Abstract
Over a relatively short period of time, the clinical geneticist’s “toolbox” has been expanded by machine-learning algorithms for image analysis, which can be applied to the task of syndrome identification on the basis of facial photographs, but these technologies harbor potential beyond the recognition of established phenotypes. Here, we comprehensively characterized two individuals with a hitherto unknown genetic disorder caused by the same de novo mutation in LEMD2 (c.1436C>T;p.Ser479Phe), the gene which encodes the nuclear envelope protein LEM domain-containing protein 2 (LEMD2). Despite different ages and ethnic backgrounds, both individuals share a progeria-like facial phenotype and a distinct combination of physical and neurologic anomalies, such as growth retardation; hypoplastic jaws crowded with multiple supernumerary, yet unerupted, teeth; and cerebellar intention tremor. Immunofluorescence analyses of patient fibroblasts revealed mutation-induced disturbance of nuclear architecture, recapitulating previously published data in LEMD2-deficient cell lines, and additional experiments suggested mislocalization of mutant LEMD2 protein within the nuclear lamina. Computational analysis of facial features with two different deep neural networks showed phenotypic proximity to other nuclear envelopathies. One of the algorithms, when trained to recognize syndromic similarity (rather than specific syndromes) in an unsupervised approach, clustered both individuals closely together, providing hypothesis-free hints for a common genetic etiology. We show that a recurrent de novo mutation in LEMD2 causes a nuclear envelopathy whose prognosis in adolescence is relatively good in comparison to that of classical Hutchinson-Gilford progeria syndrome, and we suggest that the application of artificial intelligence to the analysis of patient images can facilitate the discovery of new genetic disorders.
Keywords: LEM domain-containing protein 2”, “nuclear envelopathy”, “progeria-like”, “next-generation phenotyping”, “deep neuronal network” and “intra-syndromal similarity
Main Text
Nuclear envelopathies are rare disorders caused by mutations in genes encoding proteins of the nuclear envelope (NE); these mutations result in a variety of phenotypes, some of which mimic aspects of human aging.1 A prototypical example is Hutchinson-Gilford progeria syndrome (HGPS, MIM: 176670), which is typically accompanied by early-onset atherosclerosis, leading to cardiovascular complications and mortality in the late second decade of life (see Web Resources).2 It is caused by a recurrent de novo synonymous splicing mutation in LMNA (MIM: 150330), which encodes the splicing isoforms lamin A and C (A-type lamins), the main components of the nuclear lamina in somatic cells.3 Many other proteins are “anchored” within the meshwork created by lamins, and some of the respective genes, such as LEMD3 (also known as MAN1 [MIM: 607844]), BANF1 (MIM: 603811), and EMD (MIM: 300384), are associated with nuclear envelopathies (see Web Resources).4, 5 At the cellular level, mutations in these genes cause disruption of the nuclear architecture and gene regulation. The integral NE protein LEMD2 (LEM domain-containing protein 2) is functionally and physically linked to A-type lamins, and experimental data suggest it has a role in the structural organization of the NE6 as well as chromatin binding and distribution.7, 8 Although a single LEMD2 founder mutation has been implicated in an autosomal-recessive form of congenital cataracts (MIM: 212500),9 LEMD2 has not been associated with a complex syndromic phenotype to date.
We performed molecular analyses and artificial-intelligence-driven facial phenotyping on two individuals who are of different ethnicities and share similar facial features and physical anomalies. Both individuals and their families gave their written informed consent to participate in this study and to undergo facial analysis by specialized software (University of Cologne). Genetic analyses were approved by the ethics committees of the University of Cologne (individual 1) and the Regional Committee for Medical and Health Research Ethics in Western Norway (individual 2).
Individuals 1 and 2 presented at different university hospitals (Bologna, Italy and Oslo, Norway). Because of the individuals’ early progeria-like appearance with little subcutaneous fat and triangular facies, clinicians suspected HGPS during the first years of life in both cases. However, the individuals’ further development was encouraging, and the initial suspected diagnosis was discarded because neither developed joint contractures, alopecia, or clinical signs of atherosclerosis. Psychomotor development was normal, but poor linear growth was noted. Both had severely delayed dentition with crowded, mostly unerupted teeth in the upper and lower jaw (Figure 1C). Oral food intake was perturbed in both cases, necessitating supplementation via gastrostomy for individual 2 from the age of 3 years onward.
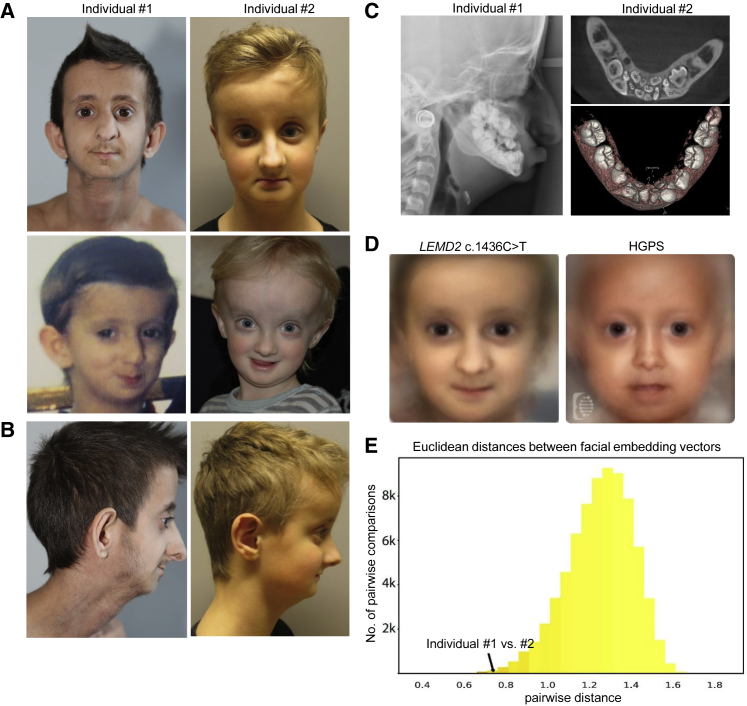
Figure 1.
Clinical Phenotype and Image Analysis of Individuals 1 and 2
(A) Facial phenotypes of individuals 1 (left) and 2 (right). Individual 1 is shown at the ages of 16 years (upper panel) and 3 years (lower panel); individual 2 is shown at the ages of 10 years and 2 years (upper and lower panel, respectively). Note the protruding eyes; small mandibula and oral aperture; and triangular face of both individuals; this latter feature is more pronounced at a younger age.
(B) (Left to right) The profiles of individuals 1 and 2 as seen at the ages of 16 and 11 years, respectively.
(C) Dental features of individuals 1 and 2. Left panel: A cranial X-ray of individual 1 shows crowded, unerupted teeth in the upper and lower jaw at the age of 13 years. Right panels: Cranial CBCT scans of individual 2 revealed complete non-erupted primary dentition and agenesis of multiple teeth in the non-erupted permanent dentition in the mandible at the age of 9 years.
(D) Composite images for LEMD2 and HGPS were averaged from photos of both individuals (left) and HGPS patients (right), as described in the Supplemental Material and Methods, and manually retouched.
(E) Histogram of the pairwise distances among all cases of the cohort. Comparisons of the same disease entity were used for adding a red blend to the respective bins according to their proportion. At the left side of the distribution, where the two individuals with the LEMD2 mutation are also posed, the percentage of pairs with the same disease-causing gene increases.
At the time of their last clinical evaluations at the ages of 16 and 10 years, respectively, individuals 1 and 2 had microcephaly, protruding eyes, and mandibular hypoplasia along with hypodontia (Figures 1A–1C). Prominent veins and a paucity of subcutaneous fat were notable in both, although these characteristics were more pronounced in individual 1. Skeletal features included wormian bones and hypoplastic clavicles, as well as reduced bone mineral density in the case of individual 1. Both had normal cognition but cerebellar intention tremor and mildly reduced muscle strength. Cranial MRIs of individual 1 had revealed white-matter hyperintensities of the peritrigonal area at the age of 5 and diffuse hyperintensities of the cerebral white matter at the age of 14, indicating progressive hypomyelination (Figure S1A). A cranial MRI of individual 2 at the age of 22 months did not show white-matter irregularities. Whereas, in the case of individual 2, growth hormone (GH) therapy initiated at age 4 substantially improved linear growth, GH therapy beginning at age 11 seemed to be ineffective for individual #1 and was discontinued after 19 months because of newly occurring insulin resistance. Repeated ophthalmological examination of both individuals revealed no significant pathologies.
Family history of both individuals was negative for similar symptomatology, and there was no history of consanguinity. More detailed descriptions of the clinical histories of individuals 1 and 2 can be found in the Supplemental Data. A compilation of the phenotypes in comparison to those associated with HGPS,2 the segmental progeroid disorders known as Néstor-Guillermo progeria syndrome (MIM: 614008),4 and PYCR1-related autosomal-recessive cutis laxa (MIM: 612940),10 to which high facial similarity to individual 1 and 2 was calculated by FaceNet (see Figure S2B), is shown in Table1. Of note, PYCR1-related autosomal-recessive cutis laxa has recently been reported to display significant clinical similarities to HGPS, predominantly in the first years of life.11
Table 1.
Compilation of Phenotypic Features of Both Individuals in Comparison to Those of Classical HGPS, NGPS, and PYCR1-Related Autosomal-Recessive Cutis Laxa
| Disorder | LEMD2-Associated Nuclear Envelopathy, Individual 1 | LEMD2-Associated Nuclear Envelopathy, Individual 2 | Hutchinson-Gilford Progeria Syndrome | Néstor-Guillermo Progeria Syndrome | PYCR1-Related Autosomal Recessive Cutis Laxa | |
|---|---|---|---|---|---|---|
| Gene, Protein(s) | LEMD2, LEM domain-containing protein 2 | LEMD2, LEM domain-containing protein 2 | LMNA, lamin A and C | BANF1, barrier-to-autointegration factor | PYCR1, pyrroline-5-carboxylate reductase 1 | |
| Facial Features | Triangular face | +++ | +++ | ++ | + | +++ |
| Prominent eyes | +++ | +++ | +++ | +++ | ++ | |
| Crooked nose, septum deviation | +++ | + | ++ | +++ | - | |
| Mandibular hypoplasia | +++ | ++ | ++ | +++ | - | |
| Skin, Hair, Nails, and | Juvenile alopezia | - | - | +++ | ++ | - |
| Subcutaneous Tissue | Loss of eyebrows | - | - | +++ | - | - |
| Thin skin, prominent veins | + | ++ | +++ | +++ | ++ | |
| Cutis laxa | - | - | - | - | +++ | |
| Patchy hyperpigmentation | - | - | + | + | - | |
| Dystrophic nails | - | - | + | + | - | |
| Generalized lipoatrophy | ++ | + | +++ | ++ | - | |
| Skull and Dentures | Microcephaly | +++ | ++ | ++ | - | + |
| Wormian bones | +++ | +++ | - | - | + | |
| Open cranial sutures | - | - | - | +++ | - | |
| Dental crowding | +++ | +++ | - | +++ | - | |
| Supernumerary teeth | +++ | +++ | - | - | - | |
| Delayed dentition | +++ | +++ | + | - | - | |
| Skeletal Features | Short stature | +++ | -a | +++ | +++ | + |
| Low bone density | ++ | - | ++ | +++ | ++ | |
| Hypoplastic clavicles | ++ | ++ | ++ | +++ | - | |
| Osteolytic foci | - | - | ++ | +++ | - | |
| Joint hyperlaxity | - | - | - | - | ++ | |
| Limited joint mobility | - | - | ++ | +++ | - | |
| Other Features | IUGR | + | - | - | - | ++ |
| Atherosclerosis | - | - | +++ | - | - | |
| Intention tremor | ++ | ++ | - | - | - | |
| Developmental delay | - | - | - | - | ++ |
Compilation of phenotypic features of both individuals in comparison to those of classical Hutchinson-Gilford Progeria Syndrome (HGPS), the phenotypically overlapping Néstor-Guillermo Progeria Syndrome (NGPS), and PYCR1-related autosomal-recessive cutis laxa, to which a high facial similarity was demonstrated upon image analysis. The novel LEMD2-associated phenotype has a characteristic and distinctive pattern of features involving several organ systems but still displaying the highest similarity to the nuclear envelopathies NGPS and HGPS.
Normal height (32th percentile) after GH-Treatment beginning at the age of 4 years.
Individual 1 had undergone molecular genetic testing of LMNA and ZMPSTE24 (MIM: 606480) prior to recruitment for the research study, and individual 2 had received testing for LMNA and an SNP chromosomal microarray. Because these tests could not provide an explanation for the unusual phenotypes, trio-exome sequencing of both individuals and their parents was performed (see Supplemental Materials and Methods for details). The German and Norwegian research groups independently identified the de novo missense variant c.1436C>T, (p. Ser479Phe) (GenBank: NM_181336.3) in LEMD2, which results in the substitution of serine with phenylalanine at position 479 (Figure 2A), as potentially causative. When both groups came into contact through GeneMatcher, a web-based tool for researchers and clinicians working on the same genes,12 the fact that both individuals carried the identical de novo variant as well as striking similarities in appearance and medical history became apparent. Given the rarity of de novo mutations in the protein-coding part of the genome, the causal role of the LEMD2 variant was established at this stage; the likelihood of finding the same de novo variant—by chance—in two individuals with a distinct and similar phenotype is negligibly low.13 We estimate this likelihood to be approximately 1 in 60 million (see Supplemental Note 2 for further details). As is to be expected for the causal variant in an ultra-rare disease, the variant c.1436C>T was absent from the ExAC and gnomAD databases. The algorithms MutationTaster, PolyPhen-2, and SIFT predicted the variant to be disease causing, probably damaging, and damaging, respectively. The CADD score was calculated as 32.000, a value strongly suggesting deleteriousness of a variant. The fact that neither affected individuals carried a second rare variant in LEMD2 suggests an autosomal-dominant mode of inheritance of the phenotype. Of note, the Hutterite founder mutation p.Leu13Arg (c.38T>G), which causes an autosomal-recessive type of juvenile-onset cataract (accompanied by sudden cardiac death at an early age in some individuals), affects an amino acid within the N-terminal LEM domain, whereas the variant p.Ser479Phe (c.1436C>T) is located at the C terminus of LEMD2 (Figure 2A). Viewed from a more global perspective, the number of observed LEMD2 missense variants (irrespective of their allele frequency) in the ExAC and gnomAD databases is lower than the number of theoretically expected variants14 for this gene: the missense Z score taken from the constraint metric for LEMD2 is 3.65 in ExAC (203.3 expected and 97 observed missense variants) and 1.98 in gnomAD (226.2 expected and 144 observed), indicating a relatively high degree of evolutionary intolerance to missense variation.
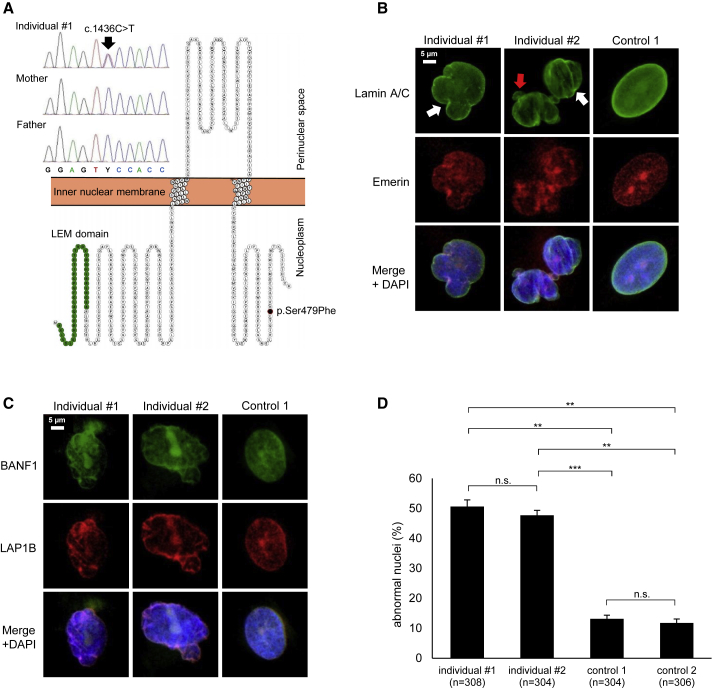
Figure 2.
Cellular Phenotype of Patient Fibroblasts
(A) Sanger Sequencing of exon 9 of LEMD2 in individual 1 and his parents shows the c.1436C>T de novo mutation in a heterozygous state. The resulting amino acid change p.Ser479Phe is indicated within the amino acid sequence of LEMD2. The N-terminal LEM domain (IPR011015), which mediates DNA binding through barrier-to-autointegration factor (BAF) proteins, and the mutated amino acid position within the C-terminal nucleoplasmic domain are highlighted. The image was generated with the Protter website (see Web Resources).
(B and C) Nuclear abnormalities in patient fibroblasts compared to control fibroblasts can be seen across multiple different IF stainings of NE proteins: stainings of lamin A and C, Emerin, BANF1, and LAP1B (TOR1AIP1) reveal irregular shapes, “blebbing” (red arrow), and invaginations (white arrows) of the nuclear membrane in patient fibroblasts (representative images).
(D) Quantification of nuclear anomalies in primary fibroblasts shows a significantly higher percentage of abnormal nuclei in both individuals’ fibroblasts as compared to cells from healthy controls. The combined results (mean ± SEM) of three separate experiments are shown (∗∗ = p ≤ 0.01, ∗∗∗ = p ≤ 0.001).
Considering the well-established role of LEMD2 in the maintenance of nuclear membrane morphology15 and LEMD2’s previously reported association with lamin A and emerin,6, 16 we performed immunofluorescence analysis of patient-derived fibroblasts and fibroblasts of two healthy control individuals (see Supplemental Material and Methods for experimental details). This analysis, through the use of four well-established integral members of the NE (lamin A and C, emerin, BANF1, and LAP1B), revealed significantly increased numbers of abnormal nuclei in both patient cell lines when compared to the control cell lines (Figures 2B–2D), although all four investigated proteins showed correct intracellular localization. These results were similar to previous findings in HGPS cells.17 Notably, although we observed some nuclear blebs and complex nuclear abnormalities, the gross majority (∼80%–90%) of nuclear abnormalities in fibroblasts from both individuals were invaginations of the NE (Figure 2B, white arrows). These data recapitulate findings in LEMD2-deficient cell lines,15 providing additional evidence for the pathogenic nature of the here identified LEMD2 variant.
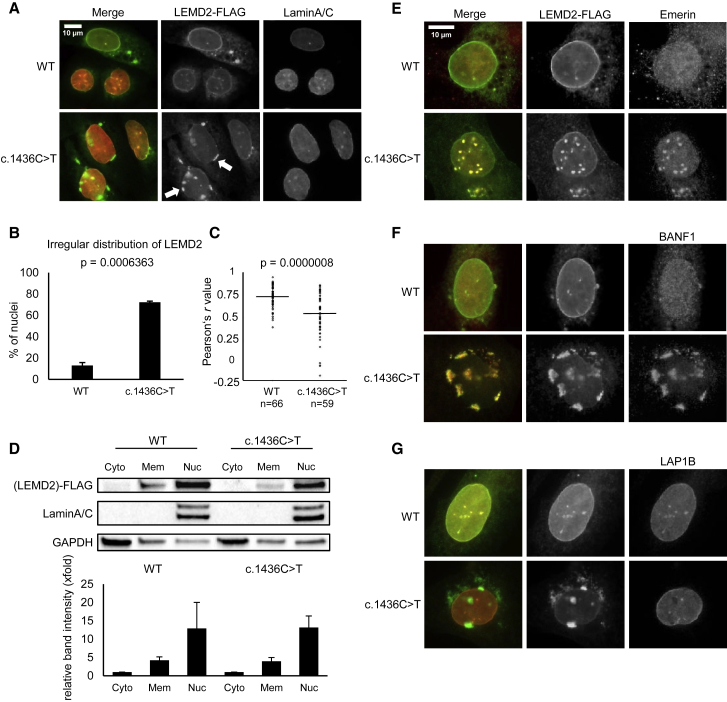
We further tried to evaluate the localization of LEMD2 in patient fibroblasts compared to control fibroblasts. However, both tested antibodies gave unspecific staining (data not shown). We therefore turned to a U2OS transient transfection system. Overexpression of wild-type (WT) and mutant LEMD2-FLAG constructs in U2OS cells led to frequent formation of “patches” of LEMD2 within the nuclear periphery, as previously described.6 However, transfection with small amounts of plasmid DNA (125 ng/mL) resulted in smooth distribution and good overall co-localization with lamin A and C in the case of WT LEMD2, whereas patchy accumulations remained predominant in cells transfected with mutant LEMD2 (Figures 3A and 3B). This visual impression was validated by quantitative co-localization analysis (Figure 3C). Both WT and mutant LEMD2-FLAG constructs were largely restricted to the nucleus, and immunoblotting showed no differences in spatial distribution within the cell (Figure 3D). The notion that mutant LEMD2 accumulates in direct contact with the nuclear lamina was further supported by co-localization of mutant and WT LEMD2-FLAG constructs with the lamina-associated proteins emerin and BANF1 (Figures 3E–3G). The preferential formation of “patches” suggests a mislocalization of mutant LEMD2 within the NE, which is in accordance with the observation of invaginations of the nuclear membrane in patient fibroblasts.18 Taken together, these data suggest that this disorder is a nuclear envelopathy on the cellular level.
Figure 3.
Immunofluorescence Analysis of Mutant LEMD2 in U2OS Cells
(A) Irregular nuclear localization of mutant LEMD2 during transient overexpression with pCMV6-LEMD2-FLAG constructs containing either the LEMD2 WT or mutant (c.1436C>T) sequence in U2OS cells. 48 h after transfection, immunofluorescence staining of FLAG (green) and lamin A and C (red) reveals patchy accumulations of mutant LEMD2 (white arrows), whereas the WT construct shows more even distribution within the nuclear lamina.
(B) Manual comparison of cells transfected with WT and mutant LEMD2 constructs shows a higher percentage of abnormal distribution in cells transfected with mutant LEMD2. Nuclear distribution of LEMD2 was characterized as normal or irregular, and the percentage of nuclei with irregular distribution was calculated. Combined results (mean ± SEM) of three separate experiments are shown. (A total of 225 cells transfected with WT and 209 cells transfected with mutant LEMD2 were analyzed.)
(C) Quantitative colocalization analysis of WT and mutant LEMD2 demonstrates disturbed colocalization of mutant LEMD2 and lamin A and C. Nuclei of cells transfected with both constructs (pCMV6-LEMD2-FLAG WT and mutant) were delimited and analyzed with the ImageJ software (Coloc2 plugin). The Pearson correlation coefficients of LEMD2-FLAG and lamin A and C of each individual nucleus, as well as averages and p values (according to the Wilcoxon rank sum test with continuity correction), are shown.
(D) WT and mutant LEMD2 are both predominantly localized within the nucleus. The Subcellular Protein Fractionation Kit (Thermo Fisher) was used to collect fractionated cell lysates 48 h after transfection with WT and mutant LEMD2 constructs. Immunoblot analysis revealed similar distribution patterns of both WT and mutant LEMD2 within cytoplasm (Cyto), membranes (Mem), and nuclear (Nuc) fractions. Comparison of band intensity normalized for total protein (shown as x-fold change with regard to the normalized band intensity of the cytoplasm fraction) from three separate experiments (mean ± SEM) did not reveal significant differences between WT and mutant constructs.
(E–G) Proteins of the nuclear lamina display different localization patterns relative to LEMD2-FLAG constructs. U2OS cells were transfected through the use of pCMV6-LEMD2-FLAG constructs (WT or mutant), and immunofluorescence stainings were performed 48 h after transfection. Whereas emerin and BANF1 colocalize with irregular nuclear accumulations of mutant LEMD2 (E and F), LAP1B does not (G).
When the German group analyzed frontal photographs of individual 1 in early 2017 by using Face2Gene, the well-established nuclear envelopathy HGPS was listed among the most likely diagnoses, and later image analysis of individual 2 produced similar results. We considered it an intriguing finding that DeepGestalt,19 this website’s deep convolutional neural network, which is commonly used as an aid to diagnosis in patients with known disorders, correctly assigned a novel syndrome to a group of syndromes for which facial similarity suggested an overlapping pathogenesis. Both individuals’ DeepGestalt similarity scores for known disorders, as well as the overlap between the individual DeepGestalt similarity scores with regard to each disorder, are shown on a radar plot in Figure S2A. DeepGestalt was trained via a supervised learning process to assign patient pictures to a set of 216 syndromes it had “learned” to recognize beforehand. In order to visualize the results of the DeepGestalt analysis, we generated composite masks of both syndromes. These masks demonstrate facial similarities and differences between individuals 1 and 2 and patients with HGPS (Figure 1D).
Because the genetic disorder of individual 1 and 2 was previously unknown, we decided to also investigate the similarity between both cases in an unsupervised, data-driven way. To do this, we clustered patient photos by using the neural network FaceNet,20 which was initially trained to recognize intra-personal similarity in a large dataset of unconstrained facial photos (i.e., in a non-medical context). After being trained to recognize intra-syndromal similarity in a group of 265 individuals with facial abnormalities from 66 different monogenic syndromes that had been selected from the PEDIA cohort (T.-C. H. et al., unpublished data), FaceNet found individuals 1 and 2 to be not only almost the most similar among 34,980 random pairwise comparisons but also more similar than most individuals inside other disease entities, including some related individuals (Figure 1E). A close-up of the dendrogram of the cophenetic distances of the clustering analysis is shown in Figure S2B. We concluded that FaceNet could determine, solely on the basis of the facial phenotype, that individuals 1 and 2 were probably affected by the same syndrome and that DeepGestalt recognized that this particular phenotype was similar to that of the family of early-onset segmental progeroid syndromes. More details about the techniques, including the cluster analysis, can be found in the Supplemental Data.
The LEMD2-associated disorder presented in this report displays a distinct combination of symptoms and has phenotypic overlap with other established segmental progeroid syndromes, such as HGPS, mandibuloacral dysplasia (MIM: 248370, 608612), and Néstor-Guillermo Progeria syndrome. However, it is important to note the better overall prognosis of the LEMD2-associated disorder when compared to HGPS: individual 1 is now 19 years of age and without clinical signs of atherosclerosis, and neither patient currently presents with life-limiting symptoms.
The available data on LEMD2,6, 7, 8, 15, 16, 21 combined with the individuals’ phenotypes and our own observations in vitro, strongly support our hypothesis of an LEMD2-associated syndromic nuclear envelopathy. The maintenance of nuclear morphology is a well-documented function of LEMD2 in vitro: it is required for reformation and closure of the NE during anaphase in mitotic yeast and HeLa cells,21 high-level overexpression of LEMD2 led to the formation of finger-type intrusions in the nuclear membrane of HeLa cells,6 and invaginations and lobulations of the NE have been reported upon siRNA-mediated depletion of LEMD2 in HeLa and U2OS cells.15 Thus, the nuclear anomalies in patient fibroblasts (Figure 2B) recapitulate the findings in the above-mentioned LEMD2-deficient cell lines,15 and these anomalies might be attributable to the observed irregular accumulations of mutant LEMD2 in the NE. Besides having an established role in the maintenance of NE integrity, LEMD2 has additionally been implicated in several other mutually non-exclusive biological processes and signaling pathways, including telomere anchoring and heterochromatin silencing,22 DNA replication,23 and MAPK cascade and ERK signaling.24 However, elucidation of the impact of the p.Ser479Phe variant in these processes as well as those linked to other progeroid syndromes will require further work and will be the main focus of our future studies.
Traditionally, recognition and classification of facial dysmorphology are among the key skills of a clinical geneticist. What used to require decades of training and clinical experience is now being supplemented by the introduction of image-analysis algorithms into clinical practice. For diagnoses with sufficiently distinct facial phenotypes, algorithms employing machine learning generally fare well in prioritizing potential diagnoses, sometimes with even better accuracy than that of seasoned geneticists.19, 25, 26 Another not-yet-exploited utility of these technologies is their potential to detect similarities of novel phenotypes to known disorders and to thereby identify a potential molecular etiology, such as the disruption of a signaling pathway or a specific cellular compartment. In our case, DeepGestalt recognized the facial similarity of individuals with the LEMD2 mutation c.1436C>T to individuals with another nuclear envelopathy, HGPS. Furthermore, FaceNet, when trained accordingly to detect intra-syndromal similarity, correctly perceived that these two individuals have highly similar facial dysmorphology. Both neural networks thus provided supporting evidence for gene identification in our particular case. We suggest that, in particular, FaceNet's ability to form groups within a cohort of syndromic patients constitutes a hypothesis-free approach to syndrome identification and patient matching and that this approach could even help to identify individuals with ultra-rare or previously unknown disorders (e.g. via a web-based patient-matching tool). Such image-analysis algorithms could be employed in the detection of syndromic “kinship” between non-related individuals and in the delineation of syndrome families to facilitate the discovery of new genetic disorders and underlying mutations, a concept that was suggested almost 15 years ago.27
In summary, we show that a recurrent de novo mutation in LEMD2 causes a syndromic nuclear envelopathy with a relatively good prognosis, and we suggest that the use of artificial intelligence could facilitate the discovery of new genetic disorders.
Declaration of Interests
Peter Krawitz works as a consultant for FDNA (Boston, MA, USA). Itmar Jorbani was formerly employed by FDNA. All other authors declare no competing interests.
Acknowledgments
We are very grateful to the individuals and their families for making this study possible. This work was supported by grants from the German Research Foundation (DFG; LE 4223/1) (Davor Lessel), and Bergen Research Foundation (BFS2016-genom) (Tomasz Stokowy). Christian Netzer and Felix Marbach thank Bernd Wollnik and Christian P. Schaaf for support of this study and helpful comments and Eike Strathmann for help in creating Figure 3C. We dedicate this work to the memory of Torunn Fiskerstrand.
Published: March 21, 2019
Footnotes
Supplemental Data can be found with this article online at https://doi.org/10.1016/j.ajhg.2019.02.021.
Accession Numbers
The identified LEMD2 variant has been deposited into the Leiden Open (Source) Variation Database (LOVD: 00222778, 00222781). The raw whole-exome sequencing data that support the findings in affected individuals cannot be made publicly available for reasons of affected individual’s confidentiality. Qualified researchers may apply for access to these data, pending approval of the institutional review board. All other data generated or analyzed during this study are included in this published article (and its Supplemental Data).
Web Resources
CADD: Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu/
Face2Gene, https://www.face2gene.com/
GeneReviews, Bonne, G., Leturcq, F., Ben Yaou, R. (2004). Emery-Dreifuss Muscular Dystrophy, https://www.ncbi.nlm.nih.gov/books/NBK1436/
GeneReviews, Gordon, L.B., Brown, W.T., Collins, F.S. (2003). Hutchinson-Gilford Progeria Syndrome, https://www.ncbi.nlm.nih.gov/books/NBK1121/
Leiden Open (Source) Variation Database (LOVD), http://www.lovd.nl/3.0/home
MutationTaster, http://www.mutationtaster.org/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protter, http://wlab.ethz.ch/protter
Supplemental Data
References
- 1.Somech R., Shaklai S., Amariglio N., Rechavi G., Simon A.J. Nuclear envelopathies--raising the nuclear veil. Pediatr. Res. 2005;57:8R–15R. doi: 10.1203/01.PDR.0000159566.54287.6C. [DOI] [PubMed] [Google Scholar]
- 2.Merideth M.A., Gordon L.B., Clauss S., Sachdev V., Smith A.C., Perry M.B., Brewer C.C., Zalewski C., Kim H.J., Solomon B. Phenotype and course of Hutchinson-Gilford progeria syndrome. N. Engl. J. Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson P.M., Lammerding J. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014;24:247–256. doi: 10.1016/j.tcb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabanillas R., Cadiñanos J., Villameytide J.A., Pérez M., Longo J., Richard J.M., Alvarez R., Durán N.S., Illán R., González D.J., López-Otín C. Néstor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. A. 2011;155A:2617–2625. doi: 10.1002/ajmg.a.34249. [DOI] [PubMed] [Google Scholar]
- 5.Hellemans J., Preobrazhenska O., Willaert A., Debeer P., Verdonk P.C., Costa T., Janssens K., Menten B., Van Roy N., Vermeulen S.J. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 2004;36:1213–1218. doi: 10.1038/ng1453. [DOI] [PubMed] [Google Scholar]
- 6.Brachner A., Reipert S., Foisner R., Gotzmann J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 2005;118:5797–5810. doi: 10.1242/jcs.02701. [DOI] [PubMed] [Google Scholar]
- 7.Barrales R.R., Braun S. Chromatin binding and silencing: Two roles of the same protein Lem2. Microb. Cell. 2016;3:185–188. doi: 10.15698/mic2016.04.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanisch K., Song C., Engelkamp D., Koch J., Wang A., Hallberg E., Foisner R., Leonhardt H., Stewart C.L., Joffe B., Solovei I. Nuclear envelope localization of LEMD2 is developmentally dynamic and lamin A/C dependent yet insufficient for heterochromatin tethering. Differentiation. 2017;94:58–70. doi: 10.1016/j.diff.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Boone P.M., Yuan B., Gu S., Ma Z., Gambin T., Gonzaga-Jauregui C., Jain M., Murdock T.J., White J.J., Jhangiani S.N. Hutterite-type cataract maps to chromosome 6p21.32-p21.31, cosegregates with a homozygous mutation in LEMD2, and is associated with sudden cardiac death. Mol. Genet. Genomic Med. 2015;4:77–94. doi: 10.1002/mgg3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimopoulou A., Fischer B., Gardeitchik T., Schröter P., Kayserili H., Schlack C., Li Y., Brum J.M., Barisic I., Castori M. Genotype-phenotype spectrum of PYCR1-related autosomal recessive cutis laxa. Mol. Genet. Metab. 2013;110:352–361. doi: 10.1016/j.ymgme.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Lessel D., Ozel A.B., Campbell S.E., Saadi A., Arlt M.F., McSweeney K.M., Plaiasu V., Szakszon K., Szőllős A., Rusu C. Analyses of LMNA-negative juvenile progeroid cases confirms biallelic POLR3A mutations in Wiedemann-Rautenstrauch-like syndrome and expands the phenotypic spectrum of PYCR1 mutations. Hum. Genet. 2018;137:921–939. doi: 10.1007/s00439-018-1957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad D.F., Keebler J.E., DePristo M.A., Lindsay S.J., Zhang Y., Casals F., Idaghdour Y., Hartl C.L., Torroja C., Garimella K.V., 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulbert S., Antonin W., Platani M., Mattaj I.W. The inner nuclear membrane protein Lem2 is critical for normal nuclear envelope morphology. FEBS Lett. 2006;580:6435–6441. doi: 10.1016/j.febslet.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 16.Huber M.D., Guan T., Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vigouroux C., Auclair M., Dubosclard E., Pouchelet M., Capeau J., Courvalin J.C., Buendia B. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 2001;114:4459–4468. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 19.Gurovich Y., Hanani Y., Bar O., Nadav G., Fleischer N., Gelbman D., Basel-Salmon L., Krawitz P.M., Kamphausen S.B., Zenker M. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019;25:60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 20.Schroff F., Kalenichenko D., Philbin J. FaceNet: A Unified Embedding for Face Recognition and Clustering. arXiv. 2015 https://arxiv.org/abs/1503.03832 arXiv:1503.03832. [Google Scholar]
- 21.Gu M., LaJoie D., Chen O.S., von Appen A., Ladinsky M.S., Redd M.J., Nikolova L., Bjorkman P.J., Sundquist W.I., Ullman K.S., Frost A. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA. 2017;114:E2166–E2175. doi: 10.1073/pnas.1613916114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrales R.R., Forn M., Georgescu P.R., Sarkadi Z., Braun S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 2016;30:133–148. doi: 10.1101/gad.271288.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y.J. Inner nuclear membrane protein Lem2 facilitates Rad3-mediated checkpoint signaling under replication stress induced by nucleotide depletion in fission yeast. Cell. Signal. 2016;28:235–245. doi: 10.1016/j.cellsig.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tapia O., Fong L.G., Huber M.D., Young S.G., Gerace L. Nuclear envelope protein Lem2 is required for mouse development and regulates MAP and AKT kinases. PLoS ONE. 2015;10:e0116196. doi: 10.1371/journal.pone.0116196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudding-Byth T., Baxter A., Holliday E.G., Hackett A., O’Donnell S., White S.M., Attia J., Brunner H., de Vries B., Koolen D. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 2017;17:90. doi: 10.1186/s12896-017-0410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferry Q., Steinberg J., Webber C., FitzPatrick D.R., Ponting C.P., Zisserman A., Nellåker C. Diagnostically relevant facial gestalt information from ordinary photos. eLife. 2014;3:e02020. doi: 10.7554/eLife.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunner H.G., van Driel M.A. From syndrome families to functional genomics. Nat. Rev. Genet. 2004;5:545–551. doi: 10.1038/nrg1383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.