ABSTRACT
An adaptive visual system is essential for organisms inhabiting new or changing light environments. The Panama Canal exhibits such variable environments owing to its anthropogenic origin and current human activities. Within the Panama Canal, Lake Gatun harbors several exotic fish species including the invasive peacock bass (Cichla monoculus), a predatory Amazonian cichlid. In this research, through spectral measurements and molecular and physiological experiments, we studied the visual system of C. monoculus and its adaptive capabilities. Our results suggest that (1) Lake Gatun is a highly variable environment, where light transmission changes throughout the canal waterway, and that (2) C. monoculus has several visual adaptations suited for this red-shifted light environment. Cichla monoculus filters short wavelengths (∼400 nm) from the environment through its ocular media and tunes its visual sensitivities to the available light through opsin gene expression. More importantly, based on shifts in spectral sensitivities of photoreceptors alone, and on transcriptome analysis, C. monoculus exhibits extreme intraspecific variation in the use of vitamin A1/A2 chromophore in their photoreceptors. Fish living in turbid water had higher proportions of vitamin A2, shifting sensitivities to longer wavelengths, than fish living in clear water. Furthermore, we also found variation in retinal transcriptomes, where fish from turbid and clear waters exhibited differentially expressed genes that vary greatly in their function. We suggest that this phenotypic plasticity has been key in the invasion success of C. monoculus.
KEY WORDS: Fish vision, Cichlids, Chromophore, Invasive species, Transcriptomics, Phenotypic plasticity
Summary: Examination of the visual system plasticity of the peacock bass, Cichla monoculus, shows that this species adapts to a changing environment by alternating the use of chromophores A1 and A2, and through differential gene expression in the retina.
INTRODUCTION
The ability of an animal to survive in a changing environment is often determined by its adaptive capabilities. Phenotypic plasticity is a key mechanism by which organisms can respond to short-term environmental changes or fluctuations. Aquatic habitats exhibit substantial spatial and temporal variation in light intensity, spectral composition, angular distribution and degree of polarized light (Warrant and Johnsen, 2013), suggesting that the eyes of aquatic organisms might be particularly promising for studies of phenotypic plasticity.
Animals are believed to adapt to their visual environment by matching their photoreceptor sensitivities to the available light. This ‘sensitivity hypothesis’ explains how modulating the visual system spectral sensitivities to a specific wavelength facilitates certain visual tasks (Lythgoe, 1979; Marshall et al., 2015). Shifts in visual sensitivities can be driven by ecological factors such as water turbidity and foraging targets, or social factors important for communication (Ehlman et al., 2015; Hofmann et al., 2009; Losey et al., 1999).
Studying adaptation of retinal visual sensitivities is possible because of a well-defined genotype-to-phenotype map. This map results from our genetic understanding of how visual sensitivities are tuned (Davies et al., 2012). Fish visual perception is a product of lens transmission and photoreceptor absorbances across the wavelength spectrum. The specific spectral sensitivity of a photoreceptor cell is largely determined by the visual pigment(s) it contains, which, in turn, consist of a transmembrane apoprotein, the opsin, bound to the photosensitive chromophore (an aldehyde of vitamin A). Photoreceptor absorbance can be tuned by opsin gene differential expression, chromophore usage (the aldehyde of either vitamin A1 or A2), opsin gene sequence variation and regional opsin co-expression in the retina (Carleton, 2009; Carleton and Kocher, 2001; Dalton et al., 2017; Hofmann et al., 2009, 2010a). Therefore, several mechanisms can act separately or in concert, giving rise to a diverse palette of visual phenotypes. Unraveling these mechanisms requires techniques such as microspectrophotometry (MSP) of individual photoreceptor cells and molecular experiments in order to characterize opsin gene sequence and expression.
Invasive species (introduced species that have successfully established and spread outside of their native range), are suitable for the study of adaptation because they can exhibit rapid responses to novel biotic and abiotic conditions (Zenni et al., 2014; Hoffmann and Sgrò, 2011; Prentis et al., 2008; Whitney and Gabler, 2008). Hence, invasive species can potentially provide insights into the mechanisms underlying adaptive phenotypic variation. In this study, we examine the peacock bass (Cichla monoculus), a diurnal predatory cichlid native to the Amazon basin (Kullander and Ferreira, 2006). Cichla monoculus was introduced to the Chagres River basin in 1967 for sport fishing, from which it dispersed and subsequently colonized Lake Gatun in the Panama Canal watershed (Zaret and Paine, 1973). The introduction of C. monoculus has had a dramatically negative effect on the native ichthyofauna. Long-term studies have shown how the introduction of C. monoculus has altered the composition of fish communities in Lake Gatun, decreasing both abundance and biomass of several local species while completely extirpating others (Sharpe et al., 2016; Zaret and Paine, 1973).
Almost half a century after its introduction, C. monoculus remains the most common fish in Lake Gatun (Sharpe et al., 2016), suggesting it has successfully adapted to the complex environment of the Panama Canal watershed. Lake Gatun was created in 1913 in order to build the Panama Canal by building a dam across the reaches of the Rio Chagres. At least half of the Panama Canal watershed is protected and covered in mature tropical lowland forest; however, there remain several threats to water quality in Lake Gatun (Ibáñez et al., 2002). First, deforestation in parts of the catchment has increased surface runoff, increasing the level of sedimentation and turbidity in the lake (Ibáñez et al., 2002). Second, the continuous transit of boats through the canal generates turbulence and wave action that also stir up sediments. Third, operation of the canal requires continual dredging, which has significantly expanded in the past 5 years as part of the recently completed expansion of the Panama Canal (Wang, 2017). Together, these factors have resulted in a strong turbidity gradient in Lake Gatun, with increasingly turbid waters as you approach the navigable sections of the waterway. The goal of our study was to analyze the visual system of C. monoculus and its adaptability to the variable light environment of Lake Gatun. Through genetic and physiological experiments we evaluated whether the visual system of C. monoculus changes across the abovementioned turbidity gradient.
MATERIALS AND METHODS
Study site and sample collection
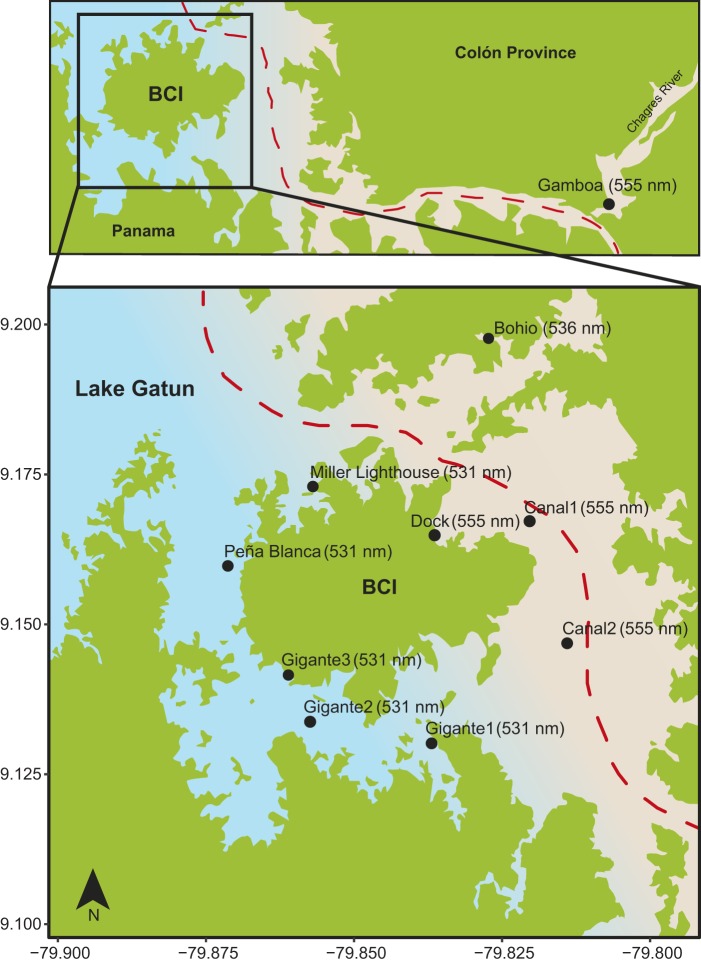
Our sampling was conducted around Barro Colorado Island (BCI) in Lake Gatun (Fig. 1). One side of the island abuts the navigable channel of the Panama Canal and the water is very turbid, whereas the other side of the island lies within the protected Barro Colorado Natural Monument, and the water is very clear (D.M.T.S., D.E.-C., personal observation). Turbidity affects light intensity but does not necessarily shift spectral wavelengths. However, the different types of suspended particles present in the Panama Canal absorb much of the short-wavelength light, causing turbid waters to be long-wavelength shifted. Throughout the article, we will refer to turbid waters as environments with decreased light intensity shifted to longer wavelengths.
Fig. 1.
Map showing the light sampling localities around Barro Colorado Island (BCI) in Lake Gatun, Panama Canal. Black circles indicate each light sampling locality, and the dashed line indicates the waterway of boats in the Panama Canal. Numbers next to locality names indicate the wavelength of peak intensity (λmax) for each locality at 1 m depth.
Fish were captured from July to August of 2016 using fishing lines with non-live bait. We collected 25 specimens of Cichla monoculus Agassiz 1831 from different sites around BCI in Lake Gatun (Table S1A,B). All fish were brought back to the BCI Smithsonian Tropical Research Institute (STRI) station and euthanized using a lethal dose of buffered MS-222 to ensure minimal suffering. Light measurements were taken at several sites (including the ones where fish were captured) at depths down to 3 m.
Sampling permits were in accordance with the Panamanian laws of environmental protection (wild collection permits from Ministerio de Ambiente de Panamá, MiAmbiente, permit nos SE/A-99-16 and SC/A-19-17). Fish were handled following the STRI IACUC protocol (2014-0901-2017-A3).
Experimental set-up
All fish were brought back to the BCI research station, and then used in one of the following ways. First, five individuals (three from clear water, two from turbid water) were killed immediately for RNA sampling. Second, 16 fish (nine from clear water, and seven from turbid water) were used for MSP. In addition, four fish captured in turbid waters were kept in clear outdoor glass tanks at Naos Marine Laboratories, STRI (Panama), for 6 months, after which their retinas were analyzed by MSP (Table S1B). This longer-term acclimation experiment was performed because cichlid visual systems have been shown to be plastic (Härer et al., 2017; Hofmann et al., 2010b; Kröger et al., 1999; Nandamuri et al., 2017). In this study, we classified our samples as clear, turbid and 6-month treated fish.
Spectral measurements
To characterize environmental light, we measured downward light intensity in nine localities around BCI (Fig. 1), which were selected a priori to cover a suspected turbidity gradient. Downward light intensity was measured in each locality with a 1000 μm fiber with a fiber-optic spectrometer based on an Ocean Optics USB2000 (Dunedin, FL, USA). We took five replicate measurements at the subsurface and at 1, 2 and 3 m depth on days of bright sunshine. In addition, we also collected measurements of sidewelling irradiance and radiance in two localities (a turbid and a clear site). For irradiance measurements, the fiber was fitted with a cosine corrector (CC-3).
We also characterized the light transmission of C. monoculus' ocular media. Lens and cornea transmission were measured by placing the isolated cichlid lens or cornea on a UV-transparent cover slip, which was illuminated from above by a fiber-optic cable attached to a pulsed xenon light source (PX-2, Ocean Optics). Another fiber-optic cable was placed directly under the specimen and delivered the signal to the spectrometer. Five replicate measurements were made. The resulting spectral scans were normalized to 100% transmission at 700 nm. Finally, we quantified the T50 values (i.e. the wavelength at which 50% transmission is reached).
Microspectrophotometry
In order to identify the peak of maximum sensitivity of C. monoculus' photopigments under different light conditions, we performed MSP on wild-caught fish from three different light conditions: clear water, turbid water and 6-month treated fish.
Each fish was dark-adapted for at least 2 h, after which it was killed with an overdose of buffered MS-222. Eyes were enucleated under a dissecting scope in dim deep red light. The retina was removed and transferred to a PBS solution containing 6.0% sucrose (Sigma-Aldrich). A small piece of retina was cut out and placed on a glass cover slip in a drop of solution, then delicately macerated with razor blades. The preparation was covered with a second glass cover slip and sealed with high-vacuum silicone grease (Dow Corning).
Spectral absorbance was measured with a computer-controlled single-beam micro-spectrophotometer fitted with quartz optics and a 100 W quartz-halogen lamp. Baseline records were taken by averaging a scan from 750 to 350 nm and a second in the opposite direction, through a clear area of the preparation and in proximity to the photoreceptor of interest. A record of the visual cell was then obtained by scanning with the MSP beam through the photoreceptor outer segment. Finally, the cell's absorption spectrum was obtained by subtracting the baseline record. A custom-designed spectral analysis program (E. R. Loew and W. Elmore, unpublished) was used to determine λmax from absorbance records using existing templates (Dartnall, 1953; Munz and Schwanzara, 1967). Individual spectra were smoothed with a nine-point adjacent averaging function and the resulting curves were differentiated to obtain a preliminary maximum value. This was used to normalize curves to zero at the baseline on the long wavelength limb and to one at the maximum value. Whitmore and Bowmaker's (1989) relationship (Eqn 1) was used to recursively fit the observed (normalized) absorption spectra to curves resulting from combinations of different proportions of pure vitamin A1 and corresponding pure vitamin A2 nomograms (Whitmore and Bowmaker, 1989) (Fig. S1):
| (1) |
Retina preparation for MSP proved to be challenging because of a particularly dense vitreous humor and thick pigment epithelium that required extensive careful manipulation, often causing detachment of the cones' outer segments. For this reason, we could not collect a sufficient number of cone records per individual to allow a satisfactory between-environment comparison. We were, nevertheless, able to characterize cone sensitivities for each spectral class and numerous rod records for analysis. A non-parametric ANOVA on ranks (Kruskal–Wallis test) followed by a pairwise Wilcoxon test was used to compare rod spectral absorbances (λmax) between fish from different groups.
RNA sequencing and opsin gene expression analysis
For RNA sequencing (RNA-Seq), fish were euthanized with buffered MS-222. Three individuals from clear water and two fish from turbid water were used for RNA-Seq. Fish eyes were enucleated and the retinas were dissected out and preserved in RNAlater. For RNA-Seq, we used both eyes from two of the five fish, resulting in a total of seven samples for transcriptome analysis. Total RNA was extracted with an RNeasy kit (Qiagen), and RNA quality was verified on an Agilent Bioanalyzer. RNA-Seq libraries were prepared using the Illumina TruSeq RNA library preparation kit (Illumina Inc., San Diego, CA, USA) by the University of Maryland Institute for Bioscience & Biotechnology Research sequencing core, obtaining 100-bp paired-end reads with all samples multiplexed in one lane of an Illumina HiSeq1500 sequencer. The data were quality-checked using FastQC version 0.10.1 to remove over-represented sequences and to retain sequences with a minimum quality score of 20 and a minimum length of 80 bp. The seven transcriptomes were combined in order to obtain a single de novo assembly. This was performed with Trinity version r20140413 (Haas et al., 2013) using only paired sequences with a minimum coverage of two to join contigs.
For estimating gene expression, reads from each sample were mapped back to the de novo assembled transcriptome using RSEM, part of the Trinity package (Haas et al., 2013). We wanted to examine whether there were differences in gene expression that might alter the visual sensitivities between clear and turbid water fish, including opsins or genes responsible for A2 chromophore synthesis. In order to analyze the opsins, read counts from each opsin class were extracted from the RSEM output (quantified as fragments per kilobase of transcript per million reads, FPKM) and then normalized to the β-actin gene. Because the RH2A duplicates are a product of gene duplication followed by gene conversion (Escobar-Camacho et al., 2017), we mapped back the reads only to the first 140 bp of the first exon in order to obtain an estimate of expression of each RH2A paralog.
To examine other genes differentially expressed between samples, we performed a differential gene expression analysis using the RSEM output from each sample. This analysis was done with the Bioconductor package DESeq2 implemented in R. We used a one-factor design (levels: clear versus turbid) to analyze all samples. We only considered genes with a sufficient number of mapped reads (FPKM>10) for the analysis.
Opsin sequence analysis
In order to expand our understanding of opsin genes in C. monoculus, we analyzed the opsin gene complement in the genome of Cichla vazzoleri. For this species, the samples were analyzed under the guidelines of Sao Paulo State University IACUC (protocol no. 34/08-CEEA/IBB/UNESP). See methods in Escobar-Camacho et al. (2017) for genome analysis.
Putative opsin sequences were identified from transcriptome-assembled FASTA files of C. monoculus. We searched for opsin sequences using Tblastx and queried with cichlid opsin genes from Oreochromis niloticus. If sequences were not found in C. monoculus' transcriptome, we looked in the genome of C. vazzoleri. Opsin class identity was confirmed based on phylogenetic relationships of the opsin sequences from other teleost lineages obtained from GenBank. We used MAFFT to align nuleotide sequences and PartitionFinder to find the best partioning scheme and molecular evolution model (Lanfear et al., 2017). Maximum likelihood analyses were conducted using Garli (version 2.0). These included a best tree search with 40 replicates and 2000 bootstrap replicates to evaluate nodal support.
We also aligned C. monoculus opsin sequences of each opsin class with bovine rhodopsin, tilapia (O. niloticus) and four other neotropical cichlids [Astronostus ocellatus, Symphysodon discus and Pterophylum scalare (Escobar-Camacho et al., 2017), and Crenicichla frenata (Weadick et al., 2012)] to identify potential spectral tuning sites. The alignments were analyzed to identify amino acids substitutions that fell in putative transmembrane regions and in the retinal binding pocket facing the chromophore (see Escobar-Camacho et al., 2017). We excluded opsin sequences from C. vazzoleri in this analysis because they were almost identical to those of C. monoculus and because sequences such as RH2A opsin were incomplete owing to genome mis-assemblies.
RESULTS
Light environment in Lake Gatun
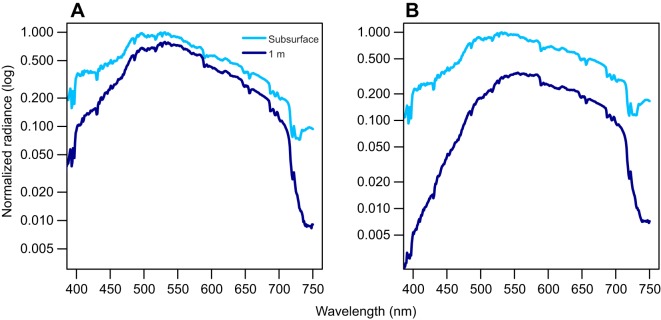
We measured downward light intensity from nine localities around BCI (Table S1C). Light diffuse attenuation was greater at sites close to the Panama Canal waterway than in clear water sites away from the canal. For example, light is best transmitted in localities near Gigante (clear) and more strongly attenuated in localities near the canal (turbid) (Fig. 2), where light intensity decreases greatly with depth. In general, the dominant wavelengths in Lake Gatun lie between 500 and 600 nm in both turbid and clear sites (Fig. 2). Short wavelengths around 400 nm appear to be transmitted only in clear water sites towards the western part of Barro Colorado Island reserve. The island seems to partially shelter these locations from turbid plumes from the main canal waterway.
Fig. 2.
Underwater light environment variation around Barro Colorado Island. Downward light intensity at (A) Gigante1 (a clear site) and (B) Canal2 (a turbid site).
Ocular media
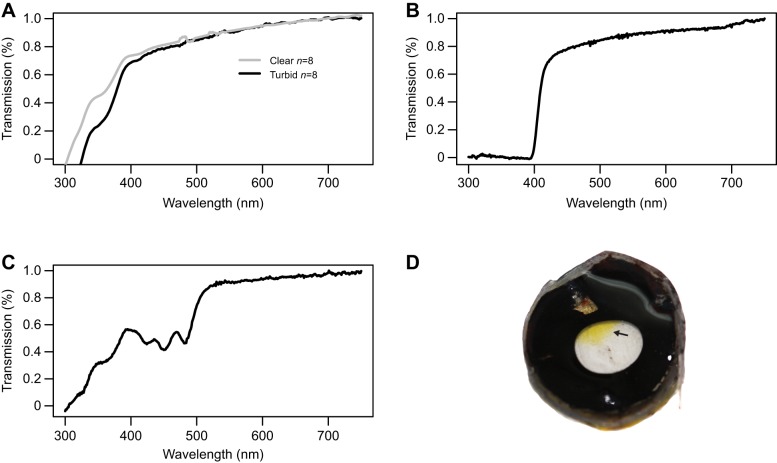
The cornea of C. monoculus is UV transmissive with a T50 cut-off around 378 nm (mean=377.56 nm, range 368–383 nm, n=8). This cut-off differed slightly between clear and turbid water sites (376 and 379.5 nm, respectively; Fig. 3A) but the difference was not significant (t5,4=0.303, d.f.=7, two-tailed P=0.334). Cichla monoculus exhibits UV blocking lenses with transmission yielding a T50 cut-off of approximately 409 nm (clear sites individuals: mean=408.94 nm; s.d.=1.055, range=406–410 nm; turbid sites individuals: mean=410.1 nm; s.d.=2.15, range=407–414 nm; Fig. 3B, Table S1D). There was no difference in lens transmission between individuals caught in turbid versus clear light environments.
Fig. 3.
Ocular media transmission measurements in Cichla monoculus. (A) Cornea transmission spectrum of C. monoculus sampled in clear and turbid waters. (B) UV-blocking lens transmission spectrum from C. monoculus (n=16). (C) Transmission spectrum of the yellow region (n=4). (D) Dissected cornea of C. monoculus. Note the yellow pigmentation in the top region.
During field work, we realized that the temporal region of the cornea was yellow (Fig. 3C,D). Transmission measurements showed that this region transmits light differently than other regions of the cornea, with three cut-off peaks at 385, 462 and 486 nm (Fig. 3C). This reduces the amount of blue light reaching the eye and perhaps acts as a sun shade to reduce scattered light.
Visual sensitivities of Cichla monoculus
MSP of rods in C. monoculus revealed remarkable variation in peak absorbances (λmax), ranging from 496 to 531 nm (n=271 records) (Table 1, Fig. 4A, Fig. S2A), suggesting variable mixtures of vitamin A1 and A2. Fish from clear water sites had greater variation in rod spectral sensitivities whereas turbid water and six-month treatment fish exhibited less variation (Fig. 4A). Rod spectral sensitivities were significantly different between fish from different locations (Kruskal–Wallis rank sum test, P<2.2e-16; Wilcoxon rank sum test, P<2e-16 and P<2e-16, respectively) (Fig. 4C). Because vitamin A2 shifts visual sensitivity to longer regions of the spectrum, our results suggest that turbid water fish had higher proportions of vitamin A2 (Fig. 4B). In addition, individuals collected at a turbid water site and kept in clear water under natural light conditions for 6 months exhibited significantly shorter rod peak absorbances (λmax) than non-treated individuals from the same site, a shift consistent with higher levels of vitamin A1 (Fig. 4A,B).
Table 1.
Range of cone and rod visual pigment peak sensitivities (λmax) measured in Cichla monoculus with microspectrophotometry (MSP)
Fig. 4.
Rod spectral sensitivity variation. (A) Maximum absorbance of rods from 20 individuals from sites with different underwater light spectra. (B) Proportion of vitamin A1 estimated from rod λmax in 20 individuals. Error bars denote 1 s.d. of all the recordings. Filled squares, circles and triangles denote samples from clear water, turbid water and the 6-month treatment, respectively. (C) Boxplots showing the rod spectral sensitivities of samples from different groups.
We identified five different photoreceptor classes (Table 1), which is consistent with the number of opsin genes C. monoculus expresses: SWS2B, SWS2A, RH2Aβ, RH2Aα and LWS (see below). Within individuals, their vitamin A1 content was consistent with the A1 content of the corresponding rod.
For single-cone photoreceptors, MSP identified two types based on pure vitamin A1, one sensitive to the violet (λmax=419–431 nm, N=2), with the other sensitive to the blue/blue-green (λmax=480–491 nm, N=5). For double-cone photoreceptors we identified three types, a short-green (λmax=524–543 nm, N=9), a long-green (λmax=530–575 nm, N=10) and a yellow-red (λmax=575–605 nm, N=3) visual pigment (Fig. S2B). The variation in the cones' MSP λmax is indicative of the presence of A1/A2 mixtures.
Opsin gene characterization and sequence analysis
RNA samples showed good quality based on the Agilent Bioanalyzer RIN (RNA integrity number varied between 8 and 9.20). The retinal transcriptomes obtained by multiplexing all samples in one lane provided sufficient data to assemble and quantify opsin transcripts. Total reads per sample varied from 24.09 to 29.33 mol l−1, and after trimming from 18.21 to 22.04 mol l−1.
We isolated five cone opsins from the retinal transcripts of C. monoculus – SWS2B, SWS2A, RH2Aβ, RH2Aα and LWS – as well as a rod opsin, RH1. Most of the opsins were complete except for SWS2B, which had lower transcript abundance than the others. Cichlids are known to have a single SWS2B copy, and we were able to generate a complete SWS2B sequence by assembling individual transcripts into a consensus SWS2B sequence. We further isolated six opsins from the genome assembly of C. vazzoleri (SWS1, SWS2B, SWS2A, RH2Aβ, RH2Aα and LWS).
The maximum likelihood tree with other neotropical cichlid species confirmed the opsin classes of Cichla spp.: SWS1, SWS2B, SWS2A, RH2A, LWS and RH1 (Fig. S3A). The cichlid opsin lineages are reciprocally monophyletic between New World and African lineages, with neotropical cichlid opsins placed as sister group to the respective African cichlid orthologs in all opsin classes (Fig. S3A). We did not find SWS1 and RH2B opsins in the expressed transcripts for C. monoculus, hence we do not know whether those genes are functional or whether they have been lost in the genome. In addition, we did not recover RH2B in C. vazzoleri, but we found what seems to be a SWS1 pseudogene in this species, as suggested by the presence of several indels.
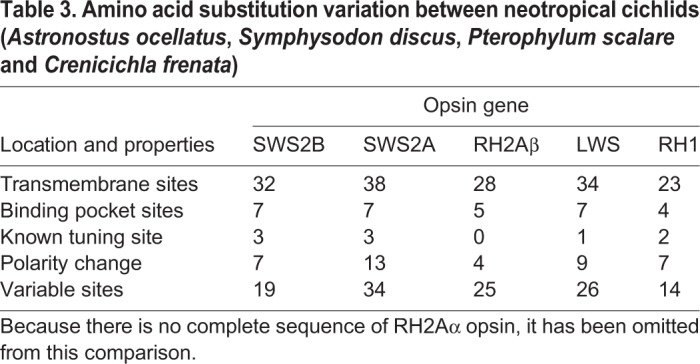
Comparisons between O. niloticus and C. monoculus opsin sequences (Table S1E) revealed evidence of amino acid changes that might shift visual pigment absorbance. Within the transmembrane region of the opsin molecule, the greatest diversity was observed in the SWS2B opsin class, with 23 variable transmembrane sites, with seven of these occurring in the retinal binding pocket and two (V46F, G109A) at known tuning sites (Table 2, Fig. S3B, Table S1F). Other opsins also showed variable diversity, including LWS, RH1 and SWS2A with 17, 17 and 12 variable transmembrane sites, respectively (Table 2). However, when we combine C. monoculus sequences with other neotropical cichlid species, the SWS2A opsin shows the highest variability followed closely by SWS2B (Table 3, Fig. S4C).
Table 2.
Amino acid substitutions between C. monoculus and Oreochromis niloticus
Table 3.
Amino acid substitution variation between neotropical cichlids (Astronostus ocellatus, Symphysodon discus, Pterophylum scalare and Crenicichla frenata)
Gene expression
Color vision in C. monoculus is based mainly on the expression of SWS2A, RH2Aβ and LWS, at 7.6, 8.2 and 82.3%, respectively (Fig. 5A,B). The most expressed single-cone pigment was SWS2A while the most expressed double-cone pigment was the LWS opsin. SWS2B and RH2Aα accounted for <5% of expressed cone opsins in all samples. RH1 was the most highly expressed visual pigment in all samples (Fig. 5C). All samples expressed the same opsins.
Fig. 5.
Opsin and Cyp27c1 expression. (A) Relative cone opsin expression profile of C. monoculus samples normalized to the β-actin gene. (B) Relative cone opsin expression of C. monoculus based on FPKM values (fragments per kilobase of transcript per million reads). (C) Normalized opsin expression as total FPKM values. (D) Opsin expression of seven C. monoculus samples. Opsin expression values were normalized to the β-actin gene. Each opsin class is specified (colored circles). (E) Gene expression of Cyp27c1 based on FPKM counts and (F) Cyp27c1 expression normalized to β-actin gene (gray circles). Dashed line separates samples from different sites (clear versus turbid). Light circles denote samples from clear waters whereas dark circles denote samples from turbid water sites. Black borders denote duplicates from a single fish and bigger circles are used to reveal overlapping data points.
Opsin expression determined from RNA-Seq data showed considerable variation amongst individuals. This remained true even when gene expression was normalized to a housekeeping gene such as β-actin. Considering the individual variation, there was no evidence for differences in opsin gene expression between samples from different light environments (Fig. 5D). The substantial differences in visual pigment peak sensitivities highlighted by the MSP analysis suggest variability in chromophore use, with the relative A1/A2 proportion potentially changing rapidly in response to underwater light conditions. Therefore, we examined the retinal transcripts for expression of Cyp27c1, a zebrafish gene involved in the synthesis of vitamin A2 from A1 (Enright et al., 2015). However, this gene was not differentially expressed between localities (Fig. 5E,F).
We performed a more global analysis of differential gene expression across all genes in the retinal transcriptomes and compared individuals sampled from clear versus turbid sites. A DESeq2 two-factor analysis detected 36 genes as being differentially expressed between clear and turbid water fish, with more genes (31) being upregulated in turbid water samples and just five being upregulated in clear water samples (Table S1G).
DISCUSSION
Visual system adaptation
We have shown that the visual system of C. monoculus exhibits remarkable plasticity, which appears to be mediated predominantly by variation in chromophore content. Overall, the visual system of C. monoculus adapts to Lake Gatun's environment by four main mechanisms: (1) filtering wavelength scattered light (∼400 nm) with ocular filters, (2) tuning visual sensitivities to the available light throughout opsin gene expression, (3) increasing the proportion of vitamin A2 in its photoreceptors, making sensitivities more red-shifted as necessary, and (4) changing gene expression in the retina as a response to environmental stressors such as turbidity. The plasticity of their visual system has probably enabled fast adaptation to this changing environment because C. monoculus is a voracious predator that relies primarily on vision. Indeed, it has been suggested that their hunting strategies are among the traits that make Cichla spp. successful invaders (Carvalho et al., 2014). Furthermore, C. monoculus might benefit from changes in gene expression as these changes provide intraspecific phenotypic variation, facilitating visual function across the diverse light environments found in the lake.
Spectral measurements
Light environment in the Panama Canal
Through spectral measurements, we describe for the first time the light environment of Lake Gatun. There is great variability in light transmission throughout Lake Gatun, which is greatly influenced by activities in the canal. There are high concentrations of suspended sediments in Lake Gatun owing to the daily transit of all types of boats, rainwater runoff and water incursion from the locks. The amount of suspended particles seems to follow the Panama Canal transit path because in our measurements we saw a pattern consistent with this trend (Figs 1 and 2). Attenuation increases at sites close to the canal waterway including Canal2 and 3, Miller, Peña Blanca and the BCI Dock, whereas it is lower at sites that remain undisturbed by the canal activity, such as Gigante1, 2 and 3 (Table S1C). Although our measurements were taken at a specific time of year and so might be subject to temporal variation, these measurements follow a turbidity gradient pattern that has been characterized by Secchi disc measurements around Lake Gatun (D. M. T. Sharpe, L. F. De Leon, R. Gonzalez and M. E. Torchin, unpublished results). Overall, our spectral measurements suggest that fish inhabit a wide range of light environments within Lake Gatun. Some of these are strongly impacted by turbidity, which could negatively affect visual perception. Particularly, turbid sites seem to be detrimental for contrast detection owing to the increased light scattering and light absorption by suspended particles, which causes light to become very dim as depth increases.
Although Lake Gatun has an artificial origin, our measurements are in agreement with previous studies that have quantified the light environment of freshwater neotropical ecosystems such as Lake Managua (Nicaragua) (Torres-Dowdall et al., 2017) and rivers from the Amazon (Costa et al., 2013), which are characterized by being long-wavelength-shifted environments.
Ocular media of C. monoculus
The ocular media transmission from C. monoculus follows a pattern similar to that of other neotropical and African cichlids (Hofmann et al., 2010a; Torres-Dowdall et al., 2017). Even though the cornea is UV transmissive, all lenses proved to be UV-blocking (Fig. 3A,B). The presence of a yellow-pigmented region in the cornea and the UV-blocking nature of the lenses indicates that the visual system of C. monoculus specifically filters short wavelengths (<400 nm), reducing the loss of acuity caused by the aberration of violet and blue wavelengths, likely a major source of visual noise in the turbid waters of Lake Gatun, and thus improving visual resolution. Furthermore, the yellow region in the cornea also filters much of the blue light (<486 nm) (Fig. 3C); this might be useful for improving contrast by filtering out the ‘veiling brightness’ (diffused blue light) found in the near-surface of the water column (Heinermann, 1984). The presence of yellow filters in C. monoculus is not surprising given its Amazonian origin and the fact that similar adaptations have been reported for several Amazonian fish (Muntz, 1973, 1982; Escobar-Camacho et al., 2017). These filters are suggested to act as an adaptation to long-wavelength-shifted environments, thereby contributing to the ability of C. monoculus to adapt well to Lake Gatun.
Microspectrophotometry
Photoreceptor spectral sensitivities and vitamin A1/A2
The peak rod spectral sensitivities in C. monoculus (496–531 nm) are in concordance with those found for other neotropical cichlids (Table S1H). The range in peak absorbance implies extreme variation in A1/A2 chromophore combinations, which seem to change according to the light environment the fish live in. Chromophore shifts are plastic and respond to the light environment. Fish sampled from turbid waters exhibited long-wavelength-shifted spectral sensitivities with average peak absorbances (λmax) between 514 and 523 nm when first sampled, whereas 6-month treatment fish exhibited shorter wavelength-shifted sensitivities, with λmax between 500 and 511 nm (Fig. 4, Fig. S2A). Furthermore, turbid water and 6-month treatment fish had a somewhat narrower set of spectral sensitivities (Fig. 4A–C) when compared with individuals collected in clear water. The variation in MSP from clear waters may be explained by fish movement within Lake Gatun. It is possible that fish collected from clear water might have migrated from turbid to clear waters relatively recently before sampling took place; however, this might suggest that fish from turbid waters change their A1/A2 ratio at a slower rate when they migrate to clear water sites. Another possibility is that clear water fish MSP variation could be a product of each fish's spectral tuning mechanisms in order to optimize their visual perception owing to their location in the water column. For example, fish living below 2 m in clear waters would still need long-wavelength sensitivities because short wavelengths are lost below 1 m depth.
Cichla monoculus spectral cone classes are similar to those reported for the Midas cichlid (Torres-Dowdall et al., 2017), but both are different from what has been suggested for neotropical cichlids, as previous studies described only three visual pigments (Table S1H). The cone spectral sensitivities of C. monoculus suggest that this species could potentially be tetrachromatic or pentachromatic, as we detected five different classes of photoreceptors (Fig. S2B). Notably, given the absence of an RH2B cone class, the blue cone appears to be particularly long-wavelength-shifted compared with that in the Midas cichlid (477 versus 450 nm). However, long-wavelength-shifted blue cones have been reported for neotropical cichlids, namely in Crenicichla frenata, Pterophyllum sp. and Neetroplus nematopus (Table S1H). Similarly, the short-green (RH2Aβ) pigment differs from the one reported for the Midas cichlid (518 versus 509 nm) but is similar to the one found for African cichlids O. niloticus and Metriaclima zebra (518 versus 518/519 nm). These differences between C. monoculus and other neotropical cichlids are the complex result of variation in chromophore usage and the accumulation of amino acid substitutions in several opsin genes.
Transcriptome analysis
Opsin genes from C. monoculus
Through transcriptome analysis we have isolated the opsin complement of C. monoculus. The opsin complement of this species is similar to that found for other South American cichlids (Escobar-Camacho et al., 2017). The absence of RH2B and SWS1 opsins in the transcriptome indicates that the inactivation of these genes has happened repeatedly among neotropical cichlids (Escobar-Camacho et al., 2017; Weadick et al., 2012). This inactivation may occur by suppressing their expression, but may eventually result in pseudogenization. This was confirmed by analysis of the genome of C. vazzoleri, which revealed a SWS1 pseudogene and the loss of the RH2B opsin.
When building phylogenetic trees of opsin genes, we observed that the opsins of C. monoculus were placed in the neotropical cichlid clade as expected, yet their position in the neotropical cichlid clade varied (Fig. S3A). For the SWS2B opsin class, Cichla spp. held the most basal position, which is consistent with previous research that showed that Cichlini (Cichla) diverges basally before Astronotini (Astronotus) and Heroini (Pterophyllum and Symphysodon) (López-Fernández et al., 2010). However, there is phylogenetic discordance between the position of the other Cichla opsin classes (SWS1, SWS2A, RH2A, LWS and RH1) and neotropical cichlid phylogenetic relationships. One possibility is that this disagreement could be the product of the mutational saturation of amino acid sequences. Furthermore, several of the opsin class clades exhibit low support.
Analysis of opsin amino acid sequence variation revealed that SWS2B is the opsin gene with the most substitution variation in C. monoculus (Table 2, Fig. S3B). Amino acid changes in the SWS2B sequence could be the result of selection changing the spectral and/or nonspectral aspect of SWS2B function. This gene has been suggested to be under positive selection in the Trinidadian pike (Crenicichla frenata) (Weadick et al., 2012). However, when amino acid variation was analyzed between the opsin set of five neotropical cichlids, SWS2A showed the highest variation, similar to previous findings (Escobar-Camacho et al., 2017) (Table 3, Fig. S3C). Both SWS2B and SWS2A in neotropical cichlids may be under strong selection to optimize visual perception in long-wavelength-shifted environments (Escobar-Camacho et al., 2017; van Nynatten et al., 2015; Schott et al., 2014; Torres-Dowdall et al., 2015; Weadick et al., 2012). In addition, it has been shown that opsin amino acid substitutions in neotropical cichlids improve opsin kinetics. For example, RH1 in C. monoculus has a D83N (Table S1E) substitution, which has been suggested to slow down the decay rate of the rhodopsin, which improves visual sensitivity in dim light conditions (Hauser et al., 2017).
Opsin gene and Cyp27c1 expression
Cone opsin expression of C. monoculus is dominated by long-wavelength-sensitive pigments and, to a lesser extent by SWS2A and RH2Aβ, a pattern similar to that of other neotropical cichlids (Escobar-Camacho et al., 2017; Torres-Dowdall et al., 2017; Weadick et al., 2012). This opsin expression profile is optimal for red-shifted light environments because the double cone spectral sensitivities match the available light (Fig. S2C). Although C. monoculus exhibited individual variation in opsin expression, we did not detect any significant correlation with the underwater light conditions at collection sites (Fig. 5D). Thus, we cannot conclude whether there is opsin expression plasticity in C. monoculus. More studies analyzing changes of opsin expression in adults and through developmental series are necessary. To date, different cichlid species have been found to exhibit gene expression plasticity at the adult stage or no adult plasticity at all (Carleton et al., 2008; Härer et al., 2017; Hofmann et al., 2010b; Nandamuri et al., 2017). Our results could also be masked by sampling effort because we do not have a large set of samples.
Similarly, although we measured large differences in spectral sensitivity between individuals from clear and turbid sites, and successfully experimentally manipulated such sensitivities by treating turbid water individuals in clear water conditions, we did not observe covariation between turbidity and the expression of Cyp27c1. This enzyme has been shown to convert vitamin A1 into vitamin A2 in the retinal pigment epithelium of zebrafish, where the proportion of A2 is positively related with its expression (Enright et al., 2015). In C. monoculus, we found a functional copy of Cyp27c1; however, turbid water fish exhibited low levels of Cyp27c1 expression when compared with clear water fish samples (Fig. 5E,F). This is surprising in light of our rod MSP results, which support more A2-based visual pigments in turbid water individuals (Fig. 4). We hypothesize two possible explanations. First, our samples in particular may not show high levels of Cyp27c1 expression owing to a sampling problem. Notably, the samples used for transcriptomes were not the same samples analyzed for MSP. Second, the conversion of A1 to A2 could be controlled by another gene in cichlids. We found other genes highly similar to Cyp27c1 in the transcriptomes. All of these genes belong to the cytochrome P450 superfamily in teleosts (Fig. S4A). Although their expression also varied between samples (Fig. S4B), it was very low and did not show any correlation with habitat lighting. Hence, more experiments are needed to unravel the relationship between Cyp27c1 expression and proportions of A1:A2 in the cichlid retina.
Differential gene expression in the C. monoculus retina
Considering more global analyses of differential retinal gene expression, our results show that there are differences in gene expression of a number of genes in the C. monoculus retina. DESeq analysis suggests that fish differentially expressed genes between clear and turbid water, including genes that vary greatly in function. Genes that were upregulated in clear water samples are involved in the oxidation of cortisol, preventing apoptosis, neutrofil regulation and intracellular trafficking (see Table S1G). Interestingly, RDH8, a key enzyme in visual pigment regeneration, was found to be upregulated in clear water samples (Table S1G). This suggests that fish from clear environments may exhibit a faster regeneration rate of visual pigment owing to greater light exposure. Furthermore, individuals from turbid waters exhibited upregulation of genes involved in transposition, anti-inflammatory processes and production of collagen. The pattern that seems to emerge is one of elevated stress in fish living in turbid sites in Lake Gatun. Individuals overexpressed genes involved in optimal immune and anti-inflammatory response (ORM1 and ANXA1, respectively); genes involved in neuroblast differentiation (AHNAK), and genes favoring bloodstream circulation (HBB, HBAA, HBAB and PDFGFRL), perhaps in response to low dissolved oxygen conditions. Interestingly, the gene TCB1, known to be involved in the transposition of transposable elements (TEs), was also found to be upregulated in turbid water samples, perhaps indicating that turbid water fish are under risk of developing mutations as a result of TE insertions. While generally detrimental, TEs might contribute to the generation of the standing variation, facilitating invasions as suggested for other species (Schrader et al., 2014; Stapley et al., 2015).
Differentially expressed genes may be relevant to the success of C. monoculus as an invasive species in Lake Gatun. Indeed, it has been suggested that invasive species can have shifted expression of genes that provide physiological advantages in different environments, enabling their adaptation and, hence, invasion success (Lockwood and Somero, 2011; Pearce et al., 2017; Ye et al., 2014; Zerebecki and Sorte, 2011).
Invasive species will only be successful if they have mechanisms for rapidly adapting to novel habitats. Here, we have documented several mechanisms that enable the visual system of C. monoculus to be particularly efficient in the temporally and spatially fluctuating turbidity of Lake Gatun. Its long-wavelength-shifted opsin palette combined with variable chromophore usage pushes its photoreceptor sensitivity to match the prevalent underwater spectrum, optimizing contrast detection in low light intensity and high scatter, even at relatively large distances. Furthermore, exceptionally large individual differences in rod spectral sensitivities correlate with local light conditions and appear to be mainly driven by plasticity in chromophore use rather than cone expression variation. Finally, these mechanisms of spectral tuning are accompanied by differential gene expression in the retina, where several genes are being upregulated and downregulated, and this varies within the same population. Overall, even though these mechanisms may occur naturally in the Amazon basin, they exhibit intraspecific variation, which is key for phenotypic plasticity and, hence, the invasive potential of C. monoculus. Unfortunately, this invasion has been at the expense of other fish as the introduction of C. monoculus has likely extirpated several native fish species in the process (Sharpe et al., 2016).
Supplementary Material
Acknowledgements
We thank the University of Maryland Institute for Bioscience & Biotechnology Research for sequencing the libraries, Matt Conte for helping with the Trinity pipeline and Noor White for help with the phylogenetic trees. We are grateful to Ellis R. Loew for generously providing his MSP machine and analysis software for this project. We thank the Panama Canal Authority (ACP) for granting us access to the canal waters. We also thank all the rangers and staff of the Smithsonian Tropical Research Institute that assisted us during our stay and sampling on Barro Colorado Island.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.E., M.E.P., V.F., D.M.S., K.L.C.; Methodology: D.E., M.E.P., V.F., E.R., C.M., K.L.C.; Software: D.E., M.E.P., E.R., C.M., K.L.C.; Validation: D.E., D.M.S., K.L.C.; Formal analysis: D.E., K.L.C.; Investigation: D.E., M.E.P., V.F., D.M.S., K.L.C.; Resources: M.E.P., K.L.C.; Data curation: D.E., M.E.P., V.F., E.R., C.M., K.L.C.; Writing - original draft: D.E., K.L.C.; Writing - review & editing: D.E., M.E.P., V.F., D.M.S., K.L.C.; Visualization: D.E., K.L.C.; Supervision: D.E., D.M.S., K.L.C.; Project administration: K.L.C.; Funding acquisition: K.L.C.
Funding
This work was funded by the National Institutes of Health (R01EY024693 to K.L.C.) and by the partnership program between the University of Maryland and Eberhard Karls Universität Tübingen for the joint course of animal communication. D.E.-C. is supported by a graduate fellowship of the Secretariat of Higher Education, Science, Technology and Innovation of Ecuador (Secretaría de Educación Superior, Ciencia, Tecnología e Innovación) (2014-AR2Q4465 to D.E.-C.). Deposited in PMC for release after 12 months.
Data availability
DNA sequences and transcriptome libraries are available from GenBank (accession nos: MK562367–MK562373, MK568303–MK568308) and the SRA database (SRR8643940–SRR8643946).
Supplementary information
Supplementary information available online at http://jeb.biologists.org/lookup/doi/10.1242/jeb.188300.supplemental
References
- Carleton K. (2009). Cichlid fish visual systems: mechanisms of spectral tuning. Integr. Zool. 4, 75-86. 10.1111/j.1749-4877.2008.00137.x [DOI] [PubMed] [Google Scholar]
- Carleton K. L. and Kocher T. D. (2001). Cone opsin genes of African cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol. Biol. Evol. 18, 1540-1550. 10.1093/oxfordjournals.molbev.a003940 [DOI] [PubMed] [Google Scholar]
- Carleton K. L., Spady T. C., Streelman J. T., Kidd M. R., McFarland W. N. and Loew E. R. (2008). Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biol. 6, 22 10.1186/1741-7007-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho D. C., Oliveira D. A. A., Sampaio I. and Beheregaray L. B. (2014). Analysis of propagule pressure and genetic diversity in the invasibility of a freshwater apex predator: the peacock bass (genus Cichla). Neotrop. Ichthyol. 12, 105-116. 10.1590/S1679-62252014000100011 [DOI] [Google Scholar]
- Costa M. P. F., Telmer K. H. and Novo E. M. L. M. (2013). Spatial and temporal variability of light attenuation in large rivers of the Amazon. Hydrobiologia 702, 171-190. 10.1007/s10750-012-1319-2 [DOI] [Google Scholar]
- Dalton B. E., de Busserolles F., Marshall N. J. and Carleton K. L. (2017). Retinal specialization through spatially varying cell densities and opsin coexpression in cichlid fish. J. Exp. Biol. 220, 266-277. 10.1242/jeb.149211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartnall H. J. A. (1953). The interpretation of spectral sensitivity curves. Br. Med. Bull. 9, 24-30. 10.1093/oxfordjournals.bmb.a074302 [DOI] [PubMed] [Google Scholar]
- Davies W. I. L., Collin S. P. and Hunt D. M. (2012). Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121-3158. 10.1111/j.1365-294X.2012.05617.x [DOI] [PubMed] [Google Scholar]
- Ehlman S. M., Sandkam B. A., Breden F. and Sih A. (2015). Developmental plasticity in vision and behavior may help guppies overcome increased turbidity. J. Comp. Physiol. A 201, 1125-1135. 10.1007/s00359-015-1041-4 [DOI] [PubMed] [Google Scholar]
- Enright J. M., Toomey M. B., Sato S., Kefalov V. J., Guengerich F. P., Corbo J. C., Enright J. M., Toomey M. B., Sato S., Temple S. E. et al. (2015). Cyp27c1 red-shifts the spectral sensitivity of photoreceptors by converting vitamin A1 into A2. Curr. Biol. 25, 3048-3057. 10.1016/j.cub.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Camacho D., Ramos E., Martins C. and Carleton K. L. (2017). The opsin genes of Amazonian cichlids. Mol. Ecol. 26, 1343-1356. 10.1111/mec.13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M. et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494-1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härer A., Torres-Dowdall J. and Meyer A. (2017). Rapid adaptation to a novel light environment: the importance of ontogeny and phenotypic plasticity in shaping the visual system of Nicaraguan Midas cichlid fish (Amphilophus citrinellus spp.). Mol. Ecol. 26, 5582-5593. 10.1111/mec.14289 [DOI] [PubMed] [Google Scholar]
- Hauser F. E., Ilves K. L., Schott R. K., Castiglione G. M., López-Fernández H. and Chang B. S. W. (2017). Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol. Biol. Evol. 34, 2650-2664. 10.1093/molbev/msx192 [DOI] [PubMed] [Google Scholar]
- Heinermann P. H. (1984). Yellow intraocular filters in fishes. Exp. Biol. 43, 127-147. [PubMed] [Google Scholar]
- Hoffmann A. A. and Sgrò C. M. (2011). Climate change and evolutionary adaptation. Nature 470, 479-485. 10.1038/nature09670 [DOI] [PubMed] [Google Scholar]
- Hofmann C. M., O'Quin K. E., Justin Marshall N., Cronin T. W., Seehausen O. and Carleton K. L. (2009). The eyes have it: regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biol. 7, e1000266 10.1371/journal.pbio.1000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C. M., O'Quin K. E., Justin Marshall N. and Carleton K. L. (2010a). The relationship between lens transmission and opsin gene expression in cichlids from Lake Malawi. Vision Res. 50, 357-363. 10.1016/j.visres.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Hofmann C. M., O'Quin K. E., Smith A. R. and Carleton K. L. (2010b). Plasticity of opsin gene expression in cichlids from Lake Malawi. Mol. Ecol. 19, 2064-2074. 10.1111/j.1365-294X.2010.04621.x [DOI] [PubMed] [Google Scholar]
- Ibáñez R., Condit R., Angehr G., Aguilar S., García T., Martínez R., Sanjur A., Stallard R., Wright S. J., Rand A. S. et al. (2002). An ecosystem report on the Panama Canal: monitoring the status of the forest communities and the watershed. Environ. Monit. Assess. 80, 65-95. 10.1023/A:1020378926399 [DOI] [PubMed] [Google Scholar]
- Kröger R. H. H., Bowmaker J. K. and Wagner H. J. (1999). Morphological changes in the retina of Aequidens pulcher (Cichlidae) after rearing in monochromatic light. Vision Res. 39, 2441-2448. 10.1016/S0042-6989(98)00256-9 [DOI] [PubMed] [Google Scholar]
- Kullander S. O. and Ferreira E. J. G. (2006). A review of the South American cichlid genus Cichla, with descriptions of nine new species (Teleostei: Cichlidae). Ichthyol. Explor. Freshwaters 17, 289-398. [Google Scholar]
- Lanfear R., Frandsen P. B., Wright A. M., Senfeld T. and Calcott B. (2017). Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772-773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Lockwood B. L. and Somero G. N. (2011). Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol. Ecol. 20, 517-529. 10.1111/j.1365-294X.2010.04973.x [DOI] [PubMed] [Google Scholar]
- López-Fernández H., Winemiller K. O. and Honeycutt R. L. (2010). Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae). Mol. Phylogenet. Evol. 55, 1070-1086. 10.1016/j.ympev.2010.02.020 [DOI] [PubMed] [Google Scholar]
- Losey G. S., Cronin T. W., Goldsmith T. H., Hydes D., Marshall N. J. and McFarland W. N. (1999). The UV visual world of fishes: a review. J. Fish Biol. 54, 921-943. 10.1111/j.1095-8649.1999.tb00848.x [DOI] [Google Scholar]
- Lythgoe J. N. (1979). The Ecology of Vision. New York: Oxford University Press. [Google Scholar]
- Marshall J., Carleton K. L. and Cronin T. (2015). Colour vision in marine organisms. Curr. Opin. Neurobiol. 34, 86-94. 10.1016/j.conb.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Muntz W. R. A. (1973). Yellow filters and the absorption of light by the visual pigments of some Amazonian fishes. Vision Res. 13, 2235-2254. 10.1016/0042-6989(73)90225-3 [DOI] [PubMed] [Google Scholar]
- Muntz W. R. A. (1982). Visual adaptations to different light environments in Amazonian fishes. Rev. Can. Biol. Exp. 41, 35-46. [PubMed] [Google Scholar]
- Munz F. W. and Schwanzara S. A. (1967). A nomogram for retinene2-based visual pigments. Vision Res. 7, 111-120. 10.1016/0042-6989(67)90078-8 [DOI] [PubMed] [Google Scholar]
- Nandamuri S. P., Yourick M. R. and Carleton K. L. (2017). Adult plasticity in African cichlids: rapid changes in opsin expression in response to environmental light differences. Mol. Ecol. 26, 6036-6052. 10.1111/mec.14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S. L., Clarke D. F., East P. D., Elfekih S., Gordon K. H. J., Jermiin L. S., Mcgaughran A., Oakeshott J. G., Papanikolaou A., Perera O. P. et al. (2017). Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 15, 1-30. 10.1186/s12915-017-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentis P. J., Wilson J. R. U., Dormontt E. E., Richardson D. M. and Lowe A. J. (2008). Adaptive evolution in invasive species. Trends Plant Sci. 13, 288-295. 10.1016/j.tplants.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Schott R. K., Refvik S. P., Hauser F. E., López-Fernández H. and Chang B. S. W. (2014). Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol. Biol. Evol. 31, 1149-1165. 10.1093/molbev/msu064 [DOI] [PubMed] [Google Scholar]
- Schrader L., Kim J. W., Ence D., Zimin A., Klein A., Wyschetzki K., Schultner E., Wurm Y., Weichselgartner T., Kemena C. et al. (2014). Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat. Commun. 5, 1-10. 10.1038/ncomms6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe D. M. T., De León L. F., González R. and Torchin M. E. (2016). Tropical fish community does not recover 45 years after predator introduction. Ecology 98, 412-424. 10.1002/ecy.1648 [DOI] [PubMed] [Google Scholar]
- Stapley J., Santure A. W. and Dennis S. R. (2015). Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol. Ecol. 24, 2241-2252. 10.1111/mec.13089 [DOI] [PubMed] [Google Scholar]
- Torres-Dowdall J., Henning F., Elmer K. R. and Meyer A. (2015). Ecological and lineage-specific factors drive the molecular evolution of rhodopsin in cichlid fishes. Mol. Biol. Evol. 32, 2876-2882. 10.1093/molbev/msv159 [DOI] [PubMed] [Google Scholar]
- Torres-Dowdall J., Pierotti M. E. R., Härer A., Karagic N., Woltering J. M., Henning F., Elmer K. R. and Meyer A. (2017). Rapid and parallel adaptive evolution of the visual system of Neotropical Midas Cichlid fishes. Mol. Biol. Evol. 34, 2469-2485. 10.1093/molbev/msx143 [DOI] [PubMed] [Google Scholar]
- van Nynatten A., Bloom D., Chang B. S. W., Lovejoy N. R. and Lovejoy N. R. (2015). Out of the blue: adaptive visual pigment evolution accompanies Amazon invasion. Biol. Lett. 11, 20150349 10.1098/rsbl.2015.0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. (2017). The role of Panama Canal in global shipping. Marit. Bus. Rev. 2, 247-260. 10.1108/MABR-07-2017-0014 [DOI] [Google Scholar]
- Warrant E. J. and Johnsen S. (2013). Vision and the light environment. Curr. Biol. 23, R990-R994. 10.1016/j.cub.2013.10.019 [DOI] [PubMed] [Google Scholar]
- Weadick C. J., Loew E. R., Rodd F. H. and Chang B. S. W. (2012). Visual pigment molecular evolution in the Trinidadian pike cichlid (Crenicichla frenata): a less colorful world for neotropical cichlids? Mol. Biol. Evol. 29, 3045-3060. 10.1093/molbev/mss115 [DOI] [PubMed] [Google Scholar]
- Whitmore A. V. and Bowmaker J. K. (1989). Seasonal variation in cone sensitivity and short-wave absorbing visual pigments in the rudd Scardinius erythrophthalmus. J. Comp. Physiol. A 166, 103-115. 10.1007/BF00190215 [DOI] [Google Scholar]
- Whitney K. D. and Gabler C. A. (2008). Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Diversity 14, 569-580. [Google Scholar]
- Ye X., Su Y., Zhao Q., Xia W., Liu S. and Wang X. (2014). Transcriptomic analyses reveal the adaptive features and biological differences of guts from two invasive whitefly species. BMC Genomics 15, 1-12. 10.1186/1471-2164-15-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret T. M. and Paine R. T. (1973). Species introduction in a tropical lake: a newly introduced piscivore can produce population changes in a wide range of trophic levels. Science 182, 449-455. 10.1126/science.182.4111.449 [DOI] [PubMed] [Google Scholar]
- Zenni R. D., Lamy J.-B., Lamarque L. J. and Porté A. J. (2014). Adaptive evolution and phenotypic plasticity during naturalization and spread of invasive species: implications for tree invasion biology. Biol. Invasions 16, 635-644. 10.1007/s10530-013-0607-8 [DOI] [Google Scholar]
- Zerebecki R. A. and Sorte C. J. B. (2011). Temperature tolerance and stress proteins as mechanisms of invasive species success. PLoS ONE 6, e14806 10.1371/journal.pone.0014806 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.