Abstract
Structural variation, composed of balanced and unbalanced genomic rearrangements, is an important contributor to human genetic diversity with prominent roles in somatic and congenital disease. At the nucleotide level, structural variants (SVs) have been shown to frequently harbor additional breakpoints and copy-number imbalances, a complexity predicted to emerge wholly as a single-cell division event. Chromothripsis, chromoplexy, and chromoanasynthesis, collectively referred to as chromoanagenesis, are three major mechanisms that explain the occurrence of complex germline and somatic SVs. While chromothripsis and chromoplexy have been shown to be key signatures of cancer, chromoanagenesis has been detected in numerous cases of developmental disease and phenotypically normal individuals. Such observations advocate for a deeper study of the polymorphic and pathogenic properties of complex germline SVs, many of which go undetected by traditional clinical molecular and cytogenetic methods. This review focuses on congenital chromoanagenesis, mechanisms leading to occurrence of these complex rearrangements, and their impact on chromosome organization and genome function. We highlight future applications of routine screening of complex and balanced SVs in the clinic, as these represent a potential and often neglected genetic disease source, a true “iceberg under water.”
Main Text
The dawn of high-resolution chromosomal microarray (CMA) and next-generation sequencing (NGS)1 has unmasked extensive submicroscopic structural variation of the human genome.2, 3, 4, 5, 6, 7, 8 Such rearrangements, collectively referred to as structural variants (SVs), include deletions, duplications, insertions, translocations, and inversions that are ≥50 base pairs (bp) in size.9 SVs represent the most variable DNA content among humans, surpassing single-nucleotide polymorphisms (SNPs).6, 8 With such prominent roles in genomic content and structure, SVs are hypothesized to be a driving force in the evolution of human phenotypes, as observed from hundreds of balanced and unbalanced differences between human and chimpanzee genomes,10 or the more recent example of positive selection of a higher copy number of the amylase gene (AMY1) in populations with high-starch diets.11
The same recombination capabilities that give rise to beneficial genomic structural variation have been extensively implicated in the generation of disease. SVs were first identified as cytogenetically detectable rearrangements (>3 Mb) associated with well-known genetic syndromes.12 Subsequent CMA and NGS publications broadly linked submicroscopic SVs to a variety of genomic disorders.13, 14, 15, 16 Similarly, an avalanche of previously undetected SVs was shown to exist in cancer genomes, many of which could directly drive tumorigenesis.17, 18 An intriguing result from the analysis of congenital and somatic SVs was the discovery that many were much more complex than previously imagined (i.e., involving more than two breakpoints with intricate rearrangement configurations). In cancer, two extremely complex categories of SVs were described, chromothripsis19 and chromoplexy,20 which involved the shattering and reshuffling of chromosome regions in a single-cell division event. In the congenital setting, a third complex SV generation mechanism was discovered, termed chromoanasynthesis, which produced localized series of germline copy-number gains and losses as well as balanced SVs.21
Currently, chromothripsis, chromoplexy, and chromoanasynthesis have been grouped under the umbrella of chromoanagenesis (from the Greek chromo for chromosome and anagenesis for rebirth) (see Table 1).22, 23 Chromoanagenesis is now regarded as an important driver of karyotypic evolution in cancer as well as a contributor to the generation of diverse developmental disorders. However, not all chromoanagenesis events are detrimental to human health, as complex SVs have also been detected surprisingly in phenotypically normal individuals.24, 25, 26, 27 In light of the observations of complex congenital SVs in apparently normal and morbidly affected individuals, we surmise that we are barely scratching the surface of congenital chromoanagenesis events, with important consequences for clinical interpretation of these rearrangements. This review provides an overview of chromoanagenesis mechanisms, reported chromoanagenesis events detected in the congenital setting, and their roles in human disease and genome architecture. We conclude the manuscript with a discussion on suggested practices to improve the detection levels of complex SVs in normal and morbid populations.
Table 1.
Definition of Chromoanagenesis Terms Included in This Review
| Term | Greek Root | Definition | Characteristics |
|||
|---|---|---|---|---|---|---|
| Chromosomes | Breakpoints | Distribution | Dosage alt | |||
| chromothripsis | chromo for chromosome and thripsis for shattering into pieces | phenomenon by which hundreds of rearrangements originate through random shattering and reshuffling of clustered chromosome regions within a single catastrophic event. The predominant method of re-assembly of the shattered pieces is c-NHEJ | typically 1 (cancer) but 1–4 observed in congenital cases | ≥5, up to 65 observed in congenital cases and hundreds in cancer | clustered | typically unbalanced (cancer) and balanced (congenital) |
| chromoplexy | chromo for chromosome and pleko for to twist or enfold | phenomenon where derivative chromosomes are generated by the chimeric joining of DNA segments from two or more chromosomes. Chromosome re-assembly is predicted to occur by c-NHEJ or alt-EJ repair | ≥2 | ≥5 to a couple dozen | interspersed | typically balanced but deletions can also be present at junctions |
| chromoanasynthesis | chromo for chromosome and anasynthesis for reconstitution | phenomenon by which multiple combinations of SVs are generated through errors in DNA replication, namely FoSTes and MMBIR | typically 1 | ≥5 to a couple dozen | clustered | unbalanced (gains and losses) |
Chromoanagenesis and Its Mechanisms of Generation
Chromothripsis
Stephens and co-authors first discovered chromothripsis (from the Greek chromo for chromosome and thripsis for shattering into pieces) in an individual with chronic lymphocytic leukemia (CLL) in 2011.19 Chromothriptic SVs were quickly identified in 2%–3% of cancer specimens, including soft tissue and central nervous system tumors, various carcinomas, and other hematological malignancies,28 and was associated with oncogene amplification through double minute formation,19, 29, 30 tumor suppressor loss,19 and promotion of gene fusions,31 all important to carcinogenesis and potential development of targeted therapies.
Chromothripsis is the localized shattering and reshuffling of tens to hundreds of chromosome segments, often restricted to one or a few chromosomal regions (Figure 1A).19 The apparent random stitching together of chromosome pieces is often observed concomitantly with DNA loss, and thus chromothripsis-derived SVs can display an oscillating copy number pattern between one and two copies with retention of allelic heterozygosity. The unexpected spatial and numeric constriction of these rearrangements led to the hypothesis that chromothripsis occurs within a single and physically isolated catastrophic event,19 contrary to the widely accepted idea of stepwise accumulation of genetic changes in cancer. Micronuclei formation was directly postulated as a source of chromothripsis, as lagging mitotic chromosomes can be encapsulated into a nuclear envelope of their own and be pulverized when failing to complete DNA replication prior to initiation of subsequent mitosis.32, 33 Zhang and co-authors tracked micronuclei formation and reincorporation into daughter cells using live-cell imaging in the RPE-1 cell line; single-cell genome sequencing of the tracked cells revealed the presence of complex chromosome rearrangements, similar to those found in chromothripsis, thus unveiling a mechanism by which complex SVs originate and are integrated back into the genome.34 While different models have been proposed to explain the origin of chromosome damage in micronuclei,23 what is clear is that double-strand breaks (DSBs) from such pulverization events could trigger DNA damage responses that would attempt their repair.35 Chromothripsis fragments have junctions sharing zero to minimal identities, which suggests their predominant re-assembly via classic non-homologous end joining (c-NHEJ). However, microhomology-mediated end-joining (MMEJ), the major form of alternative end-joining (alt-EJ), could also participate in the post-shattering stitching.19, 34, 35, 36, 37 These processes bypass the requirement of long stretches of DNA homology, allowing random fusion and loss of fragments that can explain the oscillating two copy number states without loss of heterozygosity seen in chromothripsis.
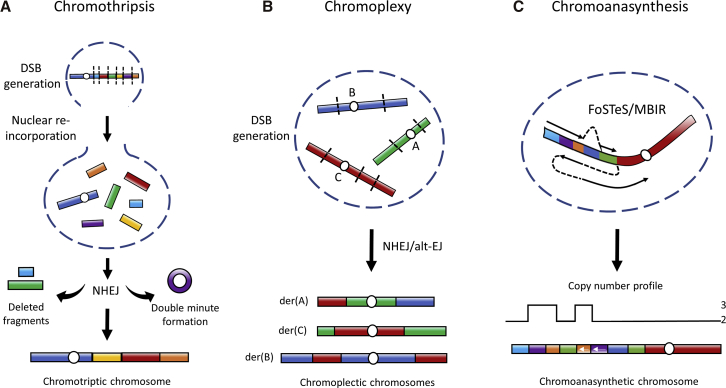
Figure 1.
Characteristics of Chromothripsis, Chromoplexy, and Chromoanasynthesis-Derived SVs
(A) A chromosome in a micronucleus can undergo massive DNA damage and result in multiple double-strand breaks (DSBs, depicted with dashed black lines). When the micronucleus is re-incorporated into the nucleus during mitosis, the DSBs undergo repair through NHEJ, where chromosome segments are randomly stitched back together, lost, or become double minutes. Functionally relevant segments could become double minutes and undergo amplification, as has been observed in MYC and other oncogene-containing segments in various cancer cases.19, 29, 30
(B) In chromoplexy, different DSBs can be repaired with or without DNA loss at the breakpoints and be arranged into various derivative configurations, as shown here by the rearrangements of example chromosomes A, B, and C.
(C) In chromoanasynthesis, a normal chromosome can undergo DNA segment re-synthesis (dashed lines to show template switches and solid arrows to show replication) mediated by replication processes such as FoSTeS and MMBIR. These mechanisms lead to templated insertions that exhibit higher copy-number and may be arranged in different orientations (depicted in purple and orange with white arrows signifying inverted sequence orientation). Notice the chromoanasynthesis chromosome has a copy-number profile exhibiting intercalating duplication-normal-duplication (dup-nml-dup) copy-number states, as seen in previous studies.21
Additional mechanisms have been proposed to explain the birth of chromothriptic events. These include localized induction of DSBs by ionizing radiation,19, 38 free radicals,19 and breakage-fusion-bridge (BFB) cycles.39 Of these, BFB is a known process leading to chromosome instability in cancer.40 In BFB, dicentric chromosomes are pulled to opposite spindle poles during mitosis, generating an anaphase bridge that matures into a chromatin bridge between daughter cells.41, 42 Rupture of the transient nuclear envelope of this bridge and physical separation of the daughter cells exposes the stretched DNA to cytoplasmic exonucleases; the resolution of these DSBs can give rise to chromothriptic-like rearrangements, and/or new unprotected chromosome ends that can initiate a new round of BFB.39
Chromoplexy
In the same year as the initial publication on chromothripsis, a second process of human genome reshuffling was described by genome sequencing of human prostate cancer.20 This process, named chromoplexy (from the Greek chromo for chromosome and pleko for to twist or enfold), involved generation of chimeric chromosomes through “closed chains” of breakage and rejoining of DNA segments (Figure 1B). Chromoplexy was subsequently identified in other fusion-driven tumors including Ewing and synovial sarcoma43 and was hypothesized to impact multiple cancer genes simultaneously, providing nascent cancer cells with proliferative advantage.
Chromoplexy events involve two to a few chromosomes, and their fusion breakpoints mostly exhibit a precise join or 2 bp microhomology; this results in chimeric chromosomes that maintain a largely balanced DNA content (i.e., minimal to no DNA gain or loss) probably due to c-NHEJ or alt-EJ repair of DSBs,20 although sizable deletions at chain fusion junctions have also been observed.44 Mechanistically, chromoplexy utilizes a different strategy for the generation of rearrangements compared to chromothripsis. Oncogenic ETS (E26 transformation-specific) family gene fusions in prostate cancer tend to be associated with a higher number of interchromosomal chromoplexy rearrangements compared to intrachromosomal ones.44 Given that androgen-driven signaling was previously shown to result in de novo TMPRSS2-ERG fusions,45 prostate cancer chromoplexy is perhaps the product of transcriptionally associated recombination at these sites. Chromoplexy-driven EWSR1-ETS fusions in Ewing sarcoma were also found to occur in transcriptionally active and early replicating genomic regions, further supporting this hypothesis.43 Such correlation is significant, as it couples the genesis of complex SVs to DNA transcription, as opposed to DNA repair/mitotic division models of chromothripsis.
In cancer chromoplexy, similar to chromothripsis, the disruption of chromosome regions is predicted to occur in a single-cell cycle event and with the presence or absence of underlying chromosome deletions.20, 43, 44 However, chromoplexy has also been shown to participate in stepwise acquisition of chromosome abnormalities, as observed from sub-clonal prostate cancer populations where driver and newly acquired rearrangements could be identified and followed-up.44 This property, in addition to the fewer number (from three and up to a couple dozen),20, 43 and generally wide spacing of rearrangements across the genome (in up to eight distinct chromosomes reported so far) can help distinguish chromoplexy from chromothripsis, where the breakpoints tend to be clustered and their numbers range from the tens to hundreds. Ultimately, both processes could arise concurrently or asynchronously within the same cell, generating different patterns of chromosome complexity with different impacts on genomic function.
Chromoanasynthesis
In characterizing individuals with various developmental and cognitive anomalies, an entirely different landscape of complex SVs was discovered, termed chromoanasynthesis.21 Chromoanasynthesis was first characterized by CMA, fluorescence in situ hybridization (FISH), and karyotyping, which brought to light complex combinations of clustered deletions, duplications, triplications, inversions, and translocations in subjects with diverse congenital anomalies. These SVs were present in different arrangements, such as duplication-normal-duplication (dup-nml-dup) copy-number states or duplication-triplication (dup-trp) configurations, among others. Such rearrangements, particularly the existence of duplications and triplications, could not be explained by the chromosome shattering and NHEJ-mediated repair distinctive of chromothripsis; for example, even if duplications or triplications were formed by shattering and re-ligation of fragments from replicated or homologous chromosomes, the reciprocal deletions associated with these fusions were never observed. It was thus that this new type of complex variation was termed chromoanasynthesis, from the Greek chromo for chromosome and anasynthesis for reconstitution, which involved the generation of multiple SVs through mechanisms other than NHEJ (Figure 1C).
Sequencing of chromoanasynthesis junctions revealed segments of extended microhomology and templated insertions, up to ∼5 kb. Such homology signatures are characteristic of errors in DNA replication, most notably fork-stalling and template switching (FoSTes)46 and microhomology-mediated break-induced replication (MMBIR).47 In the FoSTes model, stalled DNA replication forks can switch templates by using complementary stretches of microhomology from other replication fork templates. Moreover, due to the spatial arrangement of chromosomes into defined territories48 and topological associating domains (TADs),49, 50, 51 template switching and strand invasions could be constrained within adjacent intrachromosomal forks or among interchromosomal forks in close physical proximity in one or more cell division events. Re-initiation of DNA synthesis thus enables the juxtaposition of sequences from discrete genomic segments, which can grow in complexity with additional cycles of template switching. The FoSTes model was generalized to MMBIR, in which collapsed replication forks are resected and the resulting 3′ overhang is able to invade other DNA sequences; the replication fork can thus reform on different templates until it returns to the original sister chromatid.47 FoSTes/MMBIR could thus generate complex and clustered combinations of SVs, which resemble chromothripsis-like rearrangements but have additional chromosome gains or losses and other balanced SV configurations.
FoSTes/MMBIR has been reported to proceed with microhomologies as small as 2 bp.47 Given previous observations of microhomology in cancer chromothripsis, perhaps chromoanasynthesis could engage in the generation of chromothripsis events, but further research is necessary to demonstrate such involvement. Altogether, chromoanasynthesis could occur either during gametogenesis or postzygotically and could contribute to the generation of complex SVs in a non-exclusive and possibly cooperative way with other chromoanagenesis mechanisms.
Chromoanagenesis in the Congenital Setting
Chromothripsis and Chromoplexy
The identification of chromothriptic-like rearrangements in the congenital setting occurred shortly after the initial report of cancer chromothripsis.52 In this study, a child with severe developmental delay, hypertelorism, and kyphoscoliosis was studied by NGS due to the presence of a cytogenetically visible complex rearrangement involving three chromosomes (46,XY,t(1;10;4)(p32.2;q21.1;q23)dn). Genome mate-pair sequencing (MPseq) revealed the presence of 25 predicted de novo rearrangements, with 12 clustering within small regions of chromosomes 1, 4, and 10. Cardinally, the 12 SVs localized within 2–3 Mb of each other, and derivatives were observed to contain complex randomly inserted or joined fragments often in inverted orientation. Deletions between fused fragments were also observed, all of which were not found by CMA. Junctions between different chromosome fragments contained limited to no microhomology, in addition to small indels, supporting NHEJ or alt-EJ as their mechanism of repair. Collectively, the 12 rearrangements were predicted to arise simultaneously, with one rearrangement identified to have occurred in a paternal chromosome.52
Subsequent studies of constitutional chromothripsis focused on the analysis of families or cohorts of individuals presenting with various developmental phenotypes including severe growth retardation, intellectual disability, facial dysmorphism, and hypotonia, among others, and who harbored two or more cytogenetically visible and putatively balanced SVs.27, 53, 54, 55, 56, 57, 58, 59, 60, 61 The authors of these reports utilized a combination of molecular and cytogenetic techniques including MPseq, CMA, and Sanger/NGS sequencing to characterize fully the SV breakpoints; they uncovered rearrangements characterized by localized clustering of DSBs, small fragment deletions without copy number gains, and a display of microhomology-containing or blunt-ended junctions, features commonly observed in chromothripsis. While constitutional chromothripsis signatures were similar to those of cancer, notable differences between the two were observed, including largely balanced events, the number of chromosomes affected, and the number of breakpoints detected. Constitutional chromothripsis can be completely or almost perfectly balanced,27, 53, 54, 55, 56, 57, 58, 59, 60, 61 in contrast to the classic oscillating patterns of deletions observed in cancer. In addition, congenital chromothripsis has been shown to affect one or a few chromosomes simultaneously, contrary to cancer chromothripsis where many studies report the engagement of a single chromosome.19, 29, 36, 62, 63 Paradoxically, despite a higher number of chromosomes participating in constitutional chromothripsis, the number of breakpoints detected has not exceeded a couple of dozen per analyzed case, a striking disparity with cancer where chromothripsis events can contain tens to hundreds of rearrangements. A plausible explanation for these observations is the hypothesis that an extensive number of chromothriptic rearrangements and their associated fragment deletions could affect functionally relevant genomic regions and have a negative impact on fetal survival; this idea may be further supported by the multitude of chromosome rearrangements detected in products of conception (POCs, including chorionic villi samples or fetal tissue),64 many of which could arguably harbor chromothripsis-like events invisible to CMA or standard karyotyping.
In these instances, the description of congenital chromothripsis could also fit a classification of chromoplexy: ≥2 chromosomes involved, ≤30 breakpoints scattered within several Mb-sized regions, deletions present at junctions, and blunt-ended or microhomology-containing breakpoints. In reference to this observation, the case of a 10-year-old child with various developmental delays and three derivative chromosomes formed by complex translocations between chromosomes 2, 5, and 7 (with a total of 13 junctions), was classified as chromoplexy in a previous report.56 For this case, the disposition of rearrangements is similar to those observed in various published congenital chromothripsis events (Figure 2).27, 53, 54, 55, 56, 57, 58 Indubitably, these cases display characteristics of both chromothripsis (clustered breakpoints) and chromoplexy (number of chromosomes involved, distance between breakpoints), possibly arising within one or more cell division events. This observation suggests instances in which both methods could act together or in concert with other recombination mechanisms to generate such congenital complex SVs.65, 66
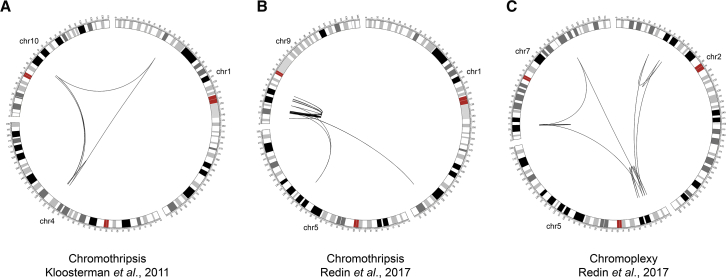
Figure 2.
Circos97 Representation and Comparison of Congenital Chromothripsis and Chromoplexy Cases
Two chromothripsis events (A, child described by Kloosterman and co-authors52 and B, Redin and co-authors [DGAP122]56) and one congenital chromoplexy instance (C, UTR20 from Redin and co-authors56) are shown. Circos diagrams look different from the original case publications as these graphs display chromoanagenesis rearrangements with whole chromosome views. Notice the presence of clustered breakpoints in all three cases, characteristic of chromothripsis events. Notice also the involvement of multiple chromosomes for all examples, similar to what is observed in chromoplexy. The occurrence of characteristic features from both mechanisms suggests that they could act sequentially or simultaneously in a cell.
Regardless of the particularities of their mechanism of generation, congenital chromothripsis/chromoplexy is usually associated with human disease, concordant with the idea that extensive chromosome rearrangements have a higher chance to disrupt gene structure, regulation, and chromosome organization (see Functional Impact of Chromoanagenesis section). Despite their described untoward clinical outcomes, the presence of chromothripsis in apparently normal individuals was incidentally encountered in phenotypically normal mothers of developmentally delayed children harboring chromothripsis-like events as well as phenotypically normal females with multiple miscarriages.25, 26, 27, 53, 54, 58 In carriers of chromothripsis/chromoplexy SVs, unbalanced segregation can result in spontaneous abortions or developmental disorders in offspring through gene dosage alterations. This is consistent with observations made by authors of these reports, where phenotypically normal women harboring apparently balanced chromosome rearrangements had multiple miscarriages25 or their children presented with various developmental abnormalities24, 27, 53, 54, 58 or syndromic-like features including cri-du-chat syndrome (MIM: 123450).26 The apparently balanced SVs of these women were found to encompass complex chromothripsis-like events truncating up to 13 protein-coding genes, which could further undergo copy-number changes in affected progeny.27 It is noteworthy to mention that, apart from the published cases of unbalanced segregation of chromothripsis/chromoplexy-derived SVs, balanced transmission without major phenotypic consequences has also been detected. Bertelsen and co-authors described stable segregation of a chromothripsis event detected as a cytogenetically visible t(3;5)(q25;q31); the event was incidentally discovered through multiple miscarriages (which were likely the product of unbalanced gametes), but the rearrangement was revealed to be paternally or maternally transmitted in a balanced fashion among 11 members of a three-generation family.25
Revision of chromothripsis/chromoplexy segregation patterns inevitably leads to a discussion of the parental origin of these events. Chromothripsis/chromoplexy SVs can be inherited from a constitutional parental event, can arise de novo in either maternal or paternal germlines, or can emerge within the early stages of embryonic development. Various studies have revealed a preferential origin of chromothripsis in paternal alleles, likely attributed to errors resulting from the higher number of cell divisions in male compared to female gametogenesis.52, 53, 58 While chromothripsis events are more likely to arise in paternal chromosomes, it is difficult to unequivocally assign the genesis of paternal chromothripsis/chromoplexy to spermatogenesis, as these events could have also occurred de novo during embryonic development. Single-cell sequencing experiments of sperm from phenotypically normal individuals would likely elucidate the burden of chromothripsis events in spermatogenesis, which could be relevant to fertility procedures such as preimplantation genetic testing. Interestingly, while the origin of these SVs was proposed to be preferentially paternal, their segregation to subsequent generations was observed to be mostly maternal,58 in agreement with previous reports of complex SV segregation where maternal transmission occurred more frequently compared to paternal.67
Chromoanasynthesis
In contrast to chromothripsis and chromoplexy, both detected in congenital and cancer cases, the identification of chromoanasynthesis has been mostly limited to the congenital setting. An explanation for this observation is that complex SVs formation in cancer mostly arises through end-joining rather than microhomology mechanisms,37 perhaps as an attempt of cancer cells to rapidly overcome extensive DNA damage.
Consistent with its initial discovery,21 additional chromoanasynthesis events have been detected in individuals presenting with various developmental and neurologic disorders.68, 69, 70, 71, 72 Thorough characterization of the SVs in these individuals revealed homology-mediated repair signatures at junctions as well as copy-number gains, primary features of chromoanasynthesis. In addition, chromoanasynthesis events were shown to occur in either maternal71 or paternal21 alleles, but no parental preference has been shown to date.
A particularly interesting chromoanasynthesis occurrence was described by Collins and co-authors, who detected a mosaic chromoanasynthesis event using a modified MPseq strategy in an individual with autism spectrum disorder (ASD [MIM: 209850]).71 The chromoanasynthesis SV occurred in chromosome 19 and involved eight duplicated loci; six of these duplications were predicted to be mosaic, while the remaining two appeared at nearly three full copies. Mosaicism was proposed to have originated from a de novo maternal duplication-inversion-duplication (dup-inv-dup) event, which was subjected to a second mutational process, thus generating duplication mosaicism. Another interesting instance of chromoanasynthesis was described by Masset and co-authors,70 who identified three derivative chromosomes with extensive copy-number gains; however, the identified junctions displayed microhomology and non-templated insertions up to 40 bp. Neither chromothripsis (c-NHEJ) nor chromoanasynthesis (microhomology) could singlehandedly explain the origin of such events. Because polymerase theta (polΘ) activity had been shown to mediate alt-NHEJ in a template-dependent and -independent manner,73 the authors proposed these events to have arisen from a combination of aberrant DNA replication and alt-NHEJ using polΘ.70 Both studies are relevant as they set the precedent that chromoanasynthesis, similarly to chromoplexy, does not necessarily have to occur in a single DNA replication/cell division event, and could act in conjunction with other DSB repair mechanisms (like chromothripsis), potentially allowing already complex SVs to acquire further structural changes and affect larger portions of the genome.
Chromoanasynthesis is hypothesized to induce significant gene dosage imbalances which can have untoward clinical consequences or result in embryonic lethality and has thus not been extensively observed in phenotypically normal individuals. However, depending on which genomic regions are included in the chromoanasynthesis event, such SVs could exist in normal individuals, as exemplified by stable inheritance of a chromoanasynthesis event involving chromosome 21 in three generations.74 This event was identified through CMA of a child with seven copy number gains in chromosome 21 and presenting with epilepsy, ataxia, and dysmorphic features; the mother and maternal grandfather harbored the unbalanced chromosome without major phenotypic consequences. A likely pathogenic SYNGAP1 variant was proposed to be responsible for the child’s clinical features. This study illustrates the point that depending on the locus of origin, complex chromoanagenesis events could represent neutral genetic variation and remain undetected in phenotypically normal individuals. However, such events may also seed further instances of complex recombination or genomic instability, as shown in the evolution of a chromothriptic rearrangement in a mother into a chromoanasynthesis rearrangement in her daughter.75
Functional Impact of Congenital Chromoanagenesis
Chromoanagenesis events can impact genome function through multiple avenues, including deletion or truncation of disease-associated genes, triplication of clinically relevant genomic segments, generation of pathogenic gene fusions, or effects on gene expression through removal/repositioning of important regulatory elements (Figure 3).
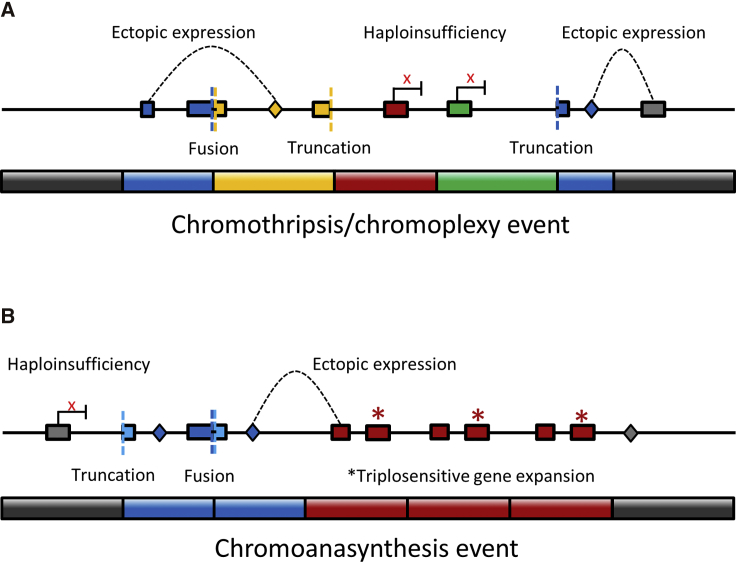
Figure 3.
Functional Consequences of Chromoanagenesis
Rearranged chromosome fragments are colored in blue, yellow, red, and green. Grey fragments represent the remainder of the chromosome, to pter (left gray fragments) and qter (right gray fragment).
(A) Chromothripsis/chromoplexy events can lead to gene truncation (colored boxes and colored dashed lines), fusions (adjacent colored boxes and colored dashed lines), gene haploinsufficiency due to removal of regulatory elements (enhancers marked as colored diamonds and haploinsufficient gene transcription marked with an x), or ectopic expression caused by position effects (enhancers marked as colored diamonds and dashed lines indicate the genes on which they are exerting their effects).
(B) Similar to chromothripsis/chromoplexy, chromoanasynthesis can lead to gene truncation, gene fusion, gene haploinsufficiency due to removal of regulatory elements or ectopic expression. In addition, expansion and transcription of triplosensitive genes could be observed in chromoanasynthesis.
Gene truncation is a common outcome of chromoanagenesis, given the number of breakpoints and small deletions generated by this process, which increase the chances of disrupting clinically relevant genes. Examples of pathogenic genes truncated after a chromoanagenesis event include PCDH15, PHIP, and MYO6, among others (Figures 3A and 3B).52, 56, 58, 71 Chromoanagenesis, specifically chromoanasynthesis, can also generate complex triplications embedded within distinct chromosome regions associated with diverse congenital phenotypes21 and can potentially induce triplosensitive gene amplification or disrupt haploinsufficient genes at the breakpoints of the duplications (Figure 3B). In addition, because chromoanagenesis SVs juxtapose segments out of their normal chromosome locations, chromoanagenesis SVs have the potential to generate pathogenic gene fusions; while chromoanagenesis-promoted fusions have been mostly observed in cancer,20, 43 there are currently no published examples of verified gain-of-function fusions due to chromoanagenesis in the congenital setting (Figures 3A and 3B). Lastly, the extensive re-shuffling of chromosome material can have an impact on underlying chromatin conformation, particularly TADs, which can impact transcriptional regulation (Figures 3A and 3B). TADs are defined as chromosome regions ranging from several hundred kb to Mb in size, with significantly increased numbers of self-interacting chromatin contacts compared to adjacent chromosome areas.49, 50 Importantly, TAD structures have been shown to be tightly linked to regulation of gene expression,50, 76, 77, 78, 79, 80 as they limit the contacts of enhancers to their specific genes within a defined 3D space. When disturbances of underlying TAD structures occur, long-range position effects of neighboring gene expression have been reported,56, 81, 82, 83, 84, 85, 86 mostly attributed to a re-wiring of local regulatory networks delimited by chromosome topology. Position effects in pathogenic genes such as FOXG1, SOX9, and SATB2 have already been observed in connection with congenital complex chromosome rearrangements,56, 84, 85 and mis-expression of TWIST1, FOXP1, and DPYD due to lost enhancer interactions upon a chromothripsis event has also been reported in an individual with craniosynostosis, facial dysmorphisms, growth retardation, and intellectual disability.59 We thus hypothesize many chromoanagenesis SVs to have important contributions to clinical phenotypes through long-range position effects, and various tools have been developed to aid in such discoveries that incorporate regulatory and chromatin conformation data.85, 87
While the majority of chromoanagenesis cases have been linked with disease, not all instances of chromoanagenesis are detrimental. Depending on the affected chromosomal regions, chromoanagenesis could be inherited without major phenotypic consequences, as demonstrated by the stable inheritance of a chromoanasynthesis event in chromosome 21 present in two healthy individuals.74 Advantageous chromoanagenesis has also been shown to exist, as exemplified by the extraordinary case of the chromothriptic cure of an individual with WHIM syndrome (WHIMS [MIM: 193670]).88 In this individual, a somatic chromothripsis event in a hematopoietic stem cell truncated the CXCR4 disease allele; this rearranged stem cell repopulated the myeloid lineage and promoted engraftment in the bone marrow, causing spontaneous remission of WHIM syndrome. Future studies on phenotypically normal control populations will eventually shed light on whether other examples of neutral and/or advantageous somatic/germinal chromoanagenesis exist, and the settings or conditions in which these complex rearrangements are selected or tolerated in the human genome.
Limitations in the Study of Congenital Chromoanagenesis
Examination of congenital chromoanagenesis has been mostly limited by identification of these complex events in the clinic, their appropriate classification, and their clinical interpretation. With each passing year, newer and more sophisticated methodologies (including high-resolution CMA, NGS, and MPseq) are utilized to question morbid and apparently normal human genomes. In practice, the majority of congenital chromoanagenesis events have been discovered through chromosome karyotyping, FISH, and CMA, where apparently balanced or unbalanced abnormalities prompted further molecular evaluation.21, 25, 27, 52, 53, 54, 55, 56, 58, 68, 69, 70 Nevertheless, characterization of chromoanagenesis junctions at the nucleotide level is not easy, given the high number of breakpoints involved and their arrangement into complex patterns. For most published congenital chromoanagenesis cases, a combination of Sanger sequencing, short-read and long-read NGS, and MPseq have been utilized to identify breakpoints and reconstruct derivatives according to the observed junctions.21, 25, 27, 52, 53, 54, 55, 56, 57, 58, 70, 71, 89 Yet, even with this battery of testing, there are instances in which junctions were predicted to exist but could not be captured, possibly due to overlaps with repetitive elements, their arrangement into highly convoluted structures, or low-level mosaicism.52, 56 Similarly, in cases where karyotyping is not performed, or the SVs do not dramatically alter chromosome banding, or the loss of genetic material is below the limit of detection, chromoanagenesis SVs can remain undetected by standard clinical testing, including CMA and WES. Several such examples exist,52, 53, 56, 71 suggesting that the frequency of congenital chromoanagenesis may be underestimated given the technical limitations in routine clinical testing. However, some of these restrictions may be alleviated through new sequencing strategies, as exemplified by Cretu Stancu and co-authors in their characterization of two congenital chromothripsis cases (associated with dysmorphic features and intellectual disability) using long-read sequencing, which proved to be a superior strategy to detect and phase complex chromothripsis SVs compared to short-read sequencing (see Discussion).89
Once complex SVs are discovered in a genome, it is important to determine if these were derived from chromoanagenesis mechanisms. While chromoanasynthesis events are easier to classify given their characteristic copy-number gains, chromothripsis and chromoplexy may be harder to distinguish from each other, as discussed earlier in this review (see Chromoanagenesis in the Congenital Setting). The clustered breakpoints together with the high number of chromosomes involved suggests instances in which both chromothripsis and chromoplexy could act in concert or with other recombination mechanisms to generate the described complex SVs. For instance, chromothriptic chromosomes could be subjected to ensuing translocations, or chromoplectic chromosomes could undergo additional chromothriptic breakage; the latter instance has been proposed as a speculative model termed “translocation-induced chromothripsis,” to explain the origin of congenital chromothripsis where multiple chromosomes are involved.65 In addition, chromoanasynthesis may display microhomologies and non-templated insertions, which also suggests a hybrid origin. Regardless of the mechanism, careful analysis should be performed before classifying a congenital complex SV as chromothripsis or chromoplexy, and diverse algorithms and rules are available to the community for appropriate designations.65, 66
The final challenge in the study of congenital chromoanagenesis is that posed by their clinical interpretation. In addition to genes amplified, fused, or disrupted by the breakpoints, one should also consider position effects associated with human disease, brought upon by disturbance of underlying chromatin organization.82, 83 Fortunately, SV position effects can now be reasonably predicted by modeling changes in chromatin topology and their effects in transcriptional regulatory networks,85, 87 which could alleviate some of the analytical workload in the laboratory and provide additional candidates to explain a phenotype. It is important to consider that various genes could participate in generating the phenotypic outcome, and the contribution of each chromoanagenesis junction should be analyzed and integrated into the final diagnosis.
Discussion
As presented in this review, constitutional chromoanagenesis has been detected in phenotypically normal and developmentally affected individuals, and these events may be more common and heterogeneous than previously appreciated. While the overall prevalence of chromoanagenesis in health and disease remains to be calculated (given the various technological and interpretative challenges that these complex events pose to the clinical community), different groups have committed great amounts of resources to the systematic profiling of complex congenital SVs at nucleotide-level resolution. For more than a decade, our laboratory has focused on the study of individuals with apparently balanced SVs and presenting with various developmental disorders through the Developmental Genome Anatomy Project (DGAP),54, 56, 84, 85, 86, 90, 91, 92 while the Talkowski laboratory has also characterized complex SVs in individuals with ASD.71 Both projects have revealed with unprecedented detail the substantial abundance and involvement of considerable structural variation at junctions of balanced and unbalanced SVs, many of them with characteristic signatures of chromoanagenesis. Collins and co-authors, for example, uncovered an overwhelming number of complex SVs at ∼5 kb resolution in subjects with ASD, with each individual averaging 14 large and newly reported complex SVs, some with minimal genomic loss missed by CMA.71 Collectively, these studies suggest that the amount of constitutional complex SVs and associated chromoanagenesis events is highly underestimated, as apparently balanced SVs are, potentially, rarely truly balanced.
Several major sequencing projects have already produced thousands of human genome sequences from phenotypically normal individuals,6, 7 which could be used to survey the frequency and complexity of chromoanagenesis with higher read-depths and/or longer read technologies. In particular, long-read single-molecule sequencing has been shown to be a suitable methodology for the comprehensive characterization of SVs, as demonstrated by the study of Cretu Stancu and co-authors in the characterization of two individuals with congenital chromothripsis events,89 as well as Nattestad and co-authors, where numerous complex SVs were captured by long reads but were mostly missed by short-read sequencing in a HER2+ breast cancer cell line.93
Because complex SVs have been observed to alter genome function by truncating or disrupting disease-associated genes and their transcriptional regulatory networks, chromoanagenesis in individuals without major clinical consequences provides an opportunity to refine further the annotation of genes, regulatory elements, and chromatin organization. Alternative and focused sources for the study of chromoanagenesis in the absence of developmental features are individuals presenting with Li-Fraumeni syndrome. TP53 is involved in DNA damage response,94 which could further increase the chances of chromoanagenesis during failed DNA replication events. Indeed, acute myeloid leukemia samples of individuals with Li-Fraumeni syndrome have been shown to be enriched in chromoanagenesis events;29 it is therefore possible that chromoanagenesis-derived SVs exist in non-cancerous cells of individuals with Li-Fraumeni, the characterization of which could help elucidate their local and long-range functional impact on gene expression in various organs and tissues.
All in all, chromoanagenesis can potentially impact the function of dozens of genes in a single event, perhaps even more if we consider long-range position effects through the deletion and/or re-shuffling of regulatory elements, which can act through vast distances of genomic space.56, 59, 82, 84, 85, 86 We speculate that the majority of chromoanagenesis events have the capacity to display position effects given their extensive impact on chromosome and TAD re-organization; such effects may explain diverse aspects of individual phenotypes, and such roles will become obvious once more chromoanagenesis cases are subjected to systematic analyses of position effects.
Lastly, it is important to remember that chromoanagenesis events can be under the detection limits of various routine clinical tests (karyotype, FISH, and CMA), so it is possible that several simple or apparently balanced SVs could actually be complex chromoanagenesis events; whenever possible, nucleotide-level resolution studies of the breakpoints should be performed in normal and morbidly affected individuals presenting with balanced or unbalanced SVs for comprehensive interpretation. We are certain to find even more convoluted examples of chromoanagenesis in seemingly simple chromosome rearrangements, in addition to those already described herein. Clarifying the frequency as well as the local and long-range functional impact of chromoanagenesis is crucial for understanding human genetics, and such knowledge will be an important asset in the identification of clinically relevant genes and genomic regions. The current utilization of clinical genome sequencing95, 96 will doubtless uncover more of the unexplored potential of complex SVs and their contribution to human health and disease.
Acknowledgments
This study was funded by the National Institutes of Health (GM061354 to C.C.M.). C.C.M. is supported by the NIHR Manchester Biomedical Research Centre.
Web Resources
OMIM, http://www.omim.org/
References
- 1.Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H. DNA sequencing at 40: past, present and future. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- 2.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 3.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weischenfeldt J., Symmons O., Spitz F., Korbel J.O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 9.Huddleston J., Chaisson M.J.P., Steinberg K.M., Warren W., Hoekzema K., Gordon D., Graves-Lindsay T.A., Munson K.M., Kronenberg Z.N., Vives L. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–685. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuk L., MacDonald J.R., Tang T., Carson A.R., Li M., Rao G., Khaja R., Scherer S.W. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tommerup N. Mendelian cytogenetics. Chromosome rearrangements associated with mendelian disorders. J. Med. Genet. 1993;30:713–727. doi: 10.1136/jmg.30.9.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupski J.R. Structural variation in the human genome. N. Engl. J. Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- 14.Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 15.Lupski J.R. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ. Mol. Mutagen. 2015;56:419–436. doi: 10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malhotra D., Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlien A., Malkin D. Copy number variations and cancer. Genome Med. 2009;1:62. doi: 10.1186/gm62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B., Conroy J.M., Morrison C.D., Odunsi A.O., Qin M., Wei L., Trump D.L., Johnson C.S., Liu S., Wang J. Structural variation discovery in the cancer genome using next generation sequencing: computational solutions and perspectives. Oncotarget. 2015;6:5477–5489. doi: 10.18632/oncotarget.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger M.F., Lawrence M.S., Demichelis F., Drier Y., Cibulskis K., Sivachenko A.Y., Sboner A., Esgueva R., Pflueger D., Sougnez C. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P., Erez A., Nagamani S.C., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland A.J., Cleveland D.W. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ly P., Cleveland D.W. Rebuilding chromosomes after catastrophe: Emerging mechanisms of chromothripsis. Trends Cell Biol. 2017;27:917–930. doi: 10.1016/j.tcb.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Binsbergen E., Hochstenbach R., Giltay J., Swinkels M. Unstable transmission of a familial complex chromosome rearrangement. Am. J. Med. Genet. A. 2012;158A:2888–2893. doi: 10.1002/ajmg.a.35580. [DOI] [PubMed] [Google Scholar]
- 25.Bertelsen B., Nazaryan-Petersen L., Sun W., Mehrjouy M.M., Xie G., Chen W., Hjermind L.E., Taschner P.E., Tümer Z. A germline chromothripsis event stably segregating in 11 individuals through three generations. Genet. Med. 2016;18:494–500. doi: 10.1038/gim.2015.112. [DOI] [PubMed] [Google Scholar]
- 26.Gu H., Jiang J.H., Li J.Y., Zhang Y.N., Dong X.S., Huang Y.Y., Son X.M., Lu X., Chen Z. A familial Cri-du-Chat/5p deletion syndrome resulted from rare maternal complex chromosomal rearrangements (CCRs) and/or possible chromosome 5p chromothripsis. PLoS ONE. 2013;8:e76985. doi: 10.1371/journal.pone.0076985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Pagter M.S., van Roosmalen M.J., Baas A.F., Renkens I., Duran K.J., van Binsbergen E., Tavakoli-Yaraki M., Hochstenbach R., van der Veken L.T., Cuppen E., Kloosterman W.P. Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am. J. Hum. Genet. 2015;96:651–656. doi: 10.1016/j.ajhg.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rode A., Maass K.K., Willmund K.V., Lichter P., Ernst A. Chromothripsis in cancer cells: An update. Int. J. Cancer. 2016;138:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 29.Rausch T., Jones D.T., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng C., Zhou Y., Li H., Xiong T., Li S., Bi Y., Kong P., Wang F., Cui H., Li Y. Whole-genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 2016;98:256–274. doi: 10.1016/j.ajhg.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker M., Mohankumar K.M., Punchihewa C., Weinlich R., Dalton J.D., Li Y., Lee R., Tatevossian R.G., Phoenix T.N., Thiruvenkatam R. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H., Sandberg A.A. Chromosome pulverization in human cells with micronuclei. J. Natl. Cancer Inst. 1968;40:165–179. [PubMed] [Google Scholar]
- 33.Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ly P., Teitz L.S., Kim D.H., Shoshani O., Skaletsky H., Fachinetti D., Page D.C., Cleveland D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017;19:68–75. doi: 10.1038/ncb3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloosterman W.P., Hoogstraat M., Paling O., Tavakoli-Yaraki M., Renkens I., Vermaat J.S., van Roosmalen M.J., van Lieshout S., Nijman I.J., Roessingh W. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malhotra A., Lindberg M., Faust G.G., Leibowitz M.L., Clark R.A., Layer R.M., Quinlan A.R., Hall I.M. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morishita M., Muramatsu T., Suto Y., Hirai M., Konishi T., Hayashi S., Shigemizu D., Tsunoda T., Moriyama K., Inazawa J. Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam irradiation system. Oncotarget. 2016;7:10182–10192. doi: 10.18632/oncotarget.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maciejowski J., Li Y., Bosco N., Campbell P.J., de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePinho R.A., Polyak K. Cancer chromosomes in crisis. Nat. Genet. 2004;36:932–934. doi: 10.1038/ng0904-932. [DOI] [PubMed] [Google Scholar]
- 41.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson N.D., de Borja R., Young M.D., Fuligni F., Rosic A., Roberts N.D., Hajjar S., Layeghifard M., Novokmet A., Kowalski P.E. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science. 2018;361:361. doi: 10.1126/science.aam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y., Park K., Kitabayashi N., MacDonald T.Y., Ghandi M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haffner M.C., Aryee M.J., Toubaji A., Esopi D.M., Albadine R., Gurel B., Isaacs W.B., Bova G.S., Liu W., Xu J. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 47.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 49.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan L., Xing D., Chang C.H., Li H., Xie X.S. Three-dimensional genome structures of single diploid human cells. Science. 2018;361:924–928. doi: 10.1126/science.aat5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kloosterman W.P., Guryev V., van Roosmalen M., Duran K.J., de Bruijn E., Bakker S.C., Letteboer T., van Nesselrooij B., Hochstenbach R., Poot M., Cuppen E. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum. Mol. Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 53.Kloosterman W.P., Tavakoli-Yaraki M., van Roosmalen M.J., van Binsbergen E., Renkens I., Duran K., Ballarati L., Vergult S., Giardino D., Hansson K. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Chiang C., Jacobsen J.C., Ernst C., Hanscom C., Heilbut A., Blumenthal I., Mills R.E., Kirby A., Lindgren A.M., Rudiger S.R. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012;44:390–397, S1. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nazaryan L., Stefanou E.G., Hansen C., Kosyakova N., Bak M., Sharkey F.H., Mantziou T., Papanastasiou A.D., Velissariou V., Liehr T. The strength of combined cytogenetic and mate-pair sequencing techniques illustrated by a germline chromothripsis rearrangement involving FOXP2. Eur. J. Hum. Genet. 2014;22:338–343. doi: 10.1038/ejhg.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Redin C., Brand H., Collins R.L., Kammin T., Mitchell E., Hodge J.C., Hanscom C., Pillalamarri V., Seabra C.M., Abbott M.A. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nazaryan-Petersen L., Bertelsen B., Bak M., Jønson L., Tommerup N., Hancks D.C., Tümer Z. Germline chromothripsis driven by L1-mediated retrotransposition and Alu/Alu homologous recombination. Hum. Mutat. 2016;37:385–395. doi: 10.1002/humu.22953. [DOI] [PubMed] [Google Scholar]
- 58.Weckselblatt B., Hermetz K.E., Rudd M.K. Unbalanced translocations arise from diverse mutational mechanisms including chromothripsis. Genome Res. 2015;25:937–947. doi: 10.1101/gr.191247.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Middelkamp S., van Heesch S., Braat A.K., de Ligt J., van Iterson M., Simonis M., van Roosmalen M.J., Kelder M.J., Kruisselbrink E., Hochstenbach R. Molecular dissection of germline chromothripsis in a developmental context using patient-derived iPS cells. Genome Med. 2017;9:9. doi: 10.1186/s13073-017-0399-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genesio R., Fontana P., Mormile A., Casertano A., Falco M., Conti A., Franzese A., Mozzillo E., Nitsch L., Melis D. Constitutional chromothripsis involving the critical region of 9q21.13 microdeletion syndrome. Mol. Cytogenet. 2015;8:96. doi: 10.1186/s13039-015-0199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamba B.F., Richieri-Costa A., Costa S., Rosenberg C., Ribeiro-Bicudo L.A. Chromothripsis with at least 12 breaks at 1p36.33-p35.3 in a boy with multiple congenital anomalies. Mol. Genet. Genomics. 2015;290:2213–2216. doi: 10.1007/s00438-015-1072-0. [DOI] [PubMed] [Google Scholar]
- 62.Magrangeas F., Avet-Loiseau H., Munshi N.C., Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molenaar J.J., Koster J., Zwijnenburg D.A., van Sluis P., Valentijn L.J., van der Ploeg I., Hamdi M., van Nes J., Westerman B.A., van Arkel J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 64.Qian Y.Q., Fu X.Y., Wang X.Q., Luo Y.Q., Chen M., Yan K., Yang Y.M., Liu B., Wang L.Y., Huang Y.Z. A feasible diagnostic approach for the translocation carrier from the indication of products of conception. Mol. Cytogenet. 2018;11:12. doi: 10.1186/s13039-018-0362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C.Z., Leibowitz M.L., Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 67.Grossmann V., Höckner M., Karmous-Benailly H., Liang D., Puttinger R., Quadrelli R., Röthlisberger B., Huber A., Wu L., Spreiz A. Parental origin of apparently balanced de novo complex chromosomal rearrangements investigated by microdissection, whole genome amplification, and microsatellite-mediated haplotype analysis. Clin. Genet. 2010;78:548–553. doi: 10.1111/j.1399-0004.2010.01419.x. [DOI] [PubMed] [Google Scholar]
- 68.Zanardo E.A., Piazzon F.B., Dutra R.L., Dias A.T., Montenegro M.M., Novo-Filho G.M., Costa T.V., Nascimento A.M., Kim C.A., Kulikowski L.D. Complex structural rearrangement features suggesting chromoanagenesis mechanism in a case of 1p36 deletion syndrome. Mol. Genet. Genomics. 2014;289:1037–1043. doi: 10.1007/s00438-014-0876-7. [DOI] [PubMed] [Google Scholar]
- 69.Plaisancié J., Kleinfinger P., Cances C., Bazin A., Julia S., Trost D., Lohmann L., Vigouroux A. Constitutional chromoanasynthesis: description of a rare chromosomal event in a patient. Eur. J. Med. Genet. 2014;57:567–570. doi: 10.1016/j.ejmg.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Masset H., Hestand M.S., Van Esch H., Kleinfinger P., Plaisancié J., Afenjar A., Molignier R., Schluth-Bolard C., Sanlaville D., Vermeesch J.R. A distinct class of chromoanagenesis events characterized by focal copy number gains. Hum. Mutat. 2016;37:661–668. doi: 10.1002/humu.22984. [DOI] [PubMed] [Google Scholar]
- 71.Collins R.L., Brand H., Redin C.E., Hanscom C., Antolik C., Stone M.R., Glessner J.T., Mason T., Pregno G., Dorrani N. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017;18:36. doi: 10.1186/s13059-017-1158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grochowski C.M., Gu S., Yuan B., Tcw J., Brennand K.J., Sebat J., Malhotra D., McCarthy S., Rudolph U., Lindstrand A. Marker chromosome genomic structure and temporal origin implicate a chromoanasynthesis event in a family with pleiotropic psychiatric phenotypes. Hum. Mutat. 2018;39:939–946. doi: 10.1002/humu.23537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabatini P.J.B., Ejaz R., Stavropoulos D.J., Mendoza-Londono R., Joseph-George A.M. Stable transmission of an unbalanced chromosome 21 derived from chromoanasynthesis in a patient with a SYNGAP1 likely pathogenic variant. Mol. Cytogenet. 2018;11:50. doi: 10.1186/s13039-018-0394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pettersson M., Eisfeldt J., Syk Lundberg E., Lundin J., Lindstrand A. Flanking complex copy number variants in the same family formed through unequal crossing-over during meiosis. Mutat. Res. 2018;812:1–4. doi: 10.1016/j.mrfmmm.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 76.Symmons O., Uslu V.V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., Ettwiller L., Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhan Y., Mariani L., Barozzi I., Schulz E.G., Blüthgen N., Stadler M., Tiana G., Giorgetti L. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27:479–490. doi: 10.1101/gr.212803.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krefting J., Andrade-Navarro M.A., Ibn-Salem J. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 2018;16 doi: 10.1186/s12915-018-0556-x. 87–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spielmann M., Lupiáñez D.G., Mundlos S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 83.Andrey G., Mundlos S. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017;144:3646–3658. doi: 10.1242/dev.148304. [DOI] [PubMed] [Google Scholar]
- 84.Ordulu Z., Kammin T., Brand H., Pillalamarri V., Redin C.E., Collins R.L., Blumenthal I., Hanscom C., Pereira S., Bradley I. Structural chromosomal rearrangements require nucleotide-level resolution: Lessons from next-generation sequencing in prenatal diagnosis. Am. J. Hum. Genet. 2016;99:1015–1033. doi: 10.1016/j.ajhg.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zepeda-Mendoza C.J., Ibn-Salem J., Kammin T., Harris D.J., Rita D., Gripp K.W., MacKenzie J.J., Gropman A., Graham B., Shaheen R. Computational prediction of position effects of apparently balanced human chromosomal rearrangements. Am. J. Hum. Genet. 2017;101:206–217. doi: 10.1016/j.ajhg.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zepeda-Mendoza C.J., Bardon A., Kammin T., Harris D.J., Cox H., Redin C., Ordulu Z., Talkowski M.E., Morton C.C. Phenotypic interpretation of complex chromosomal rearrangements informed by nucleotide-level resolution and structural organization of chromatin. Eur. J. Hum. Genet. 2018;26:374–381. doi: 10.1038/s41431-017-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bianco S., Lupiáñez D.G., Chiariello A.M., Annunziatella C., Kraft K., Schöpflin R., Wittler L., Andrey G., Vingron M., Pombo A. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat. Genet. 2018;50:662–667. doi: 10.1038/s41588-018-0098-8. [DOI] [PubMed] [Google Scholar]
- 88.McDermott D.H., Gao J.L., Liu Q., Siwicki M., Martens C., Jacobs P., Velez D., Yim E., Bryke C.R., Hsu N. Chromothriptic cure of WHIM syndrome. Cell. 2015;160:686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cretu Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Talkowski M.E., Ernst C., Heilbut A., Chiang C., Hanscom C., Lindgren A., Kirby A., Liu S., Muddukrishna B., Ohsumi T.K. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am. J. Hum. Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Talkowski M.E., Ordulu Z., Pillalamarri V., Benson C.B., Blumenthal I., Connolly S., Hanscom C., Hussain N., Pereira S., Picker J. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N. Engl. J. Med. 2012;367:2226–2232. doi: 10.1056/NEJMoa1208594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ordulu Z., Wong K.E., Currall B.B., Ivanov A.R., Pereira S., Althari S., Gusella J.F., Talkowski M.E., Morton C.C. Describing sequencing results of structural chromosome rearrangements with a suggested next-generation cytogenetic nomenclature. Am. J. Hum. Genet. 2014;94:695–709. doi: 10.1016/j.ajhg.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nattestad M., Goodwin S., Ng K., Baslan T., Sedlazeck F.J., Rescheneder P., Garvin T., Fang H., Gurtowski J., Hutton E. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018;28:1126–1135. doi: 10.1101/gr.231100.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broustas C.G., Lieberman H.B. DNA damage response genes and the development of cancer metastasis. Radiat. Res. 2014;181:111–130. doi: 10.1667/RR13515.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krier J.B., Kalia S.S., Green R.C. Genomic sequencing in clinical practice: applications, challenges, and opportunities. Dialogues Clin. Neurosci. 2016;18:299–312. doi: 10.31887/DCNS.2016.18.3/jkrier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prokop J.W., May T., Strong K., Bilinovich S.M., Bupp C., Rajasekaran S., Worthey E.A., Lazar J. Genome sequencing in the clinic: the past, present, and future of genomic medicine. Physiol. Genomics. 2018;50:563–579. doi: 10.1152/physiolgenomics.00046.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an -information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]