Abstract
Familial dysautonomia (FD) is a recessive neurodegenerative disease caused by a splice mutation in Elongator complex protein 1 (ELP1, also known as IKBKAP); this mutation leads to variable skipping of exon 20 and to a drastic reduction of ELP1 in the nervous system. Clinically, many of the debilitating aspects of the disease are related to a progressive loss of proprioception; this loss leads to severe gait ataxia, spinal deformities, and respiratory insufficiency due to neuromuscular incoordination. There is currently no effective treatment for FD, and the disease is ultimately fatal. The development of a drug that targets the underlying molecular defect provides hope that the drastic peripheral neurodegeneration characteristic of FD can be halted. We demonstrate herein that the FD mouse TgFD9;IkbkapΔ20/flox recapitulates the proprioceptive impairment observed in individuals with FD, and we provide the in vivo evidence that postnatal correction, promoted by the small molecule kinetin, of the mutant ELP1 splicing can rescue neurological phenotypes in FD. Daily administration of kinetin starting at birth improves sensory-motor coordination and prevents the onset of spinal abnormalities by stopping the loss of proprioceptive neurons. These phenotypic improvements correlate with increased amounts of full-length ELP1 mRNA and protein in multiple tissues, including in the peripheral nervous system (PNS). Our results show that postnatal correction of the underlying ELP1 splicing defect can rescue devastating disease phenotypes and is therefore a viable therapeutic approach for persons with FD.
Keywords: ELP1, familial dysautonomia, neurodegenerative disease, splicing, therapeutics, IKAP
Introduction
Familial dysautonomia (FD), also known as Riley-Day syndrome or hereditary sensory and autonomic neuropathy type III (MIM: 223900), is a congenital neurodegenerative disease caused by a splice mutation in intron 20 of ELP1 (MIM: 603722).1, 2, 3, 4, 5, 6 This mutation results in variable, tissue-specific skipping of exon 20 and a corresponding reduction of ELP1 (previously known as IKAP).7, 8 In individuals with FD, the lowest amount of ELP1 is in the nervous system.7 ELP1 is the scaffolding member of the six-subunit human Elongator complex, which is a highly conserved protein complex that participates in distinct cellular processes including transcriptional elongation, acetylation of cytoskeletal α-tubulin, and tRNA modification.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 ELP1 function has been widely investigated and has been implicated in exocytosis, cytoskeletal organization, and axonal transport, as well as cellular adhesion and migration.20, 21, 22, 23, 24 Importantly, recent in vitro and in vivo studies emphasized the role of ELP1 in neurogenesis, neuronal survival, and peripheral tissue innervation.25, 26, 27, 28, 29, 30, 31
FD occurs almost exclusively in Ashkenazi Jews and has a carrier frequency of 1 in 32 in the general Ashkenazi Jewish population and 1 in 19 in Ashkenazi Jews of Polish descent.32, 33 From birth, persons with FD display a complex neurological phenotype that is consistent with widespread loss of somatosensory feedback and that worsens over time.4, 34, 35, 36, 37 Individuals with FD have decreased sensitivity to pain and temperature sensation, visual loss, kyphoscoliosis, proprioceptive ataxia, and difficulties in regulating body temperature.3, 38, 39, 40, 41, 42, 43, 44, 45, 46 The lack of afferent baroreceptor signaling causes complete failure of blood pressure regulation and recurrent hypertensive vomiting attacks referred to as “dysautonomic crises.”2, 47, 48, 49, 50 Unexplained sudden death, aspiration pneumonias, and respiratory insufficiency remain the leading causes of death.34, 39
Many of the debilitating symptoms of the disease are due to progressive impairment of proprioception.34, 45, 46, 51, 52 A lack of afferent signaling from the muscle spindles accounts for the absence of deep tendon reflexes and for gait ataxia.46, 51 Children with FD are uncoordinated and have a tendency to fall. As they age, progressive impairment in proprioception leads to severe gait ataxia, and eventually they lose the ability to ambulate independently. This is one of the most problematic features of the disease because it severely affects their quality of life.46, 51 Proprioceptive deficits also contribute to the poor coordination of chest wall movements and explain the skeletal deformities such as early-onset kyphoscoliosis44, 45, 53, 54, 55 and abnormal craniofacial development.52 Neuropathological analysis of autopsy material from individuals with FD showed grossly reduced volume and number of neurons in the dorsal root ganglia (DRGs).35 Interestingly, recent studies conducted in Elp1-conditional-knockout mouse embryos wherein Elp1 expression was selectively deleted in the neural crest lineage, which gives rise to most of the PNS including the DRGs, showed that although Elp1 was required for the development of the pain- and temperature-receptive neurons, proprioceptors arise and differentiate normally.25, 26 Therefore, unlike nociceptors, the loss of proprioceptors in FD most likely occurs postnatally, making this subpopulation of neurons an attractive target for therapeutic intervention.
Current treatments for FD are only supportive, and they are aimed at treating symptoms rather than targeting the cause of the disease.56 Previously, we identified the small molecule kinetin (6-furfurylaminopurine) to be an orally active enhancer of ELP1 splicing.57, 58 Although in vitro and in vivo studies have demonstrated the ability of kinetin to improve ELP1 splicing,58, 59, 60, 61 until recently there has not been a way to evaluate the effect of splicing correction on disease phenotype or progression. Here, we evaluated the therapeutic potential of manipulating ELP1 splicing in vivo in the TgFD9;IkbkapΔ20/flox mouse.62 This humanized mouse model displays all the hallmarks of the human disease, and the evaluation of kinetin in this model demonstrates that splicing correction can rescue neurological phenotypes in FD. The pace of development of genetic therapies, including targeted small molecules and antisense oligonucleotides, is unprecedented. Our study shows that increasing ELP1 after birth, regardless of the therapeutic mechanism, will most likely rescue proprioceptive neurons and improve neurologic symptoms in FD.
Material and Methods
Study Design
The aim of this study was to assess the therapeutic effectiveness of the small-molecule splicing-modulator kinetin in ameliorating neurological phenotypes in vivo. In this regard, we used the recently developed FD mouse model TgFD9;IkbkapΔ20/flox because it recapitulates the same tissue-specific mis-splicing observed in individuals with FD and displays the hallmark symptoms of the disease, thus providing a powerful model for assessing the therapeutic efficacy of potential therapies. Treatment was started at birth in order to maximize the therapeutic value. At P0 (postnatal day 0), regular mouse chow was replaced with vehicle diet (LabDiet 5P00) or kinetin diet (LabDiet 5P00 with 2150 ppm kinetin), and the dam, randomly assigned, continued to be fed these diets until the time of weaning. At weaning, TgFD9;IkbkapΔ20/flox mice were genotyped and maintained in the same treatment groups. We formulated kinetin chow to provide each mouse with a dose of 200 mg/Kg/day because we have previously demonstrated that this dose was sufficient to significantly improve ELP1 splicing and protein in vivo in a phenotypically normal mouse.62
All animal experiments were designed with a commitment to minimizing both the number of mice and their suffering. We designed our preclinical animal trial on the basis of the published recommendations.63 In order to calculate appropriate sample sizes for the study, we performed a power analysis using the data generated from the phenotypic characterization of our TgFD9;IkbkapΔ20/flox mice.62 Thus, for statistical validity we used n = 4–9 mice for phenotypic assessments, n = 4–6 mice for histological analysis of the DRGs, and n = 13–14 mice for ELP1 splicing and protein analysis. All of the analyses described in this study were conducted by using animal samples from multiple litters; therefore, each unit (animal, cage, litter) represents a biologic replicate. The numbers were not altered during the course of the study. The primary endpoints were predefined in advance on the basis of our previous data.62 All data were included, and the criteria were established prospectively. Animals were assigned randomly to the vehicle- or kinetin-group via a randomization system devised by the Massachusetts General Hospital (MGH) Biostatistics Center. The system consists of a box containing cards with either “vehicle diet” or “special diet” in random order, and the animals were randomly assigned to the appropriate group by drawing a card. Investigators conducting the experiments were blind to genotype and treatment category.
Animals
Generating a mouse model for FD that recapitulated the phenotype and the tissue-specific mis-splicing seen in individuals with FD represented a significant challenge. To generate the TgFD9;IkbkapΔ20/flox mouse (C57BL/6J background) we used a breeding scheme specifically designed to increase their Mendelian ratio. Initially, we crossed the TgFD9 transgenic mouse line64 carrying the human ELP1 that had the FD major splice mutation with the mouse line heterozygous for the Ikbkapflox allele (Ikbkapflox/+)65 to obtain mice carrying both the TgFD9 transgenic and Ikbkapflox alleles (TgFD9;Ikbkapflox/+). Then, TgFD9;Ikbkapflox/+ mice were crossed with a mouse line heterozygous for the Ikbkapflox allele (Ikbkapflox/+) to obtain TgFD9;Ikbkapflox/flox mice. Finally, TgFD9;Ikbkapflox/flox male mice were crossed with female mice heterozygous for the IkbkapΔ20 allele (IkbkapΔ20/+).65 The expected Mendelian ratio of the TgFD9;IkbkapΔ20/flox mouse according to this breeding scheme was 1 in 4 (25%). However, because only about 60% of TgFD9;IkbkapΔ20/flox mice survive postnatally,62 the actual ratio was 1:8 (38/305; 12.5%).
The mice used for this study were housed in the animal facility at MGH, provided with access to food and water ad libitum, and maintained on a 12-hour light/dark cycle. All experimental protocols were approved by the MGH institutional animal care and use committee and were in accordance with National Institutes of Health (NIH) guidelines.
For the routine genotyping of progeny, genomic DNA was prepared from tail biopsies, and PCR was carried out with the forward and reverse primers 5′-TGATTGACACAGACTCTGGCCA-3′ and 5′-CTTTCACTCTGAAATTACAGGAAG-3′ to discriminate the Ikbkap alleles and the primers forward 5′-GCCATTGTACTGTTTGCGACT-3′ and reverse, 5′-TGAGTGTCACGATTCTTTCTGC-3′ to detect the TgFD9 transgene.
Behavioral Assessment in Mice
Male control and TgFD9;IkbkapΔ20/flox mice were evaluated in normal light conditions. On the day of the testing, the mice were transported in their home cages from the colony room to the behavioral testing room and allowed to acclimate to the experimental room for at least one hour.
Rotarod
Motor coordination was assessed through the use of an accelerating rotarod (Ugo Basile) and standard techniques.66 The day before the session, each animal was trained in three consecutive trials, during which the speed of the rod changed from 2 to 40 rpm (revolutions per minute) over a 5 min ramp duration with a 45 min resting interval. The day of the session, each animal was tested three times with the same acceleration scheme. The latency to fall and the terminal speed (in rpm) were recorded, and any mouse remaining on the rod for more than 5 min was removed and returned to the cage. These trials resulted in a maximum latency-to-fall value of 300 s and a speed of 40 rpm. Data from the training trial were not included in the analysis.
Open Field
Open field testing was performed on naive mice.66 Each mouse was placed in the center of a 27 × 27 cm2 Plexiglas arena, and the activity in the horizontal and vertical dimensions was recorded by the Activity Monitor program (Med Associates). The quantitative analysis of locomotor activity was measured as the total distance traveled during the first 5 min in the arena.
Measurement of the Cobb Angle
Computed tomographic (CT) images were taken on the Siemen’s Inveon system with 360 projections over 360 degrees by using an 80 kVp 500 uA X-ray tube on a 125 mm detector while the mice were under 1.5% to 2.5% isoflurane gas anesthesia. The images were reconstructed into 110 μm isotropic voxels (512 × 512 × 768 matrix) by a modified Feldkamp reconstruction algorithm (COBRA, Exxim Computer Corporation). A blinded observer evaluated the severity of the spinal deformities by using the Osirix image processing software program. The magnitude of the curvature was measured on posteroanterior radiographs by determining the Cobb angle (θ), which is the angle derived from the positions of the most-tilted vertebrae above and below the apex of the curvature.67 This angle is formed by the intersection of two lines plotted at the end vertebrae of the curve deformity. One line is parallel to the endplate of the superior end vertebra and the other is parallel to the endplate of the inferior end vertebra.62, 67
Immunohistochemistry
After euthanasia, L3 DRGs were dissected and fixed in 4% paraformaldehyde (PFA) overnight; afterward, the DRGs were washed for 24 h in PBS. The DRGs were then incubated in 30% sucrose overnight, mounted in OCT compound, and stored at −80°C. 16 μm serial cryosections that spanned the whole ganglia were performed. Proprioceptive neurons were labeled with parvalbumin (PV; Synaptic System, guinea pig, 1:200) and whole sensory neurons were labeled with fluorescent Nissl staining (NeuroTrace, Molecular Probes, 1:200). We calculated the volume of the DRG by using ImageJ to measure, in every section, the area that was occupied by neuronal cell bodies and then multiplying the area of each section by its thickness (16 μm) to find the section volume. The sum of all the section volumes provided the DRG volume, expressed in mm.3, 62 We counted the number of total proprioceptive neurons per DRG by counting the number of proprioceptive neurons in every other section and then multiplying the average by the number of sections of each DRG.62
RNA Isolation and RT-PCR Analysis of Wild-Type and Mutant ELP1 Transcripts in Mouse Tissues
The mice were euthanized, and brain, DRG, liver, lung, kidney, and heart tissues were removed and snap frozen in liquid nitrogen. The tissues were homogenized in ice-cold TRI reagent (Molecular Research Center) with a TissueLyser (QIAGEN). Total RNA was extracted via the TRI reagent procedure provided by the manufacturer. The yield, purity, and quality of the total RNA for each sample were determined with a Nanodrop ND-1000 spectrophotometer. Reverse transcription was performed with 1 μg of total RNA, Random Primers (Promega), and Superscript III reverse transcriptase (Invitrogen) according to the manufacturers’ protocols.
For RT-PCR, the cDNA equivalent of 100 ng of starting RNA in a 30 μL reaction was used with the GoTaq green master mix (Promega) and 30 amplification cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 30 s). The human-specific ELP1 forward and reverse primers 5′-CCTGAGCAG CAATCATGTG−3′ and 5′-TACATGGTCTTCGTGACATC-3′ were used for amplification of human ELP1 isoforms expressed from the transgene. The PCR products were separated on 1.5% agarose gels and stained with ethidium bromide. The relative amounts of wild-type (WT) and mutant (Δ20)-ELP1-spliced isoforms in a single PCR were determined with ImageJ, and the integrated density value for each band was determined as previously described.59, 64 The relative proportion of the WT isoform detected in a sample was calculated as a percentage.
Meso-Scale Discovery Immunoassay for ELP1 Quantification in Mouse Tissues
Tissue samples were collected in Safe-Lock tubes (Eppendorf), snap frozen in liquid nitrogen, weighed, and homogenized on the TissueLyzer II (QIAGEN) in radioimmunoprecipitation assay (RIPA) buffer (Tris-HCl 50 mM [pH 7.4]; NaCl 150 mM; NP-40 1%; sodium deoxycholate 0.5%; and SDS 0.1%) containing a cocktail of protease inhibitors (Roche) at a tissue weight-to-RIPA-buffer volume of 50 mg/mL. The samples were then centrifuged for 20 min at 14,000 × g in a microcentrifuge. The homogenates were transferred to a 96-well plate and were diluted in RIPA buffer to ∼1 mg/mL for ELP1-meso-scale discovery (MSD) and ∼0.5 mg/mL for total protein measurement with the bicinchoninic acid (BCA) protein assay (Pierce). The samples were run in duplicate and averaged. The MSD sandwich immunoassay was performed according to the manufacturer’s (Meso Scale Diagnostics) protocol. 25 μL of the diluted tissue homogenates were transferred to a 96-well standard MSD plate coated with 0.5 μg/mL of capture antibody (rabbit monoclonal anti-IKAP antibody from Abcam, #ab179437) in PBS and incubated overnight at 4°C. 0.5 μg/mL of primary detection antibody, mouse anti-IKAP (33) from Santa Cruz Biotechnology #SC-136412, was incubated for 2–3 h at room temperature. 0.5 μg/mL of Sulfo-Tag antibody, Goat anti-mouse from MSD #R32AC-1, was incubated for 1 h at room temperature. The plates were read with Sector Imager S600 (Meso Scale Diagnostics). The amount of ELP1 in the tissues from kinetin-treated mice was normalized to the ELP1 amount in the control tissues and plotted as n-fold change over controls.
RNA-Seq Experiment
Six different human fibroblast cell lines from individuals with FD were obtained from Coriell Institute (Table S1) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) with 10% fetal bovine serum (FBS; Sigma). We counted and plated the cells in order to achieve semi-confluence after 8 days. 24 h after plating, the medium was changed, and the cells were treated with kinetin or DMSO until these reached a final concentration of 200 μM and 0.5%, respectively. DMSO was used as vehicle, and the concentration of kinetin was chosen on the basis of our previous studies because at this concentration kinetin induces robust splicing changes. After seven days of treatment, cells were collected, and RNA was extracted via the TRI reagent procedure provided by the manufacturer. The Genomics and Technology Core (GTC) of MGH prepared RNA-seq libraries by using a strand-specific deoxyuridine triphosphate (dUTP) method.68 In brief, the RNA sample quality (based on the RNA integrity number, or RIN) and quantity were determined with the Agilent 2200 TapeStation, and between 100–1000 ng of total RNA were used for library preparation. Each RNA sample was spiked with 1 μl of diluted (1:100) External RNA Controls Consortium (ERCC) RNA Spike-In Mix (4456740, Thermo Fisher Scientific), alternating between mix 1 and mix 2 for each well in the batch. The samples were then enriched for mRNA via polyA capture and then subjected to stranded reverse transcription and chemical shearing to make appropriate stranded-cDNA inserts. The libraries were finished by adding Y-adapters with sample specific barcodes and then between 10–15 rounds of PCR amplification. The libraries were evaluated for final concentration and size distribution by Agilent 2200 TapeStation and/or qPCR with the Library Quantification Kit (KK4854, Kapa Biosystems), and for multiplexing, equimolar amounts of each library were pooled prior to sequencing. The pooled libraries were paired-end sequenced on the Illumina HiSeq 2500, which produced 50 bp paired reads. Real-time image analysis and base calling were performed on the HiSeq 2500 instrument with the HiSeq Sequencing Control Software (HCS), and FASTQ files were demultipled with CASAVA software version 1.8.
In order to estimate gene expression levels, we mapped paired-end RNA-seq reads to human transcriptome Ensembl GRCh37 version 75 by STAR v2.5.3a, allowing unique mapping and up to 5% mismatching (reference: PMID 23104886). To assess exon splicing changes, we first defined exon triplet structure as any three consecutive exons along each transcript annotated by Ensembl GRCh37 version 75. We then evaluated the ψ levels of the middle exon for each triplet, as described by Katz et al.69 To estimate the treatment effect on splicing, we applied a generalized linear model (GLM) on the expression of splice junctions of each triplet.69
Statistical Analysis
The analysis of the rotarod data was carried out with a generalized linear mixed model (GLMM), in which treatment and genotype were considered fixed effects and the Wald test was applied to test the significance of corresponding contrasts. For the open field data, we applied the Welch test was to assess the statistical differences between two groups. To determine the statistical differences between two groups in all the remaining analyses, we performed an unpaired Student’s t test in GraphPad Prism 7 software. When one group was compared to more than one other group, we corrected for multiple comparisons by applying the false-discovery rate (FDR) correction, and we report the FDR-adjusted p values. For differential gene expression and splicing analyses of the RNA-seq data, we used a GLM on gene counts or junction counts, respectively. We used the Wald test to estimate the raw significance of gene expression and ψ changes. We considered differentially expressed genes to be all the genes that met an FDR <0.05 and exhibited an absolute ≥2-fold change (log2 ≥1 or ≤−1). We considered significant every splicing change with ψ ≥0.2 or ≤−0.2 and FDR <0.05. For all experiments, a criterion α level was set at 0.05.
Results
Daily Consumption of Kinetin Improves Kyphosis and Motor Coordination in FD Mice
To assess the therapeutic efficacy of increasing ELP1 by splice correction, we administered kinetin orally to the phenotypic FD mouse model TgFD9;IkbkapΔ20/flox.62 Special chow was formulated so that each mouse received 200 mg/kg/day. At birth, pups were randomly assigned to vehicle- or kinetin-treated groups and were maintained in the same treatment regime for the entire trial. We started the treatment at birth in order to maximize the therapeutic value, and a preliminary study to assess correction of splicing in transgenic pups showed that kinetin can pass from dam to pups through lactation and increase the full-length ELP1 mRNA amounts in the pups (Figure S1). The treatment was well tolerated, and no weight loss or reduction in the ratio of pups with FD was observed in the kinetin-treated group (Figure S2).
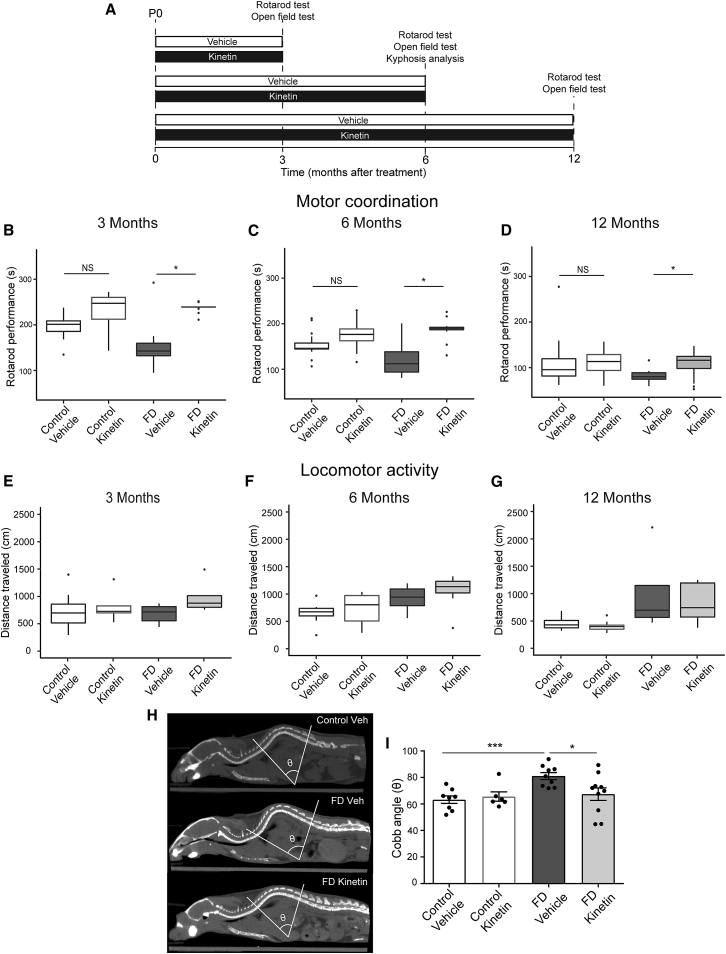
To evaluate disease progression as well as the treatment effect over time, we evaluated independent cohorts at 3, 6, and 12 months of age (Figure 1A). Because proprioceptive gait ataxia is a major morbidity in FD46, 51 and is evident in our mice,62 we measured the effect of treatment on motor coordination. Motor coordination has traditionally been assessed in mice with the rotarod test.70, 71, 72 In this analysis, increased latency to fall is indicative of better coordination.70, 71 Our results show that the kinetin-treated FD mice exhibit improvement in motor coordination at all time points tested (Figures 1B–1D). For each time point, the kinetin-treated FD mice remained on the accelerating rotarod for a significantly longer period than did the vehicle-treated FD mice. No treatment effect on coordination was observed in kinetin-treated control mice (Figures 1B–1D). As expected, we see an age-dependent decline in rotarod performance in all groups. In order to confirm that the difference in time spent on the accelerating rotarod between the treatment groups was not due to differential locomotion, we used the open field test to perform a standard evaluation of locomotor activity.73, 74 No differences were detected between vehicle-treated control and vehicle-treated FD mice or between vehicle-treated and kinetin-treated FD mice (Figures 1E–1G). These data suggest that the locomotor function of the FD mice is normal, as it is in individuals with FD,34, 37 and that the improvement in rotarod performance observed in kinetin-treated FD mice is most likely due to an improvement in coordination.
Figure 1.
Daily Consumption of Kinetin Improves Kyphosis and Motor Coordination in a Phenotypic Mouse Model of FD
(A) An experimental timeline that was used for assessing the therapeutic effect of chronic kinetin administration on disease phenotype. P0 = postnatal day 0.
(B) Quantification of motor coordination via an accelerating rotarod test at 3 months of age in vehicle-treated (n = 6) and kinetin-treated (n = 5) control mice and vehicle-treated (n = 6) and kinetin-treated (n = 5) familial dysautonomia (FD) mice. The median for each group is shown.
(C) Motor coordination assessed at 6 months of age in vehicle-treated (n = 7) and kinetin-treated (n = 8) control mice and vehicle-treated (n = 4) and kinetin-treated (n = 5) FD mice. The median for each group is shown.
(D) Motor coordination assessed at 12 months of age in vehicle-treated (n = 7) and kinetin-treated (n = 13) control mice and vehicle-treated (n = 5) and kinetin-treated (n = 9) FD mice. The median for each group is shown. In the box-and-whisker plots in B–D, for each box, the central mark shows the median, the edges of the box represent the 25th and 75th percentiles, and the whiskers extend to the most extreme data points. There was a ∗ p < 0.05 difference between vehicle-treated and kinetin-treated FD mice, but no significant difference was detected between vehicle-treated and kinetin-treated control mice (p = 0.4), determined with the Wald test.
(E) Quantification of locomotor activity as assessed by an open field test at 3 months of age in vehicle-treated (n = 8) and kinetin-treated (n = 5) control mice and in vehicle-treated (n = 7) and kinetin-treated (n = 5) FD mice. The median for each group is shown.
(F) Locomotor activity assessed at 6 months of age in vehicle-treated (n = 8) and kinetin-treated (n = 8) control mice and vehicle-treated (n = 6) and kinetin-treated (n = 7) FD mice. The median for each group is shown.
(G) Locomotor activity assessed at 12 months of age in vehicle-treated (n = 5) and kinetin-treated (n = 8) control mice and vehicle-treated (n = 4) and kinetin-treated (n = 5) FD mice. The median for each group is shown. No significant differences were detected with a two-tailed, unpaired Student’s t test.
(H) A quantification of spinal deformity in 6-month-old mice. Representative coronal CT scans and the correspondent Cobb angle are shown in vehicle-treated control mice (top) and in vehicle-treated (center) and kinetin- treated (bottom) FD mice.
(I) Cobb angle measurements in vehicle-treated (n = 8) and kinetin-treated (n = 6) control mice and vehicle-treated (n = 9) and kinetin-treated (n = 9) FD mice. Means and SEM are shown, and each data point represents an individual animal. There was a ∗∗∗ p < 0.001 difference between vehicle-treated control and vehicle-treated FD mice and a ∗ p < 0.05 difference between vehicle-treated and kinetin-treated FD mice determined via a two-tailed, unpaired Student’s t test with false discovery rate (FDR) correction. NS = not significant.
By the age of 10 years, 52% of persons with FD have scoliosis, and 21% have kyphosis. By the age of 20, 83% of them have spinal deformities.44, 53, 54, 55 To quantitatively assess the effect of kinetin treatment on spinal deformities, we performed CT to measure the magnitude of the spinal curvature via the Cobb angle.62, 67 By 6 months of age, FD mice develop severe kyphosis, confirmed by an increase in the Cobb angle (Figures 1H and 1I). Remarkably, kinetin treatment prevented the onset of these skeletal abnormalities (Figures 1H and 1I). The Cobb angle in kinetin-treated FD mice was lower than in the vehicle-treated FD mice and was comparable with that of their control littermates (Figure 1I). Taken together, these results show that kinetin prevented kyphosis and significantly improved motor coordination in our phenotypic FD mouse model, strongly suggesting that increasing production of full length ELP1 early in post-natal life can improve proprioception.
Kinetin Treatment Rescues Proprioceptive Sensory Loss in the FD Mice
In individuals with FD, fetal development and postnatal maintenance of DRG neurons is highly compromised, resulting in DRGs of grossly reduced size and significantly reduced neuronal number.35, 75 Within the DRGs, proprioceptors are the subpopulation of neurons responsible for sensory-motor coordination and posture maintenance.76, 77
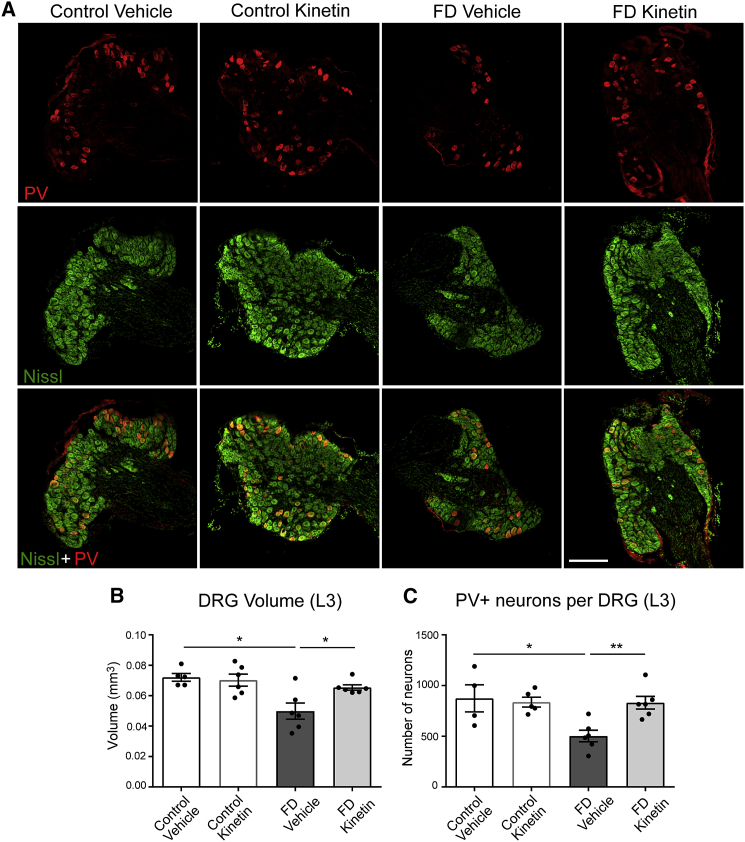
To confirm that our observed kinetin-mediated phenotypic improvement is correlated with changes in the neuropathological hallmarks of disease, we evaluated the volume of the DRGs, as well as the number of proprioceptive afferent neurons. Consistent with the observed proprioceptive deficits, FD mice showed a significant reduction in the volume of the DRG and in the number of proprioceptive neurons when compared with their control littermates (Figure 2A). The DRG volume in the FD mouse was 66% of the controls (Figure 2B) and the number of proprioceptive neurons was reduced to 57% compared with the number in their control littermates (Figure 2C). Notably, the treatment was able to rescue both neuropathological aspects of the disease. Kinetin-treated FD mice showed a normal number of proprioceptive neurons and a normal DRG volume (Figures 2B and 2C), demonstrating that starting the treatment at birth is sufficient to prevent the loss of this neuron subpopulation that plays a critical role in disease progression.
Figure 2.
Kinetin Treatment Rescues Proprioceptive Sensory Loss in the FD Mice
(A) Representative confocal images of proprioceptive (PV+) neurons (red), whole sensory (Nissl+) neurons (green), and the merged image (bottom) in L3 dorsal root ganglia (DRGs) from vehicle-treated and kinetin-treated control mice and vehicle-treated and kinetin-treated familial dysautonomia (FD) mice at 6 months of age. The scale bar represents 200 μm.
(B) The total volume of the L3 DRGs measured in vehicle-treated (n = 5) and kinetin-treated (n = 6) control mice and vehicle-treated (n = 6) and kinetin-treated (n = 6) FD mice at 6 months of age. There was a ∗ p < 0.05 difference between vehicle-treated control and vehicle-treated FD mice and a ∗ p < 0.05 difference between vehicle-treated and kinetin-treated FD mice determined by a two-tailed, unpaired Student’s t test with false discovery rate (FDR) correction.
(C) The total number of PV+ proprioceptive neurons per DRG counted in vehicle-treated (n = 4) and kinetin-treated (n = 5) control mice and vehicle-treated (n = 6) and kinetin-treated (n = 6) FD mice at 6 months of age. There was a ∗ p < 0.05 difference between vehicle-treated control and vehicle-treated FD mice and a ∗∗ p < 0.01 difference between vehicle-treated and kinetin-treated FD mice determined by a two-tailed, unpaired Student’s t test with FDR correction. In (B) and (C), the means and SEM are shown, and each data point represents an individual animal.
Kinetin Treatment Increases Full-Length ELP1 Transcript and Protein Amounts in the PNS
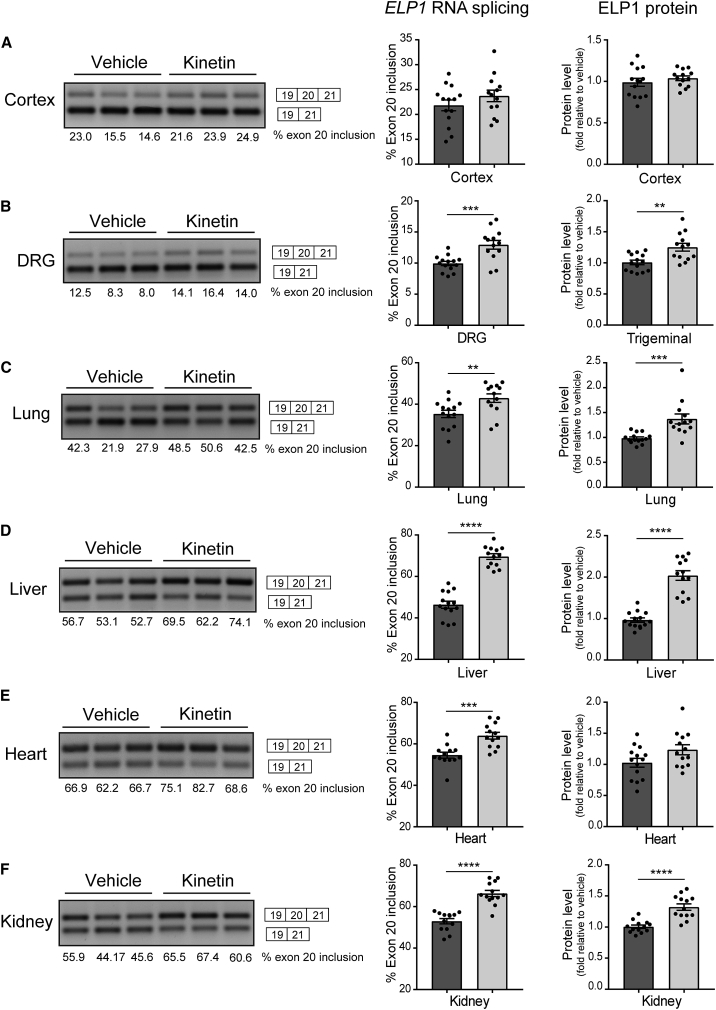
Next, we confirmed that the phenotypic and neuropathological improvement observed in the treated FD mice correlated with the correction of the underlying FD splicing defect. ELP1 splicing and protein amounts were analyzed in different tissues from vehicle- and kinetin-treated FD mice. As shown in Figure 3, kinetin treatment significantly increases ELP1 exon 20 inclusion in DRG (Figure 3B), lung (Figure 3C), liver (Figure 3D), heart (Figure 3E) and kidney tissues (Figure 3F). Importantly, the improvement of exon 20 inclusion in the ELP1 transcript results in higher protein production (Figures 3A–F). Because of the very limited amount of material available in the DRGs, the amount of ELP1 for the PNS was assessed in the trigeminal ganglia. The treatment effect on ELP1 splicing and protein amounts reflected kinetin distribution in the different tissues, and the lack of splicing correction in the cortex is consistent with the low amounts of kinetin present in the brain (Figure S3). Together, these results provide the in vivo evidence that kinetin increases the amount of ELP1 in the PNS, thereby rescuing a primary neurologic FD phenotype.
Figure 3.
Kinetin Treatment Increases Full-Length ELP1 Transcript and Protein Amounts in Several Tissues, Including in the Peripheral Nervous System, of the Phenotypic FD Mouse Model
A representative splicing analysis of human ELP1 transcripts (left), the percentage of exon 20 inclusion (middle), and amounts of ELP1 (right) from vehicle-treated (n = 13–14, dark gray) and kinetin-treated (n = 13, light gray) familial dysautonomia (FD) mice at 6 months of age in cortex (A), peripheral nervous system (PNS) (B), lung (C), liver (D), heart (E), and kidney tissues (F).
(A) In cortex tissue, no significant differences were detected in the percentage of exon 20 inclusion (p = 0.24) and the amounts of ELP1 (p = 0.39) between vehicle-treated and kinetin-treated FD mice, determined by a two-tailed, unpaired Student’s t test.
(B) In PNS tissue, dorsal root ganglia (DRGs) were used for splicing analysis, and trigeminal ganglia were used for protein quantification. There was a ∗∗∗ p < 0.001 difference in the percentage of exon 20 inclusion and a ∗∗ p < 0.01 difference in amounts of ELP1 determined by a two-tailed, unpaired Student’s t test.
(C) In lung tissue, there was a ∗∗ p < 0.01 difference in the percentage of exon 20 inclusion and a ∗∗∗ p < 0.001 difference in amounts of ELP1 determined by a two-tailed, unpaired Student’s t test.
(D) In liver tissue, there was a ∗∗∗∗ p < 0.0001 difference in the percentage of exon 20 inclusion and a ∗∗∗∗ p < 0.0001 difference in amounts of ELP1 determined by a two-tailed, unpaired Student’s t test.
(E) In heart tissue, there was a ∗∗∗ p < 0.001 difference in the percentage of exon 20 inclusion, but no significant difference detected in amounts of ELP1 (p = 0.06) determined by a two-tailed, unpaired Student’s t test.
(F) In kidney tissue, there was a ∗∗∗∗ p < 0.0001 difference in the percentage of exon 20 inclusion and a ∗∗∗∗ p < 0.0001 difference in amounts of ELP1 determined by a two-tailed, unpaired Student’s t test. The means and SEM are shown, and each data point represents an individual animal.
Kinetin Demonstrates High Selectivity when Assessed by RNA-Sequencing
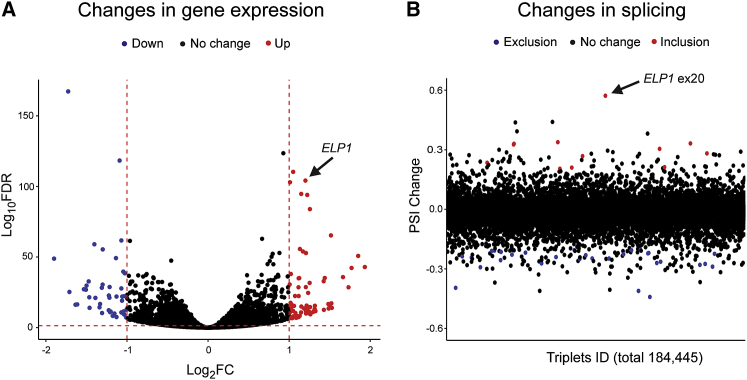
Because our ultimate goal is to move a splice-modulating therapy to the clinic, we evaluated kinetin splicing selectivity in human cell lines. We performed transcriptome profiling to compare gene expression changes as well as mRNA splicing alterations in six FD human fibroblast cell lines treated with kinetin or vehicle (DMSO) for 7 days (Table S1). We specifically chose fibroblast lines because they are the primary choice for assessing the splicing selectivity of small molecules.78, 79 Our evaluation of the gene-expression analysis conservatively restricted interpretation to genes that met an FDR <0.05 and exhibited an absolute ≥2-fold change (log2 ≥1 or ≤−1). Similar to previously reported splicing modulator compounds,78, 79, 80 kinetin treatment had minimal effect transcriptome-wide; only 118 of 18,156 (0.006%) genes were differentially expressed (DE) (Figure 4A and Table S2). We determined splicing alterations by considering the three junctions in triplets of consecutive exons and calculating the ψ levels of the middle exon, then we considered splicing changes at ψ ≥0.2 or ≤–0.2 and FDR <0.05 (Figure 4B).81, 82 As a proof-of-principle, we observed a ψ change of 0.57 for ELP1 exon 20 splicing after kinetin treatment. We detected 42 additional exon-usage differences in response to kinetin; 11 of these differences promoted inclusion of the middle exon, and 31 induced skipping of the middle exon. Given the low frequency of splice changes after treatment (0.0002%), combined with the modest changes observed in ψ when compared to the magnitude of ELP1 exon 20 inclusion post-treatment (Figure 4B and Table S3), we concluded that kinetin exhibits selective splicing modulation activity.
Figure 4.
Kinetin Demonstrates High Selectivity as Assessed by RNA-Sequencing
(A) A volcano plot showing changes in gene expression after treatment with kinetin in fibroblasts from individuals with familial dysautonomia (FD). Each dot represents one of 18,156 expressed genes in FD fibroblasts. The x axis represents log2-fold change (log2 transformed, Log2FC) of gene expression after treatment, and the y axis represents the false discovery rate (FDR) (log10 transformed, Log10FDR). The horizontal dashed line indicates the FDR = 0.05. The two vertical dashed lines indicate a 0.5-fold change and a 2-fold change, respectively. The red and blue dots represent genes with significant changes, an FDR < 0.05, and ≥2-fold changes (red) or ≤0.5-fold changes (blue), respectively.
(B) Percent-spliced-in (PSI, ψ) changes in exon triplets of kinetin-treated versus vehicle-treated fibroblasts. Each dot represents one of 184,445 expressed exon triplets. The y axis represents ψ changes. The red and blue dots represent exon triplets with significant changes, an FDR < 0.05, and ψ changes ≥ 0.2 (red) or ψ changes ≤ −0.2 (blue). See also Tables S1 and S2. See Material and Methods for details of the statistical analysis.
Discussion
FD is a devastating neurodegenerative disease, and targeted therapeutics are desperately needed. The nature of the mutation that causes FD provides a platform for the development of disease-modifying therapies that directly target the underlying genetic mechanism. The goal of such a therapy would be to halt the progressive peripheral neurodegeneration that characterizes FD—this would revolutionize patient care. Although previous studies have shown that kinetin can improve ELP1 splicing in vitro,58, 61 in vivo,59 and even in individuals with FD,60 the current study proves that postnatal ELP1 splicing correction can rescue neurological phenotypes and provides the critical proof of in vivo efficacy needed to move a splice-modulating therapy toward the clinic.
Because FD is caused by a mutation that results in aberrant splicing, small-molecule splicing modulators that specifically promote splicing correction and thereby increase the amount of full-length mRNA offer an excellent therapeutic alternative for individuals with FD. First, because FD is characterized by a complex neurological phenotype that involves multiple organs, therapies must act systemically and reach all tissues. Second, all persons with FD possess at least one copy of the major FD splice mutation, and 99.5% of them are homozygous for this mutation,8, 83 thus the development of targeted splicing therapies will benefit all individuals with FD. Third, the recent progress in the development of therapies for spinal muscular atrophy (SMA), another genetic disorder caused by a splicing alteration, has validated the utility of splicing modification as a valuable therapeutic strategy for neurologic diseases.84, 85
The loss of proprioceptors in FD accounts for many debilitating disease aspects, including gait ataxia, spinal and craniofacial deformities, and respiratory insufficiency due to neuromuscular incoordination. As individuals with FD age, progressive impairment in proprioception leads to severe gait ataxia46, 51 and worsening skeletal deformities.44, 45, 53, 54, 55 Here, we show that increasing ELP1 amounts starting at birth not only improved motor coordination but also prevented the onset of the spinal abnormalities in a phenotypically accurate FD mouse model. Kinetin-treated FD mice showed increased DRG volume and increased number of proprioceptive neurons, indicating that increasing ELP1 expression at birth is sufficient to prevent the loss of this subpopulation of neurons that play a critical role in disease progression. Finally, we confirm that the phenotypic improvement promoted by kinetin correlates with a significant increase of full-length ELP1 mRNA and protein in multiple tissues, including in the PNS, and that its activity on splicing is highly specific. Together, our results provide the critical proof of principle that increasing full-length ELP1 RNA through splicing modulation can halt the progressive neurodegeneration that characterizes this devastating disease.
Importantly, several promising therapeutic strategies that target the splice defect and increase ELP1 amounts have recently been reported for FD; these strategies include small molecules,17, 58 antisense oligonucleotides,86 and exon-specific U1 small nuclear RNAs (snRNAs).87 None, however, have demonstrated phenotypic efficacy. Our demonstration that increasing ELP1 amounts postnatally rescues proprioceptive neurons and improves phenotype clearly validates the therapeutic value of all splice-modulating therapies for FD and highlights the need for rapid translation of these targeted, disease-modifying treatments to the clinic.
Declaration of Interests
Susan A. Slaugenhaupt and Chien-Ping Ko receive research support from PTC Therapeutics to cover, in part, the data collection and analysis described in this study.
Susan A. Slaugenhaupt is an inventor on a patent entitled “Method for altering mRNA splicing and treating familial dysautonomia and other mechanistically related disorders” Patent # US9265766B2, and she is a paid consultant to PTC Therapeutics.
Jana Narasimhan, Vijayalakshmi Gabbeta, Amal Dakka, Jean Hedrick, Xin Zhao, Marla Weetall, and Nikolai A. Naryshkin are employees of PTC Therapeutics, a biotechnology company. In connection with such employment, the author receives salary, benefits, and stock-based compensation, including stock options, restricted stock, other stock-related grants, and the right to purchase discounted stock through PTC’s employee stock purchase plan. The other authors declare no competing interests.
Acknowledgments
We thank Dr. Lucy Norcliffe-Kaufmann and Dr. Horacio Kaufmann of the Dysautonomia Treatment and Evaluation Center at New York University Medical School for their long-standing collaboration and helpful discussions. We are also grateful to Dr. David Schoenfeld for his assistance in designing our animal trial and to Dr. Frances Lefcort for her comments on the manuscript. This work was supported by National Institutes of Health (NIH) grants (R37NS095640 to S.A.S.), by the Dysautonomia Foundation (to S.A.S and I.D.), and by PTC Therapeutics (to S.A.S. and C.P.K.). E.M. was the recipient of a T32 Training Grant in Genetics sponsored by NIH (5T32GM7748-37).
Published: March 21st, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.02.009.
Accession Numbers
The raw RNA-seq sequence data were deposited into NCBI GEO database and can be accessed with the accession number GEO: GSE126155 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE126155).
Web Resources
Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Axelrod F.B. Familial dysautonomia: A review of the current pharmacological treatments. Expert Opin. Pharmacother. 2005;6:561–567. doi: 10.1517/14656566.6.4.561. [DOI] [PubMed] [Google Scholar]
- 2.Axelrod F.B., Nachtigal R., Dancis J. Familial dysautonomia: Diagnosis, pathogenesis and management. Adv. Pediatr. 1974;21:75–96. [PubMed] [Google Scholar]
- 3.Pearson J. Familial dysautonomia (a brief review) J. Auton. Nerv. Syst. 1979;1:119–126. doi: 10.1016/0165-1838(79)90010-9. [DOI] [PubMed] [Google Scholar]
- 4.Pearson J., Pytel B. Quantitative studies of ciliary and sphenopalatine ganglia in familial dysautonomia. J. Neurol. Sci. 1978;39:123–130. doi: 10.1016/0022-510x(78)90193-4. [DOI] [PubMed] [Google Scholar]
- 5.Slaugenhaupt S.A. Genetics of familial dysautonomia. Tissue-specific expression of a splicing mutation in the IKBKAP gene. Clin. Auton. Res. 2002;12(Suppl 1):I15–I19. doi: 10.1007/s102860200015. [DOI] [PubMed] [Google Scholar]
- 6.Slaugenhaupt S.A., Gusella J.F. Familial dysautonomia. Curr. Opin. Genet. Dev. 2002;12:307–311. doi: 10.1016/s0959-437x(02)00303-9. [DOI] [PubMed] [Google Scholar]
- 7.Cuajungco M.P., Leyne M., Mull J., Gill S.P., Lu W., Zagzag D., Axelrod F.B., Maayan C., Gusella J.F., Slaugenhaupt S.A. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am. J. Hum. Genet. 2003;72:749–758. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkes N.A., Otero G., Winkler G.S., Marshall N., Dahmus M.E., Krappmann D., Scheidereit C., Thomas C.L., Schiavo G., Erdjument-Bromage H. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 10.Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Takagi Y., Jiang Y., Tokunaga M., Erdjument-Bromage H., Tempst P., Kornberg R.D. A multiprotein complex that interacts with RNA polymerase II elongator. J. Biol. Chem. 2001;276:29628–29631. doi: 10.1074/jbc.C100274200. [DOI] [PubMed] [Google Scholar]
- 12.Kim J.H., Lane W.S., Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pokholok D.K., Hannett N.M., Young R.A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 14.Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 15.Svejstrup J.Q. Elongator complex: How many roles does it play? Curr. Opin. Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Esberg A., Huang B., Johansson M.J., Byström A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida M., Kataoka N., Miyauchi K., Ohe K., Iida K., Yoshida S., Nojima T., Okuno Y., Onogi H., Usui T. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl. Acad. Sci. USA. 2015;112:2764–2769. doi: 10.1073/pnas.1415525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goffena J., Lefcort F., Zhang Y., Lehrmann E., Chaverra M., Felig J., Walters J., Buksch R., Becker K.G., George L. Elongator and codon bias regulate protein levels in mammalian peripheral neurons. Nat. Commun. 2018;9:889. doi: 10.1038/s41467-018-03221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsborn T., Tükenmez H., Chen C., Byström A.S. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm(5)s(2)U in tRNA. Biochem. Biophys. Res. Commun. 2014;454:441–445. doi: 10.1016/j.bbrc.2014.10.116. [DOI] [PubMed] [Google Scholar]
- 20.Rahl P.B., Chen C.Z., Collins R.N. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Johansen L.D., Naumanen T., Knudsen A., Westerlund N., Gromova I., Junttila M., Nielsen C., Bøttzauw T., Tolkovsky A., Westermarck J. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 2008;121:854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 22.Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V. Transcription impairment and cell migration defects in elongator-depleted cells: Implication for familial dysautonomia. Mol. Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Tourtellotte W.G. Axon transport and neuropathy: Relevant perspectives on the etiopathogenesis of familial dysautonomia. Am. J. Pathol. 2016;186:489–499. doi: 10.1016/j.ajpath.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naftelberg S., Abramovitch Z., Gluska S., Yannai S., Joshi Y., Donyo M., Ben-Yaakov K., Gradus T., Zonszain J., Farhy C. Phosphatidylserine ameliorates neurodegenerative symptoms and enhances axonal transport in a mouse model of familial dysautonomia. PLoS Genet. 2016;12:e1006486. doi: 10.1371/journal.pgen.1006486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson M.Z., Gruner K.A., Qin C., Tourtellotte W.G. A neuron autonomous role for the familial dysautonomia gene ELP1 in sympathetic and sensory target tissue innervation. Development. 2014;141:2452–2461. doi: 10.1242/dev.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George L., Chaverra M., Wolfe L., Thorne J., Close-Davis M., Eibs A., Riojas V., Grindeland A., Orr M., Carlson G.A., Lefcort F. Familial dysautonomia model reveals Ikbkap deletion causes apoptosis of Pax3+ progenitors and peripheral neurons. Proc. Natl. Acad. Sci. USA. 2013;110:18698–18703. doi: 10.1073/pnas.1308596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abashidze A., Gold V., Anavi Y., Greenspan H., Weil M. Involvement of IKAP in peripheral target innervation and in specific JNK and NGF signaling in developing PNS neurons. PLoS ONE. 2014;9:e113428. doi: 10.1371/journal.pone.0113428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunnicutt B.J., Chaverra M., George L., Lefcort F. IKAP/Elp1 is required in vivo for neurogenesis and neuronal survival, but not for neural crest migration. PLoS ONE. 2012;7:e32050. doi: 10.1371/journal.pone.0032050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee G., Studer L. Modelling familial dysautonomia in human induced pluripotent stem cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2286–2296. doi: 10.1098/rstb.2011.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeltner N., Fattahi F., Dubois N.C., Saurat N., Lafaille F., Shang L., Zimmer B., Tchieu J., Soliman M.A., Lee G. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 2016;22:1421–1427. doi: 10.1038/nm.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunt P.W., McKusick V.A. Familial dysautonomia. A report of genetic and clinical studies, with a review of the literature. Medicine (Baltimore) 1970;49:343–374. [PubMed] [Google Scholar]
- 33.Lehavi O., Aizenstein O., Bercovich D., Pavzner D., Shomrat R., Orr-Urtreger A., Yaron Y. Screening for familial dysautonomia in Israel: Evidence for higher carrier rate among Polish Ashkenazi Jews. Genet. Test. 2003;7:139–142. doi: 10.1089/109065703322146830. [DOI] [PubMed] [Google Scholar]
- 34.Norcliffe-Kaufmann L., Slaugenhaupt S.A., Kaufmann H. Familial dysautonomia: History, genotype, phenotype and translational research. Prog. Neurobiol. 2017;152:131–148. doi: 10.1016/j.pneurobio.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Pearson J., Pytel B.A., Grover-Johnson N., Axelrod F., Dancis J. Quantitative studies of dorsal root ganglia and neuropathologic observations on spinal cords in familial dysautonomia. J. Neurol. Sci. 1978;35:77–92. doi: 10.1016/0022-510x(78)90103-x. [DOI] [PubMed] [Google Scholar]
- 36.Geltzer A.I., Gluck L., Talner N.S., Polesky H.F. Familial dysautonomia; Studies in a newborn infant. N. Engl. J. Med. 1964;271:436–440. doi: 10.1056/NEJM196408272710903. [DOI] [PubMed] [Google Scholar]
- 37.Gutiérrez J.V., Norcliffe-Kaufmann L., Kaufmann H. Brainstem reflexes in patients with familial dysautonomia. Clin. Neurophysiol. 2015;126:626–633. doi: 10.1016/j.clinph.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahloudji M., Brunt P.W., McKusick V.A. Clinical neurological aspects of familial dysautonomia. J. Neurol. Sci. 1970;11:383–395. doi: 10.1016/0022-510x(70)90083-3. [DOI] [PubMed] [Google Scholar]
- 39.Axelrod F.B. Familial dysautonomia. Muscle Nerve. 2004;29:352–363. doi: 10.1002/mus.10499. [DOI] [PubMed] [Google Scholar]
- 40.Hilz M.J., Kolodny E.H., Neuner I., Stemper B., Axelrod F.B. Highly abnormal thermotests in familial dysautonomia suggest increased cardiac autonomic risk. J. Neurol. Neurosurg. Psychiatry. 1998;65:338–343. doi: 10.1136/jnnp.65.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunt P.W. Unusual cause of Charcot joints in early adolescence (Riley-Day syndrome) BMJ. 1967;4:277–278. doi: 10.1136/bmj.4.5574.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendoza-Santiesteban C.E., Palma J.A., Hedges T.R., 3rd, Laver N.V., Farhat N., Norcliffe-Kaufmann L., Kaufmann H. Pathological confirmation of optic neuropathy in familial dysautonomia. J. Neuropathol. Exp. Neurol. 2017;76:238–244. doi: 10.1093/jnen/nlw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendoza-Santiesteban C.E., Hedges Iii T.R., Norcliffe-Kaufmann L., Axelrod F., Kaufmann H. Selective retinal ganglion cell loss in familial dysautonomia. J. Neurol. 2014;261:702–709. doi: 10.1007/s00415-014-7258-2. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan L., Margulies J.Y., Kadari A., Floman Y., Robin G.C. Aspects of spinal deformity in familial dysautonomia (Riley-Day syndrome) Eur. Spine J. 1997;6:33–38. doi: 10.1007/BF01676572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ford D.M., Bagnall K.M., Clements C.A., McFadden K.D. Muscle spindles in the paraspinal musculature of patients with adolescent idiopathic scoliosis. Spine. 1988;13:461–465. doi: 10.1097/00007632-198805000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Macefield V.G., Norcliffe-Kaufmann L., Gutiérrez J., Axelrod F.B., Kaufmann H. Can loss of muscle spindle afferents explain the ataxic gait in Riley-Day syndrome? Brain. 2011;134:3198–3208. doi: 10.1093/brain/awr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilz M.J., Axelrod F.B., Bickel A., Stemper B., Brys M., Wendelschafer-Crabb G., Kennedy W.R. Assessing function and pathology in familial dysautonomia: Assessment of temperature perception, sweating and cutaneous innervation. Brain. 2004;127:2090–2098. doi: 10.1093/brain/awh235. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann H., Biaggioni I. Autonomic failure in neurodegenerative disorders. Semin. Neurol. 2003;23:351–363. doi: 10.1055/s-2004-817719. [DOI] [PubMed] [Google Scholar]
- 49.Norcliffe-Kaufmann L., Axelrod F., Kaufmann H. Afferent baroreflex failure in familial dysautonomia. Neurology. 2010;75:1904–1911. doi: 10.1212/WNL.0b013e3181feb283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Norcliffe-Kaufmann L., Kaufmann H. Familial dysautonomia (Riley-Day syndrome): When baroreceptor feedback fails. Auton. Neurosci. 2012;172:26–30. doi: 10.1016/j.autneu.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Macefield V.G., Norcliffe-Kaufmann L.J., Axelrod F.B., Kaufmann H. Relationship between proprioception at the knee joint and gait ataxia in HSAN III. Mov. Disord. 2013;28:823–827. doi: 10.1002/mds.25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mass E., Brin I., Belostoky L., Maayan C., Gadoth N. A cephalometric evaluation of craniofacial morphology in familial dysautonomia. Cleft Palate Craniofac. J. 1998;35:120–126. doi: 10.1597/1545-1569_1998_035_0120_aceocm_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 53.Hayek S., Laplaza F.J., Axelrod F.B., Burke S.W. Spinal deformity in familial dysautonomia. Prevalence, and results of bracing. J. Bone Joint Surg. Am. 2000;82-A:1558–1562. [PubMed] [Google Scholar]
- 54.Albanese S.A., Bobechko W.P. Spine deformity in familial dysautonomia (Riley-Day syndrome) J. Pediatr. Orthop. 1987;7:179–183. doi: 10.1097/01241398-198703000-00013. [DOI] [PubMed] [Google Scholar]
- 55.Hensinger R.N., MacEwen G.D. Spinal deformity associated with heritable neurological conditions: Spinal muscular atrophy, Friedreich’s ataxia, familial dysautonomia, and Charcot-Marie-Tooth disease. J. Bone Joint Surg. Am. 1976;58:13–24. [PubMed] [Google Scholar]
- 56.Palma J.A., Norcliffe-Kaufmann L., Fuente-Mora C., Percival L., Mendoza-Santiesteban C., Kaufmann H. Current treatments in familial dysautonomia. Expert Opin. Pharmacother. 2014;15:2653–2671. doi: 10.1517/14656566.2014.970530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heemskerk J., Tobin A.J., Bain L.J. Teaching old drugs new tricks. Meeting of the Neurodegeneration Drug Screening Consortium, 7-8 April 2002, Washington, DC, USA. Trends Neurosci. 2002;25:494–496. doi: 10.1016/s0166-2236(02)02236-1. [DOI] [PubMed] [Google Scholar]
- 58.Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004;13:429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 59.Shetty R.S., Gallagher C.S., Chen Y.T., Hims M.M., Mull J., Leyne M., Pickel J., Kwok D., Slaugenhaupt S.A. Specific correction of a splice defect in brain by nutritional supplementation. Hum. Mol. Genet. 2011;20:4093–4101. doi: 10.1093/hmg/ddr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Axelrod F.B., Liebes L., Gold-Von Simson G., Mendoza S., Mull J., Leyne M., Norcliffe-Kaufmann L., Kaufmann H., Slaugenhaupt S.A. Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr. Res. 2011;70:480–483. doi: 10.1203/PDR.0b013e31822e1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hims M.M., Ibrahim E.C., Leyne M., Mull J., Liu L., Lazaro C., Shetty R.S., Gill S., Gusella J.F., Reed R., Slaugenhaupt S.A. Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J. Mol. Med. (Berl.) 2007;85:149–161. doi: 10.1007/s00109-006-0137-2. [DOI] [PubMed] [Google Scholar]
- 62.Morini E., Dietrich P., Salani M., Downs H.M., Wojtkiewicz G.R., Alli S., Brenner A., Nilbratt M., LeClair J.W., Oaklander A.L. Sensory and autonomic deficits in a new humanized mouse model of familial dysautonomia. Hum. Mol. Genet. 2016;25:1116–1128. doi: 10.1093/hmg/ddv634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landis S.C., Amara S.G., Asadullah K., Austin C.P., Blumenstein R., Bradley E.W., Crystal R.G., Darnell R.B., Ferrante R.J., Fillit H. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490:187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hims M.M., Shetty R.S., Pickel J., Mull J., Leyne M., Liu L., Gusella J.F., Slaugenhaupt S.A. A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics. 2007;90:389–396. doi: 10.1016/j.ygeno.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dietrich P., Alli S., Shanmugasundaram R., Dragatsis I. IKAP expression levels modulate disease severity in a mouse model of familial dysautonomia. Hum. Mol. Genet. 2012;21:5078–5090. doi: 10.1093/hmg/dds354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menalled L., El-Khodor B.F., Patry M., Suárez-Fariñas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol. Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kubota K., Doi T., Murata M., Kobayakawa K., Matsumoto Y., Harimaya K., Shiba K., Hashizume M., Iwamoto Y., Okada S. Disturbance of rib cage development causes progressive thoracic scoliosis: The creation of a nonsurgical structural scoliosis model in mice. J. Bone Joint Surg. Am. 2013;95:e130. doi: 10.2106/JBJS.L.01381. [DOI] [PubMed] [Google Scholar]
- 68.Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira Fernandes M., Tourtellotte W.G. Egr3-dependent muscle spindle stretch receptor intrafusal muscle fiber differentiation and fusimotor innervation homeostasis. J. Neurosci. 2015;35:5566–5578. doi: 10.1523/JNEUROSCI.0241-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.d’Ydewalle C., Krishnan J., Chiheb D.M., Van Damme P., Irobi J., Kozikowski A.P., Vanden Berghe P., Timmerman V., Robberecht W., Van Den Bosch L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat. Med. 2011;17:968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- 72.Deacon R.M. Measuring motor coordination in mice. J. Vis. Exp. 2013;75:e2609. doi: 10.3791/2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takemiya T., Takeuchi C. Traveled distance is a sensitive and accurate marker of motor dysfunction in a mouse model of multiple sclerosis. ISRN Neurosci. 2013;2013:170316. doi: 10.1155/2013/170316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osmon K.J., Vyas M., Woodley E., Thompson P., Walia J.S. Battery of behavioral tests assessing general locomotion, muscular strength, and coordination in mice. J. Vis. Exp. 2018;(131) doi: 10.3791/55491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Axelrod F.B., Iyer K., Fish I., Pearson J., Sein M.E., Spielholz N. Progressive sensory loss in familial dysautonomia. Pediatrics. 1981;67:517–522. [PubMed] [Google Scholar]
- 76.Akay T., Tourtellotte W.G., Arber S., Jessell T.M. Degradation of mouse locomotor pattern in the absence of proprioceptive sensory feedback. Proc. Natl. Acad. Sci. USA. 2014;111:16877–16882. doi: 10.1073/pnas.1419045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takakusaki K. Functional neuroanatomy for posture and gait control. J. Mov. Disord. 2017;10:1–17. doi: 10.14802/jmd.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Naryshkin N.A., Weetall M., Dakka A., Narasimhan J., Zhao X., Feng Z., Ling K.K., Karp G.M., Qi H., Woll M.G. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 79.Palacino J., Swalley S.E., Song C., Cheung A.K., Shu L., Zhang X., Van Hoosear M., Shin Y., Chin D.N., Keller C.G. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 80.Woll M.G., Naryshkin N.A., Karp G.M. Drugging Pre-mRNA Splicing. In: Therapeutics R.N.A., Garner A.L., editors. Topics in Medicinal Chemistry. Springer International Publishing; Cham: 2018. pp. 135–176. [Google Scholar]
- 81.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakuma M., Iida K., Hagiwara M. Deciphering targeting rules of splicing modulator compounds: Case of TG003. BMC Mol. Biol. 2015;16:16. doi: 10.1186/s12867-015-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leyne M., Mull J., Gill S.P., Cuajungco M.P., Oddoux C., Blumenfeld A., Maayan C., Gusella J.F., Axelrod F.B., Slaugenhaupt S.A. Identification of the first non-Jewish mutation in familial dysautonomia. Am. J. Med. Genet. A. 2003;118A:305–308. doi: 10.1002/ajmg.a.20052. [DOI] [PubMed] [Google Scholar]
- 84.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinha R., Kim Y.J., Nomakuchi T., Sahashi K., Hua Y., Rigo F., Bennett C.F., Krainer A.R. Antisense oligonucleotides correct the familial dysautonomia splicing defect in IKBKAP transgenic mice. Nucleic Acids Res. 2018;46:4833–4844. doi: 10.1093/nar/gky249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donadon I., Pinotti M., Rajkowska K., Pianigiani G., Barbon E., Morini E., Motaln H., Rogelj B., Mingozzi F., Slaugenhaupt S.A., Pagani F. Exon-specific U1 snRNAs improve ELP1 exon 20 definition and rescue ELP1 protein expression in a familial dysautonomia mouse model. Hum. Mol. Genet. 2018;27:2466–2476. doi: 10.1093/hmg/ddy151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.