ABSTRACT
Replication stress is a common feature of cancer cells, and thus a potentially important therapeutic target. Here, we show that cyclin-dependent kinase (CDK)-induced replication stress, resulting from Wee1 inactivation, is synthetic lethal with mutations disrupting dNTP homeostasis in fission yeast. Wee1 inactivation leads to increased dNTP demand and replication stress through CDK-induced firing of dormant replication origins. Subsequent dNTP depletion leads to inefficient DNA replication, DNA damage and to genome instability. Cells respond to this replication stress by increasing dNTP supply through histone methyltransferase Set2-dependent MBF-induced expression of Cdc22, the catalytic subunit of ribonucleotide reductase (RNR). Disrupting dNTP synthesis following Wee1 inactivation, through abrogating Set2-dependent H3K36 tri-methylation or DNA integrity checkpoint inactivation results in critically low dNTP levels, replication collapse and cell death, which can be rescued by increasing dNTP levels. These findings support a ‘dNTP supply and demand’ model in which maintaining dNTP homeostasis is essential to prevent replication catastrophe in response to CDK-induced replication stress.
KEY WORDS: Wee1, Histone H3K36 modification, Schizosaccharomyces pombe, Synthetic lethality, MBF, Set2, CDK
Highlighted Article: Replication stress is a source of genome instability. In this study in fission yeast, we describe a new role for the mitotic inhibitor Wee1 in suppressing replication stress and genome instability.
INTRODUCTION
Replication stress, in which DNA replication forks stall, is a source of genome instability and a common feature of cancer cells (Gaillard et al., 2015). The ability to target such a hallmark of cancer cells is of significant therapeutic interest. Replication stress can result from multiple events including physical blockage of replication fork progression, deregulation of the replication initiation or elongation complexes or through deoxyribonucleoside triphosphate (dNTP) depletion (Dobbelstein and Sorensen, 2015; Zeman and Cimprich, 2014). Cells respond to such events by triggering checkpoint-dependent responses to facilitate DNA replication restart (Mazouzi et al., 2014). In humans, ATR and CHK1 (encoded by the CHEK1 gene) are the primary kinases responsible for replication checkpoint activity, while in fission yeast (Schizosaccharomyces pombe) the Rad3 and Cds1 kinases play a predominant role, with Cds1 being redundant with Chk1 in this response (Boddy et al., 1998; Feijoo et al., 2001; Flynn and Zou, 2011; Lindsay et al., 1998). Unresponsive stalled forks can be subject to endonucleolytic cleavage by Mus81–Eme1, generating a DNA end, which is targeted for homologous recombination (HR) (Hanada et al., 2007; Roseaulin et al., 2008).
In fission yeast, dNTP synthesis is induced in response to replication stress and DNA damage by at least two distinct mechanisms (Guarino et al., 2014). Checkpoint activation promotes ubiquitin ligase complex Ddb1–Cul4Cdt2-dependent degradation of Spd1 (Cdt2 is the substrate adapter), an inhibitor of ribonucleotide reductase (RNR) (Håkansson et al., 2006; Liu et al., 2003), thereby promoting dNTP synthesis (Holmberg et al., 2005; Liu et al., 2005, 2003). In addition, checkpoint-dependent activation of the MluI cell cycle box (MCB)-binding factor (MBF) complex promotes transcription of genes encoding one or more MCB domains within their promoter regions, including cdc22+, the catalytic subunit of RNR, thereby promoting dNTP synthesis (Dutta et al., 2008).
The chromatin state plays an important role in modulating transcriptional responses. Set2 is a histone methyltransferase required for histone H3 lysine 36 (H3K36) mono-, di- and tri-methylation in yeast (Morris et al., 2005). Various functions have been ascribed to H3K36 methylation, including DNA repair (Pai et al., 2014) and checkpoint signalling (Jha and Strahl, 2014). Furthermore, we recently described a role for Set2 in promoting dNTP synthesis in response to DNA damage and replication stress through promoting MBF-dependent transcriptional expression of cdc22+. Loss of Set2 leads to reduced Cdc22 expression, resulting in reduced dNTP levels and consequent replication stress (Pai et al., 2017). Such roles for Set2 in maintaining genome stability help explain the tumour suppressor function of the human orthologue, SETD2.
Replication stress can also arise as a result of elevated CDK activity, and cyclin E and cyclin A are frequently overexpressed in cancers (Hwang and Clurman, 2005; Yam et al., 2002). Wee1 is a negative regulator of cell cycle progression where it phosphorylates and inactivates the kinase Cdc2 (yeast) or CDK1 (mammals), thereby preventing entry into mitosis (Russell and Nurse, 1987). Inactivation of Wee1 upregulates CDK activity and promotes G2-M progression. In addition to regulating entry into mitosis, studies in mammalian cells have found that WEE1 kinase inhibition can lead to dNTP depletion through increased firing of replication origins resulting from deregulated CDK activity (Beck et al., 2012).
Synthetic lethality provides an opportunity to specifically target cancer cells (Chan and Giaccia, 2011). In this respect, previous studies using fission yeast have identified checkpoint mutants (rad1Δ, rad3Δ, rad9Δ, rad17Δ and hus1Δ) that are synthetic lethal with Wee1 inactivation by using a temperature-sensitive allele of Wee1, wee1-50 (al-Khodairy and Carr, 1992; Enoch et al., 1992). These wee1-50 checkpoint-deficient double mutants manifest a strong ‘cut’ (for ‘cell untimely torn’) phenotype in which the genetic material is mis-segregated into daughter cells, consistent with cell death arising from mitotic catastrophe (Enoch et al., 1992). Indeed, inhibitors targeting human WEE1 have been developed with the aim of promoting mitotic catastrophe in G1-S checkpoint-deficient p53 mutant cancer cells (Hirai et al., 2009). As the synthetic lethal relationship between Wee1 inactivation and loss of Chk1 is conserved in mammalian cells (Chila et al., 2015), and because inhibitors to human WEE1, ATR and CHK1 have been developed with the aim of targeting cancer cells (Dobbelstein and Sorensen, 2015; Sørensen and Syljuåsen, 2012), understanding the mechanism by which their inactivation leads to cell death is of clinical significance.
In this study, we define an evolutionarily conserved role for Wee1 in preventing replication stress through suppressing CDK-induced replication origin firing, dNTP depletion and DNA damage. Furthermore, we show that, following Wee1 inactivation, Set2-dependent histone H3K36 tri-methylation and the DNA integrity checkpoint perform an essential role in maintaining dNTP homeostasis, thus preventing replication catastrophe. These findings provide new insights into the consequences of Wee1 inactivation and its therapeutic exploitation.
RESULTS
Wee1 is required for efficient S-phase progression by limiting origin firing
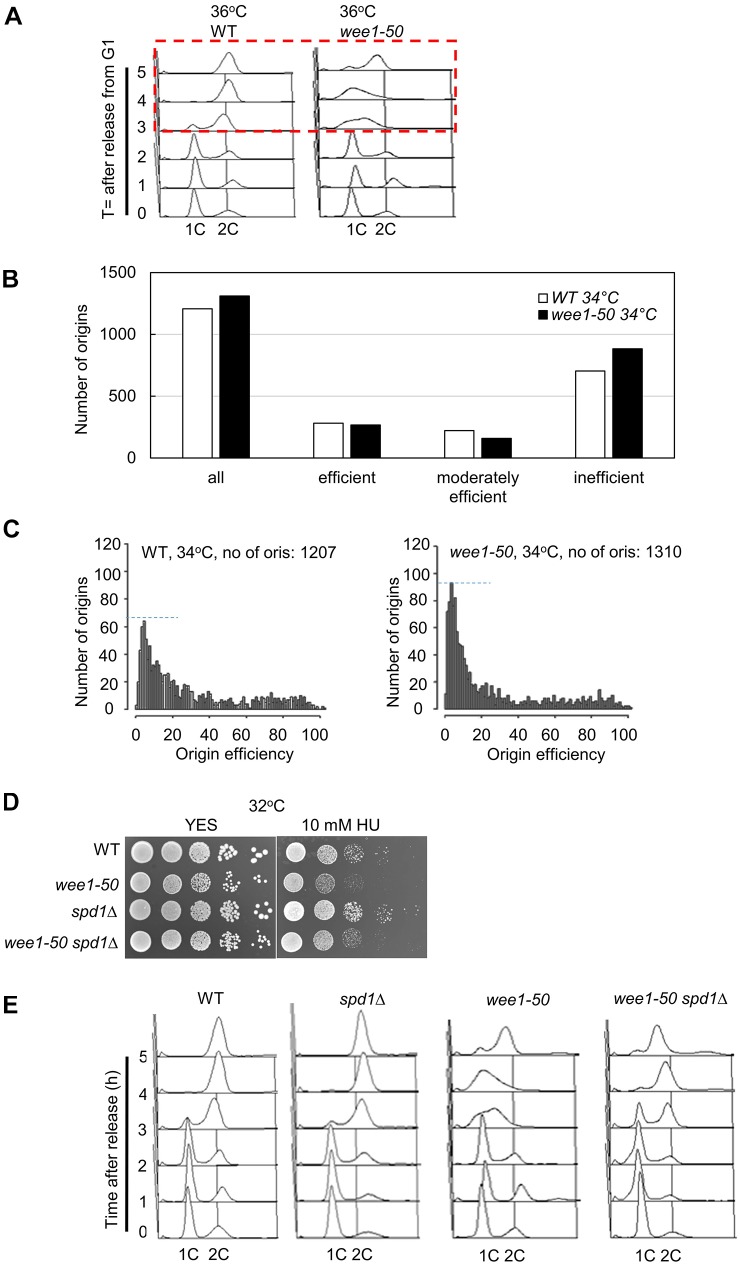
We first investigated the possible role of Wee1 in regulating S-phase progression. Nitrogen starvation was used to synchronize wee1-50 cells in G1 phase and, following re-feeding, cell cycle progression was monitored by flow cytometry. In wild-type (WT) cells, an increasing proportion of cells with a 2C DNA content was observed at 3 h following re-feeding; by 5 h, the entire population was 2C, indicating successful DNA replication (Fig. 1A). In contrast, in wee1-50 cells, at 3 h after re-feeding the population exhibited a 1C peak, and even 5 h following re-feeding there was a proportion of wee1-50 cells with a 1C peak, indicating a delay in S-phase progression (Fig. 1A, wee1-50).
Fig. 1.
Wee1 suppresses dormant origin firing. (A) Wee1 is required for efficient DNA replication. Log phase WT or wee1-50 cells were blocked in G1 phase through nitrogen starvation in EMM−N for 16 h at 25°C. Cells were released from the G1 block by re-suspending in EMM+N at 36°C. Samples were collected at the indicated time points for fluorescence-activated cell sorting (FACS) analysis. The red dashed line box indicates the delayed S-phase progression in wee1-50 cells. (B) Wee1 suppresses firing at inefficient origins. A genome-wide plot of origin usage in wee1-50 cells in comparison with WT cells at 34°C. Origin efficiencies were calculated from Pu-seq data. The sequencing experiment was performed once and therefore it is not possible to perform a statistical analysis. (C) The quantification of the frequency of origin usage (efficiency) in asynchronous WT and wee1-50 cells at 34°C. The dashed blue line indicates the higher number of low-efficiency origins used in wee1-50 cells. (D) Spd1 depletion suppresses the sensitivity of wee1-50 cells to HU. WT and wee1-50 cells were serially diluted and spotted onto YES plates containing 10 mM HU and incubated at 32°C for 2–3 days. (E) Deletion of spd1+ promotes S-phase progression in wee1-50 cells. WT, spd1Δ, wee1-50 and spd1Δ wee1-50 cells were arrested in G1 via nitrogen starvation, released and samples were taken at the time points indicated and subjected to FACS analysis.
To test whether Wee1 inactivation in fission yeast causes increased origin firing, we employed a polymerase usage sequence (Pu-seq) technique to map genome-wide origin usage as previously described (Daigaku et al., 2015). In WT cells, we identified 1207 initiation sites at 34°C including efficient (>50% usage per cell cycle), moderately efficient (25–50%) and inefficient origins (<25%) (threshold at the 20th percentile, the 99.9 percentile of all origins was set as being 100% efficient) (Fig. 1B). In the wee1-50 background, we mapped 1310 origins at 36°C (Fig. 1B). Interestingly, analysis of the distribution of origin usage in wee1-50 cells revealed a trend that an increased number of inefficient origins (dormant origins) were used compared to WT cells (Fig. 1B). There are a greater proportion of inefficient origins and less-efficient origins in wee1-50 cells compared to wild type (Fig. 1C). Taken together, these data suggest that Wee1 inactivation causes an increase in the number of DNA replication initiation sites utilized.
We tested whether the increased origin firing in wee1-50 might lead to elevated dNTP demand, thus leading to replication stress. A spot assay showed that wee1-50 cells were sensitive to hydroxyurea (HU) at the semi-restrictive temperature (Fig. 1D; Fig. S1A). Deleting the RNR inhibitor spd1+ in a wee1-50 background suppressed the sensitivity of wee1-50 cells to HU (Fig. 1D; Fig. S1A) and suppressed the delayed DNA replication of wee1-50 cells at 36°C, consistent with Wee1 inactivation having an impact on dNTP levels (Fig. 1E). Consistent with this, we showed that the dATP level (normalized to total ATP) in wee1-50 is significantly lower than in WT cells (Fig. S1B). These findings suggest that inactivation of Wee1 causes dNTP pool depletion by increased origin firing leading to replication stress.
Wee1 inactivation causes DNA damage accumulation and genome instability
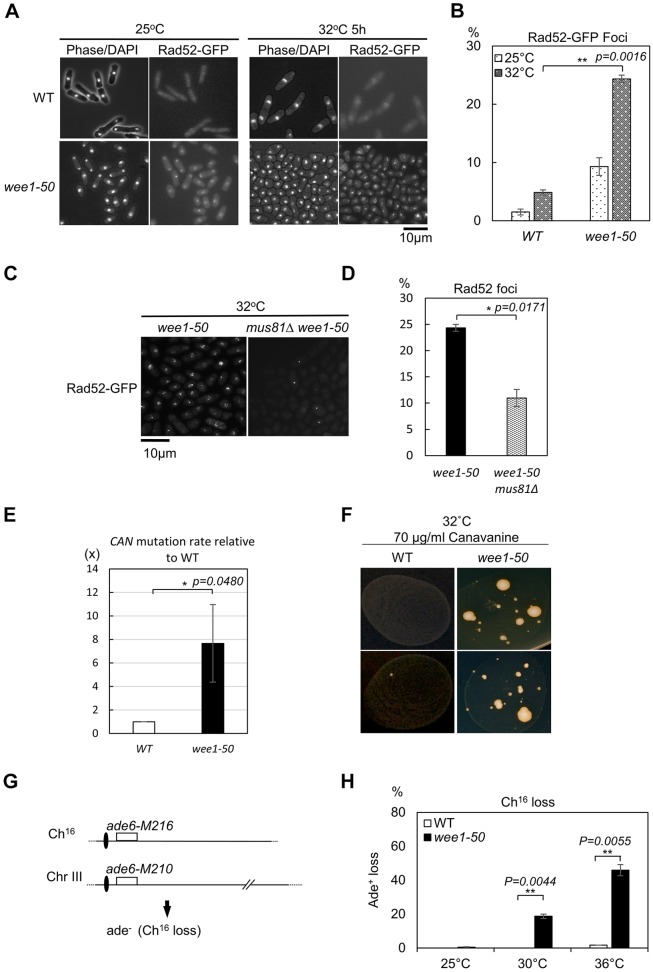
We next tested whether disrupting Wee1 could lead to DNA damage associated with replication stress. We monitored DNA damage-induced Rad52 foci in a wee1-50 mutant. Indeed, inactivation of Wee1 resulted in significantly elevated levels of DNA damage foci marked by Rad52–GFP compared to what was seen in WT cells (Fig. 2A,B). Earlier work has demonstrated that increased CDK activity promotes Mus81–Eme1 endonuclease activity (Dehé et al., 2013; Dominguez-Kelly et al., 2011). Indeed, deletion of mus81+ resulted in significantly reduced levels of Rad52–GFP DNA damage foci in a wee1-50 background (P<0.05) (Fig. 2C,D). Thus, Wee1 inactivation leads to elevated levels of Mus81-dependent DNA damage.
Fig. 2.
Wee1 inactivation causes DNA damage, increases mutation rates and leads to Ch16 loss. (A) Examination of Rad52–GFP foci in WT or wee1-50 cells at 25°C or 32°C. Cells were grown to log phase at the permissive temperature before being transferred to the semi-permissive temperature for 5 h. Samples were fixed directly in methanol/acetone and examined by fluorescence microscopy. (B) The percentage of cells containing Rad52–GFP foci in the indicated strains is shown. A total of >100 cells were counted in each experimental group from two independent experiments. **P<0.01 (t-test). (C) A similar experiment to that described in A except a wee1-50 mus81Δ rad52-GFP strain was used. (D) Quantification analysis of wee1-50 mus81Δ cells with Rad52–GFP foci compared to wee1-50 cells. A total of >100 cells were counted in each experimental group in two independent experiments *P<0.05 (t-test). (E) wee1-50 cells exhibit elevated mutation rates upon canavanine treatment compared to WT cells. Cell cultures incubated on canavanine plates at 32°C for 10 days produced Canr (canavanine-resistant) mutant colonies. Colony data were collected from 36 independent cultures. The mutation rates for WT and wee1-50 strains were calculated using the MSS statistical method. The mutation rates are shown as mean for n≥2 experiments *P<0.05 (t-test). (F) Images representative of experiments performed in E at least three times. (G) Schematic of the Ch16 strain. Centromeric regions (ovals) and complementary heteroalleles (ade6-M216 and ade6-M210) are shown for Ch16 and ChIII. (F) Elevated Ch16 loss rates associated with Wee1 inactivation. Wild-type or wee1-50 cells containing the mini-chromosome are ade+. Cells were plated on YES or adenine-limiting plates and the percentage of Ch16 loss events per division was determined (n>500 cells for each data points). The data presented are from at least two independent biological repeats. **P<0.01 (t-test). All error bars are s.e.m.
Studies in budding yeast have shown that dNTP imbalance can cause mutagenesis and induce genome instability (Kumar et al., 2011). Therefore, we tested whether Wee1 inactivation associated with DNA damage or dNTP deregulation induces mutagenesis. We used resistance to canavanine (Fraser et al., 2003; Kaur et al., 1999) to determine the mutation rate in WT and wee1-50 backgrounds. Inactivation of Wee1 led to significantly higher mutation rates (P<0.05) compared to that in WT cells (Fig. 2E,F).
It is known that either increasing or decreasing origin efficiency increases the loss of minichromosome Ch16 owing to effects on replication fork stability (Patel et al., 2008). Consistent with this, wee1-50 cells displayed high rates of minichromosome Ch16 loss at the semi-restrictive (30°C) or restrictive temperature (36°C) compared to that seen in WT cells (Fig. 2G,H). Since spd1+ deletion rescued the S-phase defect in wee1-50 cells, we expect that spd1+ deletion would also suppress genome instability in wee1-50 cells (Salguero et al., 2012). Together, these results suggest that Wee1 is essential for maintaining genome stability through suppressing replication stress, which leads to DNA damage, mutagenesis and replication fork collapse.
Loss of Set2 methyltransferase activity is synthetic lethal with wee1-50
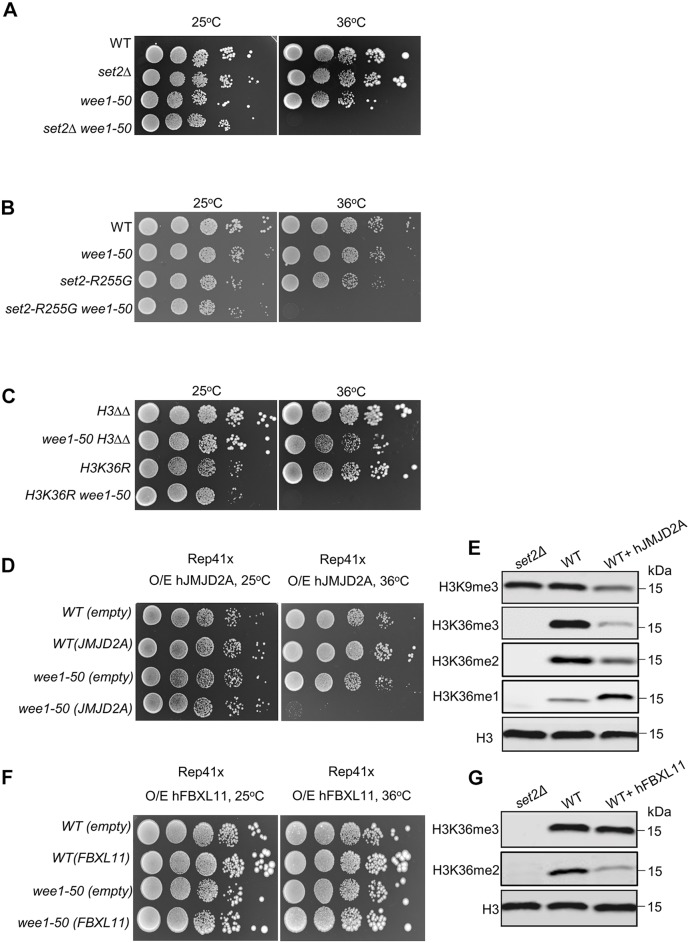
Given that the histone H3K36 methyltransferase Set2 is required for DSB repair (Pai et al., 2014) and MBF-dependent transcription in response to DNA damage (Pai et al., 2017), we tested the possibility that the double mutant set2Δ wee1-50 would show ‘sickness’ due to the accumulated DNA damage caused by Wee1 inactivation (Fig. 2A,B). Consistent with this, set2Δ and wee1-50 were synthetic lethal when grown at the restrictive temperature (36°C) (Fig. 3A). To determine whether this synthetic lethality was dependent on the histone methyltransferase activity of Set2, wee1-50 was crossed with a set2 mutant (set2-R255G) in which the methyltransferase activity was abolished (Pai et al., 2014). The set2-R255G wee1-50 double mutant was not viable at the restrictive temperature of 36°C (Fig. 3B), indicating that the methyltransferase activity of Set2 is required for viability in the absence of Wee1 kinase. Accordingly, wee1-50 was also synthetic lethal with the H3 mutant H3K36R (Fig. 3C). Taken together, these results imply that loss of Set2-dependent H3K36 methylation is synthetic lethal with Wee1 inactivation.
Fig. 3.
Mutations that cause loss of Set2-dependent H3K36 methylation are synthetic lethal with wee1-50. (A) WT, set2Δ, wee1-50 and set2Δ wee1-50 cells were serially diluted and spotted onto YES plates and incubated at the indicated temperatures for 2–3 days. (B) WT, set2-R255G, wee1-50 and set2-R255G wee1-50 cells were serially diluted and spotted onto YES plates and incubated at the indicated temperatures for 2–3 days. (C) H3ΔΔ (deletion of two of the three H3 genes), wee1-50, H3K36R, H3K36R wee1-50 cells were serially diluted and spotted onto YES plates and incubated at the indicated temperatures for 2-3 days. (D) Serial dilutions of WT cells overexpressing (O/E) empty vector pREP41x or pREP41x-JMJD2A (encoding hJMJD2A), and wee1-50 mutants expressing empty vector pREP41x or pREP41x-JMJD2A. Transformants were serially diluted and spotted onto EMM without leucine in the absence of thiamine at 25°C or 36°C. (E) Western blotting analysis of H3K9me3, H3K36me3, H3K36me2 and H3K36me1 in WT cells containing pREP41x or pREP41x-JMJD2A, and set2Δ cells. H3 is shown as a loading control. (F) Serial dilutions of WT cells overexpressing empty vector pREP41x or pREP41x-FBXL11, and wee1-50 mutants overexpressing empty vector pREP41x or pREP41x-FBXL11. Transformants were serially diluted and spotted onto EMM without leucine in the absence of thiamine at 25°C or 36°C. (G) Western blotting analysis of H3K36me3 and H3K36me2 in WT cells expressing pREP41x or pREP41x-FBXL11 and set2Δ cells. H3 is shown as a loading control.
Loss of H3K36 tri-methylation is synthetic lethal with wee1-50
In contrast to SETD2, its human homologue, Set2 in S. pombe is responsible for all three forms of H3K36 methylation (mono-, di- and tri-methylation; H3K36me1, 2 and 3) and thus its loss cannot be used to distinguish between methylation states (Morris et al., 2005). We therefore investigated the consequences of expressing human (h)JMJD2A (also known as KDM4A), the human histone demethylase that catalyses the conversion of H3K36me3/me2 into H3K36me2/me1, under the control of the thiamine repressible (nmt) promoters (Fig. S2A,C), on a plasmid in WT or wee1-50 cells at the permissive or restrictive temperatures (Hillringhaus et al., 2011; Klose et al., 2006; Shin and Janknecht, 2007; Whetstine et al., 2006). Overexpression of hJMJD2A was synthetic lethal with wee1-50 at 36°C (Fig. 3D; Fig. S2E). Consistent with previous studies, expression of hJMJD2A resulted in a reduction of H3K36me3 and H3K36me2 levels (Fig. 3E). In addition to H3K36me3 loss, expressing hJMJD2A also resulted in reduced levels of H3K9me3 (Fig. 3E; Fig. S2C). However, as we did not observe synthetic lethality between deletion of clr4+, encoding the H3K9 methyltransferase, and wee1-50 (Fig. S3A), this indicates that H3K9me3 loss is not required for cell viability in the absence of Wee1.
To distinguish between loss of H3K36me3 and H3K36me2, we expressed the WT human H3K36me2-specific demethylase human (h)FBXL11 (also known as JHDM1A and KDM2A) in WT or wee1-50 cells (Fig. S2B,D,F) (Tsukada et al., 2006). Accordingly, we found that expression of hFBXL11 in fission yeast resulted in a significant decrease in H3K36me2 but did not affect H3K36me3 levels (Fig. 3G), indicating that hFBXL11 preferentially demethylates H3K36me2 in vivo. However, expression of hFBXL11 did not induce a significant viability loss in wee1-50 cells at 36°C (Fig. 3F), and expression of hJMJD2A or hFBXL11 did not sensitize WT or wee1-50 cells at the permissive temperature (Fig. 3D,F). Collectively, these findings provide strong evidence that the histone mark H3K36me3 is required for viability in the absence of Wee1.
set2Δ synthetic lethality with wee1-50 can be suppressed by Cdc2 inactivation
We next explored whether Wee1 inactivation leads to synthetic lethality with set2Δ through elevated CDK activity or through a CDK-independent function. To test this, we investigated whether we could suppress the synthetic lethality by inhibiting CDK activity. We crossed the analogue-sensitive cdc2 mutant (cdc2-as) (Dischinger et al., 2008) with set2Δ wee1-50 to create a cdc2-as set2Δ wee1-50 triple mutant. Instead of using the ATP analogue molecule (1-NM-PP1) to inactivate Cdc2 activity, we found that the cdc2-as mutant exhibited modest temperature sensitivity. As shown in Fig. S4A, loss of CDK activity suppressed the growth defect of set2Δ wee1-50 mutants. Furthermore, the triple mutant showed a plating efficiency of 87.5±1.5% as compared to set2Δ wee1-50, which was only 0.3±0.3% (±s.e.m.) (Fig. S4B), while set2Δ and wee1-50 single mutants exhibited more than 90% plating efficiency. Collectively, these results indicate elevated CDK activity resulting from Wee1 inactivation leads to synthetic lethality in a set2Δ wee1-50 background. Since cdc2-as showed the same phenotype as cdc2-ts, we would expect that the temperature-sensitive cdc2-ts mutant would also suppresses set2Δ wee1-50 synthetic lethality.
set2Δ synthetic lethality with wee1-50 results from replication catastrophe
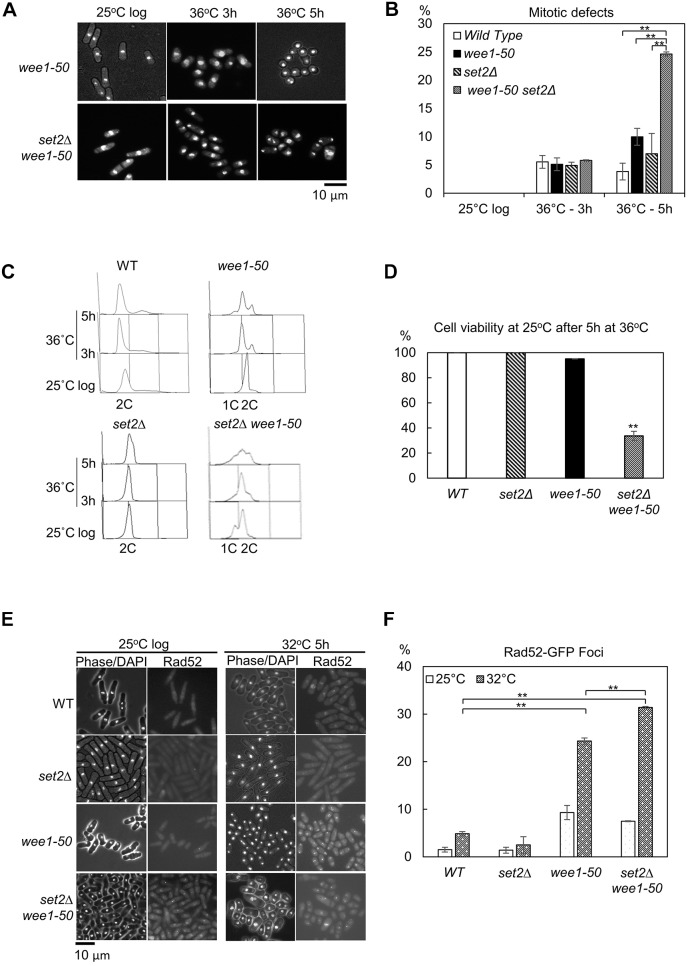
Previous studies using fission yeast found that a number of checkpoint mutants (rad1Δ, rad3Δ, rad9Δ, rad17Δ and hus1Δ) were synthetic lethal with wee1-50 at the restrictive temperature (al-Khodairy and Carr, 1992; Enoch et al., 1992). These double mutants exhibited a ‘cut’ phenotype suggesting that cell death arose through mitotic catastrophe (Enoch et al., 1992). Thus, we suspected that the synthetic lethality seen in set2Δ wee1-50 cells might be also due to premature entry into mitosis. We found that set2Δ wee1-50 cells had the ‘wee’ phenotype, and 26% of set2Δ wee1-50 cells exhibited a ‘cut’ phenotype at 36°C after 5 h incubation (Fig. 4A,B). This level of cutting in set2Δ wee1-50 cells was significantly higher than wee1-50 cells (10%) (P<0.05) (Fig. 4A,B). Surprisingly, set2Δ wee1-50 cells showed a striking S-phase delay even at the permissive temperature (Fig. 4C), suggesting inactivation of Wee1 causes more extreme DNA replication defects in set2Δ cells. Flow cytometry analysis showed that set2Δ wee1-50 cells accumulated in S-phase following a shift to 36°C for 3–5 h (Fig. 4C). This result suggests that the observed cell death might be mostly due to a permanent replication stalling in the set2Δ wee1-50 double mutant rather than through mitotic catastrophe (Fig. 4A,B). Nevertheless, 72±5% (±s.e.m.) of set2Δ wee1-50 cells exhibited a ‘cut’ phenotype following a shift to 36°C for 24 h (Fig. S5A,B), suggesting that the majority of set2Δ wee1-50 cells eventually undergo mitotic catastrophe after long-term replication stalling.
Fig. 4.
set2Δ synthetic lethality with wee1-50 results from replication catastrophe. (A) Inactivation of Wee1 in set2Δ cells results in premature entry into mitosis. wee1-50 or set2Δ wee1-50 cells were grown to log phase at the permissive temperature (25°C), then incubated at 36°C to inactivate Wee1. Samples were fixed with 70% ethanol at the indicated times. The fixed cells were stained with DAPI and examined by microscopy analysis. (B) Quantitative analysis of the percentage of cells with mitotic defect cells from experiments as in A. The data presented are from at least two independent biological repeats. **P<0.01 (t-test, n≥2 experiments for each genotype, n>200 cells for each data point). (C) WT, wee1-50, set2Δ and set2Δ wee1-50 strains were grown to log phase at the permissive temperature (25°C) then transferred to 36°C for the times shown. At the indicated times, cells were processed for FACS analysis. (D) WT, set2Δ, wee1-50 and set2Δ wee1-50 cells from the 5 h time point in (C) were collected, plated on the YES medium and incubated at 25°C for 3–4 days for viability analysis. (n≥2 experiments for each genotype, n>500 cells for each data point). **P<0.01 (t-test, between WT and set2Δ wee1-50 cells; P-value=0.0031). (E) Examination of Rad52–GFP foci in WT, set2Δ, wee1-50 and set2Δ wee1-50 cells at 25°C or 32°C. Cells were grown to log phase at the permissive temperature before being transferred to the semi-permissive temperature (32°C) for 5 h. Samples were fixed directly in methanol/acetone and examined by fluorescence microscopy. (F) The percentage of cells containing Rad52–GFP foci in the indicated strains is shown. **P<0.01 (t-test; n≥2 experiments, n≥100 cells for each data point). All error bars are s.e.m.
To investigate whether the replication arrest was the cause of synthetic lethality in set2Δ wee1-50 cells, double mutants were incubated at the restrictive temperature of 36°C for 5 h and the cell viability was examined by returning them to the permissive temperature of 25°C. The results showed that 66% of set2Δ wee1-50 cells lost viability after shifting to the restrictive temperature for 5 h (Fig. 4D), in which the majority of double mutants had arrested during DNA replication but only 26% of the double mutants underwent mitotic catastrophe (Fig. 4B), suggesting that most set2Δ wee1-50 cells were dying in S-phase.
Furthermore, we found that set2Δ wee1-50 cells exhibited elevated levels of DNA damage compared to WT cells (Fig. 4E,F), indicative of replication stress-induced DNA damage accumulation. Together, these data suggest that Wee1 inactivation in a set2Δ background leads to elevated levels of replication fork collapse and DNA damage.
set2Δ wee1-50 replication catastrophe results from nucleotide depletion
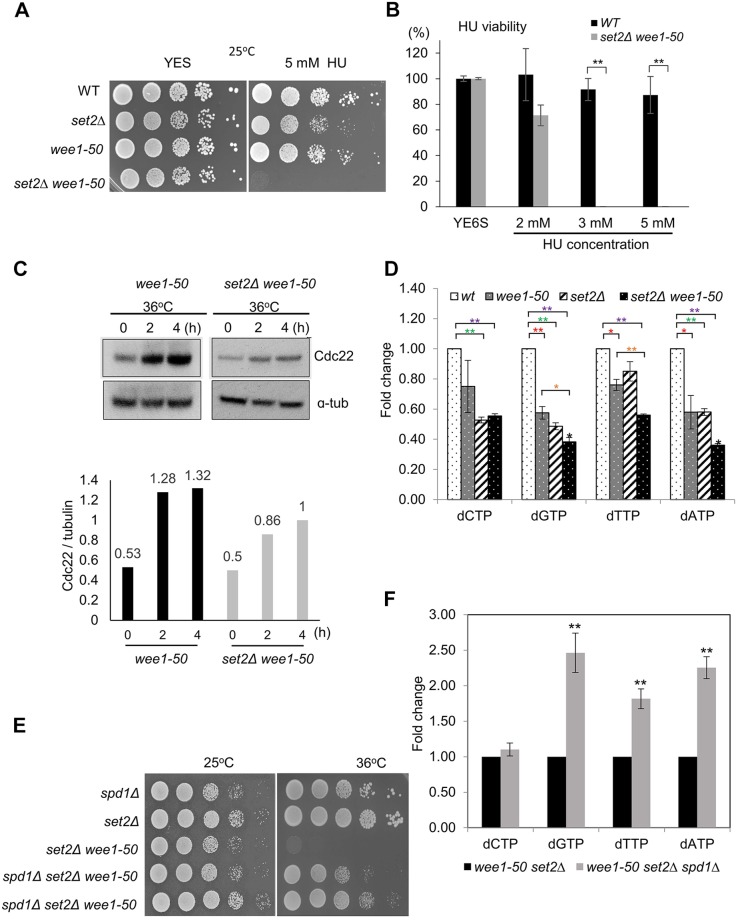
Our data indicate that Wee1 inactivation leads to nucleotide depletion. Further, we have independently identified a role for Set2 in dNTP synthesis. We showed that dNTP levels were lower in set2Δ cells compared to wild type (Pai et al., 2017). We therefore tested the possibility that the set2Δ wee1-50 synthetic lethality during S-phase was due to severe nucleotide depletion. Consistent with this, we found set2Δ wee1-50 cells to be acutely sensitive to low levels of HU (Fig. 5A,B). These cells exhibited elongated phenotypes upon HU treatment, suggesting that the lethality of the double mutant was not due to the compromised checkpoint (Fig. S5C). Instead, the lethality was more likely to be due to dNTP starvation. Consistent with this, we found that Cdc22 levels (the catalytic subunit of RNR) were reduced in response to replication stress induced in a set2Δ wee1-50 double mutant compared to a wee1-50 mutant (Fig. 5C). In accordance with this observation, dNTP levels are also significantly lower in set2Δ wee1-50 in comparison to wee1-50 cells under replication stress at 36°C (P<0.005) (Fig. 5D). Furthermore, deleting spd1+ robustly suppressed the synthetic lethality of the set2Δ wee1-50 double mutant at 36°C (Fig. 5E), indicating elevated dNTPs can suppress set2Δ wee1-50 synthetic lethality. Consistent with this observation, we found that dNTP levels are higher in the spd1Δ set2Δ wee1-50 triple mutant compared to set2Δ wee1-50 double mutants (Fig. 5F). Taken together, these findings indicate that the set2Δ wee1-50 synthetic lethality resulted from dNTP depletion, to below a critical level.
Fig. 5.
The synthetic lethality between set2Δ and wee1-50 results from dNTP depletion. (A) set2Δ wee1-50 cells are sensitive to low levels of HU. WT, set2Δ, wee1-50 and set2Δ wee1-50 cells were serially diluted and spotted onto YES plates containing 5 mM HU and incubated at the permissive temperature (25°C) for 3–4 days. (B) Quantification (mean±s.e.m.) of the viability of WT and set2Δ wee1-50 cells on YES plates at 25°C containing different concentrations of HU as indicated (n≥2 experiments, n≥500 cells for each data point). **P<0.01 (t-test). (C) The endogenous protein levels of Cdc22 were examined in wee1-50 and set2Δ wee1-50 cells following incubation at 36°C for 4 h. Samples of cells were taken at the indicated time points and cell extracts were made by using the TCA method. Cdc22 was detected using an antibody against the CFP tag. α-tubulin is shown as a loading control. The lower panel shows a quantification of Cdc22 levels in wee1-50 and set2Δ wee1-50 cells for the representative blot in the upper panel. (D) dNTP levels were measured in WT, wee1-50, set2Δ and set2Δ wee1-50 strains. Cells were grown to log phase at 25°C followed by a 5 h incubation at 36°C. Samples of cells were collected and re-suspended in 10% TCA for subsequent HPLC analysis following neutralization. Means±s.e.m. of three experiments are shown. *P<0.05, **P<0.01 (t-test). (E) spd1Δ suppresses the synthetic lethality of set2Δ wee1-50. Strains were serially diluted and spotted onto YES plates and incubated at the indicated temperatures for 2–3 days. (F) dNTP levels were measured in set2Δ wee1-50 and spd1Δ set2Δ wee1-50 strains. Cells were grown to log phase at 25°C followed by a 5 h incubation at 36°C. Samples of cells were collected and re-suspended in 10% TCA for subsequent HPLC analysis following neutralization. The mean±s.e.m. for three experiments are shown. **P<0.01 (t-test between set2Δ wee1-50 and spd1Δ set2Δ wee1-50 strains; P-values: dCTP=0.3288, dGTP=0.0065, dTTP=0.0042, dATP=0.0011). The data presented are from three independent biological repeats.
The nucleotide depletion in set2Δ wee1-50 cells is caused by downregulation of the transcription of MBF-dependent genes and increased origin firing
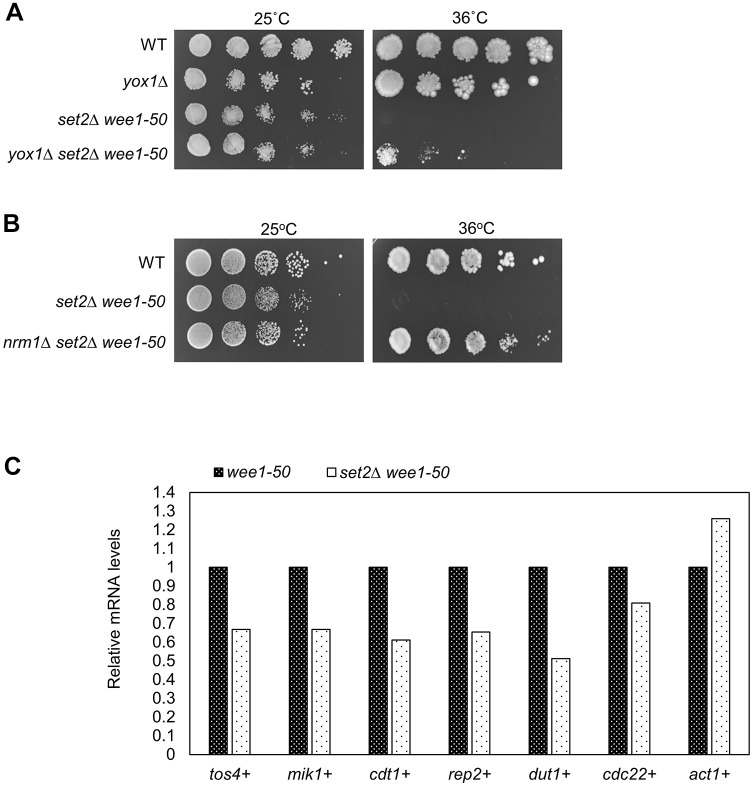
We have shown that Set2 controls dNTP synthesis through regulation of MBF transcription activity (Pai et al., 2017). As part of that study we found that deletion of MBF transcriptional repressor Yox1 suppressed the prolonged S-phase in set2Δ cells (Pai et al., 2017). Thus, we tested whether deletion of Yox1 could suppress the lethality of the set2Δ wee1-50 double mutant and found that the triple mutant exhibited an increase in viability (Fig. 6A), indicating elevated MBF transcription activity can suppress set2Δ wee1-50 synthetic lethality, presumably due to an increase in dNTP pools. Consistent with this, deletion of MBF transcriptional repressor Nrm1 also supressed the synthetic lethality of set2Δ wee1-50 cells (Fig. 6B). Furthermore, we tested whether Set2 also affected mRNA levels of MBF-dependent genes in a wee1-50 background. To do this, the set2Δ wee1-50 double and wee1-50 single mutants were grown at the restrictive temperature of 36°C for 5 h and global levels of gene expression were compared through microarray experiments. This analysis revealed that transcription of the MBF transcription factor activator rep2+ and MBF-dependent genes tos4+, cdt1+, mik1+, cdc22+ and dut1+ (Aligianni et al., 2009) was reduced following set2+ deletion in a wee1-50 background, while act1+, which is not MBF-induced, was not (Fig. 6C). Taken together, these findings support a role for Set2 in facilitating MBF transcription in response to DNA damage or replication stress resulting from Wee1 inactivation, presumably through the regulation of Rep2 function in MBF activation.
Fig. 6.
Set2 is required for MBF-dependent gene expression in wee1-50 cells. (A) yox1Δ suppresses the synthetic lethality of set2Δ wee1-50. Strains were serially diluted and spotted onto YES plates and incubated at 25°C or 36°C for 2–3 days. (B) nrm1Δ suppresses the synthetic lethality of set2Δ wee1-50 cells. Strains were serially diluted and spotted onto YES plates and incubated at 25°C or 36°C for 2–3 days. (C) tos4+, mik1+, cdt1+, rep2+, dut1+ and cdc22+ transcript levels in set2Δ wee1-50 cells relative to that in wee1-50 (the expression level in wee1-50 cells is set as 1.0). Data are the mean calculated from two biological repeats. act1+ is shown as an MBF-independent control.
Consistent with observations above (Fig. 1A), inactivation of Wee1 also leads to more origin firing in set2Δ cells (Fig. S6A). We also found that partial inactivation of replication licensing factor Cdc18 (cdc18ts at 34°C) suppressed the synthetic lethality of set2Δ wee1-50 mutants at the semi-restrictive temperature (Fig. S6B), suggesting that reducing the number of active replication origins alleviates dNTP depletion in the set2Δ wee1-50 background. No further synthetic lethality or sickness in cdc18ts wee1-50 or mcm4tdts wee1-50 cells at the semi-restrictive temperature suggesting replication stalling is due to dNTP depletion rather than defects in other steps of DNA replication (Fig. S6C). Moreover, we did not observe synthetic sickness between Polε and Wee1 inactivation, indicating that the slow S-phase in set2Δ cells is unlikely to be due to the defective polymerase function (Fig. S6D).
Disrupting checkpoint-dependent dNTP synthesis with wee1-50 results in replication catastrophe
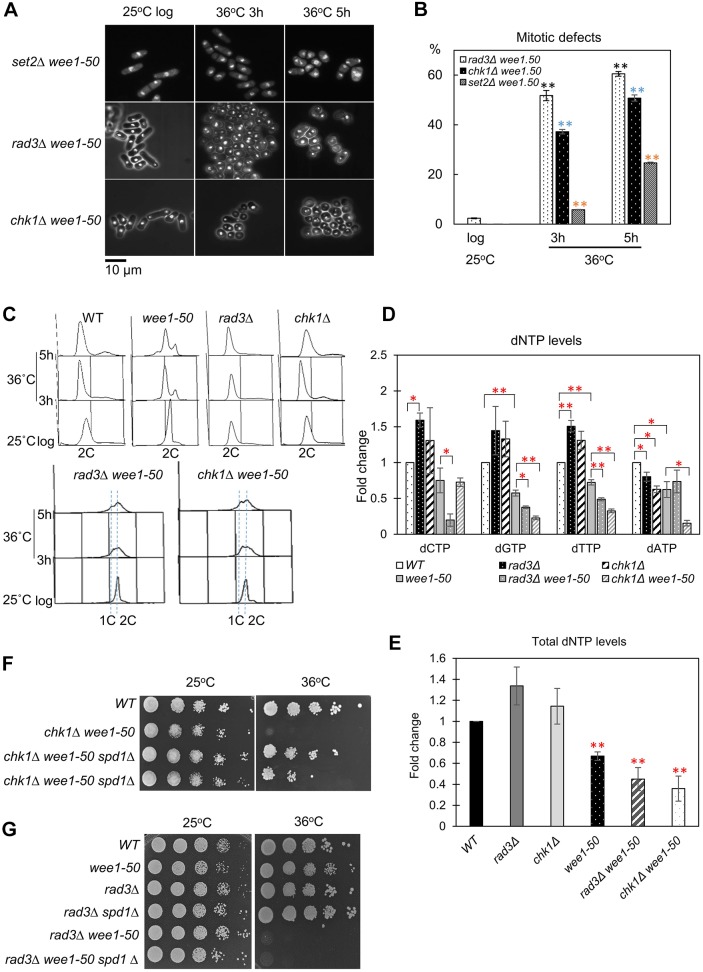
We and others have identified a role for the DNA damage checkpoint in inducing dNTP synthesis in response to genotoxic stress (Blaikley et al., 2014; Liu et al., 2003; Moss et al., 2010). We therefore compared the effects of inactivating Wee1 in set2Δ cells with those seen when Wee1 was inactivated in rad3Δ or chk1Δ cells (Fig. S7A–C). We were unable to make the cds1Δ wee1-50 double mutant as they were lethal at 25°C (Fig. S7D). We found that set2Δ wee1-50 cells phenocopied the synthetic lethality of rad3Δ wee1-50, hus1Δ wee1-50 and chk1Δ wee1-50 mutants (Fig. S7). However, we found that the percentage of cells exhibiting a ‘cut’ phenotype in set2Δ wee1-50 cells (20%) was significantly lower than for rad3Δ wee1-50 (60.5%) or chk1Δ wee1-50 (50.8%) mutants at 4 or 5 h (P<0.05) (Fig. 7A,B; Fig. S8). We also monitored cell cycle profiles of rad3Δ wee1-50 and chk1Δ wee1-50 cells following a shift to the restrictive temperature for 5 h. Surprisingly, Wee1 inactivation also caused replication stalling in rad3Δ and chk1Δ cells (Fig. 7C). In contrast, the tel1Δ wee1-50 double mutant did not exhibit synthetic lethality (Fig. S3B), consistent with the fact that tel1Δ cells exhibit normal S-phase and DNA damage checkpoints (Willis and Rhind, 2009). Taken together, the above results indicate that disrupting Wee1 causes S-phase arrest in set2Δ, rad3Δ or chk1Δ cells, consistent with Wee1 playing an important role in facilitating efficient S-phase progression in fission yeast. We also examined the dNTP levels in the single and double mutants. Unexpectedly, we found that dNTP levels were increased in rad3Δ and chk1Δ cells compared to WT cells under unstressed conditions (Fig. 7D). This may reflect a lack of DNA damage checkpoint-mediated inhibition of the MBF target genes. However, deleting rad3+ or chk1+ in a wee1-50 background resulted in a significant reduction in dNTP levels compared to WT or wee1-50 cells (Fig. 7D). Total dNTP levels were significantly reduced in wee1-50, rad3Δ wee1-50 and chk1Δ wee1-50 cells compared to WT cells (Fig. 7E). Therefore, these results suggest that Rad3 and Chk1 play an important role in maintaining dNTP levels in the absence of Wee1. Furthermore, we found that it was possible to suppress the synthetic lethality following Wee1 inactivation in a chk1Δ background by deleting spd1+ in wee1-50 chk1Δ cells (Fig. 7F). In contrast, deleting spd1+ did not suppress the synthetic lethality of rad3Δ wee1-50 double mutants (Fig. 7G), consistent with Rad3 (ATR) playing additional functions in response to replication fork stalling (Lindsay et al., 1998). These findings, together, support an essential role for Wee1 in modulating CDK-induced replication stress, and show that inactivating Wee1 together with mutations that disrupt dNTP synthesis in response to genotoxic stress results in replication catastrophe.
Fig. 7.
rad3Δ and chk1Δ are synthetic lethal with wee1-50 through replication stress. (A) set2Δ wee1-50, rad3Δ wee1-50 or chk1Δ wee1-50 result in premature entry into mitosis but the proportion of cells with the ‘cut’ phenotype in the set2Δ wee1-50 mutant is significantly lower at earlier time points. set2Δ wee1-50, rad3Δ wee1-50 and chk1Δ wee1-50 cells were grown to log phase at the permissive temperature (25°C), then incubated at 36°C to inactivate Wee1. Samples were fixed with 70% ethanol at the indicated times. The fixed cells were stained with DAPI and examined by microscopy analysis. (B) Quantitative analysis of cells in A. Means±s.e.m. are shown. **P<0.01 (t-test); black asterisks indicates statistically significant differences between rad3Δ wee1-50 cells grown at either 36°C or 25°C (P-values for rad3Δ wee1-50 cells: 4h=0.0017, 5h=0.0003; averages of n≥2 experiments, n≥100 cells for each data point); blue asterisks indicate statistically significant differences between chk1Δ wee1-50 cells grown at either 36°C or 25°C (P-values for chk1Δ wee1-50 cells: 4 h=0.0006, 5h=0.0006; averages of n≥2 experiments, n≥100 cells for each data point); orange asterisks indicate statistically significant differences between set2Δ wee1-50 cells grown at 36°C or 25°C (P-values for set2Δ wee1-50 cells: 4h=0.0002, 5h=0.0078; averages of n≥2 experiments, n≥100 cells for each data point). (C) Wee1 inactivation causes replication stress in rad3Δ or chk1Δ mutants. Flow cytometric analysis of WT, rad3Δ, chk1Δ, rad3Δ wee1-50 or chk1Δ wee1-50 cells at 25°C or 36°C at the indicated time points. (D) dNTP levels were measured in WT, wee1-50, rad3Δ wee1-50 and chk1Δ wee1-50 strains. These strains were grown to log phase at 25°C following by a 5 h incubation at 36°C. Samples of cells were collected and re-suspended in 10% TCA for HPLC analysis. Means±s.e.m. of three biological repeats are shown. *P<0.05, **P<0.01 (t-test). (E) Total dNTP levels are reduced in wee1-50, rad3Δ wee1-50 or chk1 Δ wee1-50 cells compared to WT cells. **P<0.01 (t-test; P-values: wee1-50=0.0012, rad3Δ wee1-50=0.0075, chk1Δ wee1-50=0.0059). The data presented are from three independent biological repeats. (F) spd1Δ suppresses the synthetic lethality of chk1Δ wee1-50. Strains were serially diluted and spotted onto YES plates and incubated at indicated temperatures for 2–3 days. (G) spd1Δ cannot suppress the synthetic lethality of rad3Δ wee1-50. A similar experiment was carried out as described in F.
DISCUSSION
Understanding the mechanisms that can lead to replication stress and how they can be targeted remains an important goal in cancer research. In this study, we define an evolutionarily conserved role for the CDK regulator Wee1 in suppressing replication stress through dNTP depletion, thereby maintaining genome stability. Furthermore, we demonstrate that mutations that cause dNTP homeostasis defects, resulting from either loss of Set2 or the DNA integrity checkpoint inactivation, are synthetic lethal with CDK-induced replication stress resulting from Wee1 inactivation. Taken together, our results support a ‘dNTP supply and demand’ model, which can be exploited to target replication stress.
Our data indicate that Wee1 inactivation leads to elevated levels of CDK-dependent replication origin firing, resulting in an overall increase in the total number of origins being fired. This in turn leads to dNTP depletion, replication stress-associated DNA damage and subsequent genome instability. We found that the replication stress associated with Wee1 inactivation alone, or in combination with set2Δ, resulted in DNA damage (Rad52–GFP foci formation). This DNA damage is likely to have been triggered by stalled replication forks, which present as substrates for structure-specific endonucleases (Osman et al., 2003). Consistent with this notion, elevated CDK activity has been shown to promote Mus81 activation through phosphorylation of Eme1 (Dehé et al., 2013). We hypothesize that elevated levels of CDK in the S-phase of wee1-50 cells contribute to the Mus81-dependent DNA damage. We also observed that Wee1 inactivation resulted in robust induction of Cdc22, the catalytic subunit of RNR, thus promoting dNTP synthesis. These findings are consistent with there being a major role for Wee1 in regulating CDK activity in S-phase in fission yeast (Anda et al., 2016) and support an evolutionarily conserved role for WEE1 in regulating dNTP usage and preventing DNA damage through regulating origin firing (Beck et al., 2012). Our findings further demonstrate that Wee1 inactivation has significant consequences for genome stability.
We find Wee1 inactivation together with loss of Set2-dependent histone H3K36 tri-methylation results in synthetic lethality. Our data support a key role for Set2-dependent H3K36me3 in facilitating MBF-dependent Cdc22 transcription and thus promoting dNTP synthesis in response to genotoxic stress (Pai et al., 2017). Furthermore, Set2-dependent dNTP synthesis becomes essential following Wee1 inactivation and CDK-induced dNTP depletion. In support of this, we find loss of Set2 reduces MBF-dependent expression of Cdc22, the catalytic subunit of RNR, and leads to dNTP pool depletion in response to genotoxic stress (Pai et al., 2017). Simultaneous loss of Wee1 and Set2 leads to critically low dNTP pools and a failure to induce Cdc22 expression. This in turn leads to cell death through replicative arrest and mitotic catastrophe. Consistent with this, we find that set2Δ wee1-50 synthetic lethality is associated with S-phase arrest; Cdc22 expression is significantly reduced in the double mutant compared to wee1-50 cells; dNTP levels are significantly reduced in the double mutant compared to wee1-50 cells, and the double mutant is acutely sensitive to HU at the permissive temperature. Accordingly, the synthetic lethality can be suppressed through increasing dNTP synthesis by depleting Spd1, by increasing MBF-dependent Cdc22 expression or by compromising replication origin licensing.
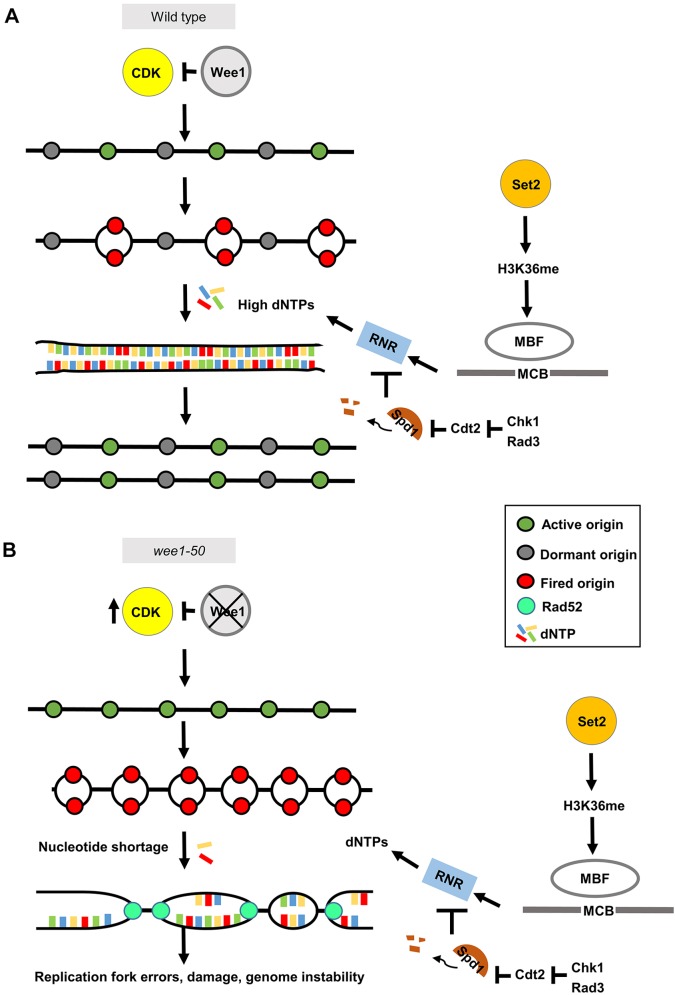
We further define a more general role for dNTP synthesis in maintaining viability in response to Wee1 inactivation. Wee1 inactivation has been previously found to be synthetic lethal with loss of Rad3 (ATR) or Chk1 in both yeast and humans (al-Khodairy and Carr, 1992; Enoch et al., 1992; Srivas et al., 2016). Synthetic lethality between Wee1 and checkpoint-deficient mutations has been proposed to be a consequence of mitotic catastrophe. However, our results demonstrate that, while mitotic catastrophe is observed in rad3Δ or chk1Δ checkpoint mutants following Wee1 inactivation, these cells undergo prior replication arrest resulting from an insufficient dNTP supply. Importantly, the Rad3 (ATR)-dependent checkpoint pathway is required to induce dNTP synthesis following replication stress and DNA damage. DNA damage checkpoint activation leads to Cul4–Ddb1Cdt2-dependent degradation of Spd1, a negative regulator of RNR to promote dNTP synthesis (Moss et al., 2010). The replication checkpoint also promotes MBF-dependent transcription of Cdc22, the catalytic subunit of RNR through Cds1-dependent phosphorylation of Yox1, which blocks the binding of this negative regulator to MBF in response to replication stress (Ivanova et al., 2011). In this respect, Set2 and the DNA integrity checkpoint function analogously to facilitate dNTP synthesis in response to both DNA damage and replication stress in fission yeast. Accordingly, we show that elevating dNTP levels through deletion of spd1+ suppressed the synthetic lethality of both the set2Δ wee1-50 and chk1Δ wee1-50 mutants. That spd1+ deletion could not suppress the synthetic lethality of rad3Δ wee1-50 mutant likely reflects the fact that Rad3 (ATR) performs additional roles in replication fork restart. Taken together, these findings support a ‘dNTP supply and demand’ model in which Set2 and DNA integrity checkpoint-dependent dNTP synthesis becomes essential following elevated CDK-induced origin firing and dNTP depletion, thereby preventing replication catastrophe. This model explains how Wee1 inactivation results in synthetic lethality with loss of Set2, sheds new light on the synthetic lethal relationship between loss of ATR, Chk1 and Wee1, and further predicts that other mutations that disrupt dNTP synthesis in response to replication stress will also be synthetic lethal with Wee1 inactivation (Fig. 8).
Fig. 8.
Wee1 limits replication stress and suppresses the synthetic lethality of Set2, Rad3 (ATR) or Chk1 loss. (A) In WT cells, Wee1, the mitotic inhibitor, controls the firing of dormant origins through limiting CDK activity during DNA replication. Set2-dependent H3K36me regulates MBF-dependent RNR expression during the cell cycle and in response to genotoxic stress. Checkpoint activation leads to degradation of Spd1 and to increased dNTP supply. Thus, Wee1, Set2 and cell cycle checkpoint proteins (Rad3 and Chk1) work together to regulate the supply of dNTP levels to limit replication stress and maintain genome stability. (B) Increased CDK activity (resulting from Wee1 inactivation) increases replication origin firing leading to increased dNTP demand. This in turn leads to replication stalling and to DNA integrity checkpoint activation. Failure to increase dNTP supply (e.g. loss of Set2, Rad3 or Chk1) when dNTP demand is high leads to replication catastrophe. See text for details.
Our findings indicate that the S-M cell cycle checkpoint is intact in set2Δ cells (Pai et al., 2017), where an elongated phenotype is seen in response to HU or bleomycin treatment. Moreover, in contrast to rad3Δ or chk1Δ, Wee1 inactivation did not lead to rapid mitotic catastrophe in set2Δ cells. While set2Δ wee1-50 cells underwent mitotic catastrophe at later time points, this may reflect a role for Set2 in promoting MBF-dependent transcription of mik1+, encoding the kinase Mik1, which negatively regulates Cdc2 and leads to mitotic catastrophe when deleted in a wee1-50 background (Christensen et al., 2000; Dutta et al., 2008; Dutta and Rhind, 2009; Lee et al., 1994; Lundgren et al., 1991; Ng et al., 2001).
In accordance with the findings described here, it was demonstrated that H3K36me3-deficient human cancers are synthetic lethal with the WEE1 inhibitor AZD1775 as a result of dNTP starvation (Pfister et al., 2015). These findings are of clinical relevance as, despite the frequent loss of histone H3K36me3 in multiple cancer types and its association with poor patient outcome, there is no therapy targeting H3K36me3-deficient cancer types (Forbes et al., 2015; Lawrence et al., 2014; Li et al., 2016). Moreover, our data suggest that inhibitors of ATR and CHK1 may have different effects in cancer therapy. As inhibitors to WEE1, ATR and CHK1 are already in clinical trials (www.clinicaltrials.gov), we anticipate that our findings described here will provide important mechanistic insights into the targeting of cancers exhibiting replication stress.
MATERIALS AND METHODS
Yeast strains, media and genetic methods
The strains used in this study are listed in Table S1. Standard media and growth conditions were used. Cultures were grown in rich media (YE6S) or Edinburgh minimal media (EMM) at 32°C with shaking, unless otherwise stated. Nitrogen starvation was carried out using EMM lacking NH4Cl (EMM−N).
Flow cytometry analysis
For flow cytometry, methanol/acetone fixed cells were rehydrated in 10 mM EDTA, pH 8.0, 0.1 mg/ml RNase A and incubated at 37°C for 2 h. Cells were then stained with 1 μM Sytox Green (ThermoFisher Scientific S7020), and analyzed using a Coulter Epics XL-MCL (Fullerton, CA).
Serial dilution assay
A dilution series for the indicated mutant cells was spotted onto YES plates. Plates were incubated at 25°C, 32°C or 36°C for 2–3 days, as indicated, before analysis.
Survival analysis
Exponential cultures were obtained in liquid YE6S medium inoculated with a single colony picked from a freshly streaked (YE6S) stock plate and grown overnight at 25°C with vigorous shaking. Exponential cells were resuspended in YE6S at a density of 2×107 cells ml−1. Serial dilutions were made, and 500 cells were plated on YE6S plates at the restrictive temperature of 36°C, as well as on a control plate incubated at 25°C. Plates were incubated for 2–3 days and colonies were then scored.
Analysis of replication origin firing
The polymerase usage sequence (Pu-seq) technique was performed as previously described (Daigaku et al., 2015). Briefly, DNA was extracted from cells grown to log phase at either 18°C or on 34°C as indicated. For ‘wt’ datasets, two strains were used, both strains containing an rnh201 deletion together with either polymerase δ (cdc6-L591G) or polymerase ε (cdc20-M630F) mutations. These strains incorporate more rNTPs on the strands synthesized by the mutant polymerase. These sites can be mapped by Pu-seq. For the wee1-50 and wee1-50 set2Δ datasets, the two strains also contained these mutations along with rnh201 and cdc6-L591G or cdc20-M630F. The isolated DNA was then subjected to alkali treatment (0.3 M NaOH for 2 h at 55°C), which digested the DNA at the positions of rNTP incorporation and also separated the double strands. The resulting single-stranded (ss)DNA fragments were size selected on agarose gel (fragments between 300–500 bp were isolated). These fragments were then used for creating strand-specific next-generation sequencing libraries and sequenced on a Next-seq Illumina platform resulting in ∼10 M reads from each strain. The Pu-Seq data has been uploaded to Gene Expression Omnibus (GEO) under accession number GSE113747. Reads were aligned to the S. pombe reference sequence (http://www.pombase.org/downloads/genome-datasets), the reads were mapped using bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), and the data was analysed and origin positions and efficiencies were determined using the tools published and described in detail in Daigaku et al., 2015 with default variables except for the ‘percentile threshold for origins’ option was set to 0.2=20th percentile. Efficient origins were determined as origins with higher than 50% efficiency and inefficient origins had less than 25% efficiency. The sequencing experiment was performed once and therefore it is not possible to perform a statistical analysis.
Mini-chromosome instability assay
The mini-chromosome loss assay was carried out as previously described (Allshire et al., 1995; Moss et al., 2010). Briefly, 500–1000 cells from individual ade+ colonies were plated on EMM plates containing low levels of adenine (5 mg/l), then incubated at 25°C, 30°C or 36°C for 3 days and stored for 48 h at 4°C before being scored for the presence of sectored colonies. The number of mini-chromosome loss events per division was determined as the number of Ade− sectored colonies divided by the sum of white and sectored colonies. The experiment was performed in triplicate.
The CanR mutation assay
To analyse mutation rates, a Luria–Delbruck fluctuation analysis was performed (Luria and Delbruck, 1943). Briefly, 1 ml cultures of WT or wee1-50 cells were grown in YES medium to saturation in 12-well plates at 32°C. 100 µl of each culture was spotted onto PMG (−Arg, −His) plates containing 70 µg/ml canavanine and incubated at 32°C for 10–12 days. Colony numbers were scored and mutation rates in culture were analysed using the FALCOR tool (Hall et al., 2009). For each strain, colony data were collected from at least 30 independent cultures. Means and standard deviations were calculated for three independent experiments.
Microscopy analysis
Asynchronous cell cultures were treated with 10 mM hydroxyurea (HU) at the indicated temperature before being fixed in methanol. Samples were rehydrated and stained with 4′,6-diamidino-2-phenylindole (DAPI) before examination using a Zeiss Axioplan 2ie microscope, Hamamatsu Orca ER camera and micromanager software. For visualization of Rad52–GFP foci, cells were incubated at 25°C or 32°C for 5 h before being fixed and visualized as above.
Protein analysis
Protein extracts were made by TCA extraction and analysed by western blotting as described previously (Pai et al., 2014). TAP-tagged proteins were detected with peroxidase-conjugated anti-peroxidase soluble complex (1:1000, P1291, Sigma). Cdc22–GFP was detected using anti-GFP antibody (1:1000, 11814460001, Roche), and α-tubulin was detected with antibody T5168 (1:10,000, Sigma).
dNTP analysis
108 cells were collected and washed with 2% glucose. Cell pellets were then lysed with 50 µl 10% trichloroacetic acid (TCA) and stored at −80°C before high-pressure liquid chromatography (HPLC) analysis. On thawing, cell extracts were spun and the supernatant diluted five-fold with water. Samples were then neutralized and analysed by HPLC as described by Moss et al. using a Waters e2695 autosampler. All peak areas were measured at 258 nm (Moss et al., 2010).
Microarray analysis
Microarray analysis was performed as previously described (Pai et al., 2014; Rallis et al., 2013). Experiments were conducted in duplicate with a dye swap. RNAs from two independent biological replicates have been utilized for cDNA production. Fig. 6C shows average expression ratios from the two repeats. Original data have been deposited in ArrayExpress under accession number E-MTAB-6795. In brief, Alexa Fluor 555- or 647-labelled cDNA was produced from the RNA, using a Superscript direct cDNA labelling system (Invitrogen) and Alexa Fluor 555 and 647 dUTP mix. cDNAs were then purified using an Invitrogen PureLink PCR Purification system and hybridized to the array using a Gene Expression Hybridization kit (Agilent). The arrays are Agilent custom-designed containing 60-mer oligonucleotides synthesized in situ containing 15,000 probes. Following hybridization for at least 17 h, the arrays were washed using a Gene Expression Wash Buffer kit (Agilent) and scanned in an Agilent Array Scanner. Signals were extracted using GenePix software.
Supplementary Material
Acknowledgements
We would like to thank members of the Humphrey lab for helpful comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.P., S.X.P., T.C.H.; Methodology: C.P.; Validation: C.P., K.H., S.C.D., S.E.K., S.E.W., N.D.L.; Formal analysis: C.P., K.H., A.K., C.R., L.K.F., N.D.L.; Investigation: C.P., K.H., S.C.D., S.E.K., R.D., S.E.W.; Writing - original draft: C.P.; Writing - review & editing: C.P., S.C.D., A.K., S.E.K., C.R., C.J.S., J.B., A.M.C., T.C.H.; Supervision: C.P., S.E.K., C.J.S., J.B., A.M.C., T.C.H.; Funding acquisition: S.E.K., C.J.S., J.B., A.M.C., T.C.H.
Funding
This research was supported by the Medical Research Council (to C.C.P., R.D., S.D. and T.C.H.); the Clarendon Scholarship (S.X.P.); L.K.F. is supported by Cancer Research UK; A.M.C.'s group is supported by European Research Council (grant 268788–SMI–DDR) and the Medical Research Council (G1100074) (A.K. and A.M.C.); J.B.’s group is supported by a Wellcome Trust Senior Investigator Award [grant number 095598/Z/11/Z] (to C.R. and J.B.); S.E.K.'s group is supported by Biotechnology and Biological Sciences Research Council (BBSRC) (grant BB/K016598/1) and the Medical Research Council (MR/L016591/1); C.J.S. thanks Cancer Research UK and the Wellcome Trust for funding. K.F.H. acknowledges scholarship support from the Ministry of National Defense-Medical Affairs Bureau, Taiwan.
Data availability
The Pu-Seq data have been uploaded to Gene Expression Omnibus (GEO) under accession number GSE113747. Original microarray data have been deposited in ArrayExpress under accession number E-MTAB-6795.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.226969.supplemental
References
- Aligianni S., Lackner D. H., Klier S., Rustici G., Wilhelm B. T., Marguerat S., Codlin S., Brazma A., de Bruin R. A. and Bähler J. (2009). The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Genet. 5, e1000626 10.1371/journal.pgen.1000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy F. and Carr A. M. (1992). DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11, 1343-1350. 10.1002/j.1460-2075.1992.tb05179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P. and Cranston G. (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218-233. 10.1101/gad.9.2.218 [DOI] [PubMed] [Google Scholar]
- Anda S., Rothe C., Boye E. and Grallert B. (2016). Consequences of abnormal CDK activity in S phase. Cell Cycle 15, 963-973. 10.1080/15384101.2016.1152423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H., Nähse-Kumpf V., Larsen M. S. Y., O'Hanlon K. A., Patzke S., Holmberg C., Mejlvang J., Groth A., Nielsen O., Syljuåsen R. G. et al. (2012). Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32, 4226-4236. 10.1128/MCB.00412-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaikley E. J., Tinline-Purvis H., Kasparek T. R., Marguerat S., Sarkar S., Hulme L., Hussey S., Wee B. Y., Deegan R. S., Walker C. A. et al. (2014). The DNA damage checkpoint pathway promotes extensive resection and nucleotide synthesis to facilitate homologous recombination repair and genome stability in fission yeast. Nucleic Acids Res. 42, 5644-5656. 10.1093/nar/gku190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Furnari B., Mondesert O. and Russell P. (1998). Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280, 909-912. 10.1126/science.280.5365.909 [DOI] [PubMed] [Google Scholar]
- Chan D. A. and Giaccia A. J. (2011). Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 10, 351-364. 10.1038/nrd3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chila R., Basana A., Lupi M., Guffanti F., Gaudio E., Rinaldi A., Cascione L., Restelli V., Tarantelli C., Bertoni F. et al. (2015). Combined inhibition of Chk1 and Wee1 as a new therapeutic strategy for mantle cell lymphoma. Oncotarget 6, 3394-3408. 10.18632/oncotarget.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P. U., Bentley N. J., Martinho R. G., Nielsen O. and Carr A. M. (2000). Mik1 levels accumulate in S phase and may mediate an intrinsic link between S phase and mitosis. Proc. Natl. Acad. Sci. USA 97, 2579-2584. 10.1073/pnas.97.6.2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y., Keszthelyi A., Müller C. A., Miyabe I., Brooks T., Retkute R., Hubank M., Nieduszynski C. A. and Carr A. M. (2015). A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22, 192-198. 10.1038/nsmb.2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P.-M., Coulon S., Scaglione S., Shanahan P., Takedachi A., Wohlschlegel J. A., Yates J. R., Llorente B., Russell P. and Gaillard P.-H. L. (2013). Regulation of Mus81–Eme1 Holliday junction resolvase in response to DNA damage. Nat. Struct. Mol. Biol. 20, 598-603. 10.1038/nsmb.2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dischinger S., Krapp A., Xie L., Paulson J. R. and Simanis V. (2008). Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J. Cell Sci. 121, 843-853. 10.1242/jcs.021584 [DOI] [PubMed] [Google Scholar]
- Dobbelstein M. and Sorensen C. S. (2015). Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 14, 405-423. 10.1038/nrd4553 [DOI] [PubMed] [Google Scholar]
- Dominguez-Kelly R., Martin Y., Koundrioukoff S., Tanenbaum M. E., Smits V. A., Medema R. H., Debatisse M. and Freire R. (2011). Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 194, 567-579. 10.1083/jcb.201101047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C. and Rhind N. (2009). The role of specific checkpoint-induced S-phase transcripts in resistance to replicative stress. PLoS ONE 4, e6944 10.1371/journal.pone.0006944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C., Patel P. K., Rosebrock A., Oliva A., Leatherwood J. and Rhind N. (2008). The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol. Cell. Biol. 28, 5977-5985. 10.1128/MCB.00596-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T., Carr A. M. and Nurse P. (1992). Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6, 2035-2046. 10.1101/gad.6.11.2035 [DOI] [PubMed] [Google Scholar]
- Feijoo C., Hall-Jackson C., Wu R., Jenkins D., Leitch J., Gilbert D. M. and Smythe C. (2001). Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154, 913-924. 10.1083/jcb.200104099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. L. and Zou L. (2011). ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 36, 133-140. 10.1016/j.tibs.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S. A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. et al. (2015). COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805-D811. 10.1093/nar/gku1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. L., Neill E. and Davey S. (2003). Fission yeast Uve1 and Apn2 function in distinct oxidative damage repair pathways in vivo. DNA Repair (Amst) 2, 1253-1267. 10.1016/j.dnarep.2003.08.005 [DOI] [PubMed] [Google Scholar]
- Gaillard H., García-Muse T. and Aguilera A. (2015). Replication stress and cancer. Nat. Rev. Cancer 15, 276-289. 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- Guarino E., Salguero I. and Kearsey S. E. (2014). Cellular regulation of ribonucleotide reductase in eukaryotes. Semin. Cell Dev. Biol. 30, 97-103. 10.1016/j.semcdb.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Håkansson P., Dahl L., Chilkova O., Domkin V. and Thelander L. (2006). The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J. Biol. Chem. 281, 1778-1783. 10.1074/jbc.M511716200 [DOI] [PubMed] [Google Scholar]
- Hall B. M., Ma C.-X., Liang P. and Singh K. K. (2009). Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25, 1564-1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S. L., van Drunen E., Onizawa H., Beverloo H. B., Maas A., Essers J., Hickson I. D. and Kanaar R. (2007). The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096-1104. 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Hillringhaus L., Yue W. W., Rose N. R., Ng S. S., Gileadi C., Loenarz C., Bello S. H., Bray J. E., Schofield C. J. and Oppermann U. (2011). Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 286, 41616-41625. 10.1074/jbc.M111.283689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Iwasawa Y., Okada M., Arai T., Nishibata T., Kobayashi M., Kimura T., Kaneko N., Ohtani J., Yamanaka K. et al. (2009). Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 8, 2992-3000. 10.1158/1535-7163.MCT-09-0463 [DOI] [PubMed] [Google Scholar]
- Holmberg C., Fleck O., Hansen H. A., Liu C., Slaaby R., Carr A. M. and Nielsen O. (2005). Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19, 853-862. 10.1101/gad.329905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H. C. and Clurman B. E. (2005). Cyclin E in normal and neoplastic cell cycles. Oncogene 24, 2776-2786. 10.1038/sj.onc.1208613 [DOI] [PubMed] [Google Scholar]
- Ivanova T., Gomez-Escoda B., Hidalgo E. and Ayte J. (2011). G1/S transcription and the DNA synthesis checkpoint: common regulatory mechanisms. Cell Cycle 10, 912-915. 10.4161/cc.10.6.14963 [DOI] [PubMed] [Google Scholar]
- Jha D. K. and Strahl B. D. (2014). An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 5, 3965 10.1038/ncomms4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B., Fraser J. L. A., Freyer G. A., Davey S. and Doetsch P. W. (1999). A Uve1p-mediated mismatch repair pathway in Schizosaccharomyces pombe. Mol. Cell. Biol. 19, 4703-4710. 10.1128/MCB.19.7.4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J. and Zhang Y. (2006). The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312-316. 10.1038/nature04853 [DOI] [PubMed] [Google Scholar]
- Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A. and Chabes A. (2011). Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39, 1360-1371. 10.1093/nar/gkq829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., Golub T. R., Meyerson M., Gabriel S. B., Lander E. S. and Getz G. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495-501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Enoch T. and Piwnica-Worms H. (1994). mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J. Biol. Chem. 269, 30530-30537. [PubMed] [Google Scholar]
- Li J., Duns G., Westers H., Sijmons R., van den Berg A. and Kok K. (2016). SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget 7, 50719 10.18632/oncotarget.9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H. D., Griffiths D. J. F., Edwards R. J., Christensen P. U., Murray J. M., Osman F., Walworth N. and Carr A. M. (1998). S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12, 382-395. 10.1101/gad.12.3.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Powell K. A., Mundt K., Wu L., Carr A. M. and Caspari T. (2003). Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17, 1130-1140. 10.1101/gad.1090803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Poitelea M., Watson A., Yoshida S. H., Shimoda C., Holmberg C., Nielsen O. and Carr A. M. (2005). Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 24, 3940-3951. 10.1038/sj.emboj.7600854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K., Walworth N., Booher R., Dembski M., Kirschner M. and Beach D. (1991). mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64, 1111-1122. 10.1016/0092-8674(91)90266-2 [DOI] [PubMed] [Google Scholar]
- Luria S. E. and Delbruck M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazouzi A., Velimezi G. and Loizou J. I. (2014). DNA replication stress: causes, resolution and disease. Exp. Cell Res. 329, 85-93. 10.1016/j.yexcr.2014.09.030 [DOI] [PubMed] [Google Scholar]
- Morris S. A., Shibata Y., Noma K., Tsukamoto Y., Warren E., Temple B., Grewal S. I. and Strahl B. D. (2005). Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot. Cell 4, 1446-1454. 10.1128/EC.4.8.1446-1454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Tinline-Purvis H., Walker C. A., Folkes L. K., Stratford M. R., Hayles J., Hoe K.-L., Kim D. U., Park H. O., Kearsey S. E. et al. (2010). Break-induced ATR and Ddb1-Cul4(Cdt)(2) ubiquitin ligase-dependent nucleotide synthesis promotes homologous recombination repair in fission yeast. Genes Dev. 24, 2705-2716. 10.1101/gad.1970810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. S., Anderson M., White S. and McInerny C. J. (2001). mik1(+) G1-S transcription regulates mitotic entry in fission yeast. FEBS Lett. 503, 131-134. 10.1016/S0014-5793(01)02720-X [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C. L. and Whitby M. C. (2003). Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12, 761-774. 10.1016/S1097-2765(03)00343-5 [DOI] [PubMed] [Google Scholar]
- Pai C. C., Deegan R. S., Subramanian L., Gal C., Sarkar S., Blaikley E. J., Walker C., Hulme L., Bernhard E., Codlin S. et al. (2014). A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 4091 10.1038/ncomms5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C. C., Kishkevich A., Deegan R. S., Keszthelyi A., Folkes L., Kearsey S. E., De Leon N., Soriano I., de Bruin R. A. M., Carr A. M. et al. (2017). Set2 methyltransferase facilitates DNA replication and promotes genotoxic stress responses through MBF-dependent transcription. Cell Rep 20, 2693-2705. 10.1016/j.celrep.2017.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. K., Kommajosyula N., Rosebrock A., Bensimon A., Leatherwood J., Bechhoefer J. and Rhind N. (2008). The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol. Biol. Cell 19, 5550-5558. 10.1091/mbc.e08-06-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S. X., Markkanen E., Jiang Y., Sarkar S., Woodcock M., Orlando G., Mavrommati I., Pai C. C., Zalmas L. P., Drobnitzky N. et al. (2015). Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28, 557-568. 10.1016/j.ccell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C., Codlin S. and Bahler J. (2013). TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 12, 563-573. 10.1111/acel.12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H. and Arcangioli B. (2008). Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 27, 1378-1387. 10.1038/emboj.2008.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. and Nurse P. (1987). Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49, 559-567. 10.1016/0092-8674(87)90458-2 [DOI] [PubMed] [Google Scholar]
- Salguero I., Guarino E., Shepherd M. E. A., Deegan T. D., Havens C. G., MacNeill S. A., Walter J. C. and Kearsey S. E. (2012). Ribonucleotide reductase activity is coupled to DNA synthesis via proliferating cell nuclear antigen. Curr. Biol. 22, 720-726. 10.1016/j.cub.2012.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. and Janknecht R. (2007). Diversity within the JMJD2 histone demethylase family. Biochem. Biophys. Res. Commun. 353, 973-977. 10.1016/j.bbrc.2006.12.147 [DOI] [PubMed] [Google Scholar]
- Sørensen C. S. and Syljuåsen R. G. (2012). Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40, 477-486. 10.1093/nar/gkr697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R., Shen J. P., Yang C. C., Sun S. M., Li J., Gross A. M., Jensen J., Licon K., Bojorquez-Gomez A., Klepper K. et al. (2016). A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell 63, 514-525. 10.1016/j.molcel.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P. and Zhang Y. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811-816. 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M. et al. (2006). Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467-481. 10.1016/j.cell.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Willis N. and Rhind N. (2009). Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-Phase DNA damage checkpoint. Mol. Biol. Cell 20, 819-833. 10.1091/mbc.e08-08-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C. H., Fung T. K. and Poon R. Y. (2002). Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 59, 1317-1326. 10.1007/s00018-002-8510-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M. K. and Cimprich K. A. (2014). Causes and consequences of replication stress. Nat. Cell Biol. 16, 2-9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.