Abstract
SAG (Sensitive to Apoptosis Gene) and ROC1 (Regulator of Cullin-1) are two family members of the RING component of CRL (Cullin RING ligase). Both members are essential for growth and survival of several types of human cancer cells; their role in renal cell carcinoma (RCC), however, remains elusive. Here we reported that compared to adjacent normal tissues, both SAG and ROC1 are overexpressed in RCC, which is positively correlated with poor patient survival, particularly for SAG. Depletion of SAG or ROC1 inhibited growth and survival of RCC cells by inducing G2/M arrest, senescence, and apoptosis likely due to accumulation of WEE1, p21, p27, NOXA, and BIM. Interestingly, simultaneous BIM knockdown in RCC cells partially rescues growth suppression triggered by depletion of SAG, but not ROC1, suggesting a differential role of BIM. Collectively, our study provides the proof-of-concept evidence that RING components of CRL are attractive candidates for targeted therapy of RCC.
Introduction
Cullin-RING Ligase (CRL) is the largest family of the E3 ubiquitin ligase that is responsible for ubiquitylation of 20% cellular proteins for degradation by proteasome system [1], [2]. CRL is a multicomponent E3, consisting of a cullin (with 8 family members), a substrate recognizing subunit (such as a F-box protein), an adaptor protein (such as SKP1), and a RING protein family member, ROC1/RBX1 or SAG/ROC2/RBX2 [3], [4], [5], [6], [7], [8]. In its founding member, also known as SCF (SKP1, Cullin-1, and F-box protein), Cullin-1 acts as a scaffold protein that at the N-terminus binds to adaptor protein SKP1 and a F-box protein and at the C-terminus binds to RING protein, ROC1 or SAG, which binds to an E2 with ubiquitin loaded, acting as an enzymatic core for ligase activity [9].
The sequence identity between human SAG and ROC1 is 50%, and both members are highly evolutionarily conserved among different species [7]. Our previous genetic studies revealed that the function of ROC1 and SAG is developmentally nonredundant since total knockout of ROC1 causes embryonic lethality at E6.5 with defective proliferation [10], whereas total knockout of SAG also causes embryonic lethality at E10.5 with defective angiogenesis and robust apoptosis [11]. ROC1 is constitutively expressed and complexes with cullins 1-4, whereas SAG is stress-inducible and complexes with cullin-5 as well as cullin-1 [12], [13], [14]. Interestingly, our recent study showed that ROC1 complexes with CDC34 or UBCH5C E2 to promote substrate polyubiquitylation via the K48 linkage, whereas SAG complexes with UBE2C/UBCH10 and UBE2S to promote substrate polyubiquitylation via the K11 linkage [15]. These unique features of two family members, leading to targeting unique sets of substrates for degradation, could explain why they are functional nonredundant.
SAG was originally cloned by us as an antioxidant protein that protects cells from apoptosis [3] and later identified as the second member of ROC/RBX RING family [6]. Both SAG and ROC1 were found to be overexpressed in human lung cancer, but only SAG overexpression was associated with poor patient survival [16]. SiRNA-based knockdown of SAG or ROC1 inhibited growth and survival of several human cancer cell lines both in vitro and in vivo [17], [18], [19]. However, whether and how these two family members regulate growth and survival of RCC cells are previously unknown.
In this study, we performed the direct comparison of SAG and ROC1 in regulation of growth and survival in RCC cells and the underlying mechanism. We found that both SAG and ROC1 are overexpressed in RCC tissues, which is correlated with poor survival of RCC patients, particularly for SAG. Depletion of SAG or ROC1 inhibited proliferation and survival of RCC cells with a greater effect seen in ROC1 depleted cells by induction of G2/M arrest, senescence, and apoptosis via accumulations of WEE1, p21, p27, NOXA, and BIM. Interestingly, simultaneous BIM knockdown partially rescued growth suppressive phenotype triggered by knockdown of SAG but not of ROC1. Collectively, our study validated that RING family proteins are attractive anticancer targets for RCC, and selective targeting ROC1 would yield a maximal growth suppression.
Materials and Methods
Cell Cultures
Human renal cell carcinoma cell lines 786-0, SLR21, RCC4, SLR20, Cak2, and ACHN were purchased from the American Type Culture Collection (Manassas, VA). The 786-0, SLR21, RCC4, and SLR20 cells were cultured in RPMI 1640 medium, whereas Cak2 and ACHN were cultured in DMEM/F12 medium, all supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C under a humidified atmosphere of 95% air and 5% CO2.
Immunohistochemistry Staining of Human RCC Tissue Array
Human RCC tissue arrays were from Biotechnology Co. LTD of Xi-An Alina (www.alenabio.com). Immunohistochemistry was performed using the ABC Vectastain kit (Vector Laboratories, Burlingame, CA) with affinity purified SAG and ROC1 specific antibody [17], [18]. The sections were developed with DAB and counterstained with hematoxylin, as described [20].
Expression of SAG and ROC1, and Survival of RCC Patients
Three subtypes of RCC—KIRC (Kidney Renal Clear Cell Carcinoma), KICH (Kidney Chromophobe), and KIRP (Kidney Renal Papillary Cell Carcinoma)—from TCGA datasets were used in this study. The mRNA expression of SAG and ROC1 in RCC tissues, and information on RCC patient survival were downloaded from the UALCAN [21]. These TCGA patient survival data were used for Kaplan-Meier survival analyses and to generate overall survival plots. For the expression of SAG and ROC1, the tumor samples in each cancer type were categorized into two groups: 1) high expression [with transcripts per million (TPM) values above upper quartile] and (2) low/medium expression (with TPM values below upper quartile). The survival curves of samples with high gene expression and low/medium gene expression were compared by log-rank test.
IB Analysis
Whole-cell lysates were prepared and subjected to immunoblotting analysis using antibodies against SAG [mAb raised against the RING domain (AA44-113)], ROC1 [18], PARP, Caspase3, and WEE1 (Cell Signaling), β-actin (Santa Cruz Biotechnology), p21, p27 (BD Transduction Laboratories), BIM (Imgenex), and NOXA (Oncogene Science).
Lentivirus-Based siRNA Knockdown
Lentivirus-based siRNAs targeting ROC1 (LT-ROC1), SAG (LT-SAG), BIM (LT-BIM), and LT-virus expressing scrambled control siRNA (LT-CONT) were constructed as described previously [18], [22], [23], [24]. For gene silencing, cells were infected with individual lenti-virus using Lipofectamine 2000 (Life Technology, Carlsbad, CA) for 48 hours in 60-mm dishes, followed by various assays for proliferation, clonal survival, and FACS profiling. For double silencing, cells were split into 60-mm dishes, after the first round of silencing, and transfected with second lenti-virus for another 48 hours before harvesting for biological assays.
ATP-Lite–Based Cell Proliferation Assay
Cells, infected with various virus, were split and seeded into 96-well plates with 3000 cells per well in quadruplicate, and cultured for up to 5 days before subjecting to the ATP-lite assay (PerkinElmer, Norwalk, CT) for proliferation [25].
Clonogenic Survival Assay
After lentivirus-based siRNA silencing, cells were split and seeded into 6-cm dish with 500 cells per dish in triplicate, followed by culture for 12 days. The colonies formed were fixed, stained, and counted under microscope [17].
Fluorescence-Activated Cell Sorting Analysis (FACS) Analysis
Cells were harvested and fixed in 70% ethanol at 20°C for 4 hours, stained with propidium iodide (18 μg/ml) containing 400 μg/ml RNaseA (Roche) with shaking for 1 hour, and analyzed by flow cytometry for apoptosis and cell cycle profile. Apoptosis was measured by the percentage of cells in sub-G1 population [17].
SA-β-gal Staining
The expression of SA-β-gal in cells was determined by SA-β-gal staining, as described [26].
Statistical Analysis
The statistical significance of differences between groups was assessed using GraphPad Prism6 software (version 4.03). Data were presented as mean ± SEM. The unpaired, two-tailed t test was used for the comparison of parameters between groups. The level of significance was set at a P value of < .05.
Results and Discussion
SAG and ROC1 Are Overexpressed in RCC Tissues with Correlation of Poor Patient Survival
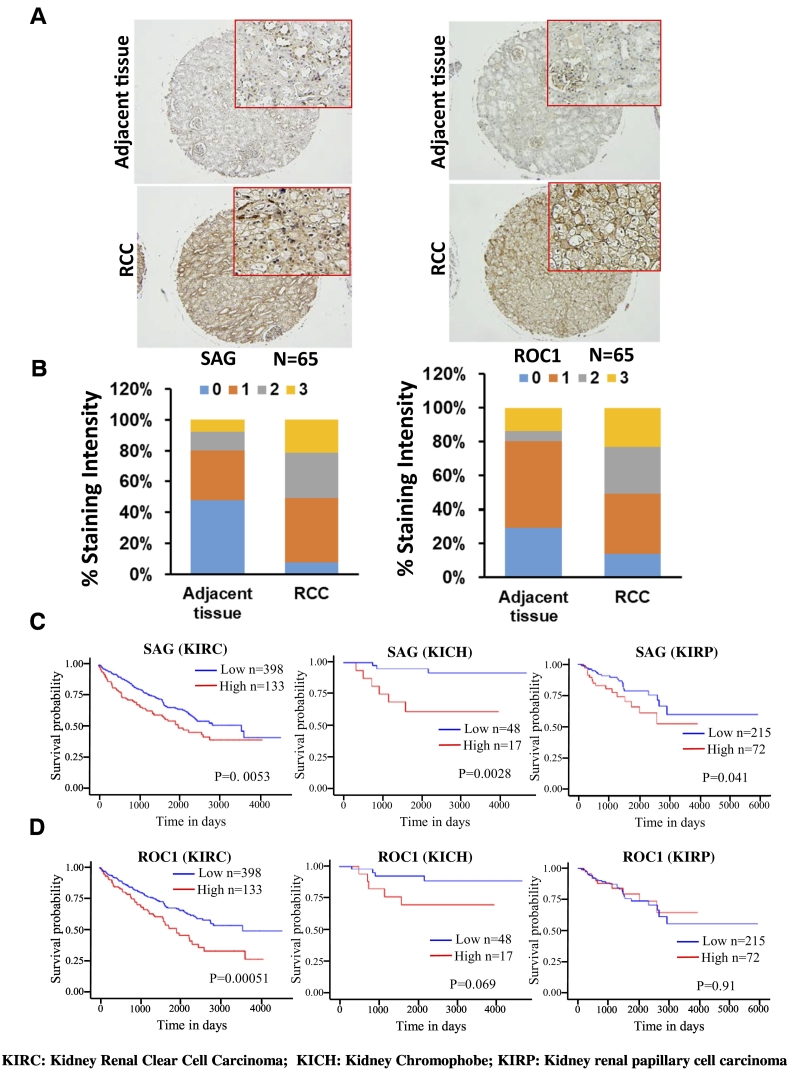
To determine potential alterations of SAG and ROC1 in RCC, we first performed immunohistochemistry staining in 65 paired RCC tissue microarrays. Based on the staining intensity, we classified the samples into four groups, with group 0 showing the least staining (+) and group 3 the highest staining (++++; Figure 1A). Compared to adjacent normal tissues with 75-80% of samples expressing under the categories of groups 0 and 1, RCC tissues had high expression of both family members with greater than 50% of cases expressing under categories of groups 2 and 3 (Figure 1, A and B). The TCGA database search revealed a rate of 0.65% of SAG amplification and 0.37% of ROC1 amplification in RCC tissues, suggesting that increased levels are likely due to increased expression.
Figure 1.
Overexpression of SAG and ROC1 in RCC tissue and its correlation with patient survival. (A) Immunostaining of SAG and ROC1 in human RCC tissues and adjacent normal tissues. Tissue microarrays of paired RCC and adjacent normal tissues from the same patient (n = 65) were stained with antibodies specifically for SAG and ROC1. Representative images are shown. (B) Percentage distribution based upon staining intensity. The staining intensity of each sample was scored and categorized into 4 groups (0-3). Percentage of distribution of these staining groups in RCC vs. adjacent tissues was plotted. (C) The Kaplan-Meier overall survival curves of high SAG expression and low SAG expression patients in three different types of RCC with sample size and P values indicated. (D) The Kaplan-Meier overall survival curves of high ROC1 expression and low ROC1 expression patients in three different types of RCC with sample size and P values indicated.
RCC represents a heterogeneous group of kidney cancer, mainly classified into three subtypes: KIRC (kidney renal clear cell carcinoma), KICH (kidney chromophobe), and KIRP (kidney renal papillary cell carcinoma) [27]. The TCGA database search for association between expression levels and patient survival revealed that high SAG expression is associated with a poor patient survival in all three types of RCC, whereas high ROC1 expression is associated with poor survival only in KIRC patients (Figure 1, C and D). Collectively, our study suggests that SAG and ROC1 may play a promoting role in renal carcinogenesis, and their overexpression could serve as the biomarkers for prognosis of kidney cancer patients.
Knockdown of SAG or ROC1 Inhibits Proliferation and Survival of RCC Cells
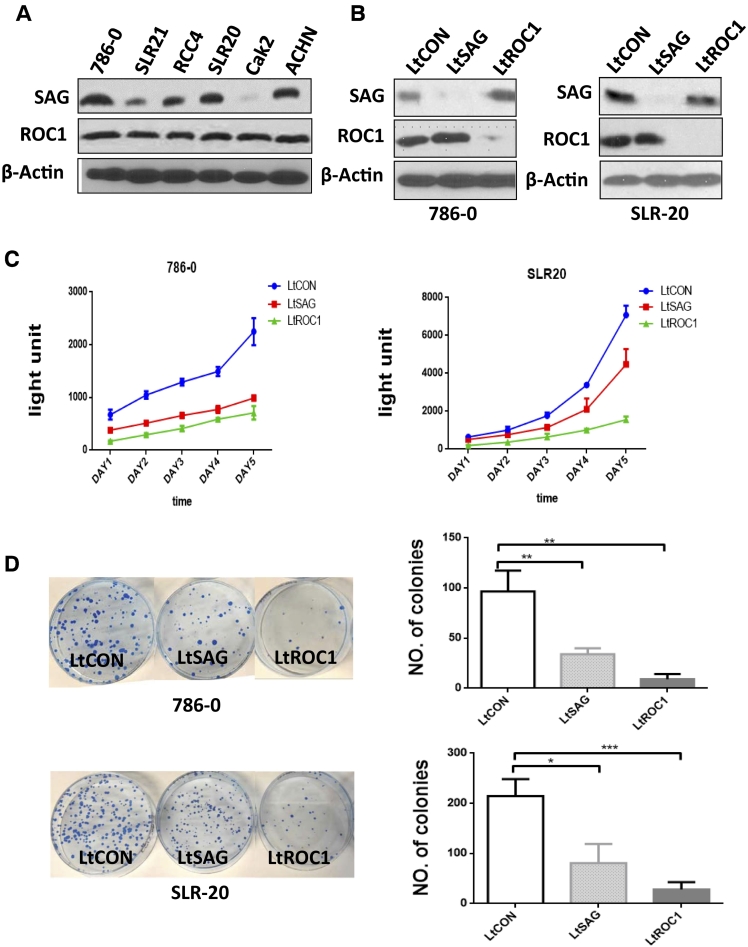
Having established that SAG and ROC1 are overexpressed in human RCC and associated with patient survival, we next used cell culture models to investigate the role of SAG and ROC1 in regulation of growth and survival of human RCC cells. We first detected the protein levels of SAG and ROC1 in six RCC cell lines and found that while ROC1 levels are similar among all lines tested, SAG levels are higher in 786-0 and SLR20 cells (Figure 2A). We then used lenti-virus–based siRNA silencing approach to deplete SAG and ROC1 in these two lines, respectively (Figure 2B), followed by ATP-lite–based cell proliferation and clonogenic based survival assays. In both assays, growth and survival of two RCC cell lines were significantly inhibited with greater effect seen in ROC1 depleted cells (Figure 2, C and D). Thus, both SAG and ROC1 are essential and required for the growth and survival of RCC cells.
Figure 2.
Knockdown of SAG or ROC1 inhibits growth and survival of RCC cells. (A) The expression levels of SAG and ROC1 in multiple RCC cell lines: Cell lysates from 6 RCC cell lines were subjected to Western blotting with SAG and ROC1 antibodies. (B-D) RCC cell lines 786-0 and SLR-20 were infected with LT-SAG or LT-ROC1 along with LT-CON for 72 hours, and cells were split for the following assays: IB for SAG and ROC1 (B), ATP-lite proliferation assay (n = 3) (C), and clonogenic survival (n = 3) (D). Shown are means ± S.E.M., *P < .05, **P < .01, ***P < .005.
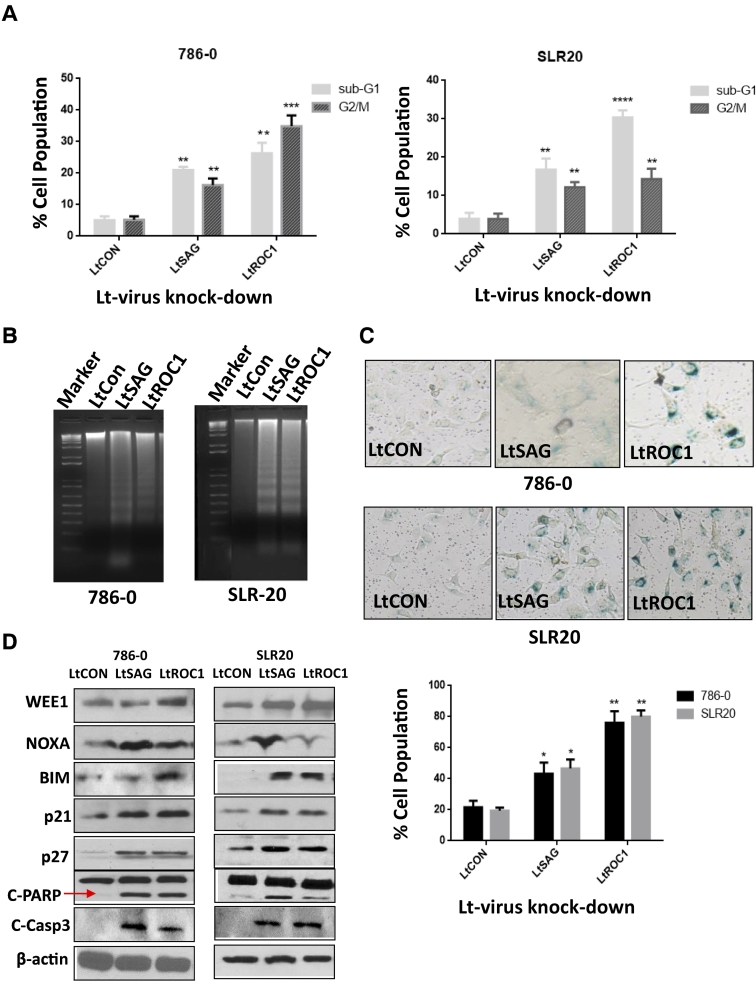
Knockdown of SAG or ROC1 Induces G2/M Arrest, Apoptosis, and Senescence of RCC Cells
To explore the nature of growth suppression, we performed FACS analysis and found that depletion of either SAG or ROC1 triggered substantial G2/M arrest and apoptosis (demonstrated as subG1 population) in both cell lines (Figure 3A). Induction of apoptosis was further confirmed by DNA fragmentation (Figure 3C) and cleavage of PARP and caspase-3 (Figure 3D) in both lines. Furthermore, we performed β-gal staining, a classic biochemical marker of senescence [26], and found that depletion of either SAG or ROC1 also triggered senescence (Figure 3C with β-Gal staining shown at the top panel and quantification at the bottom). Thus, suppression of growth and survival is caused by multiple alterations in cellular physiology. Finally, we determined potential mechanisms which mediate these biological consequences with focus on known substrates of SCF E3 ligase. Indeed, depletion of SAG or ROC1 triggered substantial accumulation of substrates, including Wee1 for G2/M arrest, NOXA and BIM for apoptosis, and p21 and p27 for senescence [16], [18], [19], [26], [28], [29], [30] (Figure 3D).
Figure 3.
Knockdown of SAG or ROC1 induced G2/M arrest, apoptosis, and senescence: RCC 786-0 and SLR20 cells were infected with LT-SAG or LT-ROC1 along with LT-CON for 72 hours, followed by FACS analysis (A), DNA fragmentation (B), β-gal staining (C), and Western blotting with indicated Abs (D). Shown are means ± S.E.M. from three independent experiments. *P < .05, **P < .01, ***P < .005, ****P < .001.
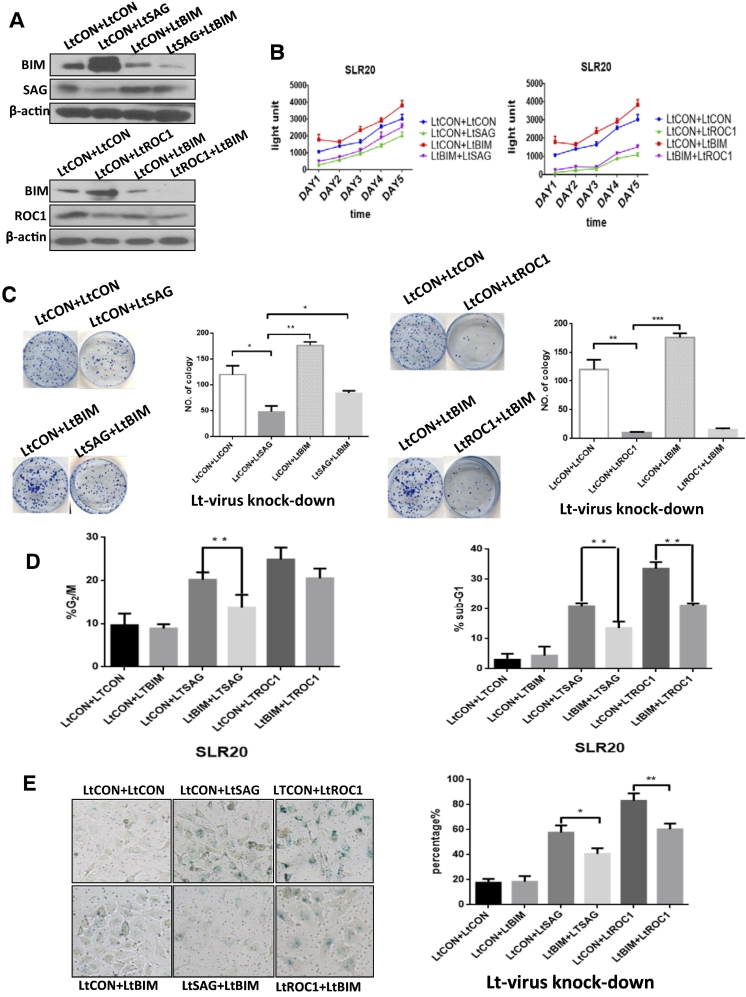
Simultaneous BIM Knockdown Partially Rescues Growth Suppression Triggered by Knockdown of SAG, But Not ROC1
Previous studies with rescue experiments have shown that NOXA plays an essential role for apoptosis, WEE1 for G2/M arrest, and p21 for senescence upon inactivation of SAG or ROC1 E3 via various approaches [29], [30], [31], [32]. We, therefore, focused our attention to the role of accumulated BIM in growth suppression triggered by SAG or ROC1 depletion. We found that single BIM knockdown moderately promoted growth and survival of SLR20 cells. Interestingly, simultaneous BIM knockdown (Figure 4A) partially but significantly rescued suppression of growth and survival triggered by SAG knockdown, but had minimal, if any, effect on growth suppression triggered by ROC1 knockdown (Figure 4, B and C). Mechanistically, BIM knockdown significantly reduced the population of G2/M arrest, apoptosis, and senescence in SAG knocked-down cells. In ROC1 knocked-down cells, BIM knockdown only rescued apoptosis and senescence, but not G2/M population (Figure 4, D and E), suggesting a key role played by G2/M arrest in manifesting growth phenotype of SLR20 RCC cells.
Figure 4.
BIM knockdown partially rescued apoptosis triggered by SAG knockdown: SLR-20 cells were first infected with LT-SAG or LT-ROC1, along with LT-CON, followed by infected LT-BIM for 72 hours. A portion of cells was harvested for Western blotting (A); the other portions for monolayer growth for 5 days, followed by ATP-lite proliferation assay (B); clonogenic assay for survival (C), FACS analysis (D), and senescence (E). Shown are means ± S.E.M. from three independent experiments. *P < .05, **P < .01, ***P < .005.
MLN4924, a small molecule inhibitor of NEDD8 activating enzyme (NAE) [1], has been shown to suppress proliferation of RCC cells by inducing G2 arrest and apoptosis in UBE2M dependent manner [33], or via accumulation of p21, p27, WEE1 [34], or NOXA [31]. Since MLN4924 inhibits neddylation E1 to block the entire neddylation pathway, whereas neddylation of cullin is required for CRL activity [2], MLN4924, therefore, inactivates ligase activity of the entire CRL family. With this broad inhibition, it is unknown whether inactivation of individual neddylation enzyme would induce growth suppression to different extent and via distinctive mechanisms. Given that SAG and ROC1 are also neddylation E3s to induce neddylation of Cullin-5 and Cullins 1-4, thus activating CRL5 and CRLs1-4, respectively [12], [14], we conducted current study via lenti-virus–based RNA silencing approach.
Here, we directly compared the role of SAG and ROC1 in regulating growth and survival of RCC cells. We demonstrated that both family members are overexpressed in RCC tissues, as compared to adjacent normal tissues, with a positive correlation of poor patient survival, particularly in the case of SAG. We further showed that both SAG and ROC1 are functionally nonredundant for growth and survival of RCC cells since depletion of either substantially suppressed proliferation. Biochemically, depletion of either SAG or ROC1 causes accumulation of few substrates, known to regulate cell growth. Biologically, accumulation of these substrates, including WEE1, NOXA, BIM, p21, and p27, triggers G2/M arrest, apoptosis, and senescence.
It is well established that BIM is a proapoptotic protein that suppresses tumor growth, while loss of BIM is associated with poor prognosis in a variety of tumor models [35], [36], [37]. Furthermore, loss of BIM was associated with chemoresistance of renal cell carcinoma [38]. We found that depletion of either SAG or ROC1 caused BIM accumulation, likely due to inactivation of SCF E3 ligase, since BIM has been reported to be the substrate of SCFβTrCP1 [39]. However, our rescue experiment showed that BIM accumulation plays a causal role at least in part for observed growth suppression in SAG-depleted cells, but not ROC1 depleted cells, further demonstrating a differential and nonredundant role of SAG vs. ROC1. It is worth noting that simultaneous BIM knockdown partially rescued apoptosis and senescence phenotypes triggered by ROC1 depletion but had no rescuing effect on G2/M arrest or on overall growth and survival (Figure 4), suggesting that extensive G2/M arrest triggered by ROC1 depletion plays the major role in growth suppression. It is also interesting to see that BIM knockdown partially rescued phenotypes of both G2/M arrest and senescence in addition to expected apoptosis in SAG-depleted cells, leading to a significant rescue of overall growth suppression. This suggests that induction of apoptosis, as a result of BIM accumulation, plays a major role in growth suppression upon SAG depletion. Nevertheless, the underlying mechanism for differential role of BIM in growth regulation of SAG vs. ROC1 depleted cells is unknown at the present time, but it is an interesting subject for future investigation. Taken together, our study demonstrated that targeting either SAG or ROC1 is sufficient to trigger growth suppression of RCC cells, and more specific inhibitors selective for SAG or ROC1 will likely have similar efficacy against RCC with anticipated less normal cells toxicity.
Authors' Contributions
Conception and design: Y. W. and Y. S.; experiment execution: Y. W. and M. T.;
data acquisition: Y. W., M. J., H. L.; data analysis and interpretation: Y. W., Haomin Li, and Y. S.; manuscript writing: Y. S.; study supervision: M. T., Y. S.
Acknowledgement
We would like to thank Dr. Wenyi Wei for providing us LT-shBIM construct. This work was supported by the fund for excellent doctor to go abroad for research and study at the first affiliated hospital of Nanchang University, and Jiangxi Educational Committee Fund (GJJ160250) to Y. W.
Footnotes
Conflicts of Interest: The authors claim no conflict of interest.
References
- 1.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate Cullin-RING ligases for anti-cancer therapy. Antioxid Redox Signal. 2014;21(17):2383–2400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 5.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 6.Swaroop M, Wang Y, Miller P, Duan H, Jatkoe T, Madore S, Sun Y. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: Chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3(4):635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr Pharm Des. 2013;19(18):3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 10.Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci U S A. 2009;106(15):6203–6208. doi: 10.1073/pnas.0812425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, Zhu Y, Sun Y. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell. 2011;21(6):1062–1076. doi: 10.1016/j.devcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33(4):483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Li H. Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase. Protein Cell. 2013;4(2):103–116. doi: 10.1007/s13238-012-2105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018;44:92–102. doi: 10.1016/j.cellsig.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuang P, Tan M, Zhou W, Zhang Q, Sun Y. SAG/RBX2 E3 ligase complexes with UBCH10 and UBE2S E2s to ubiquitylate beta-TrCP1 via K11-linkage for degradation. Sci Rep. 2016;6 doi: 10.1038/srep37441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, Xu J, Zhao L, Thomas D, Beer DG. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J Clin Invest. 2014;124(2):835–846. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16(3):814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69(12):4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235–247. doi: 10.1038/cdd.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, Liu P, Li H, Tan M, Xiong X. The beta-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67(8):3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- 23.Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1 alpha ubiquitination and degradation. Oncogene. 2008;27(10):1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- 24.Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, Liu J, Shaik S, Chen G, Deng J. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell. 2014;29(4):377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bockbrader KM, Tan M, Sun Y. A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene. 2005;24(49):7381–7388. doi: 10.1038/sj.onc.1208888. [DOI] [PubMed] [Google Scholar]
- 26.Tan M, Li H, Sun Y. Inactivation of Sag/Rbx2/Roc2 e3 ubiquitin ligase triggers senescence and inhibits kras-induced immortalization. Neoplasia. 2015;17(1):114–123. doi: 10.1016/j.neo.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F, Zhang Y, Senbabaoglu Y, Ciriello G, Yang L, Reznik E, Shuch B, Micevic G, De Velasco G, Shinbrot E. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016;14(10):2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan M, Li Y, Yang R, Xi N, Sun Y. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W, Xu J, Li H, Xu M, Chen ZJ, Wei W, Pan ZQ, Sun Y. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin Cancer Res. 2017;23:1104–1116. doi: 10.1158/1078-0432.CCR-16-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72(1):282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Wang S, Zhang W, Wang X, Liu X, Liu L, Li L, Liang Y, Yu J, Jeong LS. Targeting neddylation pathway with MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis in renal cell carcinoma. Biochem Biophys Res Commun. 2017;490(4):1183–1188. doi: 10.1016/j.bbrc.2017.06.179. [DOI] [PubMed] [Google Scholar]
- 32.Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE Inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia. 2011;13(6):561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B, Deng Y, Bi R, Guo H, Shu C, Shah NK, Chang J, Liu G, Du Y, Wei W. A first-in-class inhibitor, MLN4924 (pevonedistat), induces cell-cycle arrest, senescence, and apoptosis in human renal cell carcinoma by suppressing UBE2M-dependent neddylation modification. Cancer Chemother Pharmacol. 2018;81(6):1083–1093. doi: 10.1007/s00280-018-3582-z. [DOI] [PubMed] [Google Scholar]
- 34.Tong S, Si Y, Yu H, Zhang L, Xie P, Jiang W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci Rep. 2017;7(1):5599. doi: 10.1038/s41598-017-06098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26(9):1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2(3):183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 37.Maimaiti Y, Dong L, Aili A, Maimaitiaili M, Huang T, Abudureyimu K. Bim may be a poor prognostic biomarker in breast cancer patients especially in those with luminal A tumors. Cancer Biomark. 2017;19(4):411–418. doi: 10.3233/CBM-160377. [DOI] [PubMed] [Google Scholar]
- 38.Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, Hartmann C, Fritsch RM, Gillissen B, Daniel PT. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26(49):7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- 39.Moustafa-Kamal M, Gamache I, Lu Y, Li S, Teodoro JG. BimEL is phosphorylated at mitosis by Aurora A and targeted for degradation by betaTrCP1. Cell Death Differ. 2013;20(10):1393–1403. doi: 10.1038/cdd.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]