Abstract
High and low-density lipoproteins (HDL and LDL) are attractive targets for biomarker discovery. However, ultracentrifugation (UC), the current methodology of choice for isolating HDL and LDL is tedious, requires large sample volume, results in sample loss, and does not readily provide information on particle size. In this work, human plasma HDL and LDL are separated and collected using semi-preparative asymmetrical flow field-flow fractionation (SPAF4) and UC. The SP-AF4 and UC separation conditions, sample throughput, and liquid chromatography/mass spectrometry (LC/MS) lipidomic results are compared. Over 600 μg of total proteins are recovered in a single SP-AF4 run and Western blot results confirm apoA1 pure and apoB100 pure fractions, consistent with HDL and LDL, respectively. The SP-AF4 separation requires ~60 minutes per sample, thus providing a marked improvement over UC which can span hours to days. Lipidome analysis of SP-AF4 prepared HDL and LDL fractions are compared to UC prepared HDL and LDL samples. Over 270 lipids in positive MS mode and over 140 lipids in negative MS mode are identified by both sample preparation techniques with over 98% overlap between the lipidome. Additionally, lipoprotein size distributions are determined using analytical scale AF4 coupled with multiangle light scattering (MALS) and dynamic light scattering (DLS) detectors. These developments position SP-AF4 as a sample preparation method of choice for lipoprotein biomarker characterization and identification.
Keywords: Asymmetrical Flow Field-Flow Fractionation, Ultracentrifugation, Lipoproteins, Lipidomics, Mass Spectrometry
Graphical Abstract
INTRODUCTION
Lipoproteins are complexes of lipids and proteins responsible for transporting lipids through the blood stream. The role of lipoproteins in cardiovascular disease (CVD) has been well studied and lipoprotein blood levels are commonly used to assess cardiovascular health.[1] Lipoprotein composition, concentration, and size have been suggested or used as risk factors for CVD, but may also be useful as risk factors for other diseases such as chronic kidney disease (CKD). However, lipoproteins exist over a range of compositions and sizes making their characterization challenging.[2] Additionally, lipoprotein biomarkers may be present in low abundances further complicating screening and analysis.[3] Fast and accurate measurements of these properties and low abundance biomarkers are important for diagnosing and predicting disease. Lipoproteins have distinct subclasses based on their density: high density lipoproteins (HDL) and low density lipoproteins (LDL). Separating different subclasses from blood plasma is an important step for understanding the correlation between specific lipoprotein characteristics and disease.[4]
Ultracentrifugation (UC) is one of the primary techniques for isolating lipoproteins for clinical studies.[5] However, there are questions about the precision of the UC technique and the stability of lipoproteins during analysis.[6–9] Often UC lipoprotein separations require large sample volumes and long ultracentrifugation duration (up to 30 hours) to isolate each subclass and rely on high potassium bromide concentrations (~3 M) to create the appropriate density gradients.[6] Such a long duration of UC can be detrimental to the stability of the lipoprotein particle component proteins and lipids. Also, the high salt concentration may cause unwanted aggregation of LDL.[10] Additionally, UC studies have reported that spin rates > 30,000 rpm result in lower particle densities and increased dissociation of apoA1 from human lipoproteins.[6] Another study of human lipoproteins found that apoA1 was lost during each UC spin until one-third of the apoA1 was lost with five spins.[7] Interestingly increasing ionic strength was found to decrease apoA1 dissociation from HDL while reducing ionic strength decreased dissociation of other exchangeable apolipoproteins.[8] However, reducing ionic strength still requires high centrifugal force. Therefore, faster methods capable of separating and characterizing lipoproteins in human plasma that utilize smaller sample volumes, and do not use high centrifugal force or ionic strengths are needed to decrease analysis time and avoid potential artifacts.
Electrophoresis [11] and size exclusion chromatography (SEC) [12] have been used to separate lipoprotein subclasses. However, interactions with the chromatography packing material and restrictions on the buffer types that can be used are challenges still being addressed.[13] Analytical scale flow field-flow fractionation (FlFFF) has been shown capable of separating lipoproteins by size without significant packed column interactions.[14] It also provides flexibility in buffer composition and concentration. The capability of symmetrical FlFFF (SyFlFFF) to separate lipoproteins from human plasma was demonstrated over 20 years ago.[15] Recent work has focused on the development of two other F1FFF variants, asymmetrical flow field-flow fractionation (AF4) and hollow fiber flow field-flow fractionation (HF5). The AF4 technique provides benefits over SyF1FFF including higher resolution and faster separation speeds[16] while the HF5 variant also showed similar performance to AF4.[17, 18] Lipoprotein composition information has been obtained using a miniaturized chip type AF4 channel coupled with electrospray ionization-tandem mass (ESI-MS/MS) spectrometry [19] for top-down lipidomic analyses of lipoproteins. Lipoproteins were also used in a multivariate design of experiment approach to optimize AF4 separation conditions for coupling with LC-MS/MS.[20] These prior FFF studies typically utilized 10–80 μL of human plasma and often require the pooling of fractions collected from as many as 16 FFF separations prior to additional analyses. [21–26]
Semi-preparative flow FFF separation has been implemented by connecting six parallel HF5 modules in a multiplexed configuration (MxHF5).[21] This led to an increase of bovine serum albumin sample loading from 8 μg for a single HF5 module to ~50 μg for the MxHF5 and enabled the subsequent analysis of 50 μL of human plasma. However, two subsequent 50 μL injections of human plasma (100 μL total) were needed to produce the amount of lipoproteins suitable for bottom-up lipidomic analyses by nanoflow liquid chromatography ESI-MS/MS (nLC-ESI-MS/MS).[22, 23] The use of MxHF5 requires reproducible characteristics and performance of each individual hollow fiber. While the retention time was the same, a 56% increase in band broadening was observed over that of a single HF5 module.[21] This, along with the unavailability of commercial MxHF5, led to the recent investigation of semi-preparative AF4 (SP-AF4) channel dimensions and performance.[27, 28] In comparison to analytical scale AF4, SP-AF4 uses larger channel breadths or thicknesses to increase the channel volume allowing for larger sample quantities to be introduced into the channel without overloading. Optimized SP-AF4 channels were able to handle milligram (up to 20 mg of 50 and 100 nm silica particles) quantities and are expected to provide an important capability for studies aimed at detection of low abundance biomarkers. Additionally, increasing lipoprotein sample loadings can enable different size subpopulations within each lipoprotein subclass to be collected for future studies of size-dependent lipidomics. To date, none of the flow FFF variants have been compared to UC as sample preparation methods for lipidomics analyses. This is an important step for bringing AF4 into common use for biomarker discovery.
In this work, we develop a semi-preparative AF4 method to separate and collect lipoproteins from human plasma. Our initial focus is on HDL and LDL only because these lipoprotein classes have been most widely used for disease risk factors and existing knowledge can be used to validate SP-AF4. Objectives included 1) collection of HDL and LDL fractions under physiological conditions using SP-AF4, 2) comparison of SP-AF4 and UC separation conditions and sample recovery, and 3) comparison of lipidomics results from SP-AF4 and UC sample preparations. Additionally, analytical-scale AF4-MALS-DLS is used to separate and characterize the size distribution of LDL in human plasma.
MATERIALS AND METHODS
Standards, Samples, and AF4 Carrier Fluid
Bovine serum albumin (BSA) and thyroglobulin were from Sigma-Aldrich (St. Louis, MO), and intravenous immunoglobulin (IVIg) was provided by the School of Pharmacy and Pharmaceutical Sciences, University of Colorado Denver, Anschutz Medical Campus (Aurora, CO). Human plasma sample was obtained in mass quantity from the donors' pool of over 1000 de-identified blood donors from the American Red Cross for research purpose.
Prior to isolation of LDL and HDL fragments, the total LDL and HDL concentration of pooled human plasma samples were measured in a two-step process using the Randox (RX) Laboratories (Kearneysville, WV) protocols. In the first step, we used cholesterol esterase, cholesterol oxidase and catalase aimed at degrading other lipoproteins in the presence of specific detergents that protect either the LDL or HDL particles from degradation. This allowed the subsequent measurement of the cholesterol concentration only from the remaining LDL or HDL particles in the second step using RX Series CH 3841 protocol to measure LDL and the RX Series CH 3811 protocol to measure HDL.
Then we isolated HDL and LDL particles from pooled human plasma in to separate fractions by a multi-step UC gradient based procedure as described previously.[29, 30] After isolation of HDL and LDL fractions, the concentrations of the lipoproteins obtained from UC method were measured by Bradford protein assay using bovine serum albumin (BSA) (SigmaAldrich) and Coomassie Brilliant Blue G-250 (Sigma-Aldrich), creating standard solutions for a calibration curve, and measuring the absorbance at 595 nm.[31] These UC-prepared HDL and LDL standards of known concentrations are stored at −80° C and thawed at room temperature prior to use, analyzed within 24 hours, and thawed only once.
Pooled human plasma (HP) samples were stored at −80° C and brought to room temperature prior to use. Samples prepared for AF4 analysis were either depleted of human serum albumin (HSA) and IgG or un-depleted. Depleted pooled human plasma (HP-D) samples were prepared by removing human serum albumin (HSA) and IgG following the protocol for a ProteoPrep® Immunoaffinity albumin and IgG depletion column (PROTIA-1KT; Sigma-Aldrich, St. Louis, MO). The initial 50 μL (the largest amount recommended for the ProteoPrep® column) HP sample resulted in an HP-D sample with a total volume of 225 μL in a proprietary “low ionic strength” pH 7.4 Tris buffer. Un-depleted HP samples were prepared for SP-AF4 and analytical AF4 analysis by diluting 50 μL of human plasma to a total volume of 225 μL of AF4 carrier fluid. Samples were analyzed within 24 hours of dilution.
The carrier fluid for all AF4 separations is a 50 mM phosphate buffer prepared by mixing 4.36 g of Na2HPO4 and 2.66 g NaH2PO4 ∙ H2O in 1 L of distilled deionized 18.2 MΩ water (APS Filtration AquaMax, Van Nuys, CA) resulting in a measured pH of 7.05.
Semi-Preparative AF4
An AF4 channel (Postnova Analytics Inc., Salt Lake City, UT) with a homemade trapezoidal spacer, two LC-6A pumps (Shimadzu, Japan), and a Spectra 100 UV/Vis detector (Spectra-Physics, Santa Clara, CA) set at 280 nm formed the semi-preparative AF4 system. The spacer had a nominal thickness of 350 μm, maximum breadth of 5 cm, minimum breadth of 1 cm, and a tip-to-outlet length of 27.6 cm. A 30 kDa molecular weight cutoff regenerated cellulose membrane (Microdyn-Nadir, Wiesbaden, Germany) formed the semi-permeable accumulation wall. The outlet flow rate (⩒out) was 0.5 mL/min and the cross-flow rate (⩒c) was set to 2.0 mL/min and decreased to 0.5 mLmin at 15 minutes. The focus time was 5 minutes with an injection flow rate (⩒vnj) of 0.2 mL/min. The injected sample volume was 200 μL. Sample recovery is calculated as a ratio of the area under the peak for a retained sample to that for an unretained sample (i.e., injected into the SP-AF4 channel with no focusing or crossflow).
Analytical AF4-MALS-DLS
An AF2000 system (Postnova Analytics Inc., Salt Lake City, UT) was used for the analytical scale AF4 separations. The AF4 system was coupled to a SPD-20A UV/Vis detector (Shimadzu, Japan), a HELEOS II multi-angle light scattering (MALS) with a dynamic light scattering (DLS) detector (Wyatt Technology Corporation, Santa Barbara, CA), and an Optilab T-rEX differential refractive index detector (Wyatt Technology Corporation, Santa Barbara, CA). The AF4 spacer had a nominal thickness of 350 μm, maximum breadth of 2 cm and a minimum breadth of 0.45 cm with a tip to outlet length of 29.3 cm. The semi-permeable accumulation wall is formed using a 30 kDa molecular weight cutoff regenerated cellulose membrane (Microdyn-Nadir, Wiesbaden, Germany). The focusing time was 5 minutes, the ⩒inj was 0.2 mL/min, the sample injection volume of 20 μL, and the ⩒out was 0.5 mLmin. The ⩒c was programmed using the flow rates provided in Fig. S1 (see Electronic Supplementary Material (ESM)).
Mass Spectrometry
Details of liquid chromatography-mass spectrometry (LC-MS) based lipidomic analysis is published elsewhere.[32] In brief, the modified Bligh-Dyer method[33] is used to prepare samples. The lipoprotein fractions collected by SP-AF4 and lipoproteins isolated by UC are brought to room temperature. Lipids are extracted using water/methanol/dichloromethane (2:2:2, v/v/v), followed by spiking with 10 picomole of the MS grade lipid internal standards listed in the ESM. The dichloromethane layer is collected and dried under nitrogen. The dry extract is reconstituted using 100 μL of acetonitrile/water/isopropyl alcohol with a 10:5:85 ratio containing 10mM NH4OAc followed by injection into MS. A Shimadzu CTO-20A Nexera X2 UHPLC with Waters Acquity UPLC HSS T3 1.8 μm column (Waters, Milford, MA) and a AB SCIEX quadrupole time of flight (TOF)-5600 equipped with a Turbo VTM ion source (AB SCIEX, Concord, Canada) is used for LC/MS analyses. The mobile phase consisted of acetonitrile/water (40:60 v/v) with 10 mM ammonium acetate (solvent A) and acetonitrile/water/isopropanol (10:5:85 v/v) with 10 mM ammonium acetate (solvent B). The instrument is set to perform one time-of-flight MS survey scan (150 ms) and 15 MS/MS scans controlled by data dependent acquisition function with a total duty cycle time of 2.4 s in both positive and negative modes. The mass range in both modes was 50–1200 m/z. The mass accuracy of internal standards determined the m/z width for the chromatogram extraction and was set to 0.001 Da for positive mode and 0.005 Da for negative mode with overall mass error of less than 2 parts per million. The MS/MS data is used to facilitate identification, and MS1 for quantification. All the MS runs on all prepared samples are done in triplicate.
RESULTS AND DISCUSSION
Optimization of Semi-preparative (SP-AF4) Separation
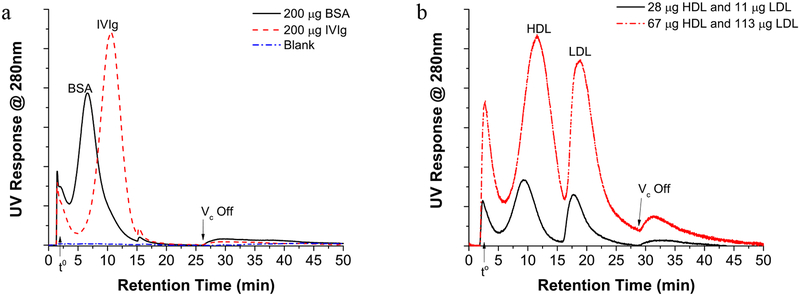
The performance of the SP-AF4 channel is evaluated and optimized by using protein standards (BSA and IVIg), and a mixture of UC prepared HDL and LDL standards (Fig. 1). Albumin protein and IVIg are chosen because they are major components of human plasma and their sizes are well suited for SP-AF4 optimization. A simple program using a single cross-flow rate ⩒c reduction from 2.0 mLmin to 0.5 mLmin at 15 minutes serves to decrease the analysis time and increase sample recovery for the LDL standard (discussed below). Fig. 1a shows fractograms obtained for individual injections of 200 μg BSA and 200 μg IVIg. Using AF4 theory and the Stokes-Einstein equation (Eqn S-1 and S-2), the retention time tr at peak maximum of 6.6 minutes (BSA) and 10.6 minutes (IVIg) minutes correspond to hydrodynamic sizes of 7 nm and 11 nm, respectively, which are in line with literature values.
Fig. 1.
Fractograms of a BSA and IVIg and b HDL and LDL standard mixtures. The void time (t0) and the time when the ⩒c is turned off are shown on each fractogram and focusing step is not shown
The purpose of using SP-AF4 is to allow for semi-preparative sample loads (>100 μg) to be processed. In human plasma, the high end concentrations of HDL and LDL are estimated to be ~0.6 μg/μL and ~2 μg/μL, respectively.[34] A 50 μL human plasma sample is thus estimated to have ~ 30 μg HDL and 100 μg LDL. Hence, SP-AF4 experiments are done with 39 μg (low) and 180 μg (high) total lipoprotein standards to assess the effects of sample loading on the separation. (A typical sample load for analytical scale AF4 and HF5 separations is <50 μg.) The amount of total lipoprotein standard injected was determined by the Bradford assay (see Materials and Methods). Figure 1b shows the separation of HDL and LDL standards mixtures at the two different injected sample loads. The HDL standard elutes at 9.3 minutes with 91% sample recovery, which is a marked improvement compared to the < 65% typically recovered by UC.[35] This tr corresponds to an AF4-calculated spherical hydrodynamic diameter of ~10 nm and is within the expected ~7 nm to 15 nm span for HDL.[36] The cross-flow rate reduction at 15 minutes serves to decrease the analysis time and increase the recovery of LDL which elutes at 18 minutes. The LDL recovery is 43% which is an improvement over UC where recoveries are often below 20%,[37–39] but further improvement in recovery should be pursued in future work. The LDL size is not readily calculated from retention time due to the programmed cross-flow rate, but the presence of apoB100 (confirmed by Western blot, see SI) strongly suggests the presence of LDL. The peak at ~27 minutes elutes after the ⩒c is turned off. No apoA1 is detected by Western blot analysis and only trace amounts of apoB100 are detected. This peak may be due to LDL that is reversibly adsorbed to the SP-AF4 membrane or more well retained intermediate density lipoproteins and very low density lipoproteins.[40] These two lipoprotein classes are not specifically investigated because the focus of this work is on the most commonly studied risk factors, HDL and LDL.
Increasing the sample load from 39 μg to 180 μg results in an increase in retention time of ~2 minutes for HDL (11.3 minutes) and ~1 minute for LDL (19 minutes) and a decrease in resolution from 1.2 to 0.8. This lower resolution still allows HDL and LDL enriched fractions to be collected as confirmed by Western blot (results not shown). The decrease in resolution is likely due to sample overloading effects, but the cause of the increase in retention time is unclear. Separation of HDL and LDL in <60 minutes is shown to be possible for a 180 μg sample load. This is a >40 times reduction in separation time than currently used UC technique and ~ 9 times increase in sample loading over analytical scale AF4. Furthermore, AF4 separations are performed in a 50 mM phosphate buffer which is preferred for biological systems (rather than the 3 M KBr used in UC lipoprotein isolation).[6, 7, 10]
Separation of Human Plasma Lipoproteins by SP-AF4
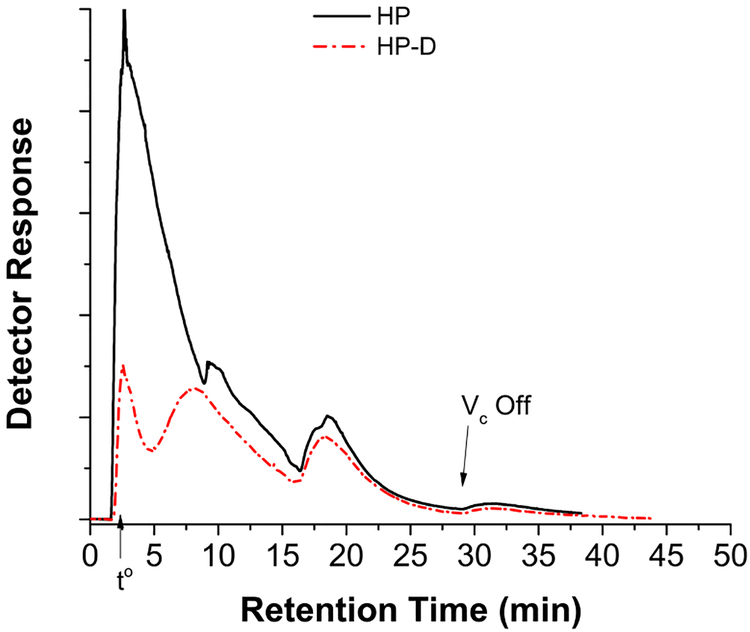
Un-depleted human plasma (HP) and HSA/IgG depleted human plasma (HP-D) samples are prepared as described in the Materials and Methods section and then fractionated by SP-AF4. The HP fractogram shown in Fig. 2 exhibits a large peak with several smaller peaks (at ~10 and 19 minutes) on the tailing edge.
Fig. 2.
Fractograms of un-depleted human plasma (HP) and HAS/IgG-depleted human plasma (HP-D) (the separation conditions are the same as Fig. 1)
The large peak is due to low molecular weight plasma proteins and/or overloaded amounts of HSA and IgG. The HSA and IgG are the major components of human plasma with concentrations of ~45 μg/μL and ~15 μg/μL, respectively. The 200 μL of 4x diluted human plasma sample separated by SP-AF4 contains ~3000 μg of HSA and IgG and ~130 μg of lipoproteins (based on apolipoprotein content). Thus, the detector response signaling the elution of this relatively small amount of lipoproteins is masked by the large amounts of co-eluting HSA and IgG. This fractogram demonstrates the need to remove HSA and IgG prior to lipoproteins separation. The ProteoPrep® column nominally removes ~95% of HSA and ~85% of IgG from a 50 μL human plasma sample. After protein depletion, the remaining amount of HSA and IgG is expected to be similar to the lipoprotein amount and more distinctive HDL and LDL peaks should be observed. This is indeed the case as demonstrated by the superimposed fractogram of a depleted plasma or HP-D sample in Fig. 2. The peak at 8 minutes is near the tr expected for HDL (9.3 minutes). The broadness and asymmetry of this peak is likely due to contributions from co-eluting HSA and IgG (trs are 6.6 and 10.6 minutes, respectively) that are not removed by the immunoaffinity column. The peak at 18.5 minutes is near the expected retention time for LDL. It is possible that other lipoprotein subclasses with larger sizes than LDL such as IDL or VLDL may partially co-elute at longer retention times or after the ⩒c is turned off.[40] The void peak at ~2.5 minutes decreases significantly after protein depletion. The peak after the ⩒c is turned off has no detectable apoA1 and contains trace levels of apoB100 (determined by Western blot).
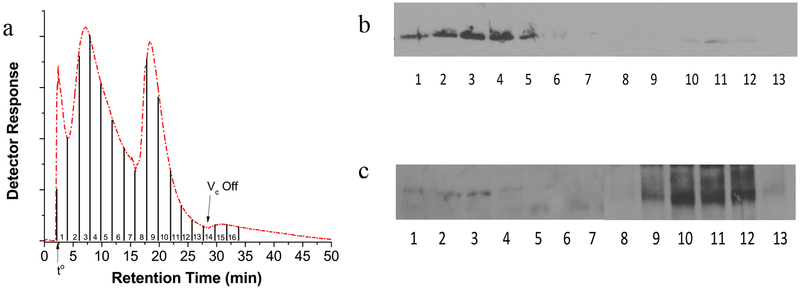
Separated HP-D fractions are collected from the detector outlet at 2 min intervals as shown in Fig. 3a. The fractions are analyzed using Western blot gels for qualitative analysis of apoA1 and apoB100 purity (Fig. 3b and 3c).
Fig. 3.
a Fractogram of HP-D separation and fraction collection by SP-AF4 and Western Blot gels for b apoA1 and c apoB100 in each fraction. Fractions 13 to 16 collected from SP-AF4 were pooled into a single sample prior to Western blot analysis and this single fraction is labeled as “13” in the apoA1 and apoB100 Western blot gels (the separation conditions are the same as Fig. 1)
Lanes 3 to 5 correspond to AF4 fractions between 6 and 12 minutes where HDL eluted (as established in Fig. 1b) and shows the presence of apoA1. Lanes 6 to 8 correspond to fractions between 12 and 18 minutes which is in-between HDL and LDL elution, so the absence of apoA1 and apoB100 is as expected. Plasma proteins not removed by the ProteoPrep® Immunoaffinity Albumin and IgG depletion column likely elute between 12 to 18 minutes. Lanes 9 to 12 correspond to AF4 fractions between 18 and 26 minutes where LDL eluted (Fig. 1b). It should be noted that apoB100 is associated with LDL, IDL, and VLDL and these classes may co-elute in the fractions 11 and 12. Further work is needed to understand the separation of these additional lipoprotein classes. Western blots also show that apoA1 is present in lanes 1 and 2 and apoB100 is present in lanes 1 to 3. The presence of both apoA1 and apoB100 in these fractions may be due to elution of smaller sized nascent preβ-HDL particles and de-lipidated apoB100 protein. It is also possible that the presence of apoA1 and apoB100 in lanes 1 to 3 is due to co-eluting small dense LDL or some HDL and LDL particles not fully equilibrating in the AF4 channel during the relaxation/focusing step resulting in early elution.
The bicinchoninic acid (BCA) assay (see SI Materials and Methods: Protein quantification) was used in determining that over 650 μg of total proteins are collected in a single SP-AF4 separation. The amount of protein in fractions 3 to 5 and 9 to 12 are ~6 to 109 μg. This is a large amount of sample compared to most AF4 studies where a total sample amount of ~20 μg is separated resulting in fractions with a maximum of only a few micrograms. Using SPAF4, the amount of protein collected from 50 μL of human plasma is similar to that of HDL and LDL obtained from UC of over 1 mL of human plasma. Additionally, the SP-AF4 separation is accomplished in under 1 hour instead of multiple hour or days, without high centrifugal force, and using buffers with lower ionic strengths reducing possible lipoprotein aggregation, damage, or apolipoprotein dissociation.
LC-MS/MS Analysis of SP-AF4 Lipoprotein Fractions
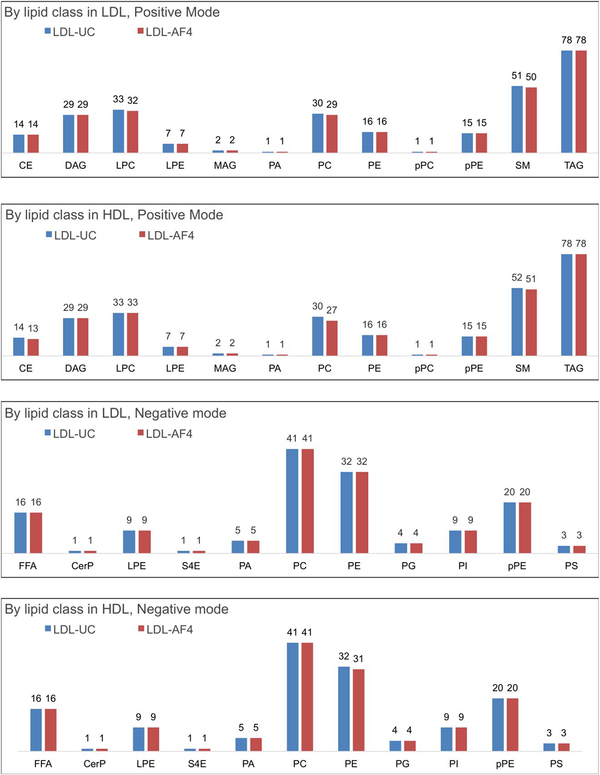
The MS determined lipidome of the UC-prepared standards and SP-AF4 prepared lipoprotein fractions are compared to show the suitability of SP-AF4 as a preparation method for HDL and LDL samples. SP-AF4 fractions 2 to 7, which are expected to contain the bulk of HDL, and fractions 9 to 13 containing the bulk of LDL are pooled prior to MS analysis to allow for a direct comparison to the UC prepared samples. The same sample amounts of SP-AF4 and UC prepared lipoprotein fractions (determined by normalizing to the protein content of each fraction) are then analyzed by LC-MS/MS. Lipid precursors identified in the first mass spectrometry (MS1), were fragmented into products in the second mass spectrometry (MS2). Specific fragmentation in MS2 allows accurate identification of unique lipids in MS1 as described previously.[32, 41] Figure 4 (ESM Table S1) shows that 277 lipids species in the LDL fraction and 278 species in the HDL fraction are identified in UC-prepared samples in positive MS mode. Similarly, 141 lipids in both LDL and HDL fractions are found in UC-prepared samples in negative mode. In comparison, over 98% of the identical lipids are found in the samples prepared by the SP-AF4 method for both the LDL and HDL fractions in positive and negative modes. The < 2% difference between UC andAF4 prepared samples is not statistically significant as this comparison is done for only one plasma sample. A head-to-head comparison of the number of lipids detected within each lipid class by lipoprotein fraction and MS mode also shows that in positive MS mode, 100% of the lipids identified in DAG, LPE, MAG, PA, PE, pPC, pPE, and TAG lipid classes of the samples prepared by the UC method are also detected in samples prepared by the SP-AF4 method. Similarly, in negative MS mode, 100% of the lipids identified in FFA, CerP, LPE, S4E, PA, PC, PG, PI, pPE, and PS lipid classes of the samples prepared by the UC method are also detected in samples prepared by the SP-AF4 method in both the LDL and HDL fractions. It should also be noted that AF4 fraction 8 which contains both HDL and LDL was not pooled and analyzed (only fractions expected to be predominantly HDL and LDL were subjected to lipidomics analysis). These lipidomics results demonstrate SP-AF4’s suitability as a preparation method for HDL and LDL from human plasma.
Fig. 4.
Comparing the number of unique lipids detected in SP-AF4 (Fractions 1 and 8 not included) and UC prepared lipoprotein fractions by tandem mass spectrometry (see ESM for definitions of lipids class acronyms)
Lipoprotein Characterization by Analytical Scale AF4-MALS-DLS
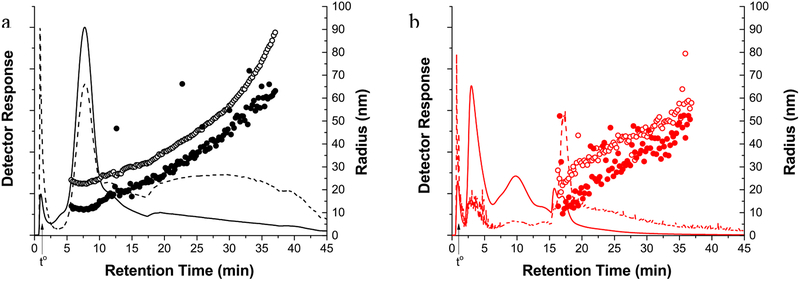
Lipoprotein size is an important physical property used to help define lipoprotein subclasses and therefore methods capable of size characterization are desirable. Analytical scale AF4 is combined with online multiangle light scattering (MALS) and dynamic light scattering (DLS) for the dual purpose of separation and size measurement of eluting components. Light scattering requires the more monodisperse samples provided by AF4 separation for accurate size measurement. The use of analytical scale AF4 reduces the amount of sample required for each injection compared to SP-AF4 while still providing acceptable light scattering intensities for size characterization. Fractograms of UC-prepared LDL standard (Fig. 5a) and HP-D samples (Fig. 5b) are superimposed with root-mean-square radius rrms values measured by MALS and hydrodynamic radius rh reported by DLS detectors.
Fig. 5.
AF4 fractograms and rrms (open circles) and rh (closed circles) distributions for a UCprepared LDL standard and b HP-D sample. The LS and UV signals are represented by solid lines and dashed lines, respectively. The ⩒out is 0.5 mL/min and initial ⩒c for the LDL and HP-D separations are 0.75 mL/min and 2.00 mL/min, respectively. The ⩒c is decreased at 15 mins using the profiles shown in Fig. S1 (see ESM)
In Fig. 5a, the peak eluting at 7.5 minutes is likely LDL based on the rh size distribution that falls in the expected LDL radius range (~11.5 to 13.5 nm). This peak represents ~55% of the UC prepared LDL sample based on the area under the UV signal. The rrms size distribution is significantly higher than the rh distribution, a difference that will be discussed in the following section. The sample components eluting between 17 to 35 minutes have an rh distribution from ~20 nm to ~60 nm. This size range is consistent with very low density lipoprotein (VLDL) which is ~20 nm to 45 nm or LDL aggregates.[10, 36] The noisy rh and rrms data is due the lower overall MALS and DLS signal intensities. The void peak at ~1 minute has a relatively high light scattering signal compared to the UV signal which is consistent with the elution of large unretained sample components.
A higher initial ⩒c is used for the separation of the HP-D sample shown in Fig. 5b so that LDL is separated from other components present in the sample. The peak at 17.5 minute is designated as LDL because the rh and rrms sizes are similar to those measured for the LDL standard in Fig. 5a. The species eluting from ~2.5 to 16 minutes are likely HSA, HDL, IgG, and other human plasma components smaller than LDL as confirmed by rh values (~3 nm to 12 nm) calculated using AF4 retention theory (see ESM Fig. S2). Determination of rh and rrms from light scattering for this rh region is not possible due to the low signal/noise ratio and because rrms cannot be determined for analytes <10 nm in diameter. In comparison to Fig. 5a, the rh in Fig. 5b increases from ~20 nm to 50 nm and is lower than the rrms over the entire distribution. The void peak at ~1 minute is most likely large analytes or other plasma components. These results show the capability of AF4-MALS-DLS to characterize the size distribution of LDL from human plasma before or after isolation by UC. Information about the particle shape can also be estimated from AF4-MALS-DLS using the ratio of rrms/rh. However, a comparison to cryoelectron microscopy and other shape sensitive methods is needed to confirm shape information obtained by AF4-MALS-DLS. This is the basis for future research and is beyond the scope of this work.
CONCLUSION
The use of SP-AF4 for isolation of human plasma HDL and LDL was demonstrated and separation conditions, sample recovery, and LC/MS/MS phospholipid profiles were compared to those obtained using a UC preparation method. Size fractionation by SP-AF4 allowed HDL and LDL fractions each containing 5 to 109 μg of total proteins to be collected in less than one hour. An approximately forty times increase in separation speed by SP-AF4 compared to the UC preparation method and nearly ten times increase in sample load compared to analytical scale AF4 was achieved. These improvements represent a significant increase in throughput and sample amount. Also, flexibility in SP-AF4 carrier fluid selection allows for the use of more biologically preferred conditions (50 mM phosphate buffer) compared to high salt concentrations (3 M KBr) used in UC that can cause unwanted protein-lipoprotein dissociation or aggregation. Analysis of LC-MS/MS lipid profiles showed that a similar number of lipids are identified in UC and SP-AF4 prepared lipoproteins, supporting SP-AF4 as a method to improve throughput for clinical studies and biomarker discovery. Analytical scale AF4-MALS-DLS was also demonstrated as a fast method for probing lipoprotein size distributions.
Supplementary Material
ACKNOWLEDGMENTS
CRMB and SKRW and the SP-AF4 and analytical AF4 work are supported by NSF-CHE1508827. FA’s time and salary is supported by the grant K08DK106523. Lipoprotein lipidomic studies, metabolomic core support, and clinical sample procurement was funded by NIH grants P30DK081943, P30DK0829503, U24DK097153 and R24DK082841 (all to SP).
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
Authors declare that they have no conflict of interest.
This research did not involve human participants or animals and therefore obtaining informed consent was not required.
Human plasma was obtained from the American Red Cross which was a pool of over 1000 deidentified plasma donors which were to be discarded due to expiration of safe window for transfusion. The study is therefore exempt from Institutional Review Board (IRB) approval.
REFERENCES
- 1.Austin MA, Rodriguez BL, McKnight B, McNeely MJ, Edwards KL, Curb JD, Sharp DS (2000) Low-density lipoprotein particle size, triglycerides, and high-density lipoprotein cholesterol as risk factors for coronary heart disease in older Japanese-American men. Am J Cardiol 86:412–416. doi: 10.1016/S0002-9149(00)00956-5 [DOI] [PubMed] [Google Scholar]
- 2.Teerlink T, Scheffer PG (2007) LDL particles are nonspherical: consequences for size determination and phenotypic classification. Clin Chem 53:361–2. doi: 10.1373/clinchem.2006.079871 [DOI] [PubMed] [Google Scholar]
- 3.Gerszten RE, Wang TJ (2008) The search for new cardiovascular biomarkers. Nature 451:949–952. doi: 10.1038/nature06802 [DOI] [PubMed] [Google Scholar]
- 4.Davidson WS, Heink A, Sexmith H, Melchior JT, Gordon SM, Kuklenyik Z, Woollett L, Barr JR, Jones JI, Toth CA, Shah AS (2016) The effects of apolipoprotein B depletion on HDL subspecies composition and function. J Lipid Res 57:674–86. doi: 10.1194/jlr.M066613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larner CD, Henriquez RR, Johnson JD, Macfarlane RD (2011) Developing High Performance Lipoprotein Density Profiling for Use in Clinical Studies Relating to Cardiovascular Disease. Anal Chem 83:8524–8530. doi: 10.1021/ac2018124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munroe WH, Phillips ML, Schumaker VN (2015) Excessive centrifugal fields damage high density lipoprotein. J Lipid Res 56:1172–81. doi: 10.1194/jlr.M058735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunitake ST, Kane JP (1982) Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. J Lipid Res 23:936–40. [PubMed] [Google Scholar]
- 8.Ståhlman M, Davidsson P, Kanmert I, Rosengren B, Borén J, Fagerberg B, Camejo G (2008) Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J Lipid Res 49:481–90. doi: 10.1194/jlr.D700025-JLR200 [DOI] [PubMed] [Google Scholar]
- 9.Jones J, Toth C, Kuklenyik Z, Schieltz D, Williamson Y, Andrews M, Parks B, Gardner M, Rees J, McWilliams L, Barr JR (2016) Effects of Ultracentrifugation on HDL and LDL Size Distribution. Pittcon Atlanta, GA [Google Scholar]
- 10.Lu M, Gursky O (2013) Aggregation and fusion of low-density lipoproteins in vivo and in vitro. Biomol Concepts 4:501–518. doi: 10.1515/bmc-2013-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh J, Chang C, Huang MT, Chang C, Chen C, Shen M-Y, Liao H-Y, Wang G, Chen C, Chen C, Yang C (2013) Biochemical and Functional Characterization of Charge-Defined Subfractions of High-Density Lipoprotein From Normal Adults. Anal Chem 85:11440–11448. doi: 10.1021/ac402516u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffer PG, Bakker SJL, Heine RJ, Teerlink T (1997) Measurement of low-density lipoprotein particle size by high-performance gel-filtration chromatography. Clin Chem 43:1904–1912. [PubMed] [Google Scholar]
- 13.Bobály B, Beck A, Fekete J, Guillarme D, Fekete S (2015) Systematic evaluation of mobile phase additives for the LC-MS characterization of therapeutic proteins. Talanta 136:60–7. doi: 10.1016/j.talanta.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 14.Williams SKR, Runyon JR, Ashames AA (2011) Field-flow fractionation: addressing the nano challenge. Anal Chem 83:634–642. doi: 10.1021/ac101759z [DOI] [PubMed] [Google Scholar]
- 15.Li P, Giddings JC (1996) Isolation and Measurement of Colloids in Human Plasma by Membrane-Selective Flow Field-Flow Fractionation: Lipoproteins and Pharmaceutical Colloids. J Pharm Sci 85:895–898. doi: 10.1021/js950335s [DOI] [PubMed] [Google Scholar]
- 16.Wahlund K (2013) Flow field-flow fractionation : Critical overview. J Chromatogr A 1287:97–112. doi: 10.1016/j.chroma.2013.02.028 [DOI] [PubMed] [Google Scholar]
- 17.Johann C, Elsenberg S, Roesch U, Rambaldi DC, Zattoni A, Reschiglian P (2011) A novel approach to improve operation and performance in flow field-flow fractionation. J Chromatogr A 1218:4126–31. doi: 10.1016/j.chroma.2010.12.077 [DOI] [PubMed] [Google Scholar]
- 18.Park I, Paeng K-J, Kang D, Moon MH (2005) Performance of hollow-fiber flow fieldflow fractionation in protein separation. J Sep Sci 28:2043–2049. doi: 10.1002/jssc.200500125 [DOI] [PubMed] [Google Scholar]
- 19.Kim KH, Moon MH (2011) Chip-Type Asymmetrical Flow Field-Flow Fractionation Channel Coupled with Mass Spectrometry for Top-Down Protein Identification. Anal Chem 83:8652–8658. doi: 10.1021/ac202098b [DOI] [PubMed] [Google Scholar]
- 20.Kuklenyik Z, Gardner M, Parks B, Schieltz D, Rees J, McWilliams L, Williamson Y, Pirkle J, Barr J (2015) Multivariate DoE Optimization of Asymmetric Flow Field Flow Fractionation Coupled to Quantitative LC-MS/MS for Analysis of Lipoprotein Subclasses. Chromatography 2:96–117. doi: 10.3390/chromatography2010096 [DOI] [Google Scholar]
- 21.Lee JY, Kim KH, Moon MH (2009) Evaluation of multiplexed hollow fiber flow fieldflow fractionation for semi-preparative purposes. J Chromatogr A 1216:6539–42. doi: 10.1016/j.chroma.2009.07.044 [DOI] [PubMed] [Google Scholar]
- 22.Lee JY, Min HK, Choi D, Moon MH (2010) Profiling of phospholipids in lipoproteins by multiplexed hollow fiber flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. J Chromatogr A 1217:1660–6. doi: 10.1016/j.chroma.2010.01.006 [DOI] [PubMed] [Google Scholar]
- 23.Byeon SK, Lee JY, Lim S, Choi D, Moon MH, Kee S, Yong J, Lim S, Choi D, Hee M, Byeon SK, Lee JY, Lim S, Choi D, Moon MH (2012) Discovery of candidate phospholipid biomarkers in human lipoproteins with coronary artery disease by flow fieldflow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. J Chromatogr A 1270:246–253. doi: 10.1016/j.chroma.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 24.Byeon SK, Kim JY, Lee JY, Chung BC, Seo HS, Moon MH (2015) Top-down and bottom-up lipidomic analysis of rabbit lipoproteins under different metabolic conditions using flow field-flow fractionation, nanoflow liquid chromatography and mass spectrometry. J Chromatogr A 1405:140–148. doi: 10.1016/j.chroma.2015.05.059 [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Byeon SK, Moon MH (2015) Profiling of Oxidized Phospholipids in Lipoproteins from Patients with Coronary Artery Disease by Hollow Fiber Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography–Tandem Mass Spectrometry. Anal Chem 87:1266–1273. doi: 10.1021/ac503973p [DOI] [PubMed] [Google Scholar]
- 26.Kuklenyik Z, Jones JI, Gardner MS, Schieltz DM, Parks BA, Toth CA, Rees JC, Andrews ML, Carter K, Lehtikoski AK, McWilliams LG, Williamson YM, Bierbaum KP, Pirkle JL, Barr JR (2018) Core lipid, surface lipid and apolipoprotein composition analysis of lipoprotein particles as a function of particle size in one workflow integrating asymmetric flow field-flow fractionation and liquid chromatography-tandem mass spectrometry. PLoS One 13:e0194797. doi: 10.1371/journal.pone.0194797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bria CRM, Skelly PW, Morse JR, Schaak RE, Williams SKR (2017) Semi-preparative asymmetrical flow field-flow fractionation: A closer look at channel dimensions and separation performance. J Chromatogr A 1499:149–157. doi: 10.1016/j.chroma.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 28.Kim SH, Yang JS, Lee JC, Lee J-Y, Lee J-Y, Kim E, Moon MH (2018) Lipidomic alterations in lipoproteins of patients with mild cognitive impairment and Alzheimer’s disease by asymmetrical flow field-flow fractionation and nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. J Chromatogr A 1568:91–100. doi: 10.1016/J.CHROMA.2018.07.018 [DOI] [PubMed] [Google Scholar]
- 29.McPherson PAC, Young IS, McKibben B, McEneny J (2007) High density lipoprotein subfractions: isolation, composition, and their duplicitous role in oxidation. J Lipid Res 48:86–95. doi: 10.1194/jlr.M600094-JLR200 [DOI] [PubMed] [Google Scholar]
- 30.Vivekanandan-Giri A, Slocum JL, Byun J, Tang C, Sands RL, Gillespie BW, Heinecke JW, Saran R, Kaplan MJ, Pennathur S (2013) High density lipoprotein is targeted for oxidation by myeloperoxidase in rheumatoid arthritis. Ann Rheum Dis 72:1725–1731. doi: 10.1136/annrheumdis-2012-202033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 32.Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, Yang W, Shafi T, Weir MR, He J, Brecklin CS, Rhee EP, Schelling JR, Ojo A, Feldman H, Michailidis G, Pennathur S, Investigators CS, Appel LJ, Go AS, Kusek JW, Lash JP, Townsend RR (2016) Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int Reports 1:256–268. doi: 10.1016/j.ekir.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. doi: 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 34.ATP III At-A-Glance: Quick Desk Reference. https://www.nhlbi.nih.gov/healthpro/guidelines/current/cholesterol-guidelines/quick-desk-reference-html. Accessed 30 Jul 2017
- 35.Mackness MI, Hallam SD, Peard T, Warner S, Walker CH (1985) The separation of sheep and human serum “A”-esterase activity into the lipoprotein fraction by ultracentrifugation. Comp Biochem Physiol - Part B Biochem Mol Biol 82:675–677. [DOI] [PubMed] [Google Scholar]
- 36.Williams PT, Zhao X-Q, Marcovina SM, Otvos JD, Brown BG, Krauss RM (2014) Comparison of four methods of analysis of lipoprotein particle subfractions for their association with angiographic progression of coronary artery disease. Atherosclerosis 233:713–720. doi: 10.1016/j.atherosclerosis.2014.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leeuwenburgh C, Rasmussen JE, Hsu FF, Mueller DM, Pennathur S, Heinecke JW (1997) Mass spectrometric quantification of markers for protein oxidation by tyrosyl radical, copper, and hydroxyl radical in low density lipoprotein isolated from human atherosclerotic plaques. J Biol Chem 272:3520–3526. doi: 10.1074/JBC.272.6.3520 [DOI] [PubMed] [Google Scholar]
- 38.Pennathur S, Wagner JD, Leeuwenburgh C, Litwak KN, Heinecke JW (2001) A hydroxyl radical-like species oxidizes cynomolgus monkey artery wall proteins in early diabetic vascular disease. J Clin Invest 107:853–860. doi: 10.1172/JCI11194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pennathur S, Ido Y, Heller JI, Byun J, Danda R, Pergola P, Williamson JR, Heinecke JW (2005) Reactive carbonyls and polyunsaturated fatty acids produce a hydroxyl radical-like species: a potential pathway for oxidative damage of retinal proteins in diabetes. J Biol Chem 280:22706–22714. doi: 10.1074/jbc.M500839200 [DOI] [PubMed] [Google Scholar]
- 40.German JB, Smilowitz JT, Zivkovic AM (2006) Lipoproteins: When size really matters. Curr Opin Colloid Interface Sci 11:171–183. doi: 10.1016/j.cocis.2005.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, Hawkins J, Bellovich K, Gipson D, Michailidis G, Pennathur S (2017) Impaired β -Oxidation and Altered Complex Lipid Fatty Acid Partitioning with Advancing CKD. J Am Soc Nephrol 29:295–306. doi: 10.1681/ASN.2017030350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.