Abstract
Exposure to environmental chemicals has been shown to have an impact on the epigenome. One example is a known human carcinogen 1,3-butadiene which acts primarily by a genotoxic mechanism, but also disrupts the chromatin structure by altering patterns of cytosine DNA methylation and histone modifications. Sex-specific differences in 1,3-butadiene-induced genotoxicity and carcinogenicity are well established; however, it remains unknown whether 1,3-butadiene-associated epigenetic alterations are also sex-dependent. Therefore, we tested the hypothesis that inhalational exposure to 1,3-butadiene will result in sex-specific epigenetic alterations. DNA damage and epigenetic effects of 1,3-butadiene were evaluated in in liver, lung, and kidney tissues of male and female mice of two inbred strains (C57BL/6J and CAST/EiJ). Mice were exposed to 0 or 425 ppm of 1,3-butadiene by inhalation (6 hr/day, 5 days/week) for 2 weeks. Strain- and tissue-specific differences in 1,3-butadiene-induced DNA adducts and crosslinks we detected in the liver, lung and kidney; however, significant sex-specific differences in DNA damage were observed in the lung of C57BL/6J mice only. In addition, we assessed expression of the DNA repair genes and observed a marked up-regulation of Mgmt in the kidney in female C57BL/6J mice. Sex-specific epigenetic effects of 1,3-butadiene exposure were evident in alterations of cytosine DNA methylation and histone modifications in the liver and lung in both strains. Specifically, we observed a loss of cytosine DNA methylation in the liver and lung of male and female 1,3-butadiene-exposed C57BL/6J mice, whereas hypermethylation was found in the liver and lung in 1,3-butadiene-exposed female CAST/EiJ mice. Our findings suggest that strain and sex-specific effects of 1,3-butadiene on the epigenome may contribute to the known differences in cancer susceptibility.
Introduction
Accumulating evidence suggests that environmental chemicals may alter the epigenome by disrupting cytosine DNA methylation patterns, chromatin structure and expression of noncoding RNAs (Baccarelli and Bollati 2009; Chappell et al. 2016; Rusyn and Pogribny 2017). Epigenetic reprogramming has been linked to genomic instability, a hallmark of cancer (Hanahan and Weinberg 2011), and induction of epigenetic alterations is one of key mechanistic characteristics of known human carcinogens (Smith et al. 2016). Epigenetic alterations may be a result of DNA damage or non-genotoxic effects of chemicals (Khobta and Epe 2012). However, even though there is an increasing interest in investigating the relationships between the states of the epigenome and chemical-induced adverse health effects (Chappell et al. 2016; Rusyn et al. 2018), few studies have considered sex-specific differences in epigenetic effects of chemicals, most of these were focused on prenatal or early life exposures (Faulk et al. 2013; Kippler et al. 2013; Kundakovic et al. 2013).
It is well acknowledged that sex is an important biological variable that now is required to be included into research designs, analyses, and reporting in vertebrate animal and human studies (Clayton and Collins 2014). Therefore, we aimed to study a chemical with well-known sex-specific differences in adverse health effects and determine whether its effects on epigenetic phenotypes are also sex-dependent. Specifically, we selected 1,3-butadiene, an industrial and environmental toxicant that is classified as a known human carcinogen (IARC 2009). In humans, exposure to 1,3-butadiene is associated with cancer in the haematolymphatic organs (Cogliano et al. 2011). In mice, liver, lung, and lymphoid tissues are the primary sites of 1,3-butadiene-induced carcinogenesis (Melnick et al. 1992).
Sex-specific differences in 1,3-butadiene-induced genotoxicity and carcinogenicity have been established in rodents and humans. Significant differences between male and female mice were reported in the levels of 1,3-butadiene DNA damage in the liver (Goggin et al. 2009). Sex-specific differences have also been identified in the occurrence of 1,3-butadiene-induced Hprt gene mutations, with female rodents exhibiting a greater susceptibility (Meng et al. 2007). In human studies, female factory workers exposed to 1,3-butadiene had lower globin adduct levels when compared to male workers, but not other markers of genotoxicity, possibly because of the sex-specific differences in toxicokinetics of 1,3-butadiene (Vacek et al. 2010). With respect to tumor development, tumor sites also vary between males and females in studies of rats and mice (Melnick et al. 1992; Owen et al. 1987). Furthermore, female mice form tumors at lower concentrations of 1,3-butadiene than male mice, further implicating sex-specific mechanisms as drivers of susceptibility to 1,3-butadiene-induced carcinogenesis (Melnick et al. 1992).
Epigenetic effects of 1,3-butadiene are well established and have been studied in multiple tissues of male mice (Chappell et al. 2017; Israel et al. 2018; Koturbash et al. 2011). These studies demonstrated that there are organ- and strain-dependent effects. Variability in 1,3-butadiene-induced chromatin alterations represents one possible mechanism for strain-specific differences in DNA damage (Chappell et al. 2017; Israel et al. 2018). Two inbred mouse strains, C57BL/6J and CAST/EiJ, have been extensively studied based on their sensitivity and resistance to 1,3-butadiene-induced DNA damage (Koturbash et al. 2011). In addition, inter-strain variability has also been identified in 1,3-butadiene-induced mitochondrial dysfunction using the Collaborative Cross mouse population model (Hartman et al. 2017). With respect to carcinogenesis-related genetic and epigenetic effects of 1,3-butadiene in mice, it was found that while exposure-induced DNA adducts are present in lung, liver, and kidney (Israel et al. 2018), epigenetic alterations (Chappell et al. 2014) and tumor formation are restricted to the liver and lung (Melnick et al. 1992; National Toxicology Program 1993). Tissue-specific epigenetic effects may provide mechanistic clues with respect to why certain organs are resistant to carcinogenesis even though they harbor appreciable genetic damage from 1,3-butadiene. In this study, we investigated sex-specific differences in DNA damage and epigenetic effects in response to 1,3-butadiene exposure in various organs in two inbred mouse strains. Overall, our results demonstrate that there are sex-specific differences in epigenetic modifications among mouse tissues and that these differences may play a role in cancer susceptibility in response to 1,3-butadiene exposures.
Methods
Animals and exposures
Male and female CAST/EiJ and C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME; 9–13 weeks old) were housed in sterilized cages in a temperature-controlled (24°C) room with a 12/12-h light/dark cycle. Mice had ad libitum access to purified water and NIH-31 pelleted diet (Purina Mills, Gray Summit, MO). These strains were selected based on previous results which indicated that C57BL/6J male mice are more susceptible to 1,3-butadiene-induced DNA damage and epigenetic effects compared to CAST/EiJ male mice, which are considered more resistant (Koturbash et al. 2011). Mice were allowed to acclimatize for 2 weeks and then were randomly assigned into control or 1,3-butadiene treatment groups. Subsequently mice were exposed to filtered air or 425 ppm 1,3-butadiene 6 hr/day, 5 days/week (Monday-Friday) for two consecutive weeks, a study dosing and duration based on previous studies of 1,3-butadiene genotoxicity(Swenberg et al. 2011). The average concentration of 1,3-butadiene in the exposure chamber in this study was 425.8+/−162.0 ppm. An in life portion of the study and tissue collection were previously described (Chappell et al. 2014) and were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
Determination of N7-guanine adducts and bis-N7-guanine crosslinks
Genomic DNA was isolated from flash-frozen liver, lung and kidney using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. Levels of N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) and 1,4-bis-(guan-7-yl)-2,3-butanediol crosslinks (bis-N7G-BD) in DNA were evaluated as described elsewhere (Goggin et al. 2009; Sangaraju et al. 2012).
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted with an RNeasy kit (Qiagen) from flash-frozen samples of the liver, lung and kidney. The High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Carlsbad, CA) was used to synthesize cDNA. Treatment-induced effects on gene expression for Mpg, Mgmt, Xrcc1, Dnmt1, Dnmt3a, and Dnmt3b were assessed by qRT-PCR. Detailed information on mouse-specific primer sequences and catalogue numbers is provided in Supplemental Table 1. Reactions were performed with two to four replicates in a 96-well plate using the 7900HT Fast Real-Time PCR system (Applied Biosystems). The housekeeping gene Gusb1 was used as a reference control.
DNA methylation of repetitive sequences
The McrBC-methylation sensitive quantitative PCR (qPCR) assay was used to determine the methylation status of short interspersed nucleotide elements B1 and B2 (SINEB1 and SINEB2) retrotransposons and minor and major satellites repetitive sequences in liver, lung, and kidney, as described in Martens et al (Martens et al. 2005). Genomic DNA was digested overnight with the restriction enzyme McrBC (New England Bio Labs, Beverly, MA) and subsequently analyzed with qPCR on the 7900 Real Time PCR System (Applied Biosystems).
Western blot analysis of histone modifications
Total histones were extracted as described elsewhere (Pogribny et al. 2006). Briefly, tissue samples were lysed with lysis buffer, incubated for one hour on ice, and centrifuged at 14,000×g for 10 minutes at 4°C. The supernatant was removed and mixed with 10 volumes of acetone for an overnight incubation. The precipitates were air-dried then dissolved in water. The levels of trimethylation of histones H3 lysine 9 (H3K9me3), H3 lysine 27 (H3K27me3), and H4 lysine 20 (H4K20me3) were evaluated by western immunoblotting using corresponding antibodies (Upstate, Charlottesville, VA) in the livers and kidneys of control and 1,3-butadiene-exposed mice as described elsewhere (Pogribny et al. 2006). Equal sample loading was confirmed by immunostaining against total H3 and H4.
Statistical Analyses
Results are presented as mean +/− SD. GraphPad Prism was used for statistical analysis of data. The student’s t test was used to evaluate differences between samples. Spearman correlation coefficients were used to evaluate correlations between phenotypes. Significance was determined when p<0.05 for all tests performed.
Results
Sex-specific differences in 1,3-butadiene-induced DNA damage and repair
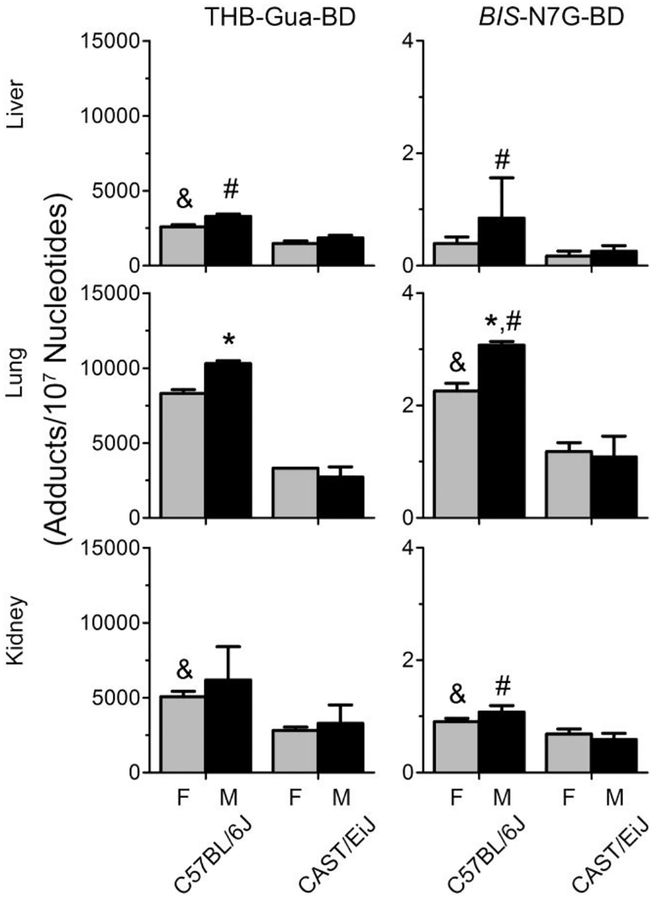
Metabolism of 1,3-butadiene yields a number of DNA-reactive moieties that form DNA adducts and cross-links in cultured cell, mice, rats, and humans (Owen et al. 1987; Swenberg et al. 2011). In the present study, we investigated THB-Gua adducts and bis-N7G crosslinks as markers of 1,3-butadiene-induced genotoxicity across several tissues in male and female mice. Figure 1 shows that THB-Gua adducts and bis-N7G crosslinks were present in all 1,3-butadiene-exposed mice. We include previously reported C57BL/6J male mice bis-N7G crosslinks results (Chappell et al. 2014) for comparison. In C57BL/6J male mice, adduct levels were similar to those previously reported at a comparable 1,3-butadiene dose (Chappell et al. 2014). Both guanine monoadducts and DNA-DNA crosslinks were 5–20% lower in female mice than in male mice across all tissues in C57BL/6J strain. Our data are also concordant with major differences in the susceptibility to 1,3-butadiene-induced DNA damage between CAST/EiJ and C57BL/6J male mice (Koturbash et al. 2011). Specifically, CAST/EiJ mice had three- to four-fold lower adduct and crosslink levels compared to C57BL/6J mice. No differences between male and female mice in adduct and crosslink levels were observed in CAST/EiJ strain. Among all tissues, in both strains and sexes, the lung had the highest levels of DNA damage, followed by the kidney and liver.
Figure 1.
Levels of THB-Gua-butadiene adducts (left panel) and bis-N7G-butadiene crosslinks (right panel) in tissues from female (gray bars) and male (black bars) mice exposed to 425 ppm of 1,3-butadiene. Data are presented as mean +/−SD. Asterisk, pound, and ampersand (*, #, and &) denote significant (p<0.05) differences in the levels of adducts or crosslinks between C57BL/6J or CAST/EiJ male and female mice, C57BL/6J and CAST/EiJ male mice, or C57BL/6J and CAST/EiJ female mice, respectively.
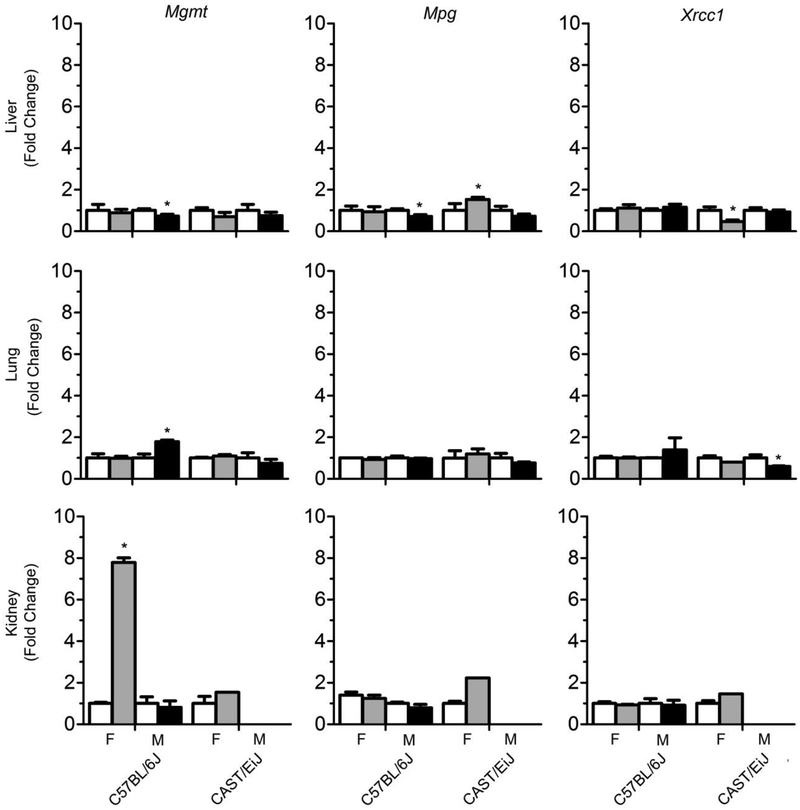
To further investigate sex-specific differences in DNA damage, we evaluated the expression of DNA repair genes that are involved in the base excision repair pathway (Mpg and Xrcc1) and removal of O6-methyl guanine (Mgmt). Figure 2 shows sex-specific changes in DNA repair enzyme expression across liver, lung, and kidney. In C57BL/6J mice, there was a marked increase in expression of Mgmt in 1,3-butadiene-exposed female mice in the kidney, while in the liver, both Mgmt and Mpg expression was significantly decreased after exposure. In CAST/EiJ mice, Mpg was induced in the liver of female mice and Xrcc1 was significantly decreased in the liver of female mice and the lung of male mice.
Figure 2.
Effects of 1,3-butadiene exposure on the expression of DNA repair genes across strains and tissues (liver – top panel, lung – middle panel, kidney – bottom panel). White bars are controls, gray bars are treated females and black bars are treated males. Results are presented as the average fold change relative to the control values for each strain and sex. Data are expressed as mean +/-SD. Asterisks (*) denote significant (p<0.05) differences from corresponding strain and sex controls. Missing samples (male kidney, CAST/EiJ) have no bars.
Sex-specific alterations in 1,3-butadiene-induced cystosine DNA methylation patterns
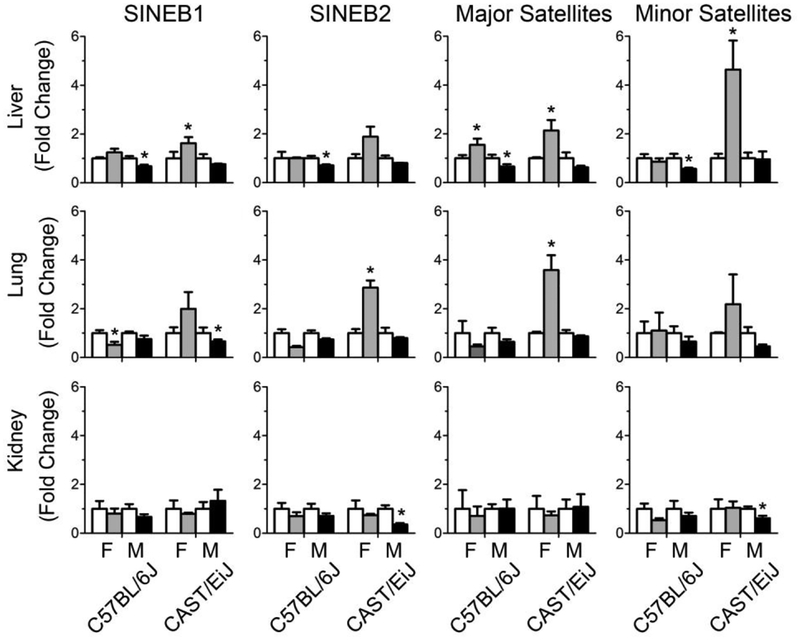
Increasing evidence suggests that disruption of cytosine DNA methylation patterns can play a role in disease (Robertson 2005). Repetitive elements that are a major component of the genome (Martens et al. 2005) are usually methylated in somatic cells (Yoder et al. 1997) and can serve as indicators of global DNA methylation status (Bae et al. 2012). To investigate sex-specific cytosine DNA methylation changes in response to 1,3-butadiene exposure, the methylation status of SINEB1 and SINEB2 retrotransposons, as well as major and minor satellites, was assessed in control and 1,3-butadiene-treated mice across tissues. It is known that 1,3-butadiene exposure resulted in strain-dependent changes in DNA methylation patterns, where CAST/EiJ remained unaffected while C57BL/6J male mice exhibited DNA hypomethylation in the liver (Koturbash et al. 2011). Previously reported DNA methylation data from male C57BL/6J mice (Chappell et al. 2014) was used here to compare to the new data for C57BL/6J female mice and male and female CAST/EiJ mice. Figure 3 shows sex-specific DNA methylation changes of SINEB1, SINEB2 retrotransposons and major and minor satellites in response to 1,3-butadiene exposure. In male C57BL/6J mice, exposure to 1,3-butadiene was associated with loss of cytosine DNA methylation in the liver, whereas in female mice significant loss of cytosine DNA methylation was observed in the lung. No treatment-related cytosine DNA methylation changes were found in the kidney in male and female C57BL/6J mice. In contrast, exposure of CAST/EiJ mice to 1,3-butadiene resulted in 2- to 4-fold increase in repetitive element cytosine DNA methylation in the liver and lung in female mice, whereas only minor and sporadic DNA hypomethylation changes were found in the liver, lung, and kidney of male CAST/EiJ mice.
Figure 3.
Effects of 1,3-butadiene exposure on the extent in DNA methylation across strains and tissues (liver – top panel, lung – middle panel, kidney – bottom panel). White bars are controls, gray bars are treated females, and black bars are treated males. The results are presented as fold change relative to the control values for each strain and sex. Data are presented as mean +/-SD. Asterisks (*) denote significant (p<0.05) difference from the corresponding strain and sex controls.
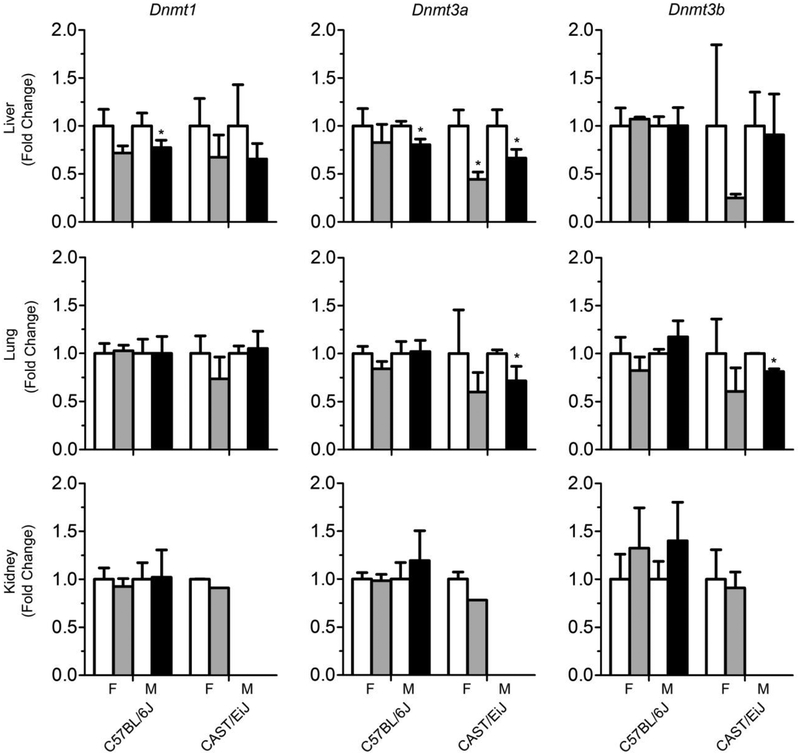
To provide mechanistic insight into 1,3-butadiene-associated sex-specific differences in DNA methylation, the expression of DNA methyltransferase genes were investigated. Figure 4 shows that overall there were few effects on expression of Dnmt1, Dnmt3a, or Dnmt3b in 1,3-butadiene-exposed mice in the liver, lung, and kidney. Specifically, in C57BL/6J mice, there was a decrease in expression in Dnmt1 and Dnmt3a in the liver in both male and female mice, albeit this effect was significant only in male mice. In CAST/EiJ mice, a significant decrease in expression of Dnmt3a was observed in the liver in both male and female mice. In the lungs, a significant decrease in expression of Dnmt3a and Dnmt3b was observed in male mice only.
Figure 4.
Effects of 1,3-butadiene exposure on the expression of DNA methyltransferase genes across strains and tissues (liver – top panel, lung – middle panel, kidney – bottom panel). White bars are controls, gray bars are treated females and black bars are treated males. The results are presented as fold change relative to the control values for each strain and sex. Data are presented as mean +/−SD. Asterisks (*) denote significant (p<0.05) differences from the corresponding strain and sex controls.
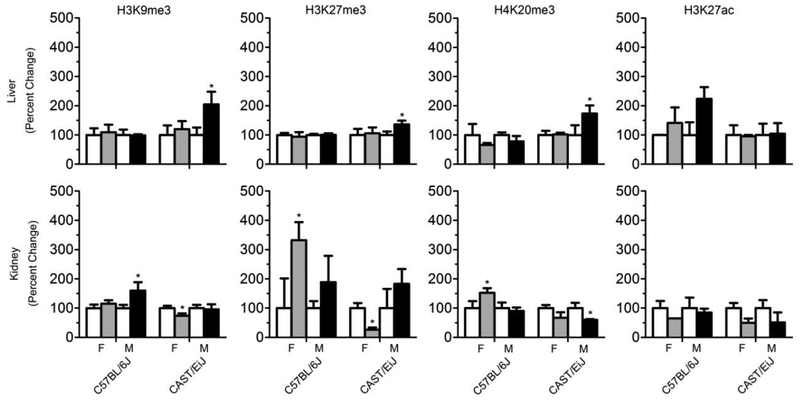
Sex-specific differences in 1,3-butadiene-induced histone modifications
Disruption of the chromatin structure has been associated with chemical carcinogen exposure (Baccarelli and Bollati 2009; Koturbash et al. 2011; Thomson et al. 2014). In the present study, histone modifications that are related to chromatin assembly and transcription repression were investigated. Figure 5 shows the levels of H3K9me3, H3K27me3, and H4K20me3 in the liver and kidney. In male C57BL/6J mice, effects of 1,3-butadiene on post-translational histone modifications were concordant with previously reported findings, which showed a significant increase in trimethylation of histones H3K9, H3K27 and H4K20 in the kidney (Chappell et al. 2014). In female C57BL/6J mice, significant increases in trimethylation of H3K27 and H4K20 were also observed in the kidney. A significant increase in H3K27ac was only observed in the liver of C57BL/6J male mice. In CAST/EiJ strain, 1,3-butadiene-exposed male mice exhibited significant increases in histone H3K9, H3K27, and H4K20 trimethylation in the liver, concordant with previous report (Koturbash et al. 2011). In contrast, there was a significant decrease in trimethylation of histone H4K20 in the kidney of male CAST/EiJ mice and decrease in trimethylation of histones H3K9 and H3K27 in in the kidney of female mice. We also evaluated histone methyltransferase genes; however, there were no significant changes (data not shown).
Figure 5.
Effects of 1,3-butadiene exposure on histone trimethylation and acetylation across strains and tissues (liver – top panel, kidney – bottom panel). All histone levels were evaluated by immunostaining using specific antibodies against trimethylated or acetylated histones. Equal sample loading was confirmed by immunostaining against total H3 or H4. Densitometry analysis of the immunostaining results is shown as percent change in methylation levels relative to control after correction for the total histone levels in each sample. White bars are controls, gray bars are treated females and black bars are treated males. The results are presented as fold change relative to the control values for each strain and sex in mouse tissues. Data are presented as mean +/-SD. Asterisks (*) denote significant (p<0.05) differences from the corresponding strain and sex controls.
Discussion
This study used 1,3-butadiene as a model genotoxic environmental chemical to investigate sex-specific differences in genotoxic and epigenetic effects. Exposure to 1,3-butadiene has been previously associated with the formation of DNA monoadducts and DNA-DNA crosslinks (Swenberg et al. 2000), as well as changes in DNA methylation and chromatin structure (Koturbash et al. 2011). Our results indicate that 1,3-butadiene-induced DNA damage and epigenome disruption are sex-dependent. In addition, these observed sex-specific 1,3-butadiene-effects are strain-specific.
It is well established that exposure to 1,3-butadiene results in the formation of DNA monoadducts and interstrand crosslinks as supported by animal and human studies (Swenberg et al. 2011). Similar to previous findings, we observed that 1,3-butadiene-induced DNA damage was highest in the lung, followed by the kidney, and liver. Our results also are in accord with previously identified inter-strain differences in DNA adduct formation between male CAST/EiJ and C57BL/6J mice (Israel et al. 2018; Koturbash et al. 2011). We found significant sex-specific differences in both monoadducts and interstrand crosslinks only in the lung of C57BL/6J mice. Sex-specific differences in 1,3-butadiene genotoxicity are well-documented. Female rats and mice have twice the DNA-DNA crosslink formation in the liver compared to male rats and mice (Goggin et al. 2009). Additionally, female rats and mice have a higher incidence of Hprt mutations (Meng et al. 2007). Human molecular epidemiology studies reported similar or lower globin adduct levels in 1,3-butadiene-exposed female factory workers compared to male workers (Vacek et al. 2010). Our results also report levels of DNA damage in C57BL/6J mice that are concordant with findings in humans. Sex-specific differences in THB-G adduct levels have not been previously reported; however, in the present study we observed significant differences between C57BL/6J male and female mice (Swenberg et al. 2011). The interplay between strain and sex may influence susceptibility to DNA damage. Strain- and sex-specific molecular events including upregulation of DNA repair enzymes and chromatin remodeling could be responsible for the observed significant differences in DNA adduct and crosslink formation between C57BL/6J male and female mice.
Investigating the expression of DNA repair enzymes may explain the variability in DNA damage observed between strains and sexes (Rusyn et al. 2004). Interestingly, the effect of exposure to 1,3-butadiene on DNA repair gene expression in the base and nucleotide excision pathways was rather muted overall. However, we observed up to an 8-fold induction of Mgmt in kidneys of female mice of C57BL/6J strain, which may be a result of their sensitivity to the adverse effects of 1,3-butadiene in comparison to other mouse strains (Koturbash et al. 2011). C57BL/6J male mice also demonstrate an increase in expression in the lung, which could be in response to the higher incidence of DNA adduct and crosslink formation. While 1,3-butadiene is not known to form O6-alkylguanine adducts, DNA cross-links at the O6 position of guanine generated by butadiene present in tobacco smoke have been reported (Arif et al. 2006). In addition, human MGMT could play a role in repair of these crosslinks (Tubbs et al. 2007). Sex-specific differences in DNA repair gene expression have been identified in acute low-dose exposure to radiation, which induced a significant upregulation of Mgmt in female mice (Kovalchuk et al. 2004). In addition, up-regulation of Mgmt, as observed in C57BL/6J female mice in the kidney, has been associated with resistance to carcinogenesis (Dumenco et al. 1993).
Chemical exposure can also result in disruption of global DNA methylation (Pogribny and Rusyn 2013). Sex-dependent changes in DNA methylation have been reported. For example, exposure to radiation and arsenic led to loss of methylation, however it was more evident in females compared to males (Hossain et al. 2017; Pogribny et al. 2004). Exposure to 1,3-butadiene is associated with strain- and tissue-specific changes in DNA methylation, with varying degrees of cytosine DNA hypomethylation or no effect (Chappell et al. 2014; Koturbash et al. 2011).
In the present study we found a sex- and strain-dependent hypomethylation of major repetitive elements in the liver in male C57BL/6J mice and in the lung in female C57BL/6J mice, two target organs for 1,3-butadiene-induced carcinogenesis in mice. In contrast, no treatment-related cytosine DNA methylation changes were found in the kidney, a non-target organ, in male and female C57BL/6J mice, even though the level of 1,3-butadiene-induced genotoxic alterations in the kidney of C57BL/6J mice was greater than in the liver. These findings provide a strong support of the importance of epigenetic alterations, in addition to genotoxic alterations, in the mechanism of chemical carcinogenesis. Additionally, the results of the present study corresponded to growing evidence of the ability of genotoxic carcinogens to induce non-genotoxic genomic alterations, e.g. trancriptomic and epigenomic, in target organ only. Specifically, it has been demonstrated that in vivo or in vitro exposure to the model genotoxic carcinogen benzo[a]pyrene resulted in the induction of both genotoxic and non-genotoxic alterations in target, but not in non-target organs (Tryndyak et al. 2018; Zuo et al. 2014).
In order to uncover the underlying mechanisms of 1,3-butadiene-induced DNA methylation effects, we investigated the expression of DNA methyltransferases, which control the status of cytosine DNA methylation. Surprisingly, we found only slight down-regulation of Dnmt1 and Dnmt3a in the livers of male C57BL/6J, indicating that inhibition of DNA methyltransferases is not the main factor that caused cytosine DNA hypomethylation in 1,3-butadiene-carcinogenesis target organs in C57BL/6J mice. It is well-known that the accurate status of the DNA methylome is maintained by the following factors; proper functioning of DNA methylation and demethylation pathways, DNA integrity, and one-carbon metabolism, which provides methyl groups for all cellular methylation reaction (Pogribny and Beland 2009). Our data indicate that neither inhibition of DNA methyltransferases, nor 1,3-butadiene-induced genotoxic alterations, except in the lung of female mice, could explain the loss of cytosine DNA methylation in the liver and lung of C57BL/6J mice. This indicates that disruption of other molecular pathways such as mitochondrial dysfunction (Hartman et al. 2017) and 1,3-butadiene-induced oxidative stress (Primavera et al. 2008; Zhang et al. 2015), known factors that cause cytosine DNA hypomethylation (Valinluck et al. 2004), or alterations in one-carbon metabolism and tricarboxylic acid cycle, may cause loss of DNA methylation in the liver and lung of C57BL/6J mice.
Another interesting finding in our study is increased cytosine DNA methylation in the lung and liver in 1,3-butadiene-exposed CAST/EiJ female mice and no methylation changes in these organs in male mice. This may be attributed, at least in part, to substantially lower levels of 1,3-butadiene-induced DNA adducts and crosslinks in CAST/EiJ mice as compared to C57BL/6J mice. Although hypermethylation has been implicated in carcinogenesis (Baylin and Herman 2000), evidence suggests that increased levels of cytosine DNA methylation at repetitive elements could also provide protection against DNA damage (Patchsung et al. 2018). Hypermethylation has also been proposed as a protective epigenetic mechanism against repeat expansion-associated pathologies (Liu et al. 2014; Yoder et al. 1997). An additional mechanism that may prevent the loss of cytosine DNA methylation in target organs is formation of compact chromatin in response to chemical exposure. Indeed, we observed an increase in trimethylation of histones H3K9, H3K27, and H4K20 in the liver of male CAST/EiJ mice. This finding reinforces previously reported observations with exposure to a higher dose of 1,3-butadiene (Koturbash et al. 2011).
In summary, this study focused on identifying sex-specific differences in DNA damage and global epigenetics. Our results demonstrate that although DNA damage was present in all tissues, changes in DNA methylation and histone modification patterns varied between strains, sexes, and tissues. The present study demonstrates the existence of sex-specific differences in response to 1,3-butadiene exposure and provides strong evidence to support NIH policy on including both sexes in experimental animal studies. Additional follow up experiments that evaluate the chromatin landscape and gene expression profiles through ATAC-seq and RNA-seq may provide additional insight to sex-dependent toxicant induced responses. Furthermore, miRNA are also key epigenetic regulators that have been shown to have sex-dependent expression. Investigating the interplay between the miRNome and epigenetic machinery could provide further mechanistic insights.
Supplementary Material
Acknowledgements
This work was supported, in part, by grants from National Institutes of Health (R01 ES023195, R01 CA095039 and P30 ES025128). The views expressed in this article are those of the authors and do not necessarily reflect the views of NIH.
Footnotes
Publisher's Disclaimer: Disclaimer: The views expressed in this manuscript do not necessarily represent those of the U.S. Food and Drug Administration.
References
- Arif JM, Dresler C, Clapper ML, et al. (2006) Lung DNA adducts detected in human smokers are unrelated to typical polyaromatic carcinogens. Chemical research in toxicology 19(2):295–9. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V (2009) Epigenetics and environmental chemicals. Curr Opin Pediatr 21(2):243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JM, Shin SH, Kwon HJ, et al. (2012) ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. International journal of cancer Journal international du cancer 131(6):1323–31. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG (2000) DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics 16(4):168–174. [DOI] [PubMed] [Google Scholar]
- Chappell G, Kobets T, O’Brien B, et al. (2014) Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice. Toxicol Sci 142(2):375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell G, Pogribny IP, Guyton KZ, Rusyn I (2016) Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: A systematic literature review. Mutat Res Rev Mutat Res 768:27–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell GA, Israel JW, Simon JM, et al. (2017) Variation in DNA-Damage Responses to an Inhalational Carcinogen (1,3-Butadiene) in Relation to Strain-Specific Differences in Chromatin Accessibility and Gene Transcription Profiles in C57BL/6J and CAST/EiJ Mice. Environ Health Perspect 125(10):107006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JA, Collins FS (2014) Policy: NIH to balance sex in cell and animal studies. Nature 509(7500):282–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliano VJ, Baan R, Straif K, et al. (2011) Preventable exposures associated with human cancers. J Natl Cancer Inst 103(24):1827–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumenco LL, Allay E, Norton K, Gerson SL (1993) The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferase. Science 259(5092):219–22. [DOI] [PubMed] [Google Scholar]
- Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC (2013) Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5(5):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin M, Swenberg JA, Walker VE, Tretyakova N (2009) Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res 69(6):2479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–74. [DOI] [PubMed] [Google Scholar]
- Hartman JH, Miller GP, Caro AA, et al. (2017) 1,3-Butadiene-induced mitochondrial dysfunction is correlated with mitochondrial CYP2E1 activity in Collaborative Cross mice. Toxicology 378:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain K, Suzuki T, Hasibuzzaman MM, et al. (2017) Chronic exposure to arsenic, LINE-1 hypomethylation, and blood pressure: a cross-sectional study in Bangladesh. Environ Health-Glob 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (2009) 1,3-Butadiene, ethylene oxide and vinyl halides (vinyl fluoride and vinyl bromide). WHO, Lyon, France. [Google Scholar]
- Israel JW, Chappell GA, Simon JM, et al. (2018) Tissue- and strain-specific effects of a genotoxic carcinogen 1,3-butadiene on chromatin and transcription. Mammalian genome: official journal of the International Mammalian Genome Society 29(1–2):153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobta A, Epe B (2012) Interactions between DNA damage, repair, and transcription. Mutation research 736(1–2):5–14. [DOI] [PubMed] [Google Scholar]
- Kippler M, Engstrom K, Mlakar SJ, et al. (2013) Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8(5):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, et al. (2011) Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol Sci 122(2):448–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk O, Burke P, Besplug J, Slovack M, Filkowski J, Pogribny I (2004) Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat Res 548(1–2):75–84. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Gudsnuk K, Franks B, et al. (2013) Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci U S A 110(24):9956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Russ J, Wu K, et al. (2014) C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 128(4):525–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, et al. (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24(4):800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick RL, Huff J, Matanoski GM (1992) Carcinogenicity of 1,3-butadiene. Lancet 340:724–725. [DOI] [PubMed] [Google Scholar]
- Meng Q, Walker DM, McDonald JD, et al. (2007) Age-, gender-, and species-dependent mutagenicity in T cells of mice and rats exposed by inhalation to 1,3-butadiene. Chem Biol Interact 166(1–3):121–31. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (1993) NTP Toxicology and Carcinogenesis Studies of 1,3-Butadiene (CAS No. 106-99-0) in B6C3F1 Mice (Inhalation Studies). NatlToxicol Program TechRepSer 434:1–389. [PubMed] [Google Scholar]
- Owen PE, Glaister JR, Gaunt IF, Pullinger DH (1987) Inhalation toxicity studies with 1,3-butadiene. 3. Two year toxicity/carcinogenicity study in rats. Am Ind Hyg Assoc J 48(5):407–13. [DOI] [PubMed] [Google Scholar]
- Patchsung M, Settayanon S, Pongpanich M, Mutirangura D, Jintarith P, Mutirangura A (2018) Alu siRNA to increase Alu element methylation and prevent DNA damage. Epigenomics 10(2):175–185. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O (2004) Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Bioph Res Co 320(4):1253–1261. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Beland FA (2009) DNA hypomethylation in the origin and pathogenesis of human diseases. Cellular and molecular life sciences: CMLS 66(14):2249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, Karpinets TV (2006) Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4–20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis 27(6):1180–1186. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Rusyn I (2013) Environmental toxicants, epigenetics, and cancer. Advances in experimental medicine and biology 754:215–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primavera A, Fustinoni S, Biroccio A, et al. (2008) Glutathione transferases and glutathionylated hemoglobin in workers exposed to low doses of 1,3-butadiene. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 17(11):3004–12. [DOI] [PubMed] [Google Scholar]
- Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6(8):597–610. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Asakura S, Pachkowski B, et al. (2004) Expression of base excision DNA repair genes is a sensitive biomarker for in vivo detection of chemical-induced chronic oxidative stress: identification of the molecular source of radicals responsible for DNA damage by peroxisome proliferators. Cancer research 64(3):1050–7. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Kleeberger SR, McAllister KA, French JE, Svenson KL (2018) Introduction to mammalian genome special issue: the combined role of genetics and environment relevant to human disease outcomes. Mammalian genome: official journal of the International Mammalian Genome Society 29(1–2):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I, Pogribny IP (2017) Editorial overview of the special issue on genomic toxicology epigenetics. Curr Opin Toxicol 6:i–iii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaraju D, Goggin M, Walker V, Swenberg J, Tretyakova N (2012) NanoHPLC-nanoESI(+)-MS/MS quantitation of bis-N7-guanine DNA-DNA cross-links in tissues of B6C3F1 mice exposed to subppm levels of 1,3-butadiene. Anal Chem 84(3):1732–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Guyton KZ, Gibbons CF, et al. (2016) Key Characteristics of Carcinogens as a Basis for Organizing Data on Mechanisms of Carcinogenesis. Environ Health Perspect 124(6):713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Bordeerat NK, Boysen G, et al. (2011) 1,3-Butadiene: Biomarkers and application to risk assessment. Chem Biol Interact 24(6):809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Ham AJ, Koc H, et al. (2000) DNA adducts: effects of low exposure to ethylene oxide, vinyl chloride and butadiene. Mutation research 464(1):77–86. [DOI] [PubMed] [Google Scholar]
- Thomson JP, Moggs JG, Wolf CR, Meehan RR (2014) Epigenetic profiles as defined signatures of xenobiotic exposure. Mutat Res Genet Toxicol Environ Mutagen 764–765:3–9. [DOI] [PubMed] [Google Scholar]
- Tryndyak V, Kindrat I, Dreval K, Churchwell MI, Beland FA, Pogribny IP (2018) Effect of aflatoxin B1, benzo[a]pyrene, and methapyrilene on transcriptomic and epigenetic alterations in human liver HepaRG cells. Food Chem Toxicol 121:214–223. [DOI] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA (2007) DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA repair 6(8):1100–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacek PM, Albertini RJ, Sram RJ, Upton P, Swenberg JA (2010) Hemoglobin adducts in 1,3-butadiene exposed Czech workers: female-male comparisons. Chem Biol Interact 188(3):668–76. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32(14):4100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics 13(8):335–340. [DOI] [PubMed] [Google Scholar]
- Zhang X, Hou H, Chen H, Liu Y, Wang A, Hu Q (2015) A column-switching LC-MS/MS method for simultaneous quantification of biomarkers for 1,3-butadiene exposure and oxidative damage in human urine. J Chromatogr B Analyt Technol Biomed Life Sci 1002:123–9. [DOI] [PubMed] [Google Scholar]
- Zuo J, Brewer DS, Arlt VM, Cooper CS, Phillips DH (2014) Benzo pyrene-induced DNA adducts and gene expression profiles in target and non-target organs for carcinogenesis in mice. BMC genomics 15:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.