Abstract
The extent to which genetic risk factors are shared between childhood-onset (COA) and adult-onset (AOA) asthma has not been estimated. On the basis of data from the UK Biobank study (n = 447,628), we found that the variance in disease liability explained by common variants is higher for COA (onset at ages between 0 and 19 years; h2g = 25.6%) than for AOA (onset at ages between 20 and 60 years; h2g = 10.6%). The genetic correlation (rg) between COA and AOA was 0.67. Variation in age of onset among COA-affected individuals had a low heritability (h2g = 5%), which we confirmed in independent studies and also among AOA-affected individuals. To identify subtype-specific genetic associations, we performed a genome-wide association study (GWAS) in the UK Biobank for COA (13,962 affected individuals) and a separate GWAS for AOA (26,582 affected individuals) by using a common set of 300,671 controls for both studies. We identified 123 independent associations for COA and 56 for AOA (37 overlapped); of these, 98 and 34, respectively, were reproducible in an independent study (n = 262,767). Collectively, 28 associations were not previously reported. For 96 COA-associated variants, including five variants that represent COA-specific risk factors, the risk allele was more common in COA- than in AOA-affected individuals. Conversely, we identified three variants that are stronger risk factors for AOA. Variants associated with obesity and smoking had a stronger contribution to the risk of AOA than to the risk of COA. Lastly, we identified 109 likely target genes of the associated variants, primarily on the basis of correlated expression quantitative trait loci (up to n = 31,684). GWAS informed by age of onset can identify subtype-specific risk variants, which can help us understand differences in pathophysiology between COA and AOA and so can be informative for drug development.
Keywords: asthma, allergy, onset, age, genetic, risk, heritability, overlap, GWAS, genome
Introduction
The age at which asthma (MIM: 600807) symptoms first develop is often used for identification of different disease subtypes,1, 2, 3, 4, 5, 6, 7 broadly separating individuals into two groups: those with childhood-onset disease and those with adult-onset disease. Perinatal factors, atopy (MIM: 147050), viral respiratory-tract infections, and the microbiome are thought to play a key role in the development of childhood-onset asthma (COA),8, 9, 10, 11 whereas adult-onset asthma (AOA) is more strongly associated with obesity (MIM: 601665), smoking, and other environmental and occupational exposures.12, 13 Such differences in etiology suggest that genetic risk factors might also be partly distinct between COA and AOA. This hypothesis, which to our knowledge has not been formally tested to date, is supported by the observations that asthma risk alleles are enriched among individuals with early-onset disease.14, 15, 16 Moreover, it has been shown that the efficacy of novel anti-cytokine asthma therapies, such as those involving anti-IL-517 and the anti-IL-5Rα,18 is greater in adult-onset disease, again pointing to age-of-onset-dependent disease mechanisms.
In this study, we used genetic data and information on asthma reported by participants from the UK Biobank study to address three main questions: To what extent do the same genetic variants influence the risk of both COA and AOA? Do genetic variants influence the specific age at which asthma first develops during childhood and during adulthood? Can we identify genetic variants that are risk factors for one disease subtype but not (or less so) for the other? Addressing these questions can potentially help us understand differences in pathophysiology between individuals with COA and individuals with AOA; this understanding could have implications for identifying age-of-onset-specific drug targets.
Subjects and Methods
Selection of Asthma-Affected Individuals from the UK Biobank Study
We first identified 53,031 individuals with self-reported doctor-diagnosed asthma among 488,365 participants from the UK Biobank study.19 We identified them on the basis of information from four data fields: field 6152 (“Has a doctor ever told you that you have had any of the following conditions?” asked on a touchscreen questionnaire), field 20002 (verbal interview), field 41202 (Diagnoses—main ICD10), and field 41204 (Diagnoses—secondary ICD10). Specifically, affected individuals had (1) either a report of “asthma” in field 6152 and a code for asthma in field 20002 or an ICD10 code for asthma in field 41202 or 41204; and (2) no report of chronic obstructive pulmonary disease (COPD; MIM: 606963) in fields 6152 or 20002, nor of other respiratory diseases in field 20002. We then excluded 9,984 individuals (leaving 43,047) on the basis of two data fields that recorded information age at asthma onset: field 3786 (“What was your age when the asthma was first diagnosed?” asked on a touchscreen questionnaire) and field 22147 (“Age you were first diagnosed by a doctor” on an online follow-up questionnaire completed only by a subset of participants). Specifically, we excluded asthma-affected individuals with (1) missing information for field 3786 (n = 7,393); (2) a diagnosis of asthma after the age of 60 (to further minimize potential confounding with COPD; n = 1,723); or (3) age at asthma onset reported in field 22147 that was >10 years apart from that reported in field 3786 (n = 868). Lastly, we excluded 2,503 asthmatics who (1) did not cluster within five standard deviations of the mean for the first and second MDS components estimated for individuals from the five European ancestry groups (CEU, GBR, FIN, IBS, and TSI) of the 1000 Genomes Project, as described previously;15 (2) had a self-reported sex different from their genetically inferred sex; (3) were outliers with regard to genotype missing rates and/or genome-wide heterozygosity levels; (4) had more than 10 third-degree relatives or were excluded from kinship inference; and/or (5) were not present in the imputed dataset released in July 2017. After these exclusions, there were 40,544 asthmatics available for analysis.
Classification of UK Biobank Asthma-Affected Individuals into Three Groups Based on Age at First Diagnosis
We used the age at first diagnosis reported in fields 3786 and 22147 (the age was averaged when values in both fields were available; see Figure S1) to group the 40,544 asthmatics into three non-overlapping groups: those first diagnosed as (1) a child or teenager, specifically at or before age 19 (n = 13,962); (2) a young or middle-aged adult, between the ages of 20 and 39 (n = 11,709); or (3) an older adult, between the ages of 40 and 60 (n = 14,873). We used a cut-off of age 19 to define COA so that those diagnosed as an adolescent or teenager (ages 13 to 19; n = 3,164) were included in the same group as those diagnosed as a child (ages 0 to 12; n = 10,798). For heritability and genetic-correlation analyses, we initially split those diagnosed as adults into two groups because data from longitudinal epidemiological studies show that asthma that presents in early to mid-adult life often has its onset in childhood.20, 21, 22, 23 As such, those diagnosed between 20 and 39 were expected to represent a more heterogeneous group of asthma-affected individuals with respect to the underlying disease course. We used a cut-off age of 39 so that both groups spanned a similar age range (∼20 years). We refer to the three affected groups as those with COA, those with young adult-onset asthma (yAOA), and those with older-adult-onset asthma (oAOA), respectively. Demographics for the three groups are summarized in Table S1. The main clinical differences between these groups were a male predominance in COA and a female predominance in AOA; affected individuals with AOA were also more likely to be obese. On the basis of results from the heritability and genetic correlation analyses, we then combined the yAOA and oAOA into a single AOA group (n = 26,582) for genome-wide association analyses.
Variation in Asthma Liability That Is Explained by Common Genetic Variants
We used the BOLT-REML24 algorithm to estimate what proportion of phenotypic variance in asthma case-control status that was explained by common single-nucleotide polymorphisms (SNPs); we call this proportion SNP heritability (h2g). This analysis was performed separately for COA, yAOA, oAOA, and AOA; controls for these analyses were all other individuals who were of European ancestry, who were included in the UK Biobank study, who did not satisfy the specific case definition for each group, and who passed the quality-control filters described above. Specifically, we identified 433,306 individuals without COA, 435,559 without yAOA, 432,395 without oAOA, and 420,686 without AOA. We did not exclude from the control groups individuals who suffered from the other asthma subtypes because this would have resulted in inflated heritability estimates (as a result of excluding controls who suffered from a genetically correlated trait).
We included as model SNPs in the BOLT-REML analysis ∼1 million autosomal SNPs from HapMap3. These had minor-allele frequencies (MAF) >1% and a <2% missing rate; discrete (i.e., hardcall) genotypes for these SNPs were derived from the ukb_imp_chr[1:22]_v3.bgen files with PLINK v. 2.00,25 with a missing genotype assigned when the inferred minor-allele dosage was >0.1 from the nearest hardcall (i.e., 0, 1 or 2). We included sex and an indicator of the array used for genotyping as covariates. The estimated SNP heritability (and its standard error) was converted to the liability scale with the formula described by Lee et al.26
Overlap in Genetic Risk Factors between COA and AOA
To determine the extent to which the same genetic risk factors contribute to the risk of COA and AOA, we used BOLT-REML as described above to estimate the pairwise genetic correlation between COA (13,962 affected individuals and 433,306 controls) and (1) yAOA (11,709 affected individuals and 435,559 controls), (2) oAOA (14,873 affected individuals and 432,395 controls), and (3) AOA (26,582 affected individuals and 420,686 controls). Pairs of genetic correlations were compared statistically with a Z test. For comparison, genetic correlations were also estimated with a different approach, linkage disequilibrium (LD)-score regression,27 based on association results from ∼1 million autosomal SNPs from HapMap3.
To understand whether the genetic etiology of COA and AOA had a comparable allergic component, we used BOLT-REML to estimate the genetic correlation between asthma-onset subtypes and a hay fever (MIM: 607154) and eczema (MIM: 603165) combined phenotype created with information provided in data field 6152, which asked, “Has a doctor ever told you that you had any of the following conditions?”. Of the individuals who were of European descent and answered this question but did not meet the exclusion filters described above, 106,782 selected the “Hayfever, allergic rhinitis or eczema” option and so were considered to be affected individuals, whereas the remaining 353,418 individuals were considered to be controls.
Contribution of Common SNPs to Variation in Age at First Diagnosis
The SNP heritability of age at first diagnosis in the UK Biobank study was estimated separately for affected individuals with COA (n = 13,962) and for those with AOA (n = 26,582) through the use of BOLT-REML as described above. Age at first diagnosis was normalized via an inverse normal transformation (ties were broken randomly) prior to the BOLT-REML analysis. We also determined whether the observed heritability of age of onset in COA-affected individuals of the UK Biobank study was consistent with that estimated on the basis of data from 3,649 COA-affected individuals identified in two independent studies described below: the Avon Longitudinal Study of Parents and Children (ALSPAC),28 described in detail in the Supplemental Data, and the Child and Adolescent Twin Study in Sweden (CATSS).28
ALSPAC
We identified 1,326 unrelated children who were of European descent and whose parent gave a positive response to the question “Did your child have asthma in the past 12 months?” in surveys completed when the child was approximately 7, 8, 9, 11, or 13 years old. Age of onset was defined as the age at which “wheeze” or “wheezing and whistling” was first reported in these surveys. The presence of wheezing was identified on the basis of a positive response to the question: “Has your child had wheezing, breathlessness, or episodes of stopping breathing in past 12 months or since he was (age at last questionnaire)?” “Wheezing and whistling” was identified on the basis of a positive response to the question: “Has your child had any periods when there was wheezing with whistling in his chest when he breathed in the past 12 months or since he was (age at last questionnaire)?” We then performed a GWAS of age of onset (which was normalized with an inverse normal transformation) by using SNPTEST with sex included as a covariate; we tested 8.4 million SNPs imputed on the basis of the 1000 Genomes Project reference panel (phase 1, version 3, released in December 2013).
CATSS
Data on asthma during childhood was collected from parental questionnaires conducted through the Swedish Twin Registry for twins born between 1992 and 1999. The questionnaires were completed when the twins were aged 9, 12, and 15. Specifically, the parents reported the age at which the twins first had asthma or wheezing or breathlessness. This information was supplemented with data from (1) the National Patient Register (NPR),29 which specifically provided the age at which asthma was first diagnosed by a doctor (ICD-10 code J45 or J46 or ICD-9 code 493), and (2) the Swedish Prescribed Drug Register (SPDG), which specifically provided the age at which the first asthma preventer medication was recorded (ATC codes R03AK, R03BA, and R03DC) for anyone with two or more prescriptions before age 20. Asthma age of onset was defined as the youngest age across all three data sources (i.e., parental questionnaires, NPR, and SPDG). After excluding individuals of non-European ancestry, we identified 2,323 twins (42% female) with asthma onset at or before age 19 (mean age of onset of 3.6). DNA samples from these twins were genotyped on the Illumina PsychChip (which includes a genome-wide association study [GWAS] backbone consisting of 265,000 tag SNPs) and, after sample and SNP quality control (described in the Supplemental Data), imputed on the basis of the 1000 Genomes phase 3 reference panel. The association between age of onset (for which quantiles were normalized) and SNP allelic dosage was tested with RAREMETALWORKER version 4.13.8; known relatedness within the sample was accounted for.
Meta-Analysis and Estimation of SNP Heritability
Age-of-onset GWAS results from the ALSPAC and CATSS studies were combined via an inverse-variance-weighted, fixed-effects meta-analysis.29 We then applied the LD-score regression approach,30 based on 1.1 million HapMap3 SNPs, to the meta-analysis results to estimate the proportion of age-of-onset variance explained by common variants.
Identification of Genetic Associations with COA and AOA
We performed a GWAS of COA (13,962 affected individuals versus 300,671 controls) and a GWAS of AOA (26,582 affected individuals versus 300,671 controls) in the UK Biobank study. We used a common set of 300,671 controls who did not suffer from any allergic disease (asthma, hay fever, eczema, or other allergies). We used this selected subset of controls for the GWAS, and not the larger set (∼430,000) used in the heritability analyses, because one can improve the power to detect associations with asthma by excluding from the control group individuals who suffer from other genetically correlated allergic diseases.31
To identify the 300,671 non-allergic controls, we used information provided in the data fields described above (6152, 20002, 41202, 41204), as well as in field 22127: “Has a doctor ever told you that you have had any of the conditions below?” which included “hay fever or allergic rhinitis” and “asthma” as possible answers. SNPs were tested for association via the linear mixed model implemented in BOLT-LMM.32 This model accounted for the presence of related individuals and for any residual population stratification among Europeans. We included as model SNPs 553,880 autosomal variants that were directly genotyped and had an MAF >1%, a call rate >95%, and a Hardy-Weinberg equilibrium p value >10−6. Age, sex, and an indicator of the genotyping array used were included as discrete covariates.
Of the 92 million variants with imputed data released by the UK Biobank, we retained results for nine million variants that (1) had an MAF >1%, (2) had matching alleles and were polymorphic in Europeans (n = 294) from the 1000 Genomes project, (3) had a unique reference sequence (rs) number and genomic position (based on hg19), and (4) had an imputation info score >0.5. For each SNP, the beta and SE were estimated on the basis of a linear model, and so both were subsequently adjusted according to the formulae adj beta = beta/(mu∗(1-mu)) and adj SE = SE/(mu∗(1-mu)), where mu is approximated by the case/control ratio. The resulting SE was then inflated by the square root of the LD-score-regression33 intercept (respectively 1.039 and 1.018 for COA and AOA), which most likely reflects inflation of test statistics as a result of unaccounted biases, and the association p value recalculated on the basis of the corrected SE. We used a p value threshold of 3 × 10−8 for genome-wide significance, as suggested for studies that analyze variants with a MAF >1%.34
For secondary analyses, we used the same approach to perform a GWAS each of (1) asthma onset type, specifically by comparing COA-affected individuals (n = 13,962, coded as “1”) with AOA-affected individuals (n = 14,873, coded as “0”), i.e., an affected-individual-only association analysis; and (2) asthma case-control status, where affected individuals were identified irrespective of age at first diagnosis (40,544 affected individuals versus 300,671 controls; SE and p value adjusted for an LD-score-regression intercept of 1.053). We performed the latter analysis to determine how many associations identified in the COA or AOA GWAS would have been identified had we instead performed an asthma GWAS that was not informed by age-of-onset information.
Identification of Variants with a Statistically Independent Association with Asthma Risk
We used the approximate joint association analysis option of GCTA35 to identify variants that remained associated with asthma risk at a p < 3 × 10−8 in the COA and AOA GWAS after accounting for the effects of nearby (<10 Mb) more strongly associated variants. We refer to these as sentinel risk variants. LD was estimated on the basis of a random subset of 5,000 individuals from the UK Biobank study.
Validation of SNP Associations
To determine whether SNP associations identified in the UK Biobank study were reproducible, we used the same age cut-offs described above to identify COA-affected individuals, AOA-affected individuals, and allergy-free controls (i.e., no asthma, eczema, rhinitis or any other allergic conditions) among research participants of the personal genetics company 23andMe (see Supplemental Data). After we restricted the analyses to unrelated individuals of confirmed European descent, sample sizes for the three association analyses performed in the replication study were as follows: (1) 31,759 COA-affected individuals versus 214,890 controls; (2) 16,297 AOA-affected individuals versus 217,711 controls; and (3) 31,002 COA-affected individuals (coded “1”) versus 16,297 AOA-affected individuals (coded “0”). The number of controls was lower in analysis (1) than in (2) because an additional 2,821 controls were relatives of COA-affected individuals and so were excluded from the analysis. Similarly, 757 COA-affected individuals were relatives of AOA-affected individuals and so were not included in analysis (3). Sentinel SNPs identified in the UK Biobank GWAS were tested for association in the 23andMe study through the use of logistic regression, for which an additive model for allelic effects was assumed and age, sex, and five ancestry-informative principal components were included as covariates. Association results from these analyses were conservatively adjusted for genomic control inflation factors of 1.101, 1.061, and 1.047, respectively.
Sentinel Risk Variants Not Previously Implicated in the Etiology of Allergic Disease
To determine whether a sentinel variant was in LD with an SNP previously reported to associate with any allergic disease, we (1) identified all SNPs in LD (r2 > 0.05) with that sentinel variant by using genotype data from European-descended individuals from the 1000 Genomes Project36 (n = 294, release 20130502_v5a); and (2) determined whether the sentinel variant or any of the correlated SNPs identified were reported to associate with any allergic disease (asthma, hay fever, eczema, food allergy, or atopy) in the NHGRI-EBI GWAS catalog database,37 which was downloaded on the 30th of September 2018.
Genetic Correlation between COA and AOA and Other Complex Diseases or Traits
To provide some insight into potential differences in genetic etiology between COA and AOA, we used the LD-score-regression approach to estimate the pairwise genetic correlation between each disease subtype and all 227 common traits or diseases with GWAS data currently available in LD Hub.38 We uploaded to LD Hub association results from ∼1.1 million HapMap3 SNPs, obtained when comparing 13,962 affected individuals with 433,306 controls for COA, and when comparing 26,582 affected individuals with 420,686 controls for AOA, as done in the BOLT-REML analysis.
Using LD with Expression Quantitative Trait Loci and Non-Synonymous SNPs to Predict Target Genes of Sentinel Variants
We performed the following steps to identify genes for which variation in gene expression and/or protein sequence was associated with sentinel SNPs identified in the COA and AOA GWAS:
First, we identified SNPs associated with variation in gene expression (i.e., expression quantitative trait loci, eQTLs) in published transcriptome studies of five broad tissue types relevant for asthma: individual immune cell types, lung, skin, spleen, and whole blood. We identified a total of 50 transcriptome studies reporting results from eQTL analyses in any one of those five tissue types (Table S2). Some studies included multiple cell types, experimental conditions, and/or eQTL types, resulting in a total of 130 separate eQTL datasets. For each eQTL dataset, we then (1) downloaded the original publication tables or files containing results for the eQTLs reported, (2) extracted the SNP identifier, gene name, association p value, and directional effect (if available; beta or z score and effect allele), (3) excluded eQTLs located >1 Mb from the respective gene (i.e., trans eQTsL) because often these are thought to be mediated by cis effects,39 (4) excluded eQTLs with an association p > 8.9 × 10−10, a conservative threshold that corrects for 55,765 genes (on the basis of GENCODE v. 19), each tested for association with 1,000 SNPs (as suggested by others40, 41, 42); and (5) for each gene, we used the --clump procedure in PLINK25 to reduce the list of eQTL identified (which often included many correlated SNPs) to a set of “sentinel eQTLs,” defined as the SNPs with the strongest association with gene expression and in low LD (r2 < 0.05, LD window of 2 Mb) with each other.
Second, we identified genes for which a sentinel eQTL reported in any of the 130 eQTL datasets described above was in high LD (r2 > 0.8) with a sentinel variant identified in the COA or AOA GWAS. That is, we only considered genes for which there was high LD between a sentinel eQTL and a sentinel asthma risk variant, which reduces the chance of spurious co-localization.
Third, we used wANNOVAR43 to identify non-synonymous SNPs among all variants in LD (r2 > 0.8) with any sentinel variants. We identified SNPs in LD with sentinel variants by using genotype data from European-descended individuals from the 1000 Genomes Project36 (n = 294; release 20130502_v5a).
It is important to note that genes identified as likely target genes on the basis of LD between risk SNPs and eQTLs and/or non-synonymous variants represent predictions that need to be confirmed by subsequent functional studies.
Ethical Approval
Appropriate informed consent was obtained from all study participants. Ethical approval for the study was obtained from the Human Ethics Committee of the QIMR Berghofer Medical Research Institute, the ALSPAC Ethics and Law Committee, and the local research ethics committees.
Results
Overlap in Genetic Risk Factors Between COA and AOA
When affected individuals in the UK Biobank study were defined as asthmatics who reported disease onset as a child (age 0 to 19, n = 13,962; demographics in Table S1) and controls as all other individuals (n = 433,306), the liability-scale SNP heritability (h2g) estimated with the BOLT-REML algorithm24 was 25.6% (SE = 0.67%; Table 1). Substantially lower SNP heritability estimates were obtained when we considered asthmatics who reported developing asthma as a young adult (age 20 to 39; 11,709 affected individuals versus 435,559 controls) or older adult (age 40 to 60; 14,873 affected individuals versus 432,395 controls): h2g = 9.8% (SE = 0.59%) and h2g = 7.8% (SE = 0.49%), respectively (Table 1).
Table 1.
Liability-Scale SNP-Based Heritability (Diagonal) of, and Genetic Correlation (Lower Diagonal) between, COA and AOA in the UK Biobank Study
|
COA (Onset: 0–19 yr) |
yAOA (Onset: 20–39 yr) |
oAOA (Onset: 40–60 yr) |
AOA (Onset: 20–60 yr) |
|
|---|---|---|---|---|
| 13,962 versus 433,306 | 11,709 versus 435,559 | 14,873 versus 432,395 | 26,582 versus 420,686 | |
| COA | 0.256 (0.007) | - | - | - |
| yAOA | 0.831 (0.032) | 0.098 (0.006) | - | - |
| oAOA | 0.469 (0.031) | 0.856 (0.050) | 0.078 (0.005) | - |
| AOA | 0.667 (0.023) | - | - | 0.106 (0.004) |
Liability-scale SNP-based heritability is shown on the diagonal, and genetic correlation is shown on the lower diagonal. Standard error of the heritability estimate is shown in parentheses.
We then estimated the extent to which the same genetic risk factors contributed to the heritability of COA and AOA. Using BOLT-REML, we observed a modest genetic correlation (rg = 0.47, SE = 0.03; Table 1) between COA and oAOA. Similar results were obtained with the LD-score-regression approach (rg = 0.42, SE = 0.06). In contrast, larger genetic correlations were observed between yAOA and both COA (rg = 0.83, SE = 0.03) and oAOA (rg = 0.86, SE = 0.05).
The large genetic correlation observed between yAOA and oAOA, together with their similar heritability estimates, suggests that most SNPs associated with developing asthma in adulthood have broadly similar allele frequencies in yAOA and oAOA cases. This is not the case between COA and yAOA, nor between COA and oAOA, given the large differences in SNP heritability and, in the case of oAOA, also the modest pairwise genetic correlation. For these reasons, we then combined the yAOA- and oAOA-affected individuals into a single group with AOA (n = 26,582). When we compared AOA-affected individuals against all other individuals as controls (n = 420,686), we observed an SNP heritability on the liability scale of 10.6% (SE = 0.38%), and the genetic correlation with COA was estimated to be 0.67 (SE = 0.023; Table 1). Similar results were obtained with LD-score regression (rg = 0.63, SE = 0.054). These results show that the genetic correlation between COA and AOA is significantly lower than 1 (p = 10−46).
To investigate whether the genetic etiology of COA had a larger allergic component than AOA, we then estimated their pairwise genetic correlation with a combined hay-fever-and-eczema phenotype measured in the same UK Biobank participants (n = 447,268). Using BOLT-REML, we found that genetic risk factors for hay fever and eczema (h2g = 15.6%, SE = 0.26%) significantly overlapped those for both COA and AOA, but the overlap was larger for the former: rg = 0.70 (SE = 0.013) versus rg = 0.57 (SE = 0.017). Similar results were obtained when we analyzed doctor-diagnosed hay fever (rg = 0.67 versus rg = 0.56); those data were available for a subset of UK Biobank individuals (n = 111,664).
Collectively, these results demonstrate that in the UK Biobank study, asthma reported to first develop in childhood has a genetic architecture that is partly similar to and partly distinct from asthma reported to first develop in adulthood.
Contribution of Common SNPs to Variation in Age at First Diagnosis among Affected Individuals with COA and AOA
Next, we used BOLT-REML to test whether common SNPs also influenced the specific age at which asthma was diagnosed during childhood and, separately, during adulthood. We first performed this analysis in the 13,962 affected individuals with COA in the UK Biobank study. We found that only 5.1% (SE = 2.2%) of the variance in self-reported asthma age of onset was explained by common SNPs; this estimate was borderline significantly greater than 0% (p = 0.011). A similarly low estimate was obtained using the GCTA-GREML approach (4.5%, SE = 2.2%, p = 0.017), and also when considering the 26,582 individuals from the UK Biobank study with AOA (onset between 20 and 60; 4.4%, SE = 1.2%, p = 0.0002). To confirm this finding, we then performed the same analysis in two independent studies conducted in children: ALSPAC (n = 1,326) and CATSS (n = 2,323) for a combined sample size of 3,649 cases with COA. Because age of onset in both studies was derived from longitudinal information provided during childhood, we expected measurement error or recall bias to have a smaller effect on this dataset than in the UK Biobank study. Using the LD-score-regression approach,27 we found that common SNPs explained a larger but still relatively modest fraction of the variation in age of onset (10.9%, SE = 12.0%, p = 0.182). Collectively, these findings indicate that common SNPs explain a relatively modest amount of variation in self-reported age of onset amongst COA and AOA cases.
Identification and Validation of Genetic Associations with COA and AOA
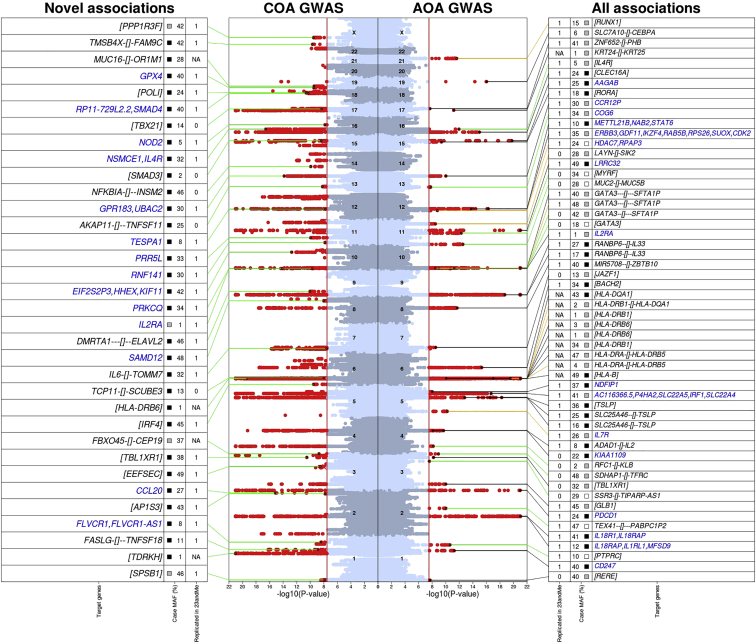
We then set out to identify genetic risk factors for COA and AOA separately. In a COA GWAS (Figure 1 and Figure S2), which included 13,962 affected individuals and 300,671 controls who did not report any allergic disease in the UK Biobank Study (i.e., these were individuals without asthma, hay fever, eczema, or other allergies), we identified 123 SNPs with an independent association with disease risk at a p < 3 × 10−8 (we call these variants sentinel SNPs; Table S3), located in 83 loci >1 Mb apart. In a separate GWAS of AOA (26,582 affected individuals and the same 300,671 controls; Figure 1 and Figure S3), we identified 56 sentinel risk SNPs (at 40 loci), including 19 that were in low LD (r2 < 0.05) with COA sentinel variants (Table S4). Consistent associations for all sentinel variants were observed when individuals with hay fever, eczema, or other allergies were not excluded from the control group (Figure S4).
Figure 1.
Summary of Association Results from the GWAS of COA and AOA in the UK Biobank Study
The middle panel shows the Manhattan plots (left for COA, based on 13,962 affected individuals and 300,671 controls; right for AOA, based on 26,582 affected individuals and 300,671 controls) with variants associated with disease risk at a p < 3 × 10−8 (red vertical line) are circled in red; associations with p < 10−21 are shown with p = 10−21. For COA, sentinel variants that were in low LD (r2 < 0.05) with previously reported associations for allergic disease are shown with a black circle (25 out of 123 in total); the green line points to additional information on the adjacent left panel. Specifically, the left panel indicates: (1) whether the association with COA was replicated in the independent 23andMe study (p < 0.05 and same direction of effect; “1” = yes, “0” = no, “NA” = results not available); (2) the minor-allele frequency (MAF; in %) in the case group; the square indicates whether the risk allele occurred at a significantly greater (in black; p < 0.05 and OR > 1 in the COA versus AOA case-case analysis, i.e., there was a stronger risk factor for COA), similar (in gray; p ≥ 0.05 in the case-case analysis, i.e., there was a similar association with COA and AOA), or lower (in white; p < 0.05 and OR < 1 in the case-case analysis; i.e., there was a stronger risk factor for AOA) frequency in COA-affected individuals than in AOA-affected individuals; and (3) the location of the sentinel risk variant relative to the nearest genes (in black font) or, for variants with an association that was replicated in the 23andMe study and with a target gene prediction, the likely target gene(s) based on LD (r2 > 0.8) with non-synonymous or sentinel eQTL (in blue font). The location of the sentinel risk variant (when shown) is indicated by “gene1–[]–gene2,” the two closest genes (upstream and downstream), when the variant was intergenic; the distance to each gene is proportional to the number of “-“ shown. Otherwise, when the risk variant was located within a gene, the respective gene name is shown between square brackets (i.e. [gene]). The right panel shows the same information for all 56 sentinel variants associated with AOA; these variants were grouped into those that (1) were in LD (r2 > 0.05) with sentinel variants identified in the COA GWAS (37 variants; highlighted by a black line); (2) were in LD (r2 > 0.05) with previous reported associations for allergic disease (eight variants; highlighted by an orange line); or (3) were not in LD (r2 < 0.05) with sentinel variants for COA, and did not have previous reported associations for allergic disease (11 variants; highlighted by a green line).
Overall, 27/123 (22%) COA and 5/56 (9%) AOA associations would not have been identified at the genome-wide significance level had we compared all asthma-affected individuals (i.e., irrespective of age of onset, n = 40,544) against the same 300,671 controls (Tables S5 and S6). For example, the COA variant rs9391997 (in IRF4 [MIM: 601900]) had only a modest association with asthma risk when all asthma-affected individuals (odds ratio [OR] = 1.02, p = 0.001) were considered.
To replicate the associations identified, we used data from research participants of the personal genetics company 23andMe. Using the same age cut-offs as for the UK Biobank Study, and after restricting the analyses to unrelated individuals of confirmed European descent, we found 31,759 COA-affected individuals and 214,890 asthma- and allergy-free controls. Of the 123 sentinel SNPs identified in the UK Biobank GWAS of COA, 108 were available for replication, and 98 of these had both a (1) reproducible association (p < 0.05 and same direction of effect) in the independent 23andMe study per se and (2) a genome-wide significant association in the meta-analysis of the UK Biobank and 23andMe studies (45,721 COA-affected individuals versus 515,561 controls; Table 2 and Table S7).
Table 2.
Independent SNP Associations with Risk of COA Discovered in the UK Biobank Study (13,962 Affected Individuals versus 300,671 Controls) and Validated in the 23andMe Study (31,759 Affected Individuals versus 214,890 Controls)
| Chr | bp | Sentinel SNP | Context | A1 |
Discovery (UK Biobank) |
Replication (23andMe) |
Meta-Analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | SE | P value | A1 freq | OR | SE | P value | OR | SE | P value | |||||
| Sentinel SNPs that Represent Novel Associations for Allergic Disease | ||||||||||||||
| 1 | 9356676 | rs67551275 | [SPSB1] | T | 1.071 | 0.012 | 1.3E-08 | 0.53 | 1.067 | 0.010 | 5.9E-11 | 1.069 | 0.008 | 3.8E-18 |
| 1 | 172715702 | rs78037977 | FASLG-[]–TNFSF18 | A | 1.116 | 0.018 | 1.4E-09 | 0.88 | 1.066 | 0.015 | 1.6E-05 | 1.086 | 0.011 | 6.2E-13 |
| 1 | 213056427 | rs12750027 | [FLVCR1] | A | 1.150 | 0.024 | 3.9E-09 | 0.07 | 1.075 | 0.022 | 1.4E-03 | 1.109 | 0.016 | 2.6E-10 |
| 2 | 224664050 | rs6755248 | [AP1S3] | G | 1.070 | 0.012 | 1.6E-08 | 0.41 | 1.046 | 0.009 | 1.3E-06 | 1.055 | 0.007 | 3.2E-13 |
| 2 | 228670437 | rs10187276 | SLC19A3-[]-CCL20 | T | 1.116 | 0.014 | 7.0E-16 | 0.25 | 1.059 | 0.011 | 6.2E-08 | 1.079 | 0.009 | 1.5E-19 |
| 3 | 127912846 | rs11412402 | [EEFSEC] | D | 1.073 | 0.012 | 3.2E-09 | 0.47 | 1.047 | 0.009 | 4.9E-07 | 1.057 | 0.007 | 3.9E-14 |
| 3 | 176852038 | rs7626218 | [TBL1XR1] | A | 1.083 | 0.012 | 3.1E-11 | 0.60 | 1.031 | 0.009 | 1.5E-03 | 1.051 | 0.007 | 3.6E-11 |
| 6 | 409119 | rs9391997 | [IRF4] | G | 1.077 | 0.012 | 3.1E-10 | 0.53 | 1.038 | 0.009 | 5.9E-05 | 1.052 | 0.007 | 3.8E-12 |
| 7 | 22780322 | rs6954667 | IL6-[]-TOMM7 | A | 1.074 | 0.013 | 2.6E-08 | 0.31 | 1.028 | 0.010 | 6.0E-03 | 1.045 | 0.008 | 3.0E-08 |
| 8 | 120063542 | rs2450083 | TNFRSF11B-[]-COLEC10 | C | 1.070 | 0.012 | 1.1E-08 | 0.50 | 1.037 | 0.009 | 1.2E-04 | 1.049 | 0.007 | 7.8E-11 |
| 9 | 23585839 | rs274943 | DMRTA1—[]–ELAVL2 | T | 1.080 | 0.012 | 9.1E-11 | 0.52 | 1.051 | 0.009 | 6.8E-08 | 1.062 | 0.007 | 2.6E-16 |
| 10 | 6093139 | rs12722502 | [IL2RA] | C | 1.326 | 0.044 | 2.0E-10 | 0.98 | 1.235 | 0.053 | 4.3E-05 | 1.288 | 0.034 | 6.8E-14 |
| 10 | 6621773 | rs943451 | [PRKCQ] | T | 1.105 | 0.013 | 7.5E-15 | 0.31 | 1.041 | 0.010 | 4.7E-05 | 1.063 | 0.008 | 3.5E-15 |
| 10 | 94384514 | rs113092121 | [KIF11] | I | 1.092 | 0.012 | 1.8E-13 | 0.56 | 1.058 | 0.009 | 1.8E-09 | 1.070 | 0.007 | 2.5E-20 |
| 11 | 10664033 | rs2052690 | [MRVI1] | T | 1.077 | 0.013 | 1.5E-08 | 0.29 | 1.043 | 0.010 | 3.2E-05 | 1.056 | 0.008 | 1.1E-11 |
| 11 | 36365253 | rs10836538 | [PRR5L] | G | 1.083 | 0.012 | 1.2E-10 | 0.65 | 1.040 | 0.010 | 5.4E-05 | 1.057 | 0.007 | 2.7E-13 |
| 12 | 55368291 | rs62623446 | [TESPA1] | T | 1.138 | 0.023 | 1.7E-08 | 0.07 | 1.103 | 0.021 | 6.2E-06 | 1.119 | 0.016 | 8.0E-13 |
| 13 | 99974492 | rs1887704 | [UBAC2] | G | 1.102 | 0.013 | 1.4E-14 | 0.68 | 1.059 | 0.010 | 4.8E-09 | 1.075 | 0.008 | 4.7E-20 |
| 16 | 27349168 | rs3785356 | [IL4R] | T | 1.116 | 0.013 | 2.7E-17 | 0.29 | 1.073 | 0.010 | 2.5E-12 | 1.089 | 0.008 | 8.5E-27 |
| 16 | 50745926 | rs2066844 | [NOD2] | T | 1.172 | 0.028 | 1.5E-08 | 0.05 | 1.086 | 0.022 | 2.5E-04 | 1.119 | 0.017 | 1.3E-10 |
| 18 | 48647640 | rs1893380 | SMAD4-[]-MEX3C | C | 1.083 | 0.012 | 5.2E-11 | 0.38 | 1.039 | 0.009 | 5.7E-05 | 1.055 | 0.007 | 3.6E-13 |
| 18 | 51816408 | rs12965763 | [POLI] | A | 1.100 | 0.014 | 2.3E-11 | 0.22 | 1.034 | 0.011 | 2.3E-03 | 1.059 | 0.009 | 4.3E-11 |
| 19 | 1152656 | rs892225 | [SBNO2] | G | 1.080 | 0.012 | 3.4E-10 | 0.38 | 1.040 | 0.010 | 9.7E-05 | 1.057 | 0.008 | 1.1E-12 |
| X | 13023741 | rs850637 | TMSB4X-[]-FAM9C | G | 1.064 | 0.010 | 2.2E-10 | 0.56 | 1.036 | 0.008 | 3.3E-06 | 1.046 | 0.006 | 8.3E-14 |
| X | 49139787 | rs5953283 | [PPP1R3F] | A | 1.064 | 0.010 | 3.1E-10 | 0.40 | 1.024 | 0.008 | 2.2E-03 | 1.038 | 0.006 | 5.6E-10 |
| Sentinel SNPs in LD (r2 > 0.05) with Variants Previously Reported to Associate with Allergic Disease | ||||||||||||||
| 1 | 151801680 | rs4845604 | [RORC] | G | 1.140 | 0.017 | 9.5E-15 | 0.86 | 1.062 | 0.014 | 9.2E-06 | 1.092 | 0.011 | 1.5E-16 |
| 1 | 152179152 | rs12123821 | RPTN-[]-HRNR | T | 1.523 | 0.028 | 4.0E-51 | 0.05 | 1.163 | 0.025 | 1.1E-09 | 1.307 | 0.018 | 7.6E-48 |
| 1 | 161185058 | rs2070901 | NDUFS2-[]-FCER1G | T | 1.093 | 0.013 | 2.4E-11 | 0.27 | 1.105 | 0.010 | 4.2E-22 | 1.100 | 0.008 | 2.8E-32 |
| 1 | 167436270 | rs1617333 | [CD247] | A | 1.098 | 0.012 | 9.9E-15 | 0.60 | 1.061 | 0.009 | 4.3E-10 | 1.075 | 0.007 | 2.5E-22 |
| 1 | 173131493 | rs10158467 | TNFSF18–[]-TNFSF4 | G | 1.116 | 0.013 | 6.3E-17 | 0.28 | 1.079 | 0.010 | 5.0E-14 | 1.093 | 0.008 | 8.3E-29 |
| 1 | 203093201 | rs12023876 | MYOG-[]-ADORA1 | G | 1.080 | 0.012 | 7.8E-10 | 0.67 | 1.071 | 0.010 | 4.3E-12 | 1.074 | 0.008 | 4.9E-21 |
| 2 | 8459404 | rs3856439 | [LINC00299] | C | 1.113 | 0.012 | 6.0E-18 | 0.66 | 1.070 | 0.010 | 7.0E-12 | 1.087 | 0.008 | 6.2E-28 |
| 2 | 102798245 | rs74180212 | IL1R1-[]-IL1RL2 | G | 1.123 | 0.012 | 1.1E-20 | 0.44 | 1.087 | 0.010 | 1.6E-17 | 1.101 | 0.008 | 5.3E-37 |
| 2 | 102882352 | rs78545931 | IL1RL2-[]-IL18R1 | A | 1.408 | 0.033 | 4.2E-25 | 0.98 | 1.391 | 0.027 | 5.9E-37 | 1.398 | 0.021 | 8.8E-59 |
| 2 | 102936159 | rs72823641 | [IL18R1] | T | 1.341 | 0.017 | 6.9E-68 | 0.86 | 1.281 | 0.014 | 5.4E-75 | 1.305 | 0.011 | 2E-136 |
| 2 | 102966906 | rs1861245 | [IL1RL1] | C | 1.194 | 0.013 | 9.3E-45 | 0.66 | 1.124 | 0.010 | 3.9E-32 | 1.150 | 0.008 | 2.3E-69 |
| 2 | 112268732 | rs143326447 | BCL2L11–[]–ANAPC1 | C | 1.108 | 0.018 | 1.8E-08 | 0.12 | 1.047 | 0.015 | 2.0E-03 | 1.071 | 0.011 | 1.8E-09 |
| 2 | 242698640 | rs34290285 | [D2HGDH] | G | 1.198 | 0.013 | 3.3E-41 | 0.74 | 1.142 | 0.012 | 5.5E-29 | 1.168 | 0.009 | 4.2E-69 |
| 3 | 33047662 | rs35570272 | [GLB1] | T | 1.106 | 0.012 | 7.8E-17 | 0.39 | 1.029 | 0.009 | 2.3E-03 | 1.057 | 0.007 | 4.1E-14 |
| 3 | 121716171 | rs1806656 | [ILDR1] | C | 1.082 | 0.013 | 6.3E-10 | 0.68 | 1.036 | 0.010 | 6.4E-04 | 1.053 | 0.008 | 1.2E-10 |
| 3 | 141150026 | rs7625643 | [ZBTB38] | G | 1.070 | 0.012 | 2.1E-08 | 0.44 | 1.029 | 0.009 | 2.7E-03 | 1.044 | 0.007 | 4.9E-09 |
| 3 | 188128979 | rs9860547 | [LPP] | A | 1.135 | 0.012 | 1.9E-26 | 0.45 | 1.087 | 0.009 | 1.5E-19 | 1.105 | 0.007 | 3.6E-42 |
| 3 | 188401138 | rs55661102 | [LPP] | A | 1.101 | 0.016 | 9.0E-10 | 0.83 | 1.093 | 0.013 | 2.4E-12 | 1.096 | 0.010 | 4.1E-20 |
| 4 | 38798648 | rs5743618 | [TLR1] | C | 1.210 | 0.014 | 2.1E-43 | 0.76 | 1.141 | 0.011 | 1.7E-36 | 1.165 | 0.008 | 3.2E-74 |
| 4 | 123353432 | rs17454584 | ADAD1-[]-IL2 | G | 1.126 | 0.014 | 1.5E-16 | 0.21 | 1.106 | 0.011 | 2.1E-20 | 1.114 | 0.009 | 5.9E-36 |
| 5 | 14610309 | rs16903574 | [FAM105A] | G | 1.191 | 0.023 | 2.1E-14 | 0.07 | 1.122 | 0.017 | 7.9E-12 | 1.145 | 0.013 | 1.0E-23 |
| 5 | 110158844 | rs7734635 | SLC25A46-[]–TSLP | G | 1.176 | 0.016 | 3.7E-23 | 0.15 | 1.084 | 0.012 | 1.8E-11 | 1.116 | 0.009 | 1.9E-30 |
| 5 | 110401872 | rs1837253 | SLC25A46–[]-TSLP | C | 1.182 | 0.013 | 1.2E-35 | 0.74 | 1.189 | 0.011 | 1.6E-59 | 1.186 | 0.008 | 7.1E-94 |
| 5 | 110470137 | rs6594499 | WDR36-[]-CAMK4 | C | 1.147 | 0.012 | 3.3E-31 | 0.51 | 1.126 | 0.009 | 5.4E-38 | 1.134 | 0.007 | 3.9E-66 |
| 5 | 131799626 | rs3749833 | [C5orf56] | C | 1.109 | 0.013 | 1.3E-14 | 0.26 | 1.057 | 0.011 | 2.2E-07 | 1.077 | 0.008 | 1.2E-19 |
| 5 | 131916940 | rs2299012 | [RAD50] | C | 1.191 | 0.015 | 1.9E-31 | 0.19 | 1.115 | 0.011 | 7.6E-23 | 1.141 | 0.009 | 1.0E-49 |
| 5 | 132105698 | rs113010607 | [SEPT8] | C | 1.172 | 0.022 | 4.1E-13 | 0.07 | 1.101 | 0.017 | 6.1E-09 | 1.126 | 0.013 | 2.0E-19 |
| 5 | 141533062 | rs449454 | [NDFIP1] | G | 1.084 | 0.012 | 3.0E-11 | 0.62 | 1.070 | 0.010 | 1.1E-12 | 1.076 | 0.007 | 2.0E-22 |
| 6 | 31303324 | rs114444221 | [HLA-B] | A | 1.231 | 0.027 | 2.3E-14 | 0.97 | 1.095 | 0.017 | 1.2E-07 | 1.133 | 0.015 | 9.0E-18 |
| 6 | 33033824 | rs111789468 | [HLA-DPA1] | T | 1.222 | 0.030 | 1.3E-11 | 0.03 | 1.069 | 0.013 | 4.1E-07 | 1.092 | 0.012 | 2.2E-13 |
| 6 | 33099538 | rs3116989 | HLA-DPB2-[]-COL11A2 | G | 1.151 | 0.017 | 5.3E-16 | 0.87 | 1.060 | 0.014 | 3.2E-05 | 1.096 | 0.011 | 2.6E-17 |
| 6 | 90976609 | rs62408233 | [BACH2] | G | 1.113 | 0.012 | 2.9E-18 | 0.64 | 1.096 | 0.010 | 3.3E-21 | 1.103 | 0.008 | 2.8E-38 |
| 6 | 106667535 | rs9372120 | [ATG5] | G | 1.106 | 0.015 | 5.5E-12 | 0.21 | 1.078 | 0.011 | 4.2E-11 | 1.088 | 0.009 | 8.8E-21 |
| 6 | 128293562 | rs55743914 | [PTPRK] | T | 1.105 | 0.014 | 4.8E-13 | 0.24 | 1.051 | 0.011 | 5.0E-06 | 1.071 | 0.009 | 1.5E-15 |
| 6 | 138002175 | rs6927172 | OLIG3–[]–LOC100130476 | C | 1.090 | 0.014 | 1.4E-09 | 0.78 | 1.051 | 0.011 | 1.1E-05 | 1.067 | 0.009 | 3.3E-13 |
| 7 | 20423923 | rs149317277 | [ITGB8] | D | 1.087 | 0.012 | 6.6E-12 | 0.61 | 1.050 | 0.009 | 3.0E-07 | 1.064 | 0.007 | 8.4E-17 |
| 7 | 20544209 | rs12531500 | ITGB8-[]–ABCB5 | A | 1.103 | 0.012 | 1.8E-16 | 0.57 | 1.056 | 0.009 | 5.3E-09 | 1.073 | 0.007 | 6.7E-22 |
| 7 | 28156606 | rs4722758 | [JAZF1] | G | 1.126 | 0.015 | 6.2E-16 | 0.20 | 1.090 | 0.011 | 6.1E-15 | 1.103 | 0.009 | 2.8E-28 |
| 8 | 81294702 | rs2221641 | MIR5708–[]–ZBTB10 | C | 1.112 | 0.012 | 2.4E-18 | 0.38 | 1.074 | 0.009 | 2.2E-14 | 1.088 | 0.007 | 1.8E-30 |
| 8 | 128777719 | rs13277355 | POU5F1B–[]-MYC | A | 1.103 | 0.013 | 1.4E-13 | 0.27 | 1.072 | 0.010 | 7.1E-12 | 1.084 | 0.008 | 9.5E-24 |
| 9 | 6081804 | rs340934 | RANBP6-[]–IL33 | T | 1.208 | 0.015 | 1.6E-36 | 0.81 | 1.142 | 0.012 | 8.7E-30 | 1.167 | 0.009 | 3.1E-62 |
| 9 | 6213468 | rs7848215 | RANBP6–[]-IL33 | T | 1.255 | 0.014 | 4.1E-63 | 0.25 | 1.181 | 0.010 | 1.3E-59 | 1.206 | 0.008 | 5E-115 |
| 10 | 8606014 | rs17144046 | GATA3–[]—SFTA1P | G | 1.102 | 0.013 | 7.0E-13 | 0.28 | 1.044 | 0.010 | 3.3E-05 | 1.066 | 0.008 | 2.9E-15 |
| 10 | 9049253 | rs12413578 | GATA3—[]—SFTA1P | C | 1.204 | 0.019 | 1.7E-22 | 0.89 | 1.160 | 0.016 | 7.7E-21 | 1.178 | 0.012 | 9.1E-41 |
| 10 | 9064716 | rs1612986 | GATA3—[]—SFTA1P | C | 1.168 | 0.015 | 4.5E-24 | 0.18 | 1.110 | 0.012 | 8.3E-19 | 1.131 | 0.009 | 8.3E-41 |
| 10 | 64391375 | rs10995245 | [ZNF365] | A | 1.073 | 0.012 | 1.3E-08 | 0.35 | 1.033 | 0.010 | 9.2E-04 | 1.048 | 0.007 | 3.6E-10 |
| 11 | 65551957 | rs479844 | AP5B1-[]-OVOL1 | G | 1.116 | 0.012 | 1.4E-20 | 0.55 | 1.052 | 0.009 | 3.9E-08 | 1.075 | 0.007 | 3.2E-23 |
| 11 | 76299431 | rs55646091 | WNT11–[]-LRRC32 | A | 1.513 | 0.028 | 3.9E-50 | 0.05 | 1.283 | 0.021 | 8.5E-30 | 1.364 | 0.017 | 5.9E-74 |
| 11 | 76299649 | rs11236797 | WNT11–[]-LRRC32 | A | 1.214 | 0.012 | 2.4E-60 | 0.45 | 1.132 | 0.009 | 1.0E-41 | 1.162 | 0.007 | 1E-93 |
| 11 | 118743286 | rs12365699 | DDX6-[]-CXCR5 | G | 1.141 | 0.016 | 6.1E-17 | 0.83 | 1.106 | 0.013 | 4.2E-16 | 1.119 | 0.010 | 4.3E-30 |
| 11 | 128172836 | rs140522418 | KIRREL3-AS3—[]–ETS1 | D | 1.095 | 0.015 | 4.8E-09 | 0.82 | 1.039 | 0.012 | 1.9E-03 | 1.062 | 0.010 | 4.3E-10 |
| 12 | 56389293 | rs705700 | [RAB5B] | C | 1.108 | 0.012 | 8.1E-18 | 0.42 | 1.060 | 0.009 | 4.3E-10 | 1.078 | 0.007 | 2.5E-24 |
| 12 | 57509102 | rs3122929 | [STAT6] | T | 1.141 | 0.012 | 7.6E-28 | 0.40 | 1.067 | 0.009 | 5.7E-12 | 1.095 | 0.007 | 2.4E-34 |
| 12 | 112059557 | rs11065979 | ATXN2-[]-BRAP | C | 1.085 | 0.012 | 6.5E-12 | 0.56 | 1.054 | 0.009 | 1.3E-08 | 1.066 | 0.007 | 4.8E-18 |
| 12 | 121365431 | rs188074962 | SPPL3-[]-HNF1A-AS1 | G | 1.112 | 0.012 | 8.8E-18 | 0.35 | 1.048 | 0.010 | 1.9E-06 | 1.073 | 0.008 | 1.6E-20 |
| 12 | 123635096 | rs1716183 | [PITPNM2] | C | 1.098 | 0.016 | 2.2E-09 | 0.83 | 1.049 | 0.013 | 1.2E-04 | 1.067 | 0.010 | 3.5E-11 |
| 14 | 68754695 | rs1885013 | [RAD51B] | G | 1.098 | 0.013 | 1.0E-12 | 0.28 | 1.078 | 0.010 | 4.4E-14 | 1.085 | 0.008 | 2.9E-25 |
| 14 | 103244070 | rs71421264 | [TRAF3] | G | 1.081 | 0.012 | 1.0E-10 | 0.59 | 1.032 | 0.009 | 7.2E-04 | 1.050 | 0.007 | 2.8E-11 |
| 15 | 41787585 | rs1655558 | [ITPKA] | G | 1.087 | 0.012 | 2.8E-12 | 0.55 | 1.056 | 0.010 | 1.7E-08 | 1.068 | 0.007 | 2.2E-18 |
| 15 | 61069988 | rs11071559 | [RORA] | C | 1.185 | 0.017 | 1.9E-22 | 0.87 | 1.121 | 0.014 | 2.5E-17 | 1.145 | 0.011 | 2.0E-37 |
| 15 | 67441750 | rs72743461 | [SMAD3] | A | 1.196 | 0.014 | 1.7E-37 | 0.23 | 1.127 | 0.011 | 3.8E-29 | 1.152 | 0.009 | 6.4E-63 |
| 15 | 67475764 | rs34445740 | [SMAD3] | D | 1.111 | 0.013 | 4.7E-16 | 0.70 | 1.055 | 0.010 | 1.8E-07 | 1.076 | 0.008 | 7.7E-20 |
| 16 | 11219041 | rs12935657 | [CLEC16A] | G | 1.154 | 0.014 | 6.1E-26 | 0.75 | 1.122 | 0.011 | 1.3E-27 | 1.133 | 0.009 | 1.0E-49 |
| 17 | 38061439 | rs4795399 | [GSDMB] | T | 1.303 | 0.012 | 3E-111 | 0.53 | 1.280 | 0.009 | 2E-155 | 1.289 | 0.007 | 1E-257 |
| 17 | 38064971 | rs117097909 | [GSDMB] | A | 1.355 | 0.026 | 1.5E-31 | 0.05 | 1.247 | 0.022 | 3.7E-22 | 1.292 | 0.017 | 1.6E-51 |
| 17 | 38755021 | rs9893132 | CCR7-[]-SMARCE1 | G | 1.095 | 0.012 | 1.4E-13 | 0.64 | 1.047 | 0.010 | 1.7E-06 | 1.065 | 0.007 | 3.0E-17 |
| 17 | 38764524 | rs112401631 | CCR7-[]-SMARCE1 | A | 1.354 | 0.043 | 1.6E-12 | 0.02 | 1.265 | 0.044 | 1.9E-07 | 1.310 | 0.031 | 2.1E-18 |
| 17 | 43457886 | rs9895436 | MAP3K14-[]-ARHGAP27 | A | 1.089 | 0.012 | 1.8E-12 | 0.40 | 1.058 | 0.009 | 1.4E-09 | 1.070 | 0.007 | 5.9E-20 |
| 17 | 47448346 | rs12952581 | ZNF652-[]-PHB | A | 1.094 | 0.012 | 2.4E-13 | 0.36 | 1.069 | 0.010 | 3.2E-12 | 1.078 | 0.007 | 4.4E-24 |
| 18 | 60009814 | rs4574025 | [TNFRSF11A] | T | 1.092 | 0.012 | 1.3E-13 | 0.53 | 1.036 | 0.010 | 2.2E-04 | 1.058 | 0.007 | 8.0E-14 |
| 18 | 61442619 | rs12964116 | [SERPINB7] | G | 1.293 | 0.031 | 3.0E-16 | 0.04 | 1.143 | 0.041 | 1.1E-03 | 1.236 | 0.025 | 8.4E-18 |
| 19 | 8785744 | rs2918302 | ADAMTS10–[]-ACTL9 | A | 1.107 | 0.016 | 5.8E-10 | 0.15 | 1.037 | 0.013 | 3.5E-03 | 1.064 | 0.010 | 4.8E-10 |
| 19 | 33726578 | rs117710327 | SLC7A10-[]-CEBPA | C | 1.205 | 0.024 | 4.7E-15 | 0.94 | 1.189 | 0.021 | 1.2E-16 | 1.196 | 0.016 | 2.8E-29 |
For AOA, data were available for 16,297 affected individuals and 217,711 controls in the 23andMe study. According to the same criteria, 34 (of 46 tested) sentinel SNPs for AOA were associated with disease risk in the 23andMe study and in the overall meta-analysis (42,879 affected individuals versus 518,382 controls; Table 3 and Table S8).
Table 3.
Independent SNP Associations with Risk of AOA Discovered in the UK Biobank Study (26,582 Affected Individuals versus 300,671 Controls) and Validated in the 23andMe Study (16,297 Affected Individuals versus 217,711 Controls)
| Chr | bp | Sentinel SNP | Context | A1 |
Discovery (UK Biobank) |
Replication (23andMe) |
Meta-Analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | SE | P value | A1 freq | OR | SE | P value | OR | SE | P value | |||||
| Sentinel SNPs that Represent Novel Associations for Allergic Disease | ||||||||||||||
| 1 | 198640488 | rs17668708 | [PTPRC] | C | 1.096 | 0.014 | 2.4E-11 | 0.90 | 1.069 | 0.020 | 6.9E-04 | 1.087 | 0.011 | 3.0E-13 |
| 2 | 146145018 | rs2381712 | TEX41–[]—PABPC1P2 | G | 1.056 | 0.008 | 9.5E-11 | 0.52 | 1.057 | 0.012 | 4.9E-06 | 1.056 | 0.007 | 2.5E-16 |
| 10 | 6093139 | rs12722502 | [IL2RA] | C | 1.237 | 0.032 | 1.9E-11 | 0.98 | 1.156 | 0.067 | 2.8E-02 | 1.222 | 0.029 | 4.3E-12 |
| 13 | 40319954 | rs28635831 | [COG6] | A | 1.051 | 0.009 | 1.7E-08 | 0.65 | 1.030 | 0.013 | 1.9E-02 | 1.044 | 0.007 | 4.3E-09 |
| 13 | 100070457 | rs4771332 | MIR548AN-[]-TM9SF2 | C | 1.061 | 0.009 | 9.8E-11 | 0.69 | 1.035 | 0.013 | 9.5E-03 | 1.053 | 0.007 | 5.5E-12 |
| 16 | 27369502 | rs3024655 | [IL4R] | G | 1.133 | 0.018 | 1.1E-12 | 0.94 | 1.051 | 0.025 | 4.9E-02 | 1.105 | 0.015 | 1.1E-11 |
| Sentinel SNPs in LD (r2 > 0.05) with Variants Previously Reported to Associate with Allergic Disease | ||||||||||||||
| 1 | 167420299 | rs2056625 | [CD247] | G | 1.060 | 0.009 | 5.6E-12 | 0.59 | 1.027 | 0.012 | 2.9E-02 | 1.048 | 0.007 | 7.6E-11 |
| 2 | 102892339 | rs60227565 | IL1RL2-[]-IL18R1 | G | 1.146 | 0.012 | 1.8E-28 | 0.87 | 1.082 | 0.018 | 7.7E-06 | 1.125 | 0.010 | 1.4E-32 |
| 2 | 102926362 | rs12470864 | IL1RL2-[]-IL18R1 | A | 1.104 | 0.009 | 2.3E-30 | 0.38 | 1.061 | 0.012 | 1.5E-06 | 1.089 | 0.007 | 1.1E-31 |
| 2 | 242698640 | rs34290285 | [D2HGDH] | G | 1.100 | 0.010 | 2.9E-23 | 0.74 | 1.091 | 0.016 | 1.8E-08 | 1.097 | 0.008 | 2.2E-28 |
| 3 | 33083985 | rs4491851 | [GLB1] | A | 1.056 | 0.008 | 1.0E-10 | 0.53 | 1.035 | 0.012 | 4.0E-03 | 1.050 | 0.007 | 3.7E-13 |
| 4 | 123359569 | rs62322662 | ADAD1-[]-IL2 | G | 1.096 | 0.016 | 8.1E-09 | 0.08 | 1.059 | 0.023 | 1.2E-02 | 1.084 | 0.013 | 7.9E-10 |
| 5 | 35881376 | rs11742240 | IL7R-[]-CAPSL | G | 1.063 | 0.009 | 6.1E-11 | 0.72 | 1.045 | 0.014 | 1.1E-03 | 1.058 | 0.007 | 9.1E-14 |
| 5 | 110161473 | rs540485182 | SLC25A46-[]–TSLP | I | 1.090 | 0.012 | 1.7E-13 | 0.15 | 1.058 | 0.016 | 4.2E-04 | 1.078 | 0.010 | 3.8E-15 |
| 5 | 110401872 | rs1837253 | SLC25A46–[]-TSLP | C | 1.084 | 0.010 | 2.9E-17 | 0.74 | 1.055 | 0.014 | 1.1E-04 | 1.074 | 0.008 | 1.4E-18 |
| 5 | 110408002 | rs1898671 | [TSLP] | T | 1.082 | 0.009 | 5.5E-19 | 0.35 | 1.060 | 0.013 | 4.9E-06 | 1.074 | 0.007 | 1.4E-22 |
| 5 | 131787137 | rs6866614 | [IRF1] | G | 1.075 | 0.009 | 2.0E-17 | 0.57 | 1.074 | 0.012 | 6.9E-09 | 1.075 | 0.007 | 4.4E-23 |
| 5 | 141518940 | rs10699671 | [NDFIP1] | I | 1.051 | 0.009 | 9.9E-09 | 0.61 | 1.055 | 0.012 | 1.6E-05 | 1.052 | 0.007 | 2.4E-12 |
| 6 | 90985198 | rs58521088 | [BACH2] | A | 1.074 | 0.009 | 5.1E-16 | 0.64 | 1.026 | 0.013 | 4.6E-02 | 1.058 | 0.007 | 2.5E-14 |
| 8 | 81302012 | rs35204956 | MIR5708–[]-ZBTB10 | D | 1.063 | 0.009 | 2.2E-12 | 0.39 | 1.057 | 0.012 | 6.9E-06 | 1.061 | 0.007 | 4.0E-16 |
| 9 | 6047765 | rs62557312 | RANBP6-[]–IL33 | C | 1.104 | 0.011 | 2.0E-19 | 0.82 | 1.049 | 0.016 | 2.1E-03 | 1.085 | 0.009 | 8.0E-20 |
| 9 | 6209697 | rs992969 | RANBP6–[]-IL33 | A | 1.115 | 0.010 | 2.4E-29 | 0.25 | 1.059 | 0.014 | 2.8E-05 | 1.095 | 0.008 | 2.0E-29 |
| 10 | 8777640 | rs10795672 | GATA3—[]—SFTA1P | A | 1.061 | 0.009 | 5.4E-12 | 0.47 | 1.040 | 0.013 | 2.5E-03 | 1.054 | 0.007 | 1.0E-12 |
| 10 | 9054340 | rs1775554 | GATA3—[]—SFTA1P | A | 1.109 | 0.008 | 3.0E-34 | 0.57 | 1.085 | 0.012 | 4.2E-11 | 1.102 | 0.007 | 3.1E-47 |
| 11 | 76293726 | rs7936312 | WNT11–[]-LRRC32 | T | 1.089 | 0.008 | 6.1E-24 | 0.47 | 1.062 | 0.012 | 5.8E-07 | 1.081 | 0.007 | 2.5E-31 |
| 12 | 48186563 | rs2544026 | [HDAC7] | T | 1.060 | 0.010 | 1.7E-09 | 0.75 | 1.066 | 0.014 | 6.2E-06 | 1.062 | 0.008 | 2.0E-13 |
| 12 | 56449435 | rs7302200 | RPS26-[]-ERBB3 | A | 1.071 | 0.009 | 1.8E-14 | 0.34 | 1.036 | 0.013 | 5.3E-03 | 1.059 | 0.007 | 4.4E-15 |
| 12 | 57489709 | rs1059513 | [STAT6] | T | 1.119 | 0.013 | 5.3E-17 | 0.89 | 1.072 | 0.020 | 3.7E-04 | 1.105 | 0.011 | 4.7E-20 |
| 15 | 61049569 | rs7183955 | [RORA] | A | 1.070 | 0.011 | 4.3E-10 | 0.81 | 1.049 | 0.015 | 1.6E-03 | 1.063 | 0.009 | 8.3E-12 |
| 15 | 67441750 | rs72743461 | [SMAD3] | A | 1.097 | 0.010 | 1.7E-20 | 0.23 | 1.067 | 0.014 | 3.8E-06 | 1.087 | 0.008 | 1.4E-24 |
| 16 | 11213021 | rs35441874 | [CLEC16A] | T | 1.081 | 0.010 | 1.1E-15 | 0.75 | 1.067 | 0.014 | 2.5E-06 | 1.076 | 0.008 | 1.2E-19 |
| 17 | 47465743 | rs10667251 | ZNF652-[]-PHB | D | 1.049 | 0.009 | 2.2E-08 | 0.58 | 1.033 | 0.012 | 9.0E-03 | 1.043 | 0.007 | 5.4E-09 |
| 19 | 33726578 | rs117710327 | SLC7A10-[]-CEBPA | C | 1.152 | 0.017 | 9.3E-17 | 0.94 | 1.089 | 0.027 | 1.5E-03 | 1.134 | 0.014 | 2.8E-18 |
| 21 | 36464631 | rs11088309 | [RUNX1] | G | 1.088 | 0.012 | 2.4E-12 | 0.14 | 1.035 | 0.017 | 4.7E-02 | 1.070 | 0.010 | 5.2E-12 |
Shared or Disease Onset-Specific SNP Associations?
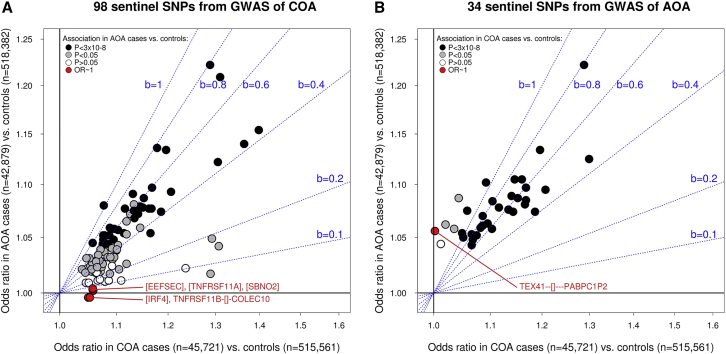
To formally test whether the 132 (98 + 34) SNP associations identified differed between COA and AOA, for each sentinel variant, we compared the frequency of the risk allele between COA- and AOA-affected individuals. Based on combined results from the UK Biobank and 23andMe studies (44,964 COA-affected individuals versus 42,879 AOA-affected individuals), we found that the risk allele was more common in individuals with COA than in those with AOA for all 98 COA sentinel variants—significantly so for 96 SNPs (p < 0.05; Table S9). Therefore, most if not all 98 COA sentinel SNPs are stronger risk factors for COA than for AOA. At least five sentinel SNPs are likely to represent COA-specific risk factors (Figure 2A) because they were not associated with AOA risk (OR ∼1 in the meta-analysis of UK Biobank and 23andMe studies; Table S9).
Figure 2.
Association between Sentinel Variants and Risk of COA and AOA
The two panels (A and B) respectively show results for COA and AOA sentinel SNPs that were identified in the UK Biobank GWAS and that were subsequently validated in the 23andMe replication study. The x axis shows SNP effects (odds ratio; “OR”) estimated in the COA-affected individuals versus control individuals meta-analysis of the UK Biobank and 23andMe studies, and the y axis shows the effect in the AOA-affected individuals versus control individuals meta-analysis of the same two studies.
In panel (A), variants for which the odds ratio in the AOA-affected individuals versus control individuals analysis was <1.005 are highlighted in red; their genomic context is also highlighted.
Similarly, in panel (B), variants in red had an odds ratio <1.005 in the COA-affected individuals versus control individuals analysis. To help interpret the correlation in odds ratios between the two disease subtypes, we have shown regression lines with increasing beta coefficients (“b,” from 0.1 to 1) in blue.
A similar pattern of results was observed for 31 of the 34 sentinel SNPs identified for AOA (Table S10), that is, the risk allele was observed at a significantly higher (22 SNPs) or similar (nine SNPs) frequency in individuals with COA compared to those with AOA. For the remaining three AOA sentinel SNPs (rs2381712 near TEX41, rs2544026 in HDAC7 [MIM: 606542], and rs28635831 in COG6 [MIM: 606977]), we observed a different pattern of results: the risk allele was significantly more common in AOA-affected individuals than in COA-affected individuals, suggesting that these SNPs are stronger risk factors for AOA. Notably, one of these SNPs is likely to represent an AOA-specific risk factor, given the lack of association with COA (OR = ∼1): rs2381712 near TEX41 (Figure 2B and Table S10).
Of the seven SNPs reported in two previously published GWASs of asthma age of onset, six were tested in our analysis, and four, those located in or near IL1RL1, HLA-DQA1, IL33, and GSDMB, had a consistent association (Table S11). The two variants for which the reported association was not supported by results from our analysis were located in or near CYLD and CRBN; neither was associated with asthma onset type (Table S11), nor with COA or AOA risk (not shown).
Sentinel Variants Not Reported in Previous GWAS of Asthma or Other Allergic Diseases
We determined which of the 132 sentinel variants identified above represented novel associations for allergic disease in general, that is, when all previously reported associations with p < 5 × 10−8 for asthma, hay fever, eczema, food allergy, and/or atopy were considered. Of the 98 sentinel variants confirmed for COA, 73 were in LD (r2 > 0.05) with variants previously reported to associate with allergic disease (Table S12), and the remaining 25 represent novel SNP associations at the genome-wide significance level (Table 1). Of the 34 sentinel associations for AOA, 28 have previously been described (Table S13), but six, including three not discovered in our GWAS of COA (in or near PTPRC [MIM: 151460], TEX41, and COG6), were novel (Table 2). Therefore, overall, we identified 28 (25 + 3) novel associations for asthma.
Genetic Correlation between COA and AOA and Other Complex Diseases or Traits
To provide some clues into the potential differences in genetic etiology between COA and AOA, we used LD Hub38 to estimate the overlap in genetic risk factors between COA and AOA and 227 human traits or diseases with publicly available GWAS results. We observed a number of genetic correlations that were significantly different from 0 (at a p < 0.05/227 = 0.0002; Table S14), and we highlight three of these groups. First, significant correlations with a similar magnitude between COA and AOA were observed for lung-function traits, for example forced expiratory volume (FEV1) and forced vital capacity (FVC) (rg = −0.35 for both). Because the SNP heritability of COA was larger than that of AOA, these results indicate that genetic variants that influence variation in lung function explain a larger proportion of variation in disease risk for COA than for AOA. Second, the genetic correlation was significant with COA but not with AOA for two traits: eczema (rg = 0.60 versus rg = 0.04) and years of schooling (rg = 0.11 versus rg = −0.09). Lastly, the genetic correlation was significant with AOA but not with COA for six sets of traits: (1) obesity-related traits (e.g., being overweight, rg = 0.08 versus rg = 0.28), (2) age when first child was born (rg = 0.07 versus rg = −0.27), (3) ever having been a smoker (rg = −0.07 versus rg = 0.23), (4) rheumatoid arthritis (rg = −0.09 versus rg = 0.18; [MIM: 180300]), (5) insomnia (rg = −0.03 versus rg = 0.21), and (6) depressive symptoms (rg = 0.02 versus rg = 0.19).
Likely Target Genes of the Sentinel Risk Variants
Finally, we found that 11 sentinel variants were in high LD (r2 ≥ 0.8) with missense or non-synonymous SNPs in 12 genes, including four with variants that were predicted by both SIFT and PolyPhen-2 to have a damaging effect on the protein: these genes were HLA-C, ITGB8, NOD2, and TESPA1 (Table S15). On the other hand, 62 sentinel variants were in high LD with a sentinel eQTL associated with gene expression in disease-relevant tissues or cell types at a conservative p < 8.9 × 10−10 (Table S2), implicating an additional 97 genes as likely targets of asthma risk variants (Tables S16 and S17). Of the 109 genes (12 + 97; Table S18), 25 were the predicted targets of novel sentinel risk SNPs for asthma (Table 4; we highlight eleven on the basis of a stricter LD threshold (r2 > 0.95): CCL20 (rs10187276; [MIM: 601960]), IL2RA (rs12722502; [MIM: 147730]), PRKCQ (rs943451; [MIM: 600448]), PRR5L (rs10836538; [MIM: 611728]), CCR12P (rs4771332), TESPA1 (rs62623446; [MIM: 615664]), NSMCE1 (rs3785356; [MIM: 617263), NOD2 (rs2066844; [MIM: 605956]), RP11-729L2.2, SMAD4 (rs1893380; [MIM: 600993]), and GPX4 (rs892225; [MIM: 138322).
Table 4.
Genes with a Sentinel eQTL and/or Non-Synonymous SNP in LD (r2 ≥ 0.8) with Sentinel Asthma Risk Variants Identified in the GWAS of COA or AOA
| Chr | bp | Sentinel SNP | Novel Association | Context |
LD between Sentinel eQTL/Non-Synonymous SNP and Sentinel GWAS SNPa |
|
|---|---|---|---|---|---|---|
| r2≥ 0.95 | 0.8 ≤ r2< 0.95 | |||||
| Sentinel SNPs Identified in the GWAS of COA | ||||||
| 1 | 151801680 | rs4845604 | No | [RORC] | RORC, TUFT1 | - |
| 1 | 161185058 | rs2070901 | No | NDUFS2-[]-FCER1G | F11R, FCER1G, TOMM40L, USF1 | - |
| 1 | 167436270 | rs1617333 | No | [CD247] | CD247 | - |
| 1 | 173131493 | rs10158467 | No | TNFSF18–[]-TNFSF4 | TNFSF4 | - |
| 1 | 203093201 | rs12023876 | No | MYOG-[]-ADORA1 | ADORA1, CHIT1, MYBPH, PPFIA4, RP11-335O13.7 | - |
| 1 | 213056427 | rs12750027 | Yes | [FLVCR1] | - | FLVCR1, FLVCR1-AS1 |
| 2 | 102882352 | rs78545931 | No | IL1RL2-[]-IL18R1 | IL18RAP, IL1RL1 | MFSD9 |
| 2 | 102936159 | rs72823641 | No | [IL18R1] | MFSD9 | - |
| 2 | 102966906 | rs1861245 | No | [IL1RL1] | - | AC007278.3, IL18R1, IL1RL1 |
| 2 | 228670437 | rs10187276 | Yes | SLC19A3-[]-CCL20 | CCL20 | - |
| 2 | 242698640 | rs34290285 | No | [D2HGDH] | PDCD1 | - |
| 3 | 141150026 | rs7625643 | No | [ZBTB38] | - | ZBTB38 |
| 3 | 188128979 | rs9860547 | No | [LPP] | BCL6 | - |
| 4 | 38798648 | rs5743618 | No | [TLR1] | TLR1∗ | - |
| 4 | 123353432 | rs17454584 | No | ADAD1-[]-IL2 | KIAA1109 | - |
| 5 | 14610309 | rs16903574 | No | [FAM105A] | FAM105A∗ | - |
| 5 | 110470137 | rs6594499 | No | WDR36-[]-CAMK4 | - | CTC-551A13.2, TSLP, WDR36 |
| 5 | 131799626 | rs3749833 | No | [C5orf56] | C5orf56,SLC22A4 | - |
| 5 | 131916940 | rs2299012 | No | [RAD50] | SLC22A5 | - |
| 5 | 141533062 | rs449454 | No | [NDFIP1] | NDFIP1 | - |
| 6 | 31303324 | rs114444221 | No | [HLA-B] | - | HLA-C∗, NOTCH4 |
| 6 | 33033824 | rs111789468 | No | [HLA-DPA1] | HLA-DPA1 | HLA-DPB1, HLA-DQB1, TAPBP |
| 7 | 20423923 | rs149317277 | No | [ITGB8] | - | ITGB8∗ |
| 7 | 28156606 | rs4722758 | No | [JAZF1] | JAZF1 | - |
| 10 | 6093139 | rs12722502 | Yes | [IL2RA] | IL2RA | - |
| 10 | 6621773 | rs943451 | Yes | [PRKCQ] | PRKCQ | - |
| 10 | 94384514 | rs113092121 | Yes | [KIF11] | - | EIF2S2P3, HHEX, KIF11 |
| 11 | 10664033 | rs2052690 | Yes | [MRVI1] | - | RNF141 |
| 11 | 36365253 | rs10836538 | Yes | [PRR5L] | PRR5L | - |
| 11 | 65551957 | rs479844 | No | AP5B1-[]-OVOL1 | OVOL1 | EFEMP2, SNX32 |
| 11 | 76299649 | rs11236797 | No | WNT11—[]-LRRC32 | LRRC32 | - |
| 11 | 128172836 | rs140522418 | No | KIRREL3-AS3—[]—ETS1 | - | ETS1 |
| 12 | 55368291 | rs62623446 | Yes | [TESPA1] | TESPA1 | - |
| 12 | 56389293 | rs705700 | No | [RAB5B] | RPS26,SUOX | - |
| 12 | 112059557 | rs11065979 | No | ATXN2-[]-BRAP | TCTN1,TRAFD1 | SH2B3∗ |
| 12 | 121365431 | rs188074962 | No | SPPL3-[]-HNF1A-AS1 | SPPL3 | - |
| 13 | 99974492 | rs1887704 | Yes | [UBAC2] | - | GPR183, UBAC2 |
| 14 | 103244070 | rs71421264 | No | [TRAF3] | TRAF3 | - |
| 15 | 41787585 | rs1655558 | No | [ITPKA] | - | D-JC17, ITPKA, RTF1, ZFYVE19 |
| 15 | 61069988 | rs11071559 | No | [RORA] | - | RP11-554D20.1 |
| 15 | 67441750 | rs72743461 | No | [SMAD3] | AAGAB | - |
| 15 | 67475764 | rs34445740 | No | [SMAD3] | SMAD3 | - |
| 16 | 11219041 | rs12935657 | No | [CLEC16A] | - | SOCS1 |
| 16 | 27349168 | rs3785356 | Yes | [IL4R] | NSMCE1 | IL4R |
| 16 | 50745926 | rs2066844 | Yes | [NOD2] | NOD2 | - |
| 17 | 38061439 | rs4795399 | No | [GSDMB] | GSDMB,ORMDL3 | GSDMA, IKZF3, RP11-94L15.2, ZPBP2 |
| 17 | 38755021 | rs9893132 | No | CCR7-[]-SMARCE1 | SMARCE1 | - |
| 17 | 43457886 | rs9895436 | No | MAP3K14-[]-ARHGAP27 | CRHR1,CRHR1-IT1,DND1P1,LRRC37A4P,RP11-105N13.4 | - |
| 17 | 47448346 | rs12952581 | No | ZNF652-[]-PHB | GNGT2 | - |
| 18 | 48647640 | rs1893380 | Yes | SMAD4-[]-MEX3C | RP11-729L2.2,SMAD4 | - |
| 18 | 60009814 | rs4574025 | No | [TNFRSF11A] | PIGN | - |
| 18 | 61442619 | rs12964116 | No | [SERPINB7] | SERPINB7 | - |
| 19 | 1152656 | rs892225 | Yes | [SBNO2] | GPX4 | - |
| Sentinel SNPs Identified in the GWAS of AOA | ||||||
| 1 | 167420299 | rs2056625 | No | [CD247] | CD247 | - |
| 2 | 102892339 | rs60227565 | No | IL1RL2-[]-IL18R1 | - | IL18RAP, IL1RL1, MFSD9 |
| 2 | 102926362 | rs12470864 | No | IL1RL2-[]-IL18R1 | IL18R1 | IL18RAP |
| 2 | 242698640 | rs34290285 | No | [D2HGDH] | PDCD1 | - |
| 4 | 123353432 | rs17454584 | No | ADAD1-[]-IL2 | KIAA1109 | - |
| 5 | 35881376 | rs11742240 | No | IL7R-[]-CAPSL | IL7R | - |
| 5 | 131787137 | rs6866614 | No | [IRF1] | AC116366.5, P4HA2, SLC22A5 | IRF1, SLC22A4 |
| 5 | 141518940 | rs10699671 | No | [NDFIP1] | NDFIP1 | - |
| 10 | 6093139 | rs12722502 | Yes | [IL2RA] | IL2RA | - |
| 11 | 76293726 | rs7936312 | No | WNT11–[]-LRRC32 | - | LRRC32 |
| 12 | 48186563 | rs2544026 | No | [HDAC7] | - | HDAC7, RPAP3 |
| 12 | 56449435 | rs7302200 | No | RPS26-[]-ERBB3 | ERBB3, GDF11, IKZF4, RAB5B, RPS26, SUOX | CDK2 |
| 12 | 57489709 | rs1059513 | No | [STAT6] | METTL21B, NAB2, STAT6 | - |
| 13 | 40319954 | rs28635831 | Yes | [COG6] | - | COG6 |
| 13 | 100070457 | rs4771332 | Yes | MIR548AN-[]-TM9SF2 | CCR12P | - |
| 15 | 67441750 | rs72743461 | No | [SMAD3] | AAGAB | - |
The pattern used for gene names is as follows. Italic only: genes implicated by an eQTL but not by a non-synonymous variant. Italic and asterisk: genes implicated by a non-synonymous variant but not an eQTL. Italic and underlined: genes implicated by both an eQTL and a non-synonymous variant.
Discussion
We found that in the UK Biobank Study, (1) common genetic variants collectively explain a larger fraction of variation in liability to COA than to AOA; (2) the genetic correlation between COA and AOA is high but significantly different from 1, and (3) variation in the specific age at which asthma first develops within each asthma subtype has a relatively low SNP heritability. We also identified and validated 98 independent genetic associations for COA and 34 for AOA, and we identified 109 likely target genes of the risk variants.
The observed difference in SNP heritabilities between COA and AOA, together with a genetic correlation that was significantly different from 1, indicates that the genetic architectures of COA and AOA are similar but not identical. Exactly how they might differ is unclear because the observed heritabilities and genetic correlation might arise under difference scenarios. For example, a genetic correlation different from 1 might arise not only when there are subtype-specific risk variants (i.e., associated with one disease subtype but not the other) but, in some situations, also when all risk variants are shared between COA and AOA, as shown by Carey et al.44 Specifically, a genetic correlation will be lower than 1 even when all risk loci are shared if there is little (or no) correlation in genetic effects between the two diseases. The latter scenario is relatively extreme and is not supported by the results of our GWAS, which found evidence for subtype-specific associations. A more realistic scenario is that for some variants, genetic effects are highly correlated between COA and AOA, and so these contribute to a higher genetic correlation. In contrast, other variants might have subtype-specific associations or genetic effects that are weakly correlated between COA and AOA; such variants would contribute to a lower genetic correlation. Under such a model, where the correlation in genetic effects differs between groups of variants, on balance, the genetic correlation between COA and AOA would be high but significantly different from 1. Association results for the sentinel SNPs identified in our GWAS suggest that this model is plausible (Figure 2).
The observation that COA and AOA have partly distinct genetic architectures is perhaps surprising given their broad similarity in clinical presentation. For comparison, the genetic overlap between COA and a combined hay fever and eczema phenotype (rg = 0.7) was similar to the genetic overlap between COA and AOA. We speculate that these results imply that there might be distinct genes or molecular pathways involved in COA but not (or less so) in AOA, or vice-versa, in which case identifying such pathways could be informative with regard to the development of drugs tailored specifically for COA or AOA. Another possibility, however, is that a particular gene contributes to the pathophysiology of both disease subtypes and that its expression is dysregulated in COA- and AOA-affected individuals by different mechanisms: inherited risk alleles in the former and acquired epigenetic modifications in the latter. Such epigenetic modifications in AOA-affected individuals could potentially result from long-term exposure to environmental risk factors, such as a low-quality diet45 or smoking.46 For example, we recently highlighted that lower expression of PITPNM2 (MIM: 608920), a gene potentially involved in neutrophil function, is independently associated with an allergy-predisposing allele and smoking-induced CpG methylation.15 Studies that carefully investigate the contribution of epigenetic modifications to the etiology of asthma, particularly with adult onset, are warranted; we suggest first investigating genes implicated by genetic studies.
Results from our heritability analysis also indicate that common SNPs explain only about 5%–10% of the variation in the specific age at which asthma was reported to be diagnosed during childhood and during adulthood. This observation, together with the widespread allele frequency differences observed between COA- and AOA-affected individuals, indicates that SNP associations discovered during analysis of the full spectrum of age of onset (i.e., spanning early childhood to older adulthood) must be interpreted with care. Specifically, one should consider whether a specific association is likely to reflect allele frequency differences between affected individuals with different asthma-onset subtypes (e.g., COA versus AOA) or between affected individuals with the same asthma subtype (e.g., COA) but with different age of onset. Our results suggest that the former scenario will be more common than the latter. By extension, our findings also suggest that measurement error and/or environmental risk factors have the largest contribution to variation in specific age of onset within each disease subtype. In childhood, candidate environmental-risk factors that might affect age of onset include the timing, frequency, and duration of chest infections, allergen exposure, pet ownership, maternal smoking, and low-quality diet during pregnancy.47, 48, 49, 50, 51, 52, 53 In adulthood, additional risk factors might include occupational exposures, active smoking, and obesity.20, 22, 54, 55, 56
To identify genetic risk factors that could be specific to COA or AOA, we performed two GWASs in the UK Biobank study. We separately compared 13,962 COA-affected individuals and 26,582 AOA-affected individuals against a common set of 300,671 asthma- and allergy-free controls. Despite a relatively small case sample size in the GWAS of COA, we identified a large number of independent SNP associations with asthma risk, 123 in total. A relatively smaller number of associations (56) were identified in the GWAS of AOA, consistent with the lower overall SNP heritability estimated for this disease subtype. For comparison, the largest number of independent associations reported to date in a single GWAS of asthma was 27 (based on 28,399 affected individuals and 128,843 controls57).
To validate the observed associations, we repeated the association analyses in the independent 23andMe study, comprising a total of 265,767 individuals. Of the 179 (123 + 56) associations discovered in the UK Biobank Study, 132 (98 + 34) were also detected in the replication study, and they remained of genome-wide significance in the combined analysis. When we took into account all associations reported to date for allergy-related traits in the GWAS catalog,58 and on the basis of a conservative LD r2 threshold of 0.05, we found that 28 of the combined 132 sentinel risk SNPs represented novel associations for asthma. Among these were the first two genome-wide-significant associations for asthma reported for variants on the X chromosome: rs850637, which overlaps a predicted transcription-factor binding site for RELA and is located between TLR7 (MIM: 300365) and TLR8 (MIM: 300366), two genes involved in anti-viral immunity;59, 60, 61 and rs5953283, located 18 kb upstream of FOXP3 (MIM: 300292), which is central to the establishment and maintenance of regulatory T cells.62
To determine whether the associations with COA and AOA were likely to be subtype specific, we then compared the frequency of the disease-predisposing allele for each sentinel risk SNP between the COA- and AOA-affected individuals. We found that for most sentinel SNPs, identified in either the COA or AOA GWAS, the predisposing allele was significantly more common in COA-affected individuals than in AOA-affected individuals, consistent with a stronger association with COA. This includes five variants that are likely to represent COA-specific risk factors because the frequency of the predisposing allele was similar in AOA-affected individuals and non-asthmatic controls (i.e., OR∼1 in the GWAS of AOA). On the basis of LD with sentinel eQTLs and non-synonymous variants, we were able to identify a likely target gene for only one of these five COA-specific risk variants (rs4574025): PIGN (MIM: 606097), a component of the glycosylphosphatidylinositol (GPI)-anchor biosynthesis pathway,63 which is involved in anchoring proteins to the cell surface64 and in ATP release.65
We also found one example of a sentinel SNP that is likely to represent an AOA-specific risk factor: rs2381712 near TEX41, which encodes a long non-coding RNA. Of note, rs2381712 is in moderate LD (r2 = 0.41; D’ = 0.75) with a variant (rs10193706) reported to associate with smoking behavior (heavy versus never smokers).66 The rs10193706:C allele that was associated with heavy smoking was associated with a lower risk of AOA, which is unexpected given that smoking is thought to be a risk factor for AOA and given the overall positive genetic correlation that we observed between smoking behavior and AOA risk. As such, it is possible that the two associations (rs2381712 with AOA and rs10193706 with smoking) do not tag the same underlying causal variant. We did not find any sentinel eQTLs or non-synonymous SNPs in high LD with rs2381712, and so the likely target gene of this risk variant remains to be identified. A nearby gene of potential interest to asthma is ZEB2 (MIM: 605802), which encodes a transcription factor that regulates T-cell and dendritic-cell development,67, 68, 69 as well as mast-cell signaling.70
For two additional sentinel SNPs, the observed difference in risk-allele frequency between the two case groups (AOA > COA) suggests that they represent stronger risk factors for AOA: rs2544026 in HDAC7 and rs28635831 in COG6. HDAC7 was identified as a likely target gene of rs2544026 on the basis of eQTL information from macrophages exposed to live bacterial pathogens (Listeria and Salmonella); results were consistent between two different eQTL studies.71, 72 Specifically, in infected macrophages but not in non-infected cells, the rs2544026:T disease-predisposing allele was associated with lower HDAC7 expression. This observation is consistent with reduced overall HDAC activity in individuals with asthma73, 74 and COPD.75 How lower HDAC7 expression in infected macrophages might then result in a higher risk of asthma needs to be determined. Because HDAC7 is thought to be a repressor of macrophage-specific genes,76 one possibility is that lower HDAC7 expression in macrophages results in a more exuberant inflammatory response against bacterial infections, which might contribute to an asthma-prone environment in the airways. Interestingly, COG6 was also identified as a likely target of rs28635831 solely on the basis of eQTLs described in macrophages stimulated with Salmonella. Variants in moderate to high LD with rs28635831 have been reported to associate with auto-immune diseases, namely psoriasis (MIM: 177900),77 rheumatoid arthritis, and lupus erythematosus (MIM: 152700).78 COG6 is part of the conserved oligomeric Golgi complex, and mutations in this gene have been described in individuals with congenital disorders of glycosylation; core clinical features include recurrent infections and hyperkeratosis.79 These observations suggest that COG6 might play a role in immune-cell function; studies that address this possibility are warranted.
Results from the genetic-correlation analysis between both COA and AOA and other complex traits or diseases provided some clues into disease mechanisms that are dysregulated by genetic variants in one disease subtype but not (or less so) in the other. First, we found a larger genetic correlation with eczema for COA than for AOA. This observation, which supported findings from the genetic-correlation analysis of hay fever that we performed in the UK Biobank study, indicates that SNPs that influence molecular pathways underlying allergies explain a larger proportion of variation in disease risk for COA than for AOA. Second, we found that alleles associated with achieving a higher education level were collectively associated with a higher risk of COA, consistent with some epidemiological studies,80, 81 but a lower risk of AOA. These opposing effects suggest that different mechanisms underlie both associations. For example, on the one hand, the development of asthma in childhood might result in an increased preference later in life for a professional career that requires a higher education. On the other hand, higher education is associated with a lower BMI in adulthood,82 which is protective for asthma. These potential causal relationships and other possible explanations (e.g., genetic pleiotropy) for the observed genetic correlations with education attainment should be explored in future studies. Lastly, we found that variants associated with three risk factors for asthma—obesity, smoking, and age at the birth of one’s first child (or puberty)—had a stronger contribution to the risk of AOA than of COA. As highlighted above, a significant genetic correlation might arise because of a causal effect of these risk factors on asthma risk.83, 84, 85, 86 At least for obesity, such a causal effect does not appear to extend to other allergies,87 which could potentially explain the difference in genetic correlation between AOA and COA.
A caveat of our analysis is that some asthma-affected individuals who were included in the UK Biobank Study and who were also included in our AOA group might not have recollected having been diagnosed with asthma when they were children. Such misclassification of true COA as AOA would tend to inflate the estimate of the genetic correlation between COA and AOA and also to decrease the power of the analysis to detect AOA-specific SNP associations.
In conclusion, we show that COA has a genetic etiology that is partly distinct from asthma that first develops in adult life. GWAS informed by age of onset can identify subtype-specific genetic risk factors, which can explain differences in pathophysiology between childhood and adult asthma.
Consortia
The members of the eQTLGen Consortium are as follows: Mawussé Agbessi, Habibul Ahsan, Isabel Alves, Anand Andiappan, Wibowo Arindrarto, Philip Awadalla, Alexis Battle, Frank Beutner, Marc Jan Bonder, Dorret Boomsma, Mark Christiansen, Annique Claringbould, Patrick Deelen, Tõnu Esko, Marie-Julie Favé, Lude Franke, Timothy Frayling, Sina A. Gharib, Gregory Gibson, Bastiaan T. Heijmans, Gibran Hemani, Rick Jansen, Mika Kähönen, Anette Kalnapenkis, Silva Kasela, Johannes Kettunen, Yungil Kim, Holger Kirsten, Peter Kovacs, Knut Krohn, Jaanika Kronberg-Guzman, Viktorija Kukushkina, Zoltan Kutalik, Bernett Lee, Terho Lehtimäki, Markus Loeffler, Urko M. Marigorta, Hailang Mei, Lili Milani, Grant W. Montgomery, Martina Müller-Nurasyid, Matthias Nauck, Michel Nivard, Brenda Penninx, Markus Perola, Natalia Pervjakova, Brandon L. Pierce, Joseph Powell, Holger Prokisch, Bruce M. Psaty, Olli T. Raitakari, Samuli Ripatti, Olaf Rotzschke, Sina Rüeger, Ashis Saha, Markus Scholz, Katharina Schramm, Ilkka Seppälä, Eline P. Slagboom, Coen D.A. Stehouwer, Michael Stumvoll, Patrick Sullivan, Peter-Bram ‘t Hoen, Alexander Teumer, Joachim Thiery, Lin Tong, Anke Tönjes, Jenny van Dongen, Maarten van Iterson, Joyce van Meurs, Jan H. Veldink, Joost Verlouw, Peter M. Visscher, Uwe Völker, Urmo Võsa, Harm-Jan Westra, Cisca Wijmenga, Hanieh Yaghootkar, Jian Yang, Biao Zeng, and Futao Zhang
The members of the BIOS (Biobank-based Integrative Omics Study) Consortium are as follows: Bastiaan T. Heijmans, Peter A.C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen MJ van Greevenbroek, Coen D.A. Stehouwer, Carla J.H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Sasha Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Bert A. Hofman, Aaron Isaacs, André G. Uitterlinden, Joyce van Meurs, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Dasha V. Zhernakova, Peter van ’t Hof, Patrick Deelen, Irene Nooren, Matthijs Moed, Martijn Vermaat, René Luijk, Marc Jan Bonder, Maarten van Iterson, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, Erik. W van Zwet, and Peter-Bram ’t Hoen, and Bastiaan T. Heijmans
Declaration of Interests
M.A.R.F. contributed to this study while employed by the QIMR Berghofer Medical Research Institute (Brisbane, Australia) but is now an employee of Regeneron Pharmaceuticals (New York, U.S.). Y.J. and D.A.H. are employed by and hold stock or stock options in 23andMe. All other authors declare no competing financial interests.
Acknowledgments
This research has been conducted through use of the UK Biobank Resource (application 10074). MARF was supported by a senior research fellowship (APP1124501) from the National Health and Medical Research Council (NHMRC) of Australia. LP was funded by a UK Medical Research Council (UK MRC) fellowship award (MR/J012165/1) and LP and RG work in a unit funded by the UK MRC (MC_UU_12013 and MC_UU_00011/1). Study-specific acknowledgments are listed in the Supplemental Data.
Published: March 28, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.02.022.
Web Resources
OMIM, https://www.omim.org/
Summary statistics from GWAS of COA and AOA in the UK Biobank, https://genepi.qimr.edu.au/staff/manuelF/gwas_results/main.html
Supplemental Data
References
- 1.Tan D.J., Walters E.H., Perret J.L., Lodge C.J., Lowe A.J., Matheson M.C., Dharmage S.C. Age-of-asthma onset as a determinant of different asthma phenotypes in adults: A systematic review and meta-analysis of the literature. Expert Rev. Respir. Med. 2015;9:109–123. doi: 10.1586/17476348.2015.1000311. [DOI] [PubMed] [Google Scholar]
- 2.Moore W.C., Meyers D.A., Wenzel S.E., Teague W.G., Li H., Li X., D’Agostino R., Jr., Castro M., Curran-Everett D., Fitzpatrick A.M., National Heart, Lung, and Blood Institute’s Severe Asthma Research Program Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deliu M., Yavuz T.S., Sperrin M., Belgrave D., Sahiner U.M., Sackesen C., Kalayci O., Custovic A. Features of asthma which provide meaningful insights for understanding the disease heterogeneity. Clin. Exp. Allergy. 2018;48:39–47. doi: 10.1111/cea.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newby C., Heaney L.G., Menzies-Gow A., Niven R.M., Mansur A., Bucknall C., Chaudhuri R., Thompson J., Burton P., Brightling C., British Thoracic Society Severe Refractory Asthma Network Statistical cluster analysis of the British Thoracic Society Severe refractory Asthma Registry: Clinical outcomes and phenotype stability. PLoS ONE. 2014;9:e102987. doi: 10.1371/journal.pone.0102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu W., Bleecker E., Moore W., Busse W.W., Castro M., Chung K.F., Calhoun W.J., Erzurum S., Gaston B., Israel E. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J. Allergy Clin. Immunol. 2014;133:1280–1288. doi: 10.1016/j.jaci.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim T.B., Jang A.S., Kwon H.S., Park J.S., Chang Y.S., Cho S.H., Choi B.W., Park J.W., Nam D.H., Yoon H.J., COREA Study Group Identification of asthma clusters in two independent Korean adult asthma cohorts. Eur. Respir. J. 2013;41:1308–1314. doi: 10.1183/09031936.00100811. [DOI] [PubMed] [Google Scholar]
- 7.Haldar P., Pavord I.D., Shaw D.E., Berry M.A., Thomas M., Brightling C.E., Wardlaw A.J., Green R.H. Cluster analysis and clinical asthma phenotypes. Am. J. Respir. Crit. Care Med. 2008;178:218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortqvist A.K., Lundholm C., Carlström E., Lichtenstein P., Cnattingius S., Almqvist C. Familial factors do not confound the association between birth weight and childhood asthma. Pediatrics. 2009;124:e737–e743. doi: 10.1542/peds.2009-0305. [DOI] [PubMed] [Google Scholar]
- 9.Tedner S.G., Örtqvist A.K., Almqvist C. Fetal growth and risk of childhood asthma and allergic disease. Clin. Exp. Allergy. 2012;42:1430–1447. doi: 10.1111/j.1365-2222.2012.03997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt P.G. The mechanism or mechanisms driving atopic asthma initiation: The infant respiratory microbiome moves to center stage. J. Allergy Clin. Immunol. 2015;136:15–22. doi: 10.1016/j.jaci.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Lynch J.P., Sikder M.A., Curren B.F., Werder R.B., Simpson J., Cuív P.O., Dennis P.G., Everard M.L., Phipps S. The influence of the microbiome on early-life severe viral lower respiratory infections and asthma—Food for thought? Front. Immunol. 2017;8:156. doi: 10.3389/fimmu.2017.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Nijs S.B., Venekamp L.N., Bel E.H. Adult-onset asthma: Is it really different? Eur. Respir. Rev. 2013;22:44–52. doi: 10.1183/09059180.00007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilmarinen P., Tuomisto L.E., Kankaanranta H. Phenotypes, risk factors, and mechanisms of adult-onset asthma. Mediators Inflamm. 2015;2015:514868. doi: 10.1155/2015/514868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarnowski C., Sugier P.E., Granell R., Jarvis D., Dizier M.H., Ege M., Imboden M., Laprise C., Khusnutdinova E.K., Freidin M.B. Identification of a new locus at 16q12 associated with time to asthma onset. J. Allergy Clin. Immunol. 2016;138:1071–1080. doi: 10.1016/j.jaci.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira M.A., Vonk J.M., Baurecht H., Marenholz I., Tian C., Hoffman J.D., Helmer Q., Tillander A., Ullemar V., van Dongen J., 23andMe Research Team. AAGC collaborators. BIOS consortium. LifeLines Cohort Study Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat. Genet. 2017;49:1752–1757. doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belsky D.W., Sears M.R., Hancox R.J., Harrington H., Houts R., Moffitt T.E., Sugden K., Williams B., Poulton R., Caspi A. Polygenic risk and the development and course of asthma: An analysis of data from a four-decade longitudinal study. Lancet Respir. Med. 2013;1:453–461. doi: 10.1016/S2213-2600(13)70101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brusselle G., Germinaro M., Weiss S., Zangrilli J. Reslizumab in patients with inadequately controlled late-onset asthma and elevated blood eosinophils. Pulm. Pharmacol. Ther. 2017;43:39–45. doi: 10.1016/j.pupt.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Bleecker E.R., Wechsler M.E., FitzGerald J.M., Menzies-Gow A., Wu Y., Hirsch I., Goldman M., Newbold P., Zangrilli J.G. Baseline patient factors impact on the clinical efficacy of benralizumab for severe asthma. Eur. Respir. J. 2018;52 doi: 10.1183/13993003.00936-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strachan D.P., Butland B.K., Anderson H.R. Incidence and prognosis of asthma and wheezing illness from early childhood to age 33 in a national British cohort. BMJ. 1996;312:1195–1199. doi: 10.1136/bmj.312.7040.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oswald H., Phelan P.D., Lanigan A., Hibbert M., Bowes G., Olinsky A. Outcome of childhood asthma in mid-adult life. BMJ. 1994;309:95–96. doi: 10.1136/bmj.309.6947.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sood A., Qualls C., Schuyler M., Arynchyn A., Alvarado J.H., Smith L.J., Jacobs D.R., Jr. Adult-onset asthma becomes the dominant phenotype among women by age 40 years. the longitudinal CARDIA study. Ann. Am. Thorac. Soc. 2013;10:188–197. doi: 10.1513/AnnalsATS.201212-115OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins M.A., Hopper J.L., Bowes G., Carlin J.B., Flander L.B., Giles G.G. Factors in childhood as predictors of asthma in adult life. BMJ. 1994;309:90–93. doi: 10.1136/bmj.309.6947.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh P.-R., Bhatia G., Gusev A., Finucane H.K., Bulik-Sullivan B.K., Pollack S.J., de Candia T.R., Lee S.H., Wray N.R., Kendler K.S., O’Donovan M.C., Neale B.M., Patterson N., Price A.L., Schizophrenia Working Group of the Psychiatric Genomics Consortium Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., Daly M.J., Price A.L., Neale B.M., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd A., Golding J., Macleod J., Lawlor D.A., Fraser A., Henderson J., Molloy L., Ness A., Ring S., Davey Smith G. Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer C.J., Li Y., Abecasis G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira M.A. Improving the power to detect risk variants for allergic disease by defining case-control status based on both asthma and hay fever. Twin Res. Hum. Genet. 2014;17:505–511. doi: 10.1017/thg.2014.59. [DOI] [PubMed] [Google Scholar]
- 32.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gazal S., Finucane H.K., Furlotte N.A., Loh P.R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fadista J., Manning A.K., Florez J.C., Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 2016;24:1202–1205. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S., Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce B.L., Tong L., Chen L.S., Rahaman R., Argos M., Jasmine F., Roy S., Paul-Brutus R., Westra H.J., Franke L. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: A genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10:e1004818. doi: 10.1371/journal.pgen.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis J.R., Fresard L., Knowles D.A., Pala M., Bustamante C.D., Battle A., Montgomery S.B. An efficient multiple-testing adjustment for eQTL studies that accounts for linkage disequilibrium between variants. Am. J. Hum. Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang X., Wang K. wANNOVAR: Annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carey G. Inference about genetic correlations. Behav. Genet. 1988;18:329–338. doi: 10.1007/BF01260933. [DOI] [PubMed] [Google Scholar]
- 45.Thorburn A.N., McKenzie C.I., Shen S., Stanley D., Macia L., Mason L.J., Roberts L.K., Wong C.H., Shim R., Robert R. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 46.Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Midodzi W.K., Rowe B.H., Majaesic C.M., Saunders L.D., Senthilselvan A. Early life factors associated with incidence of physician-diagnosed asthma in preschool children: results from the Canadian Early Childhood Development cohort study. J. Asthma. 2010;47:7–13. doi: 10.3109/02770900903380996. [DOI] [PubMed] [Google Scholar]
- 48.Ferry O.R., Duffy D.L., Ferreira M.A. Early life environmental predictors of asthma age-of-onset. Immun. Inflamm. Dis. 2014;2:141–151. doi: 10.1002/iid3.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott M., Roberts G., Kurukulaaratchy R.J., Matthews S., Nove A., Arshad S.H. Multifaceted allergen avoidance during infancy reduces asthma during childhood with the effect persisting until age 18 years. Thorax. 2012;67:1046–1051. doi: 10.1136/thoraxjnl-2012-202150. [DOI] [PubMed] [Google Scholar]
- 50.Holt P.G., Sly P.D. Viral infections and atopy in asthma pathogenesis: New rationales for asthma prevention and treatment. Nat. Med. 2012;18:726–735. doi: 10.1038/nm.2768. [DOI] [PubMed] [Google Scholar]
- 51.de Marco R., Pattaro C., Locatelli F., Svanes C., ECRHS Study Group Influence of early life exposures on incidence and remission of asthma throughout life. J. Allergy Clin. Immunol. 2004;113:845–852. doi: 10.1016/j.jaci.2004.01.780. [DOI] [PubMed] [Google Scholar]
- 52.Litonjua A.A., Milton D.K., Celedon J.C., Ryan L., Weiss S.T., Gold D.R. A longitudinal analysis of wheezing in young children: the independent effects of early life exposure to house dust endotoxin, allergens, and pets. J. Allergy Clin. Immunol. 2002;110:736–742. doi: 10.1067/mai.2002.128948. [DOI] [PubMed] [Google Scholar]
- 53.Lee-Sarwar K., Litonjua A.A. As you eat it: Effects of prenatal nutrition on asthma. J. Allergy Clin. Immunol. Pract. 2018;6:711–718. doi: 10.1016/j.jaip.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh R.E., Cullinan P., Fishwick D., Hoyle J., Warburton C.J., Strachan D.P., Butland B.K., Jarvis D. Asthma and occupation in the 1958 birth cohort. Thorax. 2013;68:365–371. doi: 10.1136/thoraxjnl-2012-202151. [DOI] [PubMed] [Google Scholar]
- 55.Karjalainen A., Martikainen R., Karjalainen J., Klaukka T., Kurppa K. Excess incidence of asthma among Finnish cleaners employed in different industries. Eur. Respir. J. 2002;19:90–95. doi: 10.1183/09031936.02.00201002. [DOI] [PubMed] [Google Scholar]
- 56.Brumpton B., Langhammer A., Romundstad P., Chen Y., Mai X.M. General and abdominal obesity and incident asthma in adults: the HUNT study. Eur. Respir. J. 2013;41:323–329. doi: 10.1183/09031936.00012112. [DOI] [PubMed] [Google Scholar]
- 57.Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clingan J.M., Matloubian M. B Cell-intrinsic TLR7 signaling is required for optimal B cell responses during chronic viral infection. J. Immunol. 2013;191:810–818. doi: 10.4049/jimmunol.1300244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson S., Kaiko G., Loh Z., Lalwani A., Zhang V., Spann K., Foo S.Y., Hansbro N., Uematsu S., Akira S. Plasmacytoid dendritic cells promote host defense against acute pneumovirus infection via the TLR7-MyD88-dependent signaling pathway. J. Immunol. 2011;186:5938–5948. doi: 10.4049/jimmunol.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cros J., Cagnard N., Woollard K., Patey N., Zhang S.Y., Senechal B., Puel A., Biswas S.K., Moshous D., Picard C. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu L., Barbi J., Pan F. The regulation of immune tolerance by FOXP3. Nat. Rev. Immunol. 2017;17:703–717. doi: 10.1038/nri.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaynor E.C., Mondésert G., Grimme S.J., Reed S.I., Orlean P., Emr S.D. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yadav U., Khan M.A. Targeting the GPI biosynthetic pathway. Pathog. Glob. Health. 2018;112:115–122. doi: 10.1080/20477724.2018.1442764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong X., Malhotra R., Guidotti G. ATP uptake in the Golgi and extracellular release require Mcd4 protein and the vacuolar H+-ATPase. J. Biol. Chem. 2003;278:33436–33444. doi: 10.1074/jbc.M305785200. [DOI] [PubMed] [Google Scholar]
- 66.Wain L.V., Shrine N., Miller S., Jackson V.E., Ntalla I., Soler Artigas M., Billington C.K., Kheirallah A.K., Allen R., Cook J.P., UK Brain Expression Consortium (UKBEC) OxGSK Consortium Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir. Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X., Briseño C.G., Grajales-Reyes G.E., Haldar M., Iwata A., Kretzer N.M., Kc W., Tussiwand R., Higashi Y., Murphy T.L., Murphy K.M. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc. Natl. Acad. Sci. USA. 2016;113:14775–14780. doi: 10.1073/pnas.1611408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scott C.L., Soen B., Martens L., Skrypek N., Saelens W., Taminau J., Blancke G., Van Isterdael G., Huylebroeck D., Haigh J. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Omilusik K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 2015;212:2027–2039. doi: 10.1084/jem.20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbu E.A., Zhang J., Berenstein E.H., Groves J.R., Parks L.M., Siraganian R.P. The transcription factor Zeb2 regulates signaling in mast cells. J. Immunol. 2012;188:6278–6286. doi: 10.4049/jimmunol.1102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A.J., Mann A.L., Kundu K., Hale C., Dougan G., Gaffney D.J., HIPSCI Consortium Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nedelec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 73.Gunawardhana L.P., Gibson P.G., Simpson J.L., Powell H., Baines K.J. Activity and expression of histone acetylases and deacetylases in inflammatory phenotypes of asthma. Clin. Exp. Allergy. 2014;44:47–57. doi: 10.1111/cea.12168. [DOI] [PubMed] [Google Scholar]
- 74.Zuccaro L., Cox A., Pray C., Radford K., Novakowski K., Dorrington M., Surette M.G., Bowdish D., Nair P. Histone deacetylase activity and recurrent bacterial bronchitis in severe eosinophilic asthma. Allergy. 2016;71:571–575. doi: 10.1111/all.12831. [DOI] [PubMed] [Google Scholar]
- 75.Ito K., Ito M., Elliott W.M., Cosio B., Caramori G., Kon O.M., Barczyk A., Hayashi S., Adcock I.M., Hogg J.C., Barnes P.J. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 76.Barneda-Zahonero B., Román-González L., Collazo O., Rafati H., Islam A.B., Bussmann L.H., di Tullio A., De Andres L., Graf T., López-Bigas N. HDAC7 is a repressor of myeloid genes whose downregulation is required for transdifferentiation of pre-B cells into macrophages. PLoS Genet. 2013;9:e1003503. doi: 10.1371/journal.pgen.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Helms C., Liao W., Zaba L.C., Duan S., Gardner J., Wise C., Miner A., Malloy M.J., Pullinger C.R. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Márquez A., Vidal-Bralo L., Rodríguez-Rodríguez L., González-Gay M.A., Balsa A., González-Álvaro I., Carreira P., Ortego-Centeno N., Ayala-Gutiérrez M.M., García-Hernández F.J. A combined large-scale meta-analysis identifies COG6 as a novel shared risk locus for rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 2017;76:286–294. doi: 10.1136/annrheumdis-2016-209436. [DOI] [PubMed] [Google Scholar]
- 79.Rymen D., Winter J., Van Hasselt P.M., Jaeken J., Kasapkara C., Gokçay G., Haijes H., Goyens P., Tokatli A., Thiel C. Key features and clinical variability of COG6-CDG. Mol. Genet. Metab. 2015;116:163–170. doi: 10.1016/j.ymgme.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 80.Mazurek J.M., Schleiff P.L., Henneberger P.K. Is childhood asthma associated with educational level and longest-held occupation? Am. J. Epidemiol. 2012;175:279–288. doi: 10.1093/aje/kwr300. [DOI] [PubMed] [Google Scholar]
- 81.Sturdy P., Bremner S., Harper G., Mayhew L., Eldridge S., Eversley J., Sheikh A., Hunter S., Boomla K., Feder G. Impact of asthma on educational attainment in a socioeconomically deprived population: A study linking health, education and social care datasets. PLoS ONE. 2012;7:e43977. doi: 10.1371/journal.pone.0043977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rohrmann S., Steinbrecher A., Linseisen J., Hermann S., May A., Luan J., Ekelund U., Overvad K., Tjønneland A., Halkjær J. The association of education with long-term weight change in the EPIC-PANACEA cohort. Eur. J. Clin. Nutr. 2012;66:957–963. doi: 10.1038/ejcn.2012.55. [DOI] [PubMed] [Google Scholar]
- 83.Granell R., Henderson A.J., Evans D.M., Smith G.D., Ness A.R., Lewis S., Palmer T.M., Sterne J.A. Effects of BMI, fat mass, and lean mass on asthma in childhood: A Mendelian randomization study. PLoS Med. 2014;11:e1001669. doi: 10.1371/journal.pmed.1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Minelli C., van der Plaat D.A., Leynaert B., Granell R., Amaral A.F.S., Pereira M., Mahmoud O., Potts J., Sheehan N.A., Bowden J. Age at puberty and risk of asthma: A Mendelian randomisation study. PLoS Med. 2018;15:e1002634. doi: 10.1371/journal.pmed.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y.C., Fan H.Y., Huang Y.T., Huang S.Y., Liou T.H., Lee Y.L. Causal relationships between adiposity and childhood asthma: bi-directional Mendelian Randomization analysis. Int. J. Obes. 2019;43:73–81. doi: 10.1038/s41366-018-0160-8. [DOI] [PubMed] [Google Scholar]
- 86.Skaaby T., Taylor A.E., Jacobsen R.K., Paternoster L., Thuesen B.H., Ahluwalia T.S., Larsen S.C., Zhou A., Wong A., Gabrielsen M.E. Investigating the causal effect of smoking on hay fever and asthma: A Mendelian randomization meta-analysis in the CARTA consortium. Sci. Rep. 2017;7:2224. doi: 10.1038/s41598-017-01977-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skaaby T., Taylor A.E., Thuesen B.H., Jacobsen R.K., Friedrich N., Møllehave L.T., Hansen S., Larsen S.C., Völker U., Nauck M. Estimating the causal effect of body mass index on hay fever, asthma and lung function using Mendelian randomization. Allergy. 2018;73:153–164. doi: 10.1111/all.13242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.