ABSTRACT
We describe a method, termed cryoAPEX, which couples chemical fixation and high-pressure freezing of cells with peroxidase tagging (APEX) to allow precise localization of membrane proteins in the context of a well-preserved subcellular membrane architecture. Further, cryoAPEX is compatible with electron tomography. As an example, we apply cryoAPEX to obtain a high-resolution three-dimensional contextual map of the human FIC (filamentation induced by cAMP) protein, HYPE (also known as FICD). HYPE is a single-pass membrane protein that localizes to the endoplasmic reticulum (ER) lumen and regulates the unfolded protein response. Alternate cellular locations for HYPE have been suggested. CryoAPEX analysis shows that, under normal and/or resting conditions, HYPE localizes robustly within the subdomains of the ER and is not detected in the secretory pathway or other organelles. CryoAPEX is broadly applicable for assessing both lumenal and cytosol-facing membrane proteins.
KEY WORDS: CryoAPEX, Electron tomography, Membrane protein, FIC AMPylation, Cryofixation
Summary: CryoAPEX couples localization of peroxidase-tagged membrane proteins at high resolution with 3D structural analysis, within an optimally preserved cellular context.
INTRODUCTION
Localization of membrane proteins via electron microscopy (EM) at high resolution is dependent on robust detection technology and on sample preparation methods that confer superior ultrastructural preservation of membranes. Unfortunately, current methods of localization of membrane-bound proteins at EM resolutions are less than optimal. Immunoelectron microscopy (IEM) to detect either an endogenous or epitope-tagged overexpressed protein using antigen-specific antibodies requires a permeabilization step that also causes degradation of cellular membranes and distortion of membrane-bound compartments (De Mey et al., 1981; Schnell et al., 2012). An alternative is to fuse enzymatic tags directly to the protein of interest in a transfection experiment, so as to avoid the necessity for introducing an antibody. A number of these enzymatic tags have been described, e.g. metallothioneine (METTEM), resorufin arsenical hairpin (ReAsH), miniSOG, horseradish peroxidase (HRP) and more recently, engineered ascorbate peroxidases (APEX and APEX2) (Hoffmann et al., 2010; Lam et al., 2015; Martell et al., 2012; Mercogliano and DeRoiser, 2007; Porstmann et al., 1985; Shu et al., 2011). While each of these technologies faces its own set of limitations when employed in conjunction with EM, the APEX2 tag, which has been used with success on mitochondrial and ER proteins, is arguably the most promising option (Martell et al., 2017). APEX2 is a monomeric 28 kDa soyabean ascorbate peroxidase that withstands strong EM fixation (Lam et al., 2015). Additionally, it is sensitive, generally straightforward in its application and, unlike horseradish peroxidase, is active in both the cytosolic and lumenal compartments (Hopkins et al., 2000; Lam et al., 2015; Martell et al., 2017). Nevertheless, the morphological damage to cellular membranes and membrane-bound organelles that occurs during conventional aldehyde fixation and alcohol dehydration protocols, even without the permeabilization required for antibodies, continues to be an impediment to obtaining optimal preservation of the subcellular architecture.
In contrast, cryofixation or high-pressure freezing (HPF) is a method for obtaining vitreous preparations of live cells and tissues up to 200 μm in thickness with minimal ice crystal formation, thus immobilizing macromolecular assemblies in their near-native state (Chan et al., 1993; Mcdonald, 1999; Zechmann et al., 2007; Studer et al., 2008). This method has become a mainstay for preparing samples for electron tomography, which employs thicker sections (McDonald and Auer, 2006). Further, HPF has been adapted in combination with freeze substitution (FS) methods, which entail organic substitution of water with acetone at low temperature, to generate plastic-embedded samples for conventional EM. However, HPF-FS has not been extensively used with protein localization methods that require chemical fixation of cells (Tsang et al., 2018).
Here, we opted to develop an EM tomography-compatible detection method to visualize the human FIC (filamentation induced by cAMP) protein, HYPE (also known as FICD). FIC proteins are a recently characterized class of enzymes that predominantly utilize ATP to attach AMP (adenosine monophosphate) to their protein targets (Casey and Orth, 2018; Truttmann et al., 2017; Worby et al., 2009). This post-translational modification is called adenylylation or AMPylation. The first FIC proteins, VopS and IbpA, were described in the pathogenic bacteria Vibrio parahemolyticus and Histophilus somni, respectively, where they serve as secreted bacterial effectors that induce toxicity in host cells by inactivating small GTPases through AMPylation (Mattoo et al., 2011; Worby et al., 2009; Xiao et al., 2010; Yarbrough et al., 2009; Zekarias et al., 2010). FIC proteins have also been implicated in bacterial cell division and persister cell formation, protein translation, cellular trafficking and neurodegeneration (Garcia-Pino et al., 2014; Harms et al., 2015; Mukherjee et al., 2011; Truttmann et al., 2018).
HYPE (huntingtin yeast interacting protein E) or FICD is the sole FIC protein encoded by the human genome (Faber et al., 1998; Sanyal et al., 2015). In humans, HYPE is expressed ubiquitously, albeit at very low levels. It is a single-pass type II membrane protein that localizes to the lumenal surface of the endoplasmic reticulum (ER) (Sanyal et al., 2015; Worby et al., 2009). HYPE plays a critical role in regulating ER homeostasis by reversibly AMPylating the Hsp70 chaperone, BiP (also known as HSPA5) (Ham et al., 2014; Sanyal et al., 2015; Preissler et al., 2015, 2017a,b). Biochemical and proteomic screens have identified additional AMPylation targets of HYPE and its orthologs, which include cytosolic chaperones, cytoskeletal proteins, transcriptional and translational regulators, and histones (Broncel et al., 2016; Truttmann et al., 2016, 2017). These data suggest that a fraction of HYPE could reside outside the ER, for example, in the nucleus or cytoplasm. Indeed, a small fraction of the HYPE homolog in Caenorhabditis elegans, FIC-1, has been shown to localize to the cytosol and AMPylate cytosolic c and Hsp40 proteins (Truttmann et al., 2017).
The low levels of HYPE expression in human cells combined with the resolution limitations of conventional immunofluorescence microscopy make obtaining definitive localization data difficult. We therefore opted to circumvent these limitations by using an electron microscopy approach. To visualize HYPE in cells, our challenge was to develop a technique that would preserve membrane ultrastructure and be compatible with transmission EM tomography methods to identify specific distribution of HYPE in three-dimensional (3D) space. We chose to visualize HYPE by genetically tagging it with APEX2. The APEX2 tag catalyzes a peroxide-based reaction that converts diaminobenzadine (DAB) into a low-diffusing precipitate that deposits at the site of the target protein (Lam et al., 2015). In our analysis of HYPE, we also included a peptide, designated endoplasmic reticulum membrane (ERM), to serve as a dual control for ER localization as well as for ER morphology. ERM consists of the N-terminal 1–27 amino acids of cytochrome P450 2C1 (CYP2C1) (Lam et al., 2015; Sandig et al., 1999). An ERM–APEX2 fusion localizes to the ER membrane such that the APEX2 tag faces the cytosol. Additionally, ERM is known to induce a reorganization of the smooth ER into distinctive ordered membrane structures called organized smooth ER (OSER) (Snapp et al., 2003; Lam et al., 2015; Sandig et al., 1999). Thus, ERM–APEX2 serves as an excellent metric for assessing both ER membrane-specific staining and ultrastructural membrane preservation.
Next, since degradation of the cellular ultrastructure in traditional aldehyde fixation and alcohol dehydration methods appears to be largely associated with the alcohol dehydration post-processing steps and not with aldehyde fixation per se, we adopted and optimized a combination method proposed by Sosinsky et al. (2008), which relies on chemical fixation prior to cryofixation and optimally preserves membrane structure. We then applied this combination approach to the detection of APEX2-tagged proteins. Using this methodology, we observed minimal lipid extraction or distortion of membrane structures, and were able to clearly detect ER membrane-bound HYPE. During submission of this manuscript, a similar conceptual approach of combining HPF and APEX tagging, albeit procedurally different and at a lower resolution, was reported for correlative light-electron microscopy (CLEM) on whole tissues (Tsang et al., 2018). We show that the addition of tannic acid, uranyl acetate and counter-staining with lead further increased the overall contrast, and made the membrane ultrastructure in the vicinity of the localized HYPE–APEX2 density easily recognizable. Such optimizations proved key to making this technology amenable to transmission EM tomography of ER membranes.
Our hybrid method – hereafter referred to as cryoAPEX – performed remarkably well for protein localization at the subcellular level. Comparison of ERM–APEX2 in cryoAPEX-treated cells versus live cryofixed (HPF alone) cells showed well-preserved OSER morphology with ER-specific staining only on the cytosolic face of the ER membrane. Similarly, we observed that HYPE localizes as periodic foci along the lumenal face of the ER membrane and, although overexpressed in the context of a transient transfection study, is retained only within the ER and the contiguous nuclear envelope. HYPE remained ER-associated even when the ER came in close proximity to the plasma membrane, mitochondria or the Golgi apparatus. Thus, it would appear unlikely that HYPE traffics via conventional means through the secretory system or is associated with other organelles under normal circumstances. Further, data collected on cryoAPEX-treated cells could be used to reconstruct a 3D representation of HYPE within the ER lumen. This ability to simultaneously assess the detection of a membrane protein in multiple cellular compartments throughout the subcellular volume of a single cell at low-nanoscale resolution is a significant advance. More broadly, we present a straightforward methodology for probing the subcellular distribution of membrane-bound proteins, with either lumen-facing or cytosol-facing topologies, that is amenable to high-resolution 3D tomographic reconstruction.
RESULTS
A combination of glutaraldehyde fixation, cryofixation and extended osmication during freeze substitution shows optimal preservation of membrane ultrastructure in HEK-293T cells
In cellular imaging, the ability to obtain 3D spatial localization of proteins in the context of well-preserved cellular structures at high resolution is highly desirable (O'Toole et al., 2018). For antibody–conjugate-based detection methods to be effective, ultrastructure-damaging permeabilization and/or technically challenging ultracryosectioning are required. At EM resolutions, the deleterious effects of such treatments, particularly upon membrane-bound compartments, become obvious. Fusion of a protein of interest to the monomeric enhanced peroxidase APEX2 avoids the need for an antibody. However, the published protocols still use chemical fixation and alcohol dehydration prior to embedding (Martell et al., 2017). We hypothesized that the alcohol dehydration step in published APEX2 protocols is limiting for ultrastructural preservation, and consequently for signal intensity and resolution. Therefore, referencing work by Sosinsky et al. (2008), we utilized cryofixation-freeze substitution (HPF-FS) of live cells as a gold standard for preservation, and combined it with chemical fixation methods to optimize ultrastructural preservation in untransfected human HEK-293T cells.
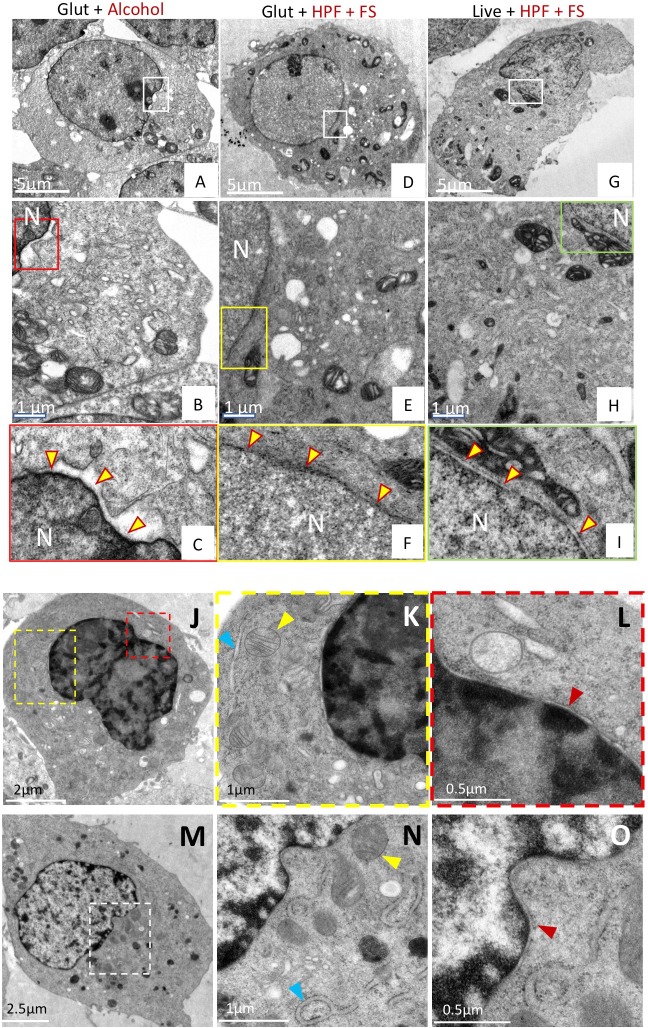
Preservation was assessed based on several criteria including membrane integrity, smoothness of intracellular membranes, densely packed cytoplasm, and maintenance of organellar structures such as mitochondria with clearly visible cristae. Smoothness of intracellular membranes is often a primary indication of good preservation, and preservation of the nuclear envelope has been used classically as a hallmark (Sosinsky et al., 2008; Tsang et al., 2018). As shown in Fig. 1A–C, traditionally used glutaraldehyde fixation and alcohol dehydration methods showed poor preservation of the nuclear membrane with intermittent ruffling and separation of the double membrane. The cytoplasm of these cells also appeared less granulated and less densely packed (Fig. S1A,B,G). By contrast, the HPF-FS-processed samples showed excellent preservation of the nuclear envelope, which was smooth and devoid of distortion (Fig. 1G–I). Additionally, the cytoplasm was densely packed and the mitochondrial membrane remained intact, with clearly visible cristae (Fig. S1E,F,I). Next, we fixed our cell samples with glutaraldehyde prior to HPF-FS (Fig. 1D–F; Fig. S1C,D,H). Morphological damage beyond that seen with HPF-FS alone was minimal, and this combination approach conferred substantially better ultrastructural preservation relative to traditional fixation techniques. This excellent membrane preservation via the combination (Glut+HPF+FS) method was further substantiated when we extended it to another cell line, BHK (baby hamster kidney) cells that are routinely used in electron microscopy studies (Hawes et al., 2007). Indeed, a comparison of ultrastructural preservation using HPF-FS (Fig. 1J–L) versus our combination method (Fig. 1M–O) showed similar results, as determined by a well-preserved nuclear membrane (red arrowheads), and comparable preservation of mitochondrial (yellow arrowheads) structure and ER (blue arrowheads) membranes.
Fig. 1.
A combination of chemical fixation and cryofixation exhibits superior ultrastructure preservation and membrane staining over traditional methods in HEK-293T cells. (A–I) HEK-293T cells. Cells prepared by conventional glutaraldehyde fixation and dehydration methods (A–C), glutaraldehyde followed by cryofixation (D–F) or cryofixation alone (G–I) were examined by thin-section EM. Panels B,E,H represent magnified views from boxed regions of panels A,D,G, respectively. Panels C,F,I represent magnified views of the boxed regions from panels B,E,H, respectively, and highlight the preservation of the nuclear membrane in each case (yellow arrowheads). The nuclear membrane appears smooth and uncompromised in samples prepared by glutaraldehyde treatment and cryofixation (Glut+HPF+FS) or cryofixation alone (Live+HPF+FS), but is ruffled and irregular in samples prepared by conventional means (Glut+Alcohol). (J–O) BHK cells. Ultrastructural preservation in cells prepared by the combined (Glut+HPF+FS) method (M–O) was comparable to that in cryofixed (Live+HPF+FS) cells (J–L), as illustrated by the presence of intact mitochondria (yellow arrowheads, panels K and N), intact ER (blue arrowheads, panels K and N), and absence of ruffled nuclear membranes (red arrowheads, panels L and O). Panels K and L are magnified views of the respective yellow and red boxes in panel J. Panel O is a magnified view of panel N, which is a magnified view of the region delineated by the white box in panel M. Glut, glutaraldehyde; HPF, high-pressure freezing; FS, freeze substitution; N, nucleus.
ERM–APEX2 induced OSER structures show that membrane preservation is improved with the cryoAPEX method
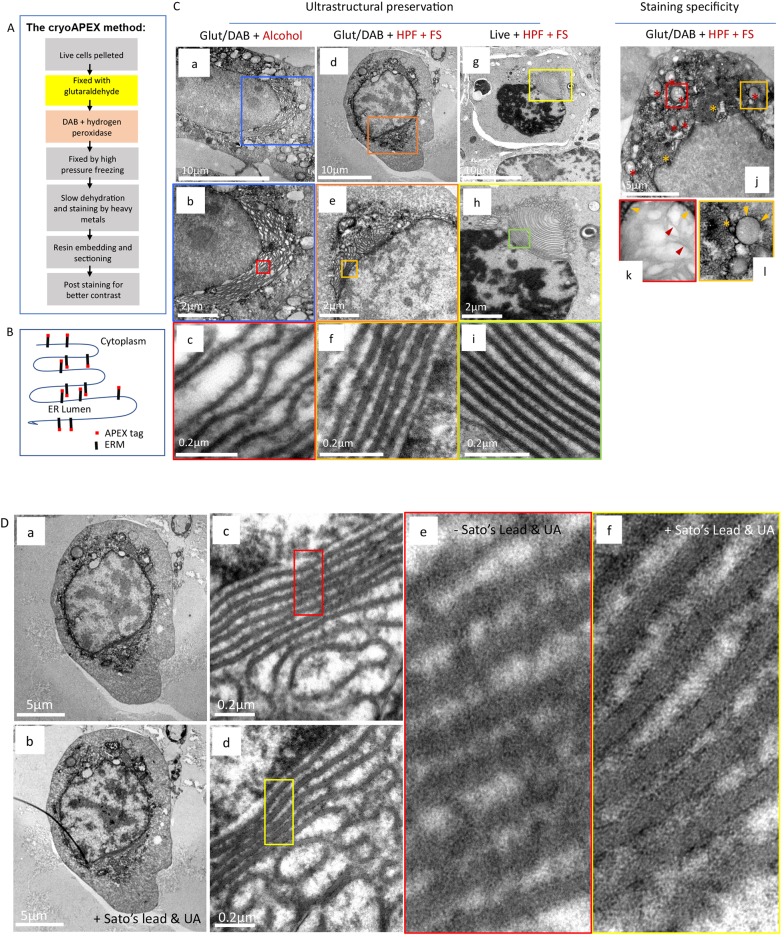
Next, we sought to test the applicability of the above combination method for ultrastructural preservation with the use of the APEX2 tag to follow a protein of interest, creating the hybrid glutaraldehyde+HPF-FS+APEX2 method we have called cryoAPEX (Fig. 2A).
Fig. 2.
OSER as a system for evaluating membrane preservation and staining specificity. (A) Flowchart describing cryoAPEX. (B) Schematic of APEX-tagged ERM expressed on the cytosolic face of the ER membrane. (C) The reorganized ER morphology in chemically fixed, DAB reacted ERM–APEX2-expressing cells that were processed via traditional chemical fixation and alcohol dehydration (a–c) or by cryoAPEX (d–f) was compared to ERM–APEX2 expressing cells that were cryofixed live and without the DAB reaction (g–i). The live cryofixed cells (g–i) represent the best attainable ultrastructural preservation and serve here as the metric for evaluating membrane preservation obtained via the two APEX-based detection protocols (a–f). The high specificity of staining obtained by cryoAPEX is exemplified in the images in panels j–l. Here, thin-section images of cells expressing ERM–APEX2 processed by cryoAPEX show preferential staining of the reorganized ER (j, orange asterisks) and the outer mitochondrial membrane (red asterisks in j and orange arrowheads in k–l, respectively) but not of the mitochondrial cristae (k, red arrowheads). (D) Post-staining with heavy metals improves definition of preferentially stained membranes. Post-staining of thin sections with heavy metals using uranyl acetate and Sato's lead solution following cryoAPEX provides additional contrast, thereby improving resolution. Shown are thin sections of the same cell imaged in Fig. 2Cd before (a,c,e) and after (b,d,f) post-staining. Comparison of panels e and f clearly shows improved definition and the resolution of membranes at high magnifications in post-stained samples (f). UA, uranyl acetate.
To serve as a proof of concept, we evaluated cryoAPEX by assessing the membrane localization of the ERM–APEX2 chimeric protein (Fig. 2B). Expression of ERM results in ER localization of the peptide as a membrane protein, such that the peroxidase tag faces the cytoplasm (Lam et al., 2015) (Fig. 2B). This topology in the ER membrane serves as an additional control for HYPE, which displays the opposite orientation in the membrane. ERM expression is known to cause reorganization of the smooth ER and increased membrane biogenesis (Lam et al., 2015; Sandig et al., 1999). Cells expressing ERM form OSERs, which are distinctive ordered labile membrane structures that can be easily visualized via electron microscopy without the need for specialized detection methods (Snapp et al., 2003). Thus, it is possible to conveniently assess the degree of membrane preservation in conjunction with various sample preparation methods, providing an excellent metric for assessing both staining specificity and ultrastructural preservation in one system.
As a morphological control, thin sections of ERM–APEX2-transfected cells were processed using HPF-FS alone and then stained using osmium tetroxide, tannic acid and uranyl acetate but without chemical fixation or the DAB reaction. Under these conditions, the typical membrane whorl pattern of reorganized ER adjacent to the nucleus was clearly visible (Fig. 2Cg–i, yellow and green boxes). OSER membranes within these were arranged in evenly spaced parallel stacks without disruption or ruffling (Fig. 2Ci). By contrast, traditional methods showed a clear disruption of the lamellar stacking of these OSER structures (Fig. 2Ca–c; Fig. S2A–D), with artefactual ruffling of the membrane stacks and presumed loss of cytoplasmic material between the stacks (compare Fig. 2Cc and Fig. S2D with Fig. 2Ci). However, the cryoAPEX method (Fig. 2Cd–f; Fig. S2E–H) resulted in well-preserved ER-derived lamellar structures comparable to the lamellar stacking in ER-derived structures observed in live HPF-FS controls (compare Fig. 2Cf and Fig. S2H with Fig. 2Ci). Peroxidase-reacted samples were also processed in parallel without uranyl acetate, as uranyl acetate stains nucleic acids and therefore the nucleus, and, to some extent, the mitochondria (Fig. 2Cj–l, red asterisks). Tannic acid was also eliminated from these samples to minimize background staining (Fig. 2Cj–l) (Huxley and Zubay, 1961; Kalina and Pease, 1977; Persi and Burnham, 1981; Schrijvers et al., 1989). Staining was not observed within the nucleus (Fig. 2Cj) or in mitochondrial cristae (Fig. 2Ck, red arrowheads within red box) but was seen in the ER-derived structures (Fig. 2Ck,Cl, yellow asterisks and yellow arrowheads within yellow box). As additional controls for the specificity of the ERM–APEX2 staining for the ER, we assessed localization of three organellar markers – namely, mito-V5–APEX2 that targets to the mitochondrial matrix (Addgene plasmid #72480; Lam et al., 2015); CAAX–APEX2 that targets to the plasma membrane (Lam et al., 2015), and ManII–APEX2 that targets to the Golgi lumen (see Materials and Methods). Fig. S3A–C show APEX2-associated staining only at the mitochondria, Golgi, and plasma membrane, respectively, confirming that APEX2-associated signals obtained in our assays are specific to the tagged protein.
To enhance the contrast of membrane staining over the background of overall osmicated DAB density (i.e. osmium-stained DAB precipitate), the same sections shown in Fig. 2Cd–f were imaged before and after post-staining with Sato's lead solution and uranyl acetate (Fig. 2D). This resulted in an improved ability to distinguish specific staining of adjacent membranes at higher magnifications (Fig. 2D, compare panels a,c and e with b,d, and f, respectively).
HYPE localizes to the ER membrane as distinct lumen-facing foci
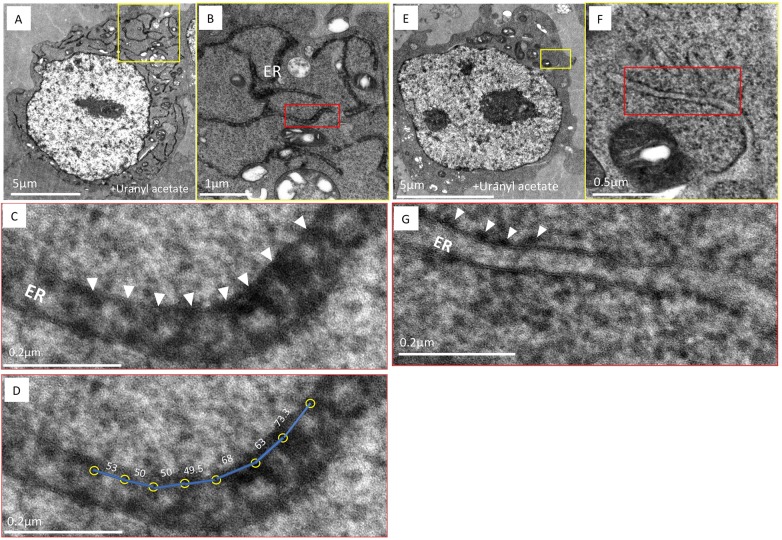
Having established the methodology to localize an APEX2-tagged ER membrane protein in an optimally preserved cell, we next applied cryoAPEX to the sole human FIC protein, HYPE. Previous immunofluorescence, cell fractionation and protease protection assays have placed HYPE as predominantly facing the ER lumen (Sanyal et al., 2015). A smaller fraction of HYPE has also been detected in the cytosol, as well as in the perinuclear space (Truttmann et al., 2015, 2017). We therefore transiently transfected HEK-293T cells with HYPE–APEX2 constructs and processed them using cryoAPEX as standardized for ERM–APEX2 above. As before, ultrastructure was well preserved and there were minimal signs of lipid extraction or membrane ruffling (Fig. 3). Osmicated DAB density was observed within the lumen of peripheral ER tubules (Fig. 3A,B, yellow and red boxes, respectively) but not in identically treated untransfected controls (Fig. 3E,F, yellow and red boxes). Remarkably, at higher magnification this density resolved into distinct foci along the length of the tubules, on the lumenal face of the ER membrane (Fig. 3C, highlighted by white arrowheads). These HYPE-specific foci averaged a distance of 61.45 nm apart (Fig. 3D). Further, this density was not an artifact of the rough ER, where such density corresponding to the presence of ribosomes is seen only on the cytosolic face of the ER membrane (Fig. 3G). Interestingly, we rarely observed ribosomal density on the outer face of HYPE–APEX2-stained ER membranes. To emphasize the merits of cryoAPEX over traditional methods, we also assessed HYPE–APEX2-transfected cells using the traditional alcohol dehydration method (Fig. S4). Analysis of thin sections using this method revealed a similar staining pattern within the tubules of the peripheral ER and nuclear envelope (Fig. S4B, white arrowheads within yellow box). However, as expected, there was poor preservation of the ER membrane and intermittent regions of membrane discontinuity (Fig. S4B, red arrowheads). Additionally, the presence of other membrane-bound structures was mostly indiscernible (compare Fig. S4A with Fig. 3A), thus making it impossible to get contextual information about the other organelles in the immediate vicinity or in contact with the ER membrane containing HYPE. Clearly, cryoAPEX offers high-resolution localization for HYPE in the ER lumen in the context of other organelles at the subcellular level.
Fig. 3.
HYPE localizes to the lumenal face of the ER membrane as periodic foci. (A) An image of a thin section of HEK-293T cells expressing HYPE–APEX2 and processed by cryoAPEX reveal staining of the ER tubules in a well-preserved (dense) cytoplasmic background. (B,C) Higher magnification images of a small section of the peripheral ER (demarcated by yellow box in A and shown in B, with further magnification of red box in B shown in C) exhibits periodic foci of APEX2-generated density (B, red box and C, white arrowheads showing periodicity between the HYPE foci). (D) Center-to-center density measurements showing the distance (in nm, blue lines) between the HYPE-specific foci (yellow circles) in C. (E) Untransfected control HEK-293T cells processed in an identical manner show the lack of APEX2-generated density within the ER lumen. (F,G) Higher magnification images of a small section of E (demarcated by yellow box and shown in F, with further magnification of red box in F shown in G) clearly shows the lack of density on the lumenal face. Additionally, density corresponding to ribosomes on the cytoplasmic face of the ER membrane is evident (G, white arrowheads). Thus, HYPE is detected only along the lumenal face of the ER membrane and never on the cytosolic face.
Assessing localization of HYPE in cellular compartments other than the ER
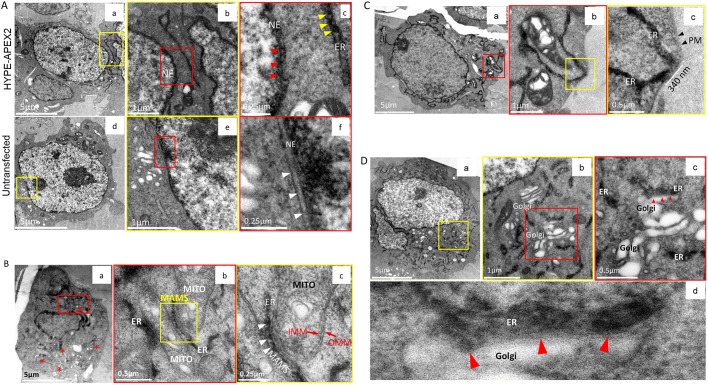
In addition to the ER, HYPE has been suggested to interact with protein targets in the cytosol and nucleus, and possibly in the secretory pathway (Broncel et al., 2016; Rahman et al., 2012; Truttmann et al., 2016, 2017). We therefore screened for the presence of HYPE in subcellular compartments other than the ER. HYPE was detected within the lumenal walls of the nuclear envelope (NE), which is expected since the nuclear membrane is contiguous with the ER (Fig. 4Aa–c, yellow and red boxes). This is in agreement with immunofluorescence data, which shows HYPE-specific staining in the perinuclear membranes (Sanyal et al., 2015; Truttmann et al., 2015). The characteristic periodic foci of osmicated DAB density were seen as well (Fig. 4Ac, red arrowheads). As a control, untransfected cells were also processed using cryoAPEX; no lumenal staining was evident, indicating the specificity of HYPE's NE localization (Fig. 4Ad–f, red box and white arrowheads in f).
Fig. 4.
Assessing localization of HYPE in subcellular compartments other than the ER. (A) Localization of HYPE to the nuclear envelope (NE). Images of thin sections from cells transfected with HYPE–APEX2 and processed by cryoAPEX show HYPE-specific density within the perinuclear space of the nuclear envelope (a–c). At higher magnification, this staining shows a pattern similar to that seen along the lumenal face of the ER membrane (c; compare red and yellow arrowheads within the NE and the ER, respectively). The same untransfected cells as used in Fig. 3E processed in this manner exhibit no membrane-associated staining within the perinuclear space of the nuclear envelope (d–f and white arrowheads in f). (B) HYPE does not localize to the mitochondria. Cells transfected with HYPE–APEX2 were processed by cryoAPEX in the presence of osmium tetroxide but without addition of uranyl acetate or tannic acid. Thin sections of these cells showed a distinct lack of mitochondrial membrane staining, making it difficult to visualize mitochondria at low magnifications (red asterisks in a). Magnified images of well-preserved ER–mitochondrial junctions (MAMS; demarcated by red box in a with further magnification of red box area shown in b) clearly show ER tubules in close contact with two adjacent mitochondria (b). A further magnified image of the MAMS shows the HYPE–APEX2 staining of the ER but no apparent staining within the inner or outer mitochondrial membranes (b, yellow box; and c, magnified image of the area within this yellow box). The periodic distribution of HYPE is retained even at the MAMS (c, white arrowheads). MITO, mitochondrion; IMM, inner mitochondrial membrane; OMM, outer mitochondrial membrane; MAMS, mitochondria-associated membranes. (C) HYPE does not localize to the plasma membrane (PM). Images of a cell expressing HYPE–APEX2 reveal absence of plasma membrane staining (a). Images of ER–PM junctions at high magnifications show well-preserved junctions where the ER makes extended contacts with the plasma membrane (red box in a; yellow box in b; white bar in c). b shows higher magnification of red box area in a, c shows higher magnification of yellow box area in b. Staining was contained within the cortical ER tubules with no apparent staining of the plasma membrane (c, black arrowheads). (D) HYPE does not enter the secretory pathway. To assess whether HYPE enters the secretory pathway, the Golgi apparatus was imaged (a–d). Images from thin sections of HYPE–APEX2-transfected cells show an area where ER tubules are interspersed within Golgi stacks (a, region within yellow box). Higher magnification of this region (b shows higher magnification of yellow box area in a, c shows higher magnification of red box area in b) shows the typical stacked morphology of the Golgi apparatus devoid of any osmicated DAB density within its lumen. Further magnification of a well-preserved ER–Golgi junction (d) shows a region of extensive contact between the two organelles where the DAB density was restricted to the ER lumen and shows no apparent staining of the Golgi apparatus (b, area within red box; red arrowheads in c and d indicate the ER–Golgi junction).
We next investigated the presence of HYPE in the mitochondria. Samples were prepared without uranyl acetate to avoid staining mitochondrial nucleic acids. Osmicated DAB density was observed at multiple junctions between the ER and mitochondria in membranes called mitochondria-associated membranes (MAMS), which are contiguous with the ER, and displayed the typical periodic HYPE foci (Fig. 4Ba–c, white arrowheads in c). Upon close examination, it was clear that although the two organelles were in close proximity, the inner and outer mitochondrial membranes were devoid of staining (Fig. 4Bc). Thus, HYPE staining remained specific for the contiguous ER and did not show any evidence for the existence of HYPE within the mitochondrion.
Similarly, we also searched for evidence of HYPE staining at the plasma membrane (PM), as the cortical ER is known to make multiple PM contacts. Even at these contact points, osmicated density was restricted to the ER lumen and was clearly absent from the adjoining PM (Fig. 4Ca–c).
Finally, we assessed the Golgi apparatus for evidence as to whether or not HYPE enters the secretory pathway. Uranyl acetate and tannic acid, in addition to osmium tetroxide, were used to enhance the contrast for the DAB density at Golgi–ER junctions (Fig. 4D, red arrowheads). The typical stacked morphology of the Golgi was clearly visible, interspersed with ER tubules (Fig. 4Da–c, yellow and red boxes). Even at the Golgi–ER junction, the HYPE-specific periodic DAB density was clearly restricted to the ER lumen and was completely absent from the Golgi (Fig. 4Dd).
Taken together, at the expression levels assessed here, our results indicate that HYPE is localized to the ER lumen and contiguous nuclear envelope, and does not appear to localize or transport to adjacent organelles or the plasma membrane even upon close contact with the ER.
CryoAPEX allows tracking of the subcellular distribution of HYPE and ERM over a large cellular volume
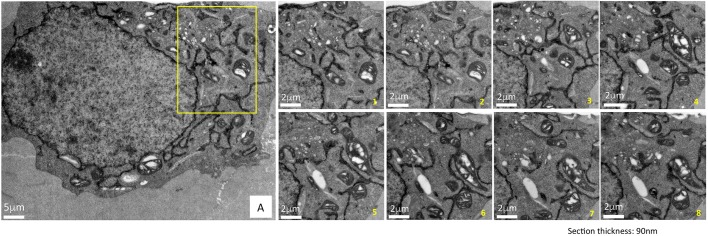
The greatest advantage of using cryofixation for sample preparation for EM is its capability to preserve membrane ultrastructure consistently throughout the cell volume. To track the distribution of HYPE over a large subcellular volume, we employed serial section EM (Fig. 5). Multiple ribbons containing between 10 and 20 serial sections, each 90 nm thick, were collected and screened. Representative images of a subset of eight serial sections selected from a larger set of images of a demarcated region from a single cell are depicted (Fig. 5A, yellow box and panels numbered 1–8). The images capture a region of a peripheral ER network showing osmicated DAB density within the ER and in close proximity to the mitochondria and Golgi apparatus. The excellent preservation of the ultrastructure of these neighboring organelles and the additional uranyl acetate staining provide contextual information within which HYPE resides. A similar analysis of ERM–APEX2 is shown in Fig. S5. Thus, such preservation and staining using cryoAPEX allows us to subsequently use EM tomography to determine the localization of membrane proteins like HYPE and ERM in 3D.
Fig. 5.
Superior ultrastructural preservation enables the tracking of HYPE's subcellular localization via serial sectioning. To demonstrate the consistency of the membrane ultrastructure preservation obtained by cryoAPEX, cells expressing HYPE–APEX2 were serially sectioned and a specific area (panel A, yellow box) was imaged. Multiple ribbons containing between 10 and 20 serial sections of 90 nm thickness were collected, screened and imaged. Representative images of eight serial sections showing ER localization of HYPE are presented (images serially numbered 1–8). Sections exhibit a dense well-preserved cytoplasm with undisrupted membrane ultrastructure of organelles such as mitochondria and Golgi complex in close proximity to the ER tubules containing HYPE–APEX2 density. Thus, we can follow HYPE localization through the volume of the cell without loss of contextual information. Scale bars: 5 μm (A); 8 μm (magnifcations 1–8).
EM tomographic reconstruction of the ER from a cell expressing HYPE–APEX2
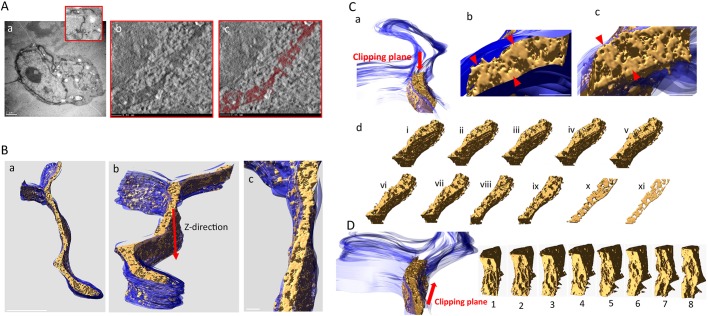
To visualize HYPE, we collected a tilt series from a single 250 nm section of HEK-293T cells expressing HYPE–APEX2 (Fig. 6Aa). The 3D reconstruction of HYPE within the ER was carried out for the region of the cell demarcated by the red box (Fig. 6Aa, red box). A movie of the 3D reconstruction for HYPE density obtained from the tilt series is shown in Movie 1, and a representative single slice from this movie is shown in Fig. 6Ab. The thresholded density for HYPE (colorized in maroon) in this movie is presented in Movie 2 and as a representative single slice in Fig. 6Ac. These reconstructions confirm the periodic distribution of HYPE density throughout the lumenal face of the ER membrane.
Fig. 6.
EM tomographic reconstruction of the ER exhibiting HYPE–APEX2 density. Tilt-series from HEK-293T cells expressing HYPE–APEX2 and processed by cryoAPEX were collected for 3D reconstruction of HYPE-specific density. (A) An image of the whole HYPE–APEX2-expressing cell showing an area containing ER tubules from where the tilt-series was collected (panel a and magnified red box). Panel b shows a snapshot from the movie (see Movie 1) showing the reconstructed tomogram of HYPE density within the ER lumen. Panel c shows a snapshot from the movie (see Movie 2) showing the reconstructed tomogram of the thresholded HYPE density (in maroon). (B) An additional view of the 3D model of the ER membrane (blue) and the HYPE density within (gold) generated and visualized with the IMOD ‘Isosurface’ tool (panels a–c). (C) Snapshots of a segment of the ER showing the HYPE–APEX2 density (gold) modeled within the lumen of the ER membrane (blue) visualized from the top of the indicated clipping plane (panels a–c). Red arrowheads in b show magnified images of different regions within this segment show HYPE's periodic density pattern along the lumenal walls. Red arrowheads in c show another view exposing the full face of the density (gold) using visualization tools that make the membrane transparent. This pattern of HYPE-specific density is more apparent when the clipping plane is moved downward in the z-direction, progressively shaving through the depths of the different layers (panel d, slices i–xi), and is most apparent when visualized in the thinnest slice (slice xi). (D) A clipping plane that moves in a head-to-tail direction shows HYPE's density pattern on the ER membrane from a front-on perspective (slices 1–8).
Next, 3D modeling by segmentation and visualization of the HYPE–APEX2 density showed that HYPE was confined within the ER membrane (Fig. 6B). We further assessed this reconstructed segment of the ER from different visual perspectives (Fig. 6Ba, whole ER segment; 6Bb, z plane; and 6Bc, top down). Finally, we used the ‘clipping planes’ tool from IMOD to incrementally trim off the HYPE-associated density from the top to the bottom (moving in the z-direction; Fig. 6C and slices i–xi in Fig. 6Cd) and in a head-to-tail direction (Fig. 6D, slices 1–8). Both clipping planes substantiated our observation of the periodic foci for HYPE on the lumenal walls of the ER (Fig. 6Cb,Cc, red arrowheads). Thinner, virtual slices of this region resembled the density pattern seen in 2D imaging of thin sections (compare Fig. 6Cd, slice xi with Fig. 3C).
DISCUSSION
FIC-mediated adenylylation (AMPylation) is an important, evolutionarily conserved mechanism of signal transduction. In humans, AMPylation mediated by HYPE regulates the unfolded protein response via reversible modification of BiP (Ham et al., 2014; Preissler, et al., 2017a,b; Sanyal et al., 2015). It is an open question as to whether HYPE functions beyond its role as a UPR regulator with additional physiological targets. Indeed, despite the fact that we and others have previously shown that HYPE is an ER membrane protein that faces the lumen (Sanyal et al., 2015; Rahman et al., 2012), a number of candidate targets have recently emerged for further evaluation that reside outside the ER (Broncel et al., 2016; Truttmann et al., 2016). For instance, the C. elegans homolog of HYPE (FIC-1) can be detected in the cytosol and is implicated in controlling the function of cytosolic chaperones (Truttmann et al., 2017). The Drosophila melanogaster HYPE homolog (dFic) is implicated in blindness and has been shown to associate with cell surface neuro–glial junctions, possibly by entering the secretory pathway (Rahman et al., 2012). Thus, a clear subcellular localization for HYPE is needed to better understand its role in the context of these new protein targets – which led us to develop a technique to determine the subcellular localization of membrane proteins like HYPE at a high (low-nanoscale) resolution.
Despite tremendous advances in light microscopy, electron microscopy still remains the technique of choice to visualize cellular ultrastructure or determine protein localization at nanoscale resolution. Here, we describe the development of an EM method, called cryoAPEX, which successfully adapts the APEX2 tag for cryofixation while simultaneously retaining membrane preservation. Additionally, we show that data obtained using cryoAPEX for visualizing ER proteins, HYPE and ERM, can be used for EM tomographic reconstruction of membranes in 3D. Applying cryoAPEX to HYPE localization, we show that HYPE appears to reside solely in the ER lumen and in the contiguous nuclear envelope, in agreement with immunofluorescence data from us and others (Sanyal et al., 2015; Truttmann et al., 2015).
CryoAPEX is designed specifically for localizing membrane-bound proteins. The methodology we present here enables sufficient resolution and membrane preservation such that even structures in close contact with the ER, such as the Golgi, mitochondria or plasma membrane, are clearly distinguishable as HYPE-negative (Fig. 4B–D). These results tend to argue against the presence of a secreted or nuclear (other than the nuclear envelope) form of HYPE. However, any trace amounts of HYPE that may potentially correspond to processed or soluble isoforms that might localize to the cytosol would not be detected unless present as immobilized complexes. We also cannot rule out the possibility that trace amounts of HYPE could exist in subcellular compartments other than the ER, or that specific stimuli could result in a dramatic reorganization of HYPE to other locations. Still, if this were the case, we predict that by overexpressing HYPE relative to endogenous levels (as we do in our study), we would see HYPE more broadly distributed throughout the cell or perhaps even mislocalized. Our data show that even upon overexpression, HYPE is most robustly observed solely within different domains of the ER, at least under resting conditions. Experiments to address the possibility of HYPE's relocalization in response to certain physiological stress signals are currently underway.
Intriguingly, our cryoAPEX analysis indicates that HYPE localizes as periodic membrane foci spanning the lumenal wall of the ER. The reasons are unknown, but this indicates that HYPE is tethered, possibly via its hydrophobic N-terminus, to repetitive structural elements along the ER, potentially as part of a larger signaling complex. The crystal structure of HYPE indicates that it exists as a dimer, interacting via its FIC domain (Bunney et al., 2014). Additionally, the activity of bacterial FIC proteins, including HYPE, has been shown to undergo regulation by transitioning between monomeric, dimeric and oligomeric states (Dedic et al., 2016; Stanger et al., 2016; Casey et al., 2017). Thus, it is possible that the HYPE-specific periodic densities that we observe may represent HYPE oligomers. Further, we were able to reconstruct this HYPE-specific density in 3D along the ER lumenal membrane using TEM tomography (Fig. 6).
In our quest for the perfect EM-based localization technology to visualize HYPE in the context of the ER and the whole cell, we were cognizant of the fact that to get a reliable picture of the presence or absence of HYPE in various subcellular compartments, we would require impeccable ultrastructural membrane preservation. Many organelles and transport vesicles within a cell are labile structures that are difficult to preserve in their native morphology. An organelle like the ER has multiple domains that make contacts with several other organelles as well as the plasma membrane. The functional relevance of these organellar contact points is of research interest and, in the case of the ER, they are known to be portals of lipid and calcium transport (English and Voeltz, 2013; Rowland and Voeltz, 2012). Thus, preservation of these structures was especially important for ascertaining the distribution of HYPE.
Next, we considered the applicability of various traditional protein localization techniques such as immunoelectron microscopy (IEM), metal-tagging EM (METTEM) and peroxidase tagging. Unfortunately, each of these techniques suffers from a variety of limitations in addition to inadequate sample preservation. Specifically, current methods of detection are based on two common processes: (1) a chemical fixation step that precedes the actual detection assay and (2) a sample preparation step involving dehydration of fixed cells via alcohol at room temperature or on ice. This combination of chemical reagents leads to poor preservation of the membrane morphology as a result of lipid extraction, and introduces artefacts. Therefore, in addition to membrane preservation, the method of choice needs to be compatible with heavy metal staining, so as to impart an adequate level of contrast between HYPE-associated membranes and other organellar membranes for contextual information about the ultrastructural environment within which HYPE resides. This ruled out METTEM tagging, as the technique is incompatible with the use of heavy metal stains (Risco et al., 2012).
Lastly, we considered the amenability of the method to 3D electron microscopic techniques. This is important as organelles or membrane structures such as the ER, Golgi, mitochondria or the plasma membrane cover a vast three-dimensional subcellular space and are in a constant state of morphological equilibrium with their surroundings. They undergo constant remodeling in their different domains in response to functional cues that can alter the localization of proteins that are associated with them (Shibata et al., 2010; Voeltz et al., 2002). To detect such changes or, alternatively, the exclusive localization of a target protein in specific domains of these large organelles, 3D information at the site of protein localization can yield critical clues about protein function. Thus, we opted to develop a method that incorporated each of the above criteria to yield HYPE's subcellular distribution in an optimally preserved and 3D EM-compatible sample.
Cryofixation of live cells under high pressure (HPF) is a method that shows the best ultrastructural preservation and is now routinely used to prepare samples for EM tomography (McDonald and Auer, 2006; O'Toole et al., 2018). It is not deemed compatible with most of the detection methods described above, however, as they require chemical fixation. Thus, to determine the subcellular distribution of HYPE, we developed cryoAPEX, a hybrid method that combines the power of APEX2 genetic tagging and HPF cryofixation. Chemically fixed cells expressing APEX2-tagged HYPE were first reacted with DAB to generate HYPE-specific density, and then cryofixed and freeze-substituted with acetone. As shown, cryoAPEX not only displays specificity of detection at high resolution for both lumen-facing (HYPE) and cytosol-facing (ERM) ER membrane proteins, but also retains ultrastructural preservation that makes cryoAPEX amenable to TEM tomography. Further, cryoAPEX can be used to assess cells grown in monolayers, making it widely applicable.
An important aspect of cryoAPEX is the robustness of the DAB byproduct that can withstand a long freeze-substitution reaction in acetone. We also noted that once chemically fixed and labeled with DAB, cells do not need to be cryofixed right away. In our hands, aldehyde-fixed, DAB-labeled cells that were cryofixed after 48 h (storage at 4°C) exhibited no deterioration of cellular ultrastructure and staining when compared to those that were cryofixed immediately. This feature could prove to be of great advantage to laboratories that do not have immediate access to HPF and freeze-substitution units. While this manuscript was being prepared, a technique with a similar goal of combining cryofixation with an APEX2-based detection method for carrying out CLEM (correlative light-EM) at the tissue (versus subcellular) level was reported. This method, called CryoChem, is procedurally different from our method. It requires immediate cryofixation of tissues and entails elaborate rehydration and dehydration steps following the initial freeze substitution in order to obtain better temporal resolution for CLEM studies (Tsang et al., 2018). Also, CryoChem is yet to be tested on cell monolayers.
In conclusion, we have developed cryoAPEX as a method for obtaining localization of a single APEX2-tagged protein at a high resolution while maintaining excellent ultrastructural preservation and compatibility with EM tomography. We applied cryoAPEX to assess the subcellular localization of the human FIC protein, HYPE, and show that it is robustly detected very specifically on the lumenal face of the ER membrane and in cellular compartments that are contiguous with the ER lumen, where it displays periodic distribution resembling possible signaling complexes. Further, we do not detect HYPE in the mitochondria, nucleus, plasma membrane or the Golgi and secretory network at the expression levels tested. Additionally, we show that cryoAPEX works equally well for cytosol-facing membrane proteins, such as ERM, and accurately reflects ultrastructural morphological changes. Finally, we demonstrate that cryoAPEX can be applied to assessing protein localization using cell monolayers and executed in basic cell biology laboratories with relative ease.
MATERIALS AND METHODS
Plasmids
The HYPE–APEX2 plasmid was ordered through Genscript. Briefly, the HYPE–APEX2 fusion was first synthesized and then inserted into pcDNA3 vector between BamHI and XhoI sites. ERM–APEX2 plasmid was obtained from Addgene (plasmid 79055). The mito-V5–APEX2 was obtained from Addgene, (plasmid 72480); CAAX–APEX2 was a kind gift from Alice Y. Ting (Stanford University, CA, USA) and the ManII–APEX2 was ordered through Epoch biosciences. Briefly, a fusion construct comprising the first 118 amino acids of the mouse isoform of α-mannosidase with the APEX2 gene in its C-terminus following a short intervening linker sequence was first synthesized and then cloned into pcDNA3.1. The complete sequence for MannII–APEX2 is provided (Table S1).
Transfection and chemical fixation
HEK-293T cells (ATCC) were grown in 10 cm dishes in Dulbecco's modified Eagle's medium (DMEM; Corning) supplemented with 10% fetal bovine serum (FBS; Corning NuSerum IV) at 37°C and 5% CO2. Cells were transfected with APEX2-tagged mammalian expression plasmids using Lipofectamine 3000 (Thermo Fisher). Cells were washed off the plate with Dulbecco's PBS 12–15 h post-transfection and then pelleted at 500 g. For those samples requiring chemical fixation, pellets were resuspended in 0.1% sodium cacodylate buffer containing 2% glutaraldehyde for 30 min, washed 3× for 5 min with 0.1% sodium cacodylate buffer and 1× with cacodylate buffer containing 1 mg/ml 3,3′-diaminobenzidine (DAB) (Sigma-Aldrich). Pellets were then incubated for 30 min in a freshly made solution of 1 mg/ml DAB and 5.88 mM hydrogen peroxide in cacodylate buffer, pelleted and washed 2× for 5 min in cacodylate buffer and once with DMEM. Finally, cell pellets were resuspended in DMEM containing 10% FBS and 15–20% BSA, then pelleted again. The supernatant was aspirated and excess media wicked off with a Kimwipe in order to remove as much liquid as possible. For BHK (ATCC) controls, cells were grown in DMEM with 10% FBS at 37°C and 5% CO2. Cells were either cryofixed directly or prefixed with glutaraldehyde prior to cryofixation. An identical freeze-substitution protocol was used for processing for both HEK-293T and BHK cells. BHK cells were post-stained with uranyl acetate and Sato's lead prior to imaging.
High-pressure freezing and freeze substitution
Cell pellets (2–3 μl) were loaded onto copper membrane carriers (1 mm×0.5 mm; Ted Pella Inc.) and cryofixed using the EM PACT2 high-pressure freezer (Leica). Cryofixed cells were then processed by freeze substitution using an AFS2 automated freeze substitution unit (Leica). An extended freeze substitution protocol was optimized for the preferential osmication of the peroxidase-DAB byproduct. Briefly, frozen pellets were incubated for 24 h at −90°C in acetone containing 0.2% tannic acid and then washed 3× for 5 min with glass-distilled acetone (EM Sciences). Pellets were resuspended in acetone containing 5% water and 1% osmium tetroxide, with or without 2% uranyl acetate (as applicable) for 72 h at −80°C. Following this extended osmication cycle, pellets were warmed to 0°C over 12–18 h. Pellets were then washed 3× for 30 min with freshly opened glass-distilled acetone. Resin exchange was carried out by infiltrating the sample with a gradually increasing concentration of Durcupan ACM resin (Sigma-Aldrich) as follows: 2%, 4% and 8% for 2 h each and then 15%, 30%, 60%, 90%, 100% and 100% plus component C (Durcupan100+C hereafter) for 4 h each. Resin-infiltrated samples in membrane carriers were then embedded in resin blocks and polymerized at 60°C for 36 h. Post-hardening, planchets were extracted by dabbing liquid nitrogen on the membrane carriers and using a razor to resect them out of the hardened resin. After extraction of membrane carriers, a thin layer of Durcupan100+C was added on top of the exposed samples and incubated in an oven at 60°C for 24–36 h to obtain the final hardened sample blocks for sectioning.
Sample preparation via conventional room temperature method
HEK-293T cells were grown on collagen-coated glass coverslips and transfected with HYPE–APEX2 or ERM–APEX2 mammalian expression plasmids for 24 h as above. Cells were then washed with DPBS and chemically fixed with 2% glutaraldehyde in 0.1% sodium cacodylate buffer for 30 min. Fixed cells were washed with cacodylate buffer and finally with 1 mg/ml of DAB in cacodylate buffer for 2 min. Following the wash, cells were incubated for 30 min in a freshly made solution of 1 mg/ml of DAB and 5.88 mM of hydrogen peroxide in cacodylate buffer at room temperature. Cells were washed 3× for 2 min each with DPBS, incubated in an aqueous solution of 1% osmium tetroxide for 10–15 min and then washed with distilled water. Dehydration was carried out using increasing concentration of 200 proof ethanol (30%, 50%, 70%, 90%, 95%, 100%) followed by resin infiltration of the cells with gradually increasing concentrations of Durcupan resin in ethanol (30%, 60%, 90%, 100%), then Durcupan100+C. Coverslips were placed on BEEM capsules filled with Durcupan100+C, cell-face-down on the resin and incubated in an oven for 48 h at 60°C. After polymerization, coverslips were extracted by dipping the coverslip face of the blocks briefly in liquid nitrogen. Serial sections were then obtained by sectioning of the blocks en face and ribbons collected on formvar-coated slot grids.
Serial sectioning, lead staining and electron microcopy
Thin (90 nm) serial sections were obtained using a UC7 ultramicrotome (Leica) and collected onto formvar-coated copper slot grids (EM sciences). Glass knives were freshly prepared from glass sticks during each sectioning exercise. Lead staining of the sections was carried out for 1 min wherever applicable with freshly made Sato's lead solution. Samples were screened on a Technai T-12 80 kV transmission electron microscope, and an average of 15–20 cells from multiple blocks were visualized for each sample.
Measurement of HYPE–APEX2 density foci
ImageJ software was used to measure the HYPE–APEX2 density foci within the ER lumen. Briefly, using the line selection tool the image scale was set up using the embedded scale bar as a yardstick. From the menu command, the ‘analyze/set scale’ was used to set up the scale by entering the dimensions of the raw image (length of the embedded scale bar) in the ‘known distance’ box. The unit of the scale bar was then set in the ‘units of the length box’. Readings were taken from multiple ER tubules and the range and average calculated.
EM tomography, data reconstruction and segmentation
Thicker (250 nm) sections were used for collecting tomographic tilt-series. Sections were coated with gold fiducials measuring 20 nm in diameter prior to collection. Tilt-series of a single 250 nm section were collected with automation using the program SerialEM (Mastronarde, 2003) on a JEOL 3200 TEM operating at 300 kV. The collected tilt-series were then aligned and tomogram generated by weighted back projection using the eTomo interface of IMOD (Kremer et al., 1996). The reconstructed tomogram was visualized in IMOD. The ER membrane was first hand-segmented and then used as a mask for thresholding of the density within the ER lumen.
Supplementary Material
Acknowledgements
We thank Dr Jason Lanman (Purdue University, IN, USA) for access to HPF, FS, and microtome units. We are also grateful to Drs Alice Ting and Jeffrey Martell (Stanford University, CA, USA) for advice and providing the CAAX–APEX2 (Lam et al., 2015) construct and to Dr Ben Giepmans (University of Groningen, The Netherlands) for providing their Golgi-FLIPPER (Kuipers et al., 2015) construct and sequence from which we cloned our mannII–APEX2 construct. HEK293T and BHK cells were a gift from Dr Richard J. Kuhn (Purdue University, IN). We are indebted to Ms Elaine Mihelc for help with IMOD segmentation and technical discussions, and to Dr Robert Stahelin for constructive and thoughtful discussions. We are also grateful to members of the Mattoo lab for feedback. Thin-section screening was carried out at the Purdue Life Sciences EM Core Facility. Tomogram collection was conducted at the Electron Microscopy Center at Indiana University – Bloomington under guidance from Dr David Morgan.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.M.; Methodology: R.S.; Software: R.S.; Validation: S.M.; Formal analysis: R.S., S.M.; Investigation: R.S., S.M.; Resources: R.S., S.M.; Data curation: R.S., S.M.; Writing - original draft: R.S., M.J.P., S.M.; Writing - review & editing: R.S., M.J.P., S.M.; Visualization: R.S., S.M.; Supervision: S.M.; Project administration: S.M.; Funding acquisition: S.M.
Funding
This work was funded in part by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM10092), an Indiana Clinical and Translational Sciences Institute Research Grant (CTSI-106564), and a Purdue University Institute for Inflammation, Immunology, and Infectious Disease Core Start Grant (PI4D-209263). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.222315.supplemental
References
- Broncel M., Serwa R. A., Bunney T. D., Katan M. and Tate E. W. (2016). Global profiling of Huntingtin-associated protein E (HYPE)-mediated AMPylation through a chemical proteomic approach. Mol. Cell. Proteomics 15, 715-725. 10.1074/mcp.O115.054429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney T. D., Cole A. R., Broncel M., Esposito D., Tate E. W. and Katan M. (2014). Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 22, 1831-1843. 10.1016/j.str.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey A. K. and Orth K. (2018). Enzymes involved in AMPylation and deAMPylation. Chem. Rev. 118, 1199-1215. 10.1021/acs.chemrev.7b00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey A. K., Moehlman A. T., Zhang J., Servage K. A., Krämer H. and Orth K. (2017). Fic-mediated deAMPylation is not dependent on homodimerization and rescues toxic AMPylation in flies. J. Biol. Chem. 292, 21193-21204. 10.1074/jbc.M117.799296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F. L., Inoue S. and Leblond C. P. (1993). The basement membranes of cryofixed or aldehyde-fixed, freeze-substituted tissues are composed of a lamina densa and do not contain a lamina lucida. Cell Tissue Res. 273, 41-52. 10.1007/BF00304610 [DOI] [PubMed] [Google Scholar]
- De Mey J., Moeremans M., Geuens G., Nuydens R. and De Brabander M. (1981). High resolution light and electron microscopic localization of tubulin with the IGS (immuno gold staining) method. Cell Biol. Int. Rep. 5, 889-899. 10.1016/0309-1651(81)90204-6 [DOI] [PubMed] [Google Scholar]
- Dedic E., Alsarraf H., Welner D. H., Østergaard O., Klychnikov O. I., Hensbergen P. J., Corver J., van Leeuwen H. C. and Jørgensen R. (2016). A novel fic (filamentation induced by cAMP) protein from Clostridium difficile reveals an inhibitory motif-independent adenylylation/AMPylation mechanism. J. Biol. Chem. 291, 13286-13300. 10.1074/jbc.M115.705491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. R. and Voeltz G. K. (2013). Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harbor Perspect. Biol. 5, a013227 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P., Barnes G. T., Srinidhi J., Chen J., Gusella J. F. and MacDonald M. E. (1998). Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 7, 1463-1474. 10.1093/hmg/7.9.1463 [DOI] [PubMed] [Google Scholar]
- Garcia-Pino A., Zenkin N. and Loris R. (2014). The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem. Sci. 39, 121-129. 10.1016/j.tibs.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Ham H., Woolery A. R., Tracy C., Stenesen D., Krämer H. and Orth K. (2014). Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059-36069. 10.1074/jbc.M114.612515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Stanger F. V., Scheu P. D., de Jong I. G., Goepfert A., Glatter T., Gerdes K., Schirmer T. and Dehio C. (2015). Adenylylation of gyrase and topo IV by FicT toxins disrupts bacterial DNA topology. Cell Rep. 12, 1497-1507. 10.1016/j.celrep.2015.07.056 [DOI] [PubMed] [Google Scholar]
- Hawes P., Netherton C. L., Mueller M., Wileman T. and Monaghan P. (2007). Rapid freeze-substitution preserves membranes in high-pressure frozen tissue culture cells. J. Microsc. 226, 182-189. 10.1111/j.1365-2818.2007.01767.x [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Gaietta G., Zürn A., Adams S. R., Terrillon S., Ellisman M. H., Tsien R. Y. and Lohse M. J. (2010). Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat. Protoc. 5, 1666-1677. 10.1038/nprot.2010.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C., Gibson A., Stinchcombe J. and Futter C. (2000). Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 327, 35-45. 10.1016/S0076-6879(00)27265-0 [DOI] [PubMed] [Google Scholar]
- Huxley H. E. and Zubay G. (1961). Preferential staining of nucleic acid-containing structures for electron microscopy. J. Cell Biol. 11, 273-296. 10.1083/jcb.11.2.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalina M. and Pease D. C. (1977). The preservation of ultrastructure in saturated phosphatidyl cholines by tannic acid in model systems and type II pneumocytes. J. Cell Biol. 74, 726-741. 10.1083/jcb.74.3.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N. and McIntosh J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kuipers J., van Ham T. J., Kalicharan R. D., Veenstra-Algra A., Sjollema K. A., Dijk F., Schnell U. and Giepmans B. N. G. (2015). FLIPPER, a combinatorial probe for correlated live imaging and electron microscopy, allows identification and quantitative analysis of various cells and organelles. Cell Tissue Res. 360, 61-70. 10.1007/s00441-015-2142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. S., Martell J. D., Kamer K. J., Deerinck T. J., Ellisman M. H., Mootha V. K. and Ting A. Y. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51-54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J. D., Deerinck T. J., Sancak Y., Poulos T. L., Mootha V. K., Sosinsky G. E., Ellisman M. H. and Ting A. (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotech. 30, 1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J. D., Deerinck T. J., Lam S. S., Ellisman M. H. and Ting A. Y. (2017). Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat. Protoc. 12, 1792-1816. 10.1038/nprot.2017.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D. (2003). SerialEM A program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182-1183. 10.1017/S1431927603445911 [DOI] [Google Scholar]
- Mattoo S., Durrant E., Chen M. J., Xiao J., Lazar C. S., Manning G., Dixon J. E. and Worby C. A. (2011). Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J. Biol. Chem. 286, 32834-32842. 10.1074/jbc.M111.227603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117, 77-97. 10.1385/1-59259-201-5:77 [DOI] [PubMed] [Google Scholar]
- McDonald K. L. and Auer M. (2006). High-pressure freezing, cellular tomography, and structural cell biology. BioTechniques 41, 137 10.2144/000112226 [DOI] [PubMed] [Google Scholar]
- Mercogliano C. and DeRoiser D. J. (2007). Concatenated metallothionein as a clonable gold label for electron microscopy. J. Struct. Biol. 160, 70-82. 10.1016/j.jsb.2007.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Liu X., Arasaki K., McDonough J., Galán J. E. and Roy C. R. (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103-106. 10.1038/nature10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E., van der Heide P., Richard McIntosh J. and Mastronarde D. (2018). Large-scale electron tomography of cells using SerialEM and IMOD. Biol. Med. Phys. Biomed. Eng. 152, 95-116. 10.1007/978-3-319-68997-5_4 [DOI] [Google Scholar]
- Persi M. A. and Burnham J. C. (1981). Use of tannic acid as a fixative-mordant to improve the ultrastructural appearance of Candida albicans blastospores. Med. Mycol. 19, 1-8. 10.1080/00362178185380021 [DOI] [PubMed] [Google Scholar]
- Porstmann B., Porstmann T., Nugel E. and Evers U. (1985). Which of the commonly used marker enzymes gives the best results in colorimetric and fluorimetric enzyme immunoassays: horseradish peroxidase, alkaline phosphatase or beta-galactosidase. J. Immunol. Methods 79, 27-37. [DOI] [PubMed] [Google Scholar]
- Preissler S., Rato C., Chen R., Antrobus R., Ding S., Fearnley I. M. and Ron D. (2015). AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. eLife 4, e12621 10.7554/eLife.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., Rato C., Perera L. A., Saudek V. and Ron D. (2017a). FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat. Struct. Mol. Biol. 24, 23-29. 10.1038/nsmb.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., Rohland L., Yan Y., Chen R., Read R. J. and Ron D. (2017b). AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation. eLife 6, e29428 10.7554/eLife.29428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Ham H., Liu X., Sugiura Y., Orth K. and Krämer H. (2012). Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 15, 871-875. 10.1038/nn.3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Sanmartín-Conesa E., Tzeng W.-P., Frey T. K., Seybold V. and de Groot R. J. (2012). Specific, sensitive, high-resolution detection of protein molecules in eukaryotic cells using metal-tagging transmission electron microscopy. Structure 20, 759-766. 10.1016/j.str.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A. A. and Voeltz G. K. (2012). Endoplasmic reticulum–mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 13, 607-615. 10.1038/nrm3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandig G., Kärgel E., Menzel R., Vogel F., Zimmer T. and Schunck W.-H. (1999). Regulation of endoplasmic reticulum biogenesis in response to cytochrome P450 overproduction. Drug Metab. Rev. 31, 393-410. 10.1081/DMR-100101926 [DOI] [PubMed] [Google Scholar]
- Sanyal A., Chen A. J., Nakayasu E. S., Lazar C. S., Zbornik E. A., Worby C. A., Koller A. and Mattoo S. (2015). A novel link between fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J. Biol. Chem. 290, 8482-8499. 10.1074/jbc.M114.618348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell U., Dijk F., Sjollema K. A. and Giepmans B. N. G. (2012). Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152-158. 10.1038/nmeth.1855 [DOI] [PubMed] [Google Scholar]
- Schrijvers A. H. G. J., Frederik P. M., Stuart M. C. A., van der Vusse G. J. and Reneman R. S. (1989). Dual effect of tannic acid on the preservation and ultrastructure of phosphatidyl choline vesicles. Mol. Cell. Biochem. 88, 91-96. 10.1007/BF00223429 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W. A., Palazzo A. F., Kozlov M. M. and Rapoport T. A. (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774-788. 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Lev-Ram V., Deerinck T. J., Qi Y., Ramko E. B., Davidson M. W., Jin Y., Ellisman M. H. and Tsien R. Y. (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041 10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N. and Lippincott-Schwartz J. (2003). Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257-269. 10.1083/jcb.200306020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky G. E., Crum J., Jones Y. Z., Lanman J., Smarr B., Terada M., Martone M. E., Deerinck T. J., Johnson J. E. and Ellisman M. H. (2008). The combination of chemical fixation procedures with high pressure freezing and freeze substitution preserves highly labile tissue ultrastructure for electron tomography applications. J. Struct. Biol. 161, 359-371. 10.1016/j.jsb.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger F. V., Burmann B. M., Harms A., Aragão H., Mazur A., Sharpe T., Dehio C., Hiller S. and Schirmer T. (2016). Intrinsic regulation of FIC-domain AMP-transferases by oligomerization and automodification. Proc. Natl. Acad. Sci. USA 113, E529-E537. 10.1073/pnas.1516930113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer D., Humbel B. M. and Chiquet M. (2008). Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem. Cell Biol. 130, 877-889. 10.1007/s00418-008-0500-1 [DOI] [PubMed] [Google Scholar]
- Truttmann M. C., Wu Q., Stiegeler S., Duarte J. N., Ingram J. and Ploegh H. L. (2015). HypE-specific nanobodies as tools to modulate HypE-mediated target AMPylation. J. Biol. Chem. 290, 9087-9100. 10.1074/jbc.M114.634287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann M. C., Cruz V. E., Guo X., Engert C., Schwartz T. U. and Ploegh H. L. (2016). The Caenorhabditis elegans protein FIC-1 is an AMPylase that covalently modifies heat-shock 70 family proteins, translation elongation factors and histones. PLoS Genet. 12, e1006023 10.1371/journal.pgen.1006023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann M. C., Zheng X., Hanke L., Damon J. R., Grootveld M., Krakowiak J., Pincus D. and Ploegh H. L. (2017). Unrestrained AMPylation targets cytosolic chaperones and activates the heat shock response. Proc. Natl Acad. Sci. USA 114, E152-E160. 10.1073/pnas.1619234114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann M. C., Pincus D. and Ploegh H. L. (2018). Chaperone AMPylation modulates aggregation and toxicity of neurodegenerative disease-associated polypeptides. Proc. Natl. Acad. Sci. USA 115, E5008-E5017. 10.1073/pnas.1801989115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang T. K., Bushong E. A., Boassa D., Hu J., Romoli B., Phan S., Su C.-Y. and Ellisman M. H. (2018). High-quality ultrastructural preservation using cryofixation for 3D electron microscopy of genetically labeled tissues. eLife 7, e35524 10.7554/eLife.35524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. K., Rolls M. M. and Rapoport T. A. (2002). Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944-950. 10.1093/embo-reports/kvf202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C. and Dixon J. E. (2009). The Fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93-103. 10.1016/j.molcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Worby C. A., Mattoo S., Sankaran B. and Dixon J. E. (2010). Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004-1010. 10.1038/nsmb.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L. and Orth K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269-272. 10.1126/science.1166382 [DOI] [PubMed] [Google Scholar]
- Zechmann B., Müller M. and Zellnig G. (2007). Membrane associated qualitative differences in cell ultrastructure of chemically and high pressure cryofixed plant cells. J. Struct. Biol. 158, 370-377. 10.1016/j.jsb.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Zekarias B., Mattoo S., Worby C., Lehmann J., Rosenbusch R. F. and Corbeil L. B. (2010). Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect. Immun. 78, 1850-1858. 10.1128/IAI.01277-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.