Abstract
Mutations in several genes encoding components of the SWI/SNF chromatin remodeling complex cause neurodevelopmental disorders (NDDs). Here, we report on five individuals with mutations in SMARCD1; the individuals present with developmental delay, intellectual disability, hypotonia, feeding difficulties, and small hands and feet. Trio exome sequencing proved the mutations to be de novo in four of the five individuals. Mutations in other SWI/SNF components cause Coffin-Siris syndrome, Nicolaides-Baraitser syndrome, or other syndromic and non-syndromic NDDs. Although the individuals presented here have dysmorphisms and some clinical overlap with these syndromes, they lack their typical facial dysmorphisms. To gain insight into the function of SMARCD1 in neurons, we investigated the Drosophila ortholog Bap60 in postmitotic memory-forming neurons of the adult Drosophila mushroom body (MB). Targeted knockdown of Bap60 in the MB of adult flies causes defects in long-term memory. Mushroom-body-specific transcriptome analysis revealed that Bap60 is required for context-dependent expression of genes involved in neuron function and development in juvenile flies when synaptic connections are actively being formed in response to experience. Taken together, we identify an NDD caused by SMARCD1 mutations and establish a role for the SMARCD1 ortholog Bap60 in the regulation of neurodevelopmental genes during a critical time window of juvenile adult brain development when neuronal circuits that are required for learning and memory are formed.
Keywords: SMARCD1, BAFopathies, SWI/SNF, intellectual disability, neurodevelopmental disorder, Drosophila mushroom body, long-term memory
Introduction
The regulation of gene expression in neurons is critical for normal brain development and for normal cognitive functioning in adults.1, 2, 3, 4 Chromatin structure is an important factor in modulating gene expression.5 The SWI/SNF chromatin remodeling complex (also known as the BAF complex in mammals) is a highly conserved protein complex that utilizes energy from ATP to alter nucleosome-DNA interactions; this alteration results in more open chromatin for transcription-factor binding.3, 6, 7, 8 In mammals, the SWI/SNF complex has multiple cell-type-specific conformations, including npBAF, specific for neuronal progenitors, and nBAF, specific for postmitotic neurons.1, 9, 10, 11, 12 Each form of the SWI/SNF complex contains 10–15 proteins encoded by 29 genes.11 The SWI/SNF complex is important for the regulation of gene-expression programs involved in neuronal differentiation and brain-region specification in mice.2, 4, 9, 13, 14, 15, 16, 17 However, the complex is also essential in mature neurons for memory formation, synaptic plasticity, and activity-responsive neurite outgrowth.3, 4, 18
The disruption of genes encoding chromatin regulators is an important cause of neurodevelopmental disorders (NDDs), which are a heterogeneous group of disorders including intellectual disability and autism.19, 20 Mutations in genes encoding several different SWI/SNF subunits cause syndromic NDDs, including Nicolaides-Baraitser syndrome (MIM: 601358) and Coffin-Siris syndrome (MIM: 135900).21, 22, 23, 24 SWI/SNF mutations are also involved in other syndromic and non-syndromic NDDs25, 26 and psychiatric disorders such as schizophrenia.27, 28, 29, 30 In total, mutations in 11 of the 28 genes encoding SWI/SNF components have been implicated in NDDs,21, 22, 23, 25, 26, 27, 28, 30, 31, 32, 33, 34 emphasizing the essential role of this complex in neuron development and function. Whether mutations in the remaining subunits are also involved in NDDs or other brain disorders remains to be determined.
Here, we characterize mutations in SMARCD1 (MIM: 601735) in individuals presenting with a syndromic NDD. SMARCD1 encodes a core SWI/SNF-complex component that has not previously been associated with NDDs. We show that the Drosophila SMARCD1 ortholog Bap60 is required in the mushroom body (MB) of adult flies for normal long-term memory. The MB is the learning and memory center of the fly brain.35, 36 We find that Bap60 has a profound effect on the expression of neurodevelopmental genes in the MB during a critical time window of juvenile brain development when synaptic connections are formed in response to early life experiences.
Subjects and Methods
Participant Enrollment
Individual 1 was enrolled in a study protocol that was approved by the institutional review boards of Yokohama City University School of Medicine. Individual 2 was enrolled in a study done by the Groupe Hospitalier Pitié-Salpêtrière and approved by the Institut national de la santé et de la recherche (INSERM) institutional review board. Individuals 3 and 4 were enrolled in the Deciphering Developmental Disorders (DDD) study.37 Contact with the clinicians was made through the DDD website, and the individuals were enrolled in a study approved by the institutional review board of the CHU (Centre Hospitalier Universitaire) Sainte-Justine. Individual 5 had exome sequencing on a clinical basis, and the family consented to the sharing of clinical information without photos. The clinicians of individuals 2 and 5 were connected with through GeneMatcher.
Exome Sequencing
For individual 1, genomic DNA was enriched for exons with the SureSelect All Human Exon kit (Agilent). Libraries were sequenced on the Illumina HiSeq, and analysis was performed as described.38 For individual 2, exome sequencing was performed as described (as for family 1) in Marsh et al..39 For the individuals 3 and 4, exome sequencing was done as part of the DDD project. The exomes were enriched with the Agilent SureSelect 55 MB Exome Plus library; this was followed by Illumina HiSeq sequencing, and analysis was performed as described.37 Exome variants passing the filtering criteria were evaluated by the DDD study’s internal clinical review team, which included two consultant clinical geneticists. For individual 5, exome sequencing was done at GeneDx. Genomic DNA from the proband and parents was used for capturing the exonic regions and flanking splice junctions of the genome were captured with the Clinical Research Exome kit (Agilent). Sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37 (UCSC hg19) and analyzed for sequence variants with a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol (e.g. Sanger) has been previously described.40 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page.
In Silico Assessment of the Variants
Secondary structure of the SMARCD1 protein (GenBank: NP_003067.3) was drawn with protein paint. The prediction of the coiled-coil domain was performed with the NPS@: Network Protein Sequence Analysis tool. A 3D model of the full SMARCD1 protein was predicted by the I-TASSER online suite under standard parameters. The model with the highest confidence was selected and modeled with Pymol.
Fly Stocks and Culture
The flies were reared on standard cornmeal-agar media with a 12 h/12 h light/dark cycle in 70% humidity. The following stocks were obtained from the Bloomington Drosophila Stock Center (Indiana University): short hairpin Bap60 RNAi lines generated by the Transgenic RNAi Project (TRiP)41 (UAS-Bap6032503 and UAS-Bap6033954); the TRiP short hairpin mCherryRNAi line (stock #35785); the mushroom-body-specific Gal4 driver line R14H06-Gal4 (stock #48667) from the Janelia Flylight42 collection; and the temperature-sensitive Gal80 (Gal80ts) driven by the tubulin promoter (stock #7019). UAS-unc84::GFP was a gift from G.L. Henry. The Bap60 RNAi lines (UAS-Bap6032503 and UAS-Bap6033954) were assessed for knockdown efficiency in a parallel study.43 When ubiquitously expressed with an Act-Gal4 driver, the UAS-Bap6032503 RNAi induces 100% lethality (no adults eclose) as expected because null mutations in Bap60 are lethal at the embryo and larval stage.44 RT-qPCR on larvae with ubiquitous knockdown shows a significant reduction of mRNA by more than 50%.43 UAS-Bap6033954 did not induce complete lethality under Act-Gal4 expression: 17% of progeny did survive to adulthood. The more potent RNAi line UAS-Bap6032503 was chosen for follow-up RNA-sequencing studies.
Courtship Conditioning
Courtship conditioning assays were performed as described previously.45 Individual males were paired with premated wild-type females for training. Long-term memory was induced through a 7 h training period and tested 24 h after training. For each fly pair, a courtship index (CI) was determined by manual observation of the percentage of time spent courting during a 10 min period. Statistically, loss of memory was identified through two complimentary methods. A reduction of the mean CI of trained (CItrained) flies compared to naïve flies (CInaive) of the same genotype was compared by a Kruskal-Wallis test and then pairwise comparisons with Dunn’s test. A learning index (LI) was also calculated (LI = [CInaive − CItrained]/CInaive). LIs were compared between genotypes through a randomization test (10,000 bootstrap replicates) performed with a custom R script,45 and the resulting p values were Bonferroni-corrected for multiple testing.
The temperature-sensitive Gal80ts system46 was used for specific knockdown of Bap60 in the adult MB. Flies of the genotype tubGal80ts; R14H06-Gal4 were crossed to Bap60 RNAi lines UAS-Bap6032503 and UAS-Bap6033954, as well as to a control UAS-mCherryRNAi that is present in the same genetic background as the Bap60 RNAi transgenes. Crosses were raised at 18°C, the permissive temperature that allows Gal80ts to repress Gal4-mediated transcription. At eclosion, male flies of the genotypes Gal80ts/+;R14H06-Gal4/UAS-Bap6032503, Gal80ts/+;R14H06-Gal4/UAS-Bap6033954, and Gal80ts/+;R14H06-Gal4/UAS-mCherryRNAi were transferred to 29°C, which causes Gal80 inactivation, allowing Gal4-mediated induction of UAS-RNAi transgenes. After 5 days, collected males were tested for long-term memory via courtship conditioning as described above.
Isolation of Nuclei in Tagged Cell Types
Isolation of nuclei in tagged cell types (INTACT) was performed as described previously.47 Fifty juvenile (0–3-hour-old) or mature (1- to 5-day-old) adult male flies of the genotype UAS-unc84::GFP/+;R14H06-Gal4/UAS-Bap6032503 (Bap60-KD) and UAS-unc84::GFP/+;R14H06-Gal4/UAS-mCherryRNAi (control) were anesthetized with CO2 and flash frozen with liquid nitrogen. Protein G Dynabeads (Invitrogen) were adsorbed to 5 μg of anti-GFP antibody (Invitrogen, G10362) in PBS 0.1% Tween-20 (PBST) for 10 min at room temperature. The beads were then isolated with a magnet and resuspended in PBST. The flies were then vortexed and placed in ice-cold sieves to separate and isolate their heads. The heads were then added to homogenization buffer (25 mM KCl [pH 7.8], 5 mM MgCl2, 20 mM Tricine, 150 nM spermine, 500 nM spermidine, 10 nM β-glycerophosphate, 250 mM sucrose, 1× Pierce protease inhibitor tablets – EDTA-free [Thermo Fisher Scientific] or 1× Halt protease inhibitor cocktail – EDTA-free [ThermoFisher Scientific]), and the suspension was homogenized for approximately 1 min with a standard tissue homogenizer at 30,000 rpm. The suspension was then placed in a Dounce homogenizer with NP-40 (Thermo Fisher Scientific) added to an end concentration of 0.3% and homogenized six times with the tight pestle. The homogenate was filtered through a 40 μm strainer and pre-cleared with non-antibody bound beads for 10 min. Antibody-bound beads were added to the homogenate for 30 min, and the beads were then washed in homogenization buffer. Total RNA was isolated from immunoprecipitated nuclei with the Arcturus PicoPure RNA isolation kit (Thermo Fisher Scientific), and DNase digestion was done with the RNase-free DNase kit (Qiagen) according to the manufacturer’s instructions. The quality of the isolated RNA was then assessed with the Bioanalyzer 2100 Pico RNA kit (Agilent) by visual examination of rRNA-peak integrity.
RNA-Sequencing and Analysis
RNA from MB-enriched samples was used for generation of an RNA-seq library with the NuGEN Ovation Drosophila RNA-Seq System (BioLynx), according to the manufacturer’s instructions. Size selection with Agencourt SPRIselect beads (Beckman-Coulter) was used for selecting library sizes of 200 bp. Library size was assessed with the Bioanalyzer 2100 DNA high-sensitivity kit (Agilent). Sequencing was performed with the Illumina NextSeq500 at the London Regional Genomics Centre (Robarts Research Institute) with the high output v2 75 cycle kit; read length was 75 bp for single-end reads.
Raw sequencing reads were trimmed with Prinseq quality trimming48 using a minimum base quality score of 30. The read quality was then assessed via FastQC. Trimmed reads were aligned to the Drosophila melanogaster reference genome (BDGP release 6)49, 50 with the STAR aligner.51 An average of 54,803,904 and 46,899,292 high-quality, uniquely aligned reads with a maximum of four mismatches were obtained from juvenile Bap60-KD MBs (n = 3) and controls (n = 2), respectively, and an average of 26,865,090 and 35,504,102 high-quality, uniquely aligned reads with a maximum of four mismatches were obtained from mature Bap60-KD MBs (n = 5) and controls (n = 5), respectively (Table S1). The number of reads per gene was quantified with HTSeq-count52 where the --type flag indicated “gene” (Table S1). Y chromosome genes, rRNA genes, and genes with no counts across all samples were excluded, leaving 12,922 and 13,440 genes for downstream analysis for juvenile and mature samples, respectively.
The raw gene counts were normalized, and differential expression analysis between Bap60-KD MBs and controls was performed with the R package DESeq2.53 Differentially expressed genes were defined as genes with a >1.5-fold or >2-fold change and a Benjamini-Hochberg adjusted p value < 0.05. Gene ontology (GO) enrichment analysis was performed on upregulated and downregulated differentially expressed genes with Panther54 (false discovery rate (FDR) < 0.05).
Classification of Tissue-Specific Genes
To generate lists of tissue-specific genes, we used normalized gene expression values from Brown et al.55 for several tissues, including adult head (nine samples), adult carcass (three samples), and adult digestive tract (three samples). The relative enrichment in gene expression levels for each of these tissues was calculated by comparing the mean bases per kilobase per million reads (bpkm) for each specific tissue to the mean bpkm across all remaining tissues. Enrichment values for each gene in each tissue type were determined by calculating the log2-fold-change in expression of that gene in each tissue compared to all other tissues. Tissue-specific and tissue depleted genes are defined as having an enrichment value outside one standard deviation of the average of all enrichment values (Table S2). We used adult heads as a representation of a neuron-enriched tissue and the adult carcass, consisting of tissues remaining after removal of the head, digestive tract, and reproductive organs, as a representation of a muscle-enriched tissue. A list of MB-specific genes (Table S2) was generated from a published MB-specific transcriptome that was obtained with the same INTACT protocol described here.47 MB-specific genes were defined as genes that were significantly upregulated more than 2-fold in MB-enriched samples compared to in biologically paired whole-head input samples. The statistical significance of over- or under-representation of tissue-specific genes in differentially upregulated and downregulated genes in Bap60-KD MBs was determined with a hypergeometric test.
Results
Human Genetic and Clinical Data
In a cohort of individuals collected because of their clinical phenotypic overlap with Coffin-Siris syndrome, de novo mutations in several genes encoding members of the SWI/SNF complex were identified.21 As an expansion of this study, an individual with a SMARCD1 variant (c.990C>G [p.Asp330Glu]; individual 1) was identified, but the de novo status of the variant could not be confirmed because the father was not available for testing. Subsequently, as part of the DDD study,37 two individuals were identified with de novo SMARCD1 variants (c.1457G>A [p.Trp486∗] and c.1483T>C [p.Phe495Leu]; individuals 3 and 4, respectively). Through GeneMatcher,56 two additional individuals were identified. One individual (individual 2) in a cohort with agenesis of the corpus callosum had a de novo c.1336A>G [p.Arg446Gly] variant, and another individual (individual 5) who had clinical exome sequencing for developmental delay and other anomalies and who was identified to have a truncating variant (c.1507C>T [p.Arg503∗]). For individuals 2–5, de novo occurrence of the variants was identified by analysis of exome trios (mother, father, and proband).
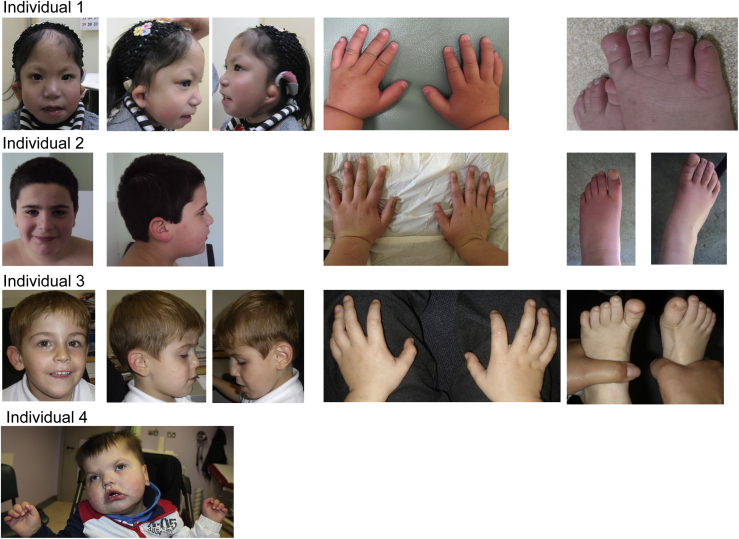
The variants and the associated clinical features are shown in Figure 1, Figure 2, Table 1, and Table S3. There is phenotypic overlap with Coffin-Siris syndrome in that two individuals have a hypoplastic 5th toenail, and all have intellectual disability or developmental delay; however, they do not have the typical facial features (a wide mouth with thick everted lips, a broad nose, thick eyebrows, and long eyelashes) (see Figure 2 for pictures). The sparse hair, thin upper vermilion, and thick lower vermilion in individuals 1 and 3 suggest overlapping facial features with Nicolaides-Baraitser syndrome, although the overall facial gestalt is quite different. All had feeding difficulties, and three had hypotonia. All individuals had developmental delay, and none had epilepsy. Three individuals had small hands and feet. The dysmorphisms were variable and were most notable in individual 4, who had the p.Phe495Leu variant, and who also had the most profound neurodevelopmental disorder (at 3 years of age, he could not talk or walk).
Figure 1.
Characterization of SMARCD1 Variants at the Protein Level
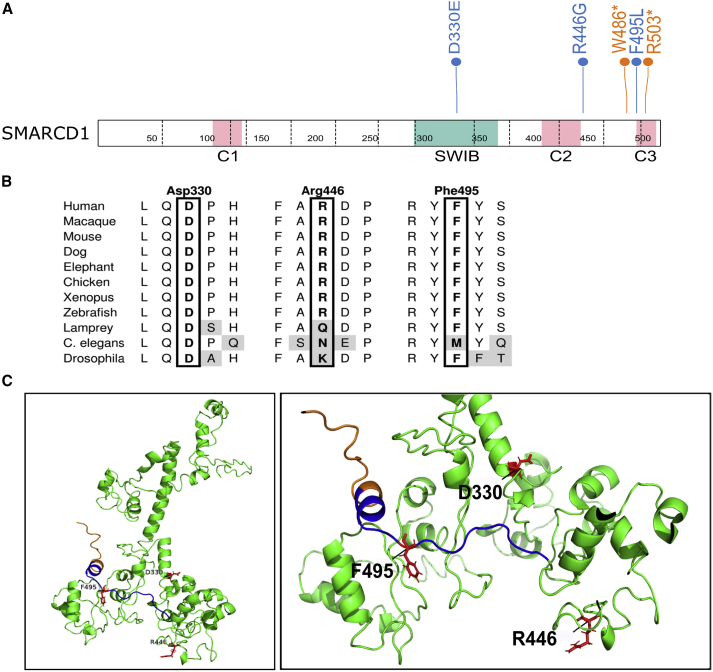
(A) Primary structure of SMARCD1. The SWIB domain is shown in green, and the predicted coiled-coil domains (C1, C2, and C3) are shown in pink. The locations of the identified variants are indicated. Missense and nonsense variants are in blue and orange, respectively.
(B) Amino acid conservation of regions around missense variants across several species.
(C) Predicted 3D model of SMARCD1. Missense variants are show in red. Residues truncated by the p.Trp486∗ variant are colored in blue, and the following residues truncated by both the p.Trp486∗ and p.Arg503∗ variants are colored in orange.
Figure 2.
Photos of Identified Individuals with SMARCD1 Mutations
Individual 1, age 7 years: note ear malformation, sparse hair, and a hypoplastic fifth toenail. Individual 2, age 11 years: note upturned, thick earlobes, a low hairline, short hands, and slender fingers. Individual 3, age 6 years: note a low hairline, long eyebrows, upturned earlobes, and small hands and feet. Individual 4, age 2 years 11 months: note a flat nasal bridge, hypertelorism, prominent philtral pillars, a broad square face with temporal narrowing, a wide mouth with downturned corners, a myopathic facial appearance, and small hands.
Table 1.
Summary of Clinical Findings
| Individual | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Mutation | GenBank: NM_003076.4; c.990C>G (p.Asp330Glu) | GenBank: NM_003076.4; c.1336A>G (p.Arg446Gly) | GenBank: NM_003076.4; c.1457G>A, (p.Trp486∗) | GenBank: NM_003076.4; c.1483T>C (p.Phe495Leu) | GenBank: NM_003076.4; c.1507C>T (p.Arg503∗) |
| Inheritance | heterozygous in child, absent in mother, father not available | de novo | de novo | de novo | de novo |
| Gender | female | male | male | male | female |
| Duration of gestation (weeks) | 37 weeks, 2 days | 38 | 40–41 | 40 | 39.4 |
| Birth weight, g | 2,598 (25th %ile) | 4,150 (>99th %ile) | 3,884 (90th %ile) | 3,480 (75th %ile) | 2,835 (20th %ile) |
| Birth length, cm | 44.5 (1st %ile) | 52 (75th %ile) | 54 (75th %ile) | NA | 48.9 (5th %ile) |
| Birth OFC, cm | 32 (3rd %ile) | 37 (>99th %ile) | NA | NA | 31.75 (10th %ile) |
| Congenital anomalies | esophageal atresia, bronchial fistula | agenesis of the corpus callosum, macrosomia | ankyloglossia (operated) | mild hydronephrosis on antenatal USS | NA |
| Post-natal age at last assessment | 7 years | 11 years, 6 months | 6 years, 3 months | 3 years | 27 months |
| Weight, kg | 17.74 (3rd %ile) | 75.5 (+3.3 SD) | 19.4 (21st %ile) | 16.5 (97th %ile) | 9.9 (−2.2 SD) |
| Height, cm | 106.2 (−2.9 SD) | 161 (+2 SD) | 105 (−2.3 SD) | 111.8 (+3.9 SD) | 84.2 (15th %ile) |
| OFC, cm | NA | 59 (+3.8 SD) | 52 (60th %ile) | 49.3 (41st %ile) | 44.3 (−2.4 SD) |
| Developmental delay | + | + | + | + | + |
| Delayed walking | + | + (17 months) | + (2 years) | not attained | − (15 months) |
| Delayed speech | NA | + (uses sentences) | not attained, some sign language | not attained | + (words at 18 months, now 2- to 3-word phrases) |
| Intellectual disability | + | + | + | + | NA |
| Hypotonia | + | − | + | + | − |
| Neuroradiology | NA | MRI age 1 month: complete agenesis of the corpus callosum | no abnormality seen | MRI at age 3 years: prominence of the lateral and third ventricles. Cavum septum pellucidum. T1 low, T2-FLAIR high in peritrigonal white matter bilaterally. Prominent perivascular spaces within the deep white matter of frontal lobes bilaterally. 4 mm rounded area of T1 low signal at pineal gland, with rim enhancement on postcontrast. Stable. |
MRI at age 22 months: nonspecific, small right frontal subcortical white matter FLAIR hyperintensity. |
| Feeding difficulties | feeding and sucking difficulties | Tube feeding during first 2 weeks. No problem after. | slow feeder, slow with milk and solids, gastroesophageal reflux | ongoing, nasogastric feeding followed by fundoplication and PEG tube, gastroesophageal reflux | gastroesophageal reflux in infancy and difficulty with regulating self-feeding |
| Hair | sparse | low hairline | low hairline | temporal deficiency | normal |
| Ears | external ear malformation | upturned, thick ear lobes | upturned earlobes | low-set, thickened, simple shape | normal |
| Hearing loss | +, malformation of ossicle | − | +, mild bilateral | − | − |
| Hands and feet | hypoplastic fifth toenails | short hands, slender fingers | small hands and feet | thick, loose palmar and plantar skin, small hands and feet, short fifth fingers | normal |
| Teeth anomalies | − | − | small, rounded teeth and pointed canines | thick, stubby teeth, delayed eruption, thick gums | − |
| Other dysmorphisms or relevant information | high palate, cleft soft palate | NA | frontal bossing, long eyebrows, plagiocephaly | broad, square face; temporal narrowing; flat nasal bridge; short nose; very prominent philtral pillars; hypertelorism, divergent squint; wide mouth with downturned corners; thick gums; high palate; "jowly", myopathic appearance; undescended testes |
prominent metopic suture, bulbous nasal tip, bifid uvula with normal palate, facial asymmetry when crying |
See Table S3 for additional details. Legend: + = present, − = absent, NA = not available, %ile = percentile, OFC = occipitofrontal circumference, USS = ultrasound scan, T2-FLAIR = T2-weighted-Fluid-Attenuated Inversion Recovery, and PEG = percutaneous endoscopic gastronomy.
Assessment of the Identified SMARCD1 Variants
SMARCD1 is 515 amino acids long and has a characteristic SWIB domain and three predicted coiled-coil domains (Figure 1A). The identified SMARCD1 variants, two nonsense and three missense, are all clustered either within or in close proximity to these domains. The two nonsense variants (p.Trp486∗ and p.Arg503∗) are located within 50 bp of the last exon junction so should escape nonsense-mediated decay. These variants are predicted to produce a truncated protein lacking the last coiled-coil domain, C3 (Figure 1). One of the missense variants (p.Asp330Glu) is located in the SWIB domain, but the two other missense mutations (p.Arg446Gly andp.Phe495Leu) are located near or in the C2 and C3 coiled-coil domains, respectively (Figure 1A). All of the missense mutations cluster close together in a 3D model of SMARCD1 (Figure 1C). The SMARCD2 SWIB domain and the coiled-coil regions have been shown to be essential for mediating SMARCD2-specific function in granulocytic development, and indeed, deletion of the coiled-coil domain located at the extreme C-terminus of the protein was shown to be sufficient to disrupt the binding between SMARCD2 and the BAF complex.57 Thus, all of the identified mutations have the potential to disrupt a putatively functional domain of SMARCD1.
Prior to the identification of the p.Arg446Gly and p.Arg503∗ variants through GeneMatcher, we had generated expression constructs for the variants p.Asp330Glu, p.Trp486∗, and p.Phe495Leu. The detection of similar amounts of ectopically expressed wild-type and mutant proteins, as visualized by immunoblot, suggests that the mutations do not dramatically affect protein stability (Figure S1). To address the question as to whether those three mutations could impair the incorporation of SMARCD1 into the SWI/SNF complex, we performed a co-immunoprecipitation (Co-IP) assay. None of the three mutations abolished the interaction between SMARCD1 and the ATPase subunit SMARCA4 or the scaffolding subunit SMARCC1 (Figure S1). Although these mutations do not appear to disrupt the interactions of SMARCD1 with SMARCC1 and SMARCA4, they might affect the function of SMARCD1 otherwise, possibly by altering interactions with other SWI/SNF proteins or transcription factors that recruit the SWI/SNF complex to specific genes.
We used several bioinformatic resources to assess the impact of the identified SMARCD1 variants in silico. SMARCD1 has an ExAC pLI score of 1 (because only one loss-of-function [LoF] variant was observed in the ExAC cohort, whereas 26 were predicted), indicating that the gene is intolerant to heterozygous LoF variants.58 It also had a positive ExAC Z score of 3.95, indicating it is also relatively intolerant to missense variants (74 were observed, but 183 were predicted). DECIPHER gives a %HI score of 16.64%, which is close to the ranks indicating that there is a high likelihood that the gene will exhibit haploinsufficiency (below 10%).59 DOMINO gives a 99.8% probability for the gene to be associated with an autosomal-dominant condition.60
We used wANNOVAR61 to predict pathogenicity scores by using tools that consider conservation (polyphen,62 LRT, MutationAssessor,63 GERP,64 PhyloP,65 phastCons,66 and SiPhy67), function (SIFT68 and PROVEAN69), or a combination thereof (FATHMM,70 MutationTaster,71 DANN,72 and CADD73). We considered the following prediction cutoffs: damaging, rank scores above 0.5, and CADD phred scores above 25 (Table S4). At these cutoffs, MutationTaster, FATHMM-MKL, and PROVEAN considered all assessed variants deleterious. LRT, GERP, PhyloP, phastCons, SyPhy, SIFT, DANN, and CADD predicted most assessed variants to be deleterious. MutationAssessor gave medium scores, and polyphen2 predicted two out of three assessed variants to be benign. p.Arg446Gly was the variant for which the most tools gave low pathogenicity predictions, mostly because of the poor conservation in animals further removed from humans (see lamprey, C. elegans, and Drosophila, Figure 1B).
The Drosophila SMARCD1 Ortholog Bap60 Is Required for Memory
Given that all of the identified SMARCD1 mutations might lead to a loss of function by affecting the SWIB or coiled-coil domains, we assessed in Drosophila the consequence of the loss of Bap60, the fly SMARCD1 ortholog. To do this, we generated Bap60 knockdown flies by using previously characterized transgenic RNAi lines (see Subjects and Methods).43 RNAi knockdown of Bap60 was targeted to the MB, the learning and memory center of the fly brain, with R14H06-Gal4. Knockdown was confined to adult flies by the temperature-sensitive Gal4 inhibitor, Gal80ts (see Subjects and Methods). In a parallel study, we have characterized the role of the SWI/SNF complex in MB development.43 Here, we decided to focus on adult flies as a model for understanding the post-natal function of SMARCD1 in the brain; this approach could be relevant toward the long-term goal of developing therapies.
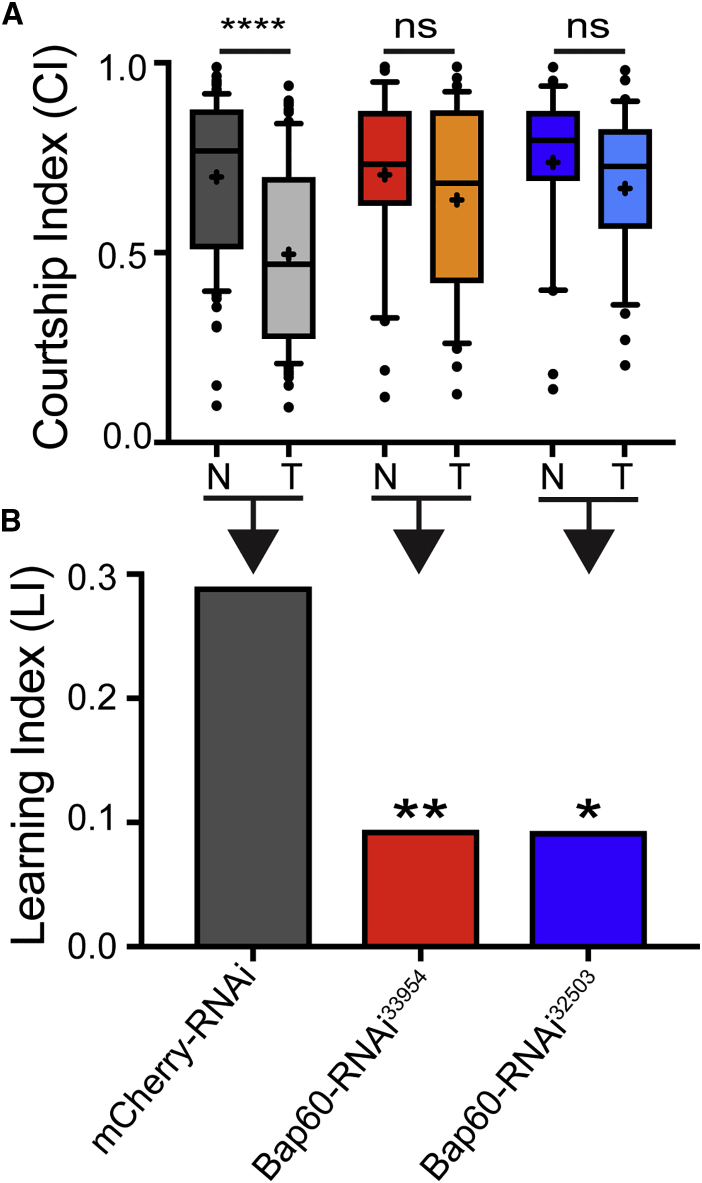
We tested memory in MB-specific Bap60-knockdown flies by using a classic paradigm known as courtship conditioning.45, 74 In this assay, male flies display a learned reduction of courtship behavior after sexual rejection by a non-receptive premated female. As expected, the control flies expressing an RNAi construct targeting mCherry showed a normal reduction in courtship behavior after being sexually rejected by a mated female (Figure 3A). However, this reduction was not seen in flies expressing RNAi transgenes targeting Bap60 (Figure 3A). Bap60-knockdown flies also showed a significantly lower learning index (LI) as compared to the controls (Figure 3B). This assay was performed with two different Bap60 RNAi lines, which have unique target sequences. For both knockdown lines, a similar reduction in memory was observed, indicating that memory defects are not a result of off-target effects. These results suggest that Bap60 is required for normal memory, post-development, in adult neurons.
Figure 3.
Adult-Specific Knockdown of Bap60 in the MB Results in Reduced Capacity for Long-Term Courtship Memory
(A) Boxplots showing courtship indices (CIs) for the mCherry RNAi control and Bap60-knockdown flies in long-term courtship conditioning assays (+ indicates the mean). Dunn's test was used for comparing the mean CIs for naïve (N) and trained (T) flies of the same genotype.
(B) Learning indices (LIs) for control and Bap60-knockdown flies. Adult-specific Bap60 knockdown resulted in a significantly reduced LI (randomization test, 10,000 bootstrap replicates) relative to control flies. ∗ p < 0.05, ∗∗ p < 0.01, and ∗∗∗∗ p < 0.0001.
Bap60 Is Required for the Expression of Neuron-Specific Genes during a Critical Period of Juvenile Adult MB Development
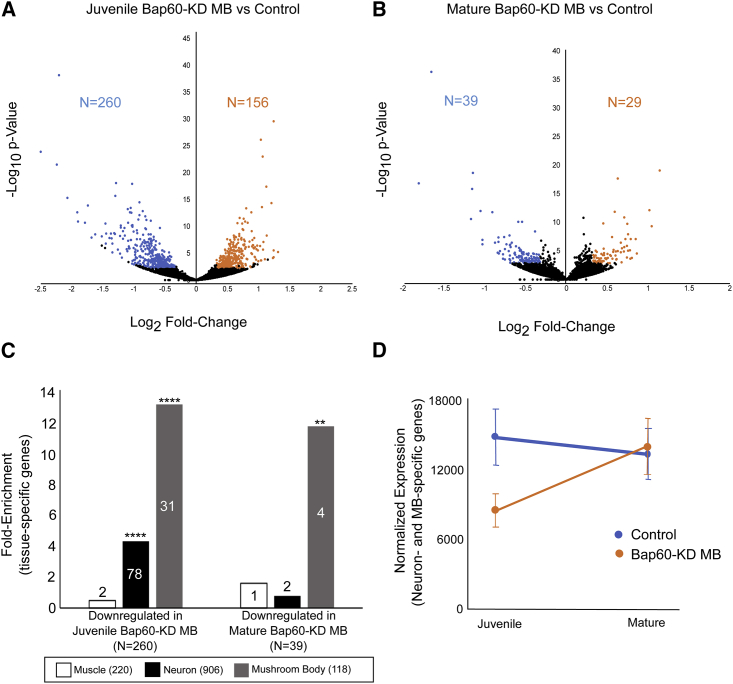
We used INTACT (see Subjects and Methods) to investigate the effect of Bap60 knockdown on gene expression in the specific MB cells that were targeted for RNAi knockdown in our learning and memory assays. In juvenile adult insects, the MB undergoes a period of development and synaptogenesis in the first hours after eclosion.75, 76, 77, 78, 79, 80, 81 During this time, neuronal connections that are critical for normal learning and memory later in life are formed.77, 79 We analyzed the MB-specific transcriptome in Bap60-knockdown flies compared to controls in early juvenile adults (0–3 hours after eclosion), and mature adults (1–5 days after eclosion). We observed a greater effect of Bap60 on gene expression at the juvenile stage than in the mature adult MB (Figures 4A and 4B). Using DESeq253 for differential expression analysis of the juvenile Bap60-KD MBs and controls yielded 416 differentially expressed genes (Padj < 0.05 and a >1.5-fold change), of which 156 were upregulated and 260 were downregulated (Figure 4A and Table S5). In contrast, only 68 differentially expressed genes (29 upregulated and 39 downregulated) were observed in mature Bap60-KD MBs (Figure 3B and Table S5). The differential expression of several genes was confirmed by RT-qPCR in independent biological replicates (Figure S2). We performed a gene ontology (GO) enrichment analysis to investigate the functions of the differentially expressed genes. Differentially expressed genes from the mature MB showed very little GO enrichment (Table S5). A GO enrichment analysis of the upregulated genes from the juvenile MB revealed many terms related to muscle, such as “myofibril assembly” and “sarcomere organization” (Table S5). A GO enrichment analysis of downregulated genes in the juvenile MB revealed neuron-related terms such as “neurotransmitter metabolic process,” “synaptic vesicle,” and “regulation of synaptic plasticity,” as well as developmental terms such as “nervous system development” and “anatomical structure development” (Table S5). This suggested an important role for Bap60 in the regulation of neurodevelopmental genes in the early juvenile adult MB.
Figure 4.
Bap60 Is Required for the Expression of Neuron-Specific Genes in the Juvenile MB
(A and B) Volcano plots showing differentially expressed genes (padj < 0.05 and >1.5-fold change) represented in blue (downregulated) or orange (upregulated) in (A) juvenile Bap60-KD MBs and (B) mature Bap60-KD MBs compared to controls of the same age.
(C) Fold enrichment of muscle-, neuron-, and mushroom-body- specific genes (Brown et al. and Jones et al.;47, 55 see Subjects and Methods) among downregulated genes in juvenile and mature Bap60-KD MBs (∗∗ p < 0.01; ∗∗∗∗ p < 0.0001; hypergeometric test). The number of genes in each category is indicated.
(D) Average normalized expression (±SEM) of neuron- and mushroom-body-specific genes in control (blue) and Bap60-KD MB (orange) flies at the juvenile and mature stages.
Having observed a downregulation of neuron-related genes and an upregulation of muscle-related genes in juvenile Bap60-KD MBs, we reasoned that Bap60 might be required at this stage to activate the expression of neuron-specific genes that contribute to cell identity. To test this, we used existing tissue-specific RNA-seq data to establish a list of 904 “neuron-specific genes” that are enriched in heads compared to other tissues,55 and 118 “MB-specific genes” that are enriched in MB-specific INTACT samples compared to whole-head samples47 (see Subjects and Methods and Table S2). Of the 260 genes that are downregulated in Bap60-KD MBs, 78 are neuron specific and 31 are MB specific; these numbers are significantly greater than those expected by chance (p < 10−25, hypergeometric test) (Figure 4C). On average, these genes are expressed at a consistent level in controls in juvenile and mature adult MBs, but in Bap60-KD MBs at the juvenile stage they have reduced expression that recovers to normal levels in mature adults (Figure 4D). These trends were validated by RT-qPCR for a selection of genes in an independent experiment (Figure S2, prt and jdp). Muscle-specific genes (enriched in the carcass compared to other tissues) were not significantly over-represented among genes that were downregulated in Bap60-KD MBs. Taken together, these results suggest that Bap60 plays a context-dependent role in activating the expression of neuron-specific genes in the MB.
Bap60 Is Required for the Expression of Developmental Genes That Are Preferentially Activated in the Juvenile MB
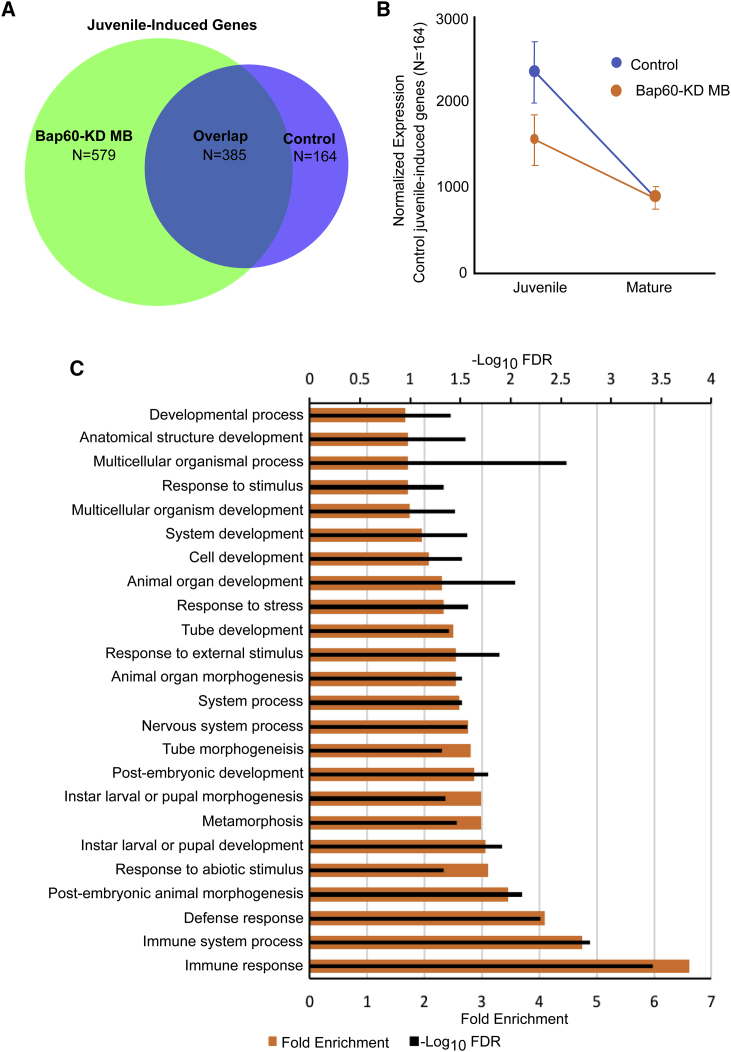
Our GO enrichment analysis of genes that are downregulated in Bap60 mutants revealed many terms related to neuronal processes and development (Table S5). We reasoned that Bap60 might be required for the activation of key genes that are involved in experience-dependent MB development in juvenile adults.79, 82 To test this, we performed differential expression analysis comparing the MB-specific transcriptome in early juvenile adults to that in mature adults (Table S6). In controls, 549 genes were significantly increased by 2-fold or more in the juvenile adult MB compared to in that of mature adults. Of these 549 juvenile enriched MB genes, 385 genes were also > 2-fold enriched in Bap60-KD MBs, but 164 were not (Figure 5A). On average, these 164 genes showed significantly lower expression in juvenile Bap60-KD MBs compared to in controls, and this difference is no longer observed in mature flies (Figure 5B). These expression trends were validated for a selection of genes in an independent RT-qPCR experiment (Figure S2). Interestingly, the 164 juvenile induced genes that are not induced in Bap60-KD MBs show a strong GO enrichment for terms related to development and immune response (Figure 5C). In contrast, the 385 genes that show juvenile enrichment in both Bap60-KD and control MBs show very little GO enrichment. This suggests that Bap60 is required for the activation of developmental genes during a critical period when the juvenile adult MB needs to establish experience-dependent synaptic connections that are required for normal learning and memory throughout life.75, 76, 77, 78, 79, 80, 81
Figure 5.
Bap60 Is Required for the Increased Expression of Developmental Genes in the Juvenile MB
(A) A Venn diagram showing overlap genes that are significantly increased in expression in the juvenile MB compared to in the mature MB (juvenile-induced genes) in control and Bap60-KD flies.
(B) Average normalized expression (±SEM) of the 164 genes that are induced in the juvenile MB in controls only.
(C) Gene ontology enrichment for biological processes of the 164 juvenile-induced genes identified in controls only. Terms with at least ten genes and an FDR < 0.05 are displayed.
Discussion
Characterization of a Neurodevelopmental Disorder Caused by SMARCD1 Mutations
In this study, we describe a genetic disorder characterized by mutations in SMARCD1, which encodes a component of the SWI/SNF chromatin remodeling complex. Mutations in several other SWI/SNF genes are implicated in syndromic NDDs that are typically characterized by intellectual disability, abnormalities of the fifth digit, and characteristic facial features.83 The five individuals described here do fit in this spectrum because they have intellectual disability and a low penetrance of fifth-digit abnormalities but lack the typical facial dysmorphisms seen in other SWI/SNF-related disorders.
The identified SMARCD1 variants are all clustered in the C-terminal end of the protein, and they are also located in close proximity in a 3D model of the protein (Figure 1). Although these missense and protein-truncating mutations are predicted to be damaging, we do not know the precise functional effect of these mutations. One missense variant is located in the highly conserved SWIB domain, and the other four variants are located in or near the C-terminal coiled-coil domains. We show here that those mutations do not disrupt the interaction with the other SWI/SNF components SMARCA4 and SMARCC1. Recently, Mashtalir et al., published an elegant study on the molecular organization of the SWI/SNF complexes.84 They demonstrated that the SWI/SNF complex is composed of an ATPase module (SMARCA4 or SMARCA2, ACTB, and ACTL6A or ACTL6B), a core scaffolding module (SMARCC1or SMARCC2; SMARCD1, SMARCD2, or SMARCD3; SMARCE1; and SMARCB1), and an ARID module that confers the specificity of the different SWI/SNF conformations (canonical BAF, PBAF, and non-canonical BAF). The minimal core module of the SWI/SNF complex is composed of the SMARCD subunit and either a homo- or heterodimer of SMARCC1 and SMARCC2. SMARCD1 can interact directly with SMARCC1 or SMARCC2 via the SWIB domain and the unstructured region of the protein toward the N-terminal side of the SWIB domain. There are minimal interactions between the C-terminal end of SMARCD1 and SMARCC1 or SMARCC2. Similarly, most interactions between SMARCA4 and SMARCD1 involve the N-terminal portion of the protein. These results are in agreement with our co-IPs showing that the tested C-terminal mutations do not disrupt the interaction between SMARCD1 and SMARCC1 or SMARCA4. Interestingly, Mashtalir et al. show that the C-terminal region located downstream of the SMARCD1 SWIB domain—where our mutations cluster—is implicated in the binding of ARID subunits (ARID1A, ARID1B, and ARID2) and necessary for the formation of the fully assembled BAF and PBAF conformations of the mammalian SWI/SNF complex.84 It will be interesting to test whether the mutations identified here affect the interaction with ARID subunits.
Bap60 in Memory and MB-specific Transcriptome Regulation in Juvenile Drosophila
Although the role of some SWI/SNF components in neuron development and function is well described,85 there was previously no work investigating the function of SMARCD1 in the nervous system. We show here that the Drosophila ortholog Bap60 is required in the adult fly MB for normal memory (Figure 3). The requirement for Bap60 in the adult fly brain is consistent with evidence from mammals suggesting the presence of a SWI/SNF complex that is only present in differentiated neurons.3, 12, 18 Indeed, it has been shown that the neuron-specific subunit BAF53b is also essential in adults for normal memory.3 Taken together, these findings suggest that SWI/SNF-related NDDs might result from defective gene regulation postnatally in differentiated neurons.
Using MB-specific transcriptome analysis, we found that Bap60 has a greater effect on gene regulation in the MB of juvenile adult flies than on that of mature flies. In particular, Bap60 seems to be important for activating the expression of neuronal genes (Figure 4) and developmental genes that normally show increased expression in juvenile MBs (Figure 5). This is interesting because the MB is known to undergo structural alterations and form new synaptic connections during the early stages of juvenile adult life.76, 77, 87, 88 These changes are dependent on sensory input, suggesting that some of the brain’s circuitry is developed in response to early life experience.75, 76, 88 This early-experience-dependent plasticity in the MB is required for normal memory at later life stages in flies76, 77, 80, 81 and bees.78, 88 Although much more complex, human brains also show periods of experience-dependent plasticity, especially during adolescence.89 So-called “environmental enrichment” therapy for autism is designed on the basis of the idea that defects in neural circuitry might be corrected by providing increased sensorimotor experience.89, 90 It will be interesting to further investigate the mechanisms of SWI/SNF-mediated gene regulation in experience-dependent brain plasticity. Understanding the role of SWI/SNF in the postnatal brain could open up possibilities for therapy, whereas prenatal developmental intervention seems unlikely.
Here we identify five mutations in SMARCD1, a subunit of the BAF complex that has not been previously associated with a neurodevelopmental disorder. Moreover, we show that its Drosophila ortholog, Bap60, regulates neurodevelopmental gene expression during a critical time window of juvenile adult brain development when neuronal circuits that are required for learning and memory are formed. Altogether, our results highlight the role of SMARCD1 in establishing proper cognitive functions.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
Thanks to the individuals and their families who participated in this study. We thank the Bloomington Drosophila Stock Center and the Transgenic RNAi Project at Harvard Medical School for creating and providing fly stocks used in this study. Thanks to David Carter at the London Regional Genomics Centre for help with sequencing. This work was funded by the Canadian Institutes of Health Research, the Canada Research Chairs program, the Canada Foundation for Innovation, the Fonds de Recherche Santé Québec, and by the Japan Agency for Medical Research and Development (AMED) under grant numbers, JP18ek0109280, JP18dm0107090, JP18ek0109301, JP18ek0109348, and JP18kk020500; Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grant numbers JP17H01539 and JP16H05160; and Takeda Science Foundation (N. Miyake and N. Matsumoto). We thank Agnès Rastetter for technical assistance and confirmation of the mutation proband 2. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health.
Published: March 14, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.02.001.
Contributor Information
Jamie M. Kramer, Email: james.kramer@schulich.uwo.ca.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Accession Numbers
RNA-seq data are available at the NCBI Gene Expression Omnibus through the accession number GEO: GSE122864.
Web Resources
Deciphering Developmental Disorders, https://decipher.sanger.ac.uk/
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc
GeneDX ClinVar Submission Page, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
I-TASSER Online Suite, https://zhanglab.ccmb.med.umich.edu/I-TASSER/
NPS@: Network Protein Analysis, https://npsa-prabi.ibcp.fr/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Protein Paint, https://pecan.stjude.cloud/proteinpaint
PyMol, https://pymol.org/2/
Supplemental Data
References
- 1.Ho L., Ronan J.L., Wu J., Staahl B.T., Chen L., Kuo A., Lessard J., Nesvizhskii A.I., Ranish J., Crabtree G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann C., Nguyen H., Rosenbusch J., Pham L., Rabe T., Patwa M., Sokpor G., Seong R.H., Ashery-Padan R., Mansouri A. mSWI/SNF (BAF) complexes are indispensable for the neurogenesis and development of embryonic olfactory epithelium. PLoS Genet. 2016;12:e1006274. doi: 10.1371/journal.pgen.1006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel-Ciernia A., Matheos D.P., Barrett R.M., Kramár E.A., Azzawi S., Chen Y., Magnan C.N., Zeller M., Sylvain A., Haettig J. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuoc T., Dere E., Radyushkin K., Pham L., Nguyen H., Tonchev A.B., Sun G., Ronnenberg A., Shi Y., Staiger J.F. Ablation of BAF170 in developing and postnatal dentate gyrus affects neural stem cell proliferation, differentiation, and learning. Mol. Neurobiol. 2017;54:4618–4635. doi: 10.1007/s12035-016-9948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett R.M., Wood M.A. Beyond transcription factors: The role of chromatin modifying enzymes in regulating transcription required for memory. Learn. Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassabov S.R., Zhang B., Persinger J., Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 7.Whitehouse I., Flaus A., Cairns B.R., White M.F., Workman J.L., Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 8.van Holde K., Yager T. Models for chromatin remodeling: A critical comparison. Biochem. Cell Biol. 2003;81:169–172. doi: 10.1139/o03-038. [DOI] [PubMed] [Google Scholar]
- 9.Lessard J., Wu J.I., Ranish J.A., Wan M., Winslow M.M., Staahl B.T., Wu H., Aebersold R., Graef I.A., Crabtree G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronan J.L., Wu W., Crabtree G.R. From neural development to cognition: Unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olave I., Wang W., Xue Y., Kuo A., Crabtree G.R. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen H., Sokpor G., Pham L., Rosenbusch J., Stoykova A., Staiger J.F., Tuoc T. Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle. 2016;15:1317–1324. doi: 10.1080/15384101.2016.1160984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayanan R., Pirouz M., Kerimoglu C., Pham L., Wagener R.J., Kiszka K.A., Rosenbusch J., Seong R.H., Kessel M., Fischer A. Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 2015;13:1842–1854. doi: 10.1016/j.celrep.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 15.Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C.D. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuoc T.C., Narayanan R., Stoykova A. BAF chromatin remodeling complex: Cortical size regulation and beyond. Cell Cycle. 2013;12:2953–2959. doi: 10.4161/cc.25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto S., Banine F., Struve J., Xing R., Adams C., Liu Y., Metzger D., Chambon P., Rao M.S., Sherman L.S. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 18.Wu J.I., Lessard J., Olave I.A., Qiu Z., Ghosh A., Graef I.A., Crabtree G.R. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Iwase S., Bérubé N.G., Zhou Z., Kasri N.N., Battaglioli E., Scandaglia M., Barco A. Epigenetic etiology of intellectual disability. J. Neurosci. 2017;37:10773–10782. doi: 10.1523/JNEUROSCI.1840-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., Kaname T., Naritomi K., Kawame H., Wakui K. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 22.Bramswig N.C., Caluseriu O., Lüdecke H.J., Bolduc F.V., Noel N.C., Wieland T., Surowy H.M., Christen H.J., Engels H., Strom T.M., Wieczorek D. Heterozygosity for ARID2 loss-of-function mutations in individuals with a Coffin-Siris syndrome-like phenotype. Hum. Genet. 2017;136:297–305. doi: 10.1007/s00439-017-1757-z. [DOI] [PubMed] [Google Scholar]
- 23.Wieczorek D., Bögershausen N., Beleggia F., Steiner-Haldenstätt S., Pohl E., Li Y., Milz E., Martin M., Thiele H., Altmüller J. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum. Mol. Genet. 2013;22:5121–5135. doi: 10.1093/hmg/ddt366. [DOI] [PubMed] [Google Scholar]
- 24.Santen G.W.E., Aten E., Sun Y., Almomani R., Gilissen C., Nielsen M., Kant S.G., Snoeck I.N., Peeters E.A., Hilhorst-Hofstee Y. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 25.Dias C., Estruch S.B., Graham S.A., McRae J., Sawiak S.J., Hurst J.A., Joss S.K., Holder S.E., Morton J.E., Turner C., DDD Study BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am. J. Hum. Genet. 2016;99:253–274. doi: 10.1016/j.ajhg.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivière J.B., van Bon B.W., Hoischen A., Kholmanskikh S.S., O’Roak B.J., Gilissen C., Gijsen S., Sullivan C.T., Christian S.L., Abdul-Rahman O.A. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012;44:440–444. doi: 10.1038/ng.1091. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleefstra T., Kramer J.M., Neveling K., Willemsen M.H., Koemans T.S., Vissers L.E., Wissink-Lindhout W., Fenckova M., van den Akker W.M., Kasri N.N. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet. 2012;91:73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koga M., Ishiguro H., Yazaki S., Horiuchi Y., Arai M., Niizato K., Iritani S., Itokawa M., Inada T., Iwata N. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 29.Loe-Mie Y., Lepagnol-Bestel A.M., Maussion G., Doron-Faigenboim A., Imbeaud S., Delacroix H., Aggerbeck L., Pupko T., Gorwood P., Simonneau M., Moalic J.M. SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet. 2010;19:2841–2857. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- 30.Nord A.S., Roeb W., Dickel D.E., Walsh T., Kusenda M., O’Connor K.L., Malhotra D., McCarthy S.E., Stray S.M., Taylor S.M., STAART Psychopharmacology Network Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur. J. Hum. Genet. 2011;19:727–731. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff D., Endele S., Azzarello-Burri S., Hoyer J., Zweier M., Schanze I., Schmitt B., Rauch A., Reis A., Zweier C. In-frame deletion and missense mutations of the C-terminal helicase domain of SMARCA2 in three patients with Nicolaides-Baraitser syndrome. Mol. Syndromol. 2012;2:237–244. doi: 10.1159/000337323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Houdt J.K., Nowakowska B.A., Sousa S.B., van Schaik B.D., Seuntjens E., Avonce N., Sifrim A., Abdul-Rahman O.A., van den Boogaard M.J., Bottani A. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat. Genet. 2012;44:445–449. doi: 10.1038/ng.1105. S1. [DOI] [PubMed] [Google Scholar]
- 33.Marom R., Jain M., Burrage L.C., Song I.W., Graham B.H., Brown C.W., Stevens S.J.C., Stegmann A.P.A., Gunter A.T., Kaplan J.D. Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum. Mutat. 2017;38:1365–1371. doi: 10.1002/humu.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machol K., Rousseau J., Ehresmann S., Garcia T., Nguyen T.T.M., Spillmann R.C., Sullivan J.A., Shashi V., Jiang Y.H., Stong N., Undiagnosed Diseases Network Expanding the spectrum of BAF-related disorders: De novo variants in SMARCC2 cause a syndrome with intellectual disability and developmental delay. Am. J. Hum. Genet. 2019;104:164–178. doi: 10.1016/j.ajhg.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heisenberg M., Borst A., Wagner S., Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 36.de Belle J.S., Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 37.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyatake S., Okamoto N., Stark Z., Nabetani M., Tsurusaki Y., Nakashima M., Miyake N., Mizuguchi T., Ohtake A., Saitsu H., Matsumoto N. ANKRD11 variants cause variable clinical features associated with KBG syndrome and Coffin-Siris-like syndrome. J. Hum. Genet. 2017;62:741–746. doi: 10.1038/jhg.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh A.P.L., Edwards T.J., Galea C., Cooper H.M., Engle E.C., Jamuar S.S., Méneret A., Moutard M.L., Nava C., Rastetter A., IRC5 Consortium DCC mutation update: Congenital mirror movements, isolated agenesis of the corpus callosum, and developmental split brain syndrome. Hum. Mutat. 2018;39:23–39. doi: 10.1002/humu.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.S. The transgenic RNAi project at Harvard medical school: Resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H.H., Kroll J.R., Lennox S.M., Ogundeyi O., Jeter J., Depasquale G., Truman J.W. A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 2014;8:897–908. doi: 10.1016/j.celrep.2014.06.065. [DOI] [PubMed] [Google Scholar]
- 43.Chubak M.C. Systematic functional characterization of the intellectual disability-associated SWI/SNF complex reveals distinct roles for the BAP and PBAP complexes in post-mitotic memory forming neurons of the Drosophila mushroom body. bioRxiv. 2018 [Google Scholar]
- 44.Möller A., Avila F.W., Erickson J.W., Jäckle H. Drosophila BAP60 is an essential component of the Brahma complex, required for gene activation and repression. J. Mol. Biol. 2005;352:329–337. doi: 10.1016/j.jmb.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Koemans T.S., Oppitz C., Donders R.A.T., van Bokhoven H., Schenck A., Keleman K., Kramer J.M. Drosophila courtship conditioning as a measure of learning and memory. J. Vis. Exp. 2017;124:1–11. doi: 10.3791/55808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;220:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 47.Jones S.G., Nixon K.C.J., Chubak M.C., Kramer J.M. Mushroom body specific transcriptome analysis reveals dynamic regulation of learning and memory genes after acquisition of long-term courtship memory in Drosophila. G3 (Bethesda) 2018;8:3433–3446. doi: 10.1534/g3.118.200560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams M.D. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 50.Clark A.G., Eisen M.B., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Priam P., Krasteva V., Rousseau P., D’Angelo G., Gaboury L., Sauvageau G., Lessard J.A. SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPɛ dependent mechanism. Nat. Genet. 2017;49:753–764. doi: 10.1038/ng.3812. [DOI] [PubMed] [Google Scholar]
- 58.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinodoz M., Royer-Bertrand B., Cisarova K., Di Gioia S.A., Superti-Furga A., Rivolta C. DOMINO: Using machine learning to predict genes associated with dominant disorders. Am. J. Hum. Genet. 2017;101:623–629. doi: 10.1016/j.ajhg.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garber M., Guttman M., Clamp M., Zody M.C., Friedman N., Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 69.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L., Edwards K.J., Day I.N., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 72.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamyshev N.G., Iliadi K.G., Bragina J.V. Drosophila conditioned courtship: Two ways of testing memory. Learn. Mem. 1999;6:1–20. [PMC free article] [PubMed] [Google Scholar]
- 75.Barth M., Hirsch H.V., Meinertzhagen I.A., Heisenberg M. Experience-dependent developmental plasticity in the optic lobe of Drosophila melanogaster. J. Neurosci. 1997;17:1493–1504. doi: 10.1523/JNEUROSCI.17-04-01493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barth M., Heisenberg M. Vision affects mushroom bodies and central complex in Drosophila melanogaster. Learn. Mem. 1997;4:219–229. doi: 10.1101/lm.4.2.219. [DOI] [PubMed] [Google Scholar]
- 77.Seugnet L., Suzuki Y., Donlea J.M., Gottschalk L., Shaw P.J. Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep. 2011;34:137–146. doi: 10.1093/sleep/34.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones B.M., Leonard A.S., Papaj D.R., Gronenberg W. Plasticity of the worker bumblebee brain in relation to age and rearing environment. Brain Behav. Evol. 2013;82:250–261. doi: 10.1159/000355845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doll C.A., Broadie K. Impaired activity-dependent neural circuit assembly and refinement in autism spectrum disorder genetic models. Front. Cell. Neurosci. 2014;8 doi: 10.3389/fncel.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doll C.A., Broadie K. Activity-dependent FMRP requirements in development of the neural circuitry of learning and memory. Development. 2015;142:1346–1356. doi: 10.1242/dev.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doll C.A., Vita D.J., Broadie K. Fragile X mental retardation protein requirements in activity-dependent critical period neural circuit refinement. Curr. Biol. 2017;27:2318–2330.e3. doi: 10.1016/j.cub.2017.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tessier C.R., Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kosho T., Miyake N., Carey J.C. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: Historical review and recent advances using next generation sequencing. Am. J. Med. Genet. C. Semin. Med. Genet. 2014;166C:241–251. doi: 10.1002/ajmg.c.31415. [DOI] [PubMed] [Google Scholar]
- 84.Mashtalir N. Modular organization and assembly of SWI / SNF family chromatin remodeling complexes. Cell. 2018;175:1272–1288e.20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sokpor G., Xie Y., Rosenbusch J., Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heisenberg M., Heusipp M., Wanke C. Structural plasticity in the Drosophila brain. J. Neurosci. 1995;15:1951–1960. doi: 10.1523/JNEUROSCI.15-03-01951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cabirol A., Brooks R., Groh C., Barron A.B., Devaud J.M. Experience during early adulthood shapes the learning capacities and the number of synaptic boutons in the mushroom bodies of honey bees (Apis mellifera) Learn. Mem. 2017;24:557–562. doi: 10.1101/lm.045492.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ismail F.Y., Fatemi A., Johnston M.V. Cerebral plasticity: Windows of opportunity in the developing brain. Eur. J. Paediatr. Neurol. 2017;21:23–48. doi: 10.1016/j.ejpn.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 90.Simpson J., Kelly J.P. The impact of environmental enrichment in laboratory rats--behavioural and neurochemical aspects. Behav. Brain Res. 2011;222:246–264. doi: 10.1016/j.bbr.2011.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.