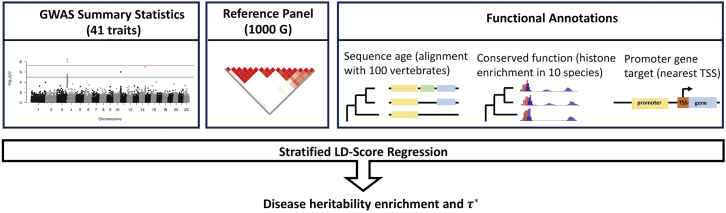
Abstract
Regulatory elements, e.g., enhancers and promoters, have been widely reported to be enriched for disease and complex trait heritability. We investigated how this enrichment varies with the age of the underlying genome sequence, the conservation of regulatory function across species, and the target gene of the regulatory element. We estimated heritability enrichment by applying stratified LD score regression to summary statistics from 41 independent diseases and complex traits (average N = 320K) and meta-analyzing results across traits. Enrichment of human putative enhancers and promoters was larger in elements with older sequence age, assessed via alignment with other species irrespective of conserved functionality: putative enhancer elements with ancient sequence age (older than the split between marsupial and placental mammals) were 8.8× enriched (versus 2.5× for all putative enhancers; p = 3e−14), and promoter elements with ancient sequence age were 13.5× enriched (versus 5.1× for all promoters; p = 5e−16). Enrichment of human putative enhancers and promoters was also larger in elements whose regulatory function was conserved across species, e.g., human putative enhancers that were enhancers in ≥5 of 9 other mammals were 4.6× enriched (p = 5e−12 versus all putative enhancers). Enrichment of human promoters was larger in promoters of loss-of-function intolerant genes: 12.0× enrichment (p = 8e−15 versus all promoters). The mean value of several measures of negative selection within these genomic annotations mirrored all of these findings. Notably, the annotations with these excess heritability enrichments were jointly significant conditional on each other and on our baseline-LD model, which includes a broad set of coding, conserved, regulatory, and LD-related annotations.
Keywords: enhancer, promoter, regulatory elements, heritability, genetic architecture
Introduction
Disease-associated variants and disease heritability have been widely reported to be concentrated in regulatory annotations, such as enhancers and promoters.1, 2, 3, 4, 5, 6, 7 These findings have motivated recent studies of how enhancers and promoters evolve across species.8, 9, 10, 11 Vierstra et al.8 analyzed DNase I hypersensitivity sites (DHSs) in humans and mice and reported that both human-specific DHSs and human DHSs that were conserved in mice were significantly enriched for disease- and trait-associated variants, despite decreased constraint within human-specific DHSs. This implies that both human-specific regulatory elements and regulatory elements that are shared across species are important for disease; however, these analyses were restricted to only one species other than humans, and do not elucidate the relative importance of these two types of regulatory elements. Villar et al.9 analyzed 20 mammalian species and reported that enhancers evolve more rapidly than promoters, and that enhancers were often species specific whereas promoters were often functionally conserved. Vermunt et al.10 and Trizzino et al.11 analyzed 3–6 primate species and reported that regulatory elements were generally functionally conserved across primates, with higher sequence and function conservation for promoters than for enhancers. However, which enhancers and promoters are most important for disease remains largely unknown. Further investigating which enhancers and promoters are most important for disease would improve our biological understanding of disease architectures.
Here, we characterize the contribution of enhancers and promoters to disease heritability based on sequence age, conserved function across species, and gene function of the target gene. We achieve this by constructing new annotations using enhancers and promoters previously identified in liver tissue using ten high-quality genomes (humans and nine other mammalian species9) and applying stratified LD score regression with the baseline-LD model6, 7 to summary association statistics from 41 independent diseases and complex traits (average N = 320K). An overview of the data sources used in our analyses is provided in Figure 1. We find that disease heritability enrichment is concentrated in putative enhancers and promoters with ancient sequence age and conserved function across species, as well as promoters of loss-of-function intolerant genes from the Exome Aggregation Consortium (ExAC).12 The mean value of several measures of negative selection within these genomic annotations mirrored all of these findings, with larger heritability enrichments for annotations under stronger negative selection. Our findings are consistent with previous studies broadly demonstrating that regions under strong negative selection are enriched for disease heritability and disease-associated variants.6, 7, 13, 14, 15, 16, 17, 18, 19, 20
Figure 1.
Data Sources Used in Analyses
New functional annotations are constructed using a variety of previous research.9, 12, 21 By applying stratified LD score regression including both these annotations and the baseline-LD model6, 7 to summary association statistics from 41 independent diseases and complex traits (average N = 320K), we can determine the disease heritability enrichment and standardized effect size for annotations of interest.
Material and Methods
Putative Enhancer and Promoter Annotations
Our goal is to understand the role of human enhancers and promoters in the genetic architecture of diseases and complex traits. We first annotated regions as putative enhancers and promoters using previously reported enhancer and promoter regions that were enriched for histone marks (H3K27ac and H3K4me3) in at least two of four human liver tissue samples.9 The study identifying these regulatory elements reported that a “sizable majority” of identified enhancers are regulatorily active (based on results of further experimental assays) and that most regions enriched for H3K4me3 (annotated as promoters) lie near transcription start sites.9 However, we conservatively refer to these enhancer regions as “putative enhancers.” We merged any overlapping annotations, resulting in putative enhancer and promoter regions with mean segment lengths of 3.4 kb and 4.3 kb, respectively. In total, 3.3% of common variants lie within putative enhancers and 1.5% within promoters (Table 1). Correlations between the putative enhancer and promoter annotations and various subsets of these annotations (described below) are reported in Figure S1 and Table S1.
Table 1.
Annotations Analyzed in Main Analyses
| Annotation |
Prop. |
Prop. Putative |
Mean Segment |
|---|---|---|---|
| SNPs | enhancer/promoter | length (BP) | |
| Putative enhancer | 0.0332 | 3,362 | |
| Promoter | 0.0152 | 4,308 | |
| Ancient putative enhancer | 0.0052 | 0.1557 | 191 |
| Ancient promoter | 0.0042 | 0.2765 | 159 |
| Conserved putative enhancer | 0.0055 | 0.1649 | 4,962 |
| Conserved promoter | 0.0080 | 0.5251 | 4,502 |
| Promoter of ExAC gene | 0.0025 | 0.1640 | 4,549 |
We report the proportion of common SNPs (MAF ≥ 0.05) and mean segment length in base pairs (BP) for each annotation. For the count annotations (putative enhancer conservation count and promoter conservation count), we report here the corresponding binary annotations, conserved putative enhancer and conserved promoter. Mean segment length is computed after merging overlapping elements. Main annotations are publicly available (see Web Resources).
Sequence Age Annotations
We constructed genomic annotations based on a previous study which classified sequence age through genome-wide alignments of 100 vertebrates.21 That study annotated each region of the human genome with an associated score between 1 and 19 based on the number of key ancestral nodes in the tree of vertebrates that it aligned to (1st root = human; 19th root = vertebrates); younger regions were assigned smaller scores, whereas older regions were assigned larger scores. Most regions were assigned a precise age (one score), but some regions were assigned an age interval (range of scores) or an inconsistent age. Regions with inconsistent age were removed.
We annotated SNPs in putative enhancer and promoter regions according to the age of the sequence in the corresponding region of the genome (start location ≤ SNP location < end location). We removed regions in which the alignment at the 19th root was uncertain and assigned the maximum sequence age for SNPs in regions with an age interval. We categorized the ages as post-eutheria split (1–11; young), eutheria (12; intermediate), and pre-eutheria split (13–19; ancient), with approximately one third of SNPs in putative enhancers and promoters falling in each of these age bins. Pre-eutheria split (or ancient sequence age) means the sequence has an age older than the split between placental and marsupial mammals (>160 million years old22, 23).
We also analyzed 24 putative regulatory annotations from the baseline model6 (Table S2). We intersected these annotations with the ancient sequence age annotation, resulting in 24 putative regulatory annotations that have ancient sequence age. Furthermore, we analyzed two chromatin marks (H3K4me3 and H3K27ac) directly measured in various tissues and cell types and defined annotations based on these marks being present in at least 1,10, or 20 tissues/cell types;5 we also intersected these annotations with the ancient sequence age annotation.
Conserved Function Annotations
We annotated human putative enhancers and promoters according to their conserved function, based on previous work specifying for each human element whether the element was functionally conserved (sequence aligned with histone mark signal conserved across species), mapped (sequence aligned), or missing in analyses of nine other mammalian species (with high-quality genomes).9 Human-specific and highly conserved putative enhancers and promoters were defined as elements with conserved function in 0 or 9 of the 9 mammals, respectively. We denote conserved putative enhancers and promoters as elements with conserved function in at least 5 of the 9 mammals.
We constructed 6 categorical annotations (3 for promoters and 3 for putative enhancers): each putative enhancer and promoter was annotated with the conservation count (CC) in other species (CC = 0,1,…9; both align and have functional conservation), the mapped count in other species (0,1,…9; align but no functional conservation) and the missing count in other species (0,1,…9). We introduced 20 binary annotations (10 for promoters and 10 for putative enhancers) reflecting the 10 possible values of CC (0,1,…9).
We computed the conservation count, mapped count, and missing count of all elements (prior to merging) and then merged information across overlapping elements. For elements that overlapped, for each of conserved, mapped, and missing count, we computed the union of each count across species; this implies that these small proportions of the genome where two or more elements overlap could get conservation, mapped, and missing counts that add up to more than 9.
Gene Function Annotations
To assess how the target gene may impact the role of a promoter in disease architecture, we annotated promoters based on whether they were a promoter of an ancient gene (P1–P10 genes24, 25, 26 which emerged before the vertebrates split 500 million years ago27), a loss-of-function intolerant gene from ExAC (ExAC gene;12 3,230 such genes; Table 1), or a gene with a mouse ortholog (identified in hg38; we assume that gene names remain consistent across builds; see Web Resources).28
We obtained the coordinates of all TSSs (see Web Resources) and associated genes.29 We calculated the mid-point of each merged promoter and determined whether the closest transcription start site (TSS) within 5 kb to the midpoint corresponded to a gene in the specified gene set.
Heritability Enrichment and Standardized Effect Size Metrics
In order to estimate the heritability enrichment of an annotation, we ran stratified LD score regression (S-LDSC)6, 7 using 1000 Genomes as the LD reference panel.30 Consider C binary or continuous-valued annotations , denote the annotation value of SNP j for annotation c, and assume that the variance of per-normalized-genotype effect sizes linearly depends on the C annotations: , where is the per-SNP contribution of one unit of the annotation c to heritability (jointly modeled with all other annotations). S-LDSC estimates using the summary statistic for a SNP j via the following equation:
| (Equation 1) |
where N is the sample size of the GWAS, b quantifies confounding biases,31 and is the LD score of SNP j to annotation c where rjk is the correlation between SNPs j and k. S-LDSC estimates two metrics quantifying the role of a functional region in diseases and complex traits. First, it estimates the heritability enrichment of binary annotations, defined as the proportion of heritability explained by SNPs in the annotation divided by the proportion of SNPs in the annotation. The enrichment of annotation c is estimated as
| (Equation 2) |
where h2(c) is the heritability causally explained by common SNPs in annotation c, h2 is the heritability causally explained by common SNPs, is the number of common SNPs that lie in the annotation, and M is the number common SNPs (in our analyses M = 5,961,159 SNPs, see below). A value greater than 1 would indicate a functional annotation is enriched for trait heritability or the proportion of heritability explained is greater than one would expect given the size of the annotation.
Standardized effect size was previously defined7 as the proportionate change in per-SNP heritability associated with a one standard deviation increase in the value of the annotation, conditional on the other annotations in the model; quantifies effects that are unique to the focal annotation, unlike heritability enrichment.6, 7, 32 In detail,
| (Equation 3) |
where sdc is the standard deviation of annotation c.
Regression SNPs (the SNPs used by S-LDSC to estimate from marginal association statistics) were obtained from the HapMap Project phase 3; these SNPs are considered to be well-imputed SNPs. SNPs with marginal association statistics larger than 80 or 0.001N and SNPs that are in the major histocompatibility complex (MHC) region were excluded from all analyses. Reference SNPs (the SNPs used by S-LDSC to compute LD scores) were defined as the set of 9,997,231 biallelic SNPs with minor allele count greater than or equal to five in the set of 489 unrelated and outbred European samples33 from phase 3 of 1000 Genomes Project (1000G).30 We note that regression SNPs tag potentially causal reference SNPs via LD scores computed using reference SNPs.6, 7 Heritability SNPs (the SNPs used by S-LDSC to compute h2, h2(c), and sdc) were defined as the 5,961,159 common variants (MAF 0.05) in the set of reference SNPs. Using the LD score for each annotation and the marginal statistics obtained from the trait phenotypes, we computed the heritability enrichment and for each annotation.
In all analyses we included the putative enhancer and promoter annotations9 as well as a broad set of 75 functional annotations from the baseline-LD (v1.1) model,7, 32 which include functional annotations from the baseline model (i.e., coding, intron, DHS, …), plus 10 MAF bins and 6 LD-related annotations (Table S2). We note that the inclusion of MAF- and LD-related annotations implies that the expected causal heritability of a SNP is a function of MAF and LD. The 75 functional annotations from the baseline-LD model are included in each analysis to account for LD-dependent architectures and to minimize the risk of model misspecification, which could bias estimates.6, 7 The 75 annotations do not all produce conditionally statistically significant signals, in part due to correlations between annotations that can compromise conditional statistical significance. However, including all of these annotations minimizes the risk of model misspecification when analyzing new annotations. We meta-analyzed results across a previously described set32 of 41 independent diseases and complex traits (average N = 320K, computed using largest dataset for each trait); for six traits we analyzed two datasets (genetic correlation > 0.9), leading to a total of 47 datasets analyzed (Table S3; see Web Resources). We performed random-effects meta-analyses across traits using the R package rmeta and the function meta.summaries() (consistent with Finucane et al.6). All models tested, including the annotations considered in each model, are listed in Table S4. Correlations between our annotations and annotations from the baseline-LD model are reported in Figure S1 and Table S1.
Reported enrichment estimates are based on a random-effects meta-analysis of enrichment estimates for each trait. The p value for enrichment is computed using a random-effect meta-analysis of across the traits (we used this quantity because enrichment is not normally distributed6) and testing the null hypothesis that this difference is 0 by computing a z-score. For enhancer conservation count and promoter conservation count, we calculate enrichment for bins of this categorical annotation.7
For each new annotation, we ran S-LDSC conditional on the putative enhancer and promoter annotations as well as the baseline-LD model. For each annotation type (sequence age, conserved function, and gene function) we derived a joint model by running S-LDSC with the full set of annotations of that type conditional on the enhancer and promoter annotations and the baseline-LD model. (For the sequence age analysis, this set consisted of four annotations: young putative enhancer/promoter and ancient putative enhancer/promoter; we removed the intermediate putative enhancer/promoter to avoid linear dependence between the annotations. For the conserved function model, this set consisted of eight annotations: human-specific putative enhancer/promoter, highly conserved putative enhancer/promoter, putative enhancer/promoter CC, and putative enhancer/promoter missing count; we removed putative enhancer/promoter mapped count to avoid linear dependence between the annotations. For the gene function model, this set consisted of three annotations: promoter of an ancient gene, promoter of ExAC gene, and promoter of a gene with a mouse ortholog.) We then iteratively removed the least statistically significant annotation (excluding annotations in the baseline-LD model as well as the enhancer and promoter annotation) until each remaining annotation was significant (after correction for multiple testing).7 To produce a combined joint model, we combined the significant sequence age, conserved function, and gene function annotations into a single model and again iteratively removed the least statistically significant annotation until each annotation remained significant (after correction for multiple testing). We note that this model selection technique may result in inflated p values, analogous to winner’s curse.34 However, our assessment of conditional significance addresses this by correcting for the total number of annotations tested. For each model, we performed a secondary analysis in which we additionally included annotations defined by 500 bp flanking regions around each of the new annotations in the model; this helps to guard against bias due to model misspecification.6
In order to determine whether a subset of putative enhancers and promoters were particularly enriched as compared to all putative enhancers or promoters, we computed the enrichment difference between an annotation A and a subset a. For each trait, we computed the difference in enrichment between the annotations and the standard error for this difference (using block-jackknife) and then meta-analyzed results across 41 traits using random-effects meta-analysis. In order to compute a p value for the difference in enrichment, we computed the normally distributed quantity , as well as its standard error for each trait (using block-jackknife), and then meta-analyzed results across traits. We then tested the null hypothesis that this difference is 0; this test assesses whether the per-SNP heritability within annotation A is different within a than outside a. This test is a natural extension of the approach used to assess statistical significance of enrichment.6
We computed the proportion of enrichment for an annotation A, attributable to a subset a. The proportion of enrichment for A attributable to a is defined as
| (Equation 4) |
If , then the proportion of enrichment for A attributable to a is just the proportion of A in subset a. We computed this quantity for each trait, used block-jackknife to compute standard errors, and meta-analyzed results across 41 traits.
Negative Selection Metrics
It has been widely reported that although regions under strong negative selection are depleted of genetic variation, these regions are enriched for disease heritability and disease-associated variants.6, 7, 13, 14, 15, 16, 17, 18, 19, 20 For example, the 2.6% of SNPs lying in regions that are conserved across 29 mammals (spanning 4.2% of the genome) were reported to explain 24%–35% of disease and complex trait heritability.6, 7, 35
We quantified the strength of negative selection within these annotations by computing the mean value of several measures of negative selection and computing the standard error using block-jackknife with 200 equally sized blocks of adjacent SNPs within the annotations (all measures are annotations in the baseline-LD model; Table S2). First, we computed the proportion of common SNPs with GERP++ rejected substitutions (RS) score ≥4 (GERP RS ≥ 4, binary annotation) within the baseline-LD annotations.7, 36 This score is equal to the difference between the neutral and observed substitution rates and reflects the intensity of constraint at a given genomic location, such that a larger score is indicative of stronger negative selection. Second, we computed the mean background selection statistic (BSS = 1-McVicker B statistic37) (at common SNPs); a BSS value close to 1 indicates that background selection resulted in near complete removal of diversity whereas a value close to 0 indicates little effect.7 Third, we computed the proportion of common SNPs conserved across mammals;6, 35 regions that are conserved across mammals are likely to be critical, as mutations were not tolerated. Fourth, we computed the mean MAF-adjusted predicted allele age (at common SNPs); on average, recent variants are more deleterious.7, 38 Fifth, we computed mean nucleotide diversity39 (at common SNPs); variants that lie in regions with low nucleotide diversity are more likely to be deleterious.7, 40
Results
Disease Enrichment Is Concentrated in Putative Regulatory Elements with Ancient Sequence Age
We focused our analyses on putative enhancer and promoter elements that were previously annotated based on H3K27ac and H3K4me3 marks assayed in human liver9 (Table 1). To assess the disease enrichment of these elements, we applied S-LDSC with the baseline-LD model6, 7 to summary statistics from 41 independent diseases and complex traits (average N = 320K; Table S3) and meta-analyzed results across traits. We observed significant heritability enrichment for both putative enhancers (2.6×, p = 3e−12) and promoters (4.6×, p = 3e−17) (Table S5A), consistent with previous studies of disease enrichment of regulatory elements.1, 2, 3, 4, 5, 6, 7 Based on significance of regression coefficients, we determined that the promoter annotation (but not the putative enhancer annotation) provides unique information conditioned on the baseline-LD model (p = 0.007; Table S5A), which includes a broad set of regulatory annotations (Table S2). Analyses of highly reproducible putative enhancer and promoter annotations (reproduced in all four tissue samples from Villar et al.9) produced similar results (Table S5B).
We annotated putative enhancer and promoter regions according to their underlying sequence age, assessed via genome-wide alignment of 100 vertebrates irrespective of conserved functionality.21 Each region of the human genome had an associated score between 1 and 19 based on the number of key ancestral nodes in the tree of vertebrates that it aligned to. We classified enhancer and promoter regions as having a young (1–11), intermediate (12), or ancient (13–19) sequence age (see Material and Methods); different regions within the same enhancer or promoter may be assigned different sequence ages, such that ancient enhancers/promoters represent the ancient parts of the enhancers/promoters rather than different enhancers/promoters. Ancient sequence age means the sequence is older than the split between marsupial and placental mammals (>160 million years old22, 23); 16% of putative enhancer SNPs were annotated as ancient putative enhancer, and 28% of promoter SNPs were annotated as ancient promoter (Table 1). We computed correlations between our annotations and 38 annotations from the baseline-LD model (Table S2): 32 functional annotations and 6 LD-related annotations. The ancient putative enhancer and ancient promoter annotations were only weakly correlated with annotations from the baseline-LD model (Figure S1 and Table S1).
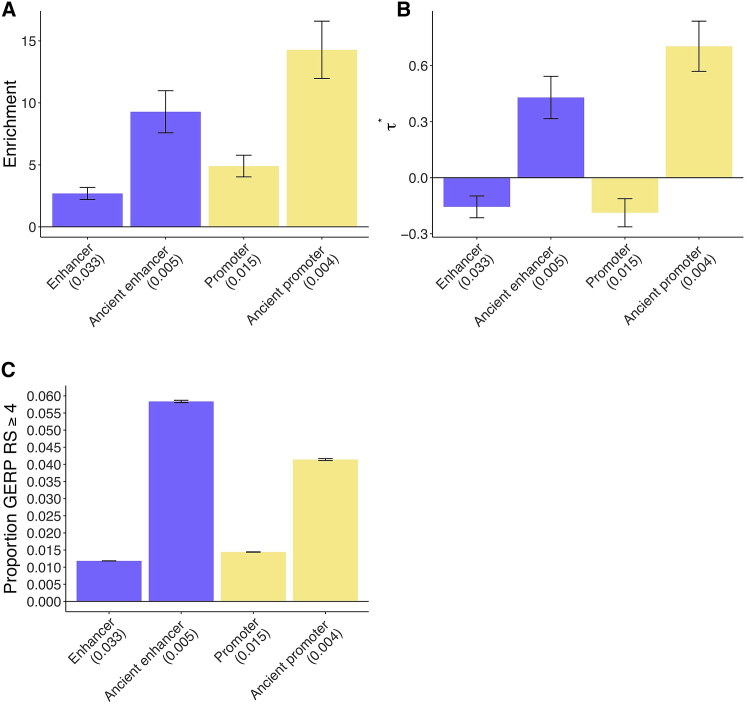
To assess how the disease enrichment of putative enhancers and promoters varies with sequence age, we repeated our S-LDSC analysis with each of the six age-specific annotations (young, intermediate, or ancient; putative enhancer or promoter) included in turn, in addition to baseline-LD + putative enhancer + promoter annotations. We observed the strongest enrichments for ancient putative enhancers and ancient promoters (Table S6). We constructed a joint sequence age model by retaining only the age-specific annotations that remained significant (after correction for multiple testing) when conditioned on the baseline-LD + putative enhancer + promoter annotations;7 only the ancient putative enhancer and ancient promoter annotations were jointly significant. Ancient putative enhancers were 9.3× enriched, compared to 2.7× for all putative enhancers (p = 4e−15 for difference), and ancient promoters were 14.3× enriched, compared to 4.9× for all promoters (p = 2e−18 for difference) (Figure 2A, Tables S7A and S8). We note that enrichment estimates - which differ from model to model - can change slightly depending on the set of annotations included in the model;6, 7 the enrichment estimates reported in the Abstract are estimates obtained using the combined joint model. Although ancient putative enhancers comprise only 16% of putative enhancers (at the level of common SNPs), they contribute 59% (SE 5%) of all putative enhancer enrichment. Analogously, although ancient promoters comprise only 28% of promoters, they contribute 82% (SE 4%) of all promoter enrichment.
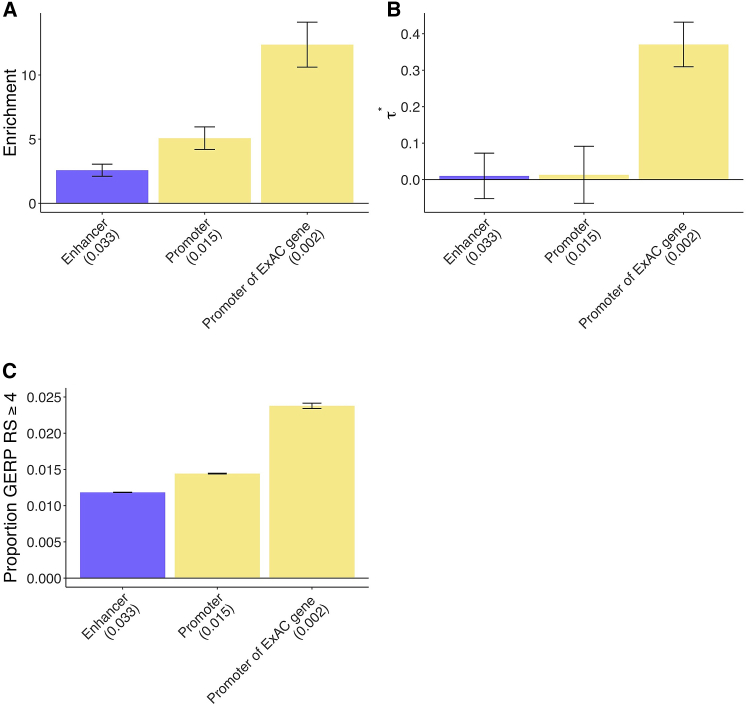
Figure 2.
Disease Enrichment of Ancient Enhancers and Ancient Promoters in Sequence Age Model
We report results for sequence age annotations that are jointly significant conditional on the baseline-LD model and putative enhancer and promoter annotations (Bonferroni p = 0.05/4 = 0.0125).
(A and B) Heritability enrichment (A) and estimates (±1.96 standard error) (B); results are meta-analyzed across 41 traits.
(C) Proportion of common SNPs within annotations with GERP RS ≥ 47, 36 (±1.96 standard error). We report the proportion of common SNPs (MAF ≥ 0.05) for each annotation. Numerical results are reported in Table S7, and results for each trait are reported in Table S8.
Both ancient putative enhancers and ancient promoters were uniquely informative for disease heritability conditional on the baseline-LD + putative enhancer + promoter annotations, as quantified by (the proportionate change in per-SNP heritability associated with an increase in the value of the annotation by one standard deviation, conditional on other annotations included in the model7) (Figure 2B and Table S7B). Specifically, we estimated large and highly significant values of for both ancient putative enhancers (, p = 1e−13) and ancient promoters (, p = 9e−25). In particular, these values were larger than the analogous values that we recently estimated for both LD-related annotations7 and molecular QTL annotations,32 implying a substantial improvement in our understanding of which regulatory elements contribute to disease heritability. The slightly but significantly negative value of for (all) putative enhancers and (all) promoters indicates conditional depletion for putative enhancers and promoters that do not have ancient sequence age (Figure 2B and Table S7B).
We quantified the mean strength of negative selection within each of the annotations from Figure 2A. We first calculated the proportion of common SNPs with GERP++ rejected substitutions (RS) score ≥ 4 (GERP RS ≥ 4).7, 36 The GERP RS score reflects the difference between the neutral and observed substitution rates and thus reflects the intensity of constraint at a given genomic location, such that a larger score is indicative of stronger negative selection. We determined that the stronger disease enrichment for ancient putative enhancers and ancient promoters is mirrored by the larger proportion of variants in these annotations with GERP RS ≥ 4, reflecting stronger negative selection (Figure 2C and Table S7C). We note that 1.2% and 1.4% of common SNPs in putative enhancer and promoter annotations (and 5.8% and 4.1% of common SNPs in ancient putative enhancer and promoter annotations) have GERP RS ≥ 4, as compared to 0.81% of all common SNPs, thus regulatory regions enter the regime of strong selection fairly frequently. We note that as sequence age is assessed via sequence conservation across species, we expected the GERP scores in ancient regions to be higher. However, we did not know in advance whether the quantitative pattern of enrichment would closely mirror the quantitative pattern of GERP++ scores. We observed similar patterns for four other measures of negative selection:7 a background selection statistic (BSS) equal to 1McVicker B statistic;37 sequence conservation across 29 mammals35; predicted allele age;7 and nucleotide diversity39 (Table S7C). However, as noted above, ancient putative enhancers and ancient promoters were uniquely informative for disease heritability conditional on the baseline-LD model, which includes all of these measures of negative selection.
We performed five secondary analyses to assess the robustness of our results. First, we repeated the analysis of Figures 2A and 2B restricting to three liver-related traits (high cholesterol, HDL, and LDL). Although this analysis is less well-powered, the conditional signal for ancient promoters remained statistically significant (p = 0.0003, Table S9). Second, we repeated the analysis of Figures 2A and 2B by adding a binary annotation defined by ancient sequence age (irrespective to enhancer or promoter status) to the model; this annotation was not conditionally informative for disease heritability as quantified by , and its addition to the model did not significantly change our results (Table S10). Third, we intersected the ancient sequence age annotation with 24 binary annotations from baseline-LD reflecting putative regulatory elements and ran S-LDSC conditional on the baseline-LD model with each of these intersected annotations included in turn. We obtained similar results, with much stronger enrichments for ancient regulatory elements (Table S11). (We used the putative enhancer and promoter annotations from Villar et al.9 in our main analyses so that we could integrate annotations based on ancient sequence age and conserved function into a combined joint model; see below.) Fourth, we repeated this analysis using H3K27ac and H3K4me3 annotations from Roadmap,5 defining annotations based on marks present in 1/10/20 tissues/cell types, respectively. As expected, we found that marks present in more tissues/cell types had higher disease enrichment (Table S12). However, we found that each of these annotations had ∼3× greater disease enrichment when restricted to regions of ancient sequence age. This shows that our finding of stronger disease enrichments for liver regulatory elements in regions of ancient sequence age is orthogonal to the number of tissues/cell types with regulatory signal. Fifth, we repeated the analysis of Figures 2A and 2B by including 500 bp flanking regions around each of the annotations from Figures 2A and 2B, to guard against bias due to model misspecification6 (see Material and Methods). We confirmed that this did not significantly change our results (Table S13).
Disease Enrichment Is Concentrated in Putative Regulatory Elements with Conserved Function
We annotated human putative enhancers and promoters according to their conserved function, assessed via how many of nine other mammalian species assayed by Villar et al.9 had shared regulatory functionality. Each putative enhancer and promoter was annotated with the conservation count (CC) in other species (CC = 0,1,…9). We constructed both integer-valued “conservation count” (value of CC) and binary “conserved” (CC ≥ 5) annotations (see Material and Methods and Table 1). A large proportion of annotated putative enhancers were functionally human specific (40% human-specific [CC = 0] versus 2% highly conserved [CC = 9]), whereas promoters were more functionally conserved (19% human-specific versus 15% highly conserved) (Table S17). Accordingly, 53% of promoters were conserved promoters, whereas only 16% of putative enhancers were conserved putative enhancers (Table 1). The putative enhancer conservation count and promoter conservation count annotations were only weakly correlated with annotations from the baseline-LD model, but moderately correlated with the ancient putative enhancer and ancient promoter annotations (Figure S1 and Table S1).
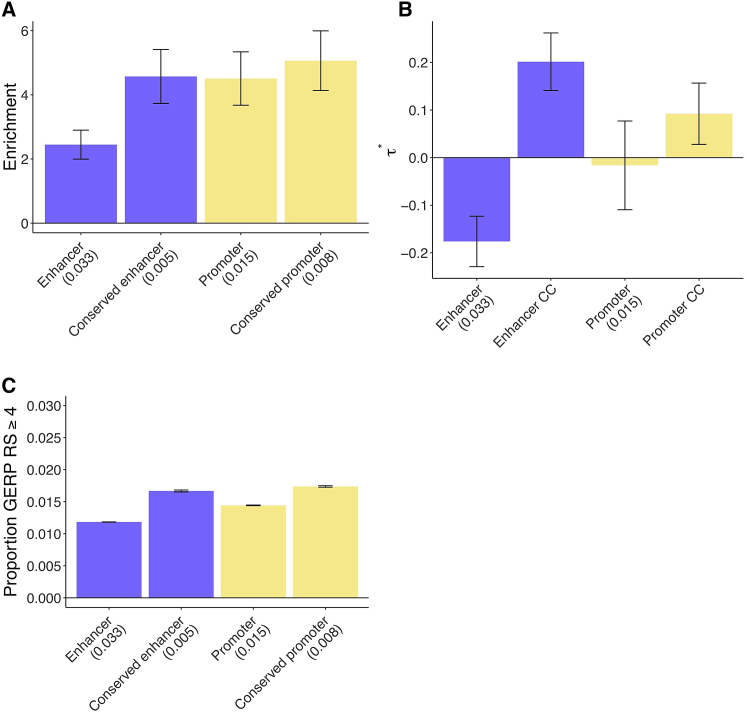
To assess how the disease enrichment of putative enhancers and promoters varies with conserved function, we performed S-LDSC analyses with each of ten conserved-function-specific annotations (conservation count, highly conserved, human-specific, mapped count, missing count [see Material and Methods]; putative enhancer or promoter) included in turn, in addition to baseline-LD + putative enhancer + promoter annotations. We observed the strongest enrichments for highly conserved putative enhancers and highly conserved promoters, and also observed that while human-specific promoters were enriched, human-specific putative enhancers were not (Table S14). We constructed a joint conserved function model by retaining only the conserved-function-specific annotations that remained significant (after correction for multiple testing) when conditioned on the baseline-LD + putative enhancer + promoter annotations;7 only the putative enhancer conservation count and promoter conservation count annotations were jointly significant. Because enrichment is not defined for annotations with value 0–9, we estimated the enrichment of the corresponding binary annotations (conserved putative enhancer and conserved promoter) in the joint model. Conserved putative enhancers were 4.6× enriched, compared to 2.4× for all putative enhancers (p = 3e−12 for difference), and conserved promoters were 5.1× enriched, compared to 4.5× for all promoters (p = 0.022 for difference) (Figure 3A, Tables S15A and S16). We note that enrichment estimates - which differ from model to model - can change slightly depending on the set of annotations included in the model;6, 7 the enrichment estimates reported in the Abstract are estimates obtained using the combined joint model. Although conserved putative enhancers comprise only 16% of putative enhancers, they contribute 35% (SE 2%) of all putative enhancer enrichment. Analogously, although conserved promoters comprise only 53% of promoters, they contribute 59% (SE 2%) of all promoter enrichment.
Figure 3.
Disease Enrichment of Conserved Enhancers and Conserved Promoters in Conserved Function Model
We report results for conserved function annotations that are jointly significant conditional on the baseline-LD model and putative enhancer and promoter annotations (Bonferroni p = 0.05/8 = 0.00625).
(A and B) Heritability enrichment (A) and estimates (±1.96 standard error) (B); results are meta-analyzed across 41 traits. CC denotes conservation count.
(C) Proportion of common SNPs within annotations with GERP RS ≥ 47, 36 (±1.96 standard error). We report the proportion of common SNPs (MAF ≥ 0.05) for each annotation. Numerical results are reported in Table S15, and results for each trait are reported in Table S16.
Both putative enhancer conservation count and promoter conservation count were uniquely informative for disease heritability conditional on the baseline-LD + putative enhancer + promoter annotations, as quantified by (Figure 3B and Table S15B). Specifically, we estimated significant values of for both putative enhancer conservation count (, p = 7e−11) and promoter conservation count (, p = 0.005). The significantly negative value of for (all) putative enhancers indicates conditional depletion for putative enhancers that are not conserved (Figure 3B, Table S15B).
We quantified the mean strength of negative selection within each of the annotations from Figure 3A. We first calculated the proportion of common SNPs with GERP RS ≥ 4.7, 36 We determined that the stronger disease enrichments for conserved putative enhancers and conserved promoters is mirrored by the larger proportion of variants in these annotations with GERP RS ≥ 4, reflecting stronger negative selection (Figure 3C and Table S15C). We observed similar patterns for four other measures of negative selection (Table S15C). However, as noted above, putative enhancer conservation count and promoter conservation count were uniquely informative for disease heritability conditional on the baseline-LD model, which includes all of these measures of negative selection.
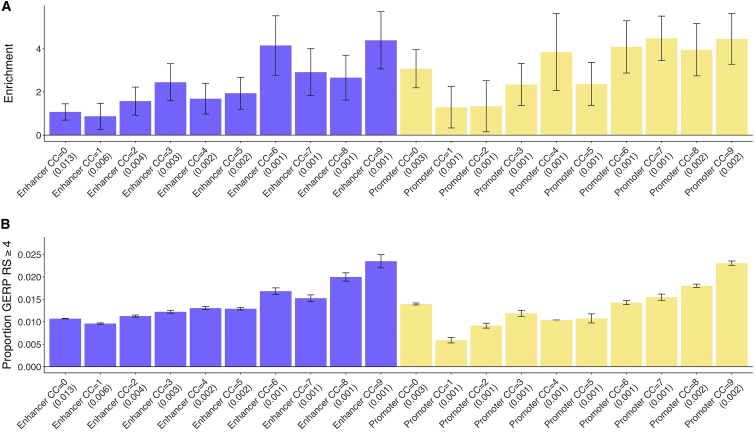
To further assess how the disease enrichment of putative enhancers and promoters varies with conserved function, we repeated our S-LDSC analysis with each of 20 binary conservation count annotations (CC = 0,1,…9; enhancer or promoter) jointly included, in addition to baseline-LD model (the putative enhancer and promoter annotations were excluded to avoid approximate colinearity of the annotations). For putative enhancers, we observed a roughly linear trend whereby putative enhancers conserved in more mammals are progressively more enriched for heritability (Figure 4A, Tables S17A and S18). For promoters, we observed a parabolic trend, similar to the linear trend but with excess heritability for human-specific promoters (Figure 4A and Table S17A).
Figure 4.
Disease Enrichment of Putative Enhancers and Promoters as a Function of Conservation Count (CC)
(A) Heritability enrichment (±1.96 standard error); results are meta-analyzed across 41 traits.
(B) Proportion of common SNPs within annotations with GERP RS ≥ 47, 36 (±1.96 standard error). We report the proportion of common SNPs (MAF ≥ 0.05) for each annotation. Numerical results are reported in Table S17, and results for each trait are reported in Table S18.
We quantified the mean strength of negative selection within each of the annotations from Figure 4A. We first calculated the proportion of common SNPs with GERP RS ≥ 4.7, 36 We determined that the linear disease enrichment trend for putative enhancers and parabolic disease enrichment trend for promoters (as conservation count increases) is mirrored by the proportion of variants in these annotations with GERP RS ≥ 4 (Figure 4B and Table S17B). We observed similar patterns for four other measures of negative selection (Table S17B).
We performed four secondary analyses. First, we repeated the analysis of Figure 3A restricting to the three liver-related traits. Although this analysis is less well powered, the conditional signal for putative enhancer conservation count remained statistically significant (p = 0.001, Table S19). Second, we repeated the analysis of Figure 3A by replacing the putative enhancer conservation count and promoter conservation count annotations in the joint model with binary conserved putative enhancer and conserved promoter annotations, and we confirmed that this did not significantly change our results (Table S20). Third, we repeated the analysis of Table S20 by including 500 bp flanking regions around each of the annotations from Figure 3A (see Material and Methods). This did not significantly change our results; the heritability enrichment for conserved putative enhancer was slightly reduced but remained highly significant (Table S21). Fourth, we repeated the analysis of Figure 3B by including human-specific promoters as an additional annotation. While this new annotation was not conditionally significant, the value of for the promoter conservation count annotation became larger and more statistically significant (Table S22), consistent with the parabolic trend for promoters in Figure 4A.
Disease Enrichment Is Concentrated in Promoters of Loss-of-Function Intolerant Genes
We annotated promoters according to the genes that they regulate (see Material and Methods). In particular, we annotated 16% of promoters as being promoters of the 3,230 ExAC LoF intolerant genes, defined as genes annotated as having a high probability of being LoF intolerant (pLI) in ExAC data12 (Table 1). The promoter of ExAC gene annotation was only weakly correlated with annotations from the baseline-LD model (Figure S1), but moderately correlated with the ancient promoter and promoter conservation count annotations (Figure S1 and Table S1).
To assess how the disease enrichment of promoters varies with the gene that it regulates, we repeated our S-LDSC analysis with the promoter of ExAC gene annotation included, in addition to baseline-LD + putative enhancer + promoter annotations. We also analyzed promoter of ancient gene and promoter of gene with mouse ortholog annotations in turn (see Material and Methods). The promoter of ExAC gene annotation produced the strongest enrichment (Table S23) and was the only gene function annotation that remained significant (after correction for multiple testing) in a joint analysis conditioned on the baseline-LD + enhancer + promoter annotations.7 Promoters of ExAC genes were 12.4× enriched, compared to 5.1× for all promoters (p = 9e−16 for the difference) (Figure 5A, Tables S24A and S25). We note that enrichment estimates - which differ from model to model - can change slightly depending on the set of annotations included in the model;6, 7 the enrichment estimates reported in the Abstract are estimates obtained using the combined joint model. Although promoters of ExAC genes comprise only 16% of promoters, they contribute 39% (SE 2%) of all promoter enrichment.
Figure 5.
Disease Enrichment of Promoters of ExAC Genes in Gene Function Model
We report results for the gene function annotation that is significant conditional on the baseline-LD model and putative enhancer and promoter annotations (Bonferroni p = 0.05/3 = 0.0167). “ExAC genes” refer to genes annotated as having high pLI in ExAC data.
(A and B) Heritability enrichment (A) and estimates (±1.96 standard error) (B); results are meta-analyzed across 41 traits.
(C) Proportion of common SNPs within annotations with GERP RS ≥ 47, 36 (±1.96 standard error). We report the proportion of common SNPs (MAF ≥ 0.05) for each annotation. Numerical results are reported in Table S24, and results for each trait are reported in Table S25.
Promoters of ExAC LoF intolerant genes were uniquely informative for disease heritability conditional on the baseline-LD + putative enhancer + promoter annotations, as quantified by . Specifically, we estimated a large and highly significant value of (, p = 2e−32) (Figure 5B and Table S24B).
We quantified the mean strength of negative selection within each of the annotations from Figures 5A and 5B. We first calculated the proportion of common SNPs with GERP RS ≥ 4.7, 36 We determined that the stronger disease enrichment for promoters of ExAC genes is mirrored by the larger proportion of variants in these annotations with GERP RS ≥ 4, reflecting stronger negative selection (Figure 5C and Table S24C). We observed similar patterns for four other measures of negative selection (Table S24C). However, as noted above, promoters of ExAC genes were uniquely informative for disease heritability conditional on the baseline-LD model, which includes all of these measures of negative selection.
We performed three secondary analyses to assess the robustness of our results. First, we repeated the analysis of Figures 5A and 5B restricting to the three liver-related traits. We observed a non-significant trend toward a conditional signal for promoters of ExAC genes (nominal p = 0.048; not significant after correcting for 3 annotations tested), consistent with the fact that this analysis is less well powered (Table S26). Second, we repeated the analysis of Figures 5A and 5B by including 500 bp flanking regions around each of the annotations from Figures 5A and 5B (see Material and Methods). We confirmed that this did not significantly change our results (Table S27). Third, we repeated the analysis of Figures 5A and 5B by including two annotations based on fine-mapped expression quantitative trait loci (eQTL): the MaxCPP annotation for all genes and the MaxCPP annotation for ExAC LoF genes only.32 Results were little changed, and promoters of ExAC genes (as well as the MaxCPP (allGenes) and MaxCPP (ExAC) annotations) were still uniquely informative for disease heritability as quantified by (Table S28).
Combined Joint Model
We constructed a combined joint model by including all jointly significant annotations involving sequence age (Figures 2A and 2B), conserved function (Figure 3B), and gene function (Figures 5A and 5B) and retaining only the annotations that remained significant (after correction for multiple testing) when conditioned both on each other and on the baseline-LD + promoter + putative enhancer annotations.7 The final joint model included ancient putative enhancer, ancient promoter, putative enhancer conservation count, and promoter of ExAC gene annotations. Because enrichment is not defined for annotations with value 0–9, we estimated the enrichment of conserved putative enhancer in lieu of putative enhancer conservation count, analogous to above.
Ancient putative enhancers were 8.8× enriched, compared to 2.5× for all putative enhancers (p = 3e−14 for difference), and ancient promoters were 13.5× enriched, compared to 5.1× for human promoters (p = 5e−16 for difference) (Figure 6A, Tables S29A and S30); these enrichments differed only slightly from the joint sequence age model (Figure 2A). Conserved putative enhancers were 4.6× enriched (p = 5e−12 for difference versus all human putative enhancers); this enrichment differed only very slightly from the joint conserved function model (Figure 3A). Promoters of ExAC genes were 12.0× enriched (p = 8e−15 for difference versus all promoters); this enrichment differed only very slightly from the joint gene function model (Figure 5A).
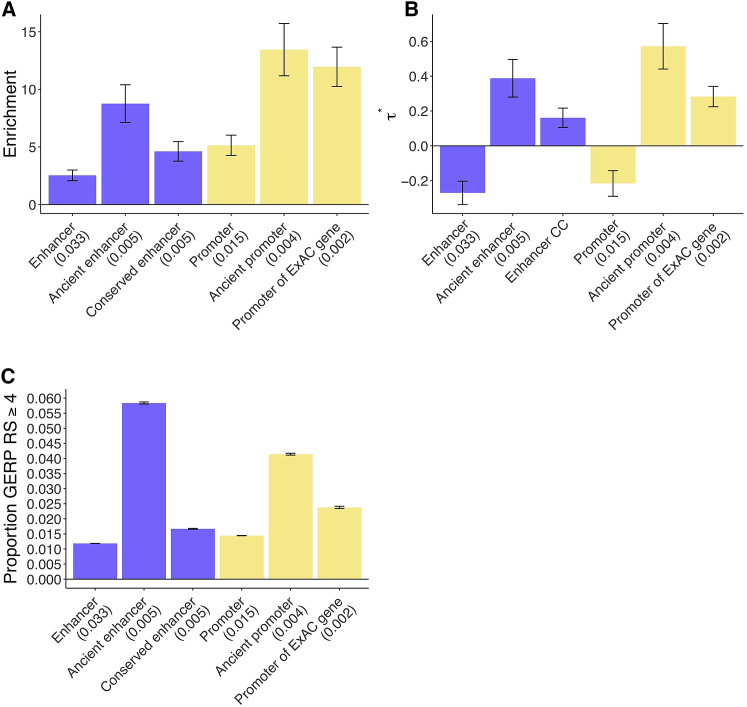
Figure 6.
Disease Enrichment of Annotations in Combined Joint Model
We report results for sequence age, conserved function, and gene function annotations that are jointly significant conditional on the baseline-LD model and putative enhancer and promoter annotations (Bonferroni p = 0.05/15 = 0.0033). (A) Heritability enrichment and (B) estimates (±1.96 standard error); results are meta-analyzed across 41 traits. CC denotes conservation count. (C) Proportion of common SNPs within annotations with GERP RS ≥ 47, 36 (±1.96 standard error). We report the proportion of common SNPs (MAF ≥ 0.05) for each annotation. Numerical results are reported in Table S29, and results for each trait are reported in Table S30.
In the combined joint model, we estimated highly significant values of for ancient putative enhancers (, p = 2e−12), ancient promoters (, p = 1e−17), putative enhancer conservation count (, p = 1e−8), and promoters of ExAC genes (, p = 2e−21) (Figure 6B and Table S29B). These estimates were slightly lower than the corresponding estimates from the joint sequence age model (Figure 2B), joint conserved function model (Figure 3B), and joint gene function model (Figure 5B), consistent with correlations between these annotations (Figure S1 and Table S1). Notably, the estimates for ancient enhancers and ancient promoters remained larger than the analogous values that we recently estimated for LD-related annotations7 and molecular QTL annotations.32
The stronger disease enrichment for ancient putative enhancers, ancient promoters, conserved putative enhancers, and promoters of ExAC genes is mirrored by the larger proportion of variants in these annotations with GERP RS ≥ 4 and four other measures of negative selection, reflecting stronger negative selection (Figure 6C and Table S29C), as we previously determined (Figure 2C and Table S7C; Figure 3C and Table S15C; Figure 5C and Table S24C). However, as noted above, all of these annotations were uniquely informative for disease heritability conditional on the baseline-LD model, which includes all of these measures of negative selection.
We performed six secondary analyses to assess the robustness of our results. First, we repeated the analysis of Figure 6A restricting to the three liver-related traits. Although this analysis is less well powered, the conditional signals for ancient promoters and enhancer conservation count remained statistically significant (Table S31). Second, we repeated the analysis of Figure 6A by replacing the putative enhancer conservation count annotation in the joint model with the binary conserved putative enhancer annotation, and confirmed that this did not significantly change our results (Table S32). Third, we repeated the analysis of Table S32 by including 500 bp flanking regions around each of the annotations from Figure 6A (see Material and Methods). We confirmed that this did not significantly change our results; the enrichment for conserved putative enhancer was slightly reduced but remained highly significant (Table S33). Fourth, we repeated the analysis of Figure 6B by including human-specific promoters and promoter conservation count as additional annotations, in order to investigate whether this might lead to a significant for the promoter conservation count annotation (as in Table S22) due to the parabolic trend for promoters in Figure 4A. However, the for both annotations was non-significant (Table S34). Fifth, we repeated the analysis of Figure 6A by including the two fine-mapped eQTL annotations: the MaxCPP annotation for all genes and the MaxCPP annotation for ExAC LoF genes only.32 Results were little changed, and our new annotations (ancient enhancer, enhancer conservation count, ancient promoter, promoter of ExAC gene) (as well as the MaxCPP [allGenes] and MaxCPP [ExAC] annotations) were still uniquely informative for disease heritability as quantified by (Table S35A). Sixth, we repeated the analysis from Table S35A, including a new annotation resulting from restricting the MaxCPP of all genes annotation to regions with ancient sequence age. We determined that the conditional eQTL signal is concentrated in the MaxCPP of all genes intersected with ancient sequence age annotation (Table S35B).
Discussion
Our results help elucidate which regulatory elements make the largest contributions to the genetic architecture of diseases and complex traits. We reached three main conclusions. First, disease heritability is concentrated in putative enhancers and promoters with ancient sequence age. Second, disease heritability is concentrated in putative enhancers and promoters with conserved function across species. Third, disease heritability is concentrated in promoters of ExAC LoF intolerant genes. These findings represent unique information about disease heritability conditional on all other available annotations, as quantified by large and highly significant values (up to 0.57 in combined joint model; Figure 6B), substantially larger than the values that we reported for other annotations in our recent work.7, 32 In addition to improving our biological understanding of disease architectures, our findings have immediate downstream applications to improve association power,3, 41, 42 fine-mapping,2, 43, 44 and genetic risk prediction,45, 46, 47 which will provide a means to validate our findings using different methods.
Promoters are known to be functionally conserved more often than enhancers;9 we determined that conserved putative enhancers, although less common than conserved promoters, are particularly strongly enriched for disease heritability (Figure 6). In addition, previous work reported that human-specific DHSs were significantly enriched for disease- and trait-associated variants, despite decreased constraint; we observed modest enrichment for human-specific promoters but no enrichment for human-specific putative enhancers (Figure 4A). The excess enrichments for putative enhancers and promoters with ancient sequence age raises the question of whether genomic regions with ancient sequence age are broadly important; however, ancient sequence age was not conditionally significant in our analyses (Table S10). Our finding of increased disease enrichment in promoters of ExAC LoF intolerant genes12 (Figure 6A) is consistent with evidence from eQTL studies;32 however, our promoter of ExAC gene annotation remains uniquely informative conditional on the fine-mapped eQTL annotations from Hormozdiari et al.32 (Table S28). We further determined that the conditional eQTL signal is concentrated in MaxCPP (allGenes) intersected with ancient sequence age, suggesting that eQTL integration studies should pay particular attention to whether an eQTL that may be linked to disease lies in a region of ancient sequence age (Table S35B). Our finding of increased disease enrichment in promoters of ancient genes (Table S23) is consistent with previous work showing that genes linked to human disease are more often ancient than recently evolved;24 however, we determined that the promoter of ancient genes annotation was not uniquely informative once the promoter of ExAC genes annotation was included in our model. Our findings are consistent with previous studies broadly demonstrating that regions under strong negative selection are enriched for disease heritability and disease-associated variants, despite being depleted for genetic variation.6, 7, 13, 14, 15, 16, 17, 18, 19, 20 (However, analogous to those studies, we are unable to make any statements about lethal mutations that preclude any genetic variation whatsoever.)
We note several limitations of our work. First, we analyzed putative enhancers that were identified using two histone marks in liver tissue,9 an approach that does not guarantee enhancer functionality. However, that study reported that the majority of the putative enhancers were regulatorily active (based on results of further experimental assays),9 implying that our finding of 3.5× stronger disease enrichment for ancient enhancers (and 1.8× stronger disease enrichment for conserved enhancers) cannot arise simply because ancient (or conserved) putative enhancers are more likely to be real enhancers. Nonetheless, the larger disease enrichment for ancient (or conserved) putative enhancers could be due to a combination of ancient (or conserved) enhancers being more strongly enriched and ancient (or conserved) putative enhancers having a higher probability of being truly functional. Second, our main analyses were restricted to putative enhancers and promoters identified in liver tissue.9 Results involving sequence age were similar for other putative regulatory annotations (Table S11). However, efforts to generalize our results for conserved function are limited by the availability of enhancer and promoter annotations across species in other tissues; one possible solution would be to predict regulatory function across species in other tissues.48, 49, 50, 51, 52, 53, 54, 55 Third, we focused our analyses on common variants by using a 1000 Genomes LD reference panel, but future work could draw inferences about low-frequency variants using larger reference panels.20 Fourth, inferences about components of heritability can potentially be biased by failure to account for LD-dependent architectures.7, 56, 57, 58 All of our analyses used the baseline-LD model, which includes six LD-related annotations.7 The baseline-LD model is supported by formal model comparisons using likelihood and polygenic prediction methods, as well as analyses using a combined model incorporating alternative approaches;59 however, there can be no guarantee that the baseline-LD model perfectly captures LD-dependent architectures. Despite these limitations, our results are highly informative for the genetic architecture of diseases and complex traits.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to P. Flicek, P. Provero, D. Marnetto, H. Finucane, J. Stamatoyannopoulos, C. Breeze, and J. Vierstra for helpful discussions. This research was funded by NIH grants U01 HG009379, R01 MH101244, R01 MH107649, and 5T32CA009337-32. This research was conducted using the UK Biobank Resource under application 16549.
Published: March 21, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.02.008.
Contributor Information
Margaux L.A. Hujoel, Email: hujoel@g.harvard.edu.
Alkes L. Price, Email: aprice@hsph.harvard.edu.
Web Resources
Ensembl hg38 build, http://useast.ensembl.org/biomart/martview
Main annotations described in this manuscript, https://data.broadinstitute.org/alkesgroup/LDSCORE/hujoel/
seqMINER, https://sourceforge.net/projects/seqminer/files/Reference%20coordinate
UK Biobank summary statistics, https://data.broadinstitute.org/alkesgroup/UKBB/
Supplemental Data
Correlation between Main Annotations and Functional Annotations of Baseline-LD Model
We report correlation of annotations with all functional annotations in baseline-LD model (among common SNPs, MAF ≥ 0.05).
List of 47 Datasets Analyzed in This Study
We meta-analyzed all results across a previously chosen collection of 47 datasets (see Hormozdiari et al.16 in Document S1). We obtained the summary statistics for each trait from previous published studies where the summary statistics are publicly available. In the case of UK Biobank traits, we computed the summary statistics using BOLT-LMM (see Loh et al.17,18 in Document S1). For some traits we have more than one dataset, thus we have 41 independent traits. However, we utilized all the 47 datasets in our meta-analyses as the number of samples that overlap is low. These traits have been selected based on a heritability z-score > 6.
List of All Models Analyzed in This Study
We report the set of annotations included in each model analyzed.
Titles and legends in Document S1.
References
- 1.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farh K.K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazal S., Finucane H.K., Furlotte N.A., Loh P.R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., Hansen R.S., Stehling-Sun S., Sabo P.J., Byron R., Humbert R. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M., Park T.J., Deaville R., Erichsen J.T., Jasinska A.J. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermunt M.W., Tan S.C., Castelijns B., Geeven G., Reinink P., de Bruijn E., Kondova I., Persengiev S., Bontrop R., Cuppen E., Netherlands Brain Bank Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat. Neurosci. 2016;19:494–503. doi: 10.1038/nn.4229. [DOI] [PubMed] [Google Scholar]
- 11.Trizzino M., Park Y., Holsbach-Beltrame M., Aracena K., Mika K., Caliskan M., Perry G.H., Lynch V.J., Brown C.D. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017;27:1623–1633. doi: 10.1101/gr.218149.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyre-Walker A. Evolution in health and medicine Sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 1):1752–1756. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwala V., Flannick J., Sunyaev S., Altshuler D., GoT2D Consortium Evaluating empirical bounds on complex disease genetic architecture. Nat. Genet. 2013;45:1418–1427. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardiñas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., Legge S.E., Bishop S., Cameron D., Hamshere M.L., GERAD1 Consortium. CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng J., de Vlaming R., Wu Y., Robinson M.R., Lloyd-Jones L.R., Yengo L., Yap C.X., Xue A., Sidorenko J., McRae A.F. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746–753. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 19.Palamara P.F., Terhorst J., Song Y.S., Price A.L. High-throughput inference of pairwise coalescence times identifies signals of selection and enriched disease heritability. Nat. Genet. 2018;50:1311–1317. doi: 10.1038/s41588-018-0177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazal S., Loh P.R., Finucane H.K., Ganna A., Schoech A., Sunyaev S., Price A.L. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 2018;50:1600–1607. doi: 10.1038/s41588-018-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marnetto D., Mantica F., Molineris I., Grassi E., Pesando I., Provero P. Evolutionary rewiring of human regulatory networks by waves of genome expansion. Am. J. Hum. Genet. 2018;102:207–218. doi: 10.1016/j.ajhg.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips M.J., Bennett T.H., Lee M.S.Y. Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proc. Natl. Acad. Sci. USA. 2009;106:17089–17094. doi: 10.1073/pnas.0904649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z.-X., Yuan C.-X., Meng Q.-J., Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 24.Domazet-Loso T., Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol. Biol. Evol. 2008;25:2699–2707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neme R., Tautz D. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics. 2013;14:117. doi: 10.1186/1471-2164-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L., Wu K., Liu Z., Yao X., Yuan S., Tao W., Yi L., Yu G., Hou Z., Fan D. Chromatin accessibility landscape in human early embryos and its association with evolution. Cell. 2018;173:248–259.e15. doi: 10.1016/j.cell.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Delsuc F., Philippe H., Tsagkogeorga G., Simion P., Tilak M.K., Turon X., López-Legentil S., Piette J., Lemaire P., Douzery E.J.P. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 2018;16:39. doi: 10.1186/s12915-018-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. Ensembl. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye T., Krebs A.R., Choukrallah M.-A., Keime C., Plewniak F., Davidson I., Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1287. e35–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J., Loh P.R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gazal S., Sahbatou M., Babron M.-C., Génin E., Leutenegger A.L. High level of inbreeding in final phase of 1000 Genomes Project. Sci. Rep. 2015;5:17453. doi: 10.1038/srep17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer C., Pe’er I. Statistical correction of the Winner’s Curse explains replication variability in quantitative trait genome-wide association studies. PLoS Genet. 2017;13:e1006916. doi: 10.1371/journal.pgen.1006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindblad-Toh K., Garber M., Zuk O., Lin M.F., Parker B.J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Broad Institute Sequencing Platform and Whole Genome Assembly Team. Baylor College of Medicine Human Genome Sequencing Center Sequencing Team. Genome Institute at Washington University A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVicker G., Gordon D., Davis C., Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruyama T. The age of a rare mutant gene in a large population. Am. J. Hum. Genet. 1974;26:669–673. [PMC free article] [PubMed] [Google Scholar]
- 39.Smith A.V., Thomas D.J., Munro H.M., Abecasis G.R. Sequence features in regions of weak and strong linkage disequilibrium. Genome Res. 2005;15:1519–1534. doi: 10.1101/gr.4421405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlesworth B., Morgan M.T., Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sveinbjornsson G., Albrechtsen A., Zink F., Gudjonsson S.A., Oddson A., Másson G., Holm H., Kong A., Thorsteinsdottir U., Sulem P. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet. 2016;48:314–317. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 42.Kichaev G., Bhatia G., Loh P.-R., Gazal S., Burch K., Freund M.K., Schoech A., Pasaniuc B., Price A.L. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kichaev G., Yang W.-Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W., McDonnell S.K., Thibodeau S.N., Tillmans L.S., Schaid D.J. Incorporating functional annotations for fine-mapping causal variants in a bayesian framework using summary statistics. Genetics. 2016;204:933–958. doi: 10.1534/genetics.116.188953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J., Park J.-H., Duan J., Berndt S.T., Moy W., Yu K., Song L., Wheeler W., Hua X., Silverman D., MGS (Molecular Genetics of Schizophrenia) GWAS Consortium. GECCO (The Genetics and Epidemiology of Colorectal Cancer Consortium) GAME-ON/TRICL (Transdisciplinary Research in Cancer of the Lung) GWAS Consortium. PRACTICAL (PRostate cancer AssoCiation group To Investigate Cancer Associated aLterations) Consortium. PanScan Consortium. GAME-ON/ELLIPSE Consortium Winner’s curse correction and variable thresholding improve performance of polygenic risk modeling based on genome- wide association study summary-level data. PLoS Genet. 2016;12:e1006493. doi: 10.1371/journal.pgen.1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y., Lu Q., Powles R., Yao X., Yang C., Fang F., Xu X., Zhao H. Leveraging functional annotations in genetic risk prediction for human complex diseases. PLoS Comput. Biol. 2017;13:e1005589. doi: 10.1371/journal.pcbi.1005589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marquez-Luna C., Gazal S., Loh P.-R., Furlotte N., Auton A., 23andMe Research Team. Price A. Modeling functional enrichment improves polygenic prediction accuracy in UK Biobank and 23andMe data sets. bioRxiv. 2018 doi: 10.1038/s41467-021-25171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee D., Karchin R., Beer M.A. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 2011;21:2167–2180. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghandi M., Lee D., Mohammad-Noori M., Beer M.A. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 2014;10:e1003711. doi: 10.1371/journal.pcbi.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitaker J.W., Chen Z., Wang W. Predicting the human epigenome from DNA motifs. Nat. Methods. 2015;12:265–272. doi: 10.1038/nmeth.3065. 7, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley D.R., Snoek J., Rinn J.L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashimoto T., Sherwood R.I., Kang D.D., Rajagopal N., Barkal A.A., Zeng H., Emons B.J., Srinivasan S., Jaakkola T., Gifford D.K. A synergistic DNA logic predicts genome-wide chromatin accessibility. Genome Res. 2016;26:1430–1440. doi: 10.1101/gr.199778.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelley D.R., Reshef Y.A., Bileschi M., Belanger D., McLean C.Y., Snoek J. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018;28:739–750. doi: 10.1101/gr.227819.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J., Theesfeld C.L., Yao K., Chen K.M., Wong A.K., Troyanskaya O.G. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 2018;50:1171–1179. doi: 10.1038/s41588-018-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speed D., Hemani G., Johnson M.R., Balding D.J. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A., Lee S.H., Robinson M.R., Perry J.R., Nolte I.M., van Vliet-Ostaptchouk J.V., LifeLines Cohort Study Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speed D., Cai N., Johnson M.R., Nejentsev S., Balding D.J., UCLEB Consortium Reevaluation of SNP heritability in complex human traits. Nat. Genet. 2017;49:986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gazal S., Marquez-Luna C., Finucane H.K., Price A.L. Reconciling S-LDSC and LDAK models and functional enrichment estimates. bioRxiv. 2018 doi: 10.1038/s41588-019-0464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between Main Annotations and Functional Annotations of Baseline-LD Model
We report correlation of annotations with all functional annotations in baseline-LD model (among common SNPs, MAF ≥ 0.05).
List of 47 Datasets Analyzed in This Study
We meta-analyzed all results across a previously chosen collection of 47 datasets (see Hormozdiari et al.16 in Document S1). We obtained the summary statistics for each trait from previous published studies where the summary statistics are publicly available. In the case of UK Biobank traits, we computed the summary statistics using BOLT-LMM (see Loh et al.17,18 in Document S1). For some traits we have more than one dataset, thus we have 41 independent traits. However, we utilized all the 47 datasets in our meta-analyses as the number of samples that overlap is low. These traits have been selected based on a heritability z-score > 6.
List of All Models Analyzed in This Study
We report the set of annotations included in each model analyzed.
Titles and legends in Document S1.