Abstract
Recent studies have revealed a role for macrophages and neutrophils in limiting chemotherapy efficacy, however the mechanisms underlying therapeutic benefit of myeloid-targeting agents in combination with chemotherapy are incompletely understood. Here, we show that targeting tumor-associated macrophages by colony-stimulating factor-1 receptor (CSF-1R) blockade in the K14cre;Cdh1F/F;Trp53F/F transgenic mouse model for breast cancer stimulates intratumoral type I interferon (IFN) signaling which enhances the anti-cancer efficacy of platinum-based chemotherapeutics. Notably, anti-CSF-1R treatment also increased intratumoral expression of type I IFN-stimulated genes in cancer patients, confirming that CSF-1R blockade is a powerful strategy to trigger an intratumoral type I IFN response. By inducing an inflamed, type I IFN-enriched tumor microenvironment and by further targeting immunosuppressive neutrophils during cisplatin therapy, anti-tumor immunity was activated in this poorly immunogenic breast cancer mouse model. These data illustrate the importance of breaching multiple layers of immunosuppression during cytotoxic therapy to successfully engage anti-tumor immunity in breast cancer.
Introduction
Poor chemotherapy response is a major obstacle to successful cancer treatment. There is a growing appreciation for the influential role of the immune system on the success of cytotoxic anti-cancer therapy1. While the adaptive immune system contributes to the therapeutic benefit of certain chemotherapeutic drugs in immunogenic tumor models2, it frequently fails to be unleashed by these same agents in less immunogenic transgenic mouse tumor models3–5, suggesting the involvement of immunosuppressive mechanisms. Indeed, macrophages and neutrophils are frequently the most abundant immune cells in tumors, and clinical studies have reported a correlation between these myeloid cells and poor chemotherapy efficacy4,6–10. Experimental animal studies confirm a causal relationship between tumor-associated myeloid cells and poor chemotherapy response4,5,11–20. For example, inhibition of macrophages in mammary tumor-bearing MMTV-PyMT mice increases paclitaxel efficacy via activation of anti-tumor immunity4,5. Notably, macrophage- and neutrophil-targeting agents are currently under clinical evaluation21–23. Although promising, the aforementioned preclinical studies only show a transient therapeutic effect of combined myeloid cell-targeting and chemotherapy. A deeper understanding of the mechanisms-of-action is needed to facilitate the rational design of therapeutic combination strategies that convert so-called ‘cold’ non-T-cell-inflamed tumors into ‘hot’ inflamed tumors, thus engaging durable anti-tumor immunity in otherwise poorly immunogenic tumors.
By combining in vivo intervention experiments and mechanistic studies in the K14cre;Cdh1F/F;Trp53F/F mouse model for spontaneous mammary tumorigenesis24 with validation studies in tumor biopsies of patients treated with anti-CSF-1R, we here demonstrate that CSF-1R inhibition synergizes with platinum-based chemotherapy by unleashing an intratumoral type I interferon response. Besides this anti-CSF-1R-mediated conversion of the tumor microenvironment (TME) into a type I interferon-enriched milieu, it takes breaching of an additional layer of immunosuppression to engage anti-tumor immunity during cytotoxic therapy.
Results
CSF-1R blockade does not affect mammary tumor growth or metastasis in K14cre;Cdh1F/F;Trp53F/F mice
We set out to assess the role of colony-stimulating factor-1 (CSF-1)/CSF-1R signaling, which is vital for macrophages25, in tumor progression in the K14cre;Cdh1F/F;Trp53F/F mouse model (KEP model), which spontaneously develops mammary tumors resembling human invasive lobular carcinomas (ILC) at 6-8 months of age24. Similar to human ILCs, KEP tumors are strongly infiltrated by macrophages (Supplementary Fig. 1a,b), Whereas in the MMTV-PyMT breast cancer model it has been reported that two distinct macrophage populations reside within the TME: CD11bhiMHCIIhiCD206hi mammary tissue macrophages (MTM) and CD11bloMHCIIhiCD206lo tumor-associated macrophages (TAMs)26, in mammary tumors of KEP mice all F4/80+ macrophages express high levels of CD11b, low levels of CD206, and only a proportion of these cells expresses MHCII (Supplementary Fig. 1c). These differences in intratumoral macrophage phenotypes between mouse tumor models underscore the complexity of macrophage plasticity in different tumor contexts. In line with the macrophage influx, CSF-1 protein levels are increased in KEP tumors versus healthy mammary glands of age-matched wild type littermates (Fig. 1a). Both cancer cells and host cells in KEP tumors express Csf-1 mRNA, while Csf-1 mRNA is barely detectable in healthy mammary glands (Fig. 1b). CSF-1R is highly expressed on TAMs and to a lesser extent on infiltrating monocytes and neutrophils (Supplementary Fig. 1d), but not on other tumor-associated immune cells or CD45- cells (Supplementary Fig. 1d).
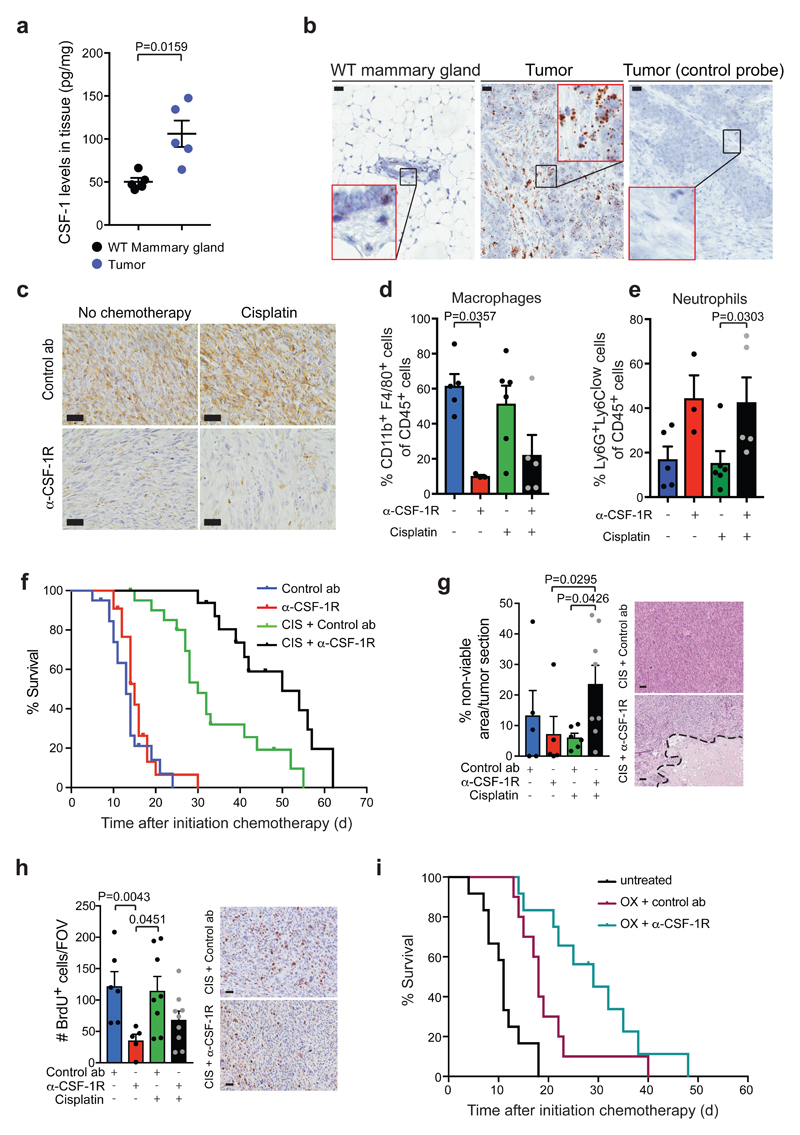
Figure 1. CSF-1R blockade improves the anti-cancer efficacy of platinum-based chemotherapeutic drugs in the K14cre;Cdh1F/F;Trp53F/F mouse model for de novo mammary tumorigenesis.
(a) CSF-1 protein levels in end-stage mammary tumors of K14cre;Cdh1F/F;Trp53F/F (KEP) mice and mammary glands of age-matched wild type (WT) mice (n=5 animals/group) measured by Luminex cytokine array. (b) Representative images of RNA in situ hybridization of CSF-1 (brown signal) in end-stage KEP tumors and normal mammary glands of age-matched WT mice. Data are representative of 3 animals/group. Scale bar=25μm. (c) Representative IHC images of F4/80+ macrophages in tumors of time point-sacrificed KEP mice treated as indicated. Data are representative of 5 animals/group. Scale bar=25μm. (d-e) Proportion of CD11b+F4/80+ macrophages (d) and Ly6G+Ly6Clow neutrophils (e) gated on CD45+ cells as determined by flow cytometry in tumors of end-stage KEP mice treated as indicated (untreated n=5 animals, anti-CSF-1R n=3 animals, cisplatin n=6 animals, cisplatin/anti-CSF-1R n=5 animals). (f) Kaplan-Meier tumor-specific survival curves of KEP mice treated with control antibody (ab) (n=20 animals), anti-CSF-1R (n=22 animals), cisplatin/control ab (n=21 animals), or cisplatin/anti-CSF-1R (n=16 animals). Cisplatin/control ab versus control ab, p=0.0001; cisplatin/control ab versus anti-CSF-1R, p=0.0001; cisplatin/anti-CSF-1R versus cisplatin/control ab, p=0.0011 (two-tailed log-rank test). (g) Percentage of non-viable area per tumor section of time point-sacrificed KEP mice quantified by digital area analysis of H&E stained sections (control ab n=5 animals, anti-CSF-1R n=5 animals, cisplatin/control ab n=6 animals, cisplatin/anti-CSF-1R n=8 animals). Representative H&E sections are shown and dashed line separates viable from non-viable area. Scale bar=50μm. (h) Quantification of BrdU+ cells in viable areas of mammary tumors of time point-sacrificed KEP mice (control ab n=6 animals, anti-CSF-1R n=5 animals, cisplatin/control ab n=8 animals, cisplatin/anti-CSF-1R n=9 animals). Values represent average number of BrdU+ cells per field of view (FOV) quantified by counting five high-power microscopic fields per tumor. Representative BrdU IHC stainings are shown. Scale bar=25μm. (i) Kaplan-Meier tumor-specific survival curves of untreated KEP mice (n=12 animals) or mice treated with oxaliplatin/control ab (n=10 animals) and oxaliplatin/anti-CSF-1R (n=12 animals). Oxaliplatin/control ab versus no treatment, p=0.0015; oxaliplatin/control ab versus oxaliplatin/anti-CSF-1R, p=0.0507 (two-tailed log-rank test). Data presented in a, d-e and g-h are mean values ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test. CIS, cisplatin, OX, oxaliplatin.
To determine whether intratumoral macrophage accumulation depends on CSF-1/CSF-1R signaling, and whether macrophages influence tumor outgrowth and dissemination, we treated tumor-bearing KEP mice with a chimeric mouse IgG1 antagonistic antibody (2G2) that binds to mouse CSF-1R with high affinity (KD=0.2 nM) or with a control antibody21. CSF-1R blockade strongly reduced the TAM population (Fig. 1c,d) and, as a result, also the total CD45+ population (Supplementary Fig. 1e). Treatment with anti-CSF-1R alone did not influence tumor-specific survival (Fig. 1f) or spontaneous metastasis formation (Supplementary Fig. 1f). We also investigated the therapeutic activity of anti-CSF-1R in the KEP-based model of spontaneous breast cancer metastasis27. In this model, after orthotopic transplantation of a KEP-derived tumor piece followed by surgical removal of the outgrown tumor, mice develop overt multi-organ metastatic disease. Anti-CSF-1R was started either after a palpable mammary tumor had developed (continuous setting) or after mastectomy (adjuvant setting) and continued until the development of metastatic disease (Supplementary Fig. 1g). Regardless of the treatment schedule, metastasis-specific survival and metastatic burden in the lungs were similar between control and anti-CSF-1R groups (Supplementary Fig. 1h,i). Thus, anti-CSF-1R monotherapy fails to affect outgrowth and dissemination of KEP mammary tumors.
CSF-1R blockade in tumor-bearing K14cre;Cdh1F/F;Trp53F/F mice enhances the anti-cancer efficacy of platinum-based chemotherapy
We next tested the anti-cancer efficacy of anti-CSF-1R in combination with two conventional chemotherapeutics with a different mode of action, cisplatin, a platinum-based anti-cancer drug that crosslinks DNA and induces apoptosis, and docetaxel an antimitotic agent that interferes with cell division through stabilization of microtubules. Successful blockade of the CSF-1R pathway during treatment of tumor-bearing KEP mice with chemotherapy and anti-CSF-1R was confirmed by the reduction of TAMs (Fig. 1c,d and Supplementary Fig. 2a,b). Interestingly, anti-CSF-1R synergized with cisplatin, resulting in prolonged survival as compared to cisplatin/control antibody-treated mice (Fig. 1f). In contrast, no therapeutic synergy was observed in docetaxel/anti-CSF-1R-treated mice (Supplementary Fig. 2c). The therapeutic synergy observed upon cisplatin/anti-CSF-1R was associated with more necrosis in KEP tumors (Fig. 1g) but not with more cleaved caspase 3+ apoptotic cells (Supplementary figure 2d). Perhaps other mechanisms of cell death are involved, or the timing of our analysis was suboptimal for this parameter. Furthermore, anti-CSF-1R monotherapy – and to a lesser extent the combination with cisplatin – decreased the number of BrdU+ proliferating cells (Fig. 1h). No significant changes in the number and pericyte coverage of CD31+ microvessels, the amount of intratumoral DNA double-strand breaks and intratumoral cisplatin-adduct formation were observed at the timepoint analyzed (Supplementary Fig. 2e-h). As expected, none of these parameters were changed in the docetaxel setting (Supplementary Fig. 2i-m).
To assess whether the anti-CSF-1R-mediated therapeutic synergy was unique to cisplatin or could be extended to drugs with a similar mechanism of action, we tested another platinum-containing drug, oxaliplatin, and found that also the survival benefit of oxaliplatin was improved by combined CSF-1R blockade (Fig. 1i and Supplementary Fig. 2n). These data demonstrate that anti-CSF-1R acts synergistically with platinum-based chemotherapeutic drugs to extend the survival of mammary tumor-bearing KEP mice.
CSF-1R inhibition alters the innate immune landscape of K14cre;Cdh1F/F;Trp53F/F tumors
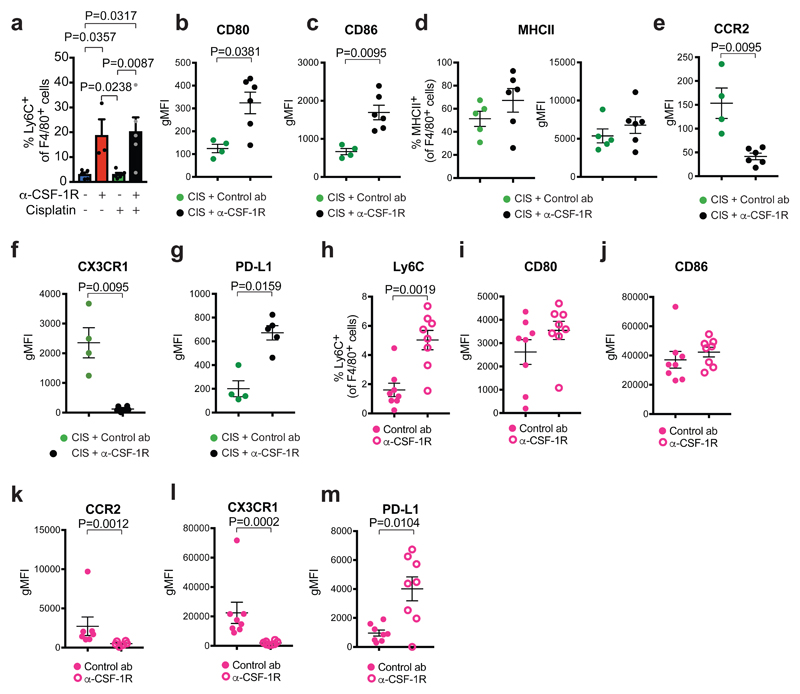
Macrophages are key orchestrators of the inflammatory TME28. We therefore set out to assess the impact of anti-CSF-1R on the innate immune landscape of KEP tumors. Despite the strong reduction of CD11b+F4/80+ TAMs upon anti-CSF-1R, up to 20% of the intratumoral CD45+ immune cells still expresses the macrophage marker F4/80 (Fig. 1d). Detailed analysis of this surviving CD11b+F4/80+ population revealed that an increased proportion of these cells expresses the inflammatory monocyte marker Ly6C as compared to CD11b+F4/80+ cells in control antibody-treated tumors (Fig. 2a and Supplementary Fig. 3a). Moreover, the surviving CD11b+F4/80+ cells in cisplatin/anti-CSF-1R-treated tumors express elevated levels of the co-stimulatory molecules CD80, CD86, slightly elevated MHCII levels, decreased levels of chemokine receptors CCR2 and CX3CR1, and increased levels of PD-L1 compared to intratumoral CD11b+F4/80+ cells in cisplatin/control antibody-treated mice (Fig. 2b-g). Also in the independent orthotopically transplanted K14cre;Trp53F/F mammary tumor model, intratumoral CD11b+F4/80+ myeloid cells remaining after CSF-1R inhibition display an altered phenotype corresponding to that in anti-CSF-1R-treated KEP tumors (Fig. 2h-m). Thus, anti-CSF-1R depletes the majority of CD11b+F4/80+ TAMs, whilst a small population of CD11b+F4/80+ cells with a distinct phenotype survives. To explore whether these surviving cells could derive from circulating monocytes, we transferred tdTomato+ monocytes into control antibody or anti-CSF-1R-treated tumor-bearing KEP mice. After 4 days, the transferred monocytes that infiltrated tumors of CSF-1R-treated, and not control antibody-treated, animals partially acquired the phenotype of the surviving intratumoral CD11b+F4/80+ cell population (i.e. loss of CX3CR1 and elevated PDL1 expression) (Supplementary Fig. 3b-d). These findings suggest that the surviving CD11b+F4/80+ cells in anti-CSF-1R-treated tumors may derive from newly recruited circulating monocytes, although other mechanisms cannot be excluded.
Figure 2. Characterization of F4/80+ cells by flow cytometry in spontaneous K14cre;Cdh1F/F;Trp53F/F tumors and in orthotopically transplanted K14cre;Trp53F/F tumors after anti-CSF-1R treatment.
(a) Percentage of CD11b+F4/80+ immune cells expressing Ly6C in end-stage KEP tumors (untreated n=5 animals, anti-CSF-1R n=3 animals, cisplatin n=6 animals, cisplatin/anti-CSF-1R n=5 animals). (b-c) Geometric Mean Fluorescence Intensity (gMFI) of CD80 (b) and CD86 (c) expression on F4/80+SiglecF- cells in KEP tumors (cisplatin/control ab n=4 animals, cisplatin/anti-CSF-1R n=6 animals). (d) Percentage of MHCII-expressing F4/80+SiglecF- cells and gMFI of MHCII on F4/80+SiglecF- in the KEP tumors (cisplatin/control ab n=5 animals, cisplatin/anti-CSF-1R n=6 animals). gMFI was calculated by subtracting the gMFI of the MHCII negative population from the gMFI of the MHCII positive population. (e-g) gMFI of CCR2 (e), CX3CR1 (f) and PD-L1 (g) expression on F4/80+Siglec F- cells in KEP tumors (CCR2 and CX3CR1: cisplatin/control ab n=4 animals, cisplatin/anti-CSF-1R n=6 animals; PD-L1: cisplatin/control ab n=4 animals, cisplatin/anti-CSF-1R n=5 animals). (h-m) K14cre;Trp53F/F (KP) tumor pieces were orthotopically transplanted in the mammary fat pad of FVB mice. (h) Percentage of CD11b+F4/80+SiglecF- immune cells expressing Ly6C in time-point-sacrificed KP tumors. (i-m) gMFI of CD80 (i), CD86 (j), CCR2 (k), CX3CR1 (l), and PD-L1 (m) expression on F4/80+SiglecF- cells in time-point-sacrificed KP tumors (n=8 animals/group, except CCR2: control ab n=7 animals, anti-CSF-1R n=6 animals). gMFI values presented in b-c, e-g and i-m are determined by subtracting gMFIs of the fluorescence minus one (FMO) staining from the gMFI of the full staining. Data presented in a-m are mean values ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test.
Whereas in treatment-naïve KEP tumors the macrophage:neutrophil ratio is approximately 3:1, in anti-CSF-1R-treated tumors, either in the presence or absence of cisplatin, this ratio is reversed (Fig. 1d,e). However, the absolute number of intratumoral neutrophils was not increased upon CSF-1R inhibition (Supplementary Fig. 3e). Anti-CSF-1R treatment induced an increase in monocytes and a modest, but non-significant and very variable increase in the number of intratumoral eosinophils and mast cells (Supplementary Fig. 3f-h). Together these data show that cisplatin/anti-CSF-1R synergy is accompanied by changes in the myeloid immune landscape of tumors. Most notably, anti-CSF-1R treatment resulted in a surviving population of CD11b+F4/80+ cells with an altered phenotype.
Macrophage blockade enhances cisplatin response by unleashing intratumoral type I interferon signaling
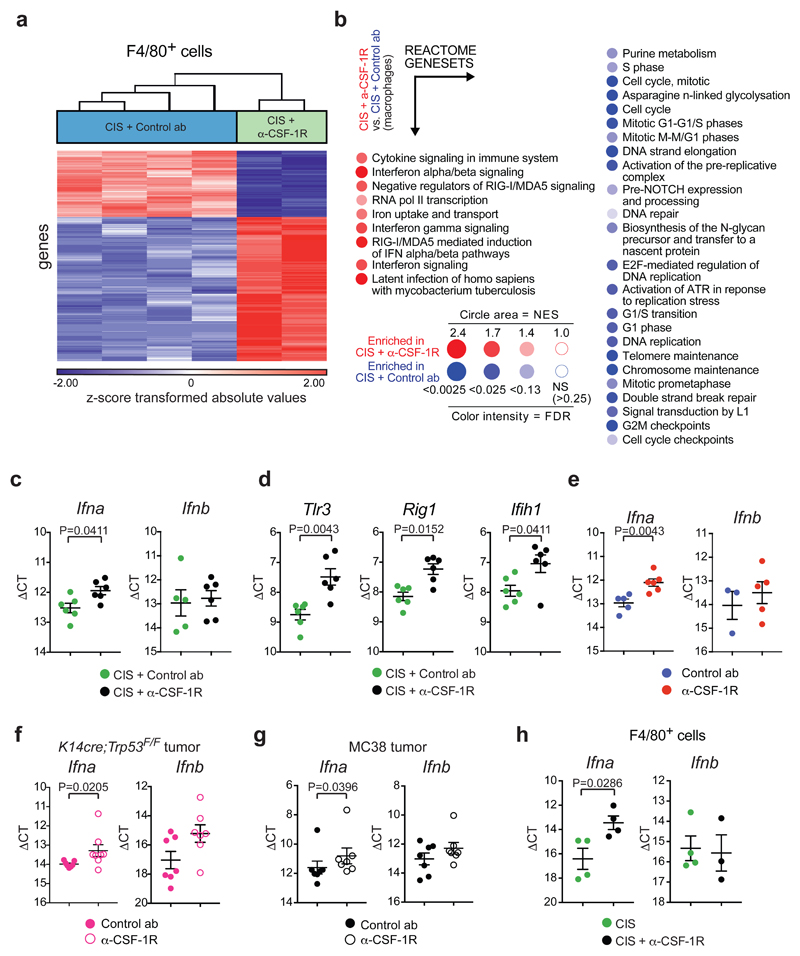
To better characterize the phenotype of the anti-CSF-1R surviving intratumoral CD11b+F4/80+ cells, next-generation RNA sequencing (RNA-Seq) analysis was performed on CD11b+F4/80+ cells sorted from cisplatin/control antibody or cisplatin/anti-CSF-1R-treated tumors. Hierarchical clustering of the top 400 variable genes revealed that CD11b+F4/80+ cells from cisplatin/anti-CSF-1R-treated tumors displayed a different transcriptome profile, mainly characterized by a strong enrichment of genes involved in type I IFN signaling and type I IFN production, while cell cycle-associated genes were reduced (Fig. 3a,b). Interestingly, CSF-1R expression levels were lower in the remaining CD11b+F4/80+ cells from cisplatin/anti-CSF-1R-treated tumors (FC:-2,04; p-value=3,63x10-5), perhaps explaining why these cells resisted anti-CSF-1R therapy. In parallel, we also performed RNA-seq on flow-sorted Ly6G+Ly6Clow neutrophils isolated from tumors of cisplatin/control antibody- and cisplatin/anti-CSF-1R-treated KEP mice. Hierarchical clustering of the top 400 variable genes within this dataset revealed that anti-CSF-1R treatment also had a significant impact on the transcriptome profile of tumor-associated neutrophils (Supplementary Fig. 4a). To ensure that these transcriptome alterations in neutrophils are not a direct effect of anti-CSF-1R on neutrophils, but rather an indirect consequence of macrophage targeting, we performed gene set enrichment analysis (GSEA) of target genes of early growth receptor 2 (Egr2), a transcription factor downstream of CSF-1R signaling29. No differences were observed in the expression of Egr2 target genes between neutrophils isolated from anti-CSF-1R and control antibody-treated tumors (Supplementary Fig. 4b), suggesting that neutrophils are not directly influenced by anti-CSF-1R. Interestingly, BiNGO analysis of the top 100 up- and down-regulated genes and Ingenuity Pathway Analysis of the differentially expressed genes revealed an enrichment in genes involved in type I IFN signaling in neutrophils isolated from cisplatin/anti-CSF-1R-treated tumors versus cisplatin/control antibody-treated tumors (Supplementary Fig. 4c,d and Supplementary Table 3). These data indicate that the therapeutic benefit of cisplatin/anti-CSF-1R is accompanied by induction of type I IFN-stimulated genes (ISGs) in both intratumoral CD11b+F4/80+ cells and neutrophils.
Figure 3. CSF-1R inhibition alters TAM phenotype and induces type I IFN signaling in the tumor microenvironment.
(a) Hierarchical clustering of the top 400 variable genes between CD11b+F4/80+ cells isolated from KEP tumors treated with cisplatin/control ab (n=4 animals) and cisplatin/anti-CSF-1R (n=2 biologically independent samples, pool of 5 mice each). FC: ≥ 1,5; p-value with FDR correction: ≤ 0,05. Statistical analysis was performed using two-way ANOVA. (b) BubbleGUM visualization of GSEA using reactome genesets comparing CD11b+F4/80+ cells from cisplatin/anti-CSF-1R-treated tumors versus macrophages from cisplatin/control ab-treated tumors. Same mice as in a. (Normalized enrichment score (NES) ≥ 1, FDR value: 0.25 ≤). Enrichments score was calculated using a weighted Kolmogorov–Smirnov-like statistic. (c) Transcripts of Ifna (n=6 animals/group) and Ifnb (n=5 animals in cisplatin/control ab, n=6 animals in cisplatin/anti-CSF-1R) and (d) Tlr3, Rig1 and Ifih1 in KEP mammary tumors isolated one day after the second cisplatin injection were determined by qPCR and normalized to β-actin (n=6 animals/group). (e-g) Transcripts of Ifna and Ifnb in KEP mammary tumors (Ifna: n=5 animals in control ab, n=6 animals in anti-CSF-1R; Ifnb: n=3 animals in control ab, n=5 animals in anti-CSF-1R) (e), orthotopically transplanted K14cre;Trp53F/F mammary tumor (Ifna: n=7 animals in control ab, n=8 animals in anti-CSF-1R; Ifnb: n=7 animals/group) (f) and subcutaneously inoculated MC38 tumors (n=7 animals/group) (g) treated with control ab and anti-CSF-1R were determined by qPCR and normalized to β-actin. Mice were analysed at a tumor size of 100mm2 (KP) or after 12 days from the start of the treatment (MC38). (h) Transcripts of Ifna (n=4 animals/group) and Ifnb (n=4 animals in cisplatin, n=3 animals in cisplatin/anti-CSF-1R) in CD11b+F4/80+ cells isolated from end-stage KEP tumors were determined by qPCR and normalized to β-actin. Graphs in c-h show the mean ± SEM in ΔCt values and statistical analysis was performed using two-tailed Mann–Whitney test.
We hypothesized that the enrichment of ISGs in these intratumoral immune populations was a consequence of increased levels of type I IFNs in KEP tumors upon CSF-1R blockade. Indeed, by using primers hybridizing to all Ifna genes, mRNA expression of Ifna, but not Ifnb, was increased in tumors of cisplatin/anti-CSF-1R-treated KEP mice compared to cisplatin/control antibody-treated mice (Fig. 3c). In line with this, mRNA levels of various intracellular pattern recognition receptors like Tlr3, Rig1 and Ifih1, whose signals induce type I IFN production, were up-regulated in cisplatin/anti-CSF-1R-treated tumors compared to cisplatin/control antibody treatment (Fig. 3d). Notably, the increase in type I IFN expression upon anti-CSF-1R was independent of chemotherapy treatment, since a similar intratumoral increase in Ifna expression was observed upon anti-CSF-1R alone (Fig. 3e) or with docetaxel/anti-CSF-1R (Supplementary Fig. 5a). We also confirmed the increase of Ifna - and of two ISGs, Isg15 and Oas1a - upon anti-CSF-1R treatment in the independent K14cre;Trp53F/F-based tumor transplantation model and in inoculated MC38 colorectal adenocarcinoma tumors21 (Fig. 3f,g and Supplementary Fig. 5b,c). Together these data demonstrate that anti-CSF-1R induces type I IFN in the TME.
To pursue the cellular source of type I IFN, we flow-sorted different cell populations from cisplatin and cisplatin/anti-CSF-1R-treated KEP tumors (Supplementary Fig. 5d) and compared Ifna and Ifnb transcript levels. Plasmacytoid dendritic cells (pDCs) are known for their ability to produce type I IFN, however, since very few pDCs – less than 0.1% of the total intratumoral immune population – are present in KEP tumors (Supplementary Fig. 5e,f), we could not recover RNA of sufficient quality. Likewise, we did not obtain RNA of sufficient quality from sorted CD31+ endothelial cells. Only the CD11b+F4/80+ immune cell population displayed elevated Ifna expression levels upon CSF-1R blockade (Fig. 3h and Supplementary Fig. 5g,h). In line with these in vivo findings, in vitro treatment of bone marrow-derived macrophages with anti-CSF-1R modestly induces Ifna levels after 24h of culture (Supplementary Fig. 5i). These analyses suggest that the surviving population of intratumoral CD11b+F4/80+ cells is an important source of IFNα in cisplatin/anti-CSF-1R-treated KEP tumors.
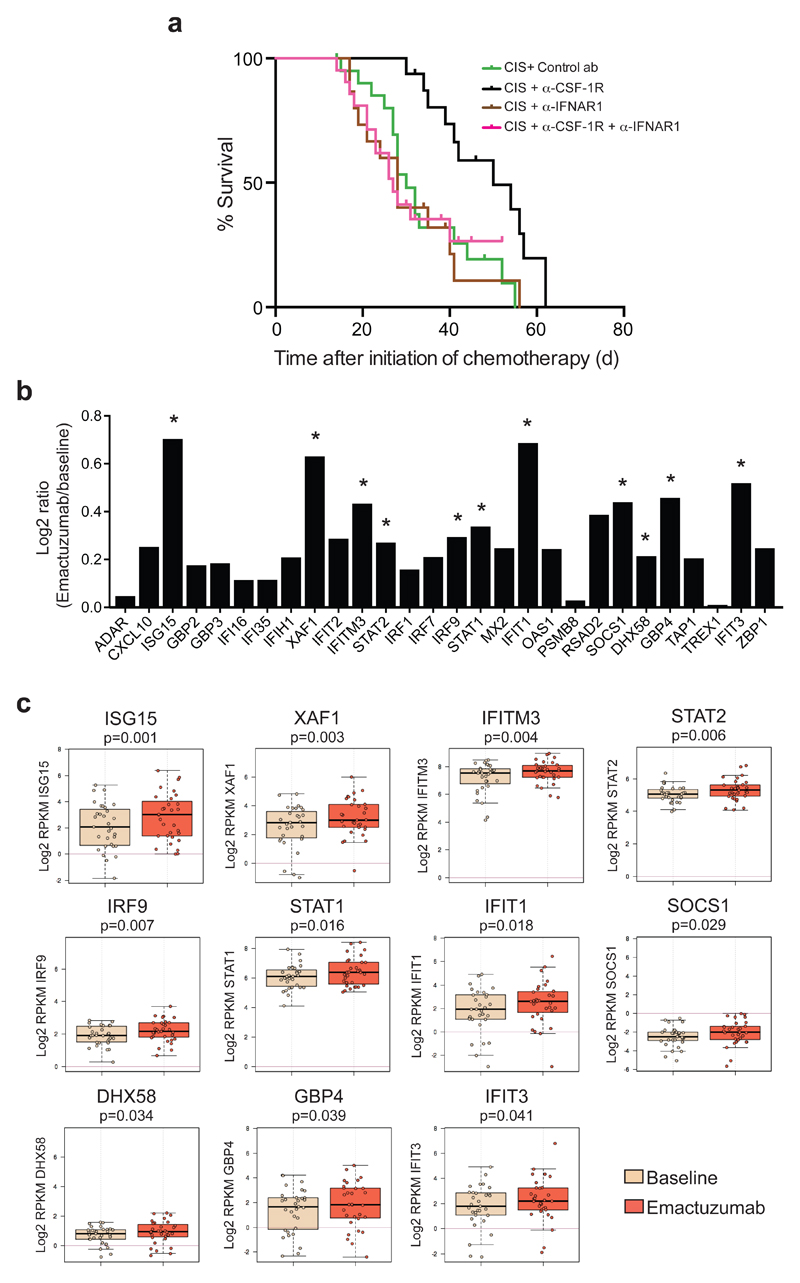
To dissect the functional significance of type I IFN signaling in the therapeutic benefit of cisplatin/anti-CSF-1R therapy, we blocked the IFN-α/β receptor subunit 1 (IFNAR1) in KEP mice. While blockade of type I IFN signaling did not influence the anti-cancer efficacy of cisplatin, anti-IFNAR1 treatment completely abrogated the synergistic effect of cisplatin/anti-CSF-1R treatment (Fig. 4a). These findings reveal that therapeutic targeting of macrophages with anti-CSF-1R in tumor-bearing KEP mice unleashes intratumoral type I IFN signaling which enhances the therapeutic efficacy of cisplatin.
Figure 4. CSF-1R blockade increases the expression of intratumoral type I IFN signaling in cancer patients treated with emactuzumab and is essential for the therapeutic synergy of cisplatin/anti-CSF-1R in the K14cre;Cdh1F/F;Trp53F/F mouse model.
(a) Kaplan-Meier tumor-specific survival curves of KEP mice treated with cisplatin/control ab, cisplatin/anti-CSF-1R (same groups as in Fig. 1f), cisplatin/anti-IFNAR1 (n=15 animals), or cisplatin/anti-CSF-1R/anti-IFNAR1 (n=21 animals). Cisplatin/anti-CSF-1R versus cisplatin/anti-CSF-1R/anti-IFNAR1, p=0.0064 (two-tailed log-rank test). (b) Log2 ratio of intra-tumoral expression levels of 28 type I interferon-stimulated genes (ISGs) in emactuzumab (anti-CSF-1R)-treated patients (n=31 patients) normalized against the pre-treatment expression levels. (c) Box plots of the expression level of the 11 statistically significant up-regulated type I ISGs in tumors of emactuzumab-treated patients (same data as in Fig. 4b). Expression levels in pre-treatment (baseline) tumor biopsies are compared to on-treatment (emactuzumab) biopsies. The topmost line is the maximum, the top of the box is the 3rd quartile, the center line is the median, the bottom of the box is the 1st quartile, the bottom-most line is the minimum. RPKM, reads per kilobase of exon per million mapped reads. P value was determined by two-tailed Student’s t-test.
Emactuzumab treatment induces intratumoral type I IFN-stimulated genes in cancer patients
To validate our preclinical findings that CSF-1R blockade unleashes intratumoral type I IFN signaling in patients, we compared ISGs expression levels in pre- and on-treatment tumor biopsies from cancer patients with advanced solid tumors treated with emactuzumab (RG7155), a humanized anti-human CSF-1R monoclonal antibody (NCT01494688)21,30. Gene expression profiling was performed on tumor biopsies taken before the start of treatment and after 4 weeks of emactuzumab therapy. We assessed the expression level of a set of 28 ISGs that was selected based on the RNA-seq results from our KEP mouse model (Fig. 3a and Supplementary Fig. 4a,c,d and Supplementary Table 3). The intratumoral expression of all 28 selected ISGs was increased in emactuzumab on-treatment biopsies versus pre-treatment biopsies, of which 11 ISGs were significantly up-regulated (Fig. 4b,c). Thus, in line with our preclinical studies, these clinical findings indicate that CSF-1R blockade is a powerful strategy to augment intratumoral type I IFN signaling.
Combined CSF-1R inhibition and neutrophil depletion engages anti-tumor immunity that further improves therapeutic benefit of cisplatin
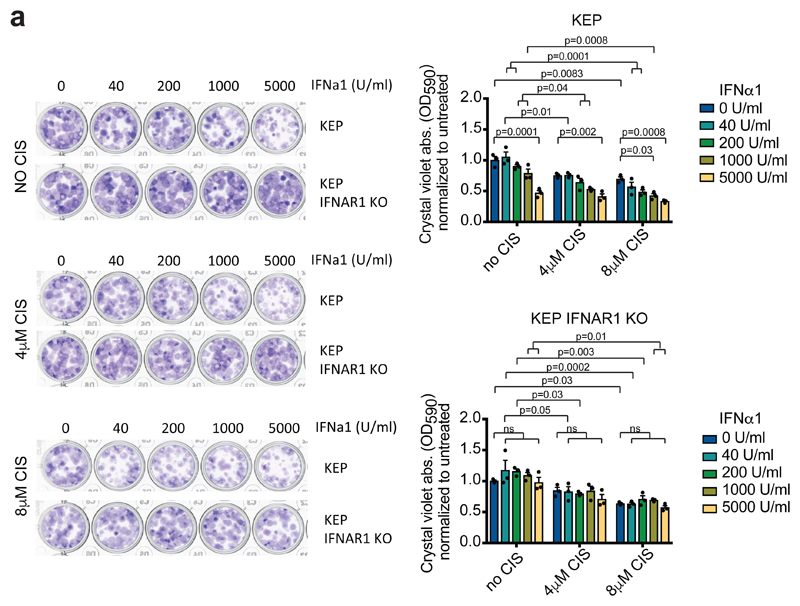
Type I IFNs are emerging as key regulators of cancer growth and therapy response31,32. Type I IFNs can affect cancer biology via different mechanisms, including induction of anti-proliferative and pro-apoptotic effects on IFNAR+ cancer cells33,34. Indeed, exposure of a cell line derived from a spontaneous KEP mammary tumor to recombinant IFNα1 results in a dose-dependent decrease in colony-forming ability, also in combination with cisplatin, suggesting that type I IFNs have a direct inhibitory impact on KEP cancer cells (Fig. 5a). Because type I IFNs are also key orchestrators of anti-tumor immunity33–35, we hypothesized that the anti-CSF-1R-induced type I IFN-enriched TME may foster anti-tumor CD8+ T cell activity. However, we observed less tumor-infiltrating CD4+ or CD8+ T cells in cisplatin/anti-CSF-1R-treated tumors compared to cisplatin/control antibody-treated tumors (Supplementary Fig. 6a-c), and the CD8/Treg ratio was not affected (Fig. 6a). More NK cells were infiltrating the cisplatin/anti-CSF-1R-treated KEP tumors, however, the number of granzyme B+ cells was not affected compared to cisplatin/control antibody treatment (Fig. 6b,c and Supplementary Fig. 6d). We previously reported that cisplatin efficacy is independent of the adaptive immune system3, and, in line with the lack of more intratumoral granzyme B+ cells and T cells, we here show that also antibody-mediated depletion of CD8+ T cells does not reduce the therapeutic efficacy of cisplatin/anti-CSF-1R therapy (Fig. 6d). In addition, genetic ablation of the entire adaptive immune system by crossing KEP mice with RAG1-/- mice did not affect therapeutic synergy (Fig. 6g). These data indicate that the anti-CSF-1R-mediated conversion of the TME into a type I IFN-enriched milieu in cisplatin-treated mice is not sufficient to successfully engage an endogenous anti-tumor T cell response.
Figure 5. Direct inhibitory effect of IFNα1 on a K14cre;Cdh1F/F;Trp53F/F-derived cancer cell line.
(a) Representative images of a colony forming assay with KEP-derived cancer cells and IFNAR1 KO KEP cancer cells treated with increased concentrations of IFNα1 and cisplatin. After 7 days, crystal violet was dissolved and absorbance was measured at 590nm. Data are representative of 3 independent experiments. Data are mean values ± SEM. P value was determined by two-way ANOVA with Tukey’s multiple comparison test. CIS, cisplatin
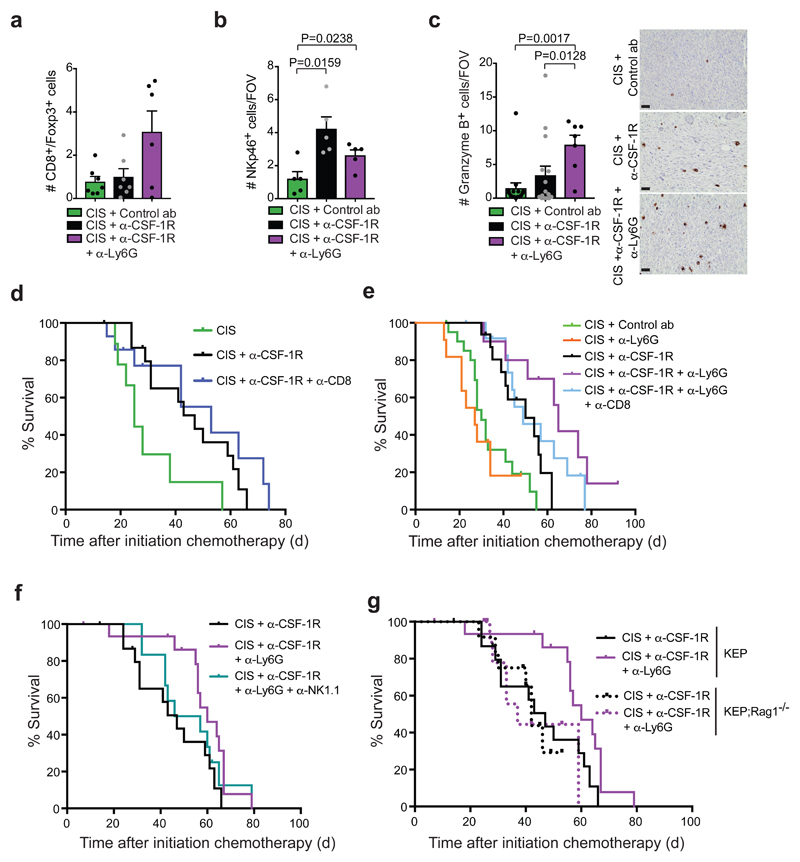
Figure 6. Neutrophil inhibition engages anti-tumor immunity and further improves cisplatin/anti-CSF-1R efficacy.
(a) CD8+ T cell/FoxP3+ T cell ratio based on IHC staining in the tumor of time point-sacrificed KEP mice (cisplatin/control ab and cisplatin/anti-CSF-1R n=7 animals; cisplatin/anti-CSF-1R/anti-Ly6G n=6 animals). (b-c) Quantification of NKp46+ cells (b) and granzyme B+ cells (c) in viable areas of mammary tumors of time point-sacrificed KEP mice treated with cisplatin/control ab (NKp46 n=5 animals; granzyme B n=15 animals), cisplatin/anti-CSF-1R (NKp46 n=5 animals; granzyme B n=15 animals) and cisplatin/anti-CSF-1R/anti-Ly6G (NKp46 n=5 animals; granzyme B n=7 animals). Values represent average number of positive cells per FOV quantified by counting five high-power microscopic fields per tumor. Representative granzyme B IHC stainings are shown. Scale bar=50μm. (d) Kaplan-Meier tumor-specific survival curves of KEP mice treated with cisplatin/control ab (n=6 animals), cisplatin/anti-CSF-1R (n=16 animals), cisplatin/anti-CSF-1R/anti-CD8 (n=14 animals). Cisplatin/anti-CSF-1R/anti-CD8-treated mice versus cisplatin/anti-CSF-1R-treated mice, p=0.3728 (two-tailed log-rank test). (e) Kaplan-Meier tumor-specific survival curves of KEP mice treated with cisplatin/control ab, cisplatin/anti-CSF-1R (same groups as Fig. 1f), cisplatin/anti-Ly6G (n=11 animals), cisplatin/anti-CSF-1R/anti-Ly6G (n=10 animals), or cisplatin/anti-CSF-1R/anti-Ly6G/anti-CD8 (n=13 animals). Cisplatin/anti-CSF-1R-treated mice versus cisplatin/anti-CSF-1R/anti-Ly6G-treated mice, p=0.0085; cisplatin/anti-CSF-1R-treated mice versus cisplatin/anti-CSF-1R/anti-Ly6G/a-CD8-treated mice, p=0.1104 (two-tailed log-rank test). (f) Kaplan-Meier tumor-specific survival curves of KEP mice treated with cisplatin/anti-CSF-1R (n=16 animals, same curve as in d), cisplatin/anti-CSF-1R/anti-Ly6G (n=17 animals of which 4 mice were treated with cisplatin/anti-CSF-1R/anti-Ly6G/IgG2a; no differences were observed between cisplatin/anti-CSF-1R/anti-Ly6G and cisplatin/anti-CSF-1R/anti-Ly6G/IgG2a), or cisplatin/anti-CSF-1R/anti-Ly6G/anti-NK1.1 (n=12 animals). Cisplatin/anti-CSF-1R-treated mice versus cisplatin/anti-CSF-1R/anti-Ly6G-treated mice, p=0.0226; cisplatin/anti-CSF-1R-treated mice versus cisplatin/anti-CSF-1R/anti-Ly6G/anti-NK1.1-treated mice, p=0.4073 (two-tailed log-rank test). (g) Kaplan-Meier tumor-specific survival curves comparing cisplatin/anti-CSF-1R and cisplatin/anti-CSF-1R/anti-Ly6G treatment in KEP (solid line; same as in f) and KEP;Rag1-/- (dotted line) mice. Cisplatin/anti-CSF-1R in KEP;Rag1-/- (n=12 animals) and cisplatin/anti-CSF-1R/anti-Ly6G in KEP;Rag1-/- (n=11 animals). Cisplatin/anti-CSF-1R/anti-Ly6G-treated KEP;Rag1-/- mice mice vs. cisplatin/anti-CSF-1R-treated KEP;Rag1-/- mice, p=0.9597 (two-tailed log-rank test). Data presented in a-c are mean values ± SEM and statistical analysis was performed using two-tailed Mann–Whitney test. CIS, cisplatin
We next hypothesized that it may be necessary to breach an additional layer of immunosuppression before anti-tumor immunity can be unleashed. The most abundant immune cell population in cisplatin/anti-CSF-1R-treated KEP tumors are neutrophils (Fig. 1e) and we have previously reported that KEP tumor-educated neutrophils are very immunosuppressive36. To address whether neutrophils impede anti-tumor immunity in cisplatin/anti-CSF-1R-treated mice, we treated tumor-bearing KEP mice with the neutrophil-specific anti-Ly6G antibody (clone 1A8). Immunohistochemistry for S100A9 confirmed a reduction in the number of neutrophils in the lungs and to a lesser extent in the tumor (Supplementary Fig. 6e,f). Cisplatin/anti-CSF-1R/anti-Ly6G treatment significantly improved tumor control and prolonged survival of KEP mice as compared to cisplatin/anti-CSF-1R therapy (Fig. 6e). While cisplatin/anti-CSF-1R temporarily stabilizes tumor ourgrowth, we observed tumor shrinkage in six out of ten mice treated with cisplatin/anti-CSF-1R/anti-Ly6G, and the mammary tumors of two of these mice regressed completely during treatment (Supplementary Fig. 6g,h). Anti-Ly6G treatment alone failed to influence primary tumor growth in KEP mice as previously shown36, neither did the combination of anti-CSF-1R/anti-Ly6G (Supplementary Fig. 6i) nor did anti-Ly6G alter the efficacy of cisplatin (Fig. 6e). Further characterization of cisplatin/anti-CSF-1R/anti-Ly6G-treated KEP tumors showed a significant reduction of the number of BrdU+ proliferating cells and γ-H2AX+ DNA damaged cells (Supplementary Fig. 6j,k). No statistically significant differences were observed in the number of apoptotic cells, CD31+ vessels and cisplatin adducts (Supplementary Fig. 6l-n). Interestingly, the CD8/Treg ratio, the absolute number of NK cells and the absolute and relative number of granzyme B+ immune cells were increased in cisplatin/anti-CSF-1R/anti-Ly6G-treated tumors compared to cisplatin/control antibody therapy (Fig. 6a-c and Supplementary Fig. 6d). Importantly, the additional therapeutic benefit obtained by anti-Ly6G treatment was partially lost after antibody-mediated depletion of CD8+ T cells or NK cells (Fig. 6e,f) and was completely abrogated when the same treatment was performed in KEP;Rag1-/- mice (Fig. 6g). Collectively, these data indicate that the combined anti-CSF-1R-mediated conversion of the tumor milieu into a type I IFN-enriched environment and the relieve of neutrophil-dependent immunosuppression fosters engagement of anti-tumor immunity in the anti-cancer effect of cisplatin in this poorly immunogenic mouse tumor model.
Discussion
There is a growing realization that immune-mediated mechanisms influence the responsiveness of tumors to chemotherapy1. Notably, macrophages actively interfere with the therapeutic efficacy of chemotherapy via several mechanisms in mouse tumor models, including suppression of anti-tumor immunity through IL-10 secretion5, secretion of chemo-protective proteases such as cathepsins12 or secretion of lysophospholipids15 that interfere with the DNA damage response. These macrophage-mediated chemotherapy resistance mechanisms are dependent on the production of soluble mediators from TAMs. Our study reveals a conceptually different mechanism of how therapeutic targeting of macrophages improves chemotherapy efficacy. Through in vivo mechanistic studies in the K14cre;Cdh1F/F;Trp53F/F transgenic mouse model for breast cancer, we demonstrate that macrophage inhibition with anti-CSF-1R induces intratumoral type I IFN signaling which acts synergistically with cisplatin to inhibit tumor outgrowth and extend survival.
There is a growing interest in the impact of type I IFNs on cancer behaviour and response to immune checkpoint inhibitors, radiotherapy and chemotherapy32,37–39. Besides being associated with an improved prognosis40–42, an intratumoral interferon signature in breast cancer patients has been correlated with improved chemotherapy response37 and preclinical studies reported that type I IFN enhanced chemotherapy efficacy37,43. However, IFN-related gene signatures have also been correlated with chemotherapy resistance44, consistent with a pleomorphic and still poorly understood role of type I IFN signaling in the tumor context. Importantly, impaired type I IFN signaling is a prominent feature of immune dysfunction in cancer patients45. Our study reveals that anti-CSF-1R represents a powerful approach to induce intratumoral IFN signaling and to sensitize tumors to cisplatin. Notably, we find that anti-CSF-1R treatment in cancer patients also results in increased intratumoral expression of ISGs, confirming our findings that anti-CSF-1R unleashes type I IFN response in tumors.
Our study shows that anti-CSF-1R depletes the majority of F4/80+ TAMs, however, a small intratumoral CD11b+F4/80+ population with a distict phenotype survives. Interestingly, these surviving cells express lower levels of Csf1r and significantly higher Ifna mRNA levels than the CD11b+F4/80+ cells in untreated tumors, likely accounting for the increased Ifna levels in the tumors. A shift in macrophage phenotype was also observed in pancreatic cancer and glioblastoma models upon interference with the CSF-1/CSF-1R pathway46–49. Similar to our model, targeting CSF-1 in the pancreatic cancer models on one hand depleted TAMs and on the other hand reprogrammed the remaining macrophages to an anti-tumor phenotype. Interestingly, type I IFN was also found increased in these macrophages49, however its effect was not functionally pursued in this study. These data, combined with our observation that IFNα is also up-regulated in anti-CSF-1R-treated MC38 colon adenocarcinoma tumors, indicate that anti-CSF-1R-mediated induction of type I IFNs is not limited to breast cancer, but extends to other cancer types.
Type I IFNs can directly affect cancer cells by inducing apoptosis or blocking proliferation, or indirectly, by stimulating anti-tumor immune responses or inhibiting angiogenesis33. In line with the observed in vivo reduction of proliferating tumor cells upon anti-CSF-1R therapy, our in vitro studies indicate that IFNα1 can directly suppress KEP cancer cells. We did not observe an impact of CSF-1R inhibition on the number of intratumoral blood vessels, or their pericyte coverage, excluding an angiogenesis effect. Despite a key role for type I IFNs in dictating anti-tumor immunity31,32, the increase in intratumoral type I IFNs was not sufficient to induce effective anti-tumor T cell responses. In line with the immunosuppressive phenotype of tumor-educated neutrophils in KEP mice36 and in other models50, the additional ablation of neutrophils stimulated anti-tumor immunity. It may be surprising that we observed therapeutic benefit of depletion of neutrophils with an IFN gene signature in cisplatin/anti-CSF-1R-treated KEP mice, whilst some studies have suggested that type I IFNs can induce anti-tumor properties in neutrophils51. However, in line with our data, a type I IFN transcriptional signature in neutrophils in malaria-infected hosts and in patients with active tuberculosis correlated with tissue damage and disease pathogenesis52,53, suggesting that in these settings type I IFN signaling in neutrophils may contribute to their harmful actions. In addition, although type I IFNs are often considered to exert anti-tumor functions, several studies on chronic viral infections show negative feedback mechanisms when persistently present in the environment by, for example, generating an immunosuppressive milieu34,54,55. Perhaps in our study a similar mechanism is involved explaining why the cytotoxic activity of platinum-based chemotherapy is enhanced by type I IFNs, but at the same time this therapeutic synergy is limited by an immunosuppressive program. Although T cell activation was implicated in controlling tumor growth upon combined macrophage and neutrophil depletion in a mouse model for pancreatic cancer20, the full mechanism in the context of chemotherapy was not completely resolved. Our in vivo data demonstrate that while the release of type I IFN is necessary for cisplatin/anti-CSF-1R therapeutic synergy, it takes further depletion of neutrophils to engage an anti-tumor immune response during cisplatin treatment.
Interestingly, the platinum-based drugs cisplatin and oxaliplatin synergized with anti-CSF-1R treatment, while docetaxel did not, despite the induction of IFNα in the docetaxel setting. It will be important to mechanistically understand how the type of chemotherapy dictates its ability to act in synergy with type I IFN signaling. These insights will facilitate the development of optimal combination therapies of CSF-1R-targeting drugs or other type I IFN-inducing agents, including STING agonists56, with chemotherapeutic agents. To maximize therapeutic benefit of cytotoxic therapy in poorly immunogenic tumor types, it will be critical to simultaneously target neutrophil-dependent immunosuppression.
Methods
Mice
The generation and characterization of K14cre;Cdh1F/F;Trp53F/F (KEP) mice have previously been described24 and are commercially available via Taconic Biosciences. KEP mice were back-crossed onto the FVB/N background and genotyping was performed by PCR analysis on tail tip DNA as described24,36. KEP mice were crossed with Rag1-/- mice (FVB/N, a gift from L. Coussens) to generate K14cre;Cdh1F/F;Trp53F/F;Rag1-/- (KEP;Rag1-/-)3. Female KEP and KEP;Rag1-/- mice were monitored twice-weekly for the spontaneous onset of mammary tumor formation by palpation starting at 4 months of age. Donor tumors from KEP and K14cre;Trp53F/F (KP)24 mice were collected in ice-cold PBS, cut in small pieces and resuspended in Dulbecco's Modified Eagle's Medium F12 containing 30% fetal calf serum and 10% dimethyl sulfoxide and stored at −150 °C. The perpendicular tumor diameters of mammary tumors were measured twice a week using a caliper. Age-matched WT littermates were used as controls. Female FVB/N mice (age 10-12 weeks) were obtained from Charles River. mTmG mice57 (back-crossed to the FVB/N background) express tdTomato ubiquitously and were used for the isolation of bone marrow monocytes. Mice were kept in individually ventilated cages at the animal laboratory facility of the Netherlands Cancer Institute. Food and water were provided ad libitum. Animal experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute and performed in accordance with institutional, national and European guidelines for Animal Care and Use. The study is compliant with all relevant ethical regulations regarding animal research.
In vivo intervention studies
KEP mice bearing spontaneous mammary tumors were randomized over the treatment groups prior to initiation of the treatment. Mice were injected intraperitoneally with the chimeric (hamster/mouse) anti-CSF-1R antibody (clone 2G2, Roche Innovation Center Munich, single loading dose of 60 mg/kg followed by 30 mg/kg once a week); control antibody (IgG1, MOPC21, Roche Innovation Center Munich, single loading dose of 60 mg/kg followed by 30 mg/kg once a week); anti-Ly6G antibody (1A8, single loading dose of 400 μg followed by 100 μg three times a week, BioXCell); anti-IFNAR1 (MAR1-5A3, 100 μg three times a week, BioXCell); anti-CD8 (2.43, single loading dose of 400 μg followed by 100 μg three times a week, BioXCell); anti-NK1.1 (PK136 single loading dose of 400 μg followed by 100 μg three times a week, BioXCell); IgG2a (C1.18.4 single loading dose of 400 μg followed by 100 μg three times a week, BioXCell). MTD dose of cisplatin (5 mg/kg, Accord Healthcare Limited) was administered intravenously every other week for a total of 4 cycles; MTD dose of docetaxel (15 mg/kg, Accord Healthcare Limited) was administered intravenously every week for a total of 4 cycles; MTD dose of oxaliplatin (6mg/kg diluted in NaCl, Fresenius Kabi) was administered intravenously every 10 days for 3 cycles.
Anti-CSF-1R, control antibodies, anti-Ly6G, anti-CD8 and anti-NK1.1 treatment started when mammary tumors reached 25mm2 anti-IFNAR1, cisplatin, docetaxel and oxaliplatin treatment started when mammary tumors reached 50mm2. For survival curve experiments and end-stage analyses, antibody treatment continued until the tumor or the cumulative tumor burden reached 225mm2. For survival curve experiments, an event is defined as an animal with a cumulative tumor size of 225 mm2. The main cause of death of censored mice was ulcerated tumors or cisplatin-induced renal toxicity for cisplatin-treated mice.
For time point analyses, mice were sacrificed one day after the second chemotherapy injection (therapy-responsive phase) or at a tumor size of 100mm2 in chemotherapy-naïve mice. To assess tumor cell proliferation, BrdU (50 mg/kg) was injected intraperitoneally 90 minutes before sacrifice.
KP tumor pieces were orthotopically transplanted in the mammary fat pad of 10-12 week-old FVB/N female mice. Before initiation of the treatment mice were randomized over the experimental groups and treated either with control antibody or anti-CSF-1R as described above. Treatment started at a tumor size of 25mm2 and continued until the tumor reached 100mm2 when the mice were sacrificed.
MC38 tumors were provided by Roche Innovation Center Munich. MC38 cells were subcutaneously injected into C57Bl6/N mice and, when the tumor volume reached 100mm3, treated with either control antibody or anti-CSF-1R as described above. Mice were sacrificed 5 days after the second treatment21.
Intervention studies in the K14cre;Cdh1F/F;Trp53F/F- based spontaneous metastasis model
The orthotopic KEP-based spontaneous metastasis model was described previously in detail27. Briefly, KEP tumor pieces (1x1 mm) were orthotopically transplanted into 10-12 week-old FVB/N female mice. Mammary tumors were surgically removed once they reached a tumor size of 225mm2, after which mice were monitored and sacrificed when they reached the humane endpoint due to clinically overt metastatic disease. Tumor-bearing recipient mice were treated either with control antibody or with anti-CSF-1R once the mammary tumors reached 5mm2 (continuous setting) or three days after mastectomy (adjuvant setting). Antibody treatment continued until recipient mice developed clinical signs of distress caused by metastatic disease (e.g. respiratory distress).
Histology, immunohistochemistry, immunofluorescence and RNA in situ hybridization
All histochemical and immunohistochemical analyses, except NKp46 IHC, were performed by the Animal Pathology facility at the NKI, Amsterdam. NKp46 immunohistochemistry was performed at the Histology core facility within the Cancer Research UK Beatson Institute, Glasgow. For histochemical analysis, formalin-fixed tissues were processed, sectioned and stained as described27. Briefly, tissues were fixed for 24 h in 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 μm and stained with heamatoxylin and eosin (H&E) for histopathological evaluation. H&E slides were digitally processed using the Aperio ScanScope (Aperio, Vista, CA). For the quantitative assessment of areas in the tumor that had lost viability, slides were analyzed with ImageJ by quantifying the percentage of non-viable areas (defined as areas that lost cellularity) over the total tumor area. Histochemistry for mast cells was performed with Toluidine blue.
For immunohistochemical analysis, paraffin sections were cut and deparaffinized. Antibodies and antigen retrieval methods are described in the Supplementary Table 1. Cisplatin adduct staining was performed on frozen tissues embedded in OCT. Quantification of positive cells was performed manually by counting five high-power (40x) fields of view (FOV) per tumor by two independent operators in a blinded fashion. Samples were visualized with a BX43 upright microscope (Olympus) and images were acquired in bright field using cellSens Entry software (Olympus).
Percentages of metastasis-bearing spontaneous KEP mice were calculated based on the microscopic presence or absence of metastatic nodules in lungs and lymph nodes. In the metastasis model, the number of metastatic nodules in the lungs was based on cytokeratin 8 expression. Mice that developed overt metastatic disease were included in the analysis and mice that were sacrificed because of local recurrence of the primary tumor were excluded.
Immunohistochemistry analysis for CD68 expression (1:2000, clone KP1, Dako) was performed by NKI-AvL Core Facility Molecular Pathology & Biobanking (CFMPB) on FFPE material of invasive lobular carcinoma breast cancer patients from the RATHER cohort58,59 enrolled at the NKI. Anonymized archival tissue was used according to national guidelines regarding the use of archival material and with approval of the NKI-AVL translational research board (TRB).
Immunofluorescence analysis was performed on FFPE material. The list of primary and secondary antibodies is list in Supplementary Table 1. Sections were counterstained with DAPI and visualized with Leica SP5 confocal microscope. Images were taken with LAS AF software (Leica) and values were obtained by counting alpha-SMA+CD31+ cells and total CD31+ cells in six field per tumor by 2 independent researches.
In situ detection of Csf-1 mRNA was performed using the RNAscope® 2.0 FFPE Assay (Advanced Cell Diagnostics, INC) and performed according to the manufacturers’ recommendations60.
Flow cytometry
KEP tumors were collected in ice-cold PBS and processed as described61. Briefly, samples were mechanically chopped using the McIlwain tissue chopper (Mickle Laboratory Engineering) and enzymatically digested with 3 mg/ml collagenase type A (Roche) and 25μg/ml of DNase I (Sigma) in serum-free medium for 1h at 37°C in a shaking water bath. After washing, cells were stained with fluorochrome-conjugated antibodies (Supplementary Table 1). For intracellular staining of granzyme B, single-cell suspensions were stimulated in IMDM containing 8% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.5% β-mercaptoethanol, 50 ng/ml PMA, 1 μM ionomycin and Golgi-Plug (1:1000; BD Biosciences) for 3 h at 37 °C. Following surface antigen staining, samples were fixed and permeabilized (BD Biosciences) and stained for intracellular proteins. Data acquisition was performed on BD LSRII or BD LSRFortessa flow cytometer using Diva software (BD Biosciences) and data analysis was performed using FlowJo software version 9.9.6.
Isolation of intratumoral cell populations
Primary mammary tumors were harvested from KEP mice one day after two cycles of chemotherapy (±100 mm2) or at end-stage (±225 mm2) and single cell suspensions were generated as described above. Enrichment of CD11b+ cells was performed using magnetic columns (Miltenyi Biotec), as described previously61. Briefly, single cell suspensions were stained with anti-CD11b-APC (1:200; clone M1/70, eBioscience) for 20 min and incubated with magnetic anti-APC MicroBeads according to manufacturer’s instructions (Miltenyi Biotec). CD11b+ cells were isolated with LS-columns (Miltenyi Biotec) according to manufacturer’s instructions. For the isolation of macrophages and neutrophils from tumors at the therapy-responsive phase, the enriched CD11b+ fraction was stained with antibodies against Ly6G-FITC (1:200; clone 1A8, BD Biosciences), F4/80-PE (1:200; clone BM8, eBioscience) and Ly6C-ef450 (1:400; clone hk1.4, eBioscience). LIVE/DEAD® fixable aqua dead cell stain (ThermoFisher Scientific) was added 1:100 in PBS to exclude dead cells. CD11b+F4/80+ macrophages and F4/80-Ly6G+Ly6Clow neutrophils were isolated with the BD FACSARIA II sorter with DIVA software (BD Biosciences).
For the isolation of cell populations from end-stage tumors, we separated intratumoral CD11b+ and CD11b- cells by magnetic cell sorting as described above. The CD11b- and CD11b+ fractions were stained as described in Supplementary Table 1. CD11b+F4/80+ macrophages, CD11b+F4/80-Ly6G-Ly6C+ monocytes, CD11b+F4/80-Ly6G+Ly6Clow neutrophils, CD11b-CD45+CD11c- lymphocytes and CD11b-CD45-CD31- tumor cells were isolated with BD FACSARIA FUSION sorter with DIVA software (BD Biosciences).
Adoptive transfer of monocytes
Front legs, hind legs and hips were collected from female mTmG mice and bone marrow was flushed out. Bone marrow cells were incubated with Fc Block (1:50, CD16/CD32, BD Biosciences), stained with anti-Ly6G-APC (1:200, clone 1A8, Biolegend) and consequently negative selection for neutrophils was performed using magnetic columns (Miltenyi Biotec) as described previously61. The Ly6G- fraction was then stained with fluorochrome-conjugated antibodies (Supplementary Table 1). After gating out Lineage+ cells (CD3, CD8, CD4, NKp46, Ter119), Siglec F+, Sca1+ and cKIT+ cells, tdTomato+CD11bintLy6G-Ly6C+ monocytes were isolated with BD FACSARIA FUSION sorter with DIVA software (BD Biosciences). Between 1,5 and 2 x 106 tdTomato+ monocytes were adoptively transfer into the tail vein of a tumor-bearing KEP mouse treated with control antibody or anti-CSF-1R. Antibody treatments started at a tumor size of 25mm2 and one day after the second antibody injection (one week apart), monocytes were transferred. KEP mice were sacrificed four days later and tumors were isolated for flow cytometry analysis. Antibodies are listed in supplementary Table 1.
CRISPR/Cas9-mediated gene disruption and colony forming assay
IFNAR1 was knocked out from a cell line derived from a spontaneous KEP mammary tumor by transient transfection with a lentiCRISPRv262 containing IFNAR1-specific sgRNA targeting exon 1 (sgRNA1: 5’- GCTCGCTGTCGTGGGCGCGG - 3’). 24 h after transfection, cells were exposed to puromycin for 48 hours. Cells were stained with IFNAR1-PE (1:200, clone MAR1-5A3, eBioscience) and IFNAR1 negative cells were sorted with BD FACSARIA FUSION sorter with DIVA software (BD Biosciences).
250 KEP and IFNAR1 KO KEP cells were seeded in triplicate in a 24 well plate and the next day cells were treated with an increasing concentration of recombinant IFNα1 (Biolegend) for 7 days. On day 5, 4μM or 8 μM of cisplatin was added. At day 7, cells were washed, fixed in ice-cold methanol and incubated with 0.05% crystal violet. For quantification, crystal violet was dissolved in 10% acetic acid for 20 min and the absorbance was measured at 590nm.
Bone marrow-derived macrophages
To generate bone marrow-derived macrophages (BMDMs), bone marrow cells were harvested from the hind legs of WT mice and cultured for 7 days in RPMI medium containing 8% FCS, 100 IU/ml penicillin, 100 mg/ml streptomycin and 20ng/ml recombinant M-CSF (Peprotech). After differentiation, BMDMs were harvested and seeded in a 24 well plate (400000 BMDMs/well) and cultured overnight. The next morning, BMDMs were exposed to conditioned medium (CM) from a KEP cancer cell line in the presence of 8μg/ml of either control antibody or anti-CSF-1R for 24 hours. CM was obtained by culturing KEP cancer cells (80-90% confluency) for 24h in RPMI containing 8% FCS, 100 IU/ml penicillin and 100 mg/ml streptomycin. RNA of BMDMs was isolated with Isolate II RNA Mini Kit (Bioline) and quantitiative RT-PCR for Ifna was performed as described below.
RNA isolation and quantitative RT-PCR
RNA from sorted cells and tumors of KEP mice was isolated using Trizol (Invitrogen). Samples were treated with DNase I (Invitrogen) followed by RNA cleanup with the Qiagen RNeasy Mini Kit according to manufacturer’s recommendation. Isolated RNA was quantified with Nanodrop (Thermo Scientific). Transformation of RNA into cDNA was performed with the Cloned AMV First-Strand cDNA synthesis Kit (Invitrogen) using Oligo(dT) primers. 20 ng per well of cDNA was analyzed by SYBR green real-time PCR with 500 nM primers (Supplementary Table 2) using a LightCycler 480 thermocycler (Roche). Samples were run in duplicate and were only further considered if the difference between the Ct values of the duplo was less than 1 cycle. β-actin was used as a reference gene.
Luminex cytokine array
Tumors and mammary glands were prepared with BioRad cell lysis buffer and protein concentration of lysates was determined using the Pierce BCA Protein Assay Kit (Thermo Fischer Scientific) according to manufacturers’ recommendations. CSF-1 concentration in protein lysates was determined using Bio-Plex Pro™ Cytokine 23-Plex Kits (BioRad) and measured according to the manufactures’ instruction. Data acquisition and analysis was performed on a Bio-Plex 200 reader, using Bio-Plex Manager 6.0 software (Bio-Rad).
RNA sequencing and data analysis
RNA isolation, library construction and deep sequencing
CD11b+F4/80+ and CD11b+F4/80-Ly6G+Ly6Clow immune cell populations were isolated as described above from KEP tumors treated with either control antibody, anti-CSF-1R, cisplatin/control antibody or cisplatin/anti-CSF-1R at the therapy-responsive phase (tumor size ±100 mm2). Some of the biological replicates consisted of pools of cells from 2-6 different mice. Total RNA was extracted using RNeasy Mini and Microkits (Qiagen). According to the Ovation RNA-Seq system V2 and Encore Rapid library systems protocols (NuGen), 10 ng of RNA was converted into cDNA libraries, subsequently sequenced on a HiSeq 1500 system and demultiplexed using CASAVA v1.8 (Illumina).
Preprocessing of sequenced data
Using default parameters, all reads were aligned against the murine mm10 reference genome by TopHat2 v2.0.1163. The data were imported into Partek Genomics Suite v6.6 (PGS) and gene and transcript information was deducted before conducting normalization utilizing statistical software R (v3.3.1) and the DESeq2 package (http://dx.doi.org/10.1101/002832). Normalized read counts were floored to a value of at least 1 thereafter and the dataset was trimmed by defining a gene as expressed if the maximum value over all group means was higher than 10.
Identification of differentially expressed genes
Using PGS, a two-way ANalysis Of Variance (ANOVA) was performed to compute the top variable genes (treatment vs. control) within the dataset, as well as differentially expressed (DE) genes present in cisplatin/anti-CSF-1R neutrophils (vs. cisplatin/control antibody neutrophils). Genes were defined to be DE when having a fold change (FC) of ≥1.5 and an unadjusted p-value of ≤0.05. Based on the ANOVA model, Hierarchical clustering (HC) was performed on the top 400 variable genes within the dataset (neutrophils and macrophages, cisplatin/anti-CSF-1R vs. cisplatin/control antibody) using default settings in PGS.
Gene ontology enrichment analysis, GO network visualization, Ingenuity Pathway Analysis and Gene Set Enrichment Analysis
To link transcriptome information to prior knowledge, we applied Gene Ontology Enrichment Analysis (GOEA) on the 100 most up- and 100 most down-regulated genes (FC: ≥1.5, unadjusted p-value: ≤0.05) extracted from neutrophils exposed to cisplatin/anti-CSF-1R treatment (vs. cisplatin/control antibody-exposed neutrophils). Subsequently, the data were visualized employing BiNGO64, EnrichmentMap65 and Word Clouding66 plug-ins in Cytoscape. In addition, all differentially expressed genes found in neutrophils were analyzed with Ingenuity Pathway analysis (QIAGEN).
Gene Set Enrichment Analysis (GSEA) was performed utilizing the BubbleGUM GSEA tool67 to find enriched pathways in macrophages from cisplatin/anti-CSF-1R-treated tumors (vs. cisplatin/control antibody macrophages). Pathways interrogated were derived from the reactome gene sets and all pathways demonstrating a significant enrichment (False discovery rate (FDR): ≤0.25) in one condition were shown. Specifically addressing enrichment of Egr2 target genes in neutrophils from anti-CSF-1R-treated tumors vs. neutrophils from control antibody-treated tumors, GSEA was employed on transcription factor target gene sets using the GSEA tool previously published68. The reactome and transcription factor target gene sets were obtained via the online available Molecular Signatures Database (MSigDB) of the Broad Institute (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
No custom codes were used in the manuscript.
Evaluation of expression of IFN-stimulated genes in patient biopsies
The selection of type I IFN-stimulated genes was based on the RNA-seq results from our KEP mouse model upon cisplatin/anti-CSF-1R. The genes belong to the biological processes listed in Supplementary Table 3. We assessed the effect of anti-CSF-1R on these selected genes in human tumors by analyzing RNA-Seq data of paired baseline and on-treatment tumor biopsies of patients enrolled in a Clinical Phase 1 Trial with emactuzumab (RG7155), a humanized anti-human CSF-1R monoclonal antibody. Biopsies were taken from a multicenter, open-label study (clinicaltrials.gov identifier NCT01494688). Patients received emactuzumab every two weeks as intravenous infusion. Tumor biopsies of 31 patients with a broad range of different solid malignancies treated with either emactuzumab alone or in combination with paclitaxel (with an overrepresentation of breast [n=13] and ovarian cancer [n=7] samples) were collected. The study was conducted in accordance with the Declaration of Helsinki, current International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) guidelines and all applicable regulatory and ethical requirements. The study is compliant with all relevant ethical regulations regarding research involving human participants. All patients provided written informed consent before study-related procedures were performed. RNA extraction, RNA-Seq and data analysis was performed as previously described69.
Statistics and reproducibility
Information on study design, sample size, number of biological replicates, number of independent experiments and statistical analysis is reported in the main text and figure legends. The survival curves of cisplatin/control ab (or cisplatin only)-, cisplatin/anti-CSF-1R- and cisplatin/anti-CSF-1R/anti-Ly6G-treated mice were repeated and confirmed in a separate animal facility (Fig. 1f, Fig. 6d-f), other in vivo interventions were performed once. In vitro experiments were repeated independently with similar results. Statistical analyses were performed using GraphPad Prism 7 and 8 (GraphPad Software Inc., La Jolla, CA). The two-tailed Mann-Whitney test was used for immunohistochemistry and flow cytometry analysis. Two-way ANOVA with Tukey’s multiple comparison test was used for the quantification of crystal violet absorbance. Two-tailed Log-Rank tests were used for Kaplan-Meier survival curves. The Fisher’s exact test (two-sided) was used for metastasis analysis. P-values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by European Union (FP7 MCA-ITN 317445 TIMCC), the Dutch Cancer Society (NKI10623), the European Research Council (ERC consolidator award INFLAMET 615300), Worldwide Cancer Research (AICR 11–0677), the Netherlands Organization for Scientific Research NWO VIDI (917.96.307), and Oncode. KK is supported by an OOA/NWO Diamond grant. KEdV is an EMBO Young Investigator. JLS is a member of the excellence cluster ImmunoSensation. JLS is in part supported by the DFG (SFB704, Excellence Cluster ImmunoSensation). We thank M.D. Wellenstein, H. Garner, S. Bissinger, J. Borst and T. Schumacher for useful discussions. We thank Michael Hauptmann for advise on statistical analyses on the mouse survival curves. We thank the clinical investigators J.-Y. Blay, C. Gomez-Roca, J.-P. Delord, M. Toulmond, C. le Tourneau and A. Italiano for running clinical trials with emactuzumab, M. Cannarile, B. Quackenbush, A. Jegg for translational medicine support at Roche. We thank the Histology core facility within the Cancer Research UK Beatson Institute for performing NKp46 immunohistochemistry on mouse tumor tissue. We thank K. Wartha, S. Klarenbeek and I. Peters Rit for technical assistance, and the researchers involved in the RATHER project for generously providing tissue sections of human ILCs. We thank the flow cytometry facility, the animal facility, the animal pathology facility and the Core Facility Molecular Pathology and Biobank (CFMPB) at the Netherlands Cancer Institute.
Footnotes
Data Availability
The RNA-Seq data derived from mouse samples that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE101881. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101881).
Source data for Fig. 1a, 1d-i, 2, 3c-h, 4, 5, 6 and Supplementary Fig. 1a-b, 1e-f,1h-i, 2, 3e-h, 5a-c, 5f-i, 6a-f, 6i-n have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Author Contributions:
C.S., M.C., C.H.R., J.J. and K.E.dV. conceived the ideas and designed the experiments. C.S., M.C., C-S.H., S.B.C., K.K., A.vW, K.V., K.K., and K.E.dV. performed the experiments and analyzed the data. C.H.R. provided the anti-CSF-1R antibody and control antibody. Preparation of samples, RNA-Seq and bioinformatics analyses on murine cells were performed by S.T., T.U. and J.L.S. Metastasis scoring was performed by J-Y.S. C-H.O., D.R. and P.A.C. were involved in collection of patients samples and bioinformatic analysis of the human data. C.S. and K.E.dV. wrote the paper.
Competing Financial Interests:
Carola H. Ries, Chia-Huey Ooi and Dominik Rüttinger are employees of F. Hoffman La Roche. Carola H. Ries is an inventor of granted and pending patent applications relating to emactuzumab and stockholder in F. Hoffman La Roche. F. Hoffman La Roche provided financial research support for the experiments with anti-CSF-1R. Philippe A. Cassier received funding from Roche for the described clinical trial and other Roche-sponsored studies. The authors declare no additional competing financial interests.
References
- 1.Coffelt SB, de Visser KE. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends in immunology. 2015;36:198–216. doi: 10.1016/j.it.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 3.Ciampricotti M, Hau CS, Doornebal CW, Jonkers J, de Visser KE. Chemotherapy response of spontaneous mammary tumors is independent of the adaptive immune system. Nat Med. 2012;18:344–346. doi: 10.1038/nm.2652. [DOI] [PubMed] [Google Scholar]
- 4.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruffell B, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell MJ, et al. The prognostic implications of macrophages expressing proliferating cell nuclear antigen in breast cancer depend on immune context. PLoS One. 2013;8:e79114. doi: 10.1371/journal.pone.0079114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua W, Charles KA, Baracos VE, Clarke SJ. Neutrophil/lymphocyte ratio predicts chemotherapy outcomes in patients with advanced colorectal cancer. Br J Cancer. 2011;104:1288–1295. doi: 10.1038/bjc.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishi Y, et al. Blood neutrophil-to-lymphocyte ratio predicts survival in patients with colorectal liver metastases treated with systemic chemotherapy. Ann Surg Oncol. 2009;16:614–622. doi: 10.1245/s10434-008-0267-6. [DOI] [PubMed] [Google Scholar]
- 9.Miao Y, Yan Q, Li S, Li B, Feng Y. Neutrophil to lymphocyte ratio and platelet to lymphocyte ratio are predictive of chemotherapeutic response and prognosis in epithelial ovarian cancer patients treated with platinum-based chemotherapy. Cancer Biomark. 2016;17:33–40. doi: 10.3233/CBM-160614. [DOI] [PubMed] [Google Scholar]
- 10.Pistelli M, et al. Pre-treatment neutrophil to lymphocyte ratio may be a useful tool in predicting survival in early triple negative breast cancer patients. BMC Cancer. 2015;15:195. doi: 10.1186/s12885-015-1204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulus P, Stanley ER, Schafer R, Abraham D, Aharinejad S. Colony-stimulating factor-1 antibody reverses chemoresistance in human MCF-7 breast cancer xenografts. Cancer Res. 2006;66:4349–4356. doi: 10.1158/0008-5472.CAN-05-3523. [DOI] [PubMed] [Google Scholar]
- 12.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchem JB, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73:1128–1141. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Mitri D, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515:134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 15.Houthuijzen JM, et al. Lysophospholipids secreted by splenic macrophages induce chemotherapy resistance via interference with the DNA damage response. Nat Commun. 2014;5 doi: 10.1038/ncomms6275. 5275. [DOI] [PubMed] [Google Scholar]
- 16.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson OC, Kim H, Quail DF, Foley EA, Joyce JA. Tumor-Associated Macrophages Suppress the Cytotoxic Activity of Antimitotic Agents. Cell Rep. 2017;19:101–113. doi: 10.1016/j.celrep.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weizman N, et al. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene. 2014;33:3812–3819. doi: 10.1038/onc.2013.357. [DOI] [PubMed] [Google Scholar]
- 19.Nakasone ES, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21:488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nywening TM, et al. Targeting both tumour-associated CXCR2(+) neutrophils and CCR2(+) macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut. 2018;67:1112–1123. doi: 10.1136/gutjnl-2017-313738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ries CH, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Quail DF, Joyce JA. Molecular Pathways: Deciphering Mechanisms of Resistance to Macrophage-Targeted Therapies. Clin Cancer Res. 2017;23:876–884. doi: 10.1158/1078-0432.CCR-16-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 24.Derksen PW, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:1810–1820. doi: 10.1182/blood-2011-09-379214. [DOI] [PubMed] [Google Scholar]
- 26.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doornebal CW, et al. A preclinical mouse model of invasive lobular breast cancer metastasis. Cancer Res. 2013;73:353–363. doi: 10.1158/0008-5472.CAN-11-4208. [DOI] [PubMed] [Google Scholar]
- 28.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley EW, Ruan MM, Oursler MJ. Novel pro-survival functions of the Kruppel-like transcription factor Egr2 in promotion of macrophage colony-stimulating factor-mediated osteoclast survival downstream of the MEK/ERK pathway. J Biol Chem. 2008;283:8055–8064. doi: 10.1074/jbc.M709500200. [DOI] [PubMed] [Google Scholar]
- 30.Gomez-Roca CA, et al. Phase I study of RG7155, a novel anti-CSF1R antibody, in patients with advanced/metastatic solid tumors. Journal of Clinical Oncology. 2015;33:3005–3005. [Google Scholar]
- 31.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 32.Vacchelli E, et al. Autocrine signaling of type 1 interferons in successful anticancer chemotherapy. Oncoimmunology. 2015;4:e988042. doi: 10.4161/2162402X.2014.988042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 34.Snell LM, McGaha TL, Brooks DG. Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 2017;38:542–557. doi: 10.1016/j.it.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuertes MB, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffelt SB, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sistigu A, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 38.Burnette BC, et al. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71:2488–2496. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bidwell BN, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18:1224–1231. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- 41.Callari M, et al. Subtype-dependent prognostic relevance of an interferon-induced pathway metagene in node-negative breast cancer. Mol Oncol. 2014;8:1278–1289. doi: 10.1016/j.molonc.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snijders AM, et al. An interferon signature identified by RNA-sequencing of mammary tissues varies across the estrous cycle and is predictive of metastasis-free survival. Oncotarget. 2014;5:4011–4025. doi: 10.18632/oncotarget.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiavoni G, et al. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res. 2011;71:768–778. doi: 10.1158/0008-5472.CAN-10-2788. [DOI] [PubMed] [Google Scholar]
- 44.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Critchley-Thorne RJ, et al. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci U S A. 2009;106:9010–9015. doi: 10.1073/pnas.0901329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quail DF, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352 doi: 10.1126/science.aad3018. aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan D, et al. Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene. 2017 doi: 10.1038/onc.2017.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 51.Pylaeva E, Lang S, Jablonska J. The Essential Role of Type I Interferons in Differentiation and Activation of Tumor-Associated Neutrophils. Front Immunol. 2016;7:629. doi: 10.3389/fimmu.2016.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rocha BC, et al. Type I Interferon Transcriptional Signature in Neutrophils and Low-Density Granulocytes Are Associated with Tissue Damage in Malaria. Cell Rep. 2015;13:2829–2841. doi: 10.1016/j.celrep.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cunningham CR, et al. Type I and Type II Interferon Coordinately Regulate Suppressive Dendritic Cell Fate and Function during Viral Persistence. PLoS Pathog. 2016;12:e1005356. doi: 10.1371/journal.ppat.1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corrales L, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 58.Michaut M, et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep. 2016;6 doi: 10.1038/srep18517. 18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schouten PC, et al. Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms. Mol Oncol. 2015;9:1274–1286. doi: 10.1016/j.molonc.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salvagno C, de Visser KE. Purification of Immune Cell Populations from Freshly Isolated Murine Tumors and Organs by Consecutive Magnetic Cell Sorting and Multi-parameter Flow Cytometry-Based Sorting. Methods Mol Biol. 2016;1458:125–135. doi: 10.1007/978-1-4939-3801-8_10. [DOI] [PubMed] [Google Scholar]
- 62.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 65.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oesper L, Merico D, Isserlin R, Bader GD. WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol Med. 2011;6:7. doi: 10.1186/1751-0473-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spinelli L, Carpentier S, Montanana Sanchis F, Dalod M, Vu Manh TP. BubbleGUM: automatic extraction of phenotype molecular signatures and comprehensive visualization of multiple Gene Set Enrichment Analyses. BMC Genomics. 2015;16:814. doi: 10.1186/s12864-015-2012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pradel LP, et al. Macrophage Susceptibility to Emactuzumab (RG7155) Treatment. Mol Cancer Ther. 2016;15:3077–3086. doi: 10.1158/1535-7163.MCT-16-0157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.