ABSTRACT
Mitochondrial DNA depletion syndromes (MDS) are a group of rare autosomal recessive disorders with early onset and no cure available. MDS are caused by mutations in nuclear genes involved in mitochondrial DNA (mtDNA) maintenance, and characterized by both a strong reduction in mtDNA content and severe mitochondrial defects in affected tissues. Mutations in MPV17, a nuclear gene encoding a mitochondrial inner membrane protein, have been associated with hepatocerebral forms of MDS. The zebrafish mpv17 null mutant lacks the guanine-based reflective skin cells named iridophores and represents a promising model to clarify the role of Mpv17. In this study, we characterized the mitochondrial phenotype of mpv17−/− larvae and found early and severe ultrastructural alterations in liver mitochondria, as well as significant impairment of the respiratory chain, leading to activation of the mitochondrial quality control. Our results provide evidence for zebrafish Mpv17 being essential for maintaining mitochondrial structure and functionality, while its effects on mtDNA copy number seem to be subordinate. Considering that a role in nucleotide availability had already been postulated for MPV17, that embryos blocked in pyrimidine synthesis do phenocopy mpv17−/− knockouts (KOs) and that mpv17−/− KOs have impaired Dihydroorotate dehydrogenase activity, we provided mpv17 mutants with the pyrimidine precursor orotic acid (OA). Treatment with OA, an easily available food supplement, significantly increased both iridophore number and mtDNA content in mpv17−/− mutants, thus linking the loss of Mpv17 to pyrimidine de novo synthesis and opening a new simple therapeutic approach for MPV17-related MDS.
KEY WORDS: Dihydroorotate dehydrogenase, Iridophore, Mitochondrial DNA depletion syndrome, Pyrimidine, Vitamin B13
Summary: The zebrafish mpv17−/− mutant shows a severe mitochondrial phenotype with ultrastructural alterations and oxidative phosphorylation impairment. The pyrimidine precursor orotic acid ameliorates mpv17−/− phenotype and increases mitochondrial DNA content, linking the loss of Mpv17 to pyrimidine de novo synthesis.
INTRODUCTION
In the past few years, the scientific scenario identified mitochondria as key players in a growing spectrum of human diseases. Beside their main functions in oxidative phosphorylation and bioenergetics, mitochondria also participate in a plethora of cell pathways, such as apoptosis, production of reactive oxygen species (ROS), heat generation and lipid metabolism (Wallace, 2005). Notably, mitochondrial functionality requires a synergistic relationship between mitochondrial and nuclear genomes (Chinnery and Hudson, 2013). In fact, mitochondrial DNA (mtDNA) encodes only 13 subunits of the respiratory chain (RC), while the remaining ∼1500 mitochondrial proteins, encoded by nuclear DNA (nDNA), are imported into the organelle (Calvo et al., 2016).
Each mitochondrion may contain hundreds of copies of mtDNA and, considering that energy demand depends on age, tissue and life style, even in physiological conditions, the amount of mtDNA differs between cell types and individuals (Suomalainen and Isohanni, 2010). What has become clear, however, is that having the correct mtDNA amount in each tissue is crucial. In fact, mtDNA depletion has been discovered to cause a continuously increasing group of severe human diseases called mtDNA depletion syndromes (MDS), first described in 1991 (Moraes et al., 1991).
MDS are a clinically heterogeneous group of autosomal-recessive mitochondrial disorders, caused by mutations in nuclear genes, such as MPV17, involved in mtDNA replication and maintenance. Symptoms of MDS typically appear during infancy and childhood, and are characterized by the reduction of mtDNA copy number in the affected tissues (Nogueira et al., 2014). MPV17 is an inner-membrane mitochondrial protein possibly involved in mitochondrial deoxynucleoside triphosphates (dNTP) pool homeostasis (Löllgen and Weiher, 2014; Krauss et al., 2013). Mutations in the MPV17 gene are known to cause Navajo neurohepatopathy (NNH) (Karadimas et al., 2006) and a hepatocerebral form of MDS (Spinazzola et al., 2006). In fact, of ∼50 MPV17 pathogenic variants reported worldwide in ∼100 patients, the c.149G>A (p.R50Q) missense mutation has been found with an unusually high frequency in NNH-affected individuals (El-Hattab et al., 2018). To date, the precise function of MPV17 is unclear, even though many studies have been carried out in different in vivo models. Among those, two spontaneous zebrafish mpv17 mutants, roy orbison (roy) and transparent (tra), have been independently identified (Krauss et al., 2013; D'Agati et al., 2017). Notably, although the genomic lesions have not been properly sequenced so far in each mutant line, they both lead to the same 19 bp deletion in the coding sequence, originating from a defective splicing of intron 2. Both roy and tra (mpv17−/−) display pigmentation abnormalities due to the lack of cells derived from the neural crest, such as iridophores, and, at later stages of development, melanophores (Krauss et al., 2013). Notably, in vertebrates, neural crest cells migrate and give rise to a large variety of complex structures, including peripheral nervous system, part of the craniofacial skeleton and pigment cells (Kelsh et al., 1996). In particular, the zebrafish pigment pattern presents three distinct types of cells: black melanophores, yellow xantophores and reflective iridophores, which strongly interact with each other and are distributed along longitudinal stripes and silvery/yellow interstripes (Kelsh et al., 2017). Several zebrafish mutants bearing recessive mutations in the de novo purine synthesis genes gart and paics show defects in iridophores, the cells containing light-reflective purine platelets, which are responsible for the shiny appearance of the striped pattern (Frohnhöfer et al., 2013; Ng et al., 2009).
In this work, we validated the functional homology between human and zebrafish Mpv17 and improved the characterization of the roy/tra phenotype, providing evidence of mitochondrial defects, RC impairment and mtDNA depletion. Our findings indicate roy and tra as valuable models for MPV17-related MDS. Moreover, because the chemical inhibition of pyrimidine de novo synthesis causes iridophore and melanophore loss in zebrafish embryos (White et al., 2011), we investigated the function of mpv17 by administrating dNTPs and pyrimidine precursors to mpv17−/− mutants, and found significant impairment of Dihydroorotate dehydrogenase (Dhodh) activity in mutants.
RESULTS
Mpv17 and its paralogue, Mpv17-like2, crosstalk in mpv17−/− larvae
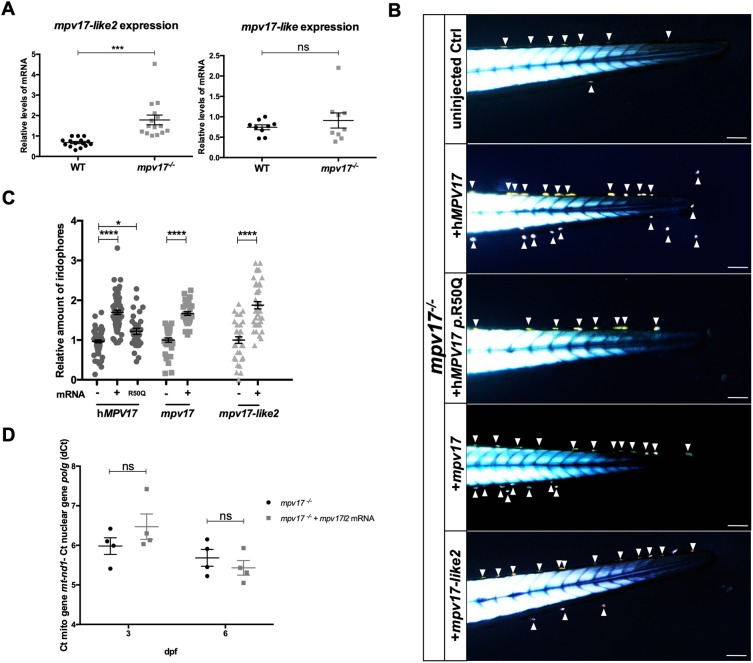
Because zebrafish mpv17 null mutants are viable and fertile, we verified whether mpv17 paralogue genes might play compensatory activities in mpv17−/− mutants, thus ameliorating their phenotype. By performing reverse transcription and quantitative polymerase chain reaction (RT-qPCR) analysis, we observed a significant overexpression of mpv17-like2 (mpv17l2), whereas the mpv17-like (mpv17l) expression level remained unchanged in both wild-type and homozygous knockout (KO) larvae at 6 days post-fertilization (dpf) (Fig. 1A). This result was also confirmed by in situ hybridization (ISH), which showed wider mpv17l2 expression in mpv17−/− mutant larvae at 3 dpf, compared with the liver-specific localization observed in wild-type controls (Fig. S1). Hence, we checked whether mpv17l2 transcripts might rescue the mpv17 null mutant phenotype, and thus whether their protein functions could overlap. To this aim, we injected one-cell-stage embryos with mRNAs encoding zebrafish mpv17 and mpv17l2, and wild-type and p.R50Q human MPV17 (Fig. 1B), and evaluated iridophore number at 3 dpf. Early phenotyping of mpv17−/− zebrafish mutants can be easily performed with the aid of a microscope equipped with two polarizing filters and using the birefringence property of iridophores. Briefly, embryos are anaesthetized, flat oriented along the lateral axis and placed above the polarizer lens in a transmitted-light-dissecting microscope. Then, according to published protocols for birefringence analysis (Smith et al., 2013), the polarization plan of the analyser lens, placed above the sample, is oriented at 90° with the polarizer. With this procedure, the light from the background is completely abolished, leaving a black surround. This application of birefringence, in contrast with Krauss et al. (2013), allows the precise counting of iridophore number independently from the orientations of light and samples (Fig. S2). In particular, iridophores are readily visible at 3 dpf; they are the first metamorphic chromatophores to become observable in the dermis of the lateral trunk, and appear only later in the anterior trunk in a segmental fashion, continuing to increase in number and to accumulate to form the first ventral interstripe (Higdon et al., 2013; Frohnhöfer et al., 2013).
Fig. 1.
Investigation of mpv17 orthologue and paralogue genes in zebrafish larvae. (A) Real-time PCR quantification of mRNA transcripts of mpv17-like2 and mpv17-like at 6 dpf (n=9). (B) Evaluation of phenotypic rescue in the tail region at 3 dpf after the injection of human MPV17 mRNAs, wild-type (WT) and p.R50Q mutated forms, and zebrafish mpv17 and mpv17-like2 mRNAs. Arrowheads point to iridophores. Scale bars: 100 µm. (C) Relative quantification of iridophore amount in controls and injected larvae (n=30). (D) mtDNA copy number analysis in mpv17−/− mutants transiently overexpressing mpv17-like2 at 3 dpf and 6 dpf. Mean dCt values were calculated as Ct of mt-nd1 (mitochondrially encoded gene) minus Ct of polg (nuclear gene) (n=4). Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ****P<0.0001; ***P<0.001; *P<0.05; ns, not significant. Experiments were performed in biological replicates.
Interestingly, in all injected samples, except for p.R50Q-injected larvae, we could observe a significant and comparable increase in the number of iridophores. Notably, injection of mRNA encoding the human p.R50Q mutation had a significantly lower effect on the amount of iridophores (Fig. 1C). Hence, we performed an mtDNA copy number analysis on mpv17l2-injected embryos and found a non-significant increase in mtDNA content at 3 dpf (Fig. 1D).
Together, these data show that mpv17 and mpv17l2 functions may partially overlap, and demonstrate a similar function for human and zebrafish mpv17, confirming roy as a sound model for investigating mpv17 roles.
Loss of Mpv17 alters mitochondrial ultrastructure and respiratory complex stability
MPV17-related MDS primarily show hepatic manifestations and patients die of liver failure between 6 and 28 months of age (Kim et al., 2016). To characterize the mpv17−/− mutant phenotype, we investigated the liver mitochondrial morphology of mpv17−/− larvae, by transmission electron microscopy (TEM) analysis. Results showed a tissue-specific disruption of mitochondrial ultrastructure in liver hepatocytes at 6 dpf, with mitochondrial ballooning and disappearance of cristae (Fig. 2A); this phenotype was absent at 3 dpf and no difference was detected in brain and muscle tissues (data not shown). In order to assess whether an impairment of mitochondrial morphology might modify the amount of mitochondria in the liver, we crossed mpv17−/− mutants with two different transgenic lines expressing, respectively, a ubiquitous mitochondrially targeted enhanced green fluorescent protein (EGFP) and a cytosolic red fluorescent protein (dsRed) under the control of a hepatic promoter. In this way, we could analyse mitochondria only where the red fluorescence was decorating the liver. When comparing heterozygous and homozygous siblings at 6 dpf and 12 dpf (Fig. 2B), we did not observe any difference in the total volume of hepatocytes (data not shown) and liver mitochondria (Fig. 2C).
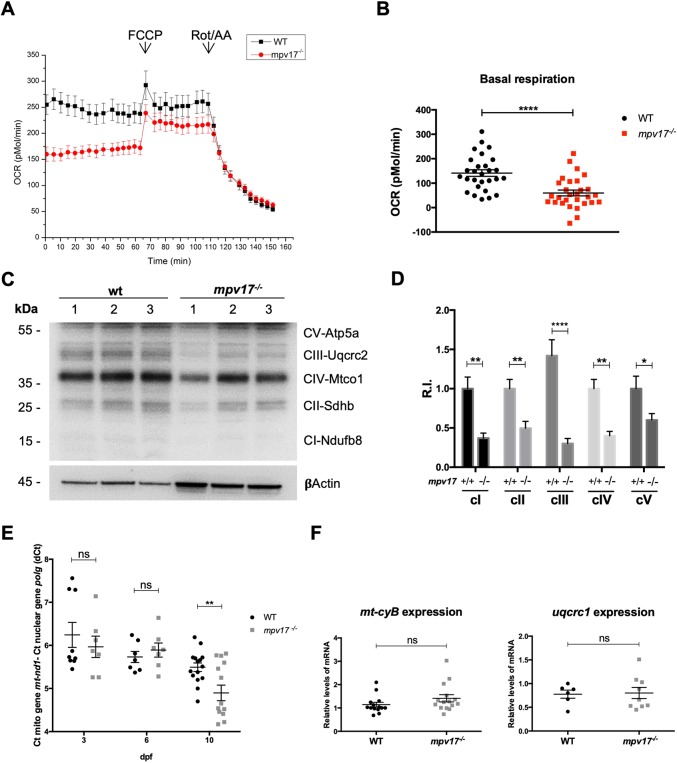
Fig. 2.
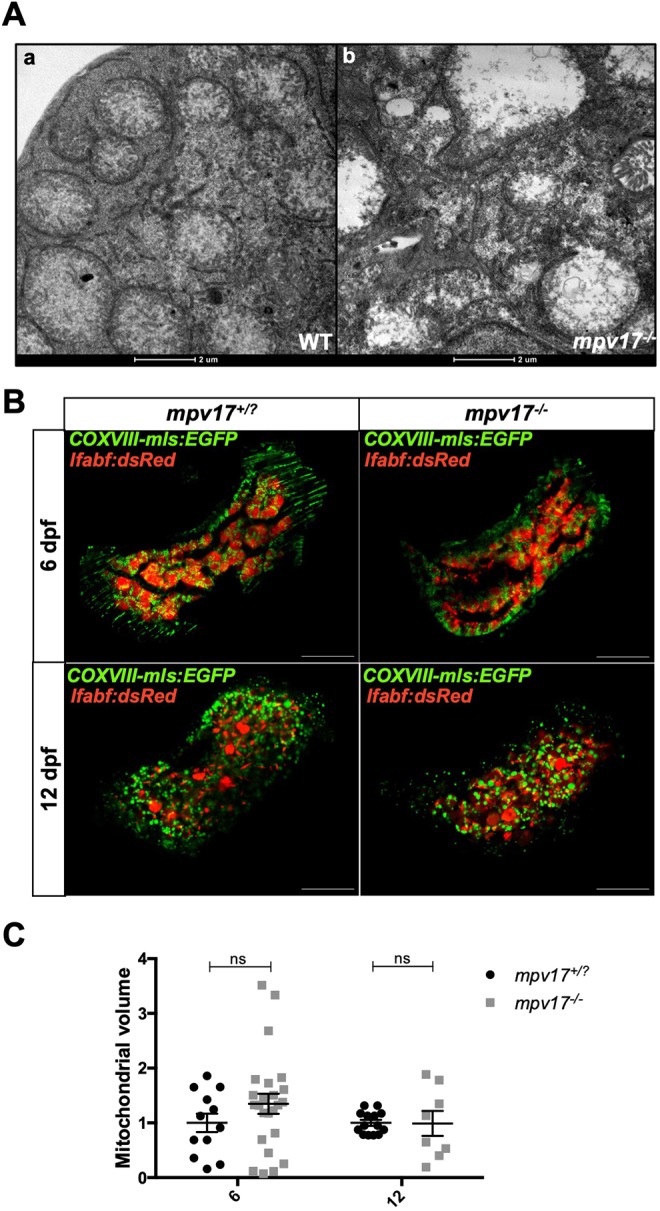
Analysis of mitochondrial ultrastructure and volume in mpv17−/− and wild-type liver at 6 dpf. (A) TEM analysis of wild-type (a) and mpv17−/− liver hepatocytes (b) (n=3). Scale bars: 2 µM. (B) Confocal images of double Tg(COXVIII-mls:EGFP); Tg(lfabp:dsRed) mpv17+/? and mpv17−/− siblings at 6 dpf (n=13) and 12 dpf (n=8). Scale bars: 50 µm. (C) Relative quantification of integrated density. Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ns, not significant. Experiments were performed in biological replicates.
To understand whether loss of mitochondrial normal morphology in mpv17 KO mutants was affecting respiratory complexes (RCs), we performed an in vivo analysis of the oxygen consumption rate (OCR) in 4 dpf larvae (Fig. 3A). We detected a significant reduction in the basal respiration level in mpv17−/− mutants compared with wild type (Fig. 3B), thus confirming the impairment of oxidative phosphorylation (OXPHOS) in larvae lacking Mpv17. To enquire whether the RC dysfunction observed was due to a depletion of OXPHOS subunits and complexes, we performed a western blot analysis (Fig. 3C). The cellular amount of all investigated subunits of RC complexes was lower in mpv17−/− mutants (Fig. 3D); the nuclear-encoded Uqcrc2b, a subunit of complex III, appeared the most affected.
Fig. 3.
Biochemical and genetic characterization of mitochondrial phenotype of mpv17 KO larvae. (A) Basal OCR was measured for ∼1 h in fish water, immediately after 4 dpf larvae were exposed to 0.5 μM FCCP and, later, to a combination of 2 μM rotenone (Rot) and 5 μM antimycin (AA). Four independent experiments were performed (n=40). (B) Quantification of basal respiration in wild-type and mpv17−/− mutants (n=40). (C) Protein blot analysis of different subunits of OXPHOS complexes. (D) Relative quantification of protein amount, using an antibody against βActin for standardization (n=3). R.I., relative intensity. (E) Relative quantification of mtDNA copy number in wild type and mpv17 homozygous mutants. Mean dCt values were calculated as Ct of mt-nd1 (mitochondrially encoded gene) minus Ct of polg (nuclear gene) and plotted with s.e.m. (n=7). (F) Real-time PCR quantification of mRNA transcripts from Ubiquinol-cytochrome c reductase complex subunits (n=6). Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ****P<0.0001; **P<0.01; *P<0.05; ns, not significant. Experiments were performed in biological replicates.
To test whether mtDNA depletion was the cause of the observed reduction in OXPHOS complexes, we assessed mtDNA copy number in mpv17 homozygous mutants by performing RT-qPCR analysis of DNA extracted from whole larvae. Surprisingly, for a MDS gene, we could not find a significant reduction in mtDNA content in mpv17−/− larvae at 6 dpf, a time point at which the morphological and respiratory phenotypes were already noticeable (Fig. 3E). However, in 10 dpf mpv17−/− larvae, we did detect a significant decrease in mtDNA content, thus validating the zebrafish mpv17 KO mutant as a model for MPV17-related MDS.
To test whether the OXPHOS reduction was due to defective gene transcription, we investigated, by RT-qPCR, the expression levels of a nuclear- and a mitochondrial-encoded subunit of complex III. No difference was observed in nuclear and mitochondrial transcription when comparing wild-type and mpv17−/− KO larvae at 6 dpf (Fig. 3F). From these analyses, we can conclude that OXPHOS complex reduction is not due to a defective transcription. Rather, mitochondrial cristae disruption impairs supercomplex localization and functionality (Stuart, 2008), leading to a defective assembly/stability or translation of respiratory complexes.
Finally, to investigate a possible activation of stress response machinery in mpv17−/− mutant mitochondria, we focused on Grp75 (also known as Hspa9), a well-studied cytoprotective factor against oxidative stress (Kaul et al., 2007). Using western blot technique, we found a significant increase in Grp75 in mpv17 KO mutants compared with wild type at 6 dpf (Fig. 4A). In addition, knowing that the mitochondrial contact site and cristae organizing system (MICOS) acts as a membrane-shaping and -connecting scaffold (Schorr and van der Laan, 2018), we tested whether the loss of mitochondrial ultrastructure might activate the quality control system of the organelle. Thus, we checked, by RT-qPCR, the expression levels of several MICOS subunits: mic13, mic19a, mic19b and mic60. As a result, we detected a specific transcriptional upregulation of mic19a in mpv17 KO larvae at 6 dpf (Fig. 4B).
Fig. 4.
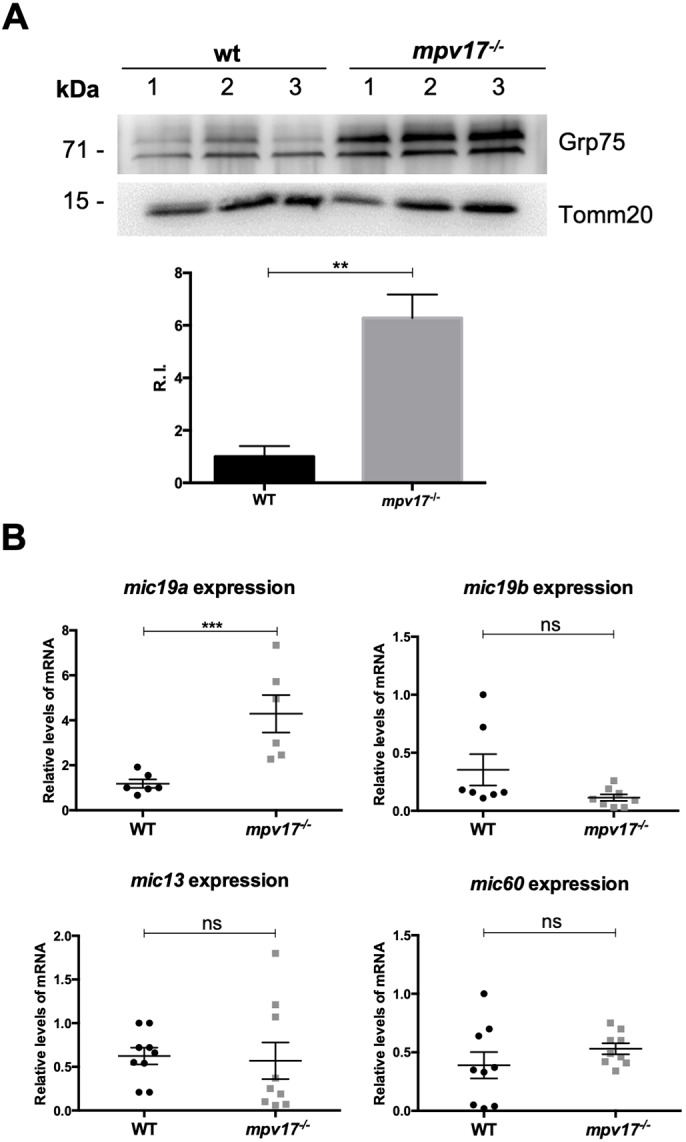
Evaluation of stress response and mitochondrial quality control system in zebrafish larvae. (A) Representative western blot analysis of Grp75 protein from 6 dpf zebrafish larvae and relative quantification, using an antibody against Tomm20 (also known as Tomm20b) for standardization (n=3). (B) Real-time PCR quantification of mRNA transcripts from different MICOS subunits at 6 dpf (n=8). Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ***P<0.001; **P<0.01; ns, not significant. Experiments were performed in biological replicates.
The pyrimidine de novo synthesis pathway is strictly linked to the iridophore phenotype in mpv17 null mutants
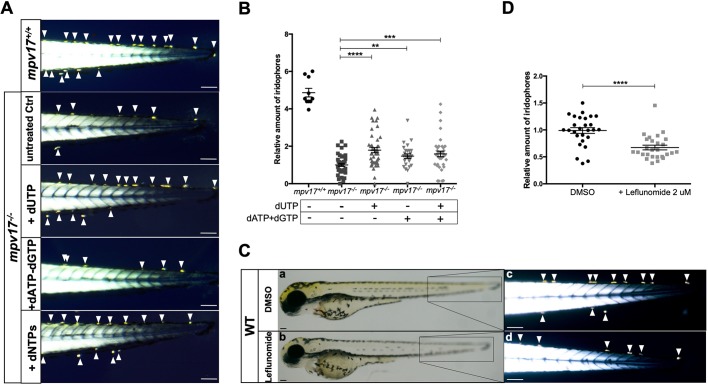
By performing a bioinformatics search on CORD platform to identify MPV17 co-regulated genes, we found that the trifunctional Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase and dihydroorotase (CAD) enzyme involved in pyrimidine production showed 66.48% of concordance in 176 experiments (Pearson coefficient 0.504 and P=1.1×10−28), among the highest scores for MPV17 co-regulated genes (Fig. S3). Remarkably, evidence of a possible connection between pyrimidine de novo pathway and the mpv17 mutant phenotype also comes from previous studies: the inhibition of Dhodh by leflunomide treatment is shown to affect iridophore and melanophore formation in zebrafish (White et al., 2011). In addition, by considering previous studies that suggest a role for Mpv17 in mitochondrial dNTP availability (Löllgen and Weiher, 2014), we decided to treat mpv17−/− embryos at 8 h post-fertilization (hpf) with different deoxynucleotide mixtures and to check the amount of iridophores at 3 dpf. Treating the embryos with a solution containing all dNTPs was able to increase iridophore number (Fig. 5A), and dUTP administration had the best effect in ameliorating the mpv17 KO phenotype (Fig. 5B).
Fig. 5.
Investigation of dNTP metabolism in zebrafish larvae. (A) Visualization of iridophores in the tail region of 3 dpf larvae treated with different mixtures of dNTPs. (B) Relative quantification of iridophore number (n=27). (C) Wild-type embryos were treated with dimethyl sulfoxide and 2 μM leflunomide, inhibitor of pyrimidine biosynthesis. Whole-body bright-field pictures (a,b) and birefringent images of the tail region (c,d) are shown. (D) Quantification of relative amounts of iridophores (n=27). Arrowheads point to iridophores. Scale bars: 100 µm. Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ****P<0.0001; ***P<0.001; **P<0.01. Experiments were performed in biological replicates.
Consequently, we decided to investigate whether the mitochondrial step of pyrimidine de novo synthesis was affected in mpv17 mutants. The conversion of dihydroorotic acid (DHOA) into orotic acid (OA) is a metabolic reaction catalysed by Dhodh, the only mitochondrial enzyme involved in pyrimidine de novo synthesis. Hence, we hypothesized that Dhodh functionality might be partially impaired in mpv17 null mutants. Notably, in accordance with the results by White et al. (2011), we found a specific effect of leflunomide on iridophore formation at low doses; melanophores correctly develop, confirming that inhibition of Dhodh significantly impairs iridophore growth at 3 dpf (Fig. 5C,D).
OA treatment restores iridophore formation and increases mtDNA copy number in mpv17 null mutants
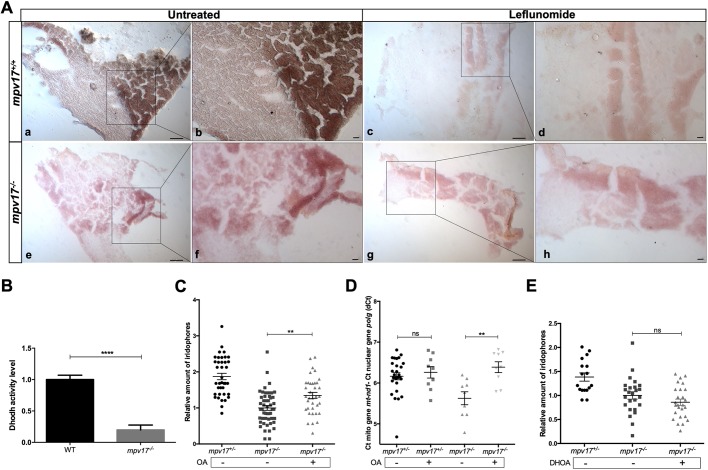
To test a connection between Dhodh impairment and the mpv17 mutant phenotype, we assessed Dhodh activity by performing a histochemistry assay on liver cryosections of wild-type and mpv17−/− adults. We decided to perform this analysis on hepatic tissue because Dhodh is highly active in liver, as reported by Löffler et al. (1996), and also because we had found a severe disruption of hepatic mitochondria in mpv17 KO larvae. Interestingly, we observed significantly lower Dhodh enzymatic activity in mpv17 null mutants compared with wild type, an activity similar to that of leflunomide-treated samples (Fig. 6A). The biochemical assay confirmed Dhodh impairment in the lysate of mpv17 KO larvae at 6 dpf (Fig. 6B). In particular, the specific activity of Dhodh in wild type (average value 4534 nmol/min per mg protein), highly similar to that measured in rat liver by Löffler et al. (1996), decreased to 20% in mpv17 null mutant larvae (average value of 1848 nmol/min per mg protein). These results highlight that the functionality of Dhodh mitochondrial enzyme is reduced at early stages of development and indicate that the impairment of Dhodh enzymatic activity might be the main cause of premature loss of iridophores in mpv17 null mutants.
Fig. 6.
Analysis of the link between Dhodh functionality and the mpv17 KO phenotype. (A) Histochemical Dhodh assay on cryosections of adult wild-type (a-d) and mpv17 KO (e-h) liver. The activity assay was performed on the same slide and a reaction mix containing 100 μM leflunomide was used as negative control (c,d,g,h). Pictures were taken at lower (a,c,e,g) and higher (b,d,f,h) magnification. Scale bars: 100 µm (n=3). (B) Dhodh biochemical assay in 6 dpf larvae. Dhodh was assessed in zebrafish homogenized through spectrophotometric recordings following the reduction of 2,6-dichlorophenolindophenol (DCPIP). Specific Dhodh activity was obtained by subtracting the leflunomide-independent fraction and measured as nmol/min per mg of protein. Values were then normalized with respect to wild-type controls. Statistical analysis was performed using paired Student's t-test (n=3). (C,D) Relative quantification of iridophore amount in 3 dpf larvae treated with 1 μM orotic acid (OA) (n=36) (C) and mtDNA copy number analysis (n=8) (D). (E) Relative evaluation of iridophore number in 3 dpf larvae treated with 100 μM dihydroorotic acid (DHOA) (n=17). Statistical analyses were performed using two-tailed Student's t-test. Statistical significance was evaluated by setting a confidence interval of 95%; data are mean±s.e.m. ****P<0.0001; **P<0.01; ns, not significant. Experiments were performed in biological replicates.
In order to examine in vivo the mitochondrial reaction of the pyrimidine metabolic pathway, we tested whether administration of OA, the product of Dhodh reaction, might rescue the mpv17−/− mutant phenotype (Fig. S4A). When OA was injected into the yolk sac of 20 hpf embryos we could observe a clear rescue of iridophores in mpv17 null larvae at 3 dpf (Fig. 6C). In addition, while mpv17 mutants displayed a constant, although non-significant, decrease in mtDNA copy number at 3 dpf, mpv17−/− larvae treated with OA exhibited increased mtDNA copy number, with no effect on heterozygotes (Fig. 6D).
Finally, we tested the effect of DHOA (the substrate of Dhodh enzyme) in mpv17 mutants. As expected, L-DHOA injected at 20 hpf into mpv17 mutant embryos had no effect on the phenotype, thus confirming Dhodh deficiency in mpv17 null mutants (Fig. 6E; Fig. S4B).
DISCUSSION
Our work validates roy as a model for investigating the role of mpv17 and a powerful tool for developing new therapeutic strategies against MPV17-related MDS. First, as already reported by D'Agati et al. (2017), we confirmed that zebrafish mpv17 mRNA can rescue the mpv17−/− mutant phenotype, and, most importantly, that human MPV17 mRNA also has similar properties in vivo, thus supporting a conserved function of the two orthologues; this finding is also in agreement with complementation assays in the yeast sym1Δ model (Trott and Morano, 2004), showing a functional link between Sym1 and mammalian MPV17. In addition, a recent work demonstrated that the growth of sym1Δ strain transformed with R51Q (hR50Q) mutant Sym1-HA allele was impaired (Gilberti et al., 2018). In agreement, also in zebrafish, the injection of the p.R50Q mutated form of human MPV17 mRNA resulted in a mild rescue of the mpv17−/− KO phenotype.
Considering that mutants showing minor phenotypical features might exhibit a compensatory upregulation of other members of the gene family (Rossi et al., 2015), we investigated possible feedback mechanisms acted by mpv17 paralogous genes, also considering that both paralogous proteins (MPV17L and MPV17L2) were previously reported to be mitochondrial proteins (Dalla Rosa et al., 2014; Krick et al., 2008). We could observe increased expression of mpv17l2 in mpv17 null mutants. Moreover, overexpression of Mpv17L2 was able to rescue the chromatophore phenotype of mpv17−/− mutants. These results support the idea of partial overlapping cellular functions for Mpv17 and Mpv17l2, at least in zebrafish. This finding contrasts with a previous work, carried out in human cellular models, that proposes different roles for MPV17, claimed to be involved in purine synthesis (Dalla Rosa et al., 2016), and MPV17L2, suggested to play a role in mitochondrial ribosome assembly (Dalla Rosa et al., 2014). On the other hand, the tissue-restricted mpv17l2 overexpression we detected was not able to prevent the strong mitochondrial phenotype assessed in mpv17 zebrafish mutants. Thus, the role of mpv17l2 in mitochondria is still an open question. In addition, we did not observe any significant effect of mpv17l2 mRNA injection on the mtDNA stability of mpv17 KO larvae. A possible explanation is the lack of significant mtDNA replication, unnecessary at early stages of development (Artuso et al., 2012), thus hampering the evaluation of the role of mpv17l2 at these time points. Because of that, stable lines overexpressing mpv17l2 in an mpv17−/− mutant background might be useful tools to follow its effect at later stages of development. Moreover, it is important to underline that the injection of mpv17l2 mRNA leads to non-physiological and ubiquitous expression of Mpv17l2, which is normally not expressed in neural-crest-derived cells, but rather restricted to liver tissue and the digestive system at later stages (Thisse and Thisse, 2004) (Fig. S1). This tissue-specific expression of mpv17l2 might be the reason why mpv17 KO mutants, although showing significant overexpression of mpv17l2, still lack iridophores, while they do not display any overt hepatic defect at the anatomical level.
In the past two decades, different MPV17-related MDS models have been developed, all showing phenotypes that are only partially representative of human clinical features. Whereas the Mpv17 KO mouse strain mainly shows glomerulosclerosis and inner ear defects (Löllgen and Weiher, 2014), the zebrafish mpv17 null mutant phenotype is mostly characterized by a specific lack of iridophores and a progressive loss of melanophores, both derived from the neural crest. This phenotype perfectly matches the Mpv17−/− mouse premature greying (Viscomi et al., 2009) and the progressive demyelinating peripheral neuropathy found in 38% of MPV17 mutant patients (El-Hattab et al., 2018), features that we can consider possible neurocristopathies.
Notably, in this work, we demonstrate that zebrafish mpv17 mutant larvae display a severe mitochondrial phenotype, characterized by disappearance of mitochondrial cristae in liver hepatocytes, impairment and reduction of RC complexes, mtDNA depletion and activation of mitochondrial stress response. Alterations of the mitochondrial ultrastructure were previously found in Mpv17 KO mouse liver (Viscomi et al., 2009) and in a yeast sym1Δ model (Dallabona et al., 2010). Moreover, Dallabona and colleagues also suggested that Sym1 is primarily involved in mitochondrial ultrastructure maintenance (Dallabona et al., 2010), taking into account that the loss of cristae architecture affects mtDNA stability and supercomplex arrangement within inner mitochondrial membrane (Stuart, 2008). Our findings strongly support this hypothesis as we were able to observe a significant reduction in mtDNA copy number only at 10 dpf, while mitochondrial ultrastructure and RC complexes were already impaired at 6 dpf. Moreover, a tissue-specific dysfunction in OXPHOS complexes is frequently found in patients affected by different mutations in the MPV17 gene (Spinazzola et al., 2006; Uusimaa et al., 2014; El-Hattab et al., 2018). Taken together, our results prove that the mpv17 null mutant perfectly resembles the human mitochondrial phenotype, thus indicating that roy is a powerful model for MPV17-related MDS.
Additionally, the overproduction of Grp75 and the overexpression of mic19a we found in mpv17−/− mutants highlight the importance of mpv17 in preserving mitochondrial homeostasis, suggesting that, in the absence of Mpv17, protective mechanisms act against oxidative stress and loss of mitochondrial morphology. Interestingly, the grp75 yeast orthologue gene, SSC1, was also found upregulated at the transcriptional level in the yeast mpv17 orthologue mutant sym1Δ (Trott and Morano, 2004). Moreover, it is indeed well-established that injured mitochondria trigger ROS production via further electron escape from impaired electron transport chain (Maharjan et al., 2014); notably, enhanced ROS levels were previously reported in mice and cell lines depleted of MPV17 (Wagner et al., 2005; Shvetsova et al., 2017).
MPV17 was discovered about 30 years ago; however, its function remains unknown. To date, the leading hypothesis is that MPV17 may represent an inner mitochondrial membrane protein involved in nucleotide supply for mtDNA synthesis (Löllgen and Weiher, 2014). As for other MDS-related mutations, MPV17 mutations might impair deoxyribonucleotide metabolism, leading to limited dNTP availability and subsequent mtDNA depletion. Indeed, administration of dNTPs is a therapeutic strategy for treatment of MDS-like mitochondrial neurogastrointestinal encephalopathy (MNGIE) syndrome (Cámara et al., 2014). It was previously reported that supplementation with different amino acids, all precursors in the synthesis of nucleotides, partially restored aerobic oxidative growth in the yeast sym1Δ strain (Dallabona et al., 2010). Coherently, we found that deoxyribonucleotide administration strongly increased iridophore number in mpv17−/− larvae. In particular, because dUTP showed the best effect, we focused on pyrimidine de novo synthesis. Serendipitously, by performing a bioinformatics analysis, we found that MPV17 and CAD, a cytosolic enzyme involved in pyrimidine production, are highly co-regulated. Moreover, we observed that a mild inhibition of Dhodh enzyme specifically impaired iridophore development and that Dhodh activity is significantly reduced in mpv17−/− mutants, in agreement with the well-established link between the Dhodh-mediated mitochondrial step of pyrimidine production and neural crest cell formation (White et al., 2011). Finally, administration of OA, but not DHOA, significantly rescued the phenotype of mpv17−/− larvae. Taken all together, our data indicate insufficient pyrimidine synthesis and impairment of Dhodh in mpv17−/− null mutants, possibly the cause of their main phenotypical trait. Whether this is due to the disruption of mitochondrial cristae, in which the enzyme is located, or to a possible interaction between Dhodh and Mpv17, needs to be further investigated. Most remarkably, the ability of OA (considered in the past vitamin B13) to increase mtDNA content in mpv17 KO mutants, the availability of this nutrient in dairy products and its current use as a food supplement open new opportunities in the treatment of MPV17-related MDS, to date a disease with poor prognosis and no cure yet available.
MATERIALS AND METHODS
Zebrafish husbandry and maintenance
All experiments were performed in accordance with the European and Italian Legislations and with permission for animal experimentation from the Local Ethics Committee of the University of Padova. Danio rerio (zebrafish) were maintained in a temperature-controlled (28.5°C) environment and fed as described by Kimmel et al. (1995). The roy strain was originally obtained from the Harvard Medical School (Boston, USA), and genotyping at early stages was performed by PCR with the following primers: forward, 5′-AACCGTTTGTCATAATGTGGC-3′; reverse-wt, 5′-CATGGGTGTTTGGCCATCAG-3′; reverse-del, 5′-ACATCAACTACATGGGTGGG-3′.
For in vivo studies, the following transgenic lines, crossed with roy, were used: Tg(lfabf:dsRed; elaA:EGFP), generated by Korzh et al. (2008); Tg(COXVIII-mls:EGFP), generated in our laboratory by N.F., using a construct kindly provided by the Seok-Yong laboratory (Chonnam National University Medical School, Gwangju, Republic of Korea), containing the mitochondrial localization sequence of subunit VIII of human cytochrome c oxidase (COXVIII; also known as COX8A).
RNA extraction and RT-qPCR
Total mRNA was isolated from 50 larvae using Trizol (Invitrogen) and reverse-transcribed using SuperScript® III First-Strand (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed using a Rotor-gene Q (Qiagen) and 5× HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis BioDyne), following the manufacturers’ protocols. The cycling parameters were 95°C for 14 min, followed by 45 cycles at 95°C for 15 s, 59°C for 25 s and 72°C for 20 s. Threshold cycles (Ct) and dissociation curves were generated automatically by Rotor-Gene Q series software. Primer sequences are listed in Table 1. Sample Ct values were normalized with Ct values from zebrafish gapdh.
Table 1.
Zebrafish gene-specific primers used for qPCR analyses
Whole-mount ISH
According to Thisse and Thisse (2004), the full-length sequence of mpv17l2 (ENSDARG00000056367) was PCR amplified from liver complementary DNA (cDNA) of adult zebrafish, using the following primers, designed by Primer3 software (http://primer3.ut.ee): mpv17l2 probe-F, 5′-CGACTCATGTTGGCTGCTC-3′; mpv17l2 probe-R, 5′-GCCCAGCGTATGTCACAAAT-3′. The PCR products were purified, then cloned into pCR2.1 TOPO vector (TOPO TA Cloning Dual Promoter Kit, Stratagene) and sequence verified. For riboprobe in vitro transcription, the constructs were linearized with the appropriate endonuclease enzyme (Promega) and transcribed with DIG-labelling mix and RNA polymerase (Roche): for antisense (working) riboprobe, HindIII/T7 RNApol; for sense (negative control) probe, NotI/SP6 RNApol. Whole-mount ISH was performed on zebrafish larvae at 3 dpf, previously fixed with 4% paraformaldehyde (PFA)/PBS and stored in 100% methanol, following standard protocols. At least 20 larvae per condition were processed in a single tube. For signal comparison, wild-type control and mpv17−/− mutant larvae were co-processed and co-stained in the same tube; controls were recognized by tail tip excision, performed after PFA fixation and before whole-mount ISH.
mRNA rescue
The full-length mpv17 and mpv17l2 cDNAs from wild-type strain and human MPV17 cDNA from pCR2.1 TOPO vector, kindly provided by the Zeviani laboratory (University of Cambridge, Cambridge, UK), were subcloned into the pCS2+ vector. The human MPV17 p.R50Q (CGG>CAG) mutated form was generated, starting from wild-type construct, using a Q5® Site-Directed Mutagenesis Kit (New England Biolabs) and the following primers: forward, 5′-CAGAGAGGCCAGACTCTGACCATG-3′; reverse, 5′-GTGTTCCTGCAGACCCCG-3′, both designed by NEBaseChanger online tool (New England Biolabs). Capped mRNA was transcribed using the mMessage mMachine® kit (Thermo Fisher Scientific) according to the manufacturer's protocol. The mRNA was resuspended in water and microinjected into one-cell mpv17−/− embryos at 70 ng/μl.
Iridophore count
Iridophore number was measured with the aid of two polarizing filters, basing on the physical property of these cells of being birefringent. Larvae at 3 dpf were anaesthetized with tricaine (0.16 mg/ml), placed on a glass, and the top filter was twisted until it was possible to see the light refracting through the striated muscle. Birefringence analysis was performed according to published methods (Smith et al., 2013). Samples were photographed using a Leica MDG41 microscope and a Nikon Digital Sight DS Fi2 camera; iridophores were manually counted in the tail region and their amount was normalized to that of controls.
Relative quantification of mtDNA content in whole larvae and sections
DNA samples, extracted from 20 pooled larvae or sections at 3, 6 and 10 dpf, were prepared as described by Rahn et al. (2015) and quantified by PicoGreen assay (Invitrogen) using the Infinite 200 PRO plate reader (Tecan). Then, 8.5 ng of each sample was used in a qPCR reaction and run in triplicate, using 5× HOT FIREPol® EvaGreen® qPCR Supermix (Solis Biodyne). mtDNA copy number was determined by the RT-qPCR standard curve method using mt-nd1 as the mitochondrial gene target and the polg gene as a reference for nDNA, following the protocol developed by Artuso et al. (2012). The delta Ct (dCt) value was calculated by subtracting the Ct of polg from the Ct of mt-nd1 for each sample.
TEM analysis
Larvae were anaesthetized and fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, then dehydrated, embedded in epoxy resin and sectioned following a standard TEM sample preparation protocol. All steps after fixation were performed by the TEM service of the Department of Biology (University of Padova).
Confocal analysis and mitochondrial volume measurements
Double transgenic fluorescence of Tg(lfabf:dsRed; COXVIII-mls:EGFP) in mpv17−/− mutant background was visualized under a Leica M165FC dissecting microscope and then with a Nikon C2 H600 L confocal microscope. Larvae were anaesthetized and mounted in 1% low-melting-point agarose gel. Mitochondrially targeted EGFP and cytosolic dsRed fluorescence was visualized by using 488 nm and 561 nm lasers, through a 40× immersion objective (Nikon). All images were analysed with Fiji software (https://fiji.sc/). Mitochondria were automatically segmented on the EGFP fluorescence image stacks, inside a dsRed fluorescent mask identifying hepatic tissue, and total mitochondrial volume was calculated, according to previous work (Facchinello et al., 2016). After confocal acquisition, heterozygous and homozygous siblings were genotyped by PCR on DNA previously extracted from a single larva, as described by Gagnon et al. (2014).
Measurement of OCR
OCR was measured in zebrafish larvae at 96 hpf with a Seahorse XF24 extracellular flux analyser. Larvae were placed into the XF24 microplate well (one larva per well) and blocked with a capture screen in the presence of 670 μl fish water (0.5 mM NaH2PO4, 0.5 mM NaHPO4, 3 mg/l instant ocean). Basal respiration was measured for 63 min at 28.5°C, while the maximal respiration was measured upon administration of 0.5 μM carbonyl cyanide-4-phenylhydrazone (FCCP). A mixture of 2 μM rotenone (Rot) and 5 μM antimycin A was added to shut down electron transport chain function, revealing the non-mitochondrial respiration. Basal respiration was obtained by subtracting non-mitochondrial respiration. Respiratory rates are the average±s.e.m. of at least 20 individual larvae per condition.
Western blot analysis
Proteins were extracted from 30 pooled larvae at 6 dpf. Lysis was performed in ice-cold RIPA buffer [125 mM NaCl, 25 mM Tris-HCl pH 7.4, 1 mM EGTA-Tris pH 7.4, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and complete EDTA-free protease inhibitor cocktail (Roche)]. Crude lysate was cleared by centrifuging for 30 min at 15,000 g and proteins in the supernatant were quantified using a BCA Protein Assay Kit (Pierce). Equal amounts of proteins (50 μg) were loaded on 4-12% Bis-Tris NuPage gels (Life Technologies) and blotted on PVDF Immobilon-P membranes (Millipore). Dried membranes were then washed with TBS buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl) with 1% (w/v) bovine serum albumin (BSA; Sigma-Aldrich) and incubated overnight with the indicated primary antibodies at 4°C: Mitoprofile® Human WB Total OXPHOS cocktail antibody (1:100; Abcam, ab110411), anti-Grp75 antibody (1:1000; Santa Cruz Biotechnology, H-155), anti-Actin antibody (1:1000; Santa Cruz Biotechnology, AC-15) or anti-Tomm20 antibody (1:10,000; Sigma-Aldrich, HPA011562). Secondary horseradish-peroxidase-conjugated antibodies (Bio-Rad) were incubated for 1 h at room temperature and protein bands were detected by chemiluminescence on a UVITEC Alliance Mini HD9. Quantitation of the signal was performed with ImageJ software (https://imagej.nih.gov/ij/).
Compound administration
Deoxynucleotide Set (SLBH8759), OA (O2750) and L-DHOA (D7128) were obtained from Sigma-Aldrich. dNTPs (12.5 µM each) were added to embryo medium at 8 hpf, and OA (1 µM) and L-DHOA (100 µM) were injected into the yolk sac of 20 hpf embryos as described by Willer et al. (2005). Controls were injected at 20 hpf, with a control solution containing NaOH or dimethylformamide, and phenotypic rescue was evaluated at 3 dpf.
Dhodh activity assays
To measure Dhodh activity, 6 dpf larvae were homogenized in a buffer composed of 100 mM HEPES pH 7.5, 150 mM NaCl, 0.01% Triton X-100, protease and phosphatase inhibitors. Cell homogenates (60 μg) were pre-incubated for 10 min with the substrate L-DHOA (10 mM) and alamethicin (2 mM), and separately with the Dhodh inhibitor leflunomide (200 μM). After the pre-incubation time, sodium azide (250 mM), antimycin A (2 mM), rotenone (2 mM), 3-nitropropionic acid (2 mM), Coenzyme Q1 (10 mM) and 2,6-dichlorophenolindophenol (DCPIP; 5 mM) were added. Dhodh activity was assessed at 30°C through spectrophotometric recordings following the reduction of DCPIP at 600 nM (ε=19.1×mM−1×cm−1). Values were normalized for protein amount.
Dhodh enzymatic activity was also histochemically assessed on frozen tissue. The sections were incubated for 30 min at 37°C with a medium containing HEPES (150 mM) pH 8, L-DHOA (10 mM), sodium azide (1 mM) and Nitro Blue Tetrazolium (NBT) (1 mM). The Dhodh inhibitor leflunomide (200 µm) was included in the test. Successively, the sections were rinsed with a wash solution containing HEPES (150 mM) pH 8 and water and covered using a mounting medium. The DHOA was oxidized to OA, producing FMNH2, and the reaction was visualized by chemically reacting the electron acceptor with the purple salt NBT. Samples were photographed using a Leica DMR microscope equipped with a Leica DFC7000T digital camera.
Statistics
Prism software (GraphPad) was used for data analysis and figure plotting. All experiments were performed at least in biological triplicate. Standard error of the mean calculations and all statistical analyses were performed using Student's t-test.
Supplementary Material
Acknowledgements
We thank Dr Carlo Viscomi for providing roy mutants; and the Zebrafish Facility of the Biology Department at the University of Padova, Martina Milanetto, Marco Schiavone, Luigi Pivotti and Andrea Vettori for technical support. We are deeply indebted to Rosario Rizzuto, Paolo Bernardi and Giorgia Pallafacchina for their kind support in this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: F.A.; Methodology: L.M., M.P., E.L., N.F., G.M., F.A.; Software: L.M., E.L.; Validation: L.M., C.L., F.A.; Formal analysis: M.P., F.A.; Investigation: L.M., M.P., C.L., E.L., N.F., G.M.; Resources: A.R., F.A.; Writing - original draft: L.M., F.A.; Writing - review & editing: L.M., N.T., D.G., F.A.; Visualization: L.M., C.L., N.F., G.M.; Supervision: E.L., A.R., D.G., F.A.; Project administration: N.T., F.A.; Funding acquisition: A.R., D.G., F.A.
Funding
This work was supported by Fondazione Telethon (GPP11011) and Fondazione Cassa di Risparmio di Padova e Rovigo. A.R. is supported by Università degli Studi di Padova, Neurofibromatosis (Therapeutic Acceleration Program) and Associazione Italiana per la Ricerca sul Cancro (IG 2017/20749).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.037226.supplemental
References
- Artuso L., Romano A., Verri T., Domenichini A., Argenton F., Santorelli F. M. and Petruzzella V. (2012). Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim. Biophys. Acta 1817, 1002-1011. 10.1016/j.bbabio.2012.03.019 [DOI] [PubMed] [Google Scholar]
- Calvo S. E., Clauser K. R. and Mootha V. K. (2016). MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251-D1257. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara Y., González-Vioque E., Scarpelli M., Torres-Torronteras J., Caballero A., Hirano M. and Martí R. (2014). Administration of deoxyribonucleosides or inhibition of their catabolism as a pharmacological approach for mitochondrial DNA depletion syndrome. Hum. Mol. Genet. 23, 2459-2467. 10.1093/hmg/ddt641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P. F. and Hudson G. (2013). Mitochondrial genetics. Br. Med. Bull. 106, 135-159. 10.1093/bmb/ldt017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agati G., Beltre R., Sessa A., Burger A., Zhou Y., Mosimann C. and White R. M. (2017). A defect in the mitochondrial protein Mpv17 underlies the transparent casper zebrafish. Dev. Biol. 430, 11-17. 10.1016/j.ydbio.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallabona C., Marsano R. M., Arzuffi P., Ghezzi D., Mancini P., Zeviani M., Ferrero I. and Donnini C. (2010). Sym1, the yeast ortholog of the MPV17 human disease protein, is a stress-induced bioenergetic and morphogenetic mitochondrial modulator. Hum. Mol. Genet. 19, 1098-1107. 10.1093/hmg/ddp581 [DOI] [PubMed] [Google Scholar]
- Dalla Rosa I., Durigon R., Pearce S. F., Rorbach J., Hirst E. M. A., Vidoni S., Reyes A., Brea-Calvo G., Minczuk M., Woellhaf M. W. et al. (2014). MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 42, 8500-8515. 10.1093/nar/gku513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Rosa I., Cámara Y., Durigon R., Moss C. F., Vidoni S., Akman G., Hunt L., Johnson M. A., Grocott S., Wang L. et al. (2016). MPV17 Loss Causes Deoxynucleotide Insufficiency and Slow DNA Replication in Mitochondria. PLoS Genet. 12: e1005779 10.1371/journal.pgen.1005779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab A. W., Wang J., Dai H., Almannai M., Staufner C., Alfadhel M., Gambello M. J., Prasun P., Raza S., Lyons H. J. et al. (2018). MPV17-related mitochondrial DNA maintenance defect: new cases and review of clinical, biochemical, and molecular aspects. Hum. Mut. 39, 461-470. 10.1002/humu.23387 [DOI] [PubMed] [Google Scholar]
- Facchinello N., Schiavone M., Vettori A., Argenton F. and Tiso N. (2016). Monitoring Wnt Signaling in Zebrafish Using Fluorescent Biosensors. Methods Mol. Biol. 1481, 81-94. 10.1007/978-1-4939-6393-5_9 [DOI] [PubMed] [Google Scholar]
- Frohnhöfer H. G., Krauss J., Maischein H. M. and Nüsslein-Volhard C. (2013). Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development 140, 2997-3007. 10.1242/dev.096719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Akhmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberti M., Baruffini E., Donnini C. and Dallabona C. (2018). Pathological alleles of MPV17 modeled in the yeast Saccharomyces cerevisiae orthologous gene SYM1 reveal their inability to take part in a high molecular weight complex. PLoS ONE 13, e0205014 10.1371/journal.pone.0205014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon C. W., Mitra R. D. and Johnson S. L. (2013). Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS ONE 8, e67801 10.1371/journal.pone.0067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadimas C. L., Vu T. H., Holve S. A., Chronopoulou P., Quinzii C., Johnsen S. D., Kurth J., Eggers E., Palenzuela L., Tanji K. et al. (2006). Navajo neurohepatopathy is caused by a mutation in the MPV17 gene. Am. J. Hum. Genet. 79, 544-548. 10.1086/506913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S. C., Deocaris C. C. and Wadhwa R. (2007). Three faces of mortalin: a housekeeper, guardian and killer. Exp. Gerontol. 42, 263-274. 10.1016/j.exger.2006.10.020 [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Brand M., Jiang Y. J., Heisenberg C. P., Lin S., Haffter P., Odenthal J., Mullins M. C., van Eeden F. J., Furutani-Seiki M. et al. (1996). Zebrafish pigmentation mutations and the processes of neural crest development. Development 123, 369-389. [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Sosa K. C., Owen J. P., Yates C. A. (2017). Zebrafish adult pigment stem cells are multipotent and form pigment cells by a progressive fate restriction process: clonal analysis identifies shared origin of all pigment cell types. BioEssays 39, 1600234 10.1002/bies.201600234 [DOI] [PubMed] [Google Scholar]
- Kim J., Kang E., Kim Y., Kim J. M., Lee B. H., Murayama K., Kim G. H., Choi I. H., Kim K. M. and Yoo H. W. (2016). MPV17 mutations in patients with hepatocerebral mitochondrial DNA depletion syndrome. Mol. Genet. Metab. Rep. 8, 74-76. 10.1016/j.ymgmr.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. and Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Korzh S., Pan X., Garcia-Lecea M., Winata C. L., Pan X., Wohland T., Korzh V. and Gong Z. (2008). Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev. Biol. 8, 84 10.1186/1471-213X-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J., Astrinidis P., Frohnhöfer H. G., Walderich B. and Nüsslein-Volhard C. (2013). transparent, a gene affecting stripe formation in Zebrafish, encodes the mitochondrial protein Mpv17 that is required for iridophore survival. Biol. Open 2, 703-710. 10.1242/bio.20135132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick S., Shi S., Ju W., Faul C., Tsai S., Mundel P. and Böttinger E. P. (2008). Mpv17l protects against mitochondrial oxidative stress and apoptosis by activation of Omi/HtrA2 protease. Proc. Natl. Acad. Sci. USA 105, 14106-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler M., Becker C. and Schuster E. W. G. (1996). Catalytic enzyme histochemistry and biochemical analysis of dihydroorotate dehydrogenase/oxidase and succinate dehydrogenase in mammalian tissues, cells and mitochondria. Histochem. Cell Biol. 105:t119-t28. 10.1007/BF01696151 [DOI] [PubMed] [Google Scholar]
- Löllgen S. and Weiher H. (2014). The role of the Mpv17 protein mutations of which cause mitochondrial DNA depletion syndrome (MDDS): lessons from homologs in different species. J. Biol. Chem. 396, 13-25. [DOI] [PubMed] [Google Scholar]
- Maharjan S., Oku M., Tsuda M., Hoseki J. and Sakai Y. (2014). Mitochondrial impairment triggers cytosolic oxidative stress and cell death following proteasome inhibition. Sci. Rep. 4, 5896 10.1038/srep05896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes C. T., Shanske S., Tritschler H. J., Aprille J. R., Andreetta F., Bonilla E., Schon E. A. and DiMauro S. (1991). mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 48, 492-501. [PMC free article] [PubMed] [Google Scholar]
- Ng A., Uribe R. A., Yieh L., Nuckels R. and Gross J. M. (2009). Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development 136, 2601-2611. 10.1242/dev.038315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira C., Almeida L. S., Nesti C., Pezzini I., Videira A., Vilarinho L. and Santorelli F. M. (2014). Syndromes associated with mitochondrial DNA depletion. Ital. J. Pediatr. 40, 34 10.1186/1824-7288-40-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn J. J., Bestman J. E., Stackley K. D. and Chan S. S. L. (2015). Zebrafish lacking functional DNA polymerase gamma survive to juvenile stage, despite rapid and sustained mitochondrial DNA depletion, altered energetics and growth. Nucleic Acids Res. 43, 10338-10352. 10.1093/nar/gkv1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M. and Stainier D. Y. R. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230-233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Schorr S. and van der Laan M. (2018). Integrative functions of the mitochondrial contact site and cristae organizing system. Semin. Cell. Dev. Biol. 76, 191-200. 10.1016/j.semcdb.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Shvetsova A. N., Mennerich D., Kerätär J. M., Kalervo Hiltunen J. and Kietzmann T. (2017). Non-electron transfer chain mitochondrial defects differently regulate HIF-1α degradation and transcription. Redox Biol. 12, 1052-1061. 10.1016/j.redox.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. L., Beggs A. H. and Gupta V. A. (2013). Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J. Vis. Exp. 13, e50925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola A., Viscomi C., Fernandez-Vizarra E., Carrara F., D'Adamo P., Calvo S., Marsano R. M., Donnini C., Weiher H., Strisciuglio P. et al. (2006). MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 38, 570-575. [DOI] [PubMed] [Google Scholar]
- Stuart R. A. (2008). Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40, 411 10.1007/s10863-008-9168-4 [DOI] [PubMed] [Google Scholar]
- Suomalainen A. and Isohanni P. (2010). Mitochondrial DNA depletion syndromes--many genes, common mechanisms. Neuromuscul. Disord. 20, 429-437. 10.1016/j.nmd.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Thisse B. and Thisse C. (2004). Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. (http://zfin.org).
- Trott A. and Morano K. A. (2004). SYM1 is the stress-induced Saccharomyces cerevisiae ortholog of the mammalian kidney disease gene Mpv17 and is required for ethanol metabolism and tolerance during heat shock. Eukaryot. Cell. 3, 620-631. 10.1128/EC.3.3.620-631.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusimaa J., Evans J., Smith C., Butterworth A., Craig K., Ashley N., Liao C., Carver J., Diot A., Macleod L. et al. (2014). Clinical, biochemical, cellular and molecular characterization of mitochondrial DNA depletion syndrome due to novel mutations in the MPV17 gene. Eur. J. Hum. Genet. 22, 184-191. 10.1038/ejhg.2013.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscomi C., Spinazzola A., Maggioni M., Fernandez-Vizarra E., Massa V., Pagano C., Vettor R., Mora M. and Zeviani M. (2009). Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum. Mol. Genet. 18, 12-26. 10.1093/hmg/ddn309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G., Stettmaier K. and Bors W. (2005). Enhanced gamma-glutamyl transpeptidase expression and superoxide production in Mpv17−/− Glomerulosclerosis Mice. J. Biol. Chem. 382, 1019-1025. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359-407 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. M., Cech J., Ratanasirintrawoot S., Lin C. Y., Rahl P. B., Burke C. J., Langdon E., Tomlinson M. L., Mosher J., Kaufman C. et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518-522. 10.1038/nature09882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer G. B., Lee V. M., Gregg R. G. and Link B. A. (2005). Analysis of the Zebrafish perplexed mutation reveals tissue-specific roles for de novo pyrimidine synthesis during development. Genetics 170, 1827-1837. 10.1534/genetics.105.041608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.