Abstract
Double-stranded breaks (DSBs) in plant organelles are repaired via genomic rearrangements characterized by microhomologous repeats. These microhomologous signatures predict the existence of an unidentified enzymatic machinery capable of repairing of DSBs via microhomology-mediated end-joining (MMEJ) in plant organelles. Here, we show that organellar DNA polymerases from Arabidopsis thaliana (AtPolIA and AtPolIB) perform MMEJ using microhomologous sequences as short as six nucleotides. AtPolIs execute MMEJ by virtue of two specialized amino acid insertions located in their thumb subdomains. Single-stranded binding proteins (SSBs) unique to plants, AtWhirly2 and organellar single-stranded binding proteins (AtOSBs), hinder MMEJ, whereas canonical mitochondrial SSBs (AtmtSSB1 and AtmtSSB2) do not interfere with MMEJ. Our data predict that organellar DNA rearrangements by MMEJ are a consequence of a competition for the 3′-OH of a DSBs. If AtWhirlies or AtOSBs gain access to the single-stranded DNA (ssDNA) region of a DSB, the reaction will shift towards high-fidelity routes like homologous recombination. Conversely MMEJ would be favored if AtPolIs or AtmtSSBs interact with the DSB. AtPolIs are not phylogenetically related to metazoan mitochondrial DNA polymerases, and the ability of AtPolIs to execute MMEJ may explain the abundance of DNA rearrangements in plant organelles in comparison to animal mitochondria.
INTRODUCTION
Plant organellar genomes encode for macromolecular assemblies indispensable for oxidative phosphorylation and photosynthesis (1–3). Thus, faithful organellar replication and regulated transcription are necessary for cell survival. Contrary to the representation of a circular structure as the predominant form of mitochondrial DNA, plant mitochondrial genomes are heterogeneous linear molecules with a strong tendency to produce rearrangements (4–7). The notion that plastid DNA exists as a circular molecule is challenged by findings of linear molecules in maize seedlings and in Medicago truncatula (4,8,9). Although the mechanisms that mediate plant organellar DNA replication are unknown, DNA rearrangements in plant organelles are common and are attributed to the repair of double-strand breaks (DSBs) (6,10–14).
Repair of DSBs occurs by two main pathways: homologous recombination (HR) and non-homologous end joining (NHEJ) (Reviewed in (15)). HR is an error-free pathway in which identical DNA segments are used as templates. In contrast, NHEJ is an error-prone pathway. NHEJ is divided into two sub-pathways: Ku-dependent classical non-homologous end joining (c-NHEJ) and alternative end joining (alt-EJ). A subset of DNA breaks repaired by alt-EJ employs a mechanism that requires minimal base pairing of microhomologous sequences, dubbed microhomology-mediated end joining (MMEJ) (16–18).
MMEJ was discovered as a sub-pathway derived from the Archaeo-Prokaryotic (AP)-NHEJ. Within this sub-pathway bacterial DNAPs promote synapsis of DSBs using short microhomologous sequences, these annealed sequences assemble the 3′-OH primers and are extended by virtue of strand-displacement synthesis by specialized DNAPs (19,20). Eukaryotic DNA polymerases θ (HsPolθ), β, λ and μ, also perform MMEJ using short microhomologous sequences. As in bacterial MMEJ, the proposed DSB repair mechanism involves annealing of microhomologous sequences in trans to create a template DNA synapse and nucleotide extension (21–24). Crystal structures of DNAPs during MMEJ illustrate that they promote the stabilization of microhomologous sequences via unique structural elements that mediate synapse formation (19,25,26).
The plant mitochondrial genome frequently recombines via homologous recombination of large size (>500 bp) or intermediate size repeats (50–500 bp). Break-Induced Replication (BIR) and Single-Strand Annealing (SSA) are postulated to drive illegitimate recombination events at intermediate size repeats (6,27,28). However, plant organellar genomes also undergo rearrangements attributed to illegitimate recombination at intermediate size repeats (>50 base pairs) (6,11,12,27,29–33). These rearrangements are associated with MMEJ, microhomology-mediated break-induced replication (MMBIR) and microhomology-mediated recombination (12,28–30). DSBs in Arabidopsis′ chloroplasts, generated by ciprofloxacin or by a homing endonuclease, are repaired using microhomologous sequences that rank between 4 and 18 nts (29,31,32). The nature of the sequences at the junctions of the DNA rearrangements indicates that DSBs are repaired by a route that involves annealing of microhomologous sequences. The length of these microhomologies resembles the length of microhomologous segments in metazoan DNA repaired by MMEJ (17) or MMBIR (34).
Flowering plants harbor two organellar family-A DNA polymerases, known as plant and protist DNA polymerases (POPs) that are not phylogenetically related to yeast or metazoan mitochondrial DNA polymerase γ and are the sole DNAPs in plant organelles (35–39). Both AtPolIs display lower nucleotide insertion fidelity that metazoan mitochondrial DNAPs, elongate RNA primers synthesized by the organellar DNA primase-helicase and harbor strand-displacement and 5′-deoxyribose phosphate lyase activities (29,40–43). From the DNAPs able to execute MMEJ, HsPolθ is the only one that belongs to the family-A of DNAPs. This enzyme harbors a polymerization domain linked to a helicase domain and seminal work demonstrated that both domains participate in MMEJ (44,45). Structure-function studies of HsPolθ indicate that a specific insertion in its polymerization domain is responsible for MMEJ (21) The ortholog of HsPolθ in plants (24), TEBICHI, is involved in T-DNA random integration by MMEJ (46,47). Although TEBICHI participates in nuclear MMEJ, a mechanism for MMEJ has not been identified in plant organelles.
AtPolIs and HsPolθ bypass DNA lesions via unique amino acid insertions in their polymerization domain (25,40). Because of this convergence, and the necessity of a unique amino acid insertion in HsPolθ to execute MMEJ (21,48), we wanted to determine whether AtPolIs could execute MMEJ. To our surprise, AtPolIs are able to efficiently mediate synapsis formation on microhomologies as short as 6-bp. We found that members of the single-stranded binding proteins unique to plants, Whirly-2 and Organellar Single-stranded DNA Binding proteins (OSBs), inhibit MMEJ by competing with the single-stranded overhang. In contrast, mtSSBs do not interfere with MMEJ formation. Our results indicate the MMEJ in plant organelles is mediated by the available concentration of single-stranded binding proteins and AtPolIs.
MATERIALS AND METHODS
Cloning of recombinant Single-Stranded DNA-Binding proteins
The coding regions corresponding to processed AtOSB2 (AT4G20010), AtOSB3 (AT5G44785), AtWhirly2 (AT1G71260), AtmtSBB1 (AT4G11060.1), AtmtSSB2 (AT3G18580.1) were PCR amplified from cDNA. The PCR products were digested and cloned into a modified pET19b or pET28b vectors. Positive clones were verified by DNA sequencing.
Expression and purification of recombinant proteins
In this study, we use recombinant AtPolIs that lack their first 257 amino acids. Those residues correspond to a dual targeting sequence (DTS) and a disordered region. Removal of those amino acids increases protein yield during heterologous purification without affecting exonucleolytic degradation or DNA polymerization (40) Thus, recombinantly expressed AtPolIs contain intact 3′-5′ exonuclease and polymerization domains, as metazoan DNAP γ and the Klenow fragment of bacterial DNAPs. The 3′-5′ exonuclease deficient variants of AtPolIs substitute two invariant carboxylates in exonuclease motif I of AtPolIA (D294 and D296) and AtPoIlB (D287 and D289) to alanine. Exonuclease deficient AtPolIB (AtPolIB exo−) was used as template to construct deletions in the unique insertions present in AtPolIs. The AtPolIB variant with a deletion in insertion 1 (AtPolIB-Δins1) eliminates residues 581 to 619, AtPolIB-Δins2 eliminates residues 648 to 712, and AtPolIB-Δins2 eliminates residues 845–869. All mutant polymerases are readily purified using three chromatographic steps (40) (Supplementary Figure S1).
AtmtSSB2 was purified following published protocols (49). Recombinant AtmtSSBs, AtOSB2, AtOSB3, and AtWhirly2 were expressed using an Escherichia coli BL21 PKJE7 strain. Cell cultures were grown until they reached an OD600 of 0.6 and were induced with 0.5 mM IPTG (isopropyl-1-thio-β-d-galactopyranoside) at 16°C during 18 h. Cells were recollected by centrifugation at 4°C and resuspended in 30 ml lysis buffer (25 mM HEPES pH 8, 600 mM NaCl, 10 mM imidazole, 10% glycerol and 1 mM PMSF). The lysate was supplemented with lysozyme 0.5 mg/ml and sonicated on ice. The clear lysate was filtrated and loaded onto a Ni-NTA column. The column was washed with lysis buffer containing 30 mM imidazole. Proteins were eluted using lysis buffer supplemented with 500 mM imidazole and dialyzed in buffer A (25 mM HEPES pH 8, 50 mM NaCl, 2 mM EDTA, 30% glycerol and 5 mM DTT). SSBs were loaded into a Hi-Trap heparin column, and further purified using a 0.1 M to 1 M NaCl gradient. Pure fractions were stored into a buffer B (25 mM HEPES pH 8.0, 100 mM NaCl, 2 mM EDTA, 50% glycerol and 5 mM DTT) at −20°C.
Substrates
5′-End oligonucleotide labeling was performed with T4 polynucleotide kinase using [γ- 32P] ATP and purified with Nucleotide Removal Kit following manufacture's protocol. Oligonucleotides used for the MMEJ assay are described in Supplementary Table S1.
Microhomology-mediated end joining (MMEJ) assays
MMEJ assays were performed at 37°C using 20 nM 5′-end radiolabeled pssDNA, in a buffer containing 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1.5 mM DTT, 10% glycerol and 0.1 mg/ml BSA. The mix was incubated with 100 nM AtPoIls for 15 min and then supplemented with 500 μM of dNTPs and 10, 6 or 2 mM MgCl2 for 30 min. The reactions were terminated using a stop buffer (95% formamide, 10 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol). The reactions were loaded on a 17% polyacrylamide denaturing gel and analyzed by phosphorimaging. In the MMEJ assay with singled-stranded DNA binding proteins, the same labeled pssDNA substrates were at 5 nM and AtSSBs were present at the concentrations described in the figure legends.
Electrophoretic-mobility shift assay (EMSA)
To investigate the binding of plant SSB to ssDNA we use a radioactively labeled poly-T of 70 nts oligonucleotide. Reactions were incubated in buffer containing 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1.5 mM DTT, 10% glycerol, 0.1 mg/ml BSA, 10 mM MgCl2, using varying concentrations of SSBs. The reactions were run on 4% polyacrylamide non-denaturing gel at 4°C and visualized by phosphorimaging.
Fluorescence anisotropy binding assay
Dissociation constant (Kd) were measured using a fluorescein-labeled ssDNA of 45 nts (ssDNA(45)) or a Cy3-labeled pssDNA-10 in 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.05 mg/ml BSA,10% glycerol and 10 mM MgCl2. Increased concentration of purified proteins (AtmtSSB1, AtOSB2 and AtWhy2) were individually evaluated. The reactions were incubated at 25°C in darkness during 20 min. Changes in anisotropy were quantified on an Infinite M1000 Microplate Reader (Tecan®) using excitation and emission wavelength (λ) of 470 and 525 nm, respectively. The data were analyzed using GraphPad Prism 6 according to the specific binding with Hill slope model following this equation: Y = Bmax*Xh/(Kdh + Xh), where Bmax indicates the maximum number of binding sites, Kd refers to ligand concentration that binds to half the receptor sites at equilibrium and h is the Hill slope.
Fluorescence resonance energy transfer (FRET) assay
For evaluation of DNA synapsis formation, oligonucleotides labeled with different fluorescence probes (Cy3 and Cy5) were annealed as described in the figure legends. Titration experiments were performed using equimolar concentration of labeled DNA (50 nM) with increasing concentrations of AtPolIs. Variations in fluorescence intensity upon incubation of AtPolIs were measured in an Infinite M1000 microplate reader using wavelengths (λ) of 530 nm for excitation and 675 nm for emission. Reactions were incubated in 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1.5 mM DTT, 10% glycerol, 0.1 mg/ml BSA and varying concentrations of AtPoIls. The reactions were incubated for 20 min in darkness at 25°C by triplicate. The data were normalized and analyzed in GraphPad Prism 6.
Gel filtration analysis
The elution profiles of plant SSBs in solution were evaluated using gel filtration. The proteins were injected into a Superdex 75 10/300 column equilibrated with 20 mM HEPES pH 8.0, 200 mM NaCl, 10% glycerol, 2 mM EDTA and 1 mM DTT. Elution peaks were analyzed using molecular weight markers.
Homology model
The homology model of AtPolIB was carried out using the homology modeling platform of the Swiss-Model Workspace (50) using as templates the Klenow fragments from the crystal structures of Thermus aquaticus, Geobacillus stearothermophilus and Geobacillus kaustophilus. From those homology models, we selected the one generated with the crystal structure of G. kaustophilus DNAP I in complex with dsDNA (PDB: 4DSE), because this model positions insertions 1 and 3 in an optimal position to interact with dsDNA.
SEC-MALS analysis
The SEC-MALS analysis was performed on a DAWN HELEOS multi-angle light scattering detector, with eighteen angles detectors and a 658.9 nm laser beam, (Wyatt Technology, Santa Barbara, CA, USA) and an Optilab T-rEX refractometer (Wyatt Technology) in-line with two coupled Superdex 200 Increase 10/300 GL size exclusion chromatography analytical columns. Experiments were performed using an isocratic pump (Agilent) with a flow of 0.5 ml/min at room temperature (25°C). Data collection was performed with ASTRA 6.1 software (Wyatt Technology). For the experiments, 300 μl at 1.3 mg/ml protein were loaded on the columns with running buffer of 25 mM HEPES pH 8, 10% glycerol, 300 mM NaCl and 2 mM EDTA. Calibration was performed with high-molecular weight markers from GE Healthcare (thyroglobulin, 669 000 Da; ferritin, 440 000 Da; aldolase, 158 000 Da; conalbumin, 75 000 Da; and ovoalbumin, 43 000 Da).
Strand-displacement assays
A 65mer DNA template (5′-CCT TGG CAC TAG CGC ACG ATG CCG CTA AGA ACC TCA GGG CCA GTT AGG TGG GCA GGT GGG CTG CG-3′) was annealed with a 5′-end radioactively labeled 24mer primer (5′-CGC AGC CCA CCT GCC CAC CTA ACT-3′) and a DNA blocking oligonucleotide (5′-GA GGT TCT TAG CGG CAT CGT GCG CTA GTG CCA AGG-3′) to create a gap of 6 nucleotides. Reactions were run at 37°C mixing 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.2 mg ml–1 BSA, 1.5 mM DTT, 2 mM MgCl2, 10% glycerol, 10 nM DNA substrate, 20 nM DNA polymerases and initiated adding 200 μM of dNTPs.
MicroScale Thermophoresis (MST)
MST analysis of the interactions between AtPolIB exo− and plant SSB’s was carried out as follows. AtPolIB was purified with a 10× histidine tag at the N terminal and labeled using NanoTemper's Monolith His-Tag Labeling Kit RED-Tris-NTA at 100 nM. SSB’s were purified and their His-tag was removed by addition of precision protease and gel filtration. SSBs were titrated against labeled AtPolIB in 16 serial dilutions from initial concentrations of 150, 200 and 250 μM for AtmtSSB1, AtOSB2 and AtWhy2, respectively. Reactions were incubated for 10 min at 25°C in PBS buffer + 0.05% Tween™ 20. The measurements were performed using a NanoTemper Monolith NT.115 pico instrument. Analyses were conducted at 10% LED power and 50% MST power on standard capillaries.
RESULTS
AtPolIs execute MMEJ on partially resected DSBs
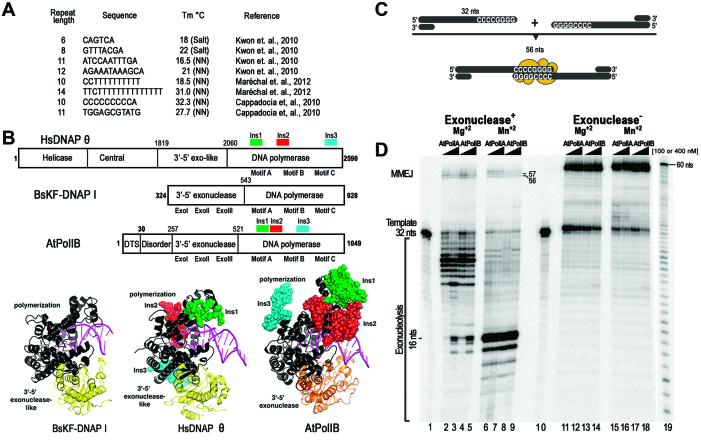
Pioneering work by Herrin and Brisson′s laboratories revealed that DSBs in chloroplast are repaired using illegitimate recombination mechanisms attributed to MMEJ (12,29–31,33). The predicted melting points for several of those microhomologies is <37°C (51) (Figure 1A), suggesting that protein factors may be involved in stabilizing the annealing of microhomologous sequences that are repaired by MMEJ during DSBs. Plant organelles in A. thaliana harbor only two DNA polymerases, AtPolIA and AtPolB that have as a distinctive feature the presence of three insertions in their polymerization domain.
Figure 1.
AtPolIs mediate MMEJ in vitro. (A) Representative microhomologous sequences present in chloroplast DNA from A. thaliana upon DSB repair (30–32). The calculated melting temperature (Tm) for single-stranded regions shorter than 8 nts were calculated using the salt-adjusted method (salt) and regions longer than 8 nts were calculated using the Nearest Neighbor (NN) method (51). (B) Structural comparison between a homology model of AtPolIs with the crystal structures of HsPolθ (25) (PDB: 4x0Q) and the Klenow fragment of Bacillus DNAP I (68) (PDB: 3BDP). The polymerization domains are colored in black, the active 3′-5′ exonuclease domain of AtPolIB in orange and the inactive 3′-5′ exonuclease domains of HsPolθ and Bacillus DNAP I in yellow. The unique amino acid insertions in HsPolθ and AtPolIB in comparison to bacterial DNAPs I are depicted in a ball-stick representation. In all DNAPs the dsDNA (magenta) is poised for nucleotide incorporation. (C) Proposed arrangement of a micromologous sequence of 8 nts showing DNA synapse formation and extension upon dNTPs incorporation by MMEJ. (D) Denaturing polyacrylamide gel showing MMEJ reactions catalyzed by wild-type and exonuclease deficient AtPolIs present at concentration of 100 or 400 nM. In the presence of Mg2+ or Mn2+. AtPolIs extend the pss-DNA(8) substrate to MMEJ products of 56, 57 and 58 nts. The migration of the MMEJ products of 56, 57 and 58 nts and the pss-DNA(8) substrate are indicated.
Given that HsPolθ performs TLS and MMEJ by a specific amino acid insertion and that unique amino acid insertions are responsible for TLS activity in AtPolIs, we hypothesize that AtPolIs may perform MMEJ via unique amino acid insertions (21,40). To investigate the putative MMEJ capabilities of AtPolIs, we used recombinant AtPolIA and AtPolIB harboring amino acid deletions of their first 257 amino acids. Those amino acids correspond to a predicted disordered protein region and their removal increases the yield of recombinant active protein (40). Thus, recombinant AtPolIs harbor both 3′-5′ exonuclease and polymerization domain activities (Figure 1B). The initial substrate used to test MMEJ, consisted of a partially resected DSB (pssDNA) with a 5′-CCCCGGGG-3′overhang sequence of 8 nts (pssDNA-8) with a predicted Tm for the microhomologous region of 22.5°C (Figure 1C). This substrate was formerly used to asses MMEJ formation by HsPolθ (21,48).
Synapse formation, followed by polymerization on the single-stranded portion of the template and strand-displacement would generate a MMEJ product of 56 nts (Figure 1C). MMEJ reactions were initially executed using 10 mM of MgCl2 and 2 mM of MnCl2, as those are optimal metal concentrations during primer extension on a canonical primer-template (data not shown) and high nucleotide concentrations (500 μM). Reaction products of 56 and 57 nts product were synthetized by exonuclease proficient AtPolIs, in the presence of Mg2+ (Figure 1D, lanes 2–5), whereas products of 56–58 nts were synthetized in reactions incubated with Mn2+ (Figure 1D, lanes 6–9). The presence of Mg2+ promotes the addition of an extra base, whereas incubation with Mn2+ promotes the addition of two extra bases, possibly due to terminal transferase activity. Thus, reactions incubated with pssDNA-8 generate MMEJ products in the presence of AtPolIs harboring an active exonuclease domain or exonuclease deficient DNAPs (Figure 1D). AtPolIs incubated with Mn2+ exhibit an increase in their 3′-5′ exonuclease activity. The exonuclease activity of AtPolIs is halted after digestion of 16 nts, at the boundary of the double-stranded DNA template (Figure 1D, lanes 2–9). Disruption of the exonuclease activity by site-directed mutagenesis in AtPolIs increases MMEJ activity, resulting in the appearance of a MMEJ product of 58 nts (Figure 1C, lanes 11–18).
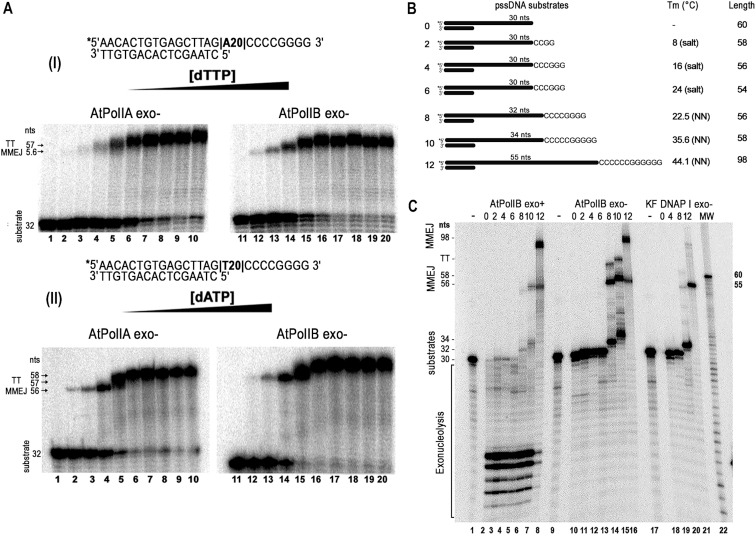
In order to asses if AtPolIs could execute MMEJ at physiological nucleotide concentrations, we used a pssDNA-8 substrate that templates for 20 consecutive thymines or adenines, after the assembly of the MMEJ substrate. In this experiment, we used nucleotide concentrations that rank from 0.0037 to 245.76 μM (Figure 2A). At low micromolar dATP or dTTP concentrations (Figure 2A and B, lanes 2–5 and 12–15) a single MMEJ product of 56 nts is observed. At higher nucleotide concentrations, a band of 57 nts is shown in reactions incubated with dTTP and a band of 58 nts is observed in reactions incubated with dATP (Figure 2A and B, lanes 7–10 and 17–20). AtPolIB exhibits a nearly 20-fold higher KM for dNTP during nucleotide incorporation on a canonical primer-template substrate with respect to AtPolIA (43). Thus, the observation that AtPolIA and AtPolIB exhibit similar MMEJ efficiency during all the assayed dNTP concentrations suggests that during MMEJ annealing the mechanism that dictates the affinity of the incoming nucleotide differs from the one during primer-template incorporation.
Figure 2.
AtPolIs performs MMEJ using microhomology sequence longer than 8 nts at high MgCl2 concentrations. (A) Exonuclease deficient AtPolIs execute MMEJ on a pss-DNA(8) substrate at physiological dNTP concentrations. Reactions incubated in the absence of dNTPs (lanes1 and 11) show the migration o the labeled substrate. MMEJ reactions were incubated using increasing dTTP (panel I) or dATP (panel II) from 0.0037 to 245.76 μM using four-fold increments (lanes 2–10 and 12–20). At low micromolar dNTP concentrations (lanes 2–5 and 12–15) a MMEJ product of 56 nts is observed. In contrast, a band of 57 nts is observed in reactions incubated with dTTP using high nucleotide concentrations and a band of 58 nts is observed in reactions incubated with high concentrations of dATP. (B) Schematic representation of the pssDNAs substrates used for MMEJ assays. Asterisks indicate the position of the 5′-radioactive label. The microhomology lengths (from 0 to 12 nts), the calculated Tm for DNA synapsis formation and the expected lengths of the MMEJ product are indicated at the right. (C) Denaturing sequencing gel showing MMEJ reactions by wild-type (lanes 2–8) and exonuclease deficient (lanes 10–16) AtPolIBs in comparison to reactions incubated with the exonuclease deficient Klenow Fragment of DNAP I (lanes 18–21). Al reactions contained 10 mM MgCl2 as a cofactor. A control labeled substrate corresponding to pss-DNA(8) is present in lanes 1, 9 and 17. MMEJ products are observed in reactions incubated with AtPolIBs on substrates with microhomologies longer than eight nucleotides (lanes 6, 7, 8, 14, 15, 16). These products correspond to bands of 56, 58 and 98 nts respectively. In contrast, Klenow Fragment is unable to efficiently use MMEJ substrates and only low amounts of products are formed with pssDNAs of 8 and 12 nts (lanes 20 and 21). TT denotes products generated by a predicted terminal transferase activity.
In order to determine the minimal microhomologous length in which AtPolls are able to execute MMEJ, we performed MMEJ reactions using substrates than contained microhomologies from 0 to 12 nts. We initially used similar reaction conditions to those previously reported for HsPolθ, but with the addition of 50 mM NaCl (21). Full nucleotide incorporations on these pssDNA substrates are designed to generate products from 54 to 98 nts, and only a pssDNA substrate containing a microhomology of 12 nts (pssDNA-12) exhibits a Tm higher than 37°C (Figure 2A).
The Klenow Fragment of E. coli DNA polymerase I (KF-DNAP I) control generates faint products of approximately 56 and 98 nts in reactions incubated with pssDNA-8 and pssDNA-12 and not with any other pssDNA substrates, indicating that transient alignment of the microhomologous sequence does not form a stable substrate for DNAPs (Figure 2B, lanes 18–21). This result contrasts with a previous report in which KF-DNAP I generates low molecular weight MMEJ products on a pssDNA containing a microhomology of 4nts (pssDNA-4) (21). Thus, exonuclease deficient KF-DNAP I is unable to efficiently use pssDNAs as substrates, including a pssDNA-12 substrate. In contrast, wild-type (AtPolIB exo+) and exonuclease deficient AtPolIB (AtPolIB exo−) efficiently use substrates with microhomologous sequences of 8 nts (pssDNA-8) to perform MMEJ (Figure 2B, lanes 1 to 16). A MMEJ product is not observed in reactions incubated with pssDNA-6, pssDNA-4, pssDNA-2, and pssDNA-0 substrates (Figure 2B, lanes 11, 12 and 13). The predicted Tms of the microhomologous regions in pssDNA-6 and pssDNA-8 are <37°C, suggesting that spontaneous template annealing is unfavorable and that MMEJ is mediated by AtPolIB. As the editing activity of AtPolIs hampers MMEJ, we subsequently utilized exonuclease deficient variants of AtPolIs to investigate their MMEJ properties.
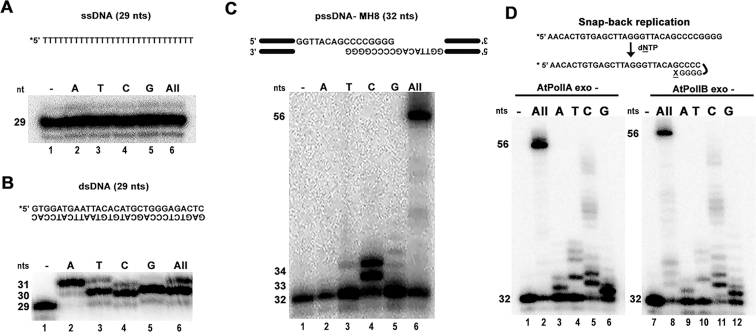
AtPolIs exhibited limited terminal deoxynucleotidyl transferase activity
In the presence of high nucleotide concentrations, AtPolIs generate MMEJ products with additional non-templated nucleotides residues at the 3′ end (Figure 2A, lanes 6–10 and 16–20). Bacterial family-A DNAPs, like Taq DNA polymerase (Taq DNAP), present limited non-templated nucleotide or terminal transferase activity. In this reaction, one or two nucleotides, preferentially dATP, are added to a 3′-OH primer (52). In contrast, HsPolθ and terminal deoxynucleotidyl transferases exhibit a robust terminal transferase activity and are able to add several nucleotides the 3′-OH primer (48,53). To test the efficiency of the predicted terminal transferase activity by AtPolIs, we used three different substrates: a homopolymeric ssDNA poly-dT, a double-stranded blunt-ended DNA substrate, and a MMEJ substrate (Figure 3). AtPolIB exo− fails to incorporate any nucleotide having as a substrate a poly-dT oligonucleotide in the presence of individual nucleotides or in the presence of all nucleotides (Figure 3A). In contrast, AtPolIB exo− exhibits limited terminal transferase activity catalyzing the addition of one or two nucleotides on a double-stranded blunt-ended DNA substrate (Figure 3B). This activity is reminiscent of the limited terminal transferase exhibited by Taq DNAP. In the presence of Mg2+ as cofactor AtPolIB exo− efficiently incorporates two non-templated dAMP molecules (Figure 3B, lane 2), whereas all other nucleotides are incorporated only once (Figure 3B). AtPolIB exo− also shows limited terminate transferase activity when MgCl2 is used as a cofactor (data not shown). In a MMEJ substrate, AtPolIB exo− synthesizes products that rank from 56 to 58 nts. This product corresponds to the annealing of the pssDNA-8 substrates plus the addition of two extra nucleotides. Using individual nucleotides, we observed an efficient nucleotide incorporation only when dCTP is present in the reaction (Figure 3C, lane 4). The pssDNA-8 substrate poises a template guanosine as the first template base after the annealing of the microhomologous CCCCGGGG sequence. The incorporation of two dCMPs on this substrate may be related to the low fidelity during nucleotide incorporation by AtPolIB (43) or to a transient misalignment of the 3′-OH (Figure 3C).
Figure 3.
AtPolIB exhibits limited terminal transferase activity on double-stranded substrates. (A) Denaturing gel showing the lack of terminal transferase activity on a poly-dT ssDNA template incubated in the presence of individual nucleotides. (B) Denaturing acrylamide gel showing AtPolIB exo- nucleotide incorporation on a ds-DNA substrate in the presence of individual nucleotides. (C) AtPolIB performs MMEJ using DNA templates in trans. Denaturing gel showing MMEJ reactions incubated in the presence of individual nucleotides on pss-DNA(8) substrate. (D) AtPolIB performs limited snap-back replication. Denaturing gel showing reactions incubated in the presence of individual nucleotides on a MH-8 substrate with AtPolIA exo- (lanes 1–6) and AtPolIB exo – (lanes 7–12).
In order to discern if the observed MMEJ products could be due to a template-dependent extension in cis (snap-back replication), we used a single-stranded pssDNA-8 substrate in the presence of individual nucleotides. In this experiment, the single-stranded oligonucleotide is predicted to assemble a primer-template by the annealing of four cytidines and four guanosines in cis (Figure 3D). This snap-back assembly is predicted to position a template guanosine for dCMP incorporation. In the presence of dCTP both AtPolIs incorporate multiple dCMPs (Figure 3D, lanes 5 and 11) possible due to a misalignment of the primer-strand. The presence of products derived from the incorporation of multiple cytidines on a single-stranded template contrast with the incorporation of two dCMPs in a pssDNA-8 substrate predicted to form a synapse (Figure 3C, lane 5). In the sole presence of dGTP, AtPolIs efficiently incorporate a dGMP molecule, possibly due to an alignment of only three cytidines with three guanosines that generate that a cytidine poised for dGTP incorporation (Figure 3C, lanes 6 and 12).
AtPolIs use substrates with microhomologies longer than 6 nts
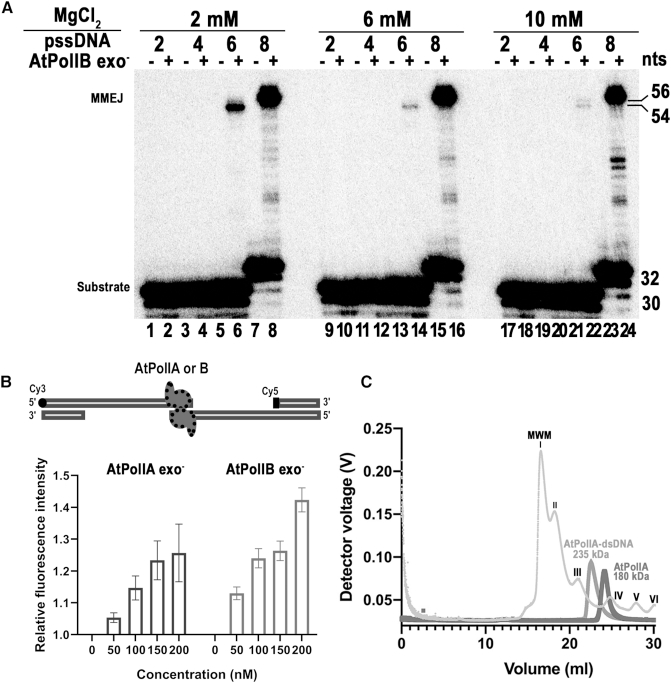
DSBs are repaired using microhomologous sequences as short as four nts in chloroplasts (30,32), suggesting that the machinery responsible of MMEJ should efficiently anneal pssDNA with short microhomologous regions. To investigate if AtPolIB exo− could perform MMEJ on these substrates, we studied the effect of three different concentrations of MgCl2 (10, 6 and 2 mM) on MMEJ. As previously shown, reactions incubated with 10 mM MgCl2 produced a faint MMEJ-product on a pssDNA-6 substrate and efficiently uses a pssDNA-8 substrate (Figure 4A, lanes 22, 24). However, in reactions incubated with 2 and 6 mM MgCl2, AtPolB exo− is able to use templates with microhomologous sequences of 6 nts (Figure 4A, lanes 6, 14).
Figure 4.
AtPolIs performs MMEJ in short homologous sequences at physiological MgCl2 concentrations and template synapse formation by AtPolIs. (A) MMEJ assays using three MgCl2 concentrations (2, 6 and 10 mM) in psDNA templates with microhomologies ranking from 2 to 8 nts. MMEJ reactions were incubated in the absence of MgCl2 (odd numbers) and with the indicated MgCl2 concentration (even numbers). A defined MMEJ product band of 54 nts is observed in reactions incubated with 2 mM MgCl2 on a pssDNA-6 substrate (lanes 6) whereas this product is hardly observed in reactions incubated with 6 and 10 mM MgCl2 (lanes 14 and 22). Reactions incubated with pssDNA-2 and pssDNA-4 do not generate a MMEJ product at any metal concentration. (B) Changes in the intensity of fluorescence indicative of the formation of a Cy3–Cy5 intermediate upon incubation of AtPolIs. The relative fluorescence increases in relation to the amount of added of exonuclease deficient AtPolIA and AtPolIB. Experiments were performed in triplicate and the graph shows the mean and the standard deviation. (C) SEC-MALS analysis of the oligomerization state of AtPolIB exo- in the absence and presence of DNA substrate. The chromatogram shows the readings of the light scattering for the molecular mass standards and AtPolIB exo- incubated in the presence and absence of DNA. The molecular mass of the standards is indicated by roman numbers: I) blue dextran (2 000 000 Da), II) thyroglobulin (669 000 Da), III) ferritin (440 000 Da), IV) aldolase (158 000 Da), V) conalbumin (75 000 Da), and ovoalbumin (43 000 Da).
We further investigated synapse formation by AtPolIs exo− using two substrates labeled with different fluorophores poised for fluorescence resonance energy transfer (FRET). Two pssDNA-8 substrates labeled with Cy3 or Cy5 dyes were incubated with AtPolIs in the absence of dNTPs. The efficiency of FRET increases in a proportional manner to the amount of AtPolIA exo− or AtPolIB exo− present in the reaction, indicating that AtPolIs drive DNA synapse formation of the fluorescent Cy3–Cy5 donor–acceptor pair (Figure 4B).
MMEJ by different DNAPs predict the formation of a synapse that is stabilized by two DNA polymerases in trans (19,21,25,26). To evaluate the possibility that AtPolIB assemblies in trans forming a dimer, we use size-exclusion chromatography (SEC)-coupled multiangle light scattering (MALS) on AtPolIBexo− incubated in the presence and absence of a DNA substrate (Figure 4C). These analyses found an average molecular weight for AtPolIB exo− of 180 kDa that corresponds to a molecular mass of two AtPolIB monomers (93 kDa). AtPolIB incubated with DNA increases its molecular mass to 235 kDa, a molecular mass that corresponds to two AtPolIB monomers interacting with two DNA molecules (27 kDa). Thus, the SEC-MALS analysis are consistent with synapse formation driven by a dimer assembly of AtPolIB and a pssDNA substrate.
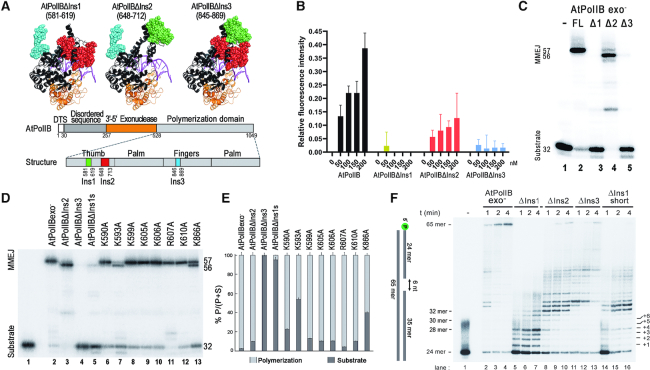
MMEJ by AtPolIB is mediated by insertions 1 and 3
AtPolIs are family-A DNA polymerases that contain three unique insertions in their polymerization domain (35,37,40) (Figure 1B). In order to investigate if MMEJ would depend on these insertions, we used mutant polymerases harboring individual deletions. Deletion mutants eliminate 38, 64, and 24 amino acids corresponding to insertions 1, 2 and 3 respectively (Figure 5A). Insertions 1 and 2 are located at the thumb subdomain and structural modeling suggests that they are poised to interact with the primer-template, whereas insertion 3 is located at the fingers subdomain in a position that is predicted to be distant from DNA (40) (Figure 5A). Mutant polymerases harboring deletions in those insertions are active on a canonical primer-template, suggesting that these mutant polymerases do not have severe alterations in their structure (40). The ability of AtPolIB exo− deletion variants to execute synapse formation was assessed by FRET on a pssDNA-8 substrate in the absence of dNTPs (Figure 1B). As previously shown, AtPolIB exo− produces a concentration-dependent increase in fluorescence. However, AtPolIB exo− harboring deletions in insertions 1 and 3 (AtPolIB-Δins1 or AtPolIB-Δins3) failed to produce a concentration-dependent change in fluorescence (Figure 5B). In contrast, a mutant AtPolIB exo− harboring a deletion in insertion 2 (AtPolIB-Δins2) showed a FRET response that is dependent on protein concentration, albeit with less efficiency compared to an unmodified AtPolIB exo− (Figure 5B). Thus, the FRET experiments suggest that AtPolIB variants harboring deletions in insertions 1 and 3 are inefficient in MMEJ (Figure 5B). To corroborate this result, we performed MMEJ reactions on a pssDNA-8 substrate. As expected, a product band of 58 nts was observed in reactions incubated with AtPolIB exo− and AtPolIB-Δins2. In contrast, no product was synthesized in samples incubated with AtPolIB exo− mutants harboring a deletion in insertions 1 and 3 (Figure 5C). Therefore, only an AtPolIB exo− mutant harboring a deletion in insertions 2 is able to execute MMEJ, although their products are different from the wild-type AtPolIB.
Figure 5.
MMEJ by AtPolIs is mediated by unique amino acid insertions in their polymerization domain. (A) Homology model showing the predicted localization of the unique insertions in the polymerization domain of AtPolIB. Insertions 1 and 2 are located at the thumb subdomain (green and red colored, respectively), whereas insertion 3 (cyan colored) is located at the palm subdomain. A representative structure of mutant polymerases showing individual deletions is present. (B) FRET assay using exonuclease deficient AtPolIB and exonuclease deficient deletion mutants. AtPolIB and AtPolIB-Δins2 generate a concentration-dependent change in the relative fluorescence, whereas incubation with AtPolIB-ΔIns1 and AtPolIB-ΔIns3 do not have an effect on the relative fluorescence. Data show the mean of three independent experiments. (C) MMEJ reaction assays using AtPolIB deletion mutants on a pssDNA-8 substrate. A MMEJ product is observed upon incubation with full-length (FL) AtPolIB (lane2), in contrast AtPolIB-ΔIns1 and ΔIns3 mutants (Δ1 and Δ3) are severely deficient in MMEJ product formation (lanes 3 and 5), AtPolIB-ΔIns2 forms the expected MMEJ product reaction products of 56 and of lower molecular weight products (lane 4) that correlate with the observed strand-displacement deficiency of this mutant (lane 4). (D and E) MMEJ reactions by point and deletion mutants in loops 1 and 3. A short deletion of 11 amino acids (599–610) in loop 1 abolishes MMEJ formation (lane 5). Point mutants K590 and K593A in loop 1 decrease MMEJ formation by 20 and 50% respectively, whereas a point mutant. K886A (loop 3) decrease MMEJ formation by 35%. MMEJ formation is quantified as the percentage of MMEJ product (P) in relation to the total amount of product and substrate (P+S). The graphical representation shows the mean of three independent experiments. (F) Denaturing polyacrylamide gel showing a time course reaction (from 1 to 4 minutes) by AtPolIB exo- and deletion mutants on a gapped-substrate. The six-nucleotides gapped DNA substrates, the length of the primer (24-mer), the expected maximal extension (30-mer) before strand-displacement and the full-length strand-displacement (65-mer) are indicated in the left side of the figure.
To further corroborate the role of insertions 1 and 3 in MMEJ, we constructed a shortened version of deletion 1 (δ599–610) and point mutations to alanine of individual lysine and arginine residues in this loop. We also constructed a point mutant in residue K866 located in loop 3. An MMEJ reaction using those mutant polymerases, show that a shorted version of loop 1 decreases MMEJ formation by ∼90% and individual K590A and K593A mutations also decrease the amount of MMEJ product to 20 and 50% of the consumed substrate (Figure 5D, lanes 5, 6, 7). Interestingly an alanine mutant of residue K866 also decreases MMEJ formation by MMEJ by nearly 40% (Figure 5D, lane 13).
AtPolIB-Δins2 primarily generates a product of approximately 42 nts that correlates with the necessity to displace the 5′-end of the primer stand corresponding to the opposite template (Figures 5C and 3C). AtPolIs harbor strong strand-displacement capabilities (40,42), mutant AtPolIs harboring a deletion in loop 1 abolish strand-displacement on a 1-nt gap, whereas mutant AtPolIB-Δins2 and AtPolIB-Δins3 severely decrease this property (42). In order to investigate if the appearance of the band of 42 nts synthesized by AtPolIB-Δins2 is related to a decrease in strand-displacement, we compare the efficiencies for strand-displacement on a 6-nt gap for all deletion mutants. Nucleotide incorporation by AtPolIB-Δins2 and AtPolIB-Δins3 is halted two and four nucleotides after the 5′-end of the blocking DNA strand (32- and 34-mer products) (Figure 5F, lanes 8 and 11). The ability of AtPolIB-Δins2 to displace two nucleotides correlates with the observed MMEJ product of approximately 42 nts (32 nts from the primer, 8 nts for extension in trans and 2 nts due to strand-displacement) show on a pssDNA-8 substrate (Figure 5C). The presence of the 42 nts band by AtPolIB-Δins2 supports the ability of AtPolIs to assemble a synapse in trans as a product mediated by snap-back replication would not be affect by the decrease in strand-displacement.
AtPolIs execute MMEJ on mismatched substrates
Analysis of microhomologous sequences found in chloroplast DNA upon DSBs contain mismatches (30,32). This suggests that in plastids MMEJ is executed by a DNAP able to extend from mismatches or that the distortions generated by a mismatch in the DNA double helix would not hamper synapse formation and its extension. We thus, investigated whether AtPoIls would use pssDNA substrates containing mismatches. We assemble six substrates using six different nucleotides annealed to the MH-8 substrate (Figure 6A and Supplementary Table S1). The first set of substrates contain three consecutive mismatches, one and two nucleotides before the 3′-OH end of the microhomologous sequence (substrate I and II, respectively). The second set of substrates (III and IV) contained one and two mismatches at their 3′-OH and the third set of substrates (V and VI) contained one and two mismatches three nucleotides before their 3′-OH (Figure 6A). To our surprise, all substrates, including a single mismatch (substrate III) and a double mismatch at the 3′-OH (substrate IV) were able to generate MMEJ products when incubated with AtPolIs exo– (Figure 6B and C). This result may relate to the fact that AtPolIs efficiently extend from a primer whose 3′-OH is confronted to an abasic site (40) and the ability of AtPolIs to extend mismatches in canonical primer-templates (43). Our data are congruent with the large repertoire of microhomologous sequences, because sequences that are not perfectly paired can be used as substrates by AtPolIs.
Figure 6.
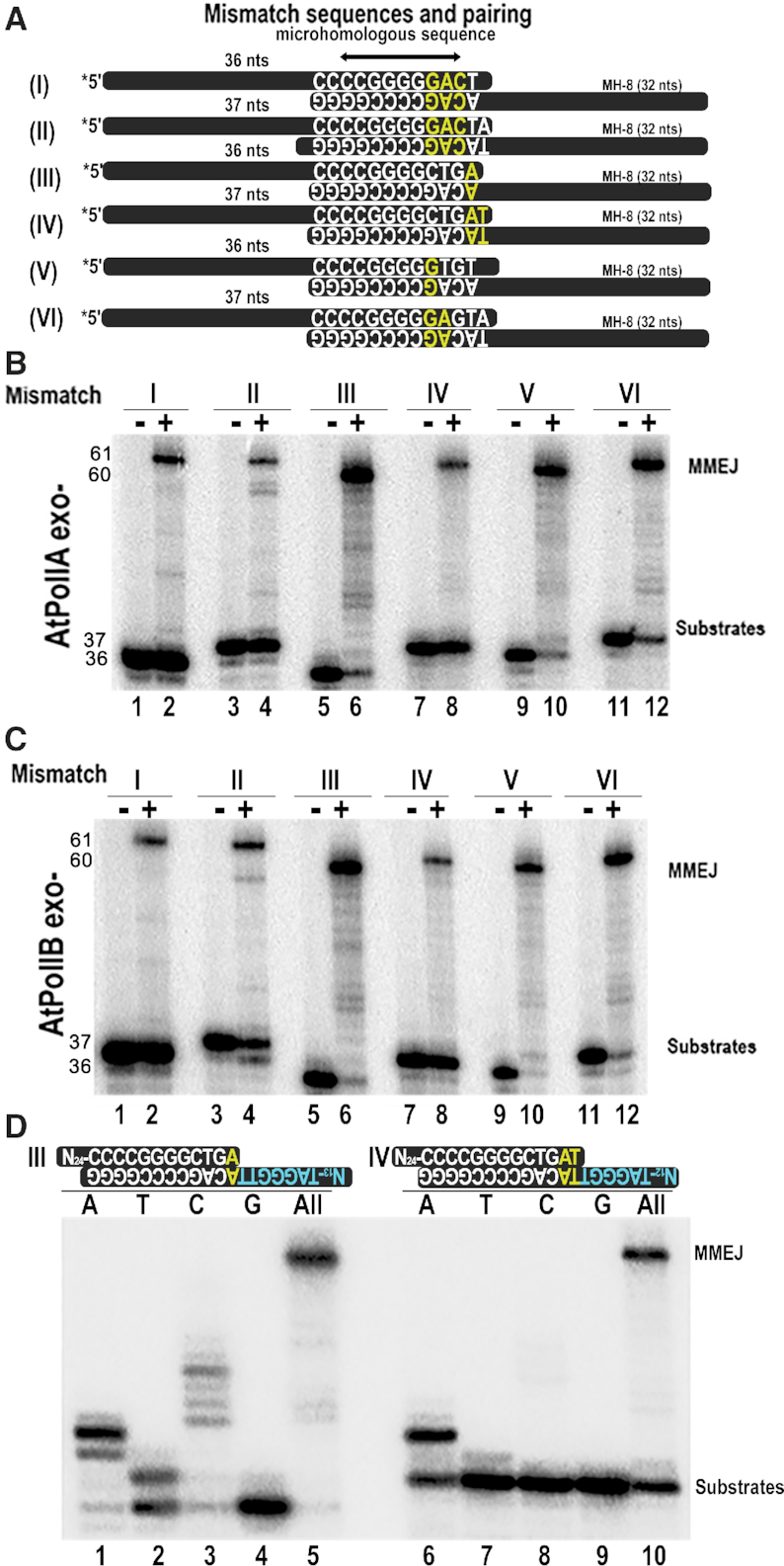
AtPolIA and AtPolIB execute MMEJ on mismatched substrates. (A) Oligonucleotide substrates containing mismatches annealed to promote MMEJ. The sequence of the microhomologous sequence of the single-stranded oligonucleotides is indicated. The mismatches are colored in red, whereas Watson-Crick pairs are colored in white. The arrangement includes mismatches located one and two nucleotides after the 3′-OH (I and II), located at the 3′-OH (III and IV), and located three and four nucleotides before the 3′-OH (V and VI). (B and C) MMEJ reactions executed by exonuclease deficient AtPolIA exo- (B) and AtPolIB exo– (C) on single-stranded substrates. The substrate without added polymerase is indicated by a negative sign (−), whereas reactions including DNA polymerase are indicated by a positive sign (+). The migration for substrates and MMEJ products are indicated. (D) Denaturing acrylamide gel showing the formation of MMEJ products by AtPolIB exo– on MMEJ with 3′-OH mismatches (substrates III and IV) annealed with the MH-8 oligonucleotide in the presence of individual nucleotides.
We decided to investigate the ability of AtPolIB exo− to incorporate individual nucleotides on substrates with one and two mismatches at their 3′-OH (substrates III and IV) (Figure 6D). On a substrate with a single nucleotide mismatch, AtPolIB exo− incorporates three dAMP nucleotides, in which two of them are expected to be template dependent and the third one due to misincorporation (Figure 6D, lane 1). AtPolIB exo− also misincoporates one adenine and several cytidines possibly due to misalignment (Figure 6D, lanes 2 and 3). On a substrate with two consecutive 3′-OH mismatches, AtPolIB exo− incorporates a dAMP molecule (Figure 6D, lane 6).
Plant specific DNA binding proteins AtmtSSBs, AtWhirlies and AtOSBs bind single-stranded DNA with different affinities
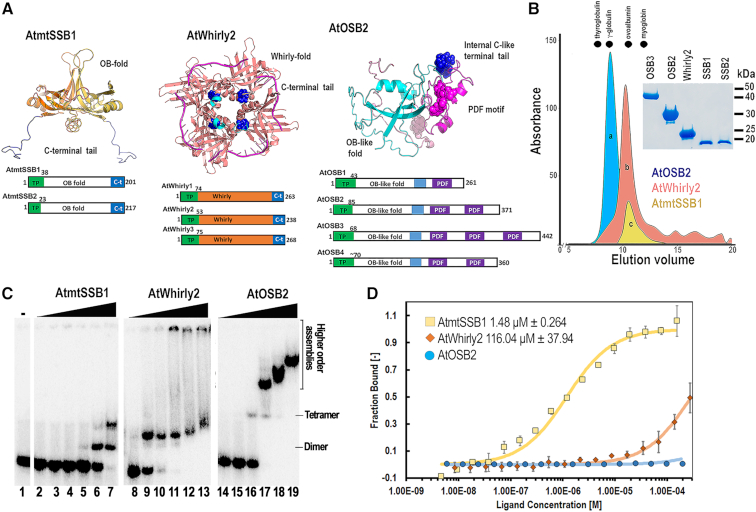
Organelles in Arabidopsis harbor three families of single-stranded DNA binding proteins (AtSSBs) and each family contains several members: A) The family of canonical mitochondrial DNA binding proteins (AtmtSSB) consisting of two members, B) Whirlies, which are composed of three members, and C) Organellar Single-stranded DNA Binding proteins (OSB), harboring four members (Figure 6A). AtmtSSBs contain an OB-domain follow by a C-terminal extension, AtOSBs, contain an N-terminal OB-like domain, followed by repeats (from one to four) of a PDF motif. This 50 amino acid motif is responsible for ssDNA binding. Whirlies assemble as tetramers and interact with ssDNA via the β-sheets of neighboring subunits (30,31,49,54–56) (Figure 7A).
Figure 7.
Arabidopsis harbors canonical and unique SSBs. (A) Structural organization of AtmtSSBs, AtWhirlies and AtOSBs. AtmtSSB1 and AtmtSSB2 are predicted to assemble as proteins with an oligonucleotide/oligosaccharide-binding (OB)-fold and a C-terminal disorder region, AtWhirlies (1–3) consists of a Whirly domain composed of two four-stranded β-sheets and two α–helices plus a disordered C-terminal extension (55,56), AtOSBs consist of an OB-like fold followed by one to three PDF motifs (54). The structural models of AtmtSSBs and AtOSBSs highlight the structural organization of the OB-fold domains and PDF motifs respectively. Structural modeling shows AtmtSSB1 and AtOSB1 as a dimer and monomer for clarity. The crystal structure of AtWhirly2 is shown in combination with the superimposed ssDNA co-crystallized in whirly2 from potato. The C-terminal amino acids of Whirlys are not present in their crystal structures and are proposed to be involved in protein-protein interactions. The AtWhirly2 structure illustrates the tetrameric organization and DNA binding of members this protein family. (B) Gel filtration profiles of purified AtmtSSB1, AtOSB2, and AtWhirly2 (inset) on a Superdex 75 column. Dots denote the elution profiles of molecular weight markers: thyrogloublin, γ globulin, ovoalbumin and myoglobin. The elution profiles of AtSSBs indicate their tetrameric assembly. (C) EMSA assay showing the binding of purified AtmtSSB1, AtOSB2, and AtWhirly2 to ssDNA. Reactions were incubated with increased concentration of the tetrameric SSBs (0, 1.15, 4.125, 14.87,53.5, 192.9, 694.2 nM) and 1 nM of a 5′-32P-labeled 70-mer oligonucleotide. Binding reactions were resolved on an 8% non-denaturing polyacrylamide gel. (D) Langmuir isotherms showing the binding of AtmtSSB1 and AtWhirly2 to AtPolIB exo− by MST.
Members of the OSB and Whirly families decrease the formation of illegitimate recombination products (31,54), whereas AtmtSSB1 promotes AtRecA strand-exchange activity (49). In order to investigate the possible role of AtSSBs in MMEJ, we directly measured their binding affinities for substrates containing ssDNAs of different lengths. In order to correctly measure their binding affinities, we first determined their oligomeric states. The elution profiles of purified AtmtSSB1, AtmtSSB2, AtOSB2, AtOSB3, and AtWhirly2 (Figure 7B, inset) on a gel filtration column revealed that all proteins assembled as tetramers in the absence of DNA (Figure 6B). In our binding studies, we used the molar concentration of the tetrameric SSBs and not the individual protomers. An EMSA on a labeled oligonucleotide of 70 nts illustrates the high binding affinity of AtWhirly2 for single-stranded DNA (Figure 7C, lanes 9–13). This binding is consistent with previous biochemical analyses of Whirly variants that bind ssDNA as tetramers with affinity constants of approximately 5 nM (30,55). In addition to binding as a tetramer, AtWhirly2 also forms high molecular weight complexes with ssDNA, as previously show by Cappadocia and coworkers (57). AtmtSSB1 forms two complexes that may correspond to dimeric and tetrameric assemblies in comparison to the tetrameric AtWhirly2 (Figure 7C, lanes 6 and 7). On the other hand, AtOSB2 initially binds ssDNA with a relative migration that corresponds to a tetrameric assembly, although high molecular assemblies are formed at higher protein concentrations (Figure 7C, lanes 14–19). An interesting point is the contrasting differences for ssDNA binding between plant SSBs. AtmtSSBs needs to be in a molar excess of 694-fold to shift all the 70-mer, whereas AtOSB2 and AtWhirly2 need to be in 53-fold excess (Figure 6C). Given these contrasting binding profiles, we measured the binding affinities for AtmtSSB1, AtOSB2 and AtWhirly2 on pssDNA-10 and ssDNA45-mer substrates using fluorescence anisotropy (Table 1). AtOSB2 and AtWhirly2 bind to a pssDNA-10 substrate with dissociation constants (Kd) of 8 and 22 nM respectively, whereas AtmtSSB1 was unable to bind to this substrate (Table 1). On the other hand, AtmtSSB1, AtOSB2, and AtWhirly2 bonded to a ssDNA 45-mer substrate with Kds of 62.8, 11.7 and 8.2 nM, respectively (Table 1). These values approximate the reported values for AtWhirly2 (30) and AtOSB2 (54). The EMSA and anisotropy experiments thus indicate that the binding affinity of AtmtSS1 for ssDNA substrates is weaker than the binding affinities of AtOSB2 and AtWhirly2.
Table 1.
Dissociation constants of plant organellar SSBs in pssDNA and ssDNA templates
| pssDNA-10 Oligomeric State in solution | ssDNA-45 mer Oligomeric State in solution | |
|---|---|---|
| Protein | K d (nM) | K d (nM) |
| AtmtSSB1 | — | 62.8 ± 3.6 (tetramer) |
| AtOSB2 | 8.6 ± 0.46 (tetramer) | 11.7 ± 2.2 (tetramer) |
| AtWhirly2 | 22.1 ± 1.3 (tetramer) | 8.2 ± 0.19 (tetramer) |
*Dissociation constant were determined by three independent experiments.
AtmtSSB1, but not AtOSB2 and AtWhirly2, interact with AtPolIB with affinity constant similar to canonical replisomes
Several SSBs bind to their cognate DNA polymerases by virtue of their exposed C-terminal amino acids. In order to investigate if AtSSBs interact with the plant organellar DNA polymerases, we evaluated a potential interaction between AtSSBs and AtPolIB exo− by MST. We found that AtPolIB exo− interacts with AtmtSSB1 with an affinity constant of 1.48 μM. This affinity constant is similar to the 1.6 μM interaction between T7 DNA polymerase and T7 SSB in the absence of thioredoxin and the 1.6 μM interaction between DNAP III and SSB from E. coli (58,59). In contrast, AtWhirly2 at a concentration of 250 μM (6 mg/ml) is not able to saturate a binding isotherm, suggesting that the observed interaction is non-specific or that is larger than 250 μM. AtOSB2 shows no observable binding to AtPolIB exo− (Figure 7D).
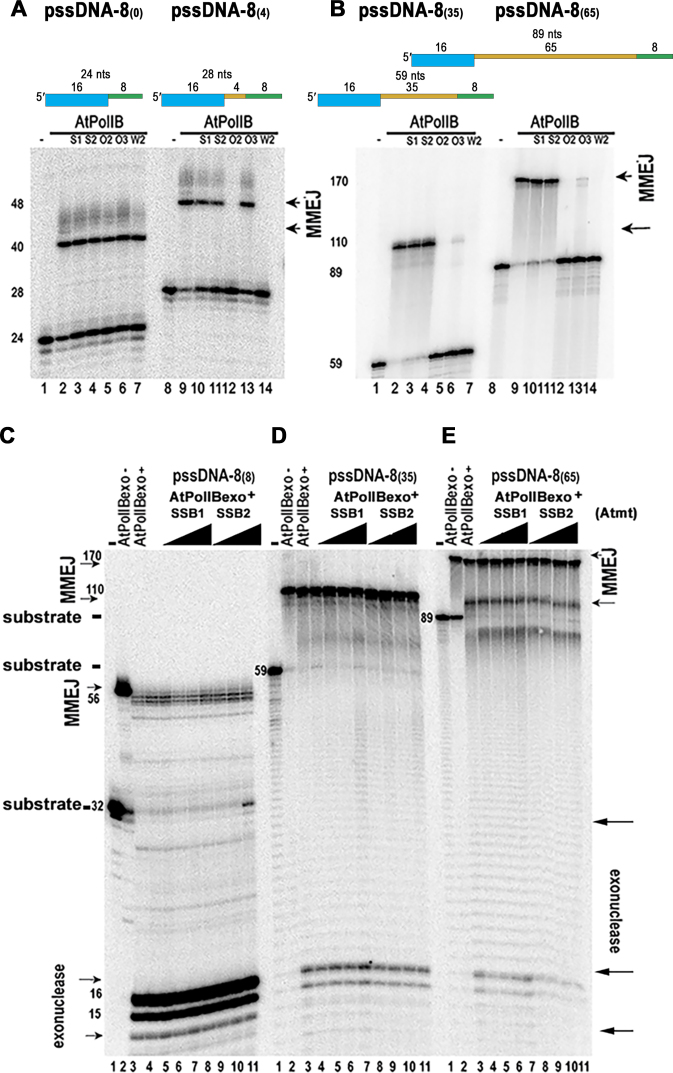
AtWhirlies and AtOSBs decrease MMEJ on long ssDNA templates
Whirlies and OSBs are proposed to compete for a partially resected DSB and prevent MMEJ in organelles (31,54). In order to obtain biochemical evidence for this hypothesis, we performed MMEJ reactions using pssDNA with microhomologies of 8 nts, but with extra single-stranded DNA sequences that ranked from 0 to 65 nts. In these reactions, we used a 50-fold molar excess of tetrameric purified SSBs (AtmtSSB1, AtmtSSB2, AtOSB2, AtOSB3, and AtWhirly2) and 5-fold AtPolIB with respect to pssDNAs (Figure 8A and B). In reactions incubated using the microhomology of 8 nts without an extra single-stranded DNA segment, pssDNA-8(0), the presence of SSBs did not have an effect on MMEJ formation by AtPolIB (Figure 8A, lanes 1–7). The lack of MMEJ inhibition on a pssDNA-8(0) correlates with the absence of binding for SSBs to this substrate (30) a (Table 1) and also indicates that the presence of SSBs does not inhibit MMEJ by AtPolIB. When 4 nts were added to the microhomologous sequence of 8 nts (pssDNA-8(4)), AtOSB2 and AtWhirly2 completely hampered MMEJ formation (Figure 8A, lanes 12 and 14). Inhibition by AtOSB2 and AtWhirly2 on a pssDNA-8(4) substrate, but not by AtmtSSB1 or AtmtSSB2, correlates with the binding of AtOSB2 and AtWhirly2 to a pssDNA-10 substrate (Table 1). The presence of AtOSB3, but not AtmtSSB1 and AtmtSSB2, in substrates with single-stranded DNA regions of 43 (pssDNA-8(35)) and 73 (pssDNA-8(65)) nucleotides decreased product formation in pssDNA-8(35) and pssDNA-8(65) substrates (Figure 8B). The lack of inhibition for AtmtSSB1 and AtmtSSB2 on longer partially single-stranded substrates was observed even with reactions incubated with 200-fold molar excess of those proteins with respect to the ppsDNA template (Supplementary Figure S2). The binding affinity of AtmtSSB1 for a 70-mer ssDNA substrate is approximately five times weaker than the binding affinity of AtOSB2 and AtWhirly2, so the lack of MMEJ product formation by AtPolIs in reactions incubated with AtmtSSBs is not explained by a lack of binding to single-stranded DNA.
Figure 8.
AtSSBs and single-stranded length of the DNA break regulate MMEJ. (A, B) Unique SSB from plant organelles hamper MMEJ on substrates with single-stranded regions longer than 8 nts. MMEJ formation by AtPolIs in the presence of plant SSBs (AtmtSSB1, S1; AtmtSSB2, S2; AtOSB2, O2; AtOSB3, O3; and AtWhirly2, W2) in MMEJ substrates with a constant microhomologous sequence of 8 nts, but varying single-stranded regions of 8 (panel A, lanes 1 to 7),12 (panel A, lanes 8–14) 43 (panel B, lanes 1–7) and 73 nts (panel B, lanes 8–14). The length of the MMEJ, single-stranded region and total oligonucleotide length of the substrates are indicated in the upper part of the panel. The length of the substrates and MMEJ products are indicated by arrows. (C–E) Wild-type AtPolIA and AtPolIB execute MMEJ with minimal exonucleolysis on substrates with single-stranded regions longer than 16 nts. (C) MMEJ reaction by AtPolIA (lanes 3–6) and AtPolIB (lanes 7–11) on a pssDNA with 8 bases of microhomologous sequence and 8 extra-bases of single-stranded DNA. The labeled substrate is indicated in lane 1 and the MMEJ product by and exonuclease deficient AtPolIB is shown in lane 2. (D and E) As in C, but with a substrate containing 35 nts or 65 nts of extra single-stranded DNA respectively.
MMEJ on long substrates is favored over exonucleolysis independently of the presence of mtSSBs
MMEJ reactions incubated with substrates containing longer single-stranded DNA regions of 43 and 73 nucleotides were more efficiently used than substrates with short regions (Figure 8A and B). Canonical SSBs recruit several enzymes to DNA replication forks via their acid C-terminal extension (60,61) and AtmtSSB1 interacts with AtPolIB with similar affinities to the interactions observed in other replisomes (Figure 8D). We therefore tested whether the presence of AtmtSSBs would promote MMEJ by wild-type AtPolIs in substrates with long single-stranded DNA regions. We used substrates with increased single-stranded DNA regions of 12, 43 and 73 nucleotides while maintaining a microhomology region of 8 nts (pssDNA-8(4), pssDNA-8(35) and pssDNA-8(65)) in the presence of increased concentrations of AtmtSSB1 and AtmtSSB2 and exonuclease proficient AtPolIB (Figure 8C–E). In a pssDNA-8(4) substrate, AtPolIB executed MMEJ in the presence and absence of mtSSBs with similar efficiencies and most of the substrate was degraded to 15- or a 16-mer (Figure 8C). A comparison between the amount of MMEJ product synthesized by exonuclease deficient AtPolIB and the exonuclease active form of the enzyme on the pssDNA-8(4) substrate shows that AtPoIlB exo- converts approximately 80% of the substrate to a MMEJ product, whereas wild-type AtPolIB converts <10% of the substrate to a MMEJ product in the absence of AtmtSSB1 (Figure 8C, lanes 2 and 3). In pssDNA-8(35) and pssDNA-8(65) substrates, the exonucleolytic degradation by wild-type AtPolIB substantially decreased independent of the presence or absence of AtmtSSBs (Figure 8D and E). MMEJ reactions incubated with a pssDNA-8(35) substrate exhibited almost no difference when incubated with exonuclease deficient or exonuclease proficient AtPolIB (Figure 8D, lanes 2 and 3) and the addition of AtmtSSBs had no effect on MMEJ (Figure 8D, lanes 4–11). Reactions incubated with wild-type AtPolIB generate a predominant MMEJ product of 110 nts and a minimal amount of exonucleolytic products that accumulate as 15 or 16-mers (Figure 8D, lanes 3–11). A similar decrease of the exonuclease degradation products by wild-type AtPolIB is also observed in reactions incubated with a pssDNA-8(65) substrate in the presence and absence of AtmtSSBs (Figure 8E).
DISCUSSION
Here we demonstrate that organellar DNA polymerases from the plant model A. thaliana, AtPolIs, efficiently perform MMEJ on partially resected DSBs longer than 6 nts, proving an explanation for DSBs repair with MMEJ signatures in plant organelles (12,29,31,32). MMEJ requires the formation of a synapse between DNA strands to join otherwise unstably paired microhomologous sequences, while creating a 3′-OH necessary for nucleotide incorporation and extension. Short microhomologous sequences could transiently pair, however these intermediates are short-lived. AtPolIs in comparison to KF DNAP I is able to stabilize these intermediates and used them as substrates. AtPolIs harbor three unique amino acid insertions in their polymerization domain (35,37) and within these insertions, insertion 1 is indispensable for MMEJ. Thus, AtPolIs and HsPolθ converged in the use of insertions for MMEJ and lesion bypass (21,40).
Mismatches are an abundant feature in DSBs repaired by MMEJ in plant organelles. For example, the Brisson group showed that 91 out of 158 induced MMEJ rearrangements in plastid DNA contain mismatches (30). Thus, the machinery responsible to execute MMEJ in plant organelles must be able to extend from mismatches. The fact that AtPolIs can efficiently anneal and extend from microhomologus sequences harboring mismatches at their 3′-OH, supports the notion that these enzymes are biochemically equipped to execute MMEJ in plant organelles. In contrast to HsPolθ, AtPolIs contain an active exonuclease domain and its modulation should be essential for MMEJ in vivo. In substrates that contain a short single-stranded region, i.e. less than 12-nts, AtPolIs exonucleolytic degradation is preferred over MMEJ. An increase in the length of the single-stranded region, however, promotes MMEJ at levels in which exonuclease deficient and wild-type AtPolIs execute MMEJ with similar efficiencies. We speculate that the presence of extra nucleotides generates a binding platform for AtPolIs that promotes synapse formation (Figure 8D and E). The ability of exonuclease proficient AtPolIs to carry out MMEJ on substrates with long single-stranded regions with the same efficiency that exonuclease deficient AtPolIs reinforces the notion of the functional relevance of these enzymes to execute MMEJ in organello.
Arabidopsis plants harboring T‐DNA insertion mutants in why and osb1 genes accumulate DSBs that are repaired via MMEJ and an increase of ectopic recombination products (30,54). As MMEJ and ectopic homologous recombination are driven by annealing of short single-stranded DNA sequences, we analyzed the effect of AtmtSSBs, At Whirly2 and AtOSBs in MMEJ by AtPolIB. Whirlies and AtOSBs are proposed to counter MMEJ and ectopic homologus recombination by binding to 3′-OH resected ends (10,30,62). This binding is predicted to hamper the annealing of the microhomologous or short-homologous sequences. We demonstrated that the presence of AtWhirly2 and AtOSBs in templates containing single-stranded regions longer than 12 nts hampers MMEJ. AtOSB3 is unable to prevent MMEJ on a pssDNA with a single-stranded region of 12 nts, but efficiently hampers MMEJ on longer substrates (Figure 8A and B). This differential correlates with the presence of an extra PDF domain, suggesting that AtOSB3 needs a larger nucleotide length for binding (10).
AtmtSSB, AtWhirly2, and AtOSBs, bind single-stranded DNA and pssDNA substrates with different affinities (10,30,31,54) (Figure 8 and Table 1). Only AtWhirly2 and AtOSBs, however, completely block MMEJ by AtPolIs. The latter suggests that the concentration of AtmtSSBs needed to block MMEJ is much higher because of its weaker affinity for ssDNA, that AtmtSSBs easily dissociate from DNA, or that AtmtSSBs physically interact with AtPolls and displaces these proteins. The weak binding for ssDNA by mtSSB is observed for other proteins from the same family that have micromolar affinities for ssDNA binding (63). A weak binding of the plant organellar OB-fold maybe related to the modular structure of AtOSBs, in which their PDF domains are responsible for binding to single-stranded DNA and their associated OB-fold domain does not contribute to ssDNA binding (54).
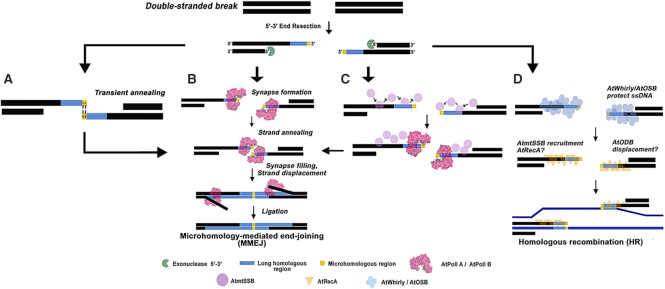
MMEJ inhibition by AtOSBs and Whirlies is in agreement with previous models explaining the fate of genomic DNA upon DSBs in plant organelles (10,30,62,64). Plant organelles harbor a bacterial RecA orthologue that mediates homologous recombination and a Rad52-like single-stranded annealing protein, dubbed organellar DNA-binding protein 1 (ODB1). ODB1 has been proposed to direct SSBs displacement from single-strand DNA regions and promotes loading of RecA (10,65,66). In plant organelles, U-turn-like rearrangements are mediated by microhomologies. The absence of whirlies and AtRecA in Arabidopsis increases the amount of U-turn-like rearrangements (67). Zampini and coworkers postulate that RecA promotes accurate fork restart suggesting that in the absence of whirlies the single-stranded 3′-OH of a collapsed replication fork anneals to an opposite strand generating U-turn-like rearrangement (67). Based upon our biochemical results and previous genetic analysis (6,11,27,28,32,54,65,67), we propose a model in which AtOSBs and AtWhirlies limit MMEJ by blocking the access of a single-stranded 3′-OH to AtPolIs or AtmtSSBs. AtmtSSBs binding to the 3′-OH may result in recruiting AtPolIs or the HR machinery, and MMEJ would be favored only if AtPolIs reach the end of partially resected 3′-OH (Figure 9). If the proposed block of the single-stranded 3′-OH is limited by the absence of whirlies and HR is impaired by lack of AtRecA, other DNA repair routes like MMEJ would mediate DSB repair.
Figure 9.
Proposed model for DSBs fate in plant organelles. Microhomology-mediated end-joining or Homologous Recombination is driven by the access of single-stranded binding proteins to a 3′-OH single-stranded end. Resected single-strand DNA regions with microhomologous (yellow) and long homologous (blue) sequences are produced upon DSBs. Plant SSBs and AtPolIs compete to bind the 3′-OH of a resected end. If single-stranded biding proteins AtOSBs or AtWhirlies bind to a resected end, they block the access to canonical AtmtSBB and AtPolIs. Binding of mtSSBs would promote MMEJ if sufficient AtPolIs is available to react with them. Conversely recruitment of RecA to a 3′-OH of a resected end would drive homologous recombination.
Both AtPolIA and AtPolB execute MMEJ, suggesting that in vivo MMEJ is executed by both polymerases. This contrasts with the role of AtPolIB in preventing MMEJ rearrangements in chloroplasts proposed by Parent and coworkers (29). A plausible explanation for the increase of MMEJ rearrangements in T-DNA insertion lines inactivating AtPolIB may lie in a differential in lesion bypass abilities between AtPolIA and AtPolIB. For instance, if AtPolA possesses less efficient lesion bypass than AtPolIB, organellar DNA replication under the sole presence of AtPolIA would have a greater tendency to collapse generating double-stranded breaks. Alternatively, it is possible that AtPolIB, but not AtPolIA, interacts with components of the HR machinery (29). Although the latter scenario is speculative, our data reveal that MMEJ in plant organelles is driven by a unique family of DNAPs. The presence of these polymerases, instead of metazoan DNAP γ, may account for the numerous MMEJ events in plant organelles. AtPolIs are functionally analogous to HsPolθ, in both lesion bypass and MMEJ. The functional similarities between AtPolIs and HsPolθ are and example of protein convergence in which nature has used amino acid insertions to decorate the active site of family-A DNA polymerases and execute novel DNA transactions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Corina Diaz-Quezada for technical support and Teri Markow for critical reading. P.L. G.-M. thanks CONACYT for her master's fellowship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CONACYT-Ciencia Básica grant [253737 to L.G.B.]; CONACYT-Infrastructure [279896]. Funding for open access charge: CONACYT-PRODEP.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gray M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012; 4:a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raven J.A., Allen J.F.. Genomics and chloroplast evolution: what did cyanobacteria do for plants. Genome Biol. 2003; 4:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D.. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oldenburg D.J., Bendich A.J.. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J. Mol. Biol. 2004; 335:953–970. [DOI] [PubMed] [Google Scholar]
- 5. Backert S., Borner T.. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 2000; 37:304–314. [DOI] [PubMed] [Google Scholar]
- 6. Davila J.I., Arrieta-Montiel M.P., Wamboldt Y., Cao J., Hagmann J., Shedge V., Xu Y.Z., Weigel D., Mackenzie S.A.. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011; 9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sloan D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013; 200:978–985. [DOI] [PubMed] [Google Scholar]
- 8. Shaver J.M., Oldenburg D.J., Bendich A.J.. The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula. Plant Physiol. 2008; 146:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bendich A.J. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004; 16:1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gualberto J.M., Newton K.J.. Plant mitochondrial Genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017; 68:225–252. [DOI] [PubMed] [Google Scholar]
- 11. Tremblay-Belzile S., Lepage E., Zampini E., Brisson N.. Short-range inversions: rethinking organelle genome stability: template switching events during DNA replication destabilize organelle genomes. Bioessays. 2015; 37:1086–1094. [DOI] [PubMed] [Google Scholar]
- 12. Marechal A., Brisson N.. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010; 186:299–317. [DOI] [PubMed] [Google Scholar]
- 13. Christensen A.C. Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biol. Evol. 2014; 6:1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christensen A.C. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013; 5:1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sfeir A., Symington L.S.. Microhomology-Mediated end Joining: A Back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chiruvella K.K., Liang Z., Wilson T.E.. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013; 5:a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brissett N.C., Pitcher R.S., Juarez R., Picher A.J., Green A.J., Dafforn T.R., Fox G.C., Blanco L., Doherty A.J.. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007; 318:456–459. [DOI] [PubMed] [Google Scholar]
- 20. Pitcher R.S., Brissett N.C., Picher A.J., Andrade P., Juarez R., Thompson D., Fox G.C., Blanco L., Doherty A.J.. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007; 366:391–405. [DOI] [PubMed] [Google Scholar]
- 21. Kent T., Chandramouly G., McDevitt S.M., Ozdemir A.Y., Pomerantz R.T.. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015; 22:230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nick McElhinny S.A., Havener J.M., Garcia-Diaz M., Juarez R., Bebenek K., Kee B.L., Blanco L., Kunkel T.A., Ramsden D.A.. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005; 19:357–366. [DOI] [PubMed] [Google Scholar]
- 23. Crespan E., Czabany T., Maga G., Hubscher U.. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases lambda and beta on normal and repetitive DNA sequences. Nucleic Acids Res. 2012; 40:5577–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublie S.. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brissett N.C., Martin M.J., Bartlett E.J., Bianchi J., Blanco L., Doherty A.J.. Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase. Cell Rep. 2013; 5:1108–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arrieta-Montiel M.P., Shedge V., Davila J., Christensen A.C., Mackenzie S.A.. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009; 183:1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller-Messmer M., Kuhn K., Bichara M., Le Ret M., Imbault P., Gualberto J.M.. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012; 159:211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parent J.S., Lepage E., Brisson N.. Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 2011; 156:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cappadocia L., Marechal A., Parent J.S., Lepage E., Sygusch J., Brisson N.. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 2010; 22:1849–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marechal A., Parent J.S., Veronneau-Lafortune F., Joyeux A., Lang B.F., Brisson N.. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwon T., Huq E., Herrin D.L.. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Odom O.W., Baek K.H., Dani R.N., Herrin D.L.. Chlamydomonas chloroplasts can use short dispersed repeats and multiple pathways to repair a double-strand break in the genome. Plant J. 2008; 53:842–853. [DOI] [PubMed] [Google Scholar]
- 34. Ottaviani D., LeCain M., Sheer D.. The role of microhomology in genomic structural variation. Trends Genet. 2014; 30:85–94. [DOI] [PubMed] [Google Scholar]
- 35. Moriyama T., Sato N.. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant Sci. 2014; 5:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moriyama T., Tajima N., Sekine K., Sato N.. Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae. Genome Biol. Evol. 2014; 6:228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moriyama T., Terasawa K., Sato N.. Conservation of POPs, the plant organellar DNA polymerases, in eukaryotes. Protist. 2011; 162:177–187. [DOI] [PubMed] [Google Scholar]
- 38. Moriyama T., Terasawa K., Fujiwara M., Sato N.. Purification and characterization of organellar DNA polymerases in the red alga Cyanidioschyzon merolae. FEBS J. 2008; 275:2899–2918. [DOI] [PubMed] [Google Scholar]
- 39. Mori Y., Kimura S., Saotome A., Kasai N., Sakaguchi N., Uchiyama Y., Ishibashi T., Yamamoto T., Chiku H., Sakaguchi K.. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005; 334:43–50. [DOI] [PubMed] [Google Scholar]
- 40. Baruch-Torres N., Brieba L.G.. Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 2017; 45:10751–10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peralta-Castro A., Baruch-Torres N., Brieba L.G.. Plant organellar DNA primase-helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence. Nucleic Acids Res. 2017; 45:10764–10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trasvina-Arenas C.H., Baruch-Torres N., Cordoba-Andrade F.J., Ayala-Garcia V.M., Garcia-Medel P.L., Diaz-Quezada C., Peralta-Castro A., Ordaz-Ortiz J.J., Brieba L.G.. Identification of a unique insertion in plant organellar DNA polymerases responsible for 5′-dRP lyase and strand-displacement activities: Implications for Base Excision Repair. DNA Repair (Amst.). 2018; 65:1–10. [DOI] [PubMed] [Google Scholar]
- 43. Ayala-Garcia V.M., Baruch-Torres N., Garcia-Medel P.L., Brieba L.G.. Plant organellar DNA polymerases paralogs exhibit dissimilar nucleotide incorporation fidelity. FEBS J. 2018; 285:4005–4018. [DOI] [PubMed] [Google Scholar]
- 44. Chan S.H., Yu A.M., McVey M.. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010; 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beagan K., Armstrong R.L., Witsell A., Roy U., Renedo N., Baker A.E., Scharer O.D., McVey M.. Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genet. 2017; 13:e1006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Kregten M., de Pater S., Romeijn R., van Schendel R., Hooykaas P.J., Tijsterman M.. T-DNA integration in plants results from polymerase-theta-mediated DNA repair. Nat. Plants. 2016; 2:16164. [DOI] [PubMed] [Google Scholar]
- 47. Inagaki S., Suzuki T., Ohto M.A., Urawa H., Horiuchi T., Nakamura K., Morikami A.. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006; 18:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Black S.J., Kashkina E., Kent T., Pomerantz R.T.. DNA polymerase theta: aunique multifunctional End-Joining machine. Genes (Basel). 2016; 7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edmondson A.C., Song D., Alvarez L.A., Wall M.K., Almond D., McClellan D.A., Maxwell A., Nielsen B.L.. Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol. Genet. Genomics. 2005; 273:115–122. [DOI] [PubMed] [Google Scholar]
- 50. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al.. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007; 35:W43–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Clark J.M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988; 16:9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kent T., Mateos-Gomez P.A., Sfeir A., Pomerantz R.T.. Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife. 2016; 5:e13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zaegel V., Guermann B., Le Ret M., Andres C., Meyer D., Erhardt M., Canaday J., Gualberto J.M., Imbault P.. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006; 18:3548–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Desveaux D., Allard J., Brisson N., Sygusch J.. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 2002; 9:512–517. [DOI] [PubMed] [Google Scholar]
- 56. Desveaux D., Marechal A., Brisson N.. Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 2005; 10:95–102. [DOI] [PubMed] [Google Scholar]
- 57. Cappadocia L., Parent J.S., Zampini E., Lepage E., Sygusch J., Brisson N.. A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res. 2012; 40:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hamdan S.M., Marintcheva B., Cook T., Lee S.J., Tabor S., Richardson C.C.. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5096–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Glover B.P., McHenry C.S.. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 1998; 273:23476–23484. [DOI] [PubMed] [Google Scholar]
- 60. Kong D., Richardson C.C.. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein-protein interactions. J. Biol. Chem. 1998; 273:6556–6564. [DOI] [PubMed] [Google Scholar]
- 61. Antony E., Weiland E., Yuan Q., Manhart C.M., Nguyen B., Kozlov A.G., McHenry C.S., Lohman T.M.. Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 2013; 425:4802–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kühn K., Gualberto J.M.. Maréchal-Drouard L. Advances in Botanical Research. 2012; 63:Elsevier; 215–252. [Google Scholar]
- 63. He Z.G., Rezende L.F., Willcox S., Griffith J.D., Richardson C.C.. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J. Biol. Chem. 2003; 278:29538–29545. [DOI] [PubMed] [Google Scholar]
- 64. Zampini E., Truche S., Lepage E., Tremblay-Belzile S., Brisson N.. Li XQ. Somatic Genome Variation in Animals, Plants, and Microorganisms. 2017; John Wiley & Sons, Inc; 119–163. [Google Scholar]
- 65. Janicka S., Kuhn K., Le Ret M., Bonnard G., Imbault P., Augustyniak H., Gualberto J.M.. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 2012; 72:423–435. [DOI] [PubMed] [Google Scholar]
- 66. Manchekar M., Scissum-Gunn K., Song D., Khazi F., McLean S.L., Nielsen B.L.. DNA recombination activity in soybean mitochondria. J. Mol. Biol. 2006; 356:288–299. [DOI] [PubMed] [Google Scholar]
- 67. Zampini E., Lepage E., Tremblay-Belzile S., Truche S., Brisson N.. Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 2015; 25:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kiefer J.R., Mao C., Braman J.C., Beese L.S.. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998; 391:304–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.