Abstract
Background
Restrictive cardiomyopathy is a rare cardiac disease, for which several genes including TNNT2, MYPN, FLNC and TNNI3 have been associated with its familial form.
Case presentation
Here we describe a female proband with a severely manifested restrictive phenotype leading to heart transplantation at the age of 41, who was found homozygous for the novel TNNI3 mutation: NM_000363.4:c.586G > C, p.(Asp196His). Her parents were third-degree cousins originating from a small village and although they were found heterozygous for the same variant they displayed no symptoms of the disease. Her older sister who was also found heterozygous was asymptomatic. Her twin sister and her brother who were homozygous for the same variant displayed a restrictive and a hypertrophic phenotype, respectively. Their children are all carriers of the mutation and remain asymptomatic until the age of 21.
Conclusion
These observations point to a recessive mode of inheritance reported for the first time for this combination of gene/disease.
Keywords: TNNI3, Restrictive cardiomyopathy, Autosomal recessive, Cardiac troponin I, Mutation, Case report
Background
Restrictive cardiomyopathy (RCM) is a rare cardiac disorder that manifests primarily as an abnormality of diastolic filling, which is also reported as a reduced ventricular compliance or increased stiffness, leading to reduced diastolic volume with preserved systolic function [1]. Several studies have confirmed that RCM can be inherited in a familial manner (MIM#PS115210) and mutations in several genes have been associated with the restrictive phenotype i.e. TNNT2 (MIM*191045) [2], MYPN (MIM*608517) [3] and FLNC (MIM*102565) [4]. In 2003, Mogensen et al. [5] showed a causal relationship between cardiac troponin I genetic abnormalities and the manifestation of the disease after studying several heterozygous patients diagnosed with idiopathic restrictive cardiomyopathy, including one large family in which affected individuals presented with either restrictive or hypertrophic cardiomyopathy (HCM).
Cardiac troponin serves as a sensor of the intracellular Ca2+ levels and regulates the interaction between the thick and thin filaments during muscle contraction. It consists of three separate subunits: troponin C, troponin T and troponin I (cTnI; TNNI3), the latter being the inhibitory subunit of the complex, primarily functioning to prevent actin and myosin from interacting in the absence of Ca2+. Mutations in the troponin complex introduce alterations in Ca2+ affinity and protein-protein interactions, which may ultimately lead to the development of cardiomyopathy.
We describe here a female patient with a severely manifested phenotype of RCM leading to heart transplantation at the age of 41, attributed to an homozygous mutation in the gene encoding the cTnI (TNNI3). In her family all heterozygous carriers were found asymptomatic, while her homozygous sister and brother displayed restrictive and hypertrophic phenotype, respectively, pointing to a recessive mode of inheritance reported for the first time for this combination of gene and disease.
Case presentation
Patients
The proband (patient II2, Fig. 1a) was referred to our hospital at the age of 41 due to progressive dyspnea on exertion over the last 3 months. Her medical history was remarkable for ischemic stroke attributed to paroxysmal atrial fibrillation. She was on oral anticoagulation (acenocumarol). Her electrocardiogram showed atrial fibrillation and her echocardiographic study revealed normal left ventricular (LV) dimensions with borderline contractile function (left ventricular ejection fraction [LVEF] 50%), severe biatrial dilatation and mild mitral regurgitation. Transmitral diastolic flow revealed monophasic flow (atrial fibrillation) with normal average maximal velocity (E) but shortened deceleration time (DT 110 ms), a finding consistent with restrictive LV filling (Table 1). Right heart catheterization demonstrated increased left atrial pressure (estimated through pulmonary capillary wedge pressure), pulmonary hypertension and restrictive diastolic filling pattern with characteristic dip-plateau morphology, confirming echocardiographic findings. Attempts to restore sinus rhythm were unsuccessful. Despite receiving appropriate treatment (metoprolol tartate, acenocoumarol, furosemide, amiloride) and achieving adequate rate control, the patient deteriorated during the following year and a new echocardiographic study revealed worsened LV systolic function (LVEF 35%, LV end-systolic diameter 34 mm), similar left and right atrial volumes, moderate mitral and tricuspid regurgitation and pericardial effusion, while right ventricular function was also impaired. Right heart catheterization confirmed hemodynamic deterioration and the patient was admitted to our center’s heart transplantation list. Due to further clinical aggravation, a pulsatile biventricular assist device was implanted as a bridge to transplant and two months later, successful heart transplantation was performed. The histology of the native heart revealed multiple foci of intestitial and pericellurar fibrosis with features of non-specific myocytic hypertrophy (Fig. 2).
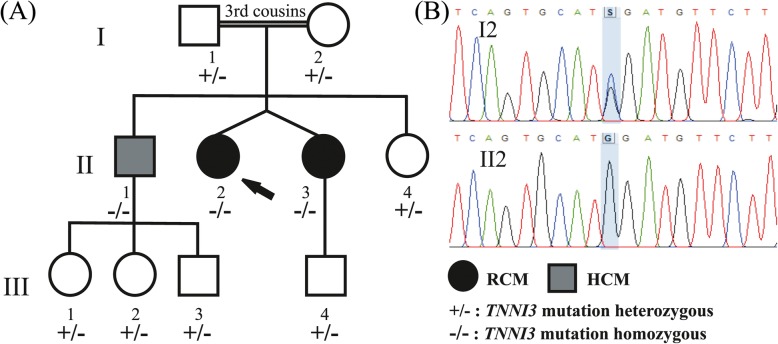
Fig. 1.
a The family pedigree is depicted. The proband (shown by arrow, II2) and her dizygotic twin sister (II3) were homozygous for the NM_000363.4:c.586G > C variant and displayed the phenotype of restrictive cardiomyopathy (RCM). Their brother (II1) also homozygous for the same variant displayed the phenotype of hypertrophic cardiomyopathy (HCM). Their children, with age range from 5 to 21 years old (III1: 5, III2: 13, III3 and III4: 21), remain unaffected along with the proband’s parents who were found heterozygous for the same mutation. Circle = female. Square = male. Filled symbols = affected individuals. b Electropherogram of the involved sequence fragment of the TNNI3 for the members of the family. The detected mutation is highlighted
Table 1.
Clinical, Electrocardiographic, and Doppler Echocardiographic Data of the proband and her family members. Approximate normal ranges of echocardiographic data according to published sources are shown in brackets
| Member of the family | TNNI3 genetic status | Current age/sex | Age at symptom onset-examination | Height (m)/ BSA (m2) | ECG, Arrhythmia | LVEDD (mm) | IVSd (mm) | PWd (mm) | LVESD (mm) | LVEF % | LAVi (ml/m2) | RAVi (ml/m2) | E (cm/s) | A (cm/s) | E/A | DT (ms) | Accompanying disease/habits (therapy) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II1 | Hom | 49 M | 42 | 1.75/2.01 | SR, incomplete RBBB, LV strain | 47 [42–58] | 12 [6–10] | 10 [6–10] | 30 [25–40] | 60 [52–72] | 53.7 [16–34] | 38.8 [11–39] | 54 [40–104] | 71 [32–92] | 0.77 [0.60–1.84] | 190 [81–293] | HCM, NYHA II, presyncope, Dyslipidemia, smoker (acetylsalicylic acid, rosuvastatin) |
| Proband II2 |
Hom | 52F | 41 | 1.60/1.49 | AF, LVH, LV strain | 45 [38–52] | 7 [6–9] | 7 [6–9] | 31 [22–35] | 50 [54–74] | 127.5 [16–34] | 100.7 [9–33] | 60 [43–111] | na (AF) | na (AF) | 110 [109–268] | RCM, NYHA III-IV (acenocumarol, metoprolol tartate, furosemide, amiloride) |
| II3 | Hom | 52F | 45 | 1.60/1.59 | SR, incomplete RBBB, LV strain | 44 [38–52] | 8 [6–9] | 8 [6–9] | 30 [22–35] | 55 [54–74] | 94.3 [16–34] | 47.2 [9–33] | 65 [43–111] | 24 [35–91] | 2.71 [0.4–2.12] | 104 [109–268] | RCM, NYHA II (furosemide) |
| II4 | Het | 55F | 1.65/1.79 | SR | 42 [38–52] | 7 [6–9] | 7 [6–9] | 26 [22–35] | 55 [54–74] | 33.5 [16–34] | 27.9 [9–33] | 63 [43–111] | 75 [35–91] | 0.87 [0.4–2.12] | 300 [109–268] | Asymptomatic | |
| I1 | Het | 75 M | 1.70/1.94 | SR | 46 [42–58] | 10 [6–10] | 10 [6–10] | 27 [25–40] | 65 [52–72] | 23.2 [16–34] | 23.7 [11–39] | 60 [37–97] | 75 [41–105] | 0.80 [0.42–1.50] | 280 [78–357] | Asymptomatic Dyslipidemia (rosuvastatin) |
|
| I2 | Het | 72F | 1.70/2.00 | SR | 41 [38–52] | 10 [6–9] | 9 [6–9] | 26 [22–35] | 55 [54–74] | 35 [16–34] | 27.9 [9–33] | 50 [38–106] | 75 [44–108] | 0.67 [0.37–1.61] | 310 [90–313] | Asymptomatic Increased apical trabeculations, AH, Dyslipidemia (olmesartan, rosuvastatin, acetylsalicylic acid) |
|
| III2 | Het | 13F | 1.61/1.45 | SR | 48 [38–52] | 7 [6–10] | 6 [6–10] | 27 [22–35] | 60 [54–74] | 33.0 [18–34] | 14.9a [9.5–19.3] | 100 [50–118] | 70 [27–75] | 1.43 [0.68–2.76] | 190 [97–257] | Asymptomatic | |
| III3 | Het | 21 M | 1.78/1.97 | SR | 50 [42–58] | 8 [6–10] | 9 [6–10] | 34 [25–40] | 55 [52–72] | 25.4 [16–34] | 22.3 [11–39] | 80 [51–107] | 45 [24–76] | 1.78 [0.65–2.73] | 195 [87–273] | Asymptomatic |
a for pediatric patient III2 Right Atrial Area is documented instead of RAVi
A: maximal velocity of the late atrial component of transmitral blood flow, AF: Atrial Fibrillation, AH: Arterial Hypertension, BSA: Body Surface Area calculated with Mosteller formula, DT: Deceleration Timeof early transmitral flow (time from maximal velocity to zero), E: maximal early transmitral blood flow velocity, ECG: ElectroCardioGram, HCM: Hypertrophic Cardiomyopathy, Het: heterozygous, Hom: homozygous, IVSd: Interventricular Septum Diameter, LAVi: Left Atrial Volume indexed to BSA, LV: Left Ventricle, LVEDD: Left Ventricular End Diastolic Diameter, LVEF: LV Ejection Fraction, LVESD: Left Ventricular End Systolic Diameter, LVH: Left Ventricular Hypertrophy, na: not applicable, NYHA: New York Heart Association class, PWd: Posterior Wall Diameter, RAVi: Right Atrial Volume indexed to BSA, RBBB: Right Bundle Branch Block, RCM: Restrictive Cardiomyopathy, SR: Sinus Rhythm
Abnormal values are shown in bold letters
Fig. 2.
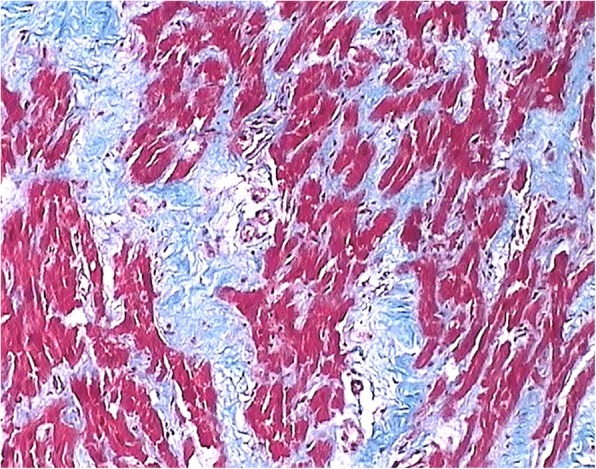
The histology of the affected myocardium revealed multiple foci of intestitial and pericellurar fibrosis with features of non-specific myocytic hypertrophy. There was no evidence of inflammation or deposition of amyloid. (trichrome stain Χ100)
The proband’s twin sister (patient II3), although initially asymptomatic, underwent screening exercise stress test and cardiac ultrasound at the age of 43. The stress test, despite LV strain findings on the resting electrocardiogram, was substantially negative for heart ischemia. The cardiac ultrasound revealed normal LV dimensions (interventricular septum 8 mm, posterior wall 8 mm, LV end-diastolic diameter 44 mm, LV end-systolic diameter 30 mm), systolic function (LVEF 55%) and left atrial enlargement (left atrial volume 94.3 ml/m2). Doppler echocardiography showed restrictive pattern of diastolic transmitral flow (early diastolic transmitral blood flow velocity E higher than late atrial contraction-induced velocity A, E/A > 2, decreased deceleration time of E: DT 120 ms), and increased right ventricular systolic pressure (55 mmHg), as well as mild mitral and tricuspid valve regurgitation. At the age of 45, she started complaining for exertional dyspnea. A new cardiac ultrasound confirmed the previously reported findings with similar E/A ratio and even lower DT (104 ms, Table 1). She underwent right heart catheterization, which confirmed restrictive diastolic filling pattern and pulmonary hypertension. As a result, the diagnosis of a form of restrictive cardiomyopathy was established and the patient was managed medically (low doses of furosemide per os) remaining stable with mild symptoms. Recently, at the age of 51, her symptoms were aggravated due to the occurrence of atrial fibrillation.
Their brother (patient II1) was admitted to hospital at the age of 42 due to recurrent pre-syncope and exertional dyspnea. His electrocardiogram revealed sinus rhythm with incomplete Right Bundle Branch Block and nonspecific repolarization abnormalities. His echocardiographic study was remarkable for asymmetric hypertrophy of the interventricular septum and papillary muscles, left atrial enlargement (milder than his affected sisters’), mild mitral regurgitation and diastolic dysfunction without restrictive pattern (E < A, normal DT), while LV size and function were normal (Table 1). Doppler velocity measurements in the LV outflow tract were normal (absence of outflow tract obstruction). The above findings were consistent with HCM. The patient showed no significant epicardial vessel stenoses, but acetylsalicylic acid and rosuvastatin were initiated. He also underwent 24-h ambulatory heart rhythm recording, which was negative for ventricular arrhythmias. The patient remains stable with minimal symptoms and his cardiac ultrasound has not changed remarkably.
His parents (I1 and I2) along with one of his sisters (II4) remain asymptomatic. The echocardiogram of the proband’s mother (patient I2) presented prominent apical trabeculations, but her electrocardiogram was essentially normal. Similarly all of the children (III1–4) of patients II1 and II3 are asymptomatic. The youngest child (III1) is 5 years old and therefore has not been clinically evaluated yet. The clinical data of children III2 (age 13) and III3 (age 21) are listed in Table 1 and for the child III4 (age 21) although reported asymptomatic no data are available as he is clinically monitored in another medical center.
Genetic analysis
The molecular basis of the disease was identified for the proband with next generation sequencing technology, using Illumina’s Trusight Cardio sequencing panel, covering 174 genes clinically relevant to cardiac diseases. Alignment, quality filtering, variant calling, and variant annotation were performed using in parallel the standard MiSeq Reporter (Illumina) and the Sophia Genetics pipeline. The variant calling files were filtered using VariantStudio Data Analysis Software (Illumina, San Diego, CA) and the Sophia Genetics DDM platform and the detected variants were characterized according to the recommendations of the American College of Medical Genetics and Genomics (ACMG) [6]. All benign or likely benign variants were filtered out and the retained variants were then evaluated according to the relevance of the gene to the observed phenotype resulting in one plausible candidate variant which was located in the TNNI3 gene: NM_000363.4:c.586G > C, p.(Asp196His). The variant (https://databases.lovd.nl/shared/individuals/00204288) was novel and characterized as likely pathogenic (according to ACMG criteria). It was absent from the population databases of Exome Sequencing Project and Genome Aggregation Database (PM2 criterion) and pathogenicity predictions were all in favor of a damaging effect of this amino acid substitution (PP3 criterion) (SIFT: 0.001, PolyPhen-2 HumVar: 0.948, Mutation Taster: 1.000 and CADD v1.3: 32). The variant affected a highly conserved nucleotide (phyloP: 4.56 [− 14.1;6.4]) and a highly conserved amino acid (Consurf: 8, [1–9]) and was located in a region where mutations of nearby residues support its functional importance [7], while missense variants in TNNI3 are a common mechanism of RCM (PP2 criterion). Additionally, another amino-acid missense variant at this position, p.Asp196Asn, is classified as likely pathogenic using ACMG criteria (PM5 criterion). Surprisingly, the proband was found homozygous for this variant but it was the second child of parents that are third cousins and originate from the same small mountainous village in Peloponnese, Greece (Fig. 1). The family of the proband underwent TNNI3 genetic testing by targeted Sanger sequencing for the detection of the variant. Her twin sister and her brother were found homozygous for the variant, while both parents and the older sister were found heterozygous (Fig. 1).
Discussion and conclusions
The C-terminal of troponin I (amino acids 184–210 in human cardiac TnI) is the most conserved structure of the molecule and interacts with tropomyosin in a calcium-regulated manner, suggesting a functional role. Despite its degree of conservation this region is highly flexible with no stable secondary structure [8] supporting the hypothesis that it is a dynamic structure in the troponin functions. The end segment of cTnI (amino acids 190–210) has been shown to play a role in the stabilization of tropomyosin in the actin filament upon Ca2+ activation [9]. In vivo data support that mutations in this region cause myofibril Ca2+ hypersensitivity and subsequent impaired relaxation, which is the main manifestation of RCM [10], suggesting that this region may constitute a mutational hotspot. Several variants located in this C-terminus end segment of the protein have been associated with HCM, while three of them (p.Asp190Gly, p.Arg192His and p.Arg204His) have been also reported to be associated with RCM as well [11]. In fact, the genetic investigation of a large family has revealed that carriers of the same variant (p.Asp190Gly) displayed considerable phenotypic heterogeneity with most of them fulfilling the HCM diagnostic criteria, while some of them were diagnosed with RCM [5]. Additionally, variants affecting the same amino acid position have been reported to confer to either the same (i.e. p.Arg192His and p.Arg192Cys are both associated with RCM) [5, 12] or different phenotypes (i.e. p.Leu144Gln is associated to RCM while p.Leu144Pro is associated to HCM) [5, 13].
Genetic analysis of our proband that presented a severe restrictive cardiomyopathy phenotype leading to heart transplantation at the age of 41, revealed a novel TNNI3 variant at homozygous state: NM_000363.4:c.586G > C, p.(Asp196His). A different variant located at the same amino acid position i.e. NM_000363.4:c.586G > A, p.(Asp196Asn) has been repeatedly associated with HCM [14–16], while recently the variant p.Asp196Gly was detected in a patient diagnosed with atrial fibrillation [17]. The parents of our proband are third-degree cousins originating from a small mountainous village in Peloponnese, Greece and are asymptomatic carriers. Homozygous TNNI3 mutations have been associated with recessive forms of DCM [18] and HCM [19] but not RCM so far.
Restrictive cardiomyopathy is considered to be inherited in an autosomal dominant way, although recently Ploski et al. [20] have reported that TNNC1 is a likely novel gene for autosomal recessive restrictive cardiomyopathy. In this family, all heterozygous carriers were asymptomatic pointing to a recessive form of the disease, but it is important to stress out that in families with restrictive cardiomyopathy inherited as dominant trait, penetrance is reduced and age dependent, so the phenotype of generation III may have not reached maturity (Fig. 1) [11]. Nevertheless, this novel variant seems less pathogenic compared to the other amino acids substitutions observed at the same position. This may reflect subtle functional differences of the different amino acids, as it has been shown for mutations at position 204, where p.Arg204His mutation affected interaction of cTnI with troponin T and troponin C, while p.Arg204Cys affected interaction with troponin C but not troponin T and both produced increased Ca2+ sensitivity [21]. Moreover, one heterozygous carrier (I2), despite being asymptomatic with normal LV systolic and diastolic function, exhibits an echocardiographic LV morphology resembling noncompaction cardiomyopathy. As TNNI3 mutations have been reported in noncompaction cardiomyopathy patients [22], a partially expressed phenotype even in heterozygous state cannot be firmly excluded. Of course, increased apical trabeculations may be a random finding unrelated to the specific mutation. Furthermore, the observed phenotypic heterogeneity in the three homozygous patients support the action of modifying genetic and/or environmental mechanisms.
Acknowledgements
We would like to thank the proband and her family for allowing us to present their results.
Funding
This work was supported by Onassis Cardiac Surgery Center and BioAnalytica Genotypes SA who provided the necessary reagents to complete the experimental part of the study.
Availability of data and materials
All data from this study that do not pertain to identifiable patient information are available and can be provided by contacting the corresponding author on reasonable request. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The identified mutation has been submitted to the corresponding LOVD database (https://databases.lovd.nl/shared/individuals/00204288).
Abbreviations
- ACMG
American College of Medical Genetics and Genomics
- HCM
Hypertrophic cardiomyopathy
- LV
Left ventricular
- LVEF
Left ventricular ejection fraction
- RCM
Restrictive cardiomyopathy
Authors’ contributions
MPP and PG participated in research design, data analysis and interpretation and writing of the manuscript, AG and IA participated in sample collection, were responsible for patient management and follow-up and provided clinical data, LK provided histological data of the native heart, CZ participated in the execution of the experiments, PC participated in research design and provided financial support, SA participated in research design and patient management, DD participated in research design and supervised the course of the project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent for molecular genetic testing was obtained from all members of the family, and the study was approved by the Ethics Committee of the Onassis Cardiac Surgery Center.
Consent for publication
Written informed consent was obtained from the patients for the publication of this report.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Malena P. Pantou, Email: pantou@ocsc.gr
Polyxeni Gourzi, Phone: 0030 210 9493023, Email: gourzi@ocsc.gr.
Aggeliki Gkouziouta, Email: agkouziouta@yahoo.gr.
Iakovos Armenis, Email: iakovosarm@hotmail.com.
Loukas Kaklamanis, Email: loukasgka@yahoo.gr.
Christianna Zygouri, Email: christianna_@hotmail.com.
Pantelis Constantoulakis, Email: pconstantoulakis@genotypos.gr.
Stamatis Adamopoulos, Email: stamatis.adamo@gmail.com.
Dimitrios Degiannis, Email: degianis@ocsc.gr.
References
- 1.Hughes SE, McKenna WJ. New insights into the pathology of inherited cardiomyopathy. Heart. 2005;91:257–264. doi: 10.1136/hrt.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menon SC, Michels VV, Pellikka PA, Ballew JD, Karst ML, Herron KJ, Nelson SM, Rodeheffer RJ, Olson TM. Cardiac troponin T mutation in familial cardiomyopathy with variable remodeling and restrictive physiology. Clin Genet. 2008;74:445–454. doi: 10.1111/j.1399-0004.2008.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huby AC, Mendsaikhan U, Takagi K, Martherus R, Wansapura J, Gong N, Osinska H, James JF, Kramer K, Saito K, Robbins J, Khuchua Z, Towbin JA, Purevjav E. Disturbance in Z-disk mechanosensitive proteins induced by a persistent mutant myopalladin causes familial restrictive cardiomyopathy. J Am Coll Cardiol. 2014;64:2765–2776. doi: 10.1016/j.jacc.2014.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodehl A, Ferrier RA, Hamilton SJ, Greenway SC, Brundler MA, Yu W, Gibson WT, McKinnon ML, McGillivray B, Alvarez N, Giuffre M, Schwartzentruber J, FORGE Canada Consortium. Gerull B. Mutations in FLNC are associated with familial restrictive cardiomyopathy. Hum Mutat. 2016;37:269–279. doi: 10.1002/humu.22942. [DOI] [PubMed] [Google Scholar]
- 5.Mogensen J, Kubo T, Duque M, Uribe W, Shaw A, Murphy R, Gimeno JR, Elliott P, McKenna WJ. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–216. doi: 10.1172/JCI200316336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akhter S, Bueltmann K, Jr, Huang X, Jin JP. Restrictive cardiomyopathy mutations demonstrate functions of the C-terminal end-segment of troponin I. Arch Biochem Biophys. 2014;552(553):3–10. doi: 10.1016/j.abb.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Julien O, Mercier P, Allen CN, Fisette O, Ramos CH, Lagüe P, Blumenschein TM, Sykes BD. Is there nascent structure in the intrinsically disordered region of troponin I? Proteins. 2011;79:1240–1250. doi: 10.1002/prot.22959. [DOI] [PubMed] [Google Scholar]
- 9.Galińska A, Hatch V, Craig R, Murphy AM, Van Eyk JE, Wang CL, Lehman W, Foster DB. The C terminus of cardiac troponin I stabilizes the Ca2+−activated state of tropomyosin on actin filaments. Circ Res. 2010;106:705–711. doi: 10.1161/CIRCRESAHA.109.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Charles PY, Nan C, Pinto JR, Wang Y, Liang J, Wu G, Tian J, Feng HZ, Potter JD, Jin JP, Huang X. Correcting diastolic dysfunction by Ca2+ desensitizing troponin in a transgenic mouse model of restrictive cardiomyopathy. J Mol Cell Cardiol. 2010;49:402–411. doi: 10.1016/j.yjmcc.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogensen J, Hey T, Lambrecht S. A systematic review of phenotypic features associated with cardiac troponin I mutations in hereditary cardiomyopathies. Can J Cardiol. 2015;31:1377–1385. doi: 10.1016/j.cjca.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 12.van den Wijngaard A, Volders P, Van Tintelen JP, Jongbloed JD, van den Berg MP, Lekanne Deprez RH, Mannens MM, Hofmann N, Slegtenhorst M, Dooijes D, Michels M, Arens Y, Jongbloed R, Smeets BJ. Recurrent and founder mutations in the Netherlands: cardiac troponin I (TNNI3) gene mutations as a cause of severe forms of hypertrophic and restrictive cardiomyopathy. Neth Heart J. 2011;19:344–351. doi: 10.1007/s12471-011-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippi G, Targher G, Franchini M, Plebani M. Genetic and biochemical heterogeneity of cardiac troponins: clinical and laboratory implications. Clin Chem Lab Med. 2009;47:1183–1194. doi: 10.1515/CCLM.2009.322. [DOI] [PubMed] [Google Scholar]
- 14.Mogensen J, Murphy RT, Kubo T, Bahl A, Moon JC, Klausen IC, Elliott PM, McKenna WJ. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:2315–2325. doi: 10.1016/j.jacc.2004.05.088. [DOI] [PubMed] [Google Scholar]
- 15.Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 16.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M, EUROGENE Heart Failure Project Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Wu M, Qian J, Li B, Tu X, Xu C, Li S, Chen S, Zhao Y, Huang Y, Shi L, Cheng X, Liao Y, Chen Q, Xia Y, Yao W, Wu G, Cheng M, Wang QK. Identification of rare variants in TNNI3 with atrial fibrillation in a Chinese GeneID population. Mol Gen Genomics. 2016;291:79–92. doi: 10.1007/s00438-015-1090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2014;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 19.Gray B, Yeates L, Medi C, Ingles J, Semsarian C. Homozygous mutation in the cardiac troponin I gene: clinical heterogeneity in hypertrophic cardiomyopathy. Int J Cardiol. 2013;168:1530–1531. doi: 10.1016/j.ijcard.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Ploski R, Rydzanicz M, Ksiazczyk TM, Franaszczyk M, Pollak A, Kosinska J, Michalak E, Stawinski P, Ziolkowska L, Bilinska ZT, Werner B. Evidence for troponin C (TNNC1) as a gene for autosomal recessive restrictive cardiomyopathy with fatal outcome in infancy. Am J Med Genet A. 2016;170:3241–3248. doi: 10.1002/ajmg.a.37860. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen S, Siu R, Dewey S, Cui Z, Gomes AV. Amino acid changes at arginine 204 of troponin I result in increased calcium sensitivity of force development. Front Physiol. 2016;7:520. doi: 10.3389/fphys.2016.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujino M, Tsuda E, Hirono K, Nakata M, Ichida F, Hata Y, Nishida N, Kurosaki K. The TNNI3 Arg192His mutation in a 13-year-old girl with left ventricular noncompaction. J Cardiol Cases. 2018;18:33–36. doi: 10.1016/j.jccase.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from this study that do not pertain to identifiable patient information are available and can be provided by contacting the corresponding author on reasonable request. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The identified mutation has been submitted to the corresponding LOVD database (https://databases.lovd.nl/shared/individuals/00204288).