Abstract
Developmental delay and intellectual disability (DD and ID) are heterogeneous phenotypes that arise in many rare monogenic disorders. Because of this rarity, developing cohorts with enough individuals to robustly identify disease-associated genes is challenging. Social-media platforms that facilitate data sharing among sequencing labs can help to address this challenge. Through one such tool, GeneMatcher, we identified nine DD- and/or ID-affected probands with a rare, heterozygous variant in the gene encoding the serine/threonine-protein kinase BRSK2. All probands have a speech delay, and most present with intellectual disability, motor delay, behavioral issues, and autism. Six of the nine variants are predicted to result in loss of function, and computational modeling predicts that the remaining three missense variants are damaging to BRSK2 structure and function. All nine variants are absent from large variant databases, and BRSK2 is, in general, relatively intolerant to protein-altering variation among humans. In all six probands for whom parents were available, the mutations were found to have arisen de novo. Five of these de novo variants were from cohorts with at least 400 sequenced probands; collectively, the cohorts span 3,429 probands, and the observed rate of de novo variation in these cohorts is significantly higher than the estimated background-mutation rate (p = 2.46 × 10−6). We also find that exome sequencing provides lower coverage and appears less sensitive to rare variation in BRSK2 than does genome sequencing; this fact most likely reduces BRSK2’s visibility in many clinical and research sequencing efforts. Altogether, our results implicate damaging variation in BRSK2 as a source of neurodevelopmental disease.
Keywords: developmental delay, intellectual disability, clinical sequencing, exome, genome, de novo, BRSK2, Mendelian disease
Main Text
Developmental delay and intellectual disability (DD and ID), attention-deficient/hyperactivity disorder (ADHD), schizophrenia, language communication disorders, autism spectrum disorders (ASDs), and motor and tic disorders lie under a more general umbrella of neurodevelopmental disorders (NDDs).1, 2 Although these are traditionally categorized into discrete disease entities, many symptoms are not unique to a single NDD. Furthermore, many genes have been associated with multiple NDDs,3 and new genetic associations continue to be discovered. This is particularly true given the recent acceleration in large-scale sequencing and cross-site genotype-phenotype “matchmaking” efforts.4, 5
Through a Clinical Sequencing Exploratory Research (CSER) project focused on sequence-driven diagnoses for probands with unexplained DD and/or ID,6 we identified variation likely to be deleterious in BRSK2 (MIM: 609236) in four unrelated probands. BRSK2 encodes a serine/threonine-protein kinase, which is involved in axonogenesis and the polarization of cortical neurons.7 BRSK2 is predicted to be relatively intolerant to protein-altering variation in the general population (%ExAC v2 residual variation intolerance score [RVIS] = 4.9462%,8 pLI score (probability that a gene is intolerant to a loss of function mutation) = 0.789). In each proband, the BRSK2 variant was prioritized, after filtering and manual curation, as the most compelling disease-candidate variant of interest (see details by Bowling and colleagues for additional information about the cohort and analytical methods6). Although these observations suggest BRKS2 as a strong candidate NDD-associated gene, we sought additional cases via GeneMatcher5 to support pathogenicity. GeneMatcher is a database developed as part of the MatchMaker Exchange and has been shown to facilitate rare-disease-gene discovery.10 Information about five additional affected probands who were found by research or diagnostic sequencing (Table 1) and who had variants likely to be deleterious in BRSK2 was independently submitted to GeneMatcher. Informed consent to publish de-identified data was obtained from all affected individuals and/or families (see Supplemental Material and Methods). Altogether, the affected probands ranged in age from 3 years and 9 months old to 19 years old and had a mean age of just under 8 years (Table 2). All probands described here display ID except for one. All probands present with developmental delays, including speech delay (9/9) and motor delay (7/9). Eight of nine probands, one of whom (proband 9) was considered borderline, present with autism, and seven were reported to have behavioral abnormalities, including stereotypies (4/7), temper tantrums (3/7), and/or ADHD (3/7). Two probands reported sleep issues that were treatable with melatonin. Although most probands were reported to have facial dysmorphism, we did not observe a consistent set of features (see Supplemental Note). Additional details of each case are provided in the supplement (See Table S1 and Supplemental Note).
Table 1.
Variant Details of Individuals with BRSK2 Variation
| Probanda | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Sequencing/analysis siteb | A | B | C | C | A | D | E | C | C |
| HGVS nomenclaturec | g.1459543G>A, c.194G>A (p.Arg65Gln) | g.1462017G>A, c.273–1G>A | g.1463837G>A, c.530+1G>A | g.1464720G>A, c.635G>A (p. Gly212Glu) | g.1464815C>T, c.730C>T (p. Gln244∗) | g.1471060_1471071del, c.1281_1287+5del | g.1471924_1471925del, c.1395_1396del (p.Ser466Glnfs∗83) | g.1472635_1472636del, c.1532_1533del (p.Glu511Valfs∗38) | g.1477839C>T, c.1861C>T (p.Arg621Cys) |
| Predicted effect | missense | splice alteration | splice alteration | missense | premature stop | splice alteration | frameshift | frameshift | missense |
| Inheritance | de novo | de novo | de novo | de novo | de novo | unknown | de novo | unknown | unknown |
| CADD_v1.4 | 25.3 | 24.8 | 26.5 | 26.9 | 38 | 32 | 34 | 34 | 29.2 |
| GERP | 2.57 | 2.65 | 3 | 3.26 | 3.27 | 4.11 | 3.91 | 3.81 | 3.8 |
| Sequencing method | exome | exome | genome | genome | exome | exome | exome | genome | genome |
Abbreviations are as follows: HGVS = Human Genome Variation Society, CADD = Combined Annotation Dependent Depletion, and GERP = genomic evolutionary rate profiling.
Probands are numbered on the basis of a 5′ to 3′ ordering of variation.
Sequencing and analysis sites are further defined in the Supplemental Note.
HGVS nomenclature applies to GenBank: NC_000011.9, GenBank: NM_001256627.1, and GenBank: NP_001243556.1.
Table 2.
Clinical Characteristics of Individuals with BRSK2 Variation
| Proband | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Female | Male | Male | Male | Male | Male | Male |
| Age at last examination | 5 years | 14 years | 5 years, 7 months | 3 years, 9 months | 19 years | 4 years | 6 years, 10 months | 10 years, 11 months | 4 years, 7 months |
| Intellectual disability | moderate | severe | mild | moderate | mild progressed to severe | none reported | mild | moderate | moderate |
| Developmental delay(s) | speech and motor | speech, including speech regression, and motor | speech and fine motor | speech and social interaction; speech regression | speech and fine motor; regression | speech and gross motor | speech | speech and gross motor | speech and gross motor |
| Autism | no | yes | yes | yes | yes | yes | yes | yes | borderline |
| Behavior | none reported | ADD and/or ADHD, stereotypic, temper tantrums, sleep issues | none reported | temper tantrums, easily cries, fidgets, hits others, prefers to play alone | ADHD, hyperactive as toddler, psychoses, schizophrenia, anxiety, hallucinations, impulsive, self-mutilation, aggression, concentration issues | stereotypies, pinching, tantrums and rigidity, sleep issues | stereotypic (mild) | ADD and/or ADHD, impulsive | stereotypic |
Abbreviations are as follows: ADD = attention deficit disorder and ADHD = attention-deficient/hyperactivity disorder.
Parents were available for testing for six of the nine probands, and in all six of these probands, the variants were found to be de novo (Table 1 and Figure 1). Six of the nine described variants, including two frameshift variants, one nonsense variant, and three variants affecting canonical splice sites, are predicted to result in loss of function. The remaining three variants were missense. All nine variants are absent from gnomAD9 and the Bravo TOPMed database. Although gnomAD does contain variant data aggregated from several disease cohorts, there has been an effort to remove any variants found in individuals with severe pediatric disease.9 All variants were computationally predicted to be deleterious and had Combined Annotation Dependent Depletion (CADD) scores11 ranging from 24.8 to 38; these scores indicate that they rank among the most highly deleterious variants possible in the human genome reference assembly, similar to most variants previously reported to cause Mendelian diseases.11
Figure 1.
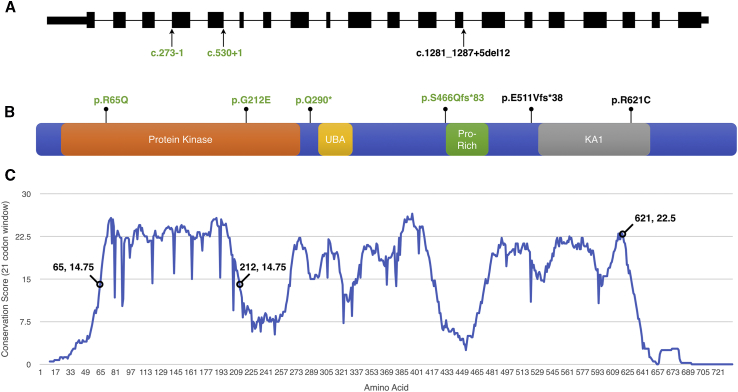
Exon and Domain Structure, Conservation, and Locations of Observed Variation in BRSK2
(A and B) Variation observed in BRSK2 is shown for (A) the canonical, 20-exon transcript, GenBank: NM_001256627.1 and (B) the 736 aa protein, GenBank: NP_001243556.1. Protein domains include protein kinase, ubiquitin-associated (UBA), proline-rich (Pro-Rich), and kinase-associated 1 (KA1) domains. Splice variants are shown below the schematic representation of the canonical transcript, and protein-altering variants are shown above the schematic representation of BRSK2. De novo variants are shown in green text, and those of unknown inheritance are shown in black.
(C) Analysis of conservation throughout BRSK2 was performed with amino acid selection scores as previously published12 and used a 21-codon sliding window. The most-selected motifs of a protein are identified as peaks. The three residues found to be affected by variation here are labeled, along with their respective conservation scores.
The canonical protein encoded by the BRSK2 locus (GenBank: NP_001243556.1, UniProt: Q8IWQ3, 736 aa) contains several domains, including a protein kinase domain (aa 19–270), a ubiquitin-associated domain (UBA; aa 297–339), a proline-rich domain (aa 424–468), and a kinase-associated domain (KA1; aa 530–653) that contains a KEN box (a degradation signal, aa 603–605) (Figure 1B). An analysis of conservation along the protein identified several regions with elevated measures of conservation (Figure 1C). Two missense variants (c.194G>A [p.Arg65Gln] and c.635G>A [p.Gly212Glu]) are located within the protein kinase domain, and one (c.1861C>T [p.Arg621Cys]) is within the KA1 domain.
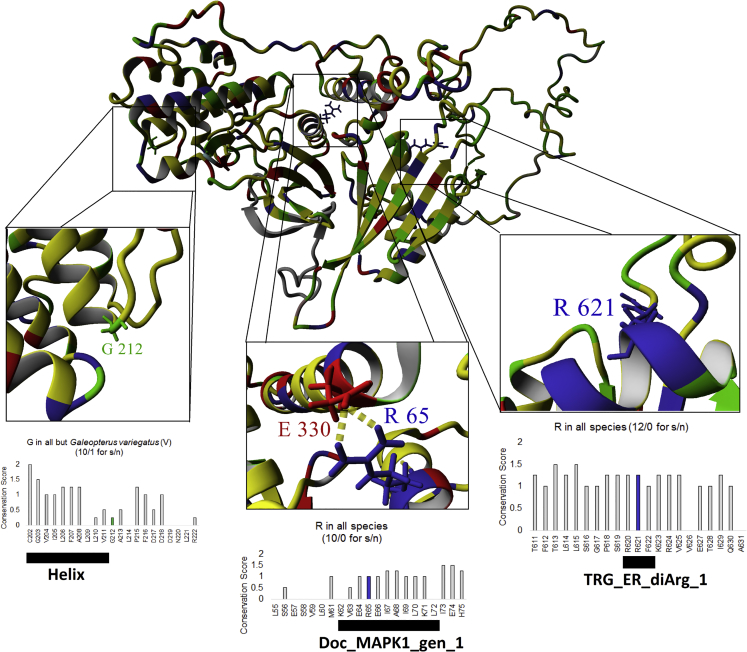
We assessed the potential structural effects of the three missense variants by performing computational modeling.12 All three missense variants lie within conserved linear motifs (Figure 1C) and affect residues that are conserved across many species (Figures 2 and S1). Arg65 lies within the protein kinase domain and has been found to coordinate intramolecularly with Glu330 to form a salt bridge.13 Arg65 also lies within a mitogen-activated protein kinase (MAPK) docking motif14 (Figure 2). Thus, p.Arg65Gln is predicted to disrupt both the structure and functional activity of BRSK2. Gly212 lies in the C-lobe of the protein kinase domain, just at the C terminus of a helix, within a flexible linker;13 thus, p.Gly212Glu might disrupt secondary protein structure. Arg620 and Arg621 comprise a di-arginine endoplasmic-reticulum (ER) retrieval-and-retention motif, and a recent publication found that Arg620 forms a salt bridge with Asp305 when the KA1 domain associates with the UBA domain.13 The authors of this paper also found that disruption of several key polar residues, including Arg620 and Arg621, in the C terminus of the protein abolishes phospholipid binding. On the basis of these observations, it is plausible that p.Arg621Cys disrupts the localization of this protein to the ER and possibly to other membranes. Given that the p.Arg621Cys variant is of unknown inheritance, it remains a variant of uncertain significance (VUS),15 and further experimental or computational analyses are needed if we are to better understand its potential molecular and disease effects.
Figure 2.
Computational Modeling of BRSK2 Missense Variants
A full model of BRSK2 was created with I-TASSER modeling using PDB: 4YOM, 4YNZ, and 4IW0. This model of BRSK2 was combined via ConSurf mapping with sequences for BRSK2 from 99 species. Amino acid coloring is as follows: gray = not conserved, yellow = conserved hydrophobic, green = conserved hydrophilic, red = conserved polar acidic, blue = conserved polar basic, and magenta = conserved human variants of interest. Zoomed-in views of the three locations are shown, along with codon usage throughout evolution. The conservation score is defined as an additive metric of amino acid conservation and codon selection as previously defined.12 For example, a conservation score of 2 indicates 100% conservation with >2 standard deviations above the mean for codon selection.12 s/n indicates synonymous mutations versus non-synonymous mutations observed at the same position in other species; differences are indicated. All three sites are under high selection and have multiple synonymous (s) amino acids in 99 open reading frames (ORFs) of BRSK2 and only a single nonsynonymous (n) change observed at G212. Linear motifs mapped with the Eukaryotic Linear Motif (ELM) tool are shown below each site.
We assessed the degree of enrichment of observed de novo variation in BRSK2 in the sequenced DD- and/or ID-affected cohorts that underlie this study. Two probands (1 and 5) were a part of a cohort of 2,418 DD- and/or ID-affected probands sequenced as trios. An additional proband (2) was sequenced as a trio among a cohort of 550 affected probands, and two others (probands 3 and 4) were among a cohort of 461. In aggregate, these cohorts include five de novo variants in 3,429 affected, sequenced individuals. We compared this observed rate to the expected rate estimated by Samocha et al.16 (2.97 × 10−5 variants per chromosome) of de novo missense, nonsense, splice, and frameshift variation in BRSK2. The observed rate of de novo variation in the DD- and/or ID-affected cohorts considered here is significantly greater than the background mutation rate (five de novo variants observed versus 0.20 expected, p = 2.46 × 10−6), and this observation remained significant even after a Bonferroni correction for 20,000 genes (p = 0.0492). We note that one proband (7) with a de novo variant was sequenced clinically as a trio, but a cohort size was not available for this proband; furthermore, one or both parents were unavailable for testing for three of the nine observed variants. Thus, although these four additional variants add to the evidence supporting a disease role for BRKS2, they are excluded from the preceding enrichment calculations.
BRSK2 and its homolog BRSK1 (MIM: 609235) encode kinases required for neuronal polarization.7 These two kinases, along with 11 other kinases, form the AMPK-related family of protein kinases.17 Although knockouts of either Brsk1 or Brsk2 alone in mice were healthy and fertile, double knockouts of Brsk1 and Brsk2 resulted in pups that exhibited reduced spontaneous movement and little response to tactile stimulation and that died within 2 h of birth.7 Expression patterns of BRSK2 also support its role in neurodevelopment. BRSK2 is most highly expressed in the brain in humans,18 and Brsk1 and Brsk2 are restricted to the nervous system in mice.7
BRSK2 interacts with several genes that are associated with NDDs, including autism, tuberous sclerosis, and DD and/or ID. For example, BRSK2 has been shown to phosphorylate TSC2 and suppress mTORC1 activity.19 The tuberous sclerosis complex (TSC) signaling pathway is one of the pathways associated with autophagy during early axonal growth,20 and TSC2, specifically, is a regulator of cellular size and growth.21, 22 BRSK2 has also been shown to interact with PTEN, which has been associated with various developmental disorders (see MIM: 60172823,24) including autism. PTEN-deficient mice display malformation of neuronal structures and autistic features that result from aberrant TSC-mTORC1 signaling.25 WDR45, also known as WIPI4, is a scaffold protein that controls autophagy and has recently been shown to be dependent on BRSK2 activity.26 Variation in WDR45 is associated with an X-linked dominant disorder: neurodegeneration with brain iron accumulation (MIM: 300894). The numerous genetic and biochemical interactions between BRSK2 and well-established NDD genes further strengthen the conclusion that damaging variation in BRSK2 underlies an NDD.
Across six recent publications reporting on de novo variation in large cohorts with DD and/or ID or autism,27, 28, 29, 30, 31, 32 two protein-altering BRSK2 variants were reported: GenBank: NM_001256627.1 (c.992_994del, [p.Lys331del]) was found in a cohort of 2,500 probands with autism,30 and GenBank: NM_001256627.1 (c.770G>A, [p.Arg257His]) was found in a cohort of 4,293 DD- and/or ID-affected probands.28 Interestingly, this second variant has been observed as a heterozygote seven times in gnomAD, suggesting it is not a highly penetrant allele contributing to DD and/or ID. These data raise an interesting question, namely as to why the frequency of observed BRSK2 variation in this study is markedly higher than that found in previous studies. This is particularly true for the HudsonAlpha CSER study,6 in which four variants were found among 581 affected probands (461 of whom were sequenced as trios). Some of the discrepancy is probably due to stochastic variability in observing a small number of rare events. However, one potential systematic explanation is that BRSK2 is less deeply covered in exomes, and the observed enrichment, in part, reflects the effects of the genome sequencing that was used for the HudsonAlpha probands described here. It has been shown previously that genome sequencing provides better coverage, in general, over coding exons than exome sequencing does,27, 31, 33, 34, 35, 36 and that some exons, including among clinically relevant genes, tend to be more poorly covered by exomes.36
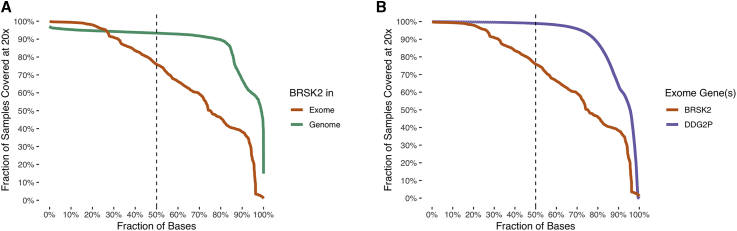
We find that BRSK2 is less well covered by exomes than by genomes in gnomAD (Figure 3). For example, when requiring a minimum depth of 20× among exonic bases (plus 10 bp on either side of each exon), we found that 76% of gnomAD exome samples, compared to 93% of genome samples, have half of all BRSK2 bases covered (Figure 3A). Furthermore, we assessed rare-variant detection rates, in particular the rate at which singletons (i.e., variants for which only one alternative allele is observed across the combined set of exomes and genomes) are observed. There are 46 singletons detected among 15,708 genomes (0.29%) in gnomAD and 189 singletons detected among 125,748 exomes (0.15%); this difference is significant (p = 1.5 × 10−4, Fisher’s exact test) and suggests increased rare-variant sensitivity in genomes relative to in exomes. Additionally, considering only exomes, we compared coverage of BRSK2 exons to exons in other disease-associated genes that are annotated in the Development Disorder Genotype-Phenotype Database (DDG2P). Although, again, only 76% of samples have at least half of BRSK2 bases covered at 20× in gnomAD exomes, 99% of samples have half or more of the bases in previously reported DDG2P genes covered in gnomAD exomes (Figure 3B). Thus, we find it likely that the lower rates of BRSK2 variation found in other DD and/or ID studies reflects, at least in part, reduced variant sensitivity of exome sequencing in BRSK2.
Figure 3.
Comparisons of BRSK2 Sequencing Depth across gnomAD Datasets
Fractions of gnomAD samples that attain a per-base sequencing depth of ≥20× are plotted as a function of the percentage of bases examined, ordered by a decreasing fraction of exonic-base coverage. Only autosomal positions are included. The dashed line shows the fraction of samples covered at the median-depth base.
(A) Using only BRSK2 exonic bases (exons plus 10 bp on either side), coverage is compared in gnomAD exomes (orange; 125,748 individuals) and gnomAD genomes (green; 15,708 individuals).
(B) Using only gnomAD exomes (125,748 individuals), exonic bases (exons plus 10 bp on either side) in BRSK2 (orange) are compared to exonic bases in 1,012 confirmed developmental-delay genes identified by the Developmental Disorders Genotype-Phenotype Database (DDG2P; purple).
We have identified nine individuals harboring rare, heterozygous BRSK2 variants that are likely to be deleterious, and we provide detailed clinical descriptions of the phenotypes observed in these individuals, who all present with varying degrees and manifestations of developmental disorders. We believe these observations strongly support the conclusion that damaging variation in BRSK2 is causally related to an NDD. The key points of evidence are as follows: (1) we observe a statistically significant enrichment of de novo variants in affected individuals relative to the estimated background mutation rate (p = 2.46 × 10−6); (2) although one or both parents were unavailable in three cases, none of the variants described here were found to be inherited, and all observed variants are absent from gnomAD and TopMed; (3) BRSK2 is relatively intolerant to protein-altering variation in the general population;8, 9 (4) all variants in affected probands are either predicted to result in loss of function or are missense variants at highly conserved residues; (5) all variants are computationally predicted to be evolutionarily deleterious and have, for example, CADD11 scores that are typical for mutations previously reported to underlie Mendelian disease; (6) model organism evidence suggests a role for BRSK2 in neurodevelopment; and (7) BRSK2 is known to genetically and/or biochemically interact with several genes that are robustly associated with developmental disease. In summary, these data collectively implicate BRSK2 as an NDD-related gene.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all of the families who participated in the study. This study was supported by: a grant from the National Human Genome Research Institute (NHGRI), UM1HG007301 (S.M.H., M.L.T., J.M.J.L., D.E.G., E.M.B., and G.M.C.); the Radiz-Rare Disease Initiative Zurich, Clinical Research Priority Program University of Zurich (A.R.); the French Foundation for Rare Diseases (C.N.); the National Institutes of Health (NIH) Office of the Director, K01ES025435 (J.W.P.); and the Fondation Maladies Rares (C.N., C.D., and C.M.).
Published: March 14, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.02.002.
Accession Numbers
All relevant variant data are supplied within the paper or in supporting files. Complete genome data for probands 3, 4, 8, and 9 are available via dbGAP (accession number phs001089.v3.p1). Complete exome data for other probands is not available for privacy and institutional review board (IRB) reasons.
Web Resources
Bedtools, https://github.com/arq5x/bedtools2
Bravo Variant Browser, https://bravo.sph.umich.edu/freeze3a/hg19/
Combined Annotation Dependent Depletion (CADD), https://cadd.gs.washington.edu/
Developmental Disorder Genotype-Phenotype Database (DDG2P), https://decipher.sanger.ac.uk/ddd#ddgenes
gnomAD Genome Aggregation Database, https://gnomad.broadinstitute.org/
Online Mendelian Inheritance in Man, http://www.omim.org/
The R Project for Statistical Computing, http://www.r-project.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Reiss A.L. Childhood developmental disorders: An academic and clinical convergence point for psychiatry, neurology, psychology and pediatrics. J. Child Psychol. Psychiatry. 2009;50:87–98. doi: 10.1111/j.1469-7610.2008.02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thapar A., Rutter M. Neurodevelopmental disorders. In: Thapar A., Pine D.S., Leckman J.F., Scott S., Snowling M.J., Taylor E.A., editors. Rutter’s Child and Adolescent Psychiatry. John Wiley & Sons, Ltd; 2015. pp. 31–40. [Google Scholar]
- 3.Plummer J.T., Gordon A.J., Levitt P. The genetic intersection of neurodevelopmental disorders and shared medical comorbidities - relations that translate from bench to bedside. Front. Psychiatry. 2016;7:142. doi: 10.3389/fpsyt.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: A platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowling K.M., Thompson M.L., Amaral M.D., Finnila C.R., Hiatt S.M., Engel K.L., Cochran J.N., Brothers K.B., East K.M., Gray D.E. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43. doi: 10.1186/s13073-017-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishi M., Pan Y.A., Crump J.G., Sanes J.R. Mammalian SAD kinases are required for neuronal polarization. Science. 2005;307:929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- 8.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobreira N.L.M., Arachchi H., Buske O.J., Chong J.X., Hutton B., Foreman J., Schiettecatte F., Groza T., Jacobsen J.O.B., Haendel M.A. Matchmaker Exchange. Curr. Protoc. Hum. Genet. 2017;95:9.31.1–9.31.15. doi: 10.1002/cphg.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokop J.W., Lazar J., Crapitto G., Smith D.C., Worthey E.A., Jacob H.J. Molecular modeling in the age of clinical genomics, the enterprise of the next generation. J. Mol. Model. 2017;23:75. doi: 10.1007/s00894-017-3258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J.-X., Cheng Y.-S., Wang J., Chen L., Ding M., Wu J.-W. Structural insight into the mechanism of synergistic autoinhibition of SAD kinases. Nat. Commun. 2015;6:8953. doi: 10.1038/ncomms9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharrocks A.D., Yang S.H., Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 15.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lizcano J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie D.G., Alessi D.R. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiyin H., Na N., Han X., Fang Y., Wu Y., Lou W., Yang X. BRSK2 induced by nutrient deprivation promotes Akt activity in pancreatic cancer via downregulation of mTOR activity. Oncotarget. 2017;8:44669–44681. doi: 10.18632/oncotarget.17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K.M., Hwang S.K., Lee J.A. Neuronal autophagy and neurodevelopmental disorders. Exp. Neurobiol. 2013;22:133–142. doi: 10.5607/en.2013.22.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tee A.R., Fingar D.C., Manning B.D., Kwiatkowski D.J., Cantley L.C., Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Gama A.M., Geng Y., Couto J.A., Martin B., Boyle E.A., LaCoursiere C.M., Hossain A., Hatem N.E., Barry B.J., Kwiatkowski D.J. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann. Neurol. 2015;77:720–725. doi: 10.1002/ana.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga E.A., Pastore M., Prior T., Herman G.E., McBride K.L. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 24.Kurata H., Shirai K., Saito Y., Okazaki T., Ohno K., Oguri M., Adachi K., Nanba E., Maegaki Y. Neurodevelopmental disorders in children with macrocephaly: A prevalence study and PTEN gene analysis. Brain Dev. 2018;40:36–41. doi: 10.1016/j.braindev.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Kwon C.-H., Luikart B.W., Powell C.M., Zhou J., Matheny S.A., Zhang W., Li Y., Baker S.J., Parada L.F. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakula D., Müller A.J., Zuleger T., Takacs Z., Franz-Wachtel M., Thost A.-K., Brigger D., Tschan M.P., Frickey T., Robenek H. WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017;8:15637. doi: 10.1038/ncomms15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner T.N., Coe B.P., Dickel D.E., Hoekzema K., Nelson B.J., Zody M.C., Kronenberg Z.N., Hormozdiari F., Raja A., Pennacchio L.A. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–722.e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.-J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 30.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 32.O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilfert A.B., Sulovari A., Turner T.N., Coe B.P., Eichler E.E. Recurrent de novo mutations in neurodevelopmental disorders: Properties and clinical implications. Genome Med. 2017;9:101. doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belkadi A., Bolze A., Itan Y., Cobat A., Vincent Q.B., Antipenko A., Shang L., Boisson B., Casanova J.L., Abel L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA. 2015;112:5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meienberg J., Bruggmann R., Oexle K., Matyas G. Clinical sequencing: Is WGS the better WES? Hum. Genet. 2016;135:359–362. doi: 10.1007/s00439-015-1631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.