Summary
Altered metabolism is a hallmark of cancer growth, forming the conceptual basis for development of metabolic therapies as cancer treatments. We performed in vivo metabolic profiling and molecular analysis of lung squamous cell carcinoma (SCC) to identify metabolic nodes to therapeutically target. Lung SCCs adapt to chronic mTOR inhibition and suppression of glycolysis through the GSK3α/β signaling pathway which upregulates glutaminolysis. Phospho-GSK3α/β protein levels are predictive of response to single therapy mTOR inhibition while combinatorial treatment with the glutaminase inhibitor CB-839 effectively overcomes therapy resistance. In addition, we identified a conserved metabolic signature in a broad spectrum of hypermetabolic human tumors that may be predictive of patient outcome and response to combined metabolic therapies targeting mTOR and glutaminase.
Introduction
Lung squamous cell carcinomas (SCC) are a highly aggressive subset of non-small cell lung cancer (NSCLC). Patients with lung SCC have a worse prognosis than those with lung adenocarcinomas (ADC) (Morgensztern et al., 2009). Lung SCC utilize high levels of glucose metabolism to meet the anabolic and catabolic needs required to sustain rapid tumor growth and are readily detected by 18F-fluoro-2-deoxyglucose positron emission tomography (18F-FDG PET) (Goodwin et al., 2017). Higher uptake of 18F-FDG by tumors predicts higher risk of recurrence as well as lower overall survival in patients with NSCLC regardless of histology (Berghmans et al., 2008). Moreover, lung SCC are highly resistant to conventional chemotherapy and many patients on recently approved checkpoint inhibitors eventually progress after an initial response (Brahmer et al., 2015) highlighting the need to identify druggable targets to more effectively treat this disease.
The Cancer Genome Atlas (TCGA) for lung SCC revealed frequent mutations in receptor tyrosine kinases (RTKs), AKT and PI3K that converge to activate the mechanistic target of rapamycin (mTOR) signaling pathway (Ding et al., 2008; Hammerman et al., 2012). The mTOR pathway is a key positive regulator of cell growth and metabolism (Guertin and Sabatini, 2007). Importantly, the PI3K-AKT-mTOR signaling axis is a druggable pathway as evidenced by multiple early phase clinical trials which have begun assessing targeted therapies against PI3K, AKT and mTOR in lung cancer (NCT01493843, NCT01248247, NCT01058707). However, single therapy targeted agents against the PI3K-AKT-mTOR pathway have had limited clinical efficacy suggesting the need to identify more effective combinatorial strategies.
Failure of single therapy kinase inhibitors to successfully treat lung SCC suggests that these tumors are not solely dependent on PI3K-AKT-mTOR signaling and glucose metabolism for survival. Agreeing with clinical results, we have demonstrated that chronic inhibition with the catalytic mTOR kinase inhibitor MLN128 effectively inhibited mTOR and suppressed glucose metabolism in both ADC and SCC tumor subtypes but failed to restrict tumor growth in lung SCC (Momcilovic et al., 2015). These results suggested that lung SCC may utilize alternate nutrients such as amino acids to circumvent MLN128-mediated suppression of glycolysis. Lung SCC upregulate the glucose transporter 1 (GLUT1) as well as the glutamine transporter SLC1A5 providing further evidence these tumors likely exhibit a dual reliance on glucose and glutamine (Goodwin et al., 2017; Hassanein et al., 2013). While acute inhibition of either GLUT1 or SLC1A5 transporters impaired survival of NSCLC cells (Goodwin et al., 2017) our data suggests chronic suppression of glycolysis through mTOR inhibition may elicit a metabolic adaptation through glutaminolysis that is more likely to occur in hypermetabolic tumors such as lung SCC. Recent in vivo studies have examined glucose and glutamine metabolism in human NSCLC and identified metabolic heterogeneity (Hensley et al., 2016; Sellers et al., 2015). Additionally, in vivo profiling of lung tumors in a KrasG12D;Trp53−/− (KP) mutant mouse model of NSCLC demonstrated these tumors were dependent on glycolysis for tumor cell survival but not glutamine (Davidson et al., 2016). However, these studies were restricted to lung ADC subtypes and, to the best of our knowledge, to date there have been no in vivo studies to profile either glucose or glutamine metabolism in lung SCC. In this study we investigated the molecular mechanisms by which these tumors adapt their metabolism to overcome suppression of glycolysis following chronic inhibition of mTOR.
Results
Lung SCC maintain high rates of anabolic growth following chronic suppression of glycolysis with the mTOR inhibitor MLN128
Lung SCC tumors are consistently avid for 18F-FDG suggestive of a hypermetabolic growth phenotype (Goodwin et al., 2017). To evaluate rates of proliferation in SCC we analyzed Ki67 mRNA expression levels from TCGA for lung SCC and ADC (Ding et al., 2008; Hammerman et al., 2012). We found mRNA expression of Ki67 was significantly higher in patients with SCC tumors compared to patients with ADC tumors (Figure 1A). We next analyzed Ki67 staining of both ADC and SCC lung tumors from the autochthonous KrasG12D;Lkb1−/− (KL) genetically engineered mouse model (GEMM) that develops tumors with multiple histologies including both ADC and SCC tumors as previously described (Ji et al., 2007).
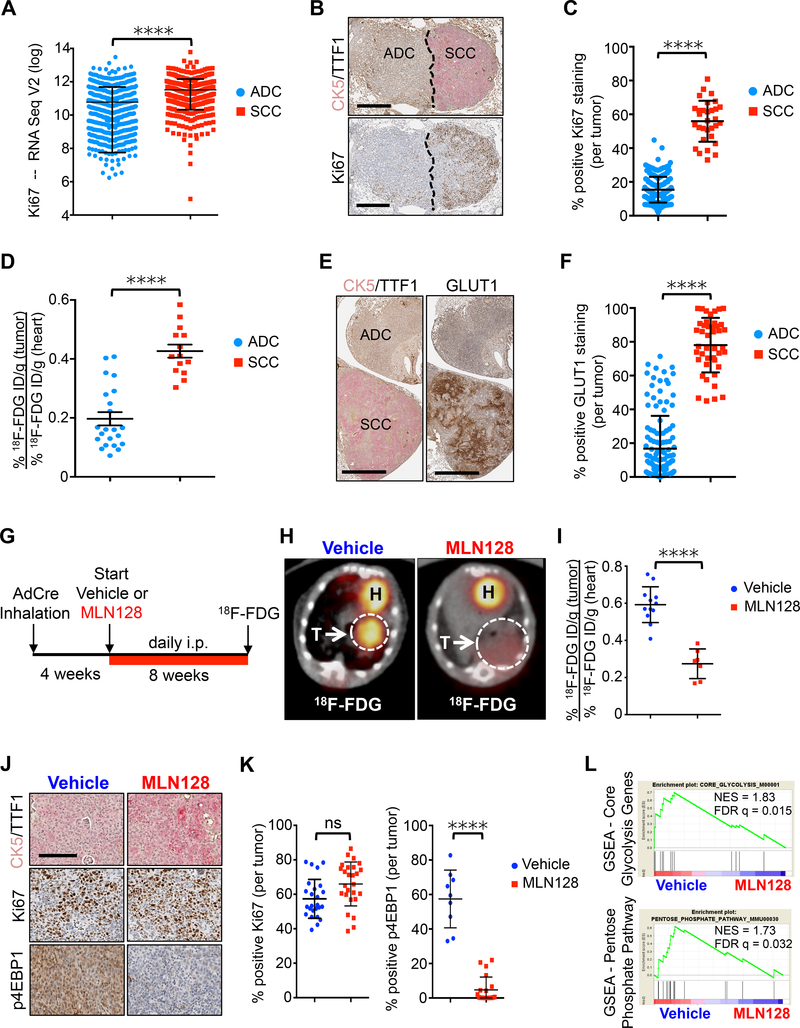
Figure 1.
Chronic MLN128 treatment reduces 18F-FDG uptake, but not tumor cell proliferation in a KL mouse model of SCC.
(A) Ki67 mRNA expression in lung ADC (n = 517) and SCC (n = 501) based on TCGA data analysis. The data are represented as the mean ± SD. Statistical significance (****p<0.0001) was calculated using two-tailed t test. (B) Representative IHC staining for CK5/TTF1 (top panel) and Ki67 (bottom panel) in tumors from KL mice. SCC – squamous cell carcinoma, ADC – adenocarcinoma. Scale bar = 200 μm. (C) Quantification of Ki67 staining from tumors in (B); n = 226 (ADC), n = 31 (SCC). (D) 18F-FDG uptake in lung SCC (n = 14) and ADC (n = 22) from KL mice in percent injected dose/gram (% ID/g). (E) Representative GLUT1 staining in lung SCC and ADC tumors from KL mice. Scale bar = 200 μm. (F) Quantification of GLUT1 from tumors in (E); n = 126 (ADC), n = 43 (SCC). (G) Dosing regiment for KL mice treated daily with Vehicle or MLN128 according to indicated scheme. (H) Representative 18F-FDG PET/CT images from KL mice treated with Vehicle or MLN128 for 8 weeks. Tumors are circled with arrow; T = tumor; H = heart. (I) Quantification of 18F-FDG signal (% ID/g) after 8 weeks of treatment; n = 12 (ADC), n = 7 (SCC). (J) Representative CK5/TTF1, Ki67 and p4EBP1 staining of lung SCC tumors from KL mice treated with Vehicle or MLN128 for 8 weeks. Scale bar = 50 μm (K) Quantification of Ki67 staining (left panel; n = 25 (Vehicle), n = 25 (MLN128)) and p4EBP1 staining (right panel; n = 9 (Vehicle), n = 20 (MLN128)) in SCC tumors from (J). (L) Gene set enrichment analysis (GSEA) of genes involved in Core Glycolysis and Pentose Phosphate Pathway from tumors isolated from KL mice treated daily with Vehicle or MLN128 for 8 weeks. NES = normalized enrichment score; FDR q = false discovery rate q-value. The data are represented as the mean ± SEM. Statistical significance (****p<0.0001; ns, not significant) was calculated using two-tailed t test. See also Figure S1.
We confirmed that Ki67 was significantly increased in the SCC compared to ADC tumor compartments (Figures 1B and 1C). We differentiated SCC and ADC tumors by staining for cytokeratin 5 (CK5) as a marker of squamous cells and thyroid transcription factor (TTF1) as a marker of ADC (Figures 1B). We reasoned the higher Ki67 index in SCC meant higher metabolic demands to sustain their accelerated anabolic growth. We took advantage of 18F-FDG PET and computed tomography (CT) imaging and quantified uptake of 18F-FDG between SCC and ADC tumors in KL mice. We confirmed that lung SCC tumors had significantly higher 18F-FDG uptake and GLUT1 staining compared to ADC (Figures 1D–1F and S1A). These results demonstrate the KL model accurately modeled the hypermetabolic glycolytic phenotype(s) of lung SCC tumors seen in patients.
We previously showed that chronic treatment of KL mice with the catalytic mTOR kinase inhibitor MLN128 significantly reduced the total number of lesions (Momcilovic et al., 2015). However, we found that in every mouse treated with MLN128 there were large SCC tumors that escaped treatment despite reduced mTOR signaling. We repeated this study using KL mice according to the scheme in Figure 1G. Mice were 18F-FDG PET imaged following 8 weeks of daily treatment with vehicle or MLN128. We again saw that SCC consistently escaped MLN128 treatment and that the 18F-FDG signal was significantly reduced in MLN128 treated SCC tumors as compared to vehicle treated ones (Figures 1H and 1I). While MLN128 induced a metabolic response in SCC tumors and effectively inhibited mTORC1 signaling as shown by reduced phosphorylation of the mTORC1 substrate 4EBP1 (p4EBP1), treatment failed to reduce the Ki67 index (Figures 1J and 1K). Gene set enrichment analysis (GSEA) performed on vehicle and MLN128 treated KL SCC tumors confirmed a decrease in expression of core genes involved in glycolysis, the pentose phosphate pathway and the mTOR pathway (Figures 1L and S1B). Together, our data suggests that hypermetabolic SCC tumors can maintain high proliferation rates despite chronic suppression of glycolysis with MLN128.
PET imaging based metabolic profiling of lung SCC identifies high influx of glucose and glutamine
We hypothesized that for lung SCC to maintain a constant Ki67 index in which majority of the tumor cells are cycling (Figure 1C) they must consume an alternate carbon source(s) if glucose metabolism is limited. Lung SCC express high levels of both GLUT1 and SLC1A5 transporters and previous studies have shown that expression of these transporters correlates with uptake of 18F-FDG and 18F- or 11C-labeled glutamine (Goodwin et al., 2017; Hassanein et al., 2016; Momcilovic et al., 2017; Venneti et al., 2015). We explored the possibility that SCC tumors may have metabolic dependencies that are distinct from ADCs, and may rely on glutamine uptake as a carbon source to supplement glucose restriction. We used PET imaging with 18F-FDG and 11C-labeled glutamine (11C-Gln) to noninvasively profile metabolic influx of glucose and glutamine in lung SCC.
We conducted serial PET imaging of KL mice with 18F-FDG and 11C-Gln within the same week as previously described (Momcilovic et al., 2017) (Figure S2). Representative images are shown in Figure 2A. In agreement with previous data, SCC tumors were highly avid for 18F-FDG (Figure 2A). We imaged the same mouse with 11C-Gln and identified this same tumor was positive for 11C-Gln (Figure 2A). Upon gross anatomical examination, we identified a large tumor that was localized below the heart, matching the position observed in PET images (Figure 2B). Immunohistochemical (IHC) examination of the tumor confirmed it was squamous based on morphology from hematoxylin and eosin (H&E) and positive CK5 staining (Figures 2C and 2D). As expected, positive GLUT1 staining in the tumor was associated with high uptake of 18F-FDG, and positive SLC1A5 staining was associated with high 11C-Gln uptake (Figures 2A and 2D).
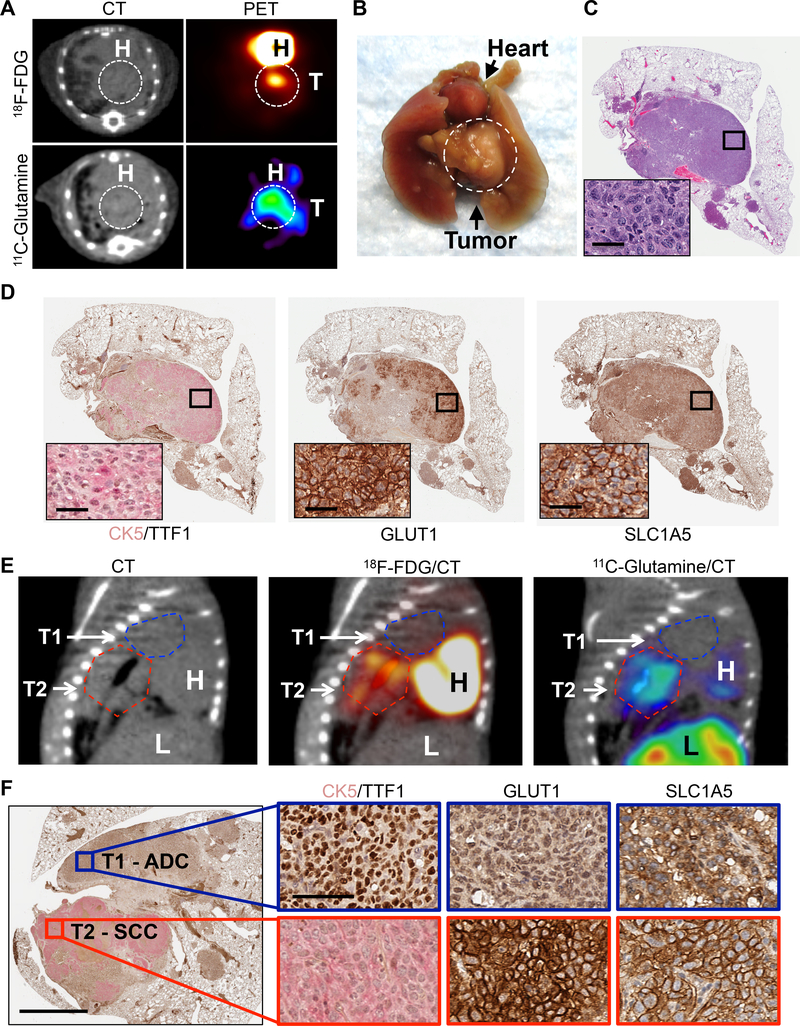
Figure 2.
High influx of glucose and glutamine in lung SCC detected by PET imaging.
(A) Representative PET and CT images of SCC tumors from KL mice imaged with 18F-FDG and 11C-Glutamine. (B) Image of whole lungs and heart from mouse imaged in (A). Tumor is circled. T = tumor. H = heart. (C) Left lung lobe and tumor from mouse imaged in (A) stained for hematoxylin and eosin (H&E). Magnified image shown in the inset; scale bar = 25 μm. (D) Serial sections of the left lung lobe stained with CK5/TTF1, GLUT1, SLC1A5 from left lobe from mouse imaged in (A). Magnified images shown in the inset; scale bar = 25 μm. (E) PET/CT images of KL mouse with both ADC and SCC tumors imaged with CT (left), 18F-FDG (middle), and 11C-Glutamine (right). H = heart, L = liver. T1 and T2 are outlined with dashed lines (T1 = the blue line, T2 = the red line). (F) PET/CT imaged ADC and SCC lung tumors from (E) were stained with antibodies against CK5/TTF1, GLUT1 and SLC1A5 (left side). Scale bar = 1 mm. Magnified images from T1 (blue box) and T2 (red box) are shown on the right side. Scale bar = 50 μm. See also Figure S2.
We next performed comparative PET analysis of 18F-FDG and 11C-Gln uptake between ADC and SCC tumor subtypes in KL mice. Figure 2E shows representative PET/CT images of a mouse with two tumors – an ADC, labeled T1, and a SCC, labeled T2 (Figure 2F). The ADC (T1) showed lower avidity for both 18F-FDG and 11C-Gln as compared to the SCC (T2). Histological analysis of the tumors confirmed T1 was an ADC and positive for TTF1 while T2 was a SCC and positive for CK5; the SCC tumor (T2) had strong plasma membrane staining for both GLUT1 and SLC1A5 as compared to the ADC (T1) (Figure 2F). These results demonstrate that in vivo, lung SCCs have high influx of both glucose and glutamine with metabolic dependencies that are distinct from lung ADCs.
In vivo metabolic tracing in lung SCC identifies adaptive glutamine metabolism following chronic MLN128 treatment.
Having established that lung SCC tumors have high influx of glutamine, we explored the possibility that glutamine supports biomass generation when glycolysis is inhibited by MLN128. We next measured glucose and glutamine consumption in cell culture media in the RH2 human SCC line following treated with MLN128. We demonstrated that both MLN128 and the allosteric mTORC1 inhibitor rapamycin suppressed glucose uptake while concomitantly inducing an increase in glutamine uptake in vitro (Figures 3A and S3A). We next stained RH2 tumor sections from mouse xenografts for GLUT1 and SLC1A5 and confirmed that both markers are highly expressed on the plasma membrane of tumor cells (Figure 3B). Treatment of RH2 xenografts with MLN128 outlined in Figure 3C induced an identical metabolic response as seen in SCC tumors from KL mice. MLN128 significantly suppressed 18F-FDG uptake in RH2 tumors (Figures 3D and S3B) and inhibited mTORC1 signaling as measured by p4EBP1 staining (Figures 3E and 3F), but it failed to reduce tumor growth or tumor cell proliferation as measured by Ki67 staining (Figures 3E–3G).
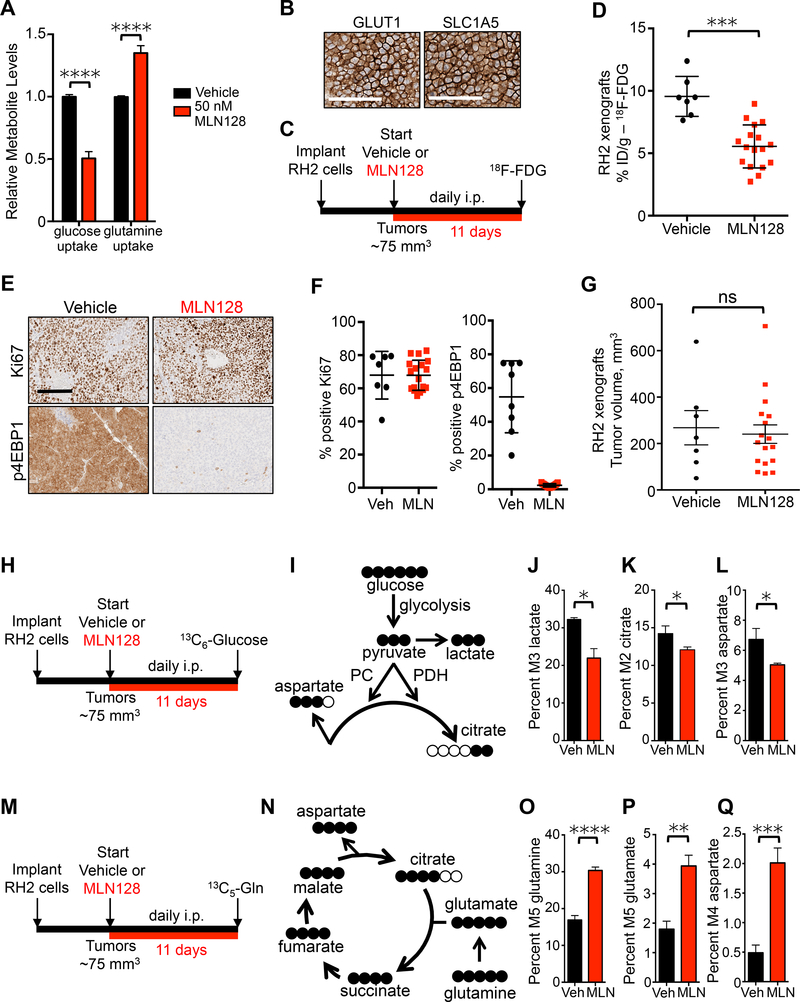
Figure 3.
Lung SCC increase glutaminolysis in vivo following treatment with MLN128.
(A) Changes in relative uptake of glucose and glutamine by RH2 cells in response to treatment with 50 nM MLN128 for 72 hr; n = 6 for Vehicle and MLN128. (B) RH2 tumor xenografts stained for GLUT1 and SLC1A5. Scale bar = 50 μm. (C) Dosing and PET imaging regimen for RH2 xenografts treated with Vehicle or MLN128 (D) 18F-FDG uptake measured by % ID/g in RH2 xenografts following treatment as described in (C); n = 7 (Vehicle), n= 17 (MLN128). (E) Representative images from RH2 tumors stained for Ki67 or p4EBP1 from mice treated daily with Vehicle or MLN128 as described in (C). Scale bar = 100 μm. (F) Quantification of Ki67 (left) and p4EBP1 (right) from Vehicle (n = 7) and MLN128 (n = 18) treatment groups. (G) Tumor volumes of RH2 xenografts following treatment with Vehicle (n = 7) or MLN128 (n = 17). (H) Overview of dosing and 13C6-Glucose infusion of RH2 xenografts. (I) Schematic of isotopomer conversion from fully labeled glucose. (J, K, and L) Graphs representing percent M3 labeled lactate (J), M2 labeled citrate (K) and M3 labeled aspartate (L) in RH2 tumors from mice treated with Vehicle (n = 4) or MLN128 (n = 6) and infused with 13C6-Glucose. (M) Overview of dosing and 13C5-Glutamine infusion of RH2 xenografts. (N). Schematic of isotopomer conversion from fully labeled glutamine. (O, P, and Q) Graphs representing percent M5 labeled glutamine (O), M5 labeled glutamate (P) and M4 labeled aspartate (Q) in RH2 tumors from mice treated with Vehicle (n = 5) or MLN128 (n = 6) and infused with 13C5-Glutamine. The data are represented as the mean ± SEM. Statistical significance (*p<0.05; **p<0.01; ***<0.001; ****p<0.0001; ns, not significant) was calculated using two-tailed t test. See also Figure S3.
We next directly measured contribution of glucose to TCA cycle intermediates by perfusing RH2 mouse xenografts with 13C6-Glucose represented in Figure 3H. The expected distribution of labeled carbons as glucose enters glycolysis and the TCA cycle is shown in Figure 3I. Glucose can contribute two carbons to citrate via acetyl-coA and three carbons to both lactate and aspartate. Plasma glucose enrichment reached 63% (± 9.7%) in mice infused with 13C6-Glucose for 120 min (Figure S3C). We detected significantly lower levels of lactate (M3 labeled) derived from 13C6-glucose in MLN128 treated mice compared with vehicle treated mice (Figure 3J). We also detected significantly lower levels of citrate (M2 labeled) and aspartate (M3 labeled) derived from 13C6-Glucose in MLN128 group, suggesting that contribution of glucose to TCA cycle was lowered by MLN128 (Figures 3K and 3L). The reduced glucose metabolism in MLN128 treated tumors is not due to differential glucose uptake as both unlabeled (M0) and fully labeled (M6) glucose were present at similar levels in vehicle and MLN128 treated tumors (Figure S3D). We also did not detect a decrease in ATP or AMP levels in MLN128 treated tumors (Figure S3E).
Next, we repeated the RH2 xenograft experiment and perfused mice with 13C5-Glutamine (Figure 3M). The expected distribution of labeled carbons from fully labeled glutamine is shown in Figure 3N. We detected significantly higher uptake of fully labeled (M5) glutamine in tumors in the MLN128 treated group compared to the vehicle group, (Figure 3O and S3F) suggesting that glutamine uptake is enhanced when glycolysis is restricted. We also detected significantly higher levels of fully labeled (M5) glutamate derived from 13C5-glutamine in MLN128 treated tumors (Figure 3P and S3F). Aspartate is a critical amino acid that supports proliferation (Sullivan et al., 2015). We detected a four-fold increase in levels of labeled aspartate (M4) derived from 13C5-glutamine in MLN128 treated tumors compared to vehicle treated tumors (Figure 3Q and S3F), suggesting that glutamine undergoes oxidative decarboxylation (Metallo et al., 2012; Mullen et al., 2012) and is used to replenish TCA cycle intermediates depleted by treatment with MLN128 and subsequent reduced glycolysis. Overall, our data describes a metabolic reliance of lung SCC on both glucose and glutamine in vivo that enables tumors to rapidly adapt to MLN128-mediated suppression of glycolysis.
The GSK3α/β pathway upregulates cMYC and cJUN to drive adaptive glutamine metabolism in lung SCC following chronic mTOR inhibition.
We next sought to identify the molecular mechanism(s) regulating metabolic adaptation in lung SCC that enabled them to escape MLN128 treatment. Tumor cells can adapt to prolonged treatment with rapamycin or catalytic mTOR inhibitors by upregulating receptor tyrosine kinase(s) and AKT activity (Momcilovic et al., 2015; Rodrik-Outmezguine et al., 2011). We previously found that SCC tumors from KL mice that escaped MLN128 treatment had increased levels of phospho-AKT Thr308 (pAKTT308) as well as increased levels of the phosphorylated form of its downstream substrate glycogen synthase kinase 3 (GSK3α/β), a regulator of cell growth and proliferation, which AKT phosphorylates at Ser21/Ser9 (pGSK3α/βS21/9). Inhibition of AKT had additive effects when combined with mTOR inhibitors MLN128 or rapamycin in vitro (Momcilovic et al., 2015). We sought to investigate the pathways downstream of pGSK3α/βS21/9 to better understand how the AKT and GSK3α/β pathways may regulate metabolic adaptation in SCC tumors.
Active GSK3α/β modulates the protein stability of the transcription factors cMYC and cJUN by facilitating their degradation by E3 ubiquitin ligases (Xu et al., 2009) as represented in Figure 4A. Both cMYC and cJUN regulate the gene expression of glutaminase (GLS) (Gao et al., 2009; Lukey et al., 2016), which is an enzyme that catalyzes the conversion of glutamine to glutamate. Phosphorylation of GSK3α/β at serine 21 and 9 inhibits activity and results in increased protein stability of both cMYC and cJUN (Gregory et al., 2003; Sears et al., 2000; Wei et al., 2005) (Figure 4A). These studies suggest that GSK3α/β may serve as a key node that upregulates GLS expression and glutamine metabolism following mTOR inhibition.
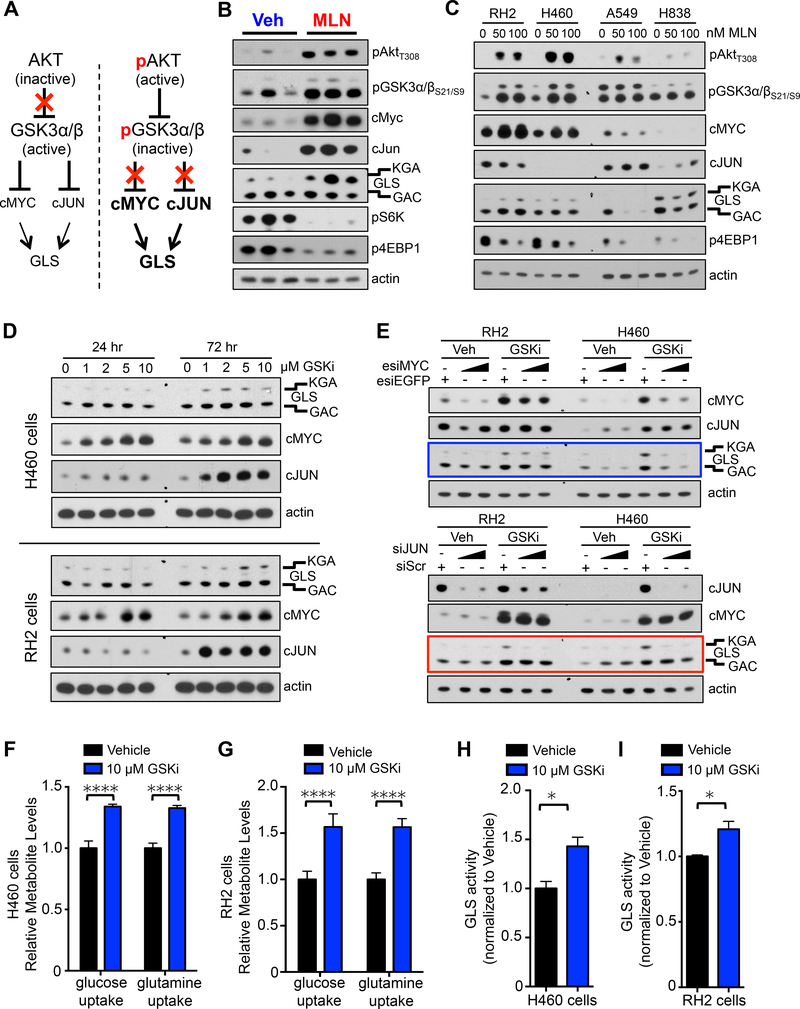
Figure 4.
GSK3α/β regulates cMYC and cJUN protein levels and glutaminase activity.
(A) Model of GSK3α/β-mediated regulation of glutaminase (GLS). (B) Lung SCC tumor lysates from KL mice treated daily with Vehicle or MLN128 for 8 weeks were immunoblotted with the indicated antibodies. (C) Human SCC/LCC cell lines (RH2 and H460) and ADC cell lines (A549 and H838) were treated with 50 nM or 100 nM MLN128 for 72 hr. Lysates were probed with indicated antibodies. (D) Human cell lines H460 (top) and RH2 (bottom) were treated with increasing doses of GSK3 inhibitor CHIR99021 (GSK3i) for indicated time. Lysates were probed with indicated antibodies. (E) Humans cell lines RH2 and H460 were treated with Vehicle or GSK inhibitor CHIR99021 (GSKi) in the presence of either esiEGFP or increasing doses of esiMyc (top panel) or either siScr or increasing doses of siJun (bottom panel) for 72 hr. Lysates were probed with indicated antibodies. (F and G) Relative uptake of glucose and glutamine in human cell line H460 (F) or RH2 (G) following 72 hr treatment with 10 μM GSKi; n = 6 for Vehicle and GSKi. (H and I) GLS activity measured in H460 (H) and RH2 (I) cells following treatment with Vehicle or 10 μM GSKi for 72 hr; n = 3 for Vehicle and GSKi. The data are represented as the mean ± SEM. Statistical significance (*p<0.05; ****p<0.0001) was calculated using two-tailed t test. See also Figure S4.
The model in Figure 4A proposes that inhibition of AKT should prevent GSK3α/β mediated upregulation of GLS in the presence of MLN128. Interestingly, we found that nanomolar doses of the pan AKT inhibitor MK2206 induced cMYC expression (Figure S4A), agreeing with a recent study that showed Lapatinib mediated AKT inhibition resulted in cMYC upregulation in breast cancer cells (Matkar et al., 2015). The combination of low dose MLN128 and MK2206 failed to fully suppress upregulation of GLS (Figure S4A) suggesting that MLN128+MK2206 may not be an effective treatment combination for lung SCC. This prompted us to identify druggable targets downstream of GSK3α/β that would overcome resistance to MLN128.
Examination of GSK3α/β signaling in lung SCC tumor lysates isolated from KL mice showed increased phosphorylation of AKTT308 and GSK3α/βS21/9 in MLN128 treated tumors as compared to vehicle (Figure 4B). We also observed an increase in total levels of PDK1, a protein that relays signal from RTKs to AKT while total levels of GSK3α/β were not changed (Figure S4B). We identified that MLN128 treated SCCs had higher levels of nuclear localized cMYC and cJUN as well as the KGA isoform of GLS (Figures 4B, and S4C). The GLS antibody correctly identified both KGA and GAC isoforms and was validated using shRNAs against GLS (Figure S4D). We found cJUN was phosphorylated at Ser73 (a JNK target site) (Figures S4B and S4C), marking transcriptionally active cJUN (Lukey et al., 2016). Both mTORC1 substrates pS6K and p4EBP1 were reduced in MLN128 treated tumors confirming mTORC1 was sufficiently inhibited in these tumors (Figure 4B). Interestingly, protein levels of GLUT1 and SLC1A5 did not change significantly (Figure S4B).
We next measured response to MLN128 in human lung SCC and large cell carcinoma (LCC) lines RH2 and H460, respectively. Nanomolar doses of MLN128 inhibited mTORC1 as shown by reduced p4EBP1 (Figure 4C). Both cell lines showed an increase in pAKTT308, pGSK3α/βS21/9 and cMYC in response to MLN128 treatment (Figure 4C). Interestingly, we observed a modest increase in GLS in RH2 but not H460 cells. The differential response to MLN128 between RH2 and H460 predicts that in vivo H460 tumor xenografts may be responsive to MLN128 while RH2 tumors are expected to be refractory to MLN128.
Additionally, we examined lung ADC cell lines to determine if the molecular response to MLN128 was unique to SCC tumors or conserved across both tumor histologies. Nanomolar doses of MLN128 inhibited mTORC1 in lung ADC cell lines A549 and H838 as measured by reduced levels of p4EBP1 (Figure 4C). Of note, we observed higher p4EBP1 levels in SCC than ADC (Figure 4C). While A549 cells showed a modest increase in pAKTT308 levels following MLN128 treatment, both ADC lines showed no change in pGSK3α/βS21/9, cMYC or cJUN levels (Figure 4C). Furthermore, MLN128 induced a reduction in GLS in both ADC cell lines opposite that which was observed in SCC cells.
We next tested whether inhibition of GSK3α/β activity in human NSCLC cell lines would lead to increased cMYC, cJUN and GLS protein levels. We treated RH2 and H460 cell lines with increasing doses of the GSK3α/β inhibitor (GSK3i) CHIR99021 and evaluated levels of GLS, cMYC and cJUN. GSK3i induced a dose dependent increase in the protein levels of GLS, cMYC and cJUN (Figure 4D) in both cell lines. This agrees with data from KL GEMMs shown in Figure 4B. Interestingly, while cMYC protein levels increased within 24 hr, both cJUN and GLS had more pronounced increase in protein levels at 72 hr (Figure 4D). Similar to tumor lysates from KL mice, cJUN was phosphorylated at Ser73 (a JNK target site) in H460 and RH2 cells (Figures S4E and S4F).
We next performed siRNA-mediated knockdown of cMYC or cJUN in RH2 and H460 cells in the presence of GSK3i and measured GLS protein levels. We showed that following cJUN knockdown, both RH2 and H460 cells are unable to upregulate GLS in the presence of GSK3i thus demonstrating a reliance on cJUN (Figure 4E bottom panel red box). In contrast, cMYC knockdown did not hinder GLS upregulation in RH2 cells while H460 cells were unable to upregulate GLS following cMYC knockdown (Figure 4E top panel blue box). These results suggest H460 cells but not RH2 cells may be reliant on cMYC to upregulate glutamine metabolism. Of note, we did observe that RH2 cells have higher basal expression of cMYC and are more resistant to cMYC knockdown than H460 cells. In addition, inhibition of the Jun N-terminal Kinase (JNK) using the JNK inhibitor JNK-IN-8 (Derijard et al., 1994) reduced phosphorylation of cJUN at Ser73 and cooperated with MLN128 to reduce cell viability in lung SCC cell lines (Figures S4G–J). Collectively, these results suggest cJUN may be a conserved regulator of GLS while cMYC regulation of GLS was cell line dependent.
Lastly, we tested whether the GSK3i-mediated increased GLS protein resulted in increased glutamine consumption in tumor cells. H460 and RH2 cells that were treated with 10 μM of GSK3i for 72 hr showed increased consumption of glutamine (Figure 4F and 4G) compared to vehicle treated cells. GSK3α/β inhibition also resulted in a concomitant increase in glucose consumption (Figures 4F and 4G). This is explained by increased protein levels of hexokinase II (HKII) (Figures S4E and S4F). HKII is a MYC target gene that is suppressed by MLN128 resulting in reduced 18F-FDG signal in MLN128 treated tumors (Momcilovic et al., 2015). Next, we directly evaluated GLS activity in H460 and RH2 cells that were treated with GSKi for 72 hr. Cells treated with 10 μM GSKi showed increased activity of GLS relative to cells treated with vehicle (Figures 4H and 4I). These results suggest that GSK3α/β modulates adaptive glutamine metabolism through regulation of cMYC and cJUN, leading to increased uptake of glutamine and increased GLS activity.
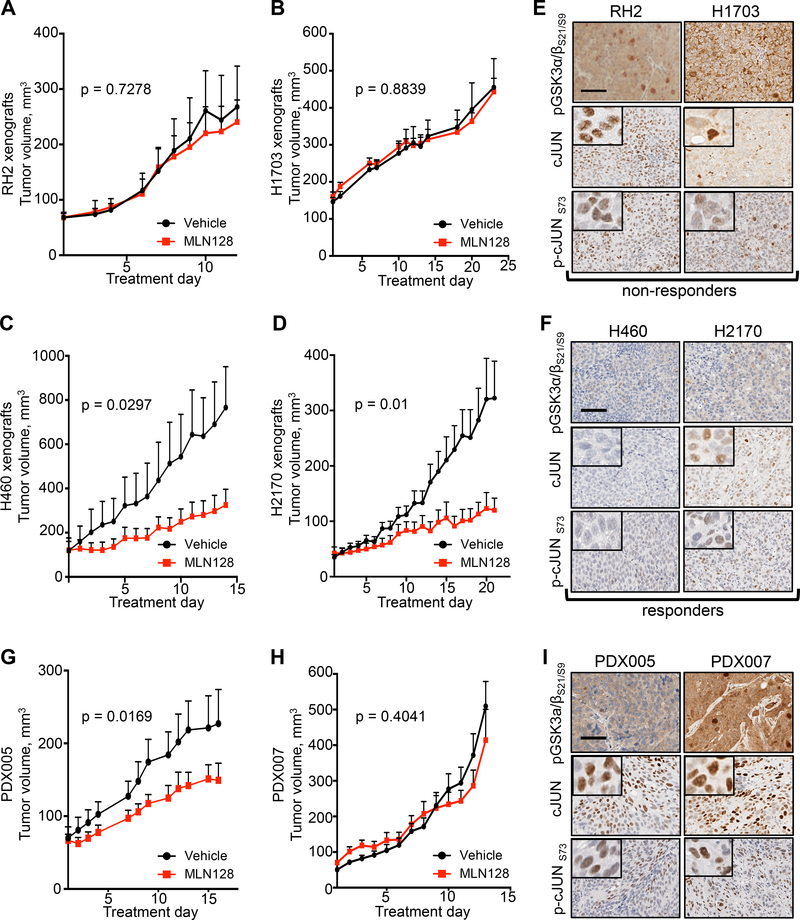
GSK3α/β activity is a predictor of either response or resistance to single therapy MLN128 in human lung SCC.
Our data in Figure 4 showed that inactivation of GSK3α/β confers resistance to MLN128 through upregulation of GLS. The data also suggest that the phosphorylation state of GSK3α/βS21/9 is predictive of differentiating responders and non-responders to single therapy MLN128. To test this we implanted mice with human lung SCC/LCC tumor lines RH2, H1703, H460 and H2170 and began daily treatment with vehicle or MLN128 when tumors reached a volume of 50 mm3-150 mm3. We identified that RH2 and H1703 tumor xenografts were unresponsive to single therapy MLN128 while H460 and H2170 tumors showed significant response to MLN128 (Figures 5A–5D). We classified these tumors as non-responders (RH2, H1703) and responders (H460, H2170). Examination of phospho-GSK3α/βS21/9 levels in tumors confirmed strong pGSK3α/βS21/9 staining in non-responders and weak pGSK3α/βS21/9 staining in responders (comparing Figures 5E to 5F). Examination of cJUN and transcriptionally active phospho-cJUN Ser73 (p-cJUNS73) showed that unresponsive tumors with strong pGSK3α/βS21/9 staining also had elevated nuclear cJUN and p-cJUNS73 levels as compared to responsive tumors (Figures 5E and 5F). While cJUN and p-cJUNS73 expression consistently delineated MLN128 responders from non-responders, cMYC staining was variable in both responders and non-responders (Figure S5A and S5B). This may be due to frequent MYC amplification, which is a common event in lung SCC tumors (Hammerman et al., 2012; Malchers et al., 2014).
Figure 5.
GSK3α/β phosphorylation state predicts therapeutic response to MLN128 in vivo.
(A and B) Tumor volumes for mouse xenografts implanted with human lung SCC cell lines RH2 (A) and H1703 (B) and treated daily with Vehicle (n = 8) or MLN128 (n = 16 (RH2), n = 10 (H1703)). (C and D) Tumor volumes for mouse xenografts implanted with human lung LCC/SCC cell lines H460 (D) and H2170 (E) and treated daily with Vehicle (n = 6 (H460), n = 10 (H2170)) or MLN128 (n = 8 (H460), n = 10 (H2170)). (E) Representative images of RH2 (left) and H1703 (right) xenografts stained with antibodies against pGSK3α/βS21/S9, cJUN or p-cJUNS73. Scale bar = 50 μm. (F) Representative images of H460 (left) and H2170 (right) xenografts stained with antibodies against pGSK3α/βS21/S9, cJUN or p-cJUNS73. Scale bar = 50 μm. (G and H) PDX005 and PDX007 were treated daily with Vehicle (n = 8 (PDX005), n = 9 (PDX007)) or MLN128 (n = 11 (PDX005), n = 9 (PDX007)) for 16 days (PDX005) or 13 days (PDX007). (I) Representative images of PDX005 and PDX007 tumors stained with antibodies against pGSK3α/βS21/S9, cJUN or p-cJUNS73. Scale bar = 50 μm. The tumor volumes are represented as the mean ± SEM. Statistical significance for final tumor volumes was calculated using a two-tailed t test. See also Figure S5.
We next sought to test whether pGSK3α/βS21/9 was a predictive marker of MLN128 response in human PDXs of lung SCC. Before treating PDXs with MLN128, we functionally characterized metabolism and protein biomarkers in lung SCC PDXs to confirm these tumors retained a hypermetabolic and glycolytic phenotype similar to the patients’ primary tumors (Figure S5C–S5E). 18F-FDG PET imaging confirmed that PDX005 and PDX007 retained a hypermetabolic, 18F-FDG avid and glycolytic tumor phenotype (Figure S5D and S5E), which was identical to the patient’s primary tumor phenotype (comparing Figures S5C to S5D). Importantly, PDXs had high activation of the mTOR pathway as indicated by positive staining for p4EBP1 (Figure S5F), thus making these PDXs logical candidates for MLN128 therapy. In addition, both PDXs had positive staining GLUT1 and SLC1A5 (Figure S5F), correlating with high 18F-FDG uptake and suggesting high influx of glutamine agreeing with data from KL mice in Figures 2 and 3. These results confirm that lung SCC tumors have a conserved hypermetabolic phenotype that can be accurately modeled in not only xenografts and GEMMs but also PDXs.
We treated NSG mice with vehicle or MLN128 for 16 days (PDX005) or 13 days (PDX007) and identified PDX005 as partial responder to MLN128, while PDX007 was completely non-responsive to MLN128 (Figures 5G and 5H). We identified differential pGSK3α/βS21/9 staining between PDXs, with PDX007 having a stronger pGSK3α/βS21/9 staining signal compared to PDX005 (Figure 5I). In addition, we identified that PDX007 had a modest increase in cJUN, p-cJUNS73 and cMYC staining compared to PDX005 (Figures 5I and S5G). Together, our data supports pGSK3α/βS21/9 as a predictive biomarker that correlates with response to treatment with MLN128.
CB-839 overcomes metabolic adaptation to MLN128 in human lung SCC.
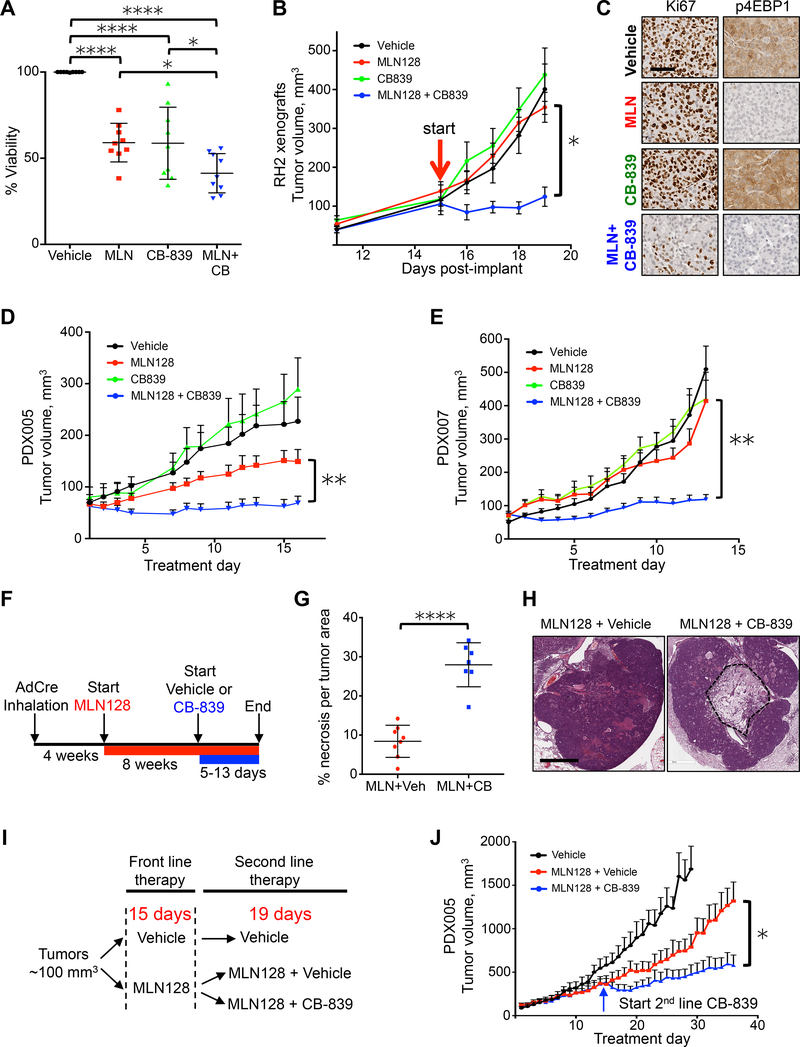
The ability of both lung SCC xenografts and PDXs to rapidly adapt their metabolism to single MLN128 treatment via upregulation of glutaminolysis suggested that selective inhibition of GLS may successfully overcome therapy resistance. We examined whether combining MLN128 with GLS inhibitor CB-839 would result in an improved therapy response in lung SCC. We first tested combinatorial MLN128 and CB-839 treatment in a panel of 9 human lung SCC cell lines. Single and combinatorial treatment with MLN128 or CB-839 significantly reduced cell viability compared to vehicle, and combinatorial MLN128+CB-839 treatment further reduced viability compared single therapy MLN128 and CB-839 (Figure 6A). Additionally, we tested rapamycin in combination with CB-839 and found the two compounds cooperated to significantly reduce cell viability in lung SCC cell lines (Figure S6A).
Figure 6.
GLS inhibition overcomes resistance to MLN128 in lung SCC tumors.
(A) Cell viability of a panel of 9 lung SCC/LCC cell lines (RH2, H460, HCC15, SW900, H1703, H226, H520, H596, H2170) that were treated with Vehicle, 20 nM MLN128 (MLN), 1 μM CB-839 (CB-839) or 20 nM MLN128+1 μM CB-839 (MLN+CB) for 72 hr. n = 3 for each cell line. (B) Tumor volumes of RH2 tumor xenografts treated with Vehicle (black; n = 8), MLN128 (red; n = 8), CB-839 (green; n = 8) or MLN128+CB-839 (blue; n = 8) for 5 days. Start of treatment is indicated with red arrow. (C) Representative images of RH2 tumors treated with Vehicle, MLN128 (MLN), CB-839 or MLN+CB-839 and stained for Ki67 (left) or p4EBP1 (right). Scale bar = 25 μm. (D and E) Tumor volumes of PDX005 (D) and PDX007 (E) treated with Vehicle (black; n = 10 (PDX005), n = 9 (PDX007)), MLN128 (red; n = 11 (PDX005), n = 9 (PDX007)), CB-839 (green; n = 8 (PDX005), n = 9 (PDX007)) or MLN128+CB-839 (blue; n = 13 (PDX005), n = 12 (PDX007)) for 16 days (for PDX005) or 13 days (for PDX007). (F) Overview of delivery of CB-839 as a second line therapy to KL mice treated daily with MLN128 treatment. (G) Percent necrosis per tumor area in KL mice treated with MLN128+vehicle (n = 8) or MLN128+CB-839 (n = 7) (H) Representative H&E slides with necrosis from tumors quantified in (G). Scale bar = 1 mm. (I) Overview of delivery of CB-839 as a second line therapy to PDX005. (J) Tumor volumes for PDX005 treated with Vehicle (black; n = 10) or MLN128 (red; n = 11) or MLN128+CB-839 second line therapy (blue; n = 8). Start of second line therapy is indicated with the blue arrow. The data are represented as the mean ± SEM. Statistical significance (*p<0.05; **p<0.01; ****p<0.0001; ns, not significant) was calculated using a non-parametric one-way ANOVA (Tukey test) for panel (A) and a two-tailed t-test for panels (B, D, E, G, J). Statistical significance for panels B, D, E and J was calculated between final tumor volumes in MLN128 group and MLN+CB839 group. See also Figure S6.
Next, we tested MLN128 in combination with CB-839 in vivo using RH2 xenografts, which were non-responsive to single MLN128 therapy (Figure 5A), in order to determine if CB-839 could overcome therapy resistance to MLN128. Single therapy MLN128 and CB-839 treatments showed no reduction in tumor volume compared to the vehicle group, however, combinatorial MLN128+CB-839 treatment significantly slowed tumor growth (Figure 6B) and significantly reduced tumor cell proliferation measured by Ki67 (Figure 6C). As expected, MLN128 effectively inhibited mTORC1 as measured by reduced p4EBP1 (Figure 6C).
We repeated combination therapy using PDX models PDX005 and PDX007. In both PDXs CB-839 treatment alone had no effect on tumor growth and MLN128 alone slowed tumor growth only in PDX005 but not PDX007 as shown in Figure 4. In both PDXs the combination of MLN128 and CB-839 significantly reduced tumor growth (Figures 6D and 6E). We also tested rapamycin alone or in combination with CB-839 in PDX005 and demonstrated that combination therapy significantly reduced tumor volume compared to vehicle or single treatments (Figure S6B).
To more closely mirror therapeutic regimens used in a clinical setting, we tested CB-839 as a second line therapy to treat tumors progressing on single therapy mTOR inhibitors. We first modeled this treatment regiment in KL GEMMs, which were treated with MLN128 for 8 weeks followed by acute treatment with CB-839 (Figure 6F). Treatment with second line therapy lasted between 5 and 13 days at which point lung tumors were analyzed by histology. We identified significant tumor cell death in mice treated with second line CB-839 compared to those receiving only MLN128, with a nearly 3 fold increase in tumor necrosis in tumors receiving CB-839 (Figures 6G and 6H). We also tested second line CB-839 treatment in PDX005 as outlined in Figure 6I. Identical to KL GEMMs, we discovered that second line CB-839 treatment cooperated with MLN128 to significantly reduce tumor growth (Figure 6J). Collectively, these results demonstrate that CB-839 overcame acquired metabolic resistance to MLN128. CB-839 was highly effective when used as either a first or second line therapy with MLN128 to significantly reduce growth of human lung SCC.
Hypermetabolic tumors share a conserved metabolic signature that predicts patient outcome and therapeutic response to mTOR and GLS inhibition.
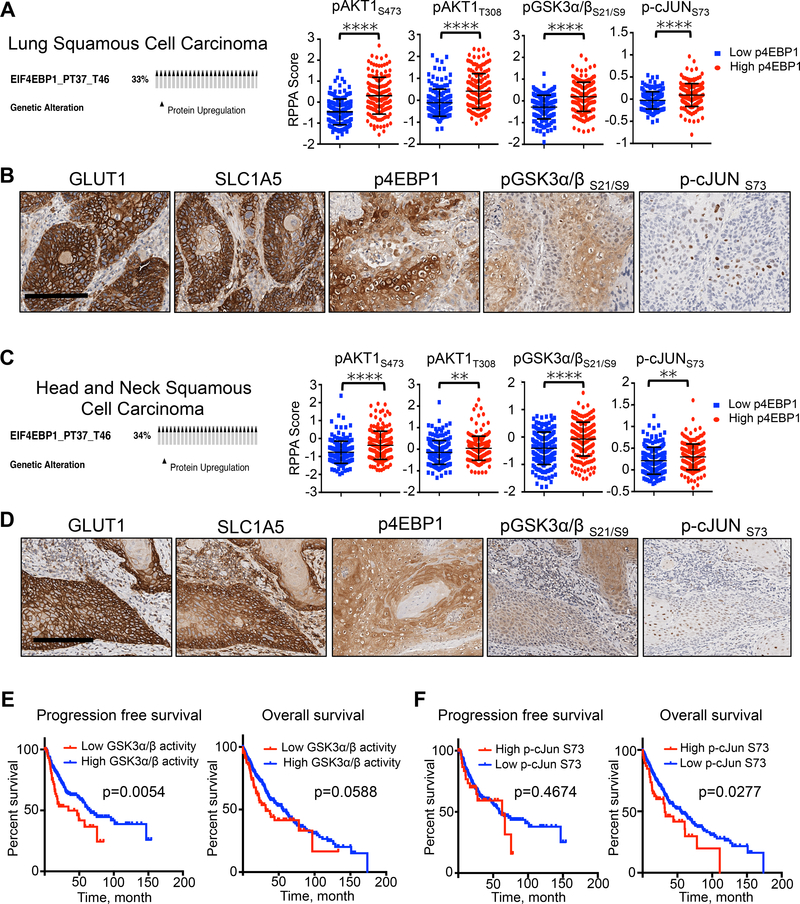
Due to the lack of defined genetic mutations in the human lung SCC that are predictive of therapeutic response to targeted therapies, we reasoned that stratification of human lung SCCs by metabolic phenotypes represents a means to identify patients who would respond to therapies targeting metabolism. We identified five proteins whose expression was consistently amplified in 18F-FDG and 11C-Glutamine positive SCC lung tumors. These proteins include the glucose and glutamine transporters GLUT1 and SLC1A5, the mTORC1 substrate p4EBP1 as well as pGSK3α/βS21/S9 and p-cJUNS73 and represent the core components of what we defined as a metabolic signature in lung SCC based on the data from this study.
We next mined data sets from TCGA for lung SCC and examined reverse phase protein assay (RPPA) analysis performed on patient biopsies. We selected patients that have higher p4EBP1 protein levels (33% of total patients) (Figure 7A) and evaluated levels of signaling proteins within the AKT–GSK3α/β–cJUN pathway that we mapped in Figure 4. We discovered that lung SCC patients with p4EBP1 positive tumors had significantly higher levels of phosphorylated AKT at Ser473 (pAKTS473) and Thr308 (pAKTT308), GSK3α/β at Ser21 and Ser9 (pGSK3α/βS21/9), and cJUN at Ser73 (p-cJUNS73) compared to patients with lower p4EBP1 levels (Figure 7A). We performed IHC staining on human lung SCC tumors and identified that these tumors were consistently marked by amplified levels of the proteins GLUT1 and SLC1A5, p4EBP1, pGSK3α/βS21/9 and nuclear p-cJUNS73 staining (Figure 7B), which constitute our metabolic signature.
Figure 7.
Human squamous cell carcinomas of the lung, head and neck share a conserved metabolic signature that is predictive of survival and therapy response to mTOR and GLS inhibition.
(A) Oncoprint from TCGA on Lung SCC showing percent of tumors with increased p4EBP1 staining in RPPA data set (left). RPPA data comparing status of indicated proteins in tumors with increased p4EBP1 staining (red; n = 162) to tumors with decreased p4EBP1 staining (blue; n = 166). The data are represented as the mean ± SD. Statistical significance (****p<0.0001) was calculated using two-tailed t test. (B) Representative images from human lung SCC tumors stained with GLUT1, SLC1A5, pGSK3α/βS21/S9, p-cJUNS73. Scale bar = 50 μm. (C) Oncoprint from TCGA on Head and Neck SCC showing 34% of tumors with increased p4EBP1 staining in RPPA data set (left). RPPA data comparing status of indicated proteins in tumors with increased p4EBP1 staining (red; n = 182) to tumors with decreased p4EBP1 staining (blue; n = 175). The data are represented as the mean ± SD. Statistical significance (**p<0.01; ****p<0.0001) was calculated using two-tailed t test. (D) Representative images from human HNSCC tumors stained with GLUT1, SLC1A5, pGSK3α/βS21/S9, p-cJUN S73. Scale bar = 50 μm. (E) Progression free survival (left) and overall survival (right) analysis of Lung SCC patients (TCGA data set) stratified for GSK3α/β activity based on the ratio of pGSK3α/βS21/S9 to total GSK3α/β. Statistical significance was calculated using log-rank test (Mantel-Cox); n = 65 (low GSK3α/β activity) and n = 310 (high GSK3α/β activity). (F) Progression free survival (left) and overall survival (right) analysis of Lung SCC patients (TCGA data set) stratified for p-cJUNS73 levels. Statistical significance was calculated using log-rank test (Mantel-Cox); n = 48 (high p-cJunS73) and n = 448 (low p-cJunS73). See also Figure S7.
We next sought to expand the discovery we made in lung SCC and identify additional tumor types that may share a similar metabolic signature. We focused on highly aggressive, therapy resistant tumors that are consistently positive for 18F-FDG PET similar to lung SCCs. Head and neck squamous cell carcinomas (HNSCC) and osteosarcomas (OS) met these criteria since these tumors are highly glycolytic, 18F-FDG positive and show poor response to conventional therapies in the clinic (Jabour et al., 1993; Walter et al., 2011). We performed RPPA analysis of TCGA data sets for HNSCC and OS identical to our analysis of lung SCC and selected tumors with elevated p4EBP1 levels (Figures 7C and S7A). We found that human HNSCC and OS patient biopsies with active mTOR signaling do share a conserved metabolic signature similar to lung SCC as defined by positive staining for GLUT1, SLC1A5, p4EBP1, pGSK3α/βS21/9 and nuclear p-cJUNS73 (compare Figure 7B to Figures 7D and S7B).
We then tested MLN128+CB-839 in mouse xenografts implanted with the HNSCC tumor line Tu686 and a PDX implanted with an OS pulmonary metastasis (OSPM). Both the HNSCC xenografts and OS PDXs significantly responded to the combination of MLN128+CB-839 (Figures S7C and S7D). Tumors from Tu686 xenografts and PDX004-OS were positive for GLUT1, SLC1A5, p4EBP1, pGSK3α/βS21/9 and nuclear p-cJUNS73 staining (Figure S7E). Positive GLUT1 staining associated with high 18F-FDG uptake in the patient’s OS tumor (shown in Figure S7D). These results demonstrate that HNSCC and OS share a similar metabolic signature with that of lung SCC tumors and are responsive to MLN128+CB-839 treatment.
Lastly, we show that patients with low activity of GSK3α/β and high p-cJUNS73 protein expression levels had significantly reduced progression free survival (PFS) or overall survival, respectively (Figures 7E and 7F). Interestingly, TCGA data suggests that lung SCC patients with high expression of SLC1A5 had significantly worse PFS and overall survival compared with patients with low expression of SLC1A5 (Figure S7F). At the same time, SLC1A5 expression did not significantly correlate with PFS or overall survival in ADC (Figure S7G) emphasizing importance of glutamine transport in SCC tumor subtype. Collectively, our data identified a conserved metabolic signature in hypermetabolic lung SCC, HNSCC and OS that predict positive response to combined mTOR and GLS inhibition. In addition, pGSK3α/βS21/9 and p-cJUNS73 may have utility as functional biomarkers that can be used to stratify patients for metabolic therapies.
Discussion
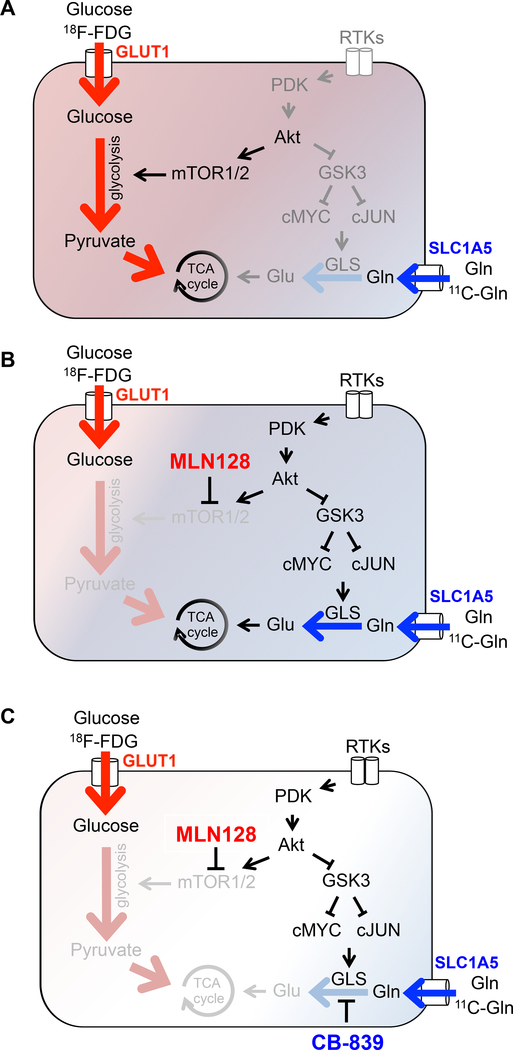
We present a proof-of-concept study where we describe a mechanism by which lung SCCs circumvent mTOR inhibition and suppression of glycolysis by upregulation of glutamine metabolism as summarized in Figures 8A–8C. Our use of metabolic profiling of lung SCC by PET and LC-MS identified a dual reliance on both glutamine and glucose metabolism. We identified a function of the GSK3α/β pathway as a central regulator of adaptive glutamine metabolism that enables lung SCC tumors to escape mTOR inhibition by upregulation of GLS. Importantly, we discovered that the phosphorylation state of GSK3α/β serves as a predictive biomarker of response or resistance to MLN128. We confirmed that CB-839 together with MLN128 was highly effective in reducing tumor growth in a series of lung SCC models that included: human cell lines, mouse xenografts, GEMMs and PDXs. Lastly, we decoded a metabolic signature within hypermetabolic human lung SCC, HNSCC and OS defined by positive staining for GLUT1, SLC1A5, p4EBP1, pGSK3α/βS21/9 and nuclear p-cJUNS73 that were responsive to combined MLN128 and CB-839 treatment.
Figure 8.
Model of adaptive glutamine metabolism in lung SCC tumors.
(A) Untreated lung SCC tumors. (B) Adaptation following chronic mTOR inhibition with MLN128. (C) Overcoming acquired resistance to MLN128 through GLS inhibition.
The mutational landscape of lung SCC is diverse (Cancer Genome Atlas Research, 2012) and SCC have high mutational burden (Alexandrov et al., 2013). Therefore unlike in ADCs there is a lack of readily targetable driver mutations in SCCs. This was the basis for our rationale to define a conserved metabolic phenotype in lung SCC that could be therapeutically targeted independent of defined genetic driver mutations. MLN128 potently inhibited mTORC1 as well as glucose metabolism in lung SCC yet tumors continued to grow. We hypothesized that lung SCCs escaped mTOR inhibition not because the drug failed to reach its target but rather because tumors upregulated alternative nutrient sources that enabled escape of MLN128 treatment.
We identified that SCC tumors share a tightly conserved metabolic phenotype that is defined in part by reliance on glucose and glutamine. Serial PET imaging of KL GEMMs with 11C-Gln and 18F-FDG identified high influx of both glutamine and glucose that was consistently elevated and associated with intense protein staining for GLUT1 and SLC1A5 transporters consistent with previously published studies (Goodwin et al., 2017; Hassanein et al., 2013; Momcilovic et al., 2017). Following up on PET imaging, we performed in vivo metabolic tracing of lung SCC tumors that successfully defined a metabolic route by which lung SCC tumors adapt to MLN128 through upregulation of glutamine. In the future it will be important to decipher cell intrinsic and extrinsic factors that contribute to the differences seen in cellular metabolism between lung SCC and ADC tumor histologies. Our work supports previous work that accurately defined human lung cancer as a metabolically heterogeneous disease (Hensley et al., 2016). Importantly, our results serve to build a more complete model of cancer metabolism in NSCLC that encompasses both ADC and SCC tumor subtypes.
By mapping the GSK3α/β as a regulator of GLS and glutaminolysis we have begun to identify a mechanism(s) underlying adaptive glutamine metabolism in lung SCC. Both, pGSK3α/βS21/9 and p-cJUNS73 have potential to be developed as surrogate biomarkers of response or resistance to MLN128 treatment. We show that tumors with low levels of pGSK3α/βS21/9 and p-cJUNS73 were sensitive to MLN128 as a single therapy while tumors with high levels of pGSK3α/βS21/9 and p-cJUNS73 were refractory to MLN128. If we extrapolate our results to clinical applications then patients with low phosphorylation of GSK3α/βS21/9 and cJUNS73 represent a patient population who may be responsive to single agent MLN128 while those with tumor(s) having elevated levels of pGSK3α/βS21/9 and p-cJUNS73 would require MLN128 in combination with CB-839.
This became our rationale for dual targeting both mTOR and GLS in lung SCC tumors. The combination of MLN128 and CB-839 was highly effective in halting growth of aggressive lung SCC tumors. In addition to testing upfront combination therapies, it was equally important we test clinically relevant drug delivery strategies. Therefore, we tested single dose delivery of MLN128 followed by second line delivery of CB-839 after tumors became non-responsive to MLN128. CB-839 effectively overcame acquired resistance to MLN128 in GEMMs and PDXs with tumors rapidly undergoing necrosis and cell death in a little as 5 days.
In a clinical setting, biomarkers will be needed to identify metabolically active tumors that will be responsive to MLN128 and CB-839 treatment. Our discovery of a conserved metabolic signature in lung SCC, HNSCC and OS suggest that hypermetabolic, 18F-FDG avid tumors may be responsive to combinatorial treatment with MLN128 and CB-839. In addition to NSCLC, high 18F-FDG uptake correlates with significantly worse survival in number of cancers, including HNSCC (Minn et al., 1997) and OS (Hwang et al., 2016). Here, PET imaging becomes not only a prognostic marker of aggressive cancer but also the first of a series of biomarkers to guide therapy. While our data demonstrate 11C-Gln and 18F-FDG pair well to identify hypermetabolic tumors with influx of glucose and glutamine, future studies coupling mass spectrometry based approaches are needed to validate 11C-Gln as a bona fide probe of glutamine dependent metabolism in lung SCC, HNSCC and OS tumors.
While genetically disparate in their mutation spectrums, lung SCC, HNSCC and OS all shared a conserved metabolic signature and showed significant response to MLN128 and CB-839 treatment. We demonstrate that metabolic profiling of tumor biomarkers by both PET imaging and histology can accurately stratify patients and delineate responders from non-responders. Targeted therapies such as MLN128 and CB-839 have an important role in oncology but only when used in the correct context and combination. Importantly, combinatorial use of MLN128 and CB-839 may provide a much needed therapeutic benefit for lung SCC, HNSCC and OS patients who are frequently refractory to conventional therapies.
The STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David B. Shackelford (DShackelford@mednet.ucla.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Cells were maintained at 37°C in a humidified incubator with 5% CO2. H460, H520, H596, H1703, H226, H2170 and SW900 cells were obtained from ATCC. RH2 lung cell line was a kind gift from Dr. Steven Dubinett (UCLA); HCC15 cell line was a kind gift from Dr. John Minna (UT Southwestern); Tu686 cell line was a kind gift from Dr. Maie St.John (UCLA). All cell lines were routinely tested and confirmed to be free of Mycoplasma using the LookOut Mycoplasma PCR Detection Kit (Sigma). Cell lines were authenticated in the UCLA Genotyping and Sequencing Core utilizing Promega’s DNA IQ System and Powerplex 1.2 system, and all cells were utilized within 10 passages of genotyping. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) plus 5% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Gibco).
Patient derived xenografts
PDXs were established in our lab using 6–12 weeks old NSG mice implanted with tumor tissue from patients undergoing surgeries. All patients provided written informed consent for use of samples in research under protocols approved by Institutional Review Boards at University of California, Los Angeles IRB # 10–001096 (for SCC), IRB #10–001857 (for OS) and at Long Beach Memorial Hospital IRB #208–13 (for SCC).
Mice
Mice were housed in pathogen-free facilities at University of California Los Angeles (UCLA). All experimental procedures performed on mice were approved by the UCLA Animal Research Committee (ARC). Both male and female mice were used in all experiments and no preference in mouse gender was given for any of the studies.
KrasG12D;Lkb1−/−;Luc mice
We performed therapeutic studies with genetically engineered mouse model of lung cancer using Lox-Stop-Lox KrasG12D, Lkb1 lox/lox, Rosa26-Lox-Stop-Lox-Luciferase mice (KL). Lung tumors were induced by intranasal administration of 2.5 × 106 plaque forming units of Adenoviral Cre (Gene Transfer Vector Core, University of Iowa) when mice were between 8–18 weeks of age. Four weeks after administration of Adenoviral Cre, mice were imaged with bioluminescence imager (IVIS, PerkinElmer) following 150 mg/kg injection of luciferin. Total luminescence flux (photons/sec) was calculated for area covering mouse chest area for both ventral and dorsal sides. Mice were randomized for treatment with vehicle or MLN128 according to luminescence flux. Mice were euthanized after ~12 weeks after tumor initiation (~20–40 weeks of age).
NSG mice
NOD.Cg-Prkdcscid IL2rgtm1Wjl / SzJ (NSG) mice were purchased from Jackson Laboratory (Bar Harbor, ME). Female NSG mice between 6–12 weeks old were used for patient derived xenograft (PDX) studies.
SCID (Beige) mice
SCID (Beige) mice were purchased from the Department of Radiation Oncology’s Division of Molecular and Cellular Oncology Animal Core. Female SCID (Beige) mice between 6–12 weeks old were used for xenografts studies with established human cell lines.
METHOD DETAILS
Therapeutic studies in GEMMs
For treatment study with MLN128, following intranasal administration of Adenoviral Cre, KL mice were randomly sorted into two groups following imaging with bioluminescence: vehicle and MLN128. Treatment was initiated 4 weeks post Adenoviral Cre delivery. Following 8 weeks of daily treatment with Vehicle or 1 mg/kg MLN128, mice were subject to imaging with 18F-FDG and CT. For PET/CT imaging study with two radio-tracers, KL mice received intraperitoneal injection with 250 mg/kg Naphthalene 72 hr prior to Adenoviral Cre delivery; 10 weeks later mice were imaged with 18F-FDG and 48 hr later with 11C-Glutamine. Using Naphthalene leads to destruction of secretory Clara cells and subsequent proliferation of basal cell (Hong et al., 2004; Rawlins et al., 2007). This leads to enrichment in SCC tumors in KL mice when coupled with Adenoviral Cre delivery. For the second line therapy with CB-839 in KL mice, tumors were induced with Adenoviral Cre. Four weeks after tumor induction, mice started receiving daily treatment of MLN128. After 8 weeks of daily 1 mg/kg MLN128 treatment, mice were randomly selected to receive either 1 mg/kg MLN128 (i.p., q.d.) + Vehicle (p.o., b.i.d.) or 1 mg/kg MLN128 (i.p., q.d.) + 200 mg/kg CB-839 (p.o., b.i.d.) for additional 5–13 days.
Therapeutic studies in xenografts
White SCID (Beige) mice (6–12 weeks old) were injected subcutaneously into two lower flanks with 3X105 cells (RH2), 5X105 cells (H460), 1X106 cells (H1703 and H2170) or 2.5X106 cells (Tu686) suspended in 50% PBS, 50% Matrigel (Corning) and allowed to grow to 75–100 mm3 before treatment was started. Animals were randomly assigned to one of two groups: Vehicle or 1 mg/kg MLN128 (i.p., q.d.). For experiment with Tu686 xenografts, mice were randomly assigned to one of four treatment groups: Vehicle, 1 mg/kg MLN128 (i.p., q.d.), 200 mg/kg CB-839 (p.o., b.i.d.), 1 mg/kg MLN128 + 200 mg/kg CB-839. Body weights (g) and caliper measurements ((LxW2)÷2 = mm3) were recorded trice a week. At the end of the study animal tissue was fixed in 10 % buffered formalin 2 hr after the final dose. Additionally, some of the mice were imaged with 18F-FDG and CT at the end of the study.
Therapeutic studies in PDXs
PDX005, PDX007 and PDX004-OS were established in our lab using 6–12 weeks old NSG mice. PDXs were established by subcutaneously injecting ~2 mm3 pieces of tissue into both flanks of NSG mice. Once tumors reached ~75 mm3 mice were randomly assigned to one of four treatment groups: Vehicle, 1 mg/kg MLN128 (i.p., q.d.), 200 mg/kg CB-839 (p.o., b.i.d.), 1 mg/kg MLN128 + 200 mg/kg CB-839. Body weights (g) and caliper measurements ((LxW2)÷2 = mm3) were recorded trice a week. For the second line therapy study with CB-839 using PDX005, when tumors reached ~75 mm3 mice were randomly assigned to either Vehicle or 1 mg/kg MLN128 (i.p., q.d.) group. After 15 days, mice in MLN128 group were randomly assigned to receive 1 mg/kg MLN128 (i.p., q.d.) + Vehicle or 1 mg/kg MLN128 (i.p., q.d.) + 200 mg/kg CB-839 (p.o., b.i.d.) for additional 19 days.
Drugs used in vivo
The vehicle for CB-839 consisted of 25% (w/v) hydroxypropyl-β-cyclodextrin in 10 mmol/L citrate, pH 2. CB-839 was formulated as a solution at 20 mg/mL (w/v) in vehicle. Both were provided by Calithera Biosciences. Vehicle and CB-839 (200 mg/kg) were delivered by oral gavage twice daily (9 am and 5 pm). MLN128 (Active Biochem) was dissolved in 1-methyl-2-pyrrolidinone, then diluted in 15% PEG400 diluted in water. MLN128 was delivered at 1 mg/kg by i.p. injection once daily (9 am). Rapamycin was dissolved in 5% Tween 80, then diluted in 5% PEG400 diluted in water. Rapamycin was delivered at 1 mg/kg by i.p. injection once daily (9 am).
Tracing with 13C6-Glucose and 13C5-Glutamine
Mice were fasted for 6 hr prior to infusions. Infusions with 13C6-glucose (Cambridge Isotope Laboratories, CLM-1396-PK) and with 13C5-Glutamine (Cambridge Isotope Laboratories, CLM-1822-H-PK) were performed via tail vein catheters for 120 min. 13C6-glucose was infused at 30 mg/kg/min and 13C5-Glutamine was infused at 3 mg/kg/min. Mice were kept under 2% isoflurane on warm pads for the entire duration of the infusion. We chose to infuse animals while under isoflurane rather than using conscious free moving animals as this closely matches conditions we use for PET/CT imaging. At the end of the infusion, mice were terminally anesthetized with 100% isoflurane and tissues were dissected within 3–5 min. Tissues was snap frozen in liquid nitrogen and stored at −80°C.
Metabolite extraction and mass spectrometry
The experiments were performed as described previously (Momcilovic et al., 2017; Thai et al., 2014). Briefly, 5–10 mg of frozen tissue was homogenized in 1 ml 80% methanol (−80°C), centrifuged for 5 min at top speed (4°C) and supernatant was transferred to a separate tube. Protein content (BCA protein kit) was measured after tissue pellet was resuspended in SDS lysis buffer (10 mM Tris HCl pH 7.5, 100 nM NaCl, 1% SDS). Supernatant equivalent to 5–10 μg of protein was transferred to a glass tube, 10 nM of norvaline was added and samples were dried using EZ-2 Elite evaporator (Genevac). Dried samples were stored in −80°C freezer until further processing. For the mass spectrometry-based analysis of the sample, 5 mu were injected onto a Luna NH2 (150 mm x 2 mm, Phenomenex) column. The samples were analyzed with an UltiMate 3000RSLC (Thermo Scientific) coupled to a Q Exactive mass spectrometer (Thermo Scientific). The Q Exactive was run with polarity switching (+4.00 kV / −4.00 kV) in full scan mode with an m/z range of 65–975. Separation was achieved using A) 5 mM NH4AcO (pH 9.9) and B) ACN. The gradient started with 15% A) going to 90% A) over 18 min, followed by an isocratic step for 9 min. and reversal to the initial 15% A) for 7 min. Metabolites were quantified with TraceFinder 3.3 using accurate mass measurements (≤ 3 ppm) and retention times established by running pure standards. Relative amounts of metabolites between various conditions, as well as percentage of labeling, were calculated and corrected for naturally occurring 13C abundance (Moseley, 2010).
Metabolite measurements in media
Media was collected from tissue culture plates and analyzed for glucose and glutamine concentrations using the Bioanalyzer 4 (Nova Biomedical). Cells were seeded into 6 cm plates overnight and were subsequently treated with DMSO (Vehicle), MLN128, Rapamycin or CHIR99021 in fresh DMEM medium for 72 hr. Metabolite concentrations were normalized to cell number.
GLS activity
Activity of GLS was measured using GLS activity kit (Biomedical Research Service and Clinical Applications, NY) according to manufacturers instructions.
Western Blotting
For in vitro studies MLN128 (Active Biochem), Rapamycin (LC laboratories), CHIR99021 (Cayman Chemical), and JNK-IN-8 (Fisher Scientific) were dissolved in DMSO. After treatment, cells were quickly rinsed with PBS, and then lysed in boiling SDS lysis buffer (10 mM Tris pH 7.5, 100 nM NaCl, 1% SDS). Lysates were sheared by passing through 25 gauge needle attached to 1 ml syringe. Protein content was normalized using BCA protein kit. Samples were resolved by SDS-PAGE, transferred to PVDF membrane, and blocked in 5% non-fat dry milk. PVDF membranes were incubated with primary antibodies overnight. Antibodies from Cell Signaling Technology, Sigma and Alpha Diagnostic used for immunoblots were diluted 1:2000; antibodies from Abcam were diluted 1:5000. All antibodies are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Cytokeratin 5 (RRID: AB_869890) | Abcam | Cat# ab52635; Clone EP1601Y |
| Mouse monoclonal anti-TTF1 | Dako/Agilent | Cat# M357529–2; Clone 8G7G3/1 |
| Rabbit polyclonal anti-Glut1 | Alpha Diagnostic | Cat# GT11-A |
| Rabbit monoclonal anti-Ki67 (RRID: AB_2314700) | ThermoFisher | Cat# MA5–14520; Clone SP6 |
| Rabbit monoclonal anti-phospho S6 ribosomal protein (Ser 235/236) (RRID: AB_916156) | Cell Signaling Technology | Cat# 4858; Clone D57.2.2E |
| Rabbit polyclonal anti-SLC1A5 (RRID: AB_10670587) | Sigma | Cat# HPA035239 |
| Rabbit monoclonal anti-phospho Akt (Thr 308) (RRID: AB_331163) | Cell Signaling Technology | Cat# 4056; Clone 244F9 |
| Rabbit polyclonal anti-phospho GSK3α/β (Ser 21/9) (RRID: AB_329830) | Cell Signaling Technology | Cat# 9331 |
| Rabbit monoclonal anti-cMyc (RRID: AB_731658) | Abcam | Cat# ab32072; Clone Y69 |
| Rabbit monoclonal anti-cJun (RRID: AB_2130165) | Cell Signaling Technology | Cat# 9165; clone 60A8 |
| Rabbit monoclonal anti-Glutaminase (RRID: AB_2721038) | Abcam | Cat# ab156876; Clone EP7212 |
| Rabbit monoclonal anti-phospho p70 S6 kinase (Thr 389) (RRID: AB_2269803) | Cell Signaling Technology | Cat# 9234, Clone 108D2 |
| Rabbit monoclonal anti-b-actin (RRID: AB_2223172) | Cell Signaling Technology | Cat# 4970, Clone 13E5 |
| Rabbit monoclonal anti-phospho cJun (Ser 73) (RRID: AB_2129575) | Cell Signaling Technology | Cat# 3270, Clone D47G9 |
| Rabbit polyclonal anti-PDK1 (RRID: AB_10828201) | Cell Signaling Technology | Cat# 3062 |
| Rabbit monoclonal anti-GSK3α/β (RRID: AB_10547140) | Cell Signaling Technology | Cat# 5676, Clone D75D3 |
| Rabbit monoclonal anti-Hexokinase II (RRID: AB_2232946) | Cell Signaling Technology | Cat# 2867, Clone C64G5 |
| Bacterial and Virus Strains | ||
| Adenoviral Cre (Ad5CMVCre) | University of Iowa Viral Vector Core | Cat# VVC-U of Iowa-5 |
| Biological Samples | ||
| Patient-derived xenografts (PDX) | This study | N/A |
| Human lung cancer IHC slides | UCLA | N/A |
| Human head and neck cancer IHC slides | UCLA | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MLN128, mTOR inhibitor, catalytic | Active Biochem | Cat# A-1443; CAS # 1224844–38-5 |
| Rapamycin, mTOR inhibitor, allosteric | LC Laboratories | Cat# R-5000; CAS # 53123–88-9 |
| MK2206, AKT inhibitor | Selleckchem | Cat# S1078, CAS # 1032350–13-2 |
| CHIR99021, GSK3α/β inhibitor | Cayman Chemical | Cat# 13122; CAS # 252917–06-9 |
| JNK-in-8, JNK inhibitor | Fisher Scientific | Cat# NC0853526; CAS # 1410880–22-6 |
| CB-839, GLS inhibitor | ApexBio | Cat# B4799; CAS# 1439399–58-2 |
| CB-839, GLS inhibitor | Calithera Biosciences | N/A |
| 13C-Glucose (U-13C6, 99%) | Cambridge Isotope Laboratories | Cat# CLM-1396; CAS# 110187–42-3 |
| 13C-Glutamine (13C5, 99%) | Cambridge Isotope Laboratories | Cat# CLM-1822-H; CAS 184161–19-1 |
| DL-Norvaline | Sigma | Cat# N7502; CAS 760–78-1 |
| 18F-FDG ([18F]fluoro-2-deoxy-glucose) | UCLA | N/A |
| 11C-Glutamine | Sam Sadeghi Lab, UCLA | N/A |
| Matrigel Membrane matrix | Fisher Scientific | Cat# CB-40234C |
| Naphthalene | Sigma | Cat# 147141; CAS# 91–20-3 |
| Critical Commercial Assays | ||
| BCA protein assay kit | ThermoFisher | Cat # 23225 |
| Glutaminase Assay kit | Biomedical Research Service and Clinical Application | Cat # E-133 |
| Deposited Data | ||
| Lung adenocarcinoma expression data | TCGA | www.cbioportal.org |
| Lung squamous carcinoma expression data | TCGA | www.cbioportal.org |
| Microarray data: Vehicle vs MLN128 treated lung SCC tumors isolated from KrasG12D; Lkb1-/- mice | This study | GEO: GSE110108 |
| Experimental Models: Cell Lines | ||
| RH2 | Steven M. Dubinett lab (UCLA) | N/A |
| NCI-H460 (Male) (RRID: CVCL_0459) | ATCC | HTB-177 |
| NCI-H1703 (Male) (RRID: CVCL_1490) | ATCC | CRL-5889 |
| NCI-H2170 (Male) (RRID: CVCL_1535) | ATCC | CRL-5928 |
| HCC15 (Male) | John Minna lab (UT Southwestern) | N/A |
| SW900 (Male) (RRID: CVCL_1731) | ATCC | HTB-59 |
| NCI-H520 (Male) (RRID: CVCL_1566) | ATCC | HTB-182 |
| NCI-H596 (Male) (RRID: CVCL_1571) | ATCC | HTB-178 |
| NCI-H226 (Male) (RRID: CVCL_1544) | ATCC | CRL-5826 |
| Tu686 (RRID: CVCL_4916) | Maie St.John lab (UCLA) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse strain: NSG; NOD.Cg-Prkdcscid IL2rgtm1Wjl / SzJ (RRID: IMSR_JAX:005557) | The Jackson Laboratory | Cat # 005557 |
| Mouse strain: SCID (beige) | UCLA Animal Core – Division of Molecular and Cellular Oncology | N/A |
| Mouse strain: KLluc; Lox-stop-lox KrasG12D; LKB1fl/fl; Lox-stop-lox Luciferase | Shackelford et al, 2013 | N/A |
| Recombinant DNA | ||
| pLKO.1-puro-scrambled shRNA | Sigma | Cat# SHC002 |
| pLKO.1-puro-shRNA targeting human GLS (134) | Sigma | Cat# TRCN0000051134 |
| pLKO.1-puro-shRNA targeting human GLS (135) | Sigma | Cat# TRCN0000051135 |
| esiEGFP; control siRNA | Sigma | Cat# EHUEGFP |
| esi-cMyc | Sigma | Cat# EHU021051 |
| siSCR; control siRNA | Sigma | Cat# SIC001 |
| c-Jun siRNA #1, targeting human c-Jun | Sigma | Cat# SASI_Hs02_00333461 |
| c-Jun siRNA #2, targeting human c-Jun | Sigma | Cat# SASI_Hs01_00150279 |
| Software and Algorithms | ||
| QuPath | (Bankhead et al., 2017) | qupath.github.io |
| GraphPad Prism 6.0 | GraphPad | www.graphpad.com |
| AMIDE | (Loening and Gambhir, 2003) | amide.sourceforge.net |
| Other | ||
| NuPAGE 4–12% Bis-Tris Protein Gels, 1.5mm, 15-well | ThermoFisher | Cat # NP0336BOX |
| PVDF membrane | ThermoFisher | Cat # 88518 |
| G8 PET/CT Preclinical Imaging System | PerkinElmer | Cat# CLS139564 |
Cell Viability Assay
Cell viability was measured using trypan blue exclusion assay (ViCell XR, Beckman Coulter).
Generation of cell lines expressing shRNA and siRNA
Lentiviral vectors (pLKO.1 plasmids) were transduced into HEK293T cells using standard protocols. Lentiviral supernatants collected from HEK293T cells were filtered and used to infect RH2 cells in the presence of polybrene (8 μg/ml) for 24 hr. Selection with puromycin (1 μg/ml) started 72 hr post-infection. Stable knockdowns were used for assays.
For transfection with esiRNA or siRNA, cells were seeded in 6-well plates and esiRNAs or siRNAs combined with Lipofectamine RNAiMax (according to manufacturers protocol) were added along with either vehicle or 10 μM CHIR99021. Following 72 hr incubation, cells were collected for Western blot analysis.
Histology and immunohistochemical analysis
Tissue isolation, fixation and staining procedures were performed as previously described (Momcilovic et al., 2017; Momcilovic et al., 2015; Shackelford et al., 2013). Briefly, lungs or tumors were fixed overnight in 10% buffered formalin, and then transferred to 70% ethanol. Tissues were processed and embedded by TPCL at UCLA. The following antibodies were used: phospho-4EBP1 (Thr37/46) (Cell Signaling Technology, #2855 1:800), anti-CK5 (EP1601Y) (abcam, ab52635 1:100), anti-TTF1 (8G7G3/1) (Dako, 1:1000), anti-Ki67 (SP6) (Thermo Scientific, RM-9106-S0 1:200), phospho-S6 (Cell Signaling Technology, #4585 1:400), phospho-GSK3α/β (Ser21/9) (Cell Signaling Technology, #9331 1:50), c-Jun (Cell Signaling Technology, #9165 1:800), phospho-c-Jun (S73) (D47G9) (Cell Signaling Technology, #3270 1:200), anti-SLC1A5 (Sigma, HPA035240, 1:400), anti-GLUT1 (Alpha Diagnostic, GT11A, 1:400), anti-Myc (Y69) (abcam, ab32072, 1:100). Slides were scanned onto a ScanScope AT (Aperio Technologies, Inc., Vista, CA). Digital slides were analyzed with QuPath software in order to determine percent positive cells for Ki67, GLUT1 and p4EBP1 stains.
PET imaging
In vivo small animal imaging was conducted at the Crump Institute’s Preclinical Imaging Technology Center. Mice we injected via lateral tail vein with ratio-tracer (70 μCi for 18F-FDG and 200–400 μCi for 11C-L-glutamine), underwent 60 min uptake under 2% isoflurane anesthesia, followed by microPET (G8 PET/CT, PerkinElmer) and microCT (CrumpCAT, Arion Hadjioannou laboratory) imaging. Synthesis of 11C-L-Glutamine was performed as previously described (Momcilovic et al., 2017). Quantification of 18F-FDG uptake was done using AMIDE software by drawing region of interest (ROI) over tumor and heart and plotting maximum uptake values (SUVmax) as percent injected dose per gram (%ID/g).
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical tests were calculated using GraphPad Prism 6. Details of the specific statistical analysis are indicated in the figure legends.
DATA AND SOFTWARE AVAILABILITY
The microarray data reported in this paper has been deposited to NCBO GEO under accession number GSE110108.
Human lung cancer gene expression data generated by TCGA and utilized in this paper can be accessed via cBioPortal (www.cBioPortal).
Supplementary Material
Significance.
Lung SCC are characterized as highly glycolytic, highly aggressive and resistant conventional chemotherapy. In this study we functionally profiled metabolic dependencies and mechanisms of metabolic adaptation in lung SCC to identify therapeutic strategies to treat lung SCC. We discovered that lung SCC are reliant on both glucose and glutamine metabolism for survival and readily adapt their metabolism through the GSK3α/β signaling pathway. Analysis of a panel of glycolytic human tumors that include SCC of the lung, head and neck, as well as osteosarcomas revealed a metabolic signature that is conserved. This signature may be predictive of patient outcome, and we propose it can be used to stratify patients for metabolic based therapies.
Acknowledgments
We thank Gigi Abdelhady for genotyping mice and helping with daily dosing of mice. We thank Waldemar Ladno, Olga Sergeeva, Tove Olafsen, Charles Zamilpa at UCLA’s Crump Imaging Technology Center for assistance with PET/CT imaging of the mice and Drs. Sherief Gamie and Corey Chakarun at Long Beach Memorial Medical Center for assistance with human PET/CT images. We thank Translational Pathology Core Laboratory at UCLA’s DGSOM for assistance with tumor sample preparation and processing (UL1TR000124). We thank Dr. Abigail Fletcher for careful review of the manuscript. D.B.S. was supported by CTSI and KL2 Translational Science Award grant numbers KL2TR000122 and UL1TR000124 at the UCLA David Geffen School of Medicine, the Department of Defense LCRP grant number W81XWH-13–1-0459, American Cancer Society grant number RSG-16–234-01-TBG, the American Lung Association and Addario Lung Cancer Foundation and the STOP Cancer Foundation. S.T.B. was supported by an NIH T32 training grant HL072752 through the UCLA David Geffen School of Medicine. T.G.G is supported by the NCI/NIH (P01 CA168585), an American Cancer Society Research Scholar Award (RSG-12–257-01-TBE), and the National Center for Advancing Translational Sciences UCLA CSTI Grant UL1TR000124.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References:
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, Scherpereel A, Mascaux C, Moreau M, Roelandts M, et al. (2008). Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 3, 6–12. [DOI] [PubMed] [Google Scholar]
- Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. (2015). Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 373, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. (2016). Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, and Davis RJ (1994). JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025–1037. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. (2008). Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, and Dang CV (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J, Neugent ML, Lee SY, Choe JH, Choi H, Jenkins DMR, Ruthenborg RJ, Robinson MW, Jeong JY, Wake M, et al. (2017). The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition. Nat Commun 8, 15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, and Hann SR (2003). Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 278, 51606–51612. [DOI] [PubMed] [Google Scholar]
- Guertin DA, and Sabatini DM (2007). Defining the role of mTOR in cancer. Cancer Cell 12, 9–22. [DOI] [PubMed] [Google Scholar]
- Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, Stojanov P, McKenna A, Lander ES, Gabriel S, et al. (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein M, Hight MR, Buck JR, Tantawy MN, Nickels ML, Hoeksema MD, Harris BK, Boyd K, Massion PP, and Manning HC (2016). Preclinical Evaluation of 4-[18F]Fluoroglutamine PET to Assess ASCT2 Expression in Lung Cancer. Mol Imaging Biol 18, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, Clark JE, Alborn WE, Eisenberg R, and Massion PP (2013). SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res 19, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. (2016). Metabolic Heterogeneity in Human Lung Tumors. Cell 164, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 164, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JP, Lim I, Kong CB, Jeon DG, Byun BH, Kim BI, Choi CW, and Lim SM (2016). Prognostic Value of SUVmax Measured by Pretreatment Fluorine-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography in Patients with Ewing Sarcoma. PLoS One 11, e0153281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabour BA, Choi Y, Hoh CK, Rege SD, Soong JC, Lufkin RB, Hanafee WN, Maddahi J, Chaiken L, Bailet J, and et al. (1993). Extracranial head and neck: PET imaging with 2-[F-18]fluoro-2-deoxy-D-glucose and MR imaging correlation. Radiology 186, 27–35. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. (2007). LKB1 modulates lung cancer differentiation and metastasis. Nature 448, 807–810. [DOI] [PubMed] [Google Scholar]
- Loening AM, and Gambhir SS (2003). AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging 2, 131–137. [DOI] [PubMed] [Google Scholar]
- Lukey MJ, Greene KS, Erickson JW, Wilson KF, and Cerione RA (2016). The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat Commun 7, 11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, Fernandez-Cuesta L, Heuckmann JM, Gautschi O, Diebold J, et al. (2014). Cell-Autonomous and Non-Cell-Autonomous Mechanisms of Transformation by Amplified FGFR1 in Lung Cancer. Cancer Discov 4, 246–257. [DOI] [PubMed] [Google Scholar]
- Matkar S, Sharma P, Gao S, Gurung B, Katona BW, Liao J, Muhammad AB, Kong XC, Wang L, Jin G, et al. (2015). An Epigenetic Pathway Regulates Sensitivity of Breast Cancer Cells to HER2 Inhibition via FOXO/c-Myc Axis. Cancer Cell 28, 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. (2012). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn H, Lapela M, Klemi PJ, Grenman R, Leskinen S, Lindholm P, Bergman J, Eronen E, Haaparanta M, and Joensuu H (1997). Prediction of survival with fluorine-18-fluoro-deoxyglucose and PET in head and neck cancer. J Nucl Med 38, 1907–1911. [PubMed] [Google Scholar]
- Momcilovic M, Bailey ST, Lee JT, Fishbein MC, Magyar C, Braas D, Graeber T, Jackson NJ, Czernin J, Emberley E, et al. (2017). Targeted Inhibition of EGFR and Glutaminase Induces Metabolic Crisis in EGFR Mutant Lung Cancer. Cell Rep 18, 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M, McMickle R, Abt E, Seki A, Simko SA, Magyar C, Stout DB, Fishbein MC, Walser TC, Dubinett SM, and Shackelford DB (2015). Heightening Energetic Stress Selectively Targets LKB1-Deficient Non-Small Cell Lung Cancers. Cancer Res 75, 4910–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgensztern D, Waqar S, Subramanian J, Gao F, and Govindan R (2009). Improving survival for stage IV non-small cell lung cancer: a surveillance, epidemiology, and end results survey from 1990 to 2005. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 4, 1524–1529. [DOI] [PubMed] [Google Scholar]
- Moseley HN (2010). Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics 11, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, and DeBerardinis RJ (2012). Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, and Hogan BL (2007). Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A 104, 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, and Rosen N (2011). mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 1, 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, and Nevins JR (2000). Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. (2015). Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest 125, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, and Shaw RJ (2013). LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai M, Graham NA, Braas D, Nehil M, Komisopoulou E, Kurdistani SK, McCormick F, Graeber TG, and Christofk HR (2014). Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab 19, 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Dunphy MP, Zhang H, Pitter KL, Zanzonico P, Campos C, Carlin SD, La Rocca G, Lyashchenko S, Ploessl K, et al. (2015). Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci Transl Med 7, 274ra217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F, Federman N, Apichairuk W, Nelson S, Phelps ME, Allen-Auerbach M, Walter MA, and Czernin J (2011). 18F-fluorodeoxyglucose uptake of bone and soft tissue sarcomas in pediatric patients. Pediatr Hematol Oncol 28, 579–587. [DOI] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, and Kaelin WG Jr. (2005). The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33. [DOI] [PubMed] [Google Scholar]
- Xu C, Kim NG, and Gumbiner BM (2009). Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data reported in this paper has been deposited to NCBO GEO under accession number GSE110108.
Human lung cancer gene expression data generated by TCGA and utilized in this paper can be accessed via cBioPortal (www.cBioPortal).