Abstract
Phage therapy is attracting growing interest among clinicians as antibiotic resistance continues becoming harder to control. However, clinical trials and animal model studies on bacteriophage treatment are still scarce and results on the efficacy vary. Recent research suggests that using traditional antimicrobials in concert with phage could have desirable synergistic effects that hinder the evolution of resistance. Here, we present a novel insect gut model to study phage–antibiotic interaction in a system where antibiotic resistance initially exists in very low frequency and phage specifically targets the resistance bearing cells. We demonstrate that while phage therapy could not reduce the frequency of target bacteria in the population during positive selection by antibiotics, it alleviated the antibiotic induced blooming by lowering the overall load of resistant cells. The highly structured gut environment had pharmacokinetic effects on both phage and antibiotic dynamics compared with in vitro: antibiotics did not reduce the overall amount of bacteria, demonstrating a simple turnover of gut microbiota from non-resistant to resistant population with little cost. The results imply moderate potential for using phage as an aid to target antibiotic resistant gut infections, and question the usefulness of in vitro inferences.
Keywords: antibiotic resistance, bacteriophage, Enterobacter cloacae, gut infection, insect model, phage therapy
1. Introduction
Problems arising from antibiotic resistant infections are set to increase worldwide. As a consequence, bacteriophages (bacteria-specific viruses) have started to attract serious consideration as antimicrobial agents after this approach was largely forgotten for decades in the Western world [1,2]. However, their efficacy as therapeutic agents remains controversial [3]. Some of the in vivo work promises therapeutic potential in insect [4], mouse [5] and human infections [6], and in vitro experiments show overwhelming evidence of phages controlling bacterial population sizes without adverse effects on non-target bacteria [7]. Despite this, the few existing modern and properly controlled medical trials report varying success [3,6,8]. Key limitations of phage therapy include high specificity, ease at which bacteria can evolve resistance, and localized activity in the body.
It has been argued that phages may be of particular therapeutic value when combined with antibiotics, by constraining the emergence and spread of antibiotic resistance [9]. A good example of recent success in combination treatment comes from a difficult chronic Pseudomonas aeruginosa infection in human aortic graft [10]. Theoretically, phages may limit antibiotic resistance during treatment because of reduction in population size and synergistic costs of resistance [9]. Moreover, with cases where antibiotic resistance is carried on plasmids, phage can be used to directly target the plasmid carrying cells [11]. However, this latter approach has only been investigated in vitro [11,12], which ignores a plethora of selection pressures towards both the host bacteria and the phage, such as host immune system, spatial structures within the tissues, nutrient availability, and presence of native microbial microbiota [13].
Here, we studied the effects of antibiotic and phage treatment on bacterial load and frequency of resistance in a gnotobiotic insect gut model system, where phage target the bacteria harbouring antibiotic resistance plasmids. We compare these findings with an in vitro context to assess the usefulness of inferring in vivo dynamics from in vitro studies. A full factorial set-up of antibiotics and phage was orally administered to cabbage looper (Trichoplusia ni) larvae harbouring Enterobacter cloacae gut bacteria and a low initial frequency of antibiotic resistance plasmid. We show that targeting the tetracycline resistant cells with phage could not prevent the increase in resistance frequency in the presence of antibiotics. However, phage reduced the bacterial population size when plasmid was driven to high frequency with antibiotic selection.
2. Material and methods
Although it has been argued that lepidopterans lack a resident microbiome [14], Enterobacter cloacae forms a persistent gut association with lepidopteran larvae [15] after oral inoculation. The strain used was isolated from Plutella xylostella in the insectary of the University of Oxford. Two spontaneous antibiotic resistant mutants, ANC C2 (rifR) and 11.1B (strepR + nalR) were used in this experiment. The IncP-type plasmid RP4, coding for tetracycline resistance, is usually conjugative [16], and the lytic, plasmid dependent tectivirus PRD1 specifically infects RP4 bearing cells by recognizing the bacterial sex-apparatus [17]. However, in this system the plasmid is essentially non-conjugative and functions as a carrier of tetracycline resistance and phage receptor genes. The mechanistic basis for low RP4 conjugation in these E. cloacae strains is unclear. Given that the phage remains infective, it could be that ANC C2 rifampicin resistant mutant was an impotent recipient for the plasmid rather than that 11.1B(RP4) lacked the ability to express pili. Although rifampicin resistance mutations are most often polymerase related, known mechanisms in Enterobacteriaceae include elongated or abundant outer membrane lipopolysaccharide chains [18] that could theoretically interfere with conjugation (but, see: [19]).
The in vitro experiment used 200 µl cultures on a 96-well plate with Luria-Bertani medium (LB) and 0.1% starting frequency of plasmid. Twenty-four replicate populations were subjected to full factorial antibiotic (AB) and phage (P) treatment: AB− P−, AB− P+, AB+ P−, and AB+ P+, respectively. The antibiotic treatments were set to 12 µg ml−1 tetracycline and phage starting density to 5 × 107 phage particles. The growth in optical density (600 nm) at 30°C was recorded at 24 h with a spectrophotometer. Plasmid frequency and transconjugants were sampled from a subset (N = 44) of populations at 24 h by selective plating.
The T. ni were maintained as follows. The adults, fed with 100 mM sucrose solution, were allowed to mate and lay eggs on paper strips in flight chambers. The eggs were placed in autoclaved, single-filter vented, 305 × 203 × 100 mm Genesis™ containers for surgical instruments to maintain sterile conditions while allowing gas exchange. Hatched larvae were allowed to feed on Hoffman diet (288 g wheatgerm, 132 g casein, 117 g sucrose, 70 g agar, 57 g dried brewers yeast, 37.5 g Wesson's salts, 6 g sorbic acid, 3.75 g cholesterol, 3.75 g methyl-4-hydroxybenzoate and 2 g Vanderzant vitamin mixture in 2750 ml water) [20]. The liquid medium was poured into a sterile steel mould and solidified to diet cubes with approximately 25 mm edges. The cubes were placed on Petri dish covers inside the containers. All rearing and in vivo bacterial work was carried out at 25°C. For population maintenance, 50–60 pupae were removed from their cocoons per flight chamber and allowed to emerge and mate to start a new generation of hosts.
For inoculating the larval gut with E. cloacae, clones ANC C2 and 11.1B bearing RP4 were grown overnight in LB (30°C, 180 r.p.m. shaking), the 11.1B diluted 1 : 10 with 0.85% NaCl, and then mixed together in a 50 : 1 ratio. The frequency of the plasmid bearing cells was counted by dilution plating as 0.072% and bacterial density approximately 1.2 × 108 colony-forming units (CFU). Diet cubes were dipped in this solution, dried for 5 min, and 3rd instar larvae were allowed to feed on this bacterial diet for 30 h. The larvae were then subjected to the AB− P−, AB− P+, AB+ P−, and AB+ P+ treatments. Tetracycline (200 µg ml−1 final concentration) was added directly to the autoclaved diet mix and 30 µl of 1010 plaque-forming units (PFU) ml−1 filtered phage lysate was pipetted onto each face of the phage treatment diet cubes. The experiment was carried out in two independent batches (final N = 156 larvae). After 96 h, the larvae were surface sterilized with 70% alcohol and homogenized in 500 µl 0.85% NaCl with Qiagen Tissuelyser II™. The homogenate was serially diluted and plated on 12 µg ml−1 tetracycline, 12 µg ml−1 tetracycline + 100 µg ml−1 rifampicin, and LB plates to calculate the frequencies of plasmid carriers and transconjugants, and total density of bacteria. Twenty-four bacterial clones from the phage treatment serial dilutions on tetracycline were tested for resistance against the ancestral phage with a phage plaque overlay assay. Phage survival in the gut was confirmed by overlaying homogenized larval faecal samples on ancestral 11.1B(RP4) lawn on soft agar and observing plaque formation (N = 24).
Treatment effects on densities and frequencies in both experiments were analysed with SPSS Statistics 21.0 with antibiotics, phage, and antibiotics * phage interaction in an ANOVA-GLM model. Post hoc multiple comparisons were Bonferroni corrected.
3. Results
In vitro, antibiotics and phage had synergistic effects on the total density of bacteria at 24 h (AB: F1,96 = 3528.0, p < 0.001; P: F1,96 = 73.7, p < 0.001; AB * P: F1,96 = 97.0, p < 0.001). Multiple comparisons showed that there was no phage effect in absence of antibiotics, but other pairwise differences were highly significant (p < 0.001, figure 1a). Thus, single treatment allowed compensatory growth of the non-target strain, but this effect was reduced for the plasmid carriers in the presence of antibiotics. The plasmid went to near fixation in the presence of antibiotics (F1,44 = 8864.4, p < 0.001) regardless of the phage (P and AB * P: p > 0.05). This was contrary to previous studies with the same plasmid and phage in E. coli [12]. The increase in frequency was not the result of conjugational transfer, because unlike in E. coli, RP4 had very low conjugation rate in E. cloacae: transconjugants were present in only one of the 44 sampled in vitro populations (AB+ P− treatment at a very low frequency). We also confirmed that the phage remained infective even with low conjugation. The cost of plasmid carriage in the absence of antibiotics manifested itself as complete plasmid extinction (at least below the detection threshold of 67 c.f.u. ml−1) in all sampled populations (figure 1b). This was the case even though RP4 has been reported to be of low cost and rapidly evolve towards no cost through chromosomal compensatory mutations [21].
Figure 1.
Dynamics in vitro. The density of bacteria (a), and the frequency of plasmid bearing cells (b) in liquid culture after 24 h factorial antibiotic (AB) and phage (P) treatments. Error bars are ±1 s.e.; ** denotes p < 0.001 difference.
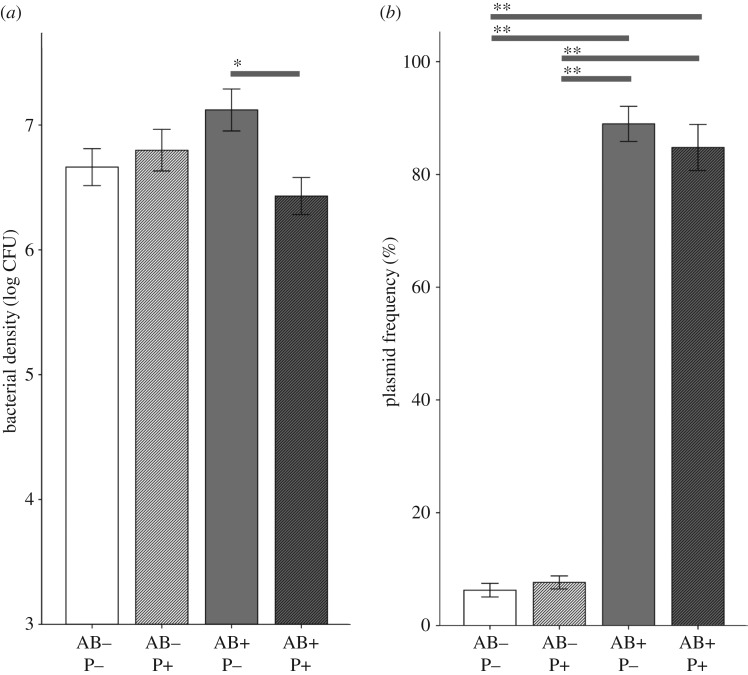
In vivo patterns of plasmid dynamics were mainly similar to that observed in vitro, but the effects smaller. The non-targeted strain could fully compensate for the reduction in density of the competing strain (AB: F1, 156 = 0.1, p = 0.77), and phage had no main effect on total bacterial load (P: F1, 156 = 3.0, p = 0.085). Phage, however, lowered bacterial density in the presence of antibiotics (interaction: F1, 156 = 6.7, p = 0.011), demonstrating that the treatment synergism was maintained in vivo (post hoc AB+ P− and AB+ P+, p = 0.020, figure 2a). Phage-susceptibility screening of evolved clones showed no emergence of resistance. Plasmid increased in frequency in all treatments, but tetracycline selection drove the majority of the population to be plasmid carriers with no effects of phage addition (effects on plasmid frequency, AB: F1, 156 = 845.0, p < 0.001; P: F1, 156 = 0.6, p = 0.43; AB * P: F1,156 = 0.4, p = 0.55 (figure 2b)). Thus, phage therapy could not restrict the spread of antibiotic resistance in the T. ni gut.
Figure 2.
Dynamics in vivo. The overall density of bacteria (a) and the frequency of plasmid bearing cells (b) in the larval guts after 96 h factorial antibiotic (AB) and phage (P) treatments. Error bars are ±1 s.e.; * and ** denote p < 0.05 and p < 0.001 differences, respectively.
4. Discussion
Here, we investigated the effect of plasmid-targeting phage on the spread of antibiotic resistance plasmid in vitro and in an insect gut model. Different selective environment in the gut made treatment effects less pronounced compared with the liquid culture: for example, the increased complexity in spatial structures may have protected the plasmid from extinction in the absence of antibiotics, and thus allowed more non-resistant bacteria to coexist in the presence of tetracycline. This happened even though the antibiotic concentration in the diet was over 16 times higher than in the liquid culture and the experiment length four times longer, allowing more time for potential fixation and/or extinction to happen. Phage driven reduction in the bacterial population size in the presence of tetracycline was also more moderate in vivo, but it is also notable that no detectable phage resistance emerged in the sampled populations. Surprisingly, antibiotics did not reduce the overall bacterial density in the gut, but rather replaced non-resistant cells with resistant ones while population size remained the same. In other words, antibiotics increased the relative fitness of the resistant strain without reducing the absolute fitness [22]. This was in contrast with liquid culture, where the plasmid went even closer to fixation in the presence of antibiotics, but resulted in lower total bacterial density, which suggests a greater growth cost associated with antibiotic resistance in vitro. We cannot attribute the inversion of fitness effects between in vivo and in vitro conditions (increase in plasmid frequency in vivo, and extinction in vitro in absence of antibiotics) directly to the plasmid, because it could also be due to unknown differences in the mutant strains. The result still highlights the limitations of making inferences from test tube dynamics.
In the absence of antibiotics neither in vitro nor in vivo regimes showed a phage effect on the bacterial population size, which was not surprising because of very low initial frequency of phage-susceptible (i.e. plasmid bearing) cells. Most importantly, when antibiotic selection increased the frequency of phage-susceptible cells by spreading antibiotic resistance, phage decreased the total density of bacteria. This effect was bigger in vitro, but also clearly visible in the insect gut. The gut lumen is a favourable environment for bacterial biofilm formation [23], which can offer non-resistant cells spatial protection from antibiotics [24] and/or from phage [25]. On the other hand, it could be that the observed phage efficacy to reduce bacterial load in vivo is partly due to their known ability to eradicate biofilms [26,27], which can be important for bacterial persistence in the insect gut [23].
One potential epidemiological implication of viable phage in the faeces is that the viral therapeutic agent could restrict ongoing pathogen transmission or be transmitted with the target pathogen [28]. The model system presented in this study allows the manipulation of structure and connectivity of the population of the treated organism and would be an interesting future endeavour towards epidemiological effects of phage therapy. Our findings have implications for therapeutic use of phage, especially considering synergistic evolutionary effects with antibiotics [9]. However, antibiotics overwhelmed the selection imposed by the phage, meaning that the relative effect remained rather small. This could be improved with using more efficient phage or phage cocktails in a way that can target wider niche space in vivo by, for example, biofilm specificity [27]. It has also been shown that the effect of phage on plasmid maintenance can depend on the type of antibiotics that are used [29]. To conclude, within-organism pharmacokinetics and the spatial, temporal, and resource-related dynamics are very different from those in a test tube, having major effects on cost of resistance, magnitude of selection, and treatment efficacy even in a highly simplified and low diversity model system.
Acknowledgements
The authors thank Alex Robinson for assistance with T. ni rearing.
Data accessibility
Data are available from the Dryad Digital Repository at: http://dx.doi.org/10.5061/dryad.sc54383 [30].
Authors' contributions
All authors conceived the study and designed the experiments. L.M. did the experimental work and analysed the data. All authors contributed to writing, approved the final version of the manuscript, and agree to be held accountable for the work performed.
Competing interests
No competing interests.
Funding
The work was funded by an MRC innovation award (MR/N013824/1), Academy of Finland grants (252411 and 297049) and Emil Aaltonen Foundation grant.
References
- 1.Reardon S. 2014. Phage therapy gets revitalized: the rise of antibiotic resistance rekindles interest in a century-old virus treatment. Nature 510, 15–17. ( 10.1038/510015a) [DOI] [PubMed] [Google Scholar]
- 2.Alavidze Z, et al. 2016. Silk route to the acceptance and re-implementation of bacteriophage therapy. Biotechnol. J. 11, 595–600. ( 10.1002/biot.201600023) [DOI] [PubMed] [Google Scholar]
- 3.Brussow H. 2017. Phage therapy for the treatment of human intestinal bacterial infections: soon to be a reality? Expert Rev. Gastroenterol. Hepatol. 11, 785–788. ( 10.1080/17474124.2017.1342534) [DOI] [PubMed] [Google Scholar]
- 4.Seed KD, Dennis JJ. 2009. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 53, 2205–2208. ( 10.1128/AAC.01166-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, Debarbieux L. 2017. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22, 38–47. ( 10.1016/j.chom.2017.06.018) [DOI] [PubMed] [Google Scholar]
- 6.Wright A, Hawkins C, Änggård E, Harper D. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357. ( 10.1111/j.1749-4486.2009.01973.x) [DOI] [PubMed] [Google Scholar]
- 7.Abedon S. 2008. Phages, ecology, evolution. In Bacteriophage ecology: population growth, evolution, and impact of bacterial viruses (ed. Abedon S.), pp. 1–28. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Sarker SA, et al. 2016. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137. ( 10.1016/j.ebiom.2015.12.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Barcelo C, Hochberg ME. 2016. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 24, 249–256. ( 10.1016/j.tim.2015.12.011) [DOI] [PubMed] [Google Scholar]
- 10.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66. ( 10.1093/emph/eoy005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalasvuori M, Friman VP, Nieminen A, Bamford JK, Buckling A. 2011. Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol. Lett. 7, 902–905. ( 10.1098/rsbl.2011.0384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ojala V, Laitalainen J, Jalasvuori M. 2013. Fight evolution with evolution: plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evol. Appl. 6, 925–932. ( 10.1111/eva.12076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne RJH, Jansen VA. 2000. Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharmacol. Ther. 68, 225–230. ( 10.1067/mcp.2000.109520) [DOI] [PubMed] [Google Scholar]
- 14.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA 114, 9641–9646. ( 10.1073/pnas.1707186114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe K, Abe K, Sato M. 2000. Biological control of an insect pest by gut-colonizing Enterobacter cloacae transformed with ice nucleation gene. J. Appl. Microbiol. 88, 90–97. ( 10.1046/j.1365-2672.2000.00904.x) [DOI] [PubMed] [Google Scholar]
- 16.Datta N, Hedges RW, Shaw EJ, Sykes RB, Richmond MH. 1971. Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108, 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen RH, Siak J-S, Grey RH. 1974. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 14, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein BP. 2014. Resistance to rifampicin: a review. J. Antibiot. 67, 625–630. ( 10.1038/ja.2014.107) [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Mendoza D, de la Cruz F. 2009. Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10, 71 ( 10.1186/1471-2164-10-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knipling E, Smith C. 1966. Semisynthetic diet used to rear Heliothis and Trichoplusia. In Insect colonisation and mass production (ed. Smith C.), pp. 479–486. New York, NY: Academic Press. [Google Scholar]
- 21.Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165, 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day T, Huijben S, Read AF. 2015. Is selection relevant in the evolutionary emergence of drug resistance? Trends Microbiol. 23, 126–133. ( 10.1016/j.tim.2015.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JK, Kwon JY, Kim SK, Han SH, Won YJ, Lee JH, Kim CH, Fukatsu T, Lee BL. 2014. Purine biosynthesis, biofilm formation, and persistence of an insect-microbe gut symbiosis. Appl. Environ. Microbiol. 80, 4374–4382. ( 10.1128/AEM.00739-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. ( 10.1016/s0140-6736(01)05321-1) [DOI] [PubMed] [Google Scholar]
- 25.Sutherland IW, Hughes KA, Skillman LC, Tait K. 2004. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232, 1–6. ( 10.1016/s0378-1097(04)00041-2) [DOI] [PubMed] [Google Scholar]
- 26.Donlan RM. 2009. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 17, 66–72. ( 10.1016/j.tim.2008.11.002) [DOI] [PubMed] [Google Scholar]
- 27.Forti F, et al. 2018. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 62, e02573-17 ( 10.1128/AAC.02573-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atad I, Zvuloni A, Loya Y, Rosenberg E. 2012. Phage therapy of the white plague-like disease of Favia favus in the Red Sea. Coral Reefs 31, 665–670. ( 10.1007/s00338-012-0900-5) [DOI] [Google Scholar]
- 29.Cairns J, et al. 2018. Black Queen evolution and trophic interactions determine plasmid survival after the disruption of the conjugation network. mSystems 3, e00104-18 ( 10.1128/mSystems.00104-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikonranta L, Buckling A, Jalasvuori M, Raymond B. 2019. Data from: Targeting antibiotic resistant bacteria with phages reduces bacterial density in an insect host Dryad Digital Repository. ( 10.5061/dryad.sc54383) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Mikonranta L, Buckling A, Jalasvuori M, Raymond B. 2019. Data from: Targeting antibiotic resistant bacteria with phages reduces bacterial density in an insect host Dryad Digital Repository. ( 10.5061/dryad.sc54383) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Data are available from the Dryad Digital Repository at: http://dx.doi.org/10.5061/dryad.sc54383 [30].