arabidopsis thaliana. PloS Genet. 2016; 12:e1006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B.. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010; 64:960–976. [DOI] [PubMed] [Google Scholar]
- 41. Sorimachi H., Toyama-Sorimachi N., Saido T.C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., Suzuki K.. Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 1993; 268:10593–10605. [PubMed] [Google Scholar]
- 42. Charton K., Sarparanta J., Vihola A., Milic A., Jonson P.H., Suel L., Luque H., Boumela I., Richard I., Udd B.. CAPN3-mediated processing of C-terminal titin replaced by pathological cleavage in titinopathy. Hum. Mol. Genet. 2015; 24:3718–3731. [DOI] [PubMed] [Google Scholar]
- 43. Zheng S., Ye K.. Purification, crystallization and preliminary X-ray diffraction analysis of Imp3 in complex with an Mpp10 peptide involved in yeast ribosome biogenesis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014; 70:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gallagher J.E.G. Two-hybrid Mpp10p interaction-defective Imp4 proteins are not interaction defective in vivo but do confer specific pre-rRNA processing defects in Saccharomyces cerevisiae. Nucleic Acids Res. 2004; 32:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.