Abstract
The mitochondrial genome is an evolutionarily persistent and cooperative component of metazoan cells that contributes to energy production and many other cellular processes. Despite sharing the same host as the nuclear genome, the multi-copy mitochondrial DNA (mtDNA) follows very different rules of replication and transmission, which translate into differences in the patterns of selection. On one hand, mtDNA is dependent on the host for its transmission, so selections would favour genomes that boost organismal fitness. On the other hand, genetic heterogeneity within an individual allows different mitochondrial genomes to compete for transmission. This intra-organismal competition could select for the best replicator, which does not necessarily give the fittest organisms, resulting in mito-nuclear conflict. In this review, we discuss the recent advances in our understanding of the mechanisms and opposing forces governing mtDNA transmission and selection in bilaterians, and what the implications of these are for mtDNA evolution and mitochondrial replacement therapy.
Keywords: mitochondrial DNA, heteroplasmy, purifying selection, selfish selection, mitochondrial replacement therapy
1. Background
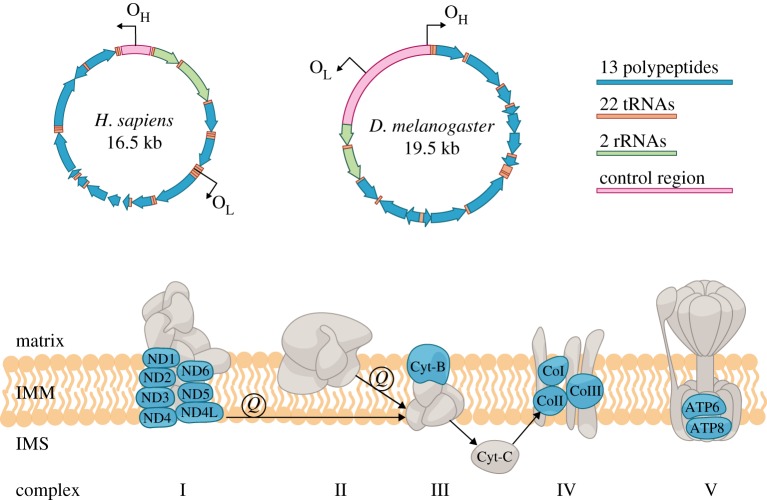
Mitochondria, the powerhouse of the cell, have attracted increasing attention because of their fascinating biology and health connections. They are thought to have evolved from free-living bacteria via symbiosis, which changed the course of eukaryotic evolution through a monumental metabolic upgrade by employing oxygen to produce energy [1,2]. While now tightly integrated into the biology of the host cell, with most proteins encoded in the nuclear genome, mitochondria still retain a reduced but vital genome of their own known as mitochondrial DNA (mtDNA). The genetic content and organization of mtDNA can vary incredibly among different species (summarized in [3,4]). For bilaterians, which are the focus of this review, mtDNA is often a compact circular DNA molecule with no introns and very few intergenic regions. It usually encodes 13 proteins of the respiratory chain complex, two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs). The genome also contains a distinct non-coding region/control region that encompasses replication origin(s) and transcription promoters (figure 1).
Figure 1.
Map of the human (Homo sapiens) and Drosophila melanogaster mtDNA, representative of the mammalian and insect genome, respectively. Both genomes have the same coding capacity, but differ in gene order, length of the control region and location of the replication origins (OL, light chain; OH, heavy chain). The 13 polypeptides (blue) form the respiratory chain complex together with the nuclear-encoded proteins (grey) [5]. In addition, a small peptide named humanin is encoded in the 16S rRNA gene of the human mtDNA. Humanin has been shown to have a role in regulating stress resistance and conferring specific protection against Alzheimer's disease [6–8]. IMM, inner mitochondrial membrane; IMS, intermembrane space; Q, the ubiquinone form of CoQ10.
Unlike the nuclear genome, which represents an assorted mixture of both maternal and paternal DNA, animal mtDNA is normally inherited exclusively from the mother. As such, the maternal genomes do not face any heredity competitors from the male parent and can safely assume their places in the next generation. Yet, not all maternal genomes are the same [9]. As most cells contain hundreds or even thousands of copies of mtDNA, spontaneous and inherited mutations can occur in a subpopulation, creating heteroplasmic organisms with genetic diversity in the mtDNA population. Theoretically, constantly occurring mutations would make heteroplasmy a default state. Even if the selection is actively removing mutant genomes, a return to homoplasmy can take time, resulting in transient heteroplasmy. Indeed, modern high-throughput sequencing provides evidence of widespread low-level heteroplasmy in many tissues of healthy individuals in humans [10–13]. Extensive heteroplasmy has also been reported in a number of other species including rabbits, horses, macaques, ferrets, cats and dogs [14–16]. In rare cases, heteroplasmy can be created by paternal leakage in animals that follow strict maternal inheritance [17–23]. In over 100 species of different bivalve orders, heteroplasmy occurs in male somatic tissues owing to doubly uniparental inheritance, where the female genome is transmitted to both male and female soma, and also female gonad, while the male genome is transmitted only to the male soma and gonad [24]. Among bilaterians, doubly uniparental inheritance is very much an exception to the rule with probably a single evolutionary origin [25].
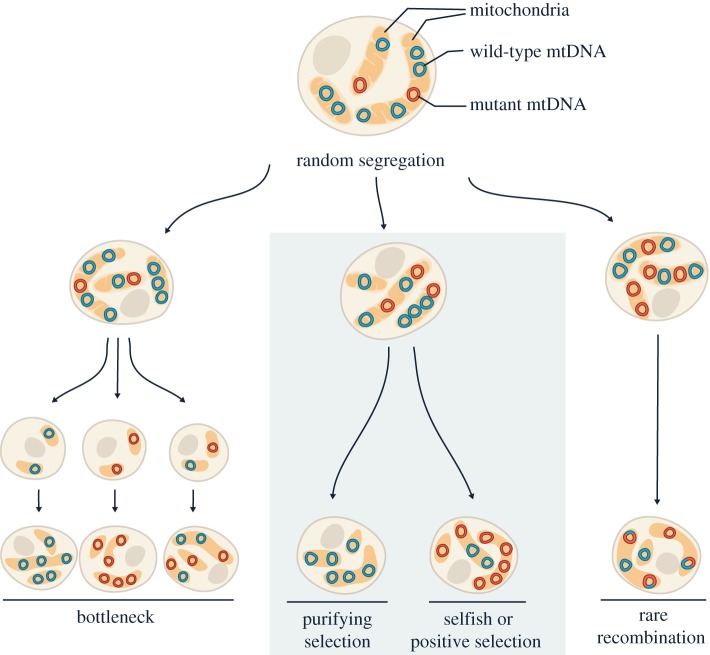
Heteroplasmy can represent a dynamic and constantly changing mtDNA population within an organism [26] (figure 2). This is because individual mtDNA molecules do not replicate in equal numbers in dividing cells, nor do they turn over at equal rates in non-dividing cells. By chance, a variant molecule may replicate more frequently than the wild-type genome and thus increase in abundance. mtDNA also lacks segregation mechanisms that ensure unbiased transmission into daughter cells, so the genome can be under the strong influence of genetic drift [27–29]. Besides random fluctuation, selection can further change heteroplasmy levels; mitochondrial genomes that provide better respiratory function might be preferentially transmitted owing to positive or purifying selection, while genomes that have a replicative advantage will increase in abundance through selfish selection (i.e. selection for selfish gains in transmission). Moreover, germline bottlenecks [30–35] and occasional recombination [36–43] can quickly shift mtDNA from one subpopulation to another within individuals and between generations.
Figure 2.
Heteroplasmy dynamics during somatic and germline transmission of mtDNA. In each cell, mitochondrial genomes are dispersed throughout the dynamic mitochondrial network and are packed in nucleoid structures, with each nucleoid containing one or more copies of mtDNA. As the cell divides, relaxed replication and random segregation of mtDNA create daughter cells with different heteroplasmy levels, while often maintaining total mtDNA copy number. The shift in the heteroplasmy level can be accelerated when there is a sharp decline in the number of transmitted mtDNA (i.e. genetic bottleneck, left panel). Besides neutral drift, selections can further alter heteroplasmy levels in a biased manner (middle panel). Very occasionally, recombination events can create hybrid genomes and alter the heteroplasmy composition (right panel).
When the abundance of pathogenic mutations reaches a threshold level, physiological consequences will become apparent (reviewed by [44,45]). To date, over 350 pathogenic mitochondrial mutations have been reported to cause a spectrum of mitochondrial diseases [46], for which there are still no cures. One emerging strategy to prevent the transmission of mitochondrial mutations to offspring is mitochondrial replacement therapy (MRT), which has been approved in the UK as part of in vitro fertilization (IVF) treatment since 2015 [47]. MRT involves the transfer of the nucleus from a fertilized or unfertilized egg which carries mitochondrial mutations into an enucleated egg of a healthy donor, producing ‘three-parent babies’. However, carryover of pathogenic mtDNA has been observed in multiple experimental trials using human or rhesus macaque eggs [48–54], and also in the first child born from MRT [55]. Even though the carried over mutants often account for less than 2% of total mtDNA, they may increase in abundance in somatic and germline tissues of those born from MRT as the individuals develop and age, and cause mitochondrial diseases later in life or in their children.
Heteroplasmy creates a battlefield for coexisting mitochondrial genomes to compete for transmission. There could be conflicts between the cooperative interest enforced by the nuclear genome and the selfish interest of the mitochondrial genome. The outcome of the competition has profound and incompletely understood impacts on the accumulation of mtDNA mutations during development and ageing, the progression and phenotypic complexity of mitochondrial disease, the inheritance of mitochondrial mutations from mother to progeny and the effectiveness of MRT. This review focuses on some of the recent efforts to investigate how different types of selection shape bilaterian mtDNA evolution within individuals and between generations, and how unexpected interactions can compromise the efficacy of MRT.
2. Selections for organismal fitness
All gene products of mtDNA are devoted to energy production via oxidative phosphorylation (OXPHOS), which is of paramount importance to the host. However, the mitochondrial genome is vulnerable to mutational meltdown because uniparental inheritance and little recombination has limited power of removing de novo mutations. A small proportion of these mutations have been shown to be adaptive and have experienced positive selection. For instance, high-altitude populations in Tibet show adaptive mtDNA haplotypes compared with low-altitude, related groups in humans [56,57], grasshoppers [58] and horses [59,60] (reviewed by Luo et al. [61]). Similarly, mtDNA haplotypes have been shown to be positively selected in populations owing to their effect on tolerance to local temperatures in humans [62] and in other animals [63–67]. However, a larger proportion of mitochondrial mutations are deleterious, and purifying selection is known to be the dominant force to purge these mutations and keep the functional integrity of mitochondrial genes.
The presence of purifying selection is reflected by the fact that mitochondrial-encoded proteins evolve much more slowly than predicted [68]. In addition, several multi-generational experiments in mouse and Drosophila have shown that purifying selection in the female germline reduced the transmission of detrimental mtDNA mutations [69–73] (also recently reviewed by [26,74–76]). In humans, two studies which analysed heteroplasmy transmission in mother–child pairs of European ancestry using blood or buccal mtDNA data showed a significant decrease in minor non-synonymous alleles in offspring mtDNA [35,77]. More recently, sequencing mtDNA of human primordial germline cells (PGCs) isolated from various embryonic stages revealed a reduced number of non-synonymous and tRNA mutations during PGC development [78]. Although pre-existing differences in the heteroplasmy level of different tissues or embryos could contribute to the observed decline in mutation load, the above studies suggest that purifying selection is likely to occur in the female germline in humans.
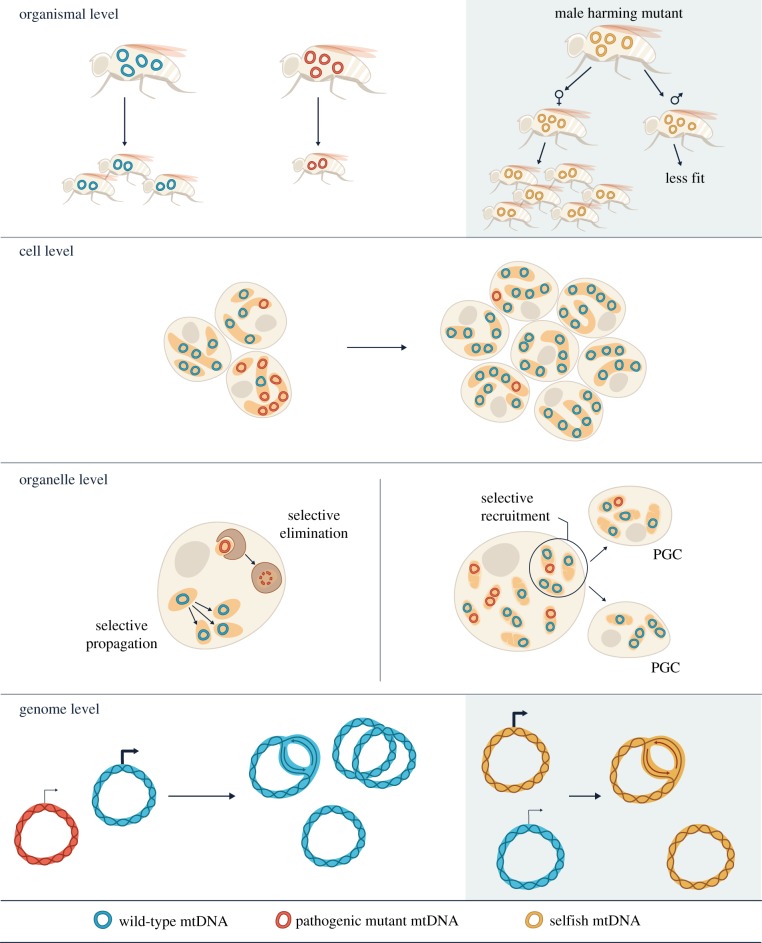
Intra-organismal purifying selection could act at the level of the cell, organelle or genome (figure 3). In some organisms, mitochondrial genetic bottlenecks (figure 2) in the germline facilitate selection at the cell level: cells that inherit more mutant mtDNA are less fit, so are less likely to propagate further. In zebrafish [79], sheep [80], mice [31,32] and humans [78], there is a dramatic decline in mtDNA copy number in PGCs. In Caenorhabditis elegans, PGCs form lobes that are removed and digested by endodermal cells, dramatically reducing the total amount of mitochondria in those cells [81]. In humans, oocytes were found to contain an average of 1.22 × 106 copies of mtDNA, while PGCs contained just 1425 copies on average, with an estimated five copies per mitochondrion [78]. This reduction in mtDNA copy number during germline development has been proposed to cause large shifts in heteroplasmy level between generations [30–32,35]. In addition, a bottleneck may result from unequal segregation or replication of mitochondrial genomes [33,34]. The bottleneck may even occur post-fertilization when there is rapid zygotic division unaccompanied by mtDNA replication, producing somatic cells with different mitochondrial mutation loads (e.g. [82]). Such bottlenecks may increase inter-cellular variation of the mtDNA pool, and thus accelerate purifying selection at the cell level.
Figure 3.
Selective transmission of mtDNA can be achieved by multiple mechanisms operating at both organismal and intra-organismal levels. Purifying selection (clear background) can occur at the organismal level owing to reduced host fitness caused by accumulation of mtDNA mutations. This mode of selection is more effective when the mutation level is high because the coexisting functional genomes can mask the physiological effects of low-abundance mutants. Within individuals, purifying selection can occur through selective propagation of more functional cells, mitochondria and mtDNA in germline and soma. For selfish gains in transmission (shaded background), mutations that are male harming, but neutral or beneficial to female fitness, can increase their abundance. This is because maternal inheritance limits the scope of purifying selection at the organismal level against such mutations among the male population. Within individuals, selfish transmission is often due to gains in replication.
Purifying selection may occur at the level of the organelle (figure 3), although it is still unclear how the OXPHOS function of individual mitochondria or mitochondrial networks is sensed to achieve selection. For example, selective recruitment or active propagation of functional mitochondria to the germplasm has been suggested by studies in zebrafish and Drosophila [83,84]. In Drosophila, there is also evidence linking mtDNA replication to OXPHOS function, such that purifying selection occurs by preferential replication of functional mtDNA [73]. The mt:CoIts genome is a temperature-sensitive lethal mutant isolated through a selection method based on expressing a restriction enzyme targeted to the mitochondria in the female germline [85]. The defect is due to a missense mutation in the coding region of cytochrome c oxidase I (CoI). When heteroplasmic flies containing mt:CoIts and wild-type mtDNA were created by cytoplasmic transfer, the level of mt:CoIts decreased over generations at the restrictive temperature, and this eventually led to its elimination [72,73]. Hill et al. [73] showed that mt:CoIts underwent reduced replication in early oogenesis and that reduced mtDNA replication also occurred when mitochondrial function was impaired by other means, such as uncoupling drugs. In order for this mechanism of selection to be effective, mtDNA must be relatively homoplasmic within an organelle when selection occurs. Indeed, increased fission of mitochondria before mtDNA replication was observed, suggesting a low mtDNA copy number per mitochondrion [73]. It is still unclear how preferential mtDNA replication is achieved during oogenesis or whether there are other mechanisms of selection acting in the germline simultaneously (e.g. [86–88]), especially since selective elimination of mt:CoIts was not observed in post-development somatic tissues in the same heteroplasmic flies [89].
As an alternative mechanism of selection at the organelle level, mitochondria with mutant mtDNA can be eliminated by mitochondrial quality control mechanisms such as mitophagy (reviewed by Pickles et al. [90]). In a study using D. melanogaster heteroplasmic for both wild-type and a deletion-bearing mtDNA variant in their post-mitotic flight muscle, overexpression of some autophagy and mitophagy proteins (e.g. Atg8a, PINK1 and Parkin) promoted selective removal of deletion-bearing molecules [91]. In addition, decreased mitofusin levels, which limit the ability of mitochondrial fragments to re-fuse with the network, enhanced the removal of deletion-bearing mtDNA in the flight muscle [91]. However, the selective elimination of mt:CoIts in the fly germline, as described earlier, did not seem to require Parkin [72]. Furthermore, knockout of Parkin did not affect the level of somatic mtDNA mutations in mutator mice, which are known to accumulate mtDNA mutations, although it did lead to more mitochondrial dysfunction in those mice [92]. These studies show that Parkin-mediated mitophagy may not always play a role in eliminating mtDNA mutations and reveals the diverse nature of purifying selection.
While there is ample evidence in favour of purifying selection, there are also examples where purifying selection was not detected. Many population data in humans find that segregation of mutations appears to follow random genetic drift without selection [93–96]. In a mouse model containing two mutant genomes among the wild-type genome, one mutant mtDNA that contained a missense mutation in ND6 was rapidly eliminated within a few generations, whereas the other mtDNA containing a missense mutation in CoI that causes myopathy and cardiopathy persisted [69]. Similarly, Freyer et al. [71] showed that mitochondrial mutations in protein-coding genes were preferentially eliminated in mice over generations, whereas mutations in tRNA genes evaded selection, despite the fact that many of these mutations are potentially pathogenic. Therefore, whether purifying selection takes place or not seems to depend on the nature of the mutation, the competing mitochondrial genomes, the tissue and the nuclear background. It is most likely that the term ‘purifying selection’ summarizes a plethora of selective phenomena that could differ completely for the underlying mechanisms, resulting in the complex dynamics of heteroplasmy observed.
3. Selections for selfish mtDNA
In addition to favouring traits that enhance organismal fitness, evolution favours selfish traits that give replication or transmission gains. Both mitochondrial and nuclear genomes are selected to maximally propagate the genes comprising its own set, independently of the effect on the other gene set or host. With few constraints on replication and segregation of mtDNA, free-wheeling intra-organismal competition is likely to select for the best replicator, regardless of its OXPHOS output.
The occurrence of selfish transmission is hard to detect in natural populations, as its consequence only becomes obvious when the selfish genome also possesses a detrimental mutation. Even if a detrimental selfish mtDNA does arise, its increase in abundance can run the risk of lowering host fitness to the point where it drives the host, and therefore itself, to extinction. Nevertheless, male harming mtDNA variants that are neutral or beneficial to female fitness can reach high frequencies in populations because males are a dead-end for mtDNA transmission (figure 3). This is known as the mother's curse and has been primarily studied in plants (summarized in [97]). A number of cases have also been recently reported in Drosophila [98,99] and in a human population in Canada [100].
Selfish mutations that reduce both male and female fitness are less common, but have been found in natural populations of Drosophila subobscura [101], C. elegans [102] and Caenorhabditis briggsae [103]. In all cases, the selfish genomes that exhibited long-term persistence in isolated strains were mtDNA variants with a large deletion. For the D. subobscura strain, the mutant genome contains a 5 kb deletion affecting 10 genes and accounts for approximately 80% of the total mtDNA copies [101]. The stable transmission of the deletion molecule is unlikely owing to physical attachment to the wild-type mtDNA because both genomes are autonomous monomers [104]. In C. elegans, the uaDf5 mitochondrial genome, which has a 3.1 kb deletion that removes 11 genes, accounts for approximately 60% of the total mtDNA copies in heteroplasmic animals [102]. uaDf5 was shown to be stably transmitted for over 100 generations, during which not a single line homoplasmic for wild-type or uaDf5 mtDNA arose [102]. In another nematode species, C. briggsae, many natural lineages are heteroplasmic for mtDNA with a 786 bp deletion in the ND5 coding region (nad5Δ) [103]. This deletion mutant is found in several geographical locations, indicating its evolutionary persistence. Clark et al. [105] investigated the inheritance patterns of nad5Δ in eight lineages and found a uniform bias towards the inheritance of a greater proportion of the nad5Δ genome, despite that high levels of nad5Δ caused reduced fertility and pharyngeal pumping rates. It is currently unclear how these deletion-bearing molecules persist in wild populations. A recent study suggested that uaDf5 can somehow run away from the copy number control mechanism that regulates the coexisting wild-type mtDNA levels because they observed a wider variation in uaDf5 copy number relative to that of wild-type [106]. An increase in the total mtDNA copy number could be an attempt to alleviate OXPHOS deficiency as proposed by previous theoretical work [106–109]. Furthermore, high levels of uaDf5 activated the mitochondrial unfolded protein response, which was suggested to help the maintenance of the uaDf5 genome [106,110].
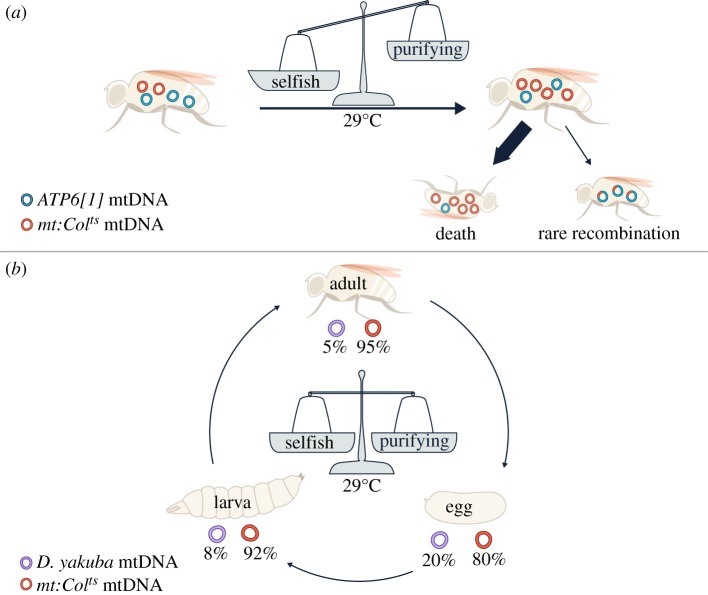
More evidence of selfish transmission has recently emerged from experimentally generated heteroplasmic lines in D. melanogaster, where diverged mitochondrial genomes from different D. melanogaster strains or even different Drosophila species were paired for competition [111]. Apparently, these diverged genomes often do not compete based on their OXPHOS function. In one example, the temperature-sensitive mutant mt:CoIts displaced a complementing genome, leading to population death after several generations at the restrictive temperature. As mentioned earlier, the mt:CoIts mutant genome is eliminated by purifying selection when paired with a closely related wild-type D. melanogaster mtDNA [72,73]. However, when it was paired with another functional but more diverged genome called ATP6[1], which is a D. melanogaster mtDNA variant that differs from mt:CoIts by multiple single nucleotide polymorphisms (SNPs) and indels in both coding and non-coding regions, the level of mt:CoIts increased from around 20% to 90% within four generations. Eventually, the ATP6[1] genome declined to the extent that it could no longer sustain the life of the flies [111] (figure 4a). In this case, the mt:CoIts mutant was considered to have a selfish drive. Interestingly, while most lines ended in lethality, a few survived [42]. In these lines, recombinant mtDNA were generated which combined the functional CoI allele from the ATP6[1] genome with the selfish drive from mt:CoIts. Once emerged, such recombinant genomes quickly outcompeted coexisting mt:CoIts because of purifying selection and the stock became healthier over time. Since all recombinant genomes retained the non-coding region of mt:CoIts, which differs significantly from the ATP6[1] genome at the sequence level, this region is believed to be responsible for the strong selfish drive. That is why the selfish drive of mt:CoIts is not apparent when paired with its closely related wild-type D. melanogaster mtDNA as they share the same non-coding region. The non-coding region contains the origins of replication, so the selfish drive in this case has been linked to replicative advantage. It is worth noting that the non-coding region of mt:CoIts is significantly longer than that of the ATP6[1] genome. This is surprising as the mitochondrial genomes with the smallest and least redundant DNA are believed to go to fixation within cells, organisms and then populations [112–114]. This example implies that other factors besides genome size can play a more important role in selfish transmission.
Figure 4.
Different selective pressures dynamically act on mtDNA in D. melanogaster [111]. (a) When the mt:CoIts mutant was paired with a diverged, functional ATP6[1] genome at the restrictive temperature, selfish selection dominated and allowed the mt:CoIts genome to take over, despite purifying selection against mt:CoIts. This led to the death of the population within a few generations. However, in three out of the 51 heteroplasmic lineages, recombination generated mtDNA containing the functional coding region of ATP6[1] and the selfish non-coding region of mt:CoIts [42]. Once emerged, these recombinants became dominant owing to purifying selection against mt:CoIts, allowing the flies to survive the selection. (b) When mt:CoIts was paired with wild-type Drosophila yakuba mtDNA at the restrictive temperature, the relative proportion of each genome remained stable over many generations. Interestingly, the heteroplasmy level differed at various developmental stages. This is likely to be due to a dynamic balance of purifying and selfish selection in different tissues and at different developmental stages.
Another example of selfish mtDNA was revealed by a number of cross-species pairings in the same study. Ma and O'Farrell [111] created D. melanogaster flies with only wild-type D. yakuba mtDNA via cytoplasmic transfer followed by expression of a mitochondrially targeted restriction enzyme that will only cut the D. melanogaster mtDNA [111]. Despite that D. melanogaster and D. yakuba diverged about 10 million years ago, the D. melanogaster (mito-D. yakuba) flies are as healthy as wild-type, indicating that D. yakuba mtDNA can provide the wild-type level of function in the D. melanogaster nuclear background. Nevertheless, when D. yakuba mtDNA was placed in competition with a number of functionally compromised D. melanogaster mtDNA variants, it was quickly outcompeted despite providing better function. In this case, the D. melanogaster mtDNA variants had a selfish advantage over D. yakuba mtDNA. Interestingly, mtDNA from Drosophila mauritiana (a species diverged ∼2 million years ago) can outcompete endogenous D. melanogaster mtDNA with no deleterious effect, indicating that the home genome is not always the winner [111,115]. De Stordeur [116] also used cytoplasmic transfer to study competition between different Drosophila simulans mtDNA haplotypes and found a hierarchy of which haplotypes could overtake which others, although it is not clear whether selfish selection plays a role in each context. Overall, these examples suggest that the mismatches in competitive strength are common among diverged genomes.
Of note, selfish selection can be neutral to the host when the selfish drive is not linked to detrimental mutations. It can even be beneficial if a more functional genome gains a replicative advantage, as it will speed up the takeover of the functional genome. For instance, Rand [117] has shown that spontaneous mutations that increase the length of the non-coding region of Drosophila mtDNA could occur in natural populations. These long variants were preferentially transmitted to the offspring, but there was no evidence for fitness difference among flies carrying mtDNA variants of different length [117]. In such cases, selfish selection results in rapid divergence of mtDNA sequence among different female lineages. This is because, during evolution, constant waves of taking over by a new mutant genome with replicative advantage will continuously select for the best replicator in a given nuclear background, especially if that mutation does not result in functional sacrifices. Uniparental inheritance limits mitochondrial variants to evolve in individual lineages, so, within each lineage, different winning mutations are likely to be fixed independently. As the non-coding region is often linked to selfish drive, selfish selection can accelerate divergence of this region. Indeed, for most mitochondrial genomes sequenced so far, their non-coding regions are highly variable [111].
4. The interplay of different types of selection at multiple levels
The outcome of mtDNA competition will depend on the relative strength of purifying and selfish selection. These two forces can oppose one another at both organismal and intra-organismal levels. In cases where deleterious mutations are linked to a strong selfish drive, they will quickly accumulate within individuals. This will eventually lower the fitness of the host and trigger purifying selection at the organismal level. In this way, a selfish element drives its own extinction. When such an element arises de novo, maternal inheritance restricts it to a single female lineage, and thus facilitates its elimination without spreading the detrimental effect to the rest of the population [118]. Diverse mechanisms have been described that eliminate paternal mtDNA before and/or after fertilization in different species to ensure maternal inheritance [119–125] (summarized by Sato & Sato [126]). Nevertheless, paternal leakage has been reported in multiple cases [17–23], and it is unclear to what extent rare leakage can affect the spread of selfish mtDNA within a species.
Within individuals, opposing types of selection may explain why sometimes purifying selection was not detected or was not always complete, resulting in the persistence of detrimental variants [69,76]. This may occur if the selfish drive only gives the linked detrimental allele a small gain in replication or transmission, which can be counterbalanced by purifying selection at the cell or organelle level. Some of the long-term heteroplasmy discussed earlier could also be due to balanced purifying and selfish selection occurring within individuals. For instance, in C. elegans, the uaDf5 genome appears to be under opposing selective pressures which have different strengths at different levels of heteroplasmy [102]. When the proportion of uaDf5 in hermaphrodites is high, the average uaDf5 content in progeny is significantly lower. Conversely, when the proportion of uaDf5 in hermaphrodites is low, the average uaDf5 contents in progeny increases significantly. These data suggest the existence of at least two opposing forces, with one force leading to the increased proportion of uaDf5 mtDNA when its levels are low, while the second force leads to decreased proportions of uaDf5 mtDNA when its levels are high [102]. In Drosophila, opposing selfish and purifying selection was shown to counterbalance in a cross-species heteroplasmic line, allowing stable transmission of the functional D. yakuba mtDNA at 5% and the selfish detrimental D. melanogaster mt:CoIts mutant at 95% in adults at the restrictive temperature [111]. How the two types of selection counteract can be complex. Selfish selection mainly operates at the genome level, whereas purifying selection can occur at genome, organelle and cell level. Furthermore, purifying selection may occur at different developmental stages and in different tissues (germline versus soma) from selfish selection, creating an oscillation in the relative levels of the two genomes when comparing different developmental stages, without changing the ratio of the genomes when comparing different generations (figure 4b) [111,127].
The nuclear background can influence the strength of various types of selection, and thus alter the outcome of mtDNA competition. This is because the nuclear genome encodes nearly all of the proteins in mitochondria, as well as external regulators of mitochondrial biogenesis, dynamics and mitophagy/turnover. Differences in the nuclear genome can re-define the functional OXPHOS capacity of mtDNA and determine whether or not a variant is a detrimental mutation: one mtDNA variant may result in deficient OXPHOS capacity in one nuclear background but not in another owing to differences in the nuclear-encoded complex proteins [128,129]. The strength of purifying selection can depend on the severity of mismatch. Selfish transmission of certain mtDNA variants may only manifest in a given nuclear background as particular isoforms of nuclear genes are required to allow them to replicate or transmit better. Tissues with different energy demands may have preferences for mitochondrial genomes with certain metabolic rates, energy expenditures or replication features [130–132]. Other changes, such as temperature and age, may also impact mtDNA competition, probably through altering nuclear transcriptional profiles [133–135].
The nuclear influence on mtDNA competition has been shown in a number of studies. In C. briggsae, the occurrence of mtDNA with large deletions varies between different strains with different nuclear genomes [103]. In D. subobscura, the abundance of the 5 kb deletion-bearing molecule was stable across generations, but changed during backcrosses to a different nuclear genotype [136]. In D. melanogaster, the level of D. yakuba mtDNA was initially stabilized at 5% when paired with mt:CoIts, but in another nuclear background it stabilized at 20% [111]. In mice, tissue-specific segregation of heteroplasmy has been reported in a number of studies [137–139]. Furthermore, certain human alleles are selected for at specific nucleotide positions in specific tissues as individuals age [12].
5. Heteroplasmy and mitochondrial replacement therapy
The last 5–10 years has been an exciting time for MRT, as fundamental scientific discoveries have significantly advanced the clinical strategies to prevent the transmission of pathogenic mitochondrial mutations. In 2016, the first ‘three-parent boy’ was born in Mexico [55], and in early 2017 the first ‘three-parent girl’ was born in Ukraine. In the UK, three-parent babies could be born this year, as two cases have been approved by the UK's Human Fertilization and Embryology Authority to take place at the Newcastle Fertility Centre.
However, MRT has raised a number of ethical and safety concerns. Although it has been argued that MRT poses no greater risk of mito-nuclear incompatibility in humans than normal reproduction [140,141], matching the parents' nuclear genome with the donor's mitochondrial genome could be considered to minimize potential effects of mito-nuclear interactions observed in cybrid studies and genetic rescues [142–144].
Another safety concern is the carryover of mutant mtDNA from the original mother's egg. Historically, embryos carrying mtDNA from both a donor and the mother were created by cytoplasmic transfer, which was developed to enable women with impaired fertility to bear children. Although mitochondrial transfer was not the primary objective at the time, 5–15% of ooplasm from unfertilized oocytes is transferred to the recipient oocyte during this process, thus creating babies with multiple mitochondrial genotypes. Analysis of the mtDNA from offspring produced using cytoplasmic transfer confirmed the presence of donor mtDNA. Between the late 1990s and early 2000s, cytoplasmic transfer resulted in over 30 live births in the USA [145]. Currently, MRT can be performed by either maternal spindle transfer before fertilization or by pro-nuclear transfer after fertilization, and both methods lead to some carryover [48–50,54]. For example, oocytes and embryos produced from maternal spindle transfer by Tachibana et al. [48] had a mean carryover of 0.5%, while embryos produced from pro-nuclear transfer by Craven et al. [49] had a mean carryover of 1.8%. In addition, the first boy born from MRT via spindle transfer contained 2–9% maternal mutant mtDNA in tissues examined (hair follicles, circumcised foreskin and umbilical blood) [55]. Recently, polar body transfer has been suggested as an alternative approach to reduce mtDNA carryover [54,146,147], as polar bodies contain few mtDNA copies. Nevertheless, an average carryover of 0.26% in blastocysts has been observed [54].
Even a small trace of carried-over mutant mtDNA could reach the disease-causing threshold level in specific tissues later in life. This can occur through genetic drift, as has been observed when passaging human pluripotent stem cell lines derived from blastocysts created by MRT [52]. Reversion to maternal haplotype can occur more rapidly when the maternal genome has a replicative advantage. For example, in a study where spindle transfers were carried out between healthy human oocytes with preselected mtDNA haplotypes, two out of 15 blastocyst-derived embryonic stem cell lines reverted to the maternal haplotype [53]. These two cell lines were created by transfer events that mixed a maternal haplotype U5a with a donor haplotype H1b (differ by 33 SNPs). Sibling cell lines generated that mixed the same maternal haplotype with a different donor haplotype V3 (also differ by 33 SNPs) did not show reversion, suggesting that reversion is specific to a certain combination of haplotypes in the maternal nuclear background. The starting abundance of the maternal mtDNA was less than 1%, but it reached 81% and 94%, respectively, after two or three passages. Further passaging resulted in a complete loss of donor mtDNA. This reversion also occurred during stem cell differentiation, raising the possibility that reversal to the mutant mtDNA may occur in some MRT children. The group identified a polymorphism within the control region of the maternal haplotype and suggested that this polymorphism could enhance replication priming and thus the proliferation of the maternal genome when paired with H1b [53]. This is very similar to the selfish selection described earlier in Drosophila, where genomes with a certain non-coding region showed a transmission advantage regardless of their OXPHOS function [111].
Although it is not known whether mtDNA shifts in embryonic stem cell lines will truly reflect those in the developing embryo, precautions ought to be taken to minimize the reversion after MRT. In humans, there are at least 25 major mitochondrial haplotypes, each containing many subclades that differ significantly for their control region [148]. If we can make sure that the donor's mitochondrial genotype not only is compatible with the nuclear genome, but also has a selective advantage in replication or transmission, it would have the advantage of fully outcompeting the maternal genome, even if the carryover level is high. For that, we need to know more about how the outcome of competition is determined in order to know which genome will have a competitive advantage. In particular, we need to identify sequences in mtDNA that can confer replicative advantage to certain mitochondrial genomes and understand how changes in the nuclear genome can influence the outcome of mtDNA competition [149]. This is relevant not only to MRT but also to cytoplasmic transfer, which, although abandoned in the USA because of uncertainties about its safety and effective benefit, is still commercially available in IVF clinics in numerous countries worldwide [150].
6. Conclusion and future perspectives
It is encouraging to see that powerful tools and animal models have been established to reveal how different forces influence the transmission of mtDNA, given that selective transmission influences mitochondrial disease and guides mtDNA evolution. Moreover, recent advances in genome editing based on mitochondrially targeted transcription activator-like effector nucleases and zinc finger nucleases have allowed selective elimination of pathogenic mutations in mouse germ cells [151] and somatic tissues [152,153], and in induced pluripotent stem cells derived from patients with mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) [154]. These technological breakthroughs will certainly widen the therapeutic options in the near future. Nevertheless, the study of mtDNA is far from exhausted and the management of mitochondrial diseases has lagged behind the genetic revolution. In the future, we need to gain a better understanding of what and how sequence differences in mtDNA give a selfish transmission advantage and how the nuclear genome modulates purifying selection to safeguard the organismal investment in mitochondrial genes. To answer these questions, we need to pursue even more basic questions such as how mtDNA replication is controlled and how the genome segregates. Furthermore, essential aspects of mitochondrial biology that were once thought fundamental and universal, such as the lack of recombination [36–43] and strict maternal inheritance [17–23], must now be revisited with new tools and systems that provide higher detection power.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
Funding for this work was provided by the Wellcome Trust (grant no. 202269/Z/16/Z).
References
- 1.Margulis L. 1970. Origin of eukaryotic cells: evidence and research implications for a theory of the origin and evolution of microbial, plant and animal cells on the precambrian Earth. New Haven, CT: Yale University Press. [Google Scholar]
- 2.Lane N. 2014. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb. Persp. Biol. 6, a015982 ( 10.1101/cshperspect.a015982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DR, Keeling PJ. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc. Natl Acad. Sci. USA 112, 10 177–10 184. ( 10.1073/pnas.1422049112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavrov DV, Pett W. 2016. Animal mitochondrial DNA as we do not know it: mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 8, 2896–2913. ( 10.1093/gbe/evw195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Signes A, Fernández-Vizarra E. 2018. Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem. 62, 255–270. ( 10.1042/EBC20170098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto Y, et al. 2001. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proc. Natl Acad. Sci. USA 98, 6336–6341. ( 10.1073/pnas.101133498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. 2003. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 423, 456–461. ( 10.1038/nature01627) [DOI] [PubMed] [Google Scholar]
- 8.Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P. 2011. Interaction between the Alzheimer's survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc. Natl Acad. Sci. USA 100, 13 042–13 047. ( 10.1073/pnas.2135111100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryaman J, Johnston IG, Jones NS. 2018. Mitochondrial heterogeneity. Front. Genet. 9, 718 ( 10.3389/fgene.2018.00718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne BAI, et al. 2013. Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet. 22, 384–390. ( 10.1093/hmg/dds435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naue J, Hörer S, Sänger T, Strobl C, Hatzer-Grubwieser P, Parson W, Lutz-Bonengel S. 2015. Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion 20, 82–94. ( 10.1016/j.mito.2014.12.002) [DOI] [PubMed] [Google Scholar]
- 12.Li M, Schröder R, Ni S, Madea B, Stoneking M. 2015. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl Acad. Sci. USA 112, 2491–2496. ( 10.1073/pnas.1419651112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding J, et al. 2015. Assessing mitochondrial DNA variation and copy number in lymphocytes of ∼2,000 sardinians using tailored sequencing analysis tools. PLoS Genet. 11, e1005306 ( 10.1371/journal.pgen.1005306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casane D, Dennebouy N, de Rochambeau H, Mounolou JC, Monnerot M. 1994. Genetic analysis of systematic mitochondrial heteroplasmy in rabbits. Genetics 138, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Arnason U. 1994. The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene 148, 357–362. ( 10.1016/0378-1119(94)90713-7) [DOI] [PubMed] [Google Scholar]
- 16.Rensch T, Villar D, Horvath J, Odom DT, Flicek P. 2016. Mitochondrial heteroplasmy in vertebrates using ChIP-sequencing data. Genome Biol. 17, 139 ( 10.1186/s13059-016-0996-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyllensten U, Wharton D, Josefsson A, Wilson AC. 1991. Paternal inheritance of mitochondrial DNA in mice. Nature 352, 255–257. ( 10.1038/352255a0) [DOI] [PubMed] [Google Scholar]
- 18.Magoulas A, Zouros E. 1993. Restriction-site heteroplasmy in anchovy (Engraulis encrasicolus) indicates incidental biparental inheritance of mitochondrial DNA. Mol. Biol. Evol. 10, 319–325. ( 10.1093/oxfordjournals.molbev.a040016) [DOI] [Google Scholar]
- 19.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. 1999. Ubiquitin tag for sperm mitochondria. Nature 402, 371–372. ( 10.1038/46466) [DOI] [PubMed] [Google Scholar]
- 20.Schwartz M, Vissing J. 2002. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 347, 576–580. ( 10.1056/NEJMoa020350) [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, Li N, Guo W, Hu X, Liu Z, Gong G, Wang A, Feng J, Wu C. 2004. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity (Edinb) 93, 399–403. ( 10.1038/sj.hdy.6800516) [DOI] [PubMed] [Google Scholar]
- 22.Ross JA, Howe DK, Coleman-Hulbert A, Denver DR, Estes S. 2016. Paternal mitochondrial transmission in intra-species Caenorhabditis briggsae hybrids. Mol. Biol. Evol. 33, 3158–3160. ( 10.1093/molbev/msw192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo S, et al. 2018. Biparental inheritance of mitochondrial DNA in humans. Proc. Natl Acad. Sci. USA 58, 201810946 ( 10.1073/pnas.1810946115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusman A, Lecomte S, Stewart DT, Passamonti M, Breton S. 2016. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ 4, e2760 ( 10.7717/peerj.2760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol. Biol. 10, 50 ( 10.1186/1471-2148-10-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart JB, Chinnery PF. 2015. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542. ( 10.1038/nrg3966) [DOI] [PubMed] [Google Scholar]
- 27.Wonnapinij P, Chinnery PF, Samuels DC. 2008. The distribution of mitochondrial DNA heteroplasmy due to random genetic drift. Am. J. Hum. Genet. 83, 582–593. ( 10.1016/j.ajhg.2008.10.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. 1996. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14, 146–151. ( 10.1038/ng1096-146) [DOI] [PubMed] [Google Scholar]
- 29.Konrad A, Thompson O, Waterston RH, Moerman DG, Keightley PD, Bergthorsson U, Katju V. 2017. Mitochondrial mutation rate, spectrum and heteroplasmy in Caenorhabditis elegans spontaneous mutation accumulation lines of differing population size. Mol. Biol. Evol. 34, 1319–1334. ( 10.1093/molbev/msx051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashley MV, Laipis PJ, Hauswirth WW. 1989. Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res. 17, 7325–7331. ( 10.1093/nar/17.18.7325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl H-HM, Chinnery PF. 2008. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 40, 249–254. ( 10.1038/ng.2007.63) [DOI] [PubMed] [Google Scholar]
- 32.Wai T, Teoli D, Shoubridge EA. 2008. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 40, 1484–1488. ( 10.1038/ng.258) [DOI] [PubMed] [Google Scholar]
- 33.Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J-I, Yonekawa H. 2007. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 39, 386–390. ( 10.1038/ng1970) [DOI] [PubMed] [Google Scholar]
- 34.Cao L, Shitara H, Sugimoto M, Hayashi J-I, Abe K, Yonekawa H. 2009. New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS Genet. 5, e1000756 ( 10.1371/journal.pgen.1000756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebolledo-Jaramillo B, et al. 2014. Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc. Natl Acad. Sci. USA 111, 15 474–15 479. ( 10.1073/pnas.1409328111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladoukakis ED, Zouros E. 2001. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18, 1168–1175. ( 10.1093/oxfordjournals.molbev.a003904) [DOI] [PubMed] [Google Scholar]
- 37.Hoarau G, Holla S, Lescasse R, Stam WT, Olsen JL. 2002. Heteroplasmy and evidence for recombination in the mitochondrial control region of the flatfish Platichthys flesus. Mol. Biol. Evol. 19, 2261–2264. ( 10.1093/oxfordjournals.molbev.a004049) [DOI] [PubMed] [Google Scholar]
- 38.Kraytsberg Y. 2004. Recombination of human mitochondrial DNA. Science 304, 981 ( 10.1126/science.1096342) [DOI] [PubMed] [Google Scholar]
- 39.Guo X, Liu S, Liu Y. 2006. Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics 172, 1745–1749. ( 10.1534/genetics.105.049841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciborowski KL, Consuegra S, García de Leániz C, Beaumont MA, Wang J, Jordan WC. 2007. Rare and fleeting: an example of interspecific recombination in animal mitochondrial DNA. Biol. Lett. 3, 554–557. ( 10.1098/rsbl.2007.0290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ujvari B, Dowton M, Madsen T. 2007. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 3, 189–192. ( 10.1098/rsbl.2006.0587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma H, O'Farrell PH. 2015. Selections that isolate recombinant mitochondrial genomes in animals. eLife 4, e07247 ( 10.7554/eLife.07247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strakova A, et al. 2016. Mitochondrial genetic diversity, selection and recombination in a canine transmissible cancer. eLife 5, 415 ( 10.7554/eLife.14552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schon EA, DiMauro S, Hirano M. 2012. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878–890. ( 10.1038/nrg3275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorman GS, et al. 2016. Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 ( 10.1038/nrdp.2016.80) [DOI] [PubMed] [Google Scholar]
- 46.2018. MITOMAP: A Human Mitochondrial Genome Database. See http://www.mitomap.org .
- 47.Herbert M, Turnbull D. 2018. Progress in mitochondrial replacement therapies. Nat. Rev. Mol. Cell Biol. 19, 71–72. ( 10.1038/nrm.2018.3) [DOI] [PubMed] [Google Scholar]
- 48.Tachibana M, et al. 2009. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature 461, 367–372. ( 10.1038/nature08368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craven L, et al. 2010. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82–85. ( 10.1038/nature08958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paull D, et al. 2013. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 493, 632–637. ( 10.1038/nature11800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachibana M, et al. 2013. Towards germline gene therapy of inherited mitochondrial diseases. Nature 493, 627–631. ( 10.1038/nature11647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada M, et al. 2016. Genetic drift can compromise mitochondrial replacement by nuclear transfer in human oocytes. Cell Stem Cell 18, 749–754. ( 10.1016/j.stem.2016.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang E, et al. 2016. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature 540, 270–275. ( 10.1038/nature20592) [DOI] [PubMed] [Google Scholar]
- 54.Wu K, et al. 2017. Polar bodies are efficient donors for reconstruction of human embryos for potential mitochondrial replacement therapy. Cell Res. 27, 1069–1072. ( 10.1038/cr.2017.67) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, et al. 2016. First live birth using human oocytes reconstituted by spindle nuclear transfer for mitochondrial DNA mutation causing Leigh syndrome. Fertil. Steril. 106, e375–e376. ( 10.1016/j.fertnstert.2016.08.004) [DOI] [Google Scholar]
- 56.Ji F, et al. 2012. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc. Natl Acad. Sci. USA 109, 7391–7396. ( 10.1073/pnas.1202484109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu M, Dong X, Shi L, Shi L, Lin K, Huang X, Chu J. 2012. Differences in mtDNA whole sequence between Tibetan and Han populations suggesting adaptive selection to high altitude. Gene 496, 37–44. ( 10.1016/j.gene.2011.12.016) [DOI] [PubMed] [Google Scholar]
- 58.Li X-D, Jiang G-F, Yan L-Y, Li R, Mu Y, Deng W-A. 2018. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 9, 605 ( 10.3389/fgene.2018.00605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu S, et al. 2007. High altitude adaptation and phylogenetic analysis of Tibetan horse based on the mitochondrial genome. J. Genet. Genomics 34, 720–729. ( 10.1016/S1673-8527(07)60081-2) [DOI] [PubMed] [Google Scholar]
- 60.Ning T, Xiao H, Li J, Hua S, Zhang YP. 2010. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. 9, 144–150. ( 10.4238/vol9-1gmr705) [DOI] [PubMed] [Google Scholar]
- 61.Luo Y, Yang X, Gao Y. 2013. Mitochondrial DNA response to high altitude: a new perspective on high-altitude adaptation. Mitochondrial DNA 24, 313–319. ( 10.3109/19401736.2012.760558) [DOI] [PubMed] [Google Scholar]
- 62.Mishmar D, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 100, 171–176. ( 10.1073/pnas.0136972100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernatchez L, Glémet H, Wilson CC, Danzmann RG. 1995. Introgression and fixation of Arctic char (Salvelinus alpinus) mitochondrial genome in an allopatric population of brook trout (Salvelinus fontinalis). Can. J. Fish. Aquat. Sci. 52, 179–185. ( 10.1139/f95-018) [DOI] [Google Scholar]
- 64.Fangue NA, Richards JG, Schulte PM. 2009. Do mitochondrial properties explain intraspecific variation in thermal tolerance? J. Exp. Biol. 212, 514–522. ( 10.1242/jeb.024034) [DOI] [PubMed] [Google Scholar]
- 65.Morales HE, Pavlova A, Amos N, Major R, Kilian A, Greening C, Sunnucks P. 2018. Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nat. Ecol. Evol. 2, 1258–1267. ( 10.1038/s41559-018-0606-3) [DOI] [PubMed] [Google Scholar]
- 66.Melo-Ferreira J, Boursot P, Suchentrunk F, Ferrand N, Alves PC. 2005. Invasion from the cold past: extensive introgression of mountain hare (Lepus timidus) mitochondrial DNA into three other hare species in northern Iberia. Mol. Ecol. 14, 2459–2464. ( 10.1111/j.1365-294X.2005.02599.x) [DOI] [PubMed] [Google Scholar]
- 67.Lajbner Z, Pnini R, Camus MF, Miller J, Dowling DK. 2018. Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci. Rep. 8, 9500 ( 10.1038/s41598-018-27805-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238, 195–209. ( 10.1016/S0378-1119(99)00270-X) [DOI] [PubMed] [Google Scholar]
- 69.Fan W, et al. 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319, 958–962. ( 10.1126/science.1147786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson N-GR. 2008. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol 6, e10 ( 10.1371/journal.pbio.0060010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freyer C, et al. 2012. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat. Genet. 44, 1282–1285. ( 10.1038/ng.2427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma H, Xu H, O'Farrell PH. 2014. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet. 46, 393–397. ( 10.1038/ng.2919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill JH, Chen Z, Xu H. 2014. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat. Genet. 46, 389–392. ( 10.1038/ng.2920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stewart JB, Larsson N-G. 2014. Keeping mtDNA in shape between generations. PLoS Genet. 10, e1004670 ( 10.1371/journal.pgen.1004670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palozzi JM, Jeedigunta SP, Hurd TR. 2018. Mitochondrial DNA purifying selection in mammals and invertebrates. J. Mol. Biol. 430, 4834–4848. ( 10.1016/j.jmb.2018.10.019) [DOI] [PubMed] [Google Scholar]
- 76.Burr SP, Pezet M, Chinnery PF. 2018. Mitochondrial DNA heteroplasmy and purifying selection in the mammalian female germ line. Develop. Growth Differ. 60, 21–32. ( 10.1111/dgd.12420) [DOI] [PubMed] [Google Scholar]
- 77.Li M, et al. 2016. Transmission of human mtDNA heteroplasmy in the genome of the Netherlands families: support for a variable-size bottleneck. Genome Res. 26, 417–426. ( 10.1101/gr.203216.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Floros VI, et al. 2018. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat. Cell Biol. 20, 144–151. ( 10.1038/s41556-017-0017-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otten ABC, et al. 2016. Differences in strength and timing of the mtDNA bottleneck between zebrafish germline and non-germline cells. Cell Rep. 16, 622–630. ( 10.1016/j.celrep.2016.06.023) [DOI] [PubMed] [Google Scholar]
- 80.Cotterill M, Harris SE, Collado Fernandez E, Lu J, Huntriss JD, Campbell BK, Picton HM. 2013. The activity and copy number of mitochondrial DNA in ovine oocytes throughout oogenesis in vivo and during oocyte maturation in vitro. MHR: Basic Sci. Reprod. Med. 19, 444–450. ( 10.1093/molehr/gat013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdu Y, Maniscalco C, Heddleston JM, Chew T-L, Nance J. 2016. Developmentally programmed germ cell remodelling by endodermal cell cannibalism. Nat. Cell Biol. 18, 1302–1310. ( 10.1038/ncb3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee H-S, et al. 2012. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell Rep. 1, 506–515. ( 10.1016/j.celrep.2012.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou RR, Wang B, Wang J, Schatten H, Zhang YZ. 2010. Is the mitochondrial cloud the selection machinery for preferentially transmitting wild-type mtDNA between generations? Rewinding Müller's ratchet efficiently. Curr. Genet. 56, 101–107. ( 10.1007/s00294-010-0291-5) [DOI] [PubMed] [Google Scholar]
- 84.Cox RT, Spradling AC. 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579–1590. ( 10.1242/dev.00365) [DOI] [PubMed] [Google Scholar]
- 85.Xu H, DeLuca SZ, O'Farrell PH. 2008. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science 321, 575–577. ( 10.1126/science.1160226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hurd TR, Herrmann B, Sauerwald J, Sanny J, Grosch M, Lehmann R. 2016. Long oskar controls mitochondrial inheritance in Drosophila melanogaster. Dev. Cell 39, 560–571. ( 10.1016/j.devcel.2016.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teixeira FK, Sanchez CG, Hurd TR, Seifert JRK, Czech B, Preall JB, Hannon GJ, Lehmann R. 2015. ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat. Cell Biol. 17, 689–696. ( 10.1038/ncb3165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Paula WBM, Agip A-NA, Missirlis F, Ashworth R, Vizcay-Barrena G, Lucas CH, Allen JF. 2013. Female and male gamete mitochondria are distinct and complementary in transcription, structure, and genome function. Genome Biol. Evol. 5, 1969–1977. ( 10.1093/gbe/evt147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Z, Qi Y, French S, Zhang G, Covian Garcia R, Balaban R, Xu H. 2015. Genetic mosaic analysis of a deleterious mitochondrial DNA mutation in Drosophila reveals novel aspects of mitochondrial regulation and function. Mol. Biol. Cell 26, 674–684. ( 10.1091/mbc.E14-11-1513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pickles S, Vigié P, Youle RJ. 2018. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185. ( 10.1016/j.cub.2018.01.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kandul NP, Zhang T, Hay BA, Guo M. 2016. Selective removal of deletion-bearing mitochondrial DNA in heteroplasmic Drosophila. Nat. Commun. 7, 13100 ( 10.1038/ncomms13100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pickrell AM, Huang C-H, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. 2015. Endogenous Parkin preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron 87, 371–381. ( 10.1016/j.neuron.2015.06.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Monnot S, et al. 2010. Segregation of mtDNA throughout human embryofetal development: m.3243A > G as a model system. Hum. Mutat. 32, 116–125. ( 10.1002/humu.21417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. 2001. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am. J. Hum. Genet. 68, 533–536. ( 10.1086/318190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson IJ, et al. 2016. Mitochondrial DNA sequence characteristics modulate the size of the genetic bottleneck. Hum. Mol. Genet. 25, 1031–1041. ( 10.1093/hmg/ddv626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holland MM, Makova KD, McElhoe JA. 2018. Deep-coverage MPS analysis of heteroplasmic variants within the mtGenome allows for frequent differentiation of maternal relatives. Genes 9, 124 ( 10.3390/genes9030124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gemmell NJ, Metcalf VJ, Allendorf FW. 2004. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol. 19, 238–244. ( 10.1016/j.tree.2004.02.002) [DOI] [PubMed] [Google Scholar]
- 98.Camus MF, Wolf JBW, Morrow EH, Dowling DK. 2015. Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr. Biol. 25, 2717–2722. ( 10.1016/j.cub.2015.09.012) [DOI] [PubMed] [Google Scholar]
- 99.Patel MR, et al. 2016. A mitochondrial DNA hypomorph of cytochrome oxidase specifically impairs male fertility in Drosophila melanogaster. eLife 5, 1144 ( 10.7554/eLife.16923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milot E, Moreau C, Gagnon A, Cohen AA, Brais B, Labuda D. 2017. Mother's curse neutralizes natural selection against a human genetic disease over three centuries. Nat. Ecol. Evol. 1, 1400–1406. ( 10.1038/s41559-017-0276-6) [DOI] [PubMed] [Google Scholar]
- 101.Beziat F, Morel F, Volz-Lingenhol A, Saint Paul N, Alziari S. 1993. Mitochondrial genome expression in a mutant strain of D. subobscura, an animal model for large scale mtDNA deletion. Nucleic Acids Res. 21, 387–392. ( 10.1093/nar/21.3.387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsang WY, Lemire BD. 2002. Stable heteroplasmy but differential inheritance of a large mitochondrial DNA deletion in nematodes. Biochem. Cell Biol. 80, 645–654. ( 10.1139/o02-135) [DOI] [PubMed] [Google Scholar]
- 103.Howe DK, Denver DR. 2008. Muller's Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 8, 62 ( 10.1186/1471-2148-8-62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petit N, Touraille S, Debise R, Morel F, Renoux M, Lécher P, Alziari S. 1998. Developmental changes in heteroplasmy level and mitochondrial gene expression in a Drosophila subobscura mitochondrial deletion mutant. Curr. Genet. 33, 330–339. ( 10.1007/s002940050344) [DOI] [PubMed] [Google Scholar]
- 105.Clark KA, Howe DK, Gafner K, Kusuma D, Ping S, Estes S, Denver DR. 2012. Selfish little circles: transmission bias and evolution of large deletion-bearing mitochondrial DNA in Caenorhabditis briggsae nematodes. PLoS ONE 7, e41433 ( 10.1371/journal.pone.0041433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, Patel MR. 2016. Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab. 24, 91–103. ( 10.1016/j.cmet.2016.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Capps GJ, Samuels DC, Chinnery PF. 2003. A model of the nuclear control of mitochondrial DNA replication. J. Theoret. Biol. 221, 565–583. ( 10.1006/jtbi.2003.3207) [DOI] [PubMed] [Google Scholar]
- 108.Chinnery PF, Samuels DC. 1999. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am. J. Hum. Genet. 64, 1158–1165. ( 10.1086/302311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tam ZY, Gruber J, Halliwell B, Gunawan R. 2015. Context-dependent role of mitochondrial fusion-fission in clonal expansion of mtDNA mutations. PLoS Comput. Biol. 11, e1004183 ( 10.1371/journal.pcbi.1004183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin Y-F, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. 2016. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533, 416–419. ( 10.1038/nature17989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma H, O'Farrell PH. 2016. Selfish drive can trump function when animal mitochondrial genomes compete. Nat. Genet. 48, 798–802. ( 10.1038/ng.3587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rand DM, Harrison RG. 1986. Mitochondrial DNA transmission genetics in crickets. Genetics 114, 955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. 1991. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 88, 10 614–10 618. ( 10.1073/pnas.88.23.10614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT. 2002. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 30, 4626–4633. ( 10.1093/nar/gkf602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Niki Y, Chigusa SI, Matsuura ET. 1989. Complete replacement of mitochondrial DNA in Drosophila. Nature 341, 551–552. ( 10.1038/341551a0) [DOI] [PubMed] [Google Scholar]
- 116.De Stordeur E. 1997. Nonrandom partition of mitochondria in heteroplasmic Drosophila. Heredity (Edinb) 79(Pt 6), 615–623. ( 10.1038/sj.hdy.6882760) [DOI] [PubMed] [Google Scholar]
- 117.Rand DM. 2011. Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica 139, 685–697. ( 10.1007/s10709-011-9576-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Greiner S, Sobanski J, Bock R. 2015. Why are most organelle genomes transmitted maternally? BioEssays 37, 80–94. ( 10.1002/bies.201400110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nishimura Y, Yoshinari T, Naruse K, Yamada T, Sumi K, Mitani H, Higashiyama T, Kuroiwa T. 2006. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc. Natl Acad. Sci. USA 103, 1382–1387. ( 10.1073/pnas.0506911103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. ( 10.1126/science.1211878) [DOI] [PubMed] [Google Scholar]
- 121.Rojansky R, Cha M-Y, Chan DC. 2016. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife 5, 1144 ( 10.7554/eLife.17896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.DeLuca SZ, O'Farrell PH. 2012. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell 22, 660–668. ( 10.1016/j.devcel.2011.12.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo S-M, Ge Z-J, Wang Z-W, Jiang Z-Z, Wang Z-B, Ouyang Y-C, Hou Y, Schatten H, Sun Q-Y. 2013. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl Acad. Sci. USA 110, 13 038–13 043. ( 10.1073/pnas.1303231110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E. 2014. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev. Cell 29, 305–320. ( 10.1016/j.devcel.2014.04.005) [DOI] [PubMed] [Google Scholar]
- 125.Yu Z, O'Farrell PH, Yakubovich N, DeLuca SZ. 2017. The mitochondrial DNA polymerase promotes elimination of paternal mitochondrial genomes. Curr. Biol. 27, 1033–1039. ( 10.1016/j.cub.2017.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sato K, Sato M. 2017. Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J. Biochem. 162, 247–253. ( 10.1093/jb/mvx052) [DOI] [PubMed] [Google Scholar]
- 127.Lécher P, Beziat F, Alziari S. 1994. Tissular distribution of heteroplasmy and ultrastructural studies of mitochondria from a Drosophila subobscura mitochondrial deletion mutant. Biol. Cell 80, 25–33. ( 10.1016/0248-4900(94)90013-2) [DOI] [PubMed] [Google Scholar]
- 128.Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Phil. Trans. R. Soc. B 369, 20130443 ( 10.1098/rstb.2013.0443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sloan DB, Havird JC, Sharbrough J. 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 26, 2212–2236. ( 10.1111/mec.13959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herbers E, Kekäläinen NJ, Hangas A, Pohjoismäki JL, Goffart S. 2018. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion 44, 85–92. ( 10.1016/j.mito.2018.01.004) [DOI] [PubMed] [Google Scholar]
- 131.Fernández-Vizarra E, Enríquez JA, Pérez-Martos A, Montoya J, Fernández-Silva P. 2011. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion 11, 207–213. ( 10.1016/j.mito.2010.09.011) [DOI] [PubMed] [Google Scholar]
- 132.Samuels DC, et al. 2013. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 9, e1003929 ( 10.1371/journal.pgen.1003929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matsuura ET. 1991. Selective transmission of mitochondrial DNA in Drosophila. Jpn. J. Genet. 66, 683–700. ( 10.1266/jjg.66.683) [DOI] [PubMed] [Google Scholar]
- 134.Matsuura ET, Tanaka YT, Yamamoto N. 1997. Effects of the nuclear genome on selective transmission of mitochondrial DNA in Drosophila. Genes Genet. Syst. 72, 119–123. ( 10.1266/ggs.72.119) [DOI] [PubMed] [Google Scholar]
- 135.Kann LM, Rosenblum EB, Rand DM. 1998. Aging, mating, and the evolution of mtDNA heteroplasmy in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 95, 2372–2377. ( 10.1073/pnas.95.5.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Farge G, Touraille S, Le Goff S, Petit N, Renoux M, Morel F, Alziari S. 2002. The nuclear genome is involved in heteroplasmy control in a mitochondrial mutant strain of Drosophila subobscura. Eur. J. Biochem. 269, 998–1005. ( 10.1046/j.0014-2956.2001.02737.x) [DOI] [PubMed] [Google Scholar]
- 137.Jenuth JP, Peterson AC, Shoubridge EA. 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16, 93–95. ( 10.1038/ng0597-93) [DOI] [PubMed] [Google Scholar]
- 138.Battersby BJ, Loredo-Osti JC, Shoubridge EA. 2003. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 33, 183–186. ( 10.1038/ng1073) [DOI] [PubMed] [Google Scholar]
- 139.Burgstaller JP, et al. 2014. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 7, 2031–2041. ( 10.1016/j.celrep.2014.05.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eyre-Walker A. 2017. Mitochondrial replacement therapy: are mito-nuclear interactions likely to be a problem? Genetics 205, 1365–1372. ( 10.1534/genetics.116.196436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rishishwar L, Jordan IK. 2017. Implications of human evolution and admixture for mitochondrial replacement therapy. BMC Genomics 18, 140 ( 10.1186/s12864-017-3539-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin T-K, et al. 2012. The creation of cybrids harboring mitochondrial haplogroups in the Taiwanese population of ethnic Chinese background: an extensive in vitro tool for the study of mitochondrial genomic variations. Oxidative Med. Cell. Longevity 2012, 1–13. ( 10.1155/2012/824275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kenney MC, et al. 2014. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum. Mol. Genet. 23, 3537–3551. ( 10.1093/hmg/ddu065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Havird JC, Fitzpatrick SW, Kronenberger J, Funk WC, Angeloni LM, Sloan DB. 2016. Sex, mitochondria, and genetic rescue. Trends Ecol. Evol. 31, 96–99. ( 10.1016/j.tree.2015.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barritt J, Willadsen S, Brenner C, Cohen J. 2001. Cytoplasmic transfer in assisted reproduction. Hum. Reprod. Update 7, 428–435. ( 10.1093/humupd/7.4.428) [DOI] [PubMed] [Google Scholar]
- 146.Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. 2014. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 157, 1591–1604. ( 10.1016/j.cell.2014.04.042) [DOI] [PubMed] [Google Scholar]
- 147.Ma H, et al. 2017. Functional human oocytes generated by transfer of polar body genomes. Cell Stem Cell 20, 112–119. ( 10.1016/j.stem.2016.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.van Oven M, Kayser M. 2009. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394. ( 10.1002/humu.20921) [DOI] [PubMed] [Google Scholar]
- 149.Dunbar DR, Moonie PA, Jacobs HT, Holt IJ. 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl Acad. Sci. USA 92, 6562–6566. ( 10.1073/pnas.92.14.6562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cree L, Loi P. 2015. Mitochondrial replacement: from basic research to assisted reproductive technology portfolio tool-technicalities and possible risks. Mol. Hum. Reprod. 21, 3–10. ( 10.1093/molehr/gau082) [DOI] [PubMed] [Google Scholar]
- 151.Reddy P, et al. 2015. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell 161, 459–469. ( 10.1016/j.cell.2015.03.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bacman SR, et al. 2018. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. 24, 1696–1700. ( 10.1038/s41591-018-0166-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gammage PA, et al. 2018. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med. 24, 1691–1695. ( 10.1038/s41591-018-0165-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang Y, et al. 2018. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell 9, 283–297. ( 10.1007/s13238-017-0499-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.