Abstract
Pheochromocytomas and paragangliomas (PPGLs) provide some of the clearest genetic evidence for the critical role of metabolism in the tumorigenesis process. Approximately 40% of PPGLs are caused by driver germline mutations in 16 known susceptibility genes, and approximately half of these genes encode members of the tricarboxylic acid (TCA) cycle. Taking as a starting point the involvement of the TCA cycle in PPGL development, we aimed to identify unreported mutations that occurred in genes involved in this key metabolic pathway and that could explain the phenotypes of additional individuals who lack mutations in known susceptibility genes. To accomplish this, we applied a targeted sequencing of 37 TCA-cycle-related genes to DNA from 104 PPGL-affected individuals with no mutations in the major known predisposing genes. We also performed omics-based analyses, TCA-related metabolite determination, and 13C5-glutamate labeling assays. We identified five germline variants affecting DLST in eight unrelated individuals (∼7%); all except one were diagnosed with multiple PPGLs. A recurrent variant, c.1121G>A (p.Gly374Glu), found in four of the eight individuals triggered accumulation of 2-hydroxyglutarate, both in tumors and in a heterologous cell-based assay designed to functionally evaluate DLST variants. p.Gly374Glu-DLST tumors exhibited loss of heterozygosity, and their methylation and expression profiles are similar to those of EPAS1-mutated PPGLs; this similarity suggests a link between DLST disruption and pseudohypoxia. Moreover, we found positive DLST immunostaining exclusively in tumors carrying TCA-cycle or EPAS1 mutations. In summary, this study reveals DLST as a PPGL-susceptibility gene and further strengthens the relevance of the TCA cycle in PPGL development.
Keywords: TCA cycle, DLST, pheochromocytoma, paraganglioma, cancer susceptibility gene
Introduction
Pheochromocytomas and paragangliomas, together called PPGLs (MIM: 171300), are rare neuroendocrine tumors derived from chromaffin cells of the adrenal medulla and extra-adrenal paraganglia, respectively. PPGLs have one of the highest heritability rates of all neoplasms in humans. Approximately 40% of individuals with this genetically heterogeneous disease carry a germline mutation in one of 16 different susceptibility genes (FH [MIM: 136850], KIF1B [MIM: 605995], MAX [MIM: 154950], MDH2 [MIM: 154100], NF1 [MIM: 613113], EGLN2 [MIM: 606424], EGLN1 [MIM: 606425], RET [MIM: 164761], SDHA [MIM: 600857], SDHAF2 [MIM: 613019], SDHB [MIM: 185470], SDHC [MIM: 602413], SDHD [MIM: 602690], SLC25A11 [MIM: 604165], TMEM127 [MIM: 613403], and VHL [MIM: 608537]).1, 2 However, an important fraction of individuals with clinical features (such as a family history of PPGL, multifocal tumors, or an early age of onset) indicative of a hereditary condition lack mutations in any of the known PPGL susceptibility genes.
On the basis of many expression and methylation profiling studies (for a review, see Dahia, 20143), PPGLs can be classified into two main clusters; one of them (cluster 1) is enriched in tumors that carry mutations in genes, such as the succinate dehydrogenase (SDH) genes FH, MDH2, and IDH1 [MIM: 147700], encoding tricarboxylic acid (TCA) cycle enzymes. These alterations lead to disruption of the TCA cycle, and that disruption results in the accumulation of “oncometabolites” like succinate, fumarate, and 2-hydroxyglutarate (2HG). These oncometabolites contribute to PPGL tumorigenesis through abrogating the function of α-ketoglutarate (αKG)-dependent dioxygenases such as Tet methylcytosine dioxygenase 2 or histone N-methyl-lysine demethylases. This leads to a hypermethylation signature, also known as the CpG-island-methylator-phenotype (CIMP) profile, similar to the ones observed in glioblastomas4 and renal cell carcinomas5 (Ricketts et al., 2016, Cancer Res., abstract) that carry metabolic alterations such as gain-of-function IDH1 or IDH2 and loss-of-function (LoF) FH or SDHB mutations, respectively. Interestingly, there are still PPGLs that exhibit a CIMP-like profile but have no alteration in any of the known TCA-cycle-related susceptibility genes.6
Most PPGLs are benign, but 10%–20% of PPGL-affected individuals develop metastases in distant, non-chromaffin tissues such as in the liver, bones, lymph nodes, and lungs.7, 8 Despite limited knowledge of the markers of malignancy, it is known that PPGLs carrying mutations in TCA-cycle-related genes, such as SDHB or FH, show particularly high rates of malignancy and lower survival rates.9 Therefore, a better understanding of the molecular pathogenesis of this disease might determine the appropriate health management or treatment strategy for affected individuals.
Considering the relevance of the TCA cycle in PPGL development, we hypothesized that other genes involved in this metabolic pathway could be responsible for the disease in genetically as-yet-uncharacterized cases. In the present study, we identified a PPGL susceptibility gene by sequencing 37 TCA-cycle-related genes in a series of affected individuals without known mutations. We found four different missense mutations and one splice-site variant in the dihydrolipoamide S-succinyltransferase (DLST [MIM: 126063]) gene in eight unrelated individuals, suggesting a pathogenic role for the disruption of the activity of the αKG dehydrogenase (OGDH) complex. The characteristic methylation profile and the accumulation of 2HG found in tumors and cells carrying the recurrent variant c.1121G>A (p.Gly374Glu) (dbSNP: rs1270341616) suggest that DLST might be a PPGL susceptibility gene and further supports the importance of TCA-cycle alterations in PPGL development.
Material and Methods
Research Subjects
104 affected index individuals without mutations in known PPGL susceptibility genes (RET, VHL, NF1, MAX, TMEM127, SDHA, SDHB, SDHC, SDHD, SDHAF2, MDH2, FH, EPAS1 [MIM: 603349], and HRAS [MIM: 190020]) were included in the study. A summary of the clinical data of the affected individuals is shown in Table S1. In order to rule out hidden mutations affecting the SDH genes, we performed immunostaining of SDHB when formalin-fixed, paraffin-embedded (FFPE) tumor tissues were available.10 Genomic DNA was extracted from peripheral-blood leukocytes with the Maxwell 16 Blood DNA-purification system (Promega). Tumor DNA was obtained with the DNeasy Blood and Tissue kit (QIAGEN) for frozen tissues and the Covaris S2 System (Covaris) for FFPE tissues according to the manufacturers’ instructions. The Instituto de Salud Carlos III (ISCIII) ethics committee (Spain) approved the study, and all individuals provided written informed consent to participate in this study.
Targeted Next-Generation Sequencing
We extracted DNA from the selected samples and sequenced it for a set of 37 genes involved in the TCA cycle (Table S2) with TruSight sequencing technology and using a previously reported NGS panel.6 In brief, the NGS panel was designed in DesignStudio software (Illumina), and the DNA samples were sequenced with a MiSeq desktop sequencer as previously described.6 Identified coding and splice-site variants were filtered by considering mapping quality, variant score, depth, strand bias, annotation quality, and predicted effect. The cutoff applied for considering a nucleotide substitution as a candidate pathogenic variant was 1.805 × 10−5, which is the highest frequency of a known pathogenic SDHB mutation in the Genome Aggregation Consortium (gnomAD) database. We used the PredictSNP consensus classifier11 to predict the effects of the substitutions that passed all filtering steps, and we used MultAlin software to study and visualize the conservation of specific residues in multiple aligned sequences from different species.
Structural Modeling of the DLST Catalytic Domain and Predictions on Variants
The full-length amino acid sequence of hDLST was submitted to the Phyre2 server for structure prediction.12 14 templates were selected for protein modeling on the basis of heuristics designed to maximize confidence, percentage identity, and alignment coverage. Of these, we used 13 templates to model the biotin-lipoyl attachment domain. We used the E. coli dihydrolipoamide succinyltransferase structure (PDB: 1SCZ, 60% identity, 74% similarity) to model the catalytic domain. Nine residues were modeled ab initio. The final model of the protein had 98% of its sequence modeled at >90% confidence. hDLST has been shown to be active as a trimer in vivo.13 We generated the trimeric structure by using the E. coli dihydrolipoamide succinyltransferase structure crystallized in a trigonal space group (PDB: 1C4T) as a template. The steric clashes that were generated during modeling and trimerization were removed with the program Chiron.14 The active site of the catalytic domain was localized with the known catalytic residues His424 and Ser372. We predicted the binding site of the substrate by using the dihydrolipoyl transacetylase structure of A. vinelandii crystallized in the presence of 6,8-dimercapto-octanoid acid amide and coenzyme A (PDB: 1EAB). Images were generated with PyMOL.15
Loss of Heterozygosity and SNP-Array Analyses
Loss-of-heterozygosity (LOH) analysis of the DLST locus was performed on samples for which tumor DNA was available. Sanger sequencing of tumor DNA with specific primers that amplified DLST regions containing the mutations identified was performed in eight tumor samples from five individuals. To characterize the mechanism of LOH, high-density SNP-array analysis was performed on DNA of sufficient quality from two tumors (#4 and #5A). A genome-wide scan was conducted on 250 ng of tumor DNA with the Illumina Human610-Quad BeadChip (Illumina) according to the manufacturer’s specifications. Image data was analyzed with the Chromosome Viewer tool contained in GenomeStudio 2010.2 (Illumina). The metric we used was the log-R ratio, which is the binary logarithm of the ratio of the observed to expected normalized R values for a given SNP.16 In addition, we estimated the allele frequency for all SNPs. Microsatellite analysis was carried out on DNA from an additional individual (#3). Primers for three polymorphic markers located in chromosome 14 and chromosome 2 (which we used as a control) were designed and labeled (5′ 6-FAM). We amplified genomic and tumor DNA samples separately by means of a multiplex PCR kit (QIAGEN), and we used PCR amplification products for fragment analysis on an ABI PRISMTM 310 capillary sequencer (Applied Biosystems) and analyzed them with Peak Scanner Software v1.0 (Applied Biosystems) as previously described.17
Lentiviral Constructs and Techniques
We overexpressed cDNA constructs by using the pLVX-EF1a-IRES-neo vector, which we derived from the pLVX-EF1a-IRES-puro vector (631988, Clontech) by exchanging the selection cassettes. All DLST cDNA constructs were untagged. All constructs were confirmed by sequencing. A catalytically dead DLST mutant was designed on the basis of the crystal structure of the E.coli DLST ortholog (PDB: 1SCZ), and His375 was selected as critical for the deprotonation of the thiol group of coenzyme A. The corresponding human residue, His424, was selected for mutagenesis. Lentivirus-based constructs were made according to the standard protocol of the RNAi Consortium from the Broad Institute. In order to evaluate the role of DLST variants, we used H838 DLST-KO cells previously generated by CRISPR-Cas9 technology.18 For infection, H838 DLST-KO cells were centrifuged for 1 h in the presence of the viruses and 8 μgg/mL Polybrene (H9268, Sigma). The medium was replaced after the spin, and drug selection was started 24 h later. Selection was carried out until all uninfected control cells were dead.
13C5-glutamate Labeling Studies in DLST-KO Cancer Cells
We conducted labeling studies and liquid chromatography-mass spectrometry (LC-MS) methods as previously described.18 In brief, cells were plated at 60%–70% confluency in triplicate in fresh Roswell Park Memorial Institute 1640 (RPMI-1640) medium and incubated overnight. The next day, the medium was replaced with medium containing 2 mM 13C5-glutamate. Labeling was done for 3 h, after which we washed the cells three times with PBS (pH 7.4), and we extracted metabolites via the addition of ice-cold 80% methanol. Cell debris was removed by centrifugation, and samples were dried under a stream of nitrogen. Internal standard (L-Glutamic acid-13C5, 15N,2,3,3,4,4-d5, 749850, Sigma-Aldrich) was added at 1 mg/mL during the extraction step. In all labeling studies, the reported amounts of labeled species were corrected for natural isotope abundance.
Immunoblotting
Immunoblotting was conducted as previously described.18 In brief, cells were harvested in 1× RIPA buffer (BP-115, Boston BioProducts) containing a phosphatase and protease inhibitor cocktail (5872S, Cell Signaling Technology). We scraped adherent cells into lysis buffer and then sonicated the mixture for a brief time to solubilize the cells. Cell lysates were cleared by centrifugation for 10 min at 4°C. Protein quantification was done with the Pierce BCA Assay Kit (23225, Life Technologies). The primary antibodies we used were anti-DLST (HPA003010; rabbit polyclonal 1:1000, Sigma) and anti-GAPDH (97166, 1:10,000, Cell Signaling Technology). The results were visualized with the Odyssey imaging system (LI-COR) and the following secondary antibodies: IRDye 680RD donkey anti-rabbit (926-68073, LI-COR) and IRDye 800CW donkey anti-mouse (926-32212, LI-COR).
Liquid Chromatographic-Tandem Mass Spectrometric Determination of TCA-Related Metabolites
Fresh-frozen or FFPE tumor tissues (5–10 mg) from four available individuals (tumors #1; #3A,B, and C; #4; and #5A and B) (Table 1) and from 51 controls (DLST-WT PPGLs) were immersed in 500 μL LC-MS/MS-grade methanol containing isotope-labeled internal standards and processed as previously described.19 An analysis of metabolites was carried out with an AB Sciex 5500 QTRAP mass spectrometer coupled to an Acquity ultra-high-performance liquid chromatographic system (Waters) as previously described.19
Table 1.
Clinical Data of the Individuals with DLST Mutations
| ID | Tumors Analyzed | Gender | Age (Years) | Tumors | Other Tumors | Biochemical Phenotype | Behavior | cDNA Variant | Protein Change | PredictSNP | LOH | DLST IHC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | #1 | m | 45 | PCC | – | NM | Mg | c.692G>A | p.Arg231Gln | deleterious | no | + |
| #2 | – | f | 63 | H&N (n = 2) | – | NS | Bg | c.910G>A | p.Asp304Asn | neutral | NA | NA |
| #3 | #3A, B, and C | f | 27 | TAP (n = 7) | uterine endometrioid carcinoma | NM | Bg | c.1121G>A | p.Gly374Glu | deleterious | yes (UPD) | +++ |
| #4 | #4 | m | 38 | TAP (n = 3) | – | NM | Bg | c.1121G>A | p.Gly374Glu | deleterious | yes (UPD) | +++ |
| #5 | #5A and B | f | 24 | TAP and PCC | – | NM | Bg | c.1121G>A | p.Gly374Glu | deleterious | yes (UPD) | NA |
| #6a | – | m | 29 | TAP (n = 4) | – | NM | Bg | c.1121G>A | p.Gly374Glu | deleterious | NA | NA |
| #7 | – | m | 29 | TAP (n = 3) | – | NM | Bg | c.1265A>G | p.Tyr422Cys | deleterious | NA | NA |
| #8 | #8 | m | 54 | PCC (n = 2) | pituitary adenoma (PRL) | NM | Bg | c.1060-3T>A | – | – | no | +++ |
Abbreviations are as follows: Bg = benign; f = female; H&N = head and neck pheochromocytomas and paragangliomas (PGLs); IHC = immunohistochemistry; LOH = loss of heterozygosity; m = male; Mg = malignant; MTC = medullary thyroid carcinoma; NA = not available; NM = normetanephrine; NS = non-secretory; PCC = pheochromocytoma; PRL = prolactinoma; TAP = thoracic-abdominal-pelvic PGL; UPD = uniparental disomy; + = weak immunostaining; +++ = strong immunostaining. a indicates an individual studied by exome sequencing. DLST cDNA mutations are numbered according to human cDNA reference sequence GenBank: NM_001933.
DNA Methylation Array
Bisulfite conversion of DNA was performed with the EZ DNA Methylation Kit (Zymo Research), and genome-wide DNA methylation was assayed with the Infinium MethylationEPIC BeadChip Kit (Illumina) at the Centro Nacional de Genotipado (CEGEN-ISCIII) as previously described.20 This BeadChip interrogates over 850,000 methylation sites per sample at single-nucleotide resolution. Beta values for interrogated CpGs were assigned with the Genome Studio Methylation module and transformed into M values by applying the formula log2[beta value/(1-beta value)]. Negative M values indicate less than 50% methylation, and positive M values indicate more than 50% methylation.21 We used the M values for statistical analyses. Hierarchical clustering of methylation data from three unrelated DLST-mutated tumors and 13 PPGLs carrying known mutations in major susceptibility genes (four SDHB-, two DNMT3A-, two HRAS-, one NF1-, two MAX-, one EPAS1-, and one RET-mutated tumor) was performed with GeneCluster 2.0.22 A second, unsupervised analysis was performed with a list of 125,112 probes corresponding to 4,662 genes with CpG sites reported as significantly hypermethylated in SDHx-mutated (M1) PPGLs.23 We used the complete linkage as a clustering method and the city-block metric as the distance measure in the unsupervised comparisons. The methylomes we used in the analysis have been deposited in the National Center for Biotechnology Information GEO database, under accession numbers GEO: GSE111336 and GSE123185.
Gene Expression Array
We used the Agilent Whole Human Genome platform (4×44K) for the competitive hybridization of labeled and amplified cDNAs obtained from Universal Human Reference RNA (Stratagene) and from RNAs extracted from PPGL tumor samples as previously described.24 To identify a transcriptional profile related to DLST alteration, we used gene expression data from two available DLST-mutated tumors and from 67 controls harboring mutations in other PPGL susceptibility genes (GEO: GSE19422). We grouped tumor samples according to their expression profiles by unsupervised clustering, and we used a supervised analysis to identify specific transcripts related to the presence of DLST mutations. For the hierarchical clustering, we used expression data from a previously reported list of 451 genes (572 probes) differentially expressed in SDHx-, VHL-, and RET-, NF1-, or TMEM127-mutated PPGLs.25 Differentially expressed genes were identified by a t test (limma) carried out with POMELO II software.26 To account for multiple hypothesis testing, the estimated significance level (P) was adjusted with the Benjamini false discovery rate (FDR) correction.27 Genes with an FDR <0.05 were considered to be differentially expressed between tumor classes. The transcriptome from tumor #5A (data from tumor #4 are already included in GEO: GSE19422) has been deposited in GEO, under accession number GEO: GSE123344.
Quantitative Real-Time PCR
We obtained total RNA from FFPE or frozen material by using the RNeasy FFPE (QIAGEN) or TriReagent (MRC) kit, respectively, according to the manufacturers’ instructions. We prepared cDNA from 1 μg of total RNA by using a mix of oligo-dT and random primers and the qScript cDNA Synthesis Kit (#95047-100, Quanta Biosciences). mRNA concentrations from genes of interest were determined by quantitative PCR on a 7500 fast real-time PCR system (Applied Biosystems) that used the Universal ProbeLibrary set; each sample was analyzed in triplicate. Relative mRNA concentrations were estimated by the ΔΔCt method28 and normalized using β-actin (ACTB) as a housekeeping gene. The results are shown as the mean ± SD, and mRNAs obtained from frozen or FFPE tumors carrying mutations in other known PPGL susceptibility genes were used as controls.
Immunohistochemistry
Immunohistochemical (IHC) staining of DLST (11954; rabbit monoclonal 1:150, Cell Signaling Technology) was performed with 3 μm FFPE sections from six DLST-mutated tumor samples available, according to standard procedures. 82 tumors carrying mutations in other PPGL susceptibility genes were used as controls.
Results
TCA-Targeted NGS Findings
In order to identify additional genes mutated in PPGL-affected individuals, we applied targeted sequencing of 37 TCA-cycle-related genes to individuals with PPGLs and without mutations in the major known predisposing genes. Targeted NGS identified nine germline LoF and non-synonymous coding variants affecting five different genes in 13 unrelated, affected individuals from the 104 cases analyzed (Table S3). All substitutions were validated by Sanger sequencing. One of the variants was found in a recently reported PPGL susceptibility gene (SLC25A11; GenBank: NM_003562 [MIM: 604165]),2 and the remaining changes affected genes that encode different subunits of TCA-cycle enzymes (DLST and SUCLG1; GenBank: NM_001933 [MIM: 126063] and NM_003849 [MIM: 611224], respectively), a cytosolic TCA-cycle-related protein (IDH1; GenBank: NM_001282387 [MIM: 147700]), and a mitochondrial carrier (SLC25A10; GenBank: NM_012140 [MIM: 606794]). The only recurrently mutated gene was DLST, in which we found five different variants in seven unrelated individuals (Table 1). In addition, upon revisiting 14 whole exomes from PPGL-affected individuals, we found another person carrying a germline DLST mutation (individual 6). We therefore focused our further research on the role of this gene in PPGL development.
DLST Is Recurrently Mutated in Individuals with Multiple PPGLs
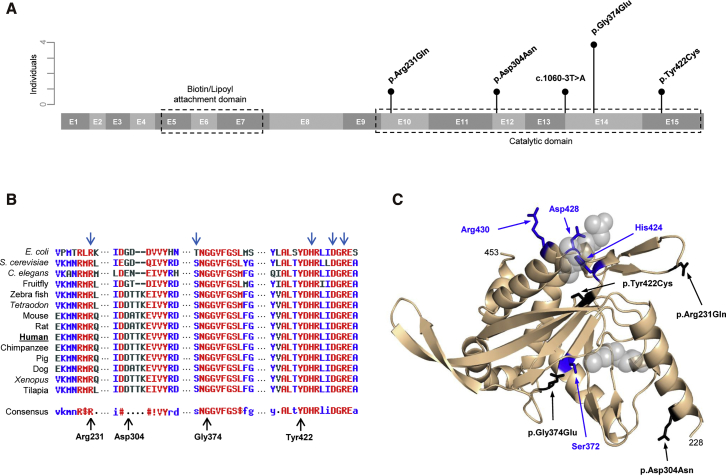
The five different DLST variants consisted of four missense mutations (c.1121G>A [p.Gly374Glu] [in four individuals]; c.692G>A [p.Arg231Gln] [dbSNP: rs771616810]; c.910G>A [p.Asp304Asn] [COSMIC: COSM957880]; and c.1265A>G [p.Tyr422Cys] [dbSNP: rs778239022]) and one intronic splice site variant (c.1060–3T>A) (dbSNP: rs769909757) (Figure 1A and Table 1). None of the individuals with a DLST mutation had a family history of the disease, and two of them also had non-PPGL tumors. Seven of the eight individuals carrying DLST mutations presented with tumors in multiple locations (significantly higher than 21/97 non-DLST-mutated cases; p < 0.001), especially in the thoracic-abdominal region (p < 0.001). One of the affected individuals developed distant metastasis, and all functional (i.e., catecholamine-producing) tumors produced normetanephrine (Table 1). Three of the four DLST missense substitutions were predicted to be deleterious by PredictSNP (Figure S1A) and affected highly-conserved protein residues located close to amino acids predicted to be critical for protein functions (Figure 1B); Gly374 and Tyr422 are part of a pocket where the succinyl group fits in E. coli, and Tyr422 is close to the active site of the protein (Figure 1C).29 All DLST variants were absent or found in at most three controls in gnomAD (out of more than 122,000 individuals) (Table S3), and two of them (p.Asp304Asn and p.Gly374Glu) were found as somatic variants in other cancers (Figure S1B). The number of heterozygous LoF mutations per coding nucleotide found in gnomAD for DLST (n = 12) was significantly lower (p < 0.05) than that observed for SDHB (n = 25), and it was similar to that observed for FH (n = 18). Assuming that disease-causing genes are more intolerant of rare LoF variants,30 this suggests that the presence of heterozygous DLST LoF variants could have phenotypic consequences.
Figure 1.
DLST Protein Structure
(A) A schematic representation of DLST (15 exons) indicating the main active sites and all variants found in this study. Vertical bars represent the number of individuals carrying each variant.
(B) A multiple-sequence alignment and DLST residue conservation across different species determined with MultAlin software; high consensus is marked in red, neutral in black, and low consensus in blue. The blue arrows = predicted active sites in E. coli DLST. The black arrows indicate the different amino acids found mutated in our affected individuals.
(C) A predicted structural model of the DLST catalytic domain. The catalytic residues are colored blue and indicated by blue arrows. The variants found in this study have been modeled, colored black, and are indicated by black arrows. The position of the putative substrate binding site is indicated with gray spheres for clarity.
The DLST Variants Locate in a Region Critical for Functional Activity
Structural analysis revealed that all detected missense variants map in regions of DLST that are involved in its catalytic activity. A structural model of human DLST was generated on the basis of a comparison with available crystal structures from prokaryotic orthologs. The structures of the N- and C-terminal domains were predicted with significant confidence because of their high conservation, but the spatial relationship between the two domains cannot be predicted accurately because there seems to be a poorly structured region connecting both domains. The structures of related complexes provide evidence of flexibility in DLST as well;13 such flexibility could be conferred by the linker connecting the catalytic and biotin-lipoyl attachment domains. The dehydrogenase multienzyme complexes in the family that includes DLST function as trimeric complexes that have a catalytic domain formed by residues from two adjacent subunits.31 Gly374 locates within the pocket where the succinyl group fits, but it also locates in the region of interaction between two monomers. In contrast to the makeup of the normal Gly-containing structure, the recurrent p. Gly374Glu substitution introduces a large and negatively charged side chain, which could interfere with the binding to the succinyl group and/or oligomerization (Figure 1C).
p.Gly374Glu-DLST Tumors Show LOH Due to Uniparental Disomy
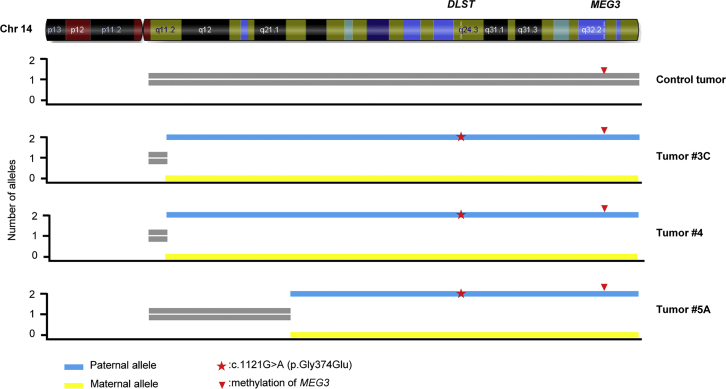
Sanger sequencing revealed LOH of the wild-type (WT) DLST allele in six tumors from three independent individuals, all of whom carried the p.Gly374Glu variant (Table 1 and Figure S2A). A genome-wide SNP array and a microsatellite analysis revealed that uniparental disomy (UPD) of chromosomal region 14q, on which DLST is located, caused LOH in tumors from three available individuals carrying the p.Gly374Glu variant (Figure 2). One of the tumors also showed loss of chromosome 11 and the long arm of chromosome 3. The other two showed clonal gains in chromosomal regions 6p and 7p and in chromosome 22, and no chromosomal alterations in PPGL-predisposing loci (i.e., loss of chromosomal region 1p and chromosomes 11 and 3 and gain of chromosomal region 2p). In addition, a paternal origin of the UPD was determined in the three individuals carrying the p.Gly374Glu variant on the basis of the methylation status (data obtained from the methylation arrays) of a maternally expressed imprinted gene, MEG3, which is located at the distal part of chromosomal region 14q (Figure 2). Collectively, the observed pattern of genetic changes at this locus suggests a tumor suppressor role for DLST. Two additional DLST-mutated tumors, carrying substitutions different from the recurrent p.Gly374Glu variant, showed no LOH (Table 1).
Figure 2.
LOH Analysis of p.Gly374Glu-DLST Tumors
A schematic representation of chromosome 14 showing the uniparental disomy (UPD) identified by SNP array analysis in three PPGLs (#3C, #4, and #5A) that carry the p.Gly374Glu-DLST variant (indicated by a red asterisk) compared to a control tumor. Yellow bars represent the alleles with the lowest copy number (n = 0), and blue bars represent the alleles with the highest copy number (n = 2). MEG3, a maternally expressed imprinted gene, was used to determine the origin of the observed UPD. The presence in the three tumors of only the methylated allele of MEG3 (indicated by a red arrowhead) indicates that the UPD has a paternal origin.
Functional Evaluation of DLST Variants
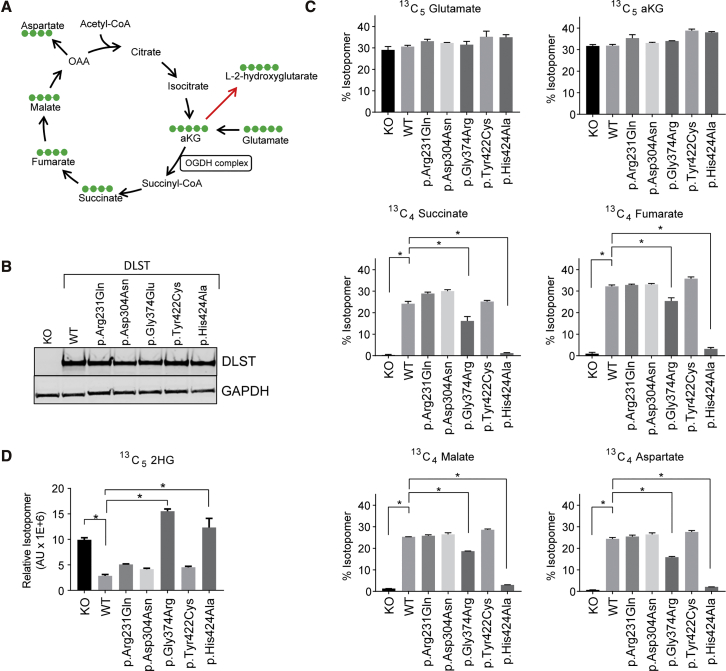
To assess the impact of the missense variants on DLST function, we conducted 13C5-glutamate labeling studies to trace the carbon flow in the TCA cycle (Figure 3A). The individual DLST mutants (see Table 1), along with the corresponding WT DLST control, were introduced in H838 DLST-KO cells. These cells do not demonstrate dramatic DLST-associated growth defects, probably because of the significant activity of compensatory metabolic pathways.18 The complete inhibition of oxidative TCA in these cells, demonstrated by the loss of 13C4-succinate, 13C4-fumarate, and 13C4-malate fractions, provides a robust assay for the evaluation of DLST variants. We also included a predicted catalytically-dead DLST mutant, p.His424Ala. The prediction was made on the basis of homology modeling of the E.coli DLST ortholog (PDB: 1SCZ) and the critical role this histidine plays in deprotonation of the coenzyme A thiol group. An equivalent protein amount was achieved for all DLST constructs (Figure 3B).
Figure 3.
Functional Analysis of DLST Variants
(A) A schematic illustration of the tricarboxylic acid (TCA) cycle. Carbon-13 is denoted by green dots in the context of a 13C5 –glutamate labeling experiment. The red arrow pointing at 2-hydroxyglutarate (2HG) represents the collective action of malate and lactate dehydrogenases.
(B) An immunoblot analysis showing equivalent protein levels of wild-type (WT) and mutant DLST in DLST-KO cells.
(C) Labeling patterns of TCA intermediates after 13C5-glutamate labeling.
(D) A pattern of 13C5-2HG labeling after 13C5-glutamate labeling.
(C and D) Error bars represent the SD from a representative experiment. ∗ p < 0.05 (Student’s t test; n = 3).
As expected, DLST-KO cells displayed a significant block in carbon flow in the TCA cycle as evidenced by negligible amounts of labeled downstream metabolites, such as 13C4-succinate, 13C4-fumarate, 13C4-malate, and 13C4-aspartate (Figure 3C). Reintroduction of WT DLST was able to effectively rescue this metabolic phenotype, and the predicted catalytically-dead mutant, p.His424Ala, had dramatically diminished activity. Of the four variants identified in our PPGLs, only p.Gly374Glu showed partially compromised activity when compared to WT DLST. Importantly, all cells displayed an equivalent amount of precursor labeling, as evidenced by 13C5-glutamate and 13C5-αKG rates. We obtained confirmatory results by using the 13C5-2HG readout, where DLST-KO cells displayed a significant amount of 2HG accumulation (L-2HG/D-2HG >25) that was decreased by the reintroduction of WT DLST. Only two mutants, p.His424Ala and p.Gly374Glu, showed a behavior consistent with diminished enzymatic activity (Figure 3D). Collectively, these results indicate that the p.Gly374Glu variant results in functionally compromised DLST. Considering this result, and because of tissue limitations, we focused our further research on the p.Gly374Glu-DLST tumors.
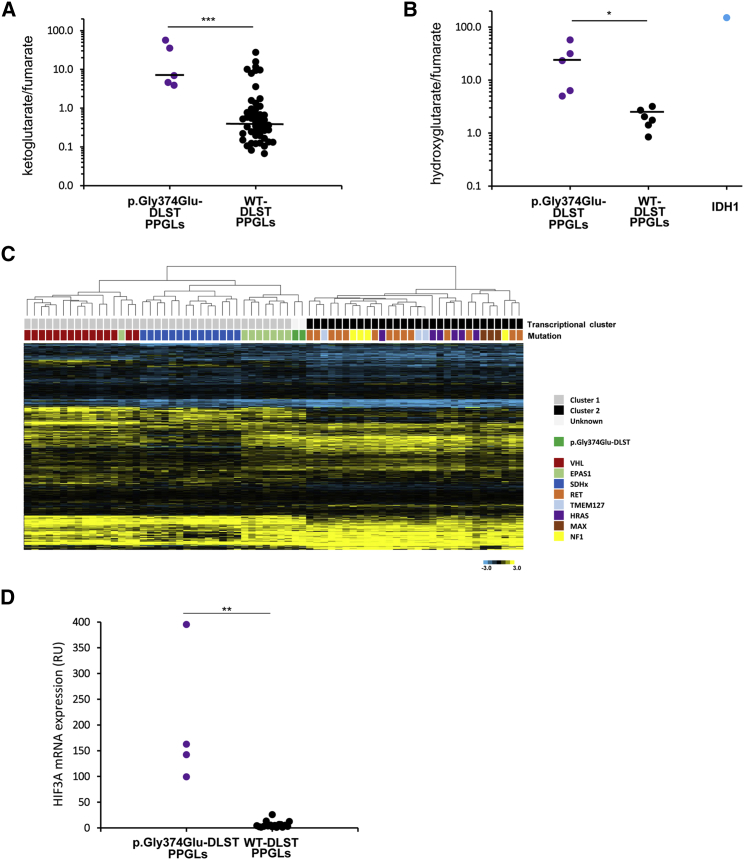
p.Gly374Glu-DLST Tumors Accumulate αKG and 2HG
Liquid chromatography tandem-mass spectrometry revealed a significantly higher αKG/fumarate ratio (21.48 ± 23.65; n = 5) in p.Gly374Glu-DLST tumors as compared to WT-DLST tumors (2.27 ± 4.99; n = 51, p < 0.001) (Figure 4A). Additionally, p.Gly374Glu-DLST tumors presented with a significantly higher 2HG/fumarate ratio (24.64 ± 21.40; n = 5) than PPGLs arising from other mutations (1.97 ± 0.87; n = 6) (p < 0.05) (Figure 4B). The amount of L-2HG, the 2HG enantiomer generated from αKG by promiscuous activity of lactate and malate dehydrogenases, was >2.3 times higher than the amount of D-2HG in three p.Gly374Glu-DLST tumors available for the measurement (data not shown). Because this ratio reflects normally found proportions of L-2HG to D-2HG, and because IDH1 and IDH2 mutations are associated with a complete reversal to many-fold higher D-2HG to L-2HG concentrations,32 the data suggest that the increase in 2HG was not related to IDH1 or IDH2 neomorphic mutations. All these data suggest that disruption of the TCA cycle in p.Gly374Glu-DLST tumors leads to aberrantly elevated αKG/fumarate and 2HG/fumarate ratios.
Figure 4.
Metabolite Assessment and Gene Expression Profiling of p.Gly374Glu-DLST Tumors
(A) α-ketoglutarate/fumarate ratios assessed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in p.Gly374Glu-DLST tumors (n = 5) compared with wild-type (WT)-DLST control PPGLs (n = 51). Black lines represent medians. A t test identified differences between means; ∗∗∗ p < 0.001.
(B) 2-Hydroxyglutarate/fumarate ratios assessed by LC-MS/MS in p.Gly374Glu-DLST tumors (n = 5) compared to WT-DLST control PPGLs (n = 6). The ratio of these metabolites in an IDH1-mutated tumor was included as a positive control of 2HG accumulation. Black lines represent medians. A t test identified differences between means; ∗ p < 0.05.
(C) A hierarchical clustering of 69 mutated tumors made on the basis of expression data for 451 genes reported as differentially expressed in PPGL-mutated samples.25 Control tumors (denoted with different colors depending on the gene mutated) were split up between the two main transcriptional clusters of PPGLs: cluster 1 (denoted in gray), which included VHL- (n = 12), SDHx- (n = 15), and EPAS1- (n = 8) mutated tumors, and cluster 2 (denoted in black), which included RET- (n = 14), HRAS- (n = 6), NF1- (n = 4), TMEM127- (n = 3), and MAX- (n = 3) mutated PPGLs. Two tumors carrying the p.Gly374Glu-DLST variant (#4 and #5A) were clustered within cluster 1 and grouped with EPAS1-mutated cases. City Block-uncentered and complete linkage characteristics were used for the analyses.
(D) HIF3A mRNA expression in p.Gly374Glu-DLST PPGLs (n = 4) versus WT-DLST control PPGLs (n = 18) by RT-qPCR. The expression level was normalized to β-actin (ACTB) and presented as a mean (n = 3). Significance was determined by a Mann-Whitney U non-parametric test; ∗∗ p < 0.01.
p.Gly374Glu-DLST Tumors Show a Characteristic Methylation Profile
To understand the impact of DLST mutations on the global methylation patterns in PPGLs tumors, we profiled three tumors from unrelated individuals carrying the p.Gly374Glu-DLST variant and 14 tumors harboring known mutations in other PPGL susceptibility genes. The three DLST-mutated cases consistently clustered together within the non-CIMP methylation cluster, together with tumors that carried EPAS1 and MAX mutations, close to tumors carrying RET, NF1, or HRAS mutations, and separated from tumors carrying SDHx and DNMT3A mutations (Figure S2B). This relatively uniform methylation profile further supports the relevance of the p.Gly374Glu DLST variant, and it is in agreement with previous studies that showed the importance of the driver mutation in PPGL methylation profiles. Hierarchical clustering that used methylation data from 125,112 probes corresponding to 4,662 genes reported as significantly hypermethylated in SDHx-mutated tumors grouped the four SDHB- and the two DNMT3A-mutated tumors together in a CIMP cluster, whereas HRAS-, NF1-, RET-, and MAX-mutated cases and the only EPAS1-mutated tumor clustered in the non-CIMP cluster (Figure S2C). This unsupervised analysis clustered the three p.Gly374Glu-DLST tumors together within the non-CIMP cluster. Thus, despite a very homogeneous methylation profile, p.Gly374Glu-DLST tumors do not show the CIMP profile associated with other TCA-cycle-related mutations. Rather, DLST-mutated tumors clustered with the only M2 tumor (i.e., PPGLs showing an intermediate methylation phenotype) included in the analysis; this clustering leads to questions about the role of the observed accumulation of metabolites as the mediators of tumorigenesis in DSLT-mutated PPGLs.
HIF3A Is Overexpressed in DLST-Mutated Tumors
Hierarchical clustering of whole-gene expression data from 69 frozen tumors grouped the two (#4 and #5A) p.Gly374Glu-DLST tumors together, suggesting a common transcriptional profile. A second unsupervised clustering that used a list of 451 genes differentially expressed in PPGLs with different genetic backgrounds grouped all VHL- and SDHx-mutated tumors in cluster 1 and all tumors carrying mutations in TMEM127, MAX, NF1, or HRAS in cluster 2. Interestingly, the two p.Gly374Glu-DLST cases (#4 and #5A) were grouped in cluster 1 together with all, except one, tumors carrying mutations in EPAS1 (Figure 4C). This result suggests a consistent impact of the potential driver mutation on gene expression and a link between the p.Gly374Glu-DLST variant and pseudohypoxia. The supervised analysis revealed 132 probes that are significantly differentially expressed in DLST-mutated tumors compared to in tumors carrying known mutations in other PPGL susceptibility genes (Table S4). The most representative probes targeted signal transduction and hematopoiesis genes such as JAK3 (MIM: 600173), ERBB4 (MIM: 600543), or LEPR (MIM: 601007); neural proteins such as MAP7D1, SPTBN5 (MIM: 605916), GABRA4 (MIM: 137141), ZIC4 (MIM: 608948), and FRMD7 (MIM: 300628); and seven nucleosome genes. Moreover, four different probes targeting HIF3A (MIM: 609976) were found differentially overexpressed and were ranked among the five most significant probes. mRNA expression of HIF3A, measured by RT-qPCR, showed an overexpression of HIF3A in p.Gly374Glu-DLST tumors (#3A and C, #4, and #5A) when compared to in controls (n = 18) (Figure 4D); this difference further suggests a link between DLST disruption and pseudohypoxia.
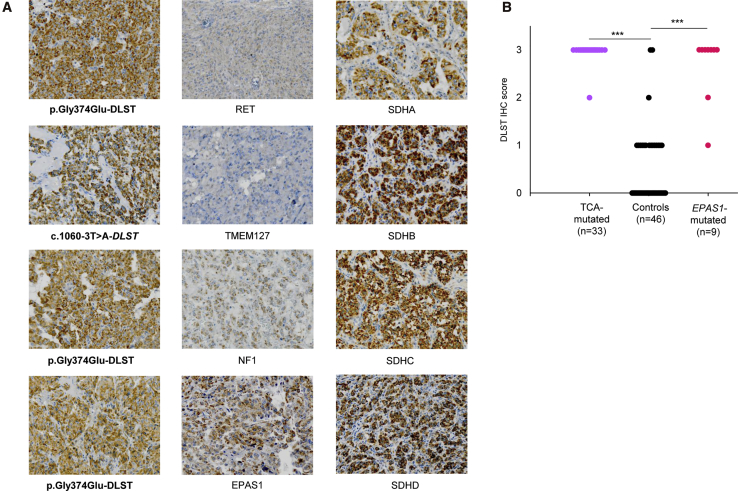
DLST- and TCA-Cycle-Mutated PPGLs Show Positive DLST Immunostaining
Immunohistochemical (IHC) staining of DLST in all available DLST-mutated tumors revealed that this enzyme was more abundant in these samples than in DLST-WT PPGLs (Figure 5A). A strong staining was also observed in all tumors carrying mutations in genes encoding TCA cycle enzymes (n = 33) compared with in control PPGLs carrying mutations in other PPGL susceptibility genes (p < 0.001) (Figure 5B). Four of the control tumors without TCA-cycle mutations showed an intense IHC staining (IHC score = 3), and two of them carried a mutation in EPAS1. Therefore, we extended the IHC study to six additional EPAS1-mutated tumors and found a positive staining in all of them (Figure 5B). These data suggest that DLST immunohistochemistry as a useful tool for identifying tumors that carry alterations either in TCA-cycle-related genes or in EPAS1 and are located in the pseudohypoxic cluster 1.
Figure 5.
Immunohistochemistry of DLST in Different Tumors
(A) Positive immunostaining (×20) with cytoplasmic aggregates was assessed in DLST- and TCA-cycle-mutated tumors (left and right columns, respectively) and compared to control tumors (middle column).
(B) Representation of DLST immunohistochemistry (IHC) score (ranging from 0 to 3) for the 88 analyzed tumors, including tumors carrying mutations in TCA-cycle-related genes (n = 33; SDHA [1], SDHB [11], SDHC [1], SDHD [6], SDHAF2 [1], GOT2 [2], MDH2 [3], IDH1 [1], IDH3B [1], and DLST [6]), tumors carrying EPAS1 mutations (n = 9), and tumors carrying other mutations as controls (n = 46; RET [24], VHL [11], HRAS [1], NF1 [7], MAX [2], and TMEM127 [1]). Significance was determined by a Fisher’s exact test; ∗∗∗ p < 0.001.
Discussion
The identification of heterozygous germline mutations in more than ten genes that are directly or indirectly involved in the TCA cycle point to this metabolic pathway as one of the main drivers for the development of PPGLs. Thus, the assessment of TCA-cycle status has become a routine part of the genetic diagnosis of PPGL and has fueled intense interest in this pathway for the purpose of identifying therapeutic targets. In the present study, we have found variants affecting DLST in more than 6% of affected individuals (most of whom had multiple tumors) without mutations in other PPGL-related genes. Moreover, we have demonstrated the deleterious effect for DLST function of one recurrently mutated residue (found in ∼3% of the whole series of affected individuals) and therefore propose DLST as a PPGL susceptibility gene.
DLST encodes the E2 subunit of the mitochondrial αKG dehydrogenase (OGDH) complex, which has two additional subunits: E1 (encoded by OGDH [MIM: 613022]) and E3 (encoded by DLD [MIM: 238331]). The OGDH complex catalyzes the overall conversion of αKG to succinyl-CoA and CO2. Depletion of any of the OGDH complex subunits leads to impaired enzymatic activity and αKG accumulation, and it has been demonstrated that DLST is a non-redundant member of the OGDH complex.33 Dlst KO mice are embryonically lethal, and although some DLST-KO cells can grow, they exhibit a complete disruption of the oxidative TCA cycle.18, 34 Any condition causing even a modest variation in the cytosolic levels of αKG might affect different pathways, promoting either oncogenic or tumor suppressive responses: an increase of fatty acid biosynthesis or promotion of aberrant mammalian target of rapamycin 1 (mTORC1) activation (Figure S3).35, 36, 37 In addition, DLST mutation causing the loss of OGDH-complex nuclear-translocation capacity might lead to altered overall gene expression (Figure S3).38 The increased αKG to fumarate ratio in p.Gly374Glu-mutated tumors suggests that this variant leads to a disruption of OGDH complex activity. Moreover, the accumulation of significant levels of L-2HG in DLST-KO cells reconstituted with p.Gly374Glu-DLST mutant protein further suggests a functional impairment of this mutant DLST because an increase in L-2HG has been previously noted in other experimental models upon OGDH complex disruption.33
In contrast to D-2HG, which can be generated by neomorphic mutations in IDH1 or IDH2 [MIM: 147650],39 the stereospecific production of L-2HG is driven by the reduction of mitochondrial αKG by both malate dehydrogenases (MDH1 and MDH2) and mainly by lactate dehydrogenase A (LDHA),41 the specific inhibition of which prevents the accumulation of L-2HG in OGDH-null cells.33 This promiscuous substrate usage of αKG occurs specifically under conditions of increased αKG as a result of TCA-cycle dysfunction, increased mitochondrial-reducing potential, or oxygen limitation.33, 40, 41 The labeling studies that used 13C5-glutamate revealed that the p.Gly374Glu-DLST variant is functionally compromised because it was not able to reconstitute WT-DLST function in the context of DLST-KO cells. The location of Gly374 close to Ser372, an amino acid described to be essential for catalysis,31 reinforces the hypothesis that distortion introduced by the Gly to Glu substitution could interfere with the catalytic activity of DLST. The remaining substitutions, although two of them affect residues that are highly conserved and were predicted to be deleterious, displayed similar behavior to WT-DLST in our cellular model. Moreover, Tyr422 is very close to the critical residue of the active site (His424) of the protein. It is important to note that we cannot exclude the possibility that these variants affect other, less-explored DLST functions, such as the biogenesis of the respiratory chain,42 or that they manifest defects in a more physiologically relevant context that is not recapitulated by our heterologous system.
As in other hereditary cancer syndromes, the presence of multiple tumors in most of the individuals with DLST mutations supports the pathological role of DLST germline variants. On the other hand, the lack of family history of the disease in these particular individuals (Figure S4) points to a de novo or low penetrance inheritance; this latter mechanism is frequently observed in PPGL.19, 43 It is interesting to note that one of the individuals also harbored a pituitary adenoma, a pathology that has been described to co-occur in some persons with PPGLs.44 However, given the singularity of this observation, further studies in a larger number of affected individuals will be needed to elucidate the functional contribution of DLST.
To the best of our knowledge, there are no DLST or OGDH complex germline mutations reported in individuals with cancer. However, mutations in DLD result in an atypical form of αKG dehydrogenase deficiency,45 recessive DHTKD1 (MIM: 614984) and OGDHL (MIM: 617513) variants that encode proteins similar to subunits of the OGDH complex have been found in individuals with neurodegenerative phenotypes,37, 46 and a decline in OGDH complex activity has also been associated with neurodegeneration.47 Moreover, it is well known that homozygous or compound heterozygous mutations in TCA-cycle-related genes (e.g., SDH genes, FH, MDH2, ACO2 [MIM: 100850], IDH3A [MIM: 601149], or SLC25A10), lead to encephalopathy and neurodegeneration.48, 49, 50, 51, 52 Thus, if one takes into account the evidences linking intermediary metabolism, tumorigenesis, and neurodegeneration, it is not surprising to find that mutations in DLST lead to cancer.
The LOH of chromosomal regions is crucial in tumor progression and has been successfully used for mapping the locations of tumor suppressor genes. Because disease-causing genes appear to be more intolerant of rare LoF variants,30 the presence of heterozygous DLST LoF variants could have phenotypic consequences. On the other hand, six tumors carrying the recurrent p.Gly374Glu variant showed LOH, suggesting a tumor suppressor role for DLST. Moreover, in all individuals carrying the p.Gly374Glu-DLST variant we were able to demonstrate that the chromosomal loss observed in their tumors was due to a UPD in paternal chromosomal-region-14q UPD, a second-hit mechanism previously reported in PPGLs carrying mutations in the susceptibility gene MAX.17 Interestingly, hierarchical genome-wide expression analysis of two DLST-mutated tumors showed that they had similar expression patterns that were also alike to those in MAX-mutated PPGLs. We studied these tumors in more detail and were able to rule out the presence of rare promoter or deep intronic MAX mutations (data not shown). The existence of a particular MAX-like expression profile has been previously reported in PPGL,53 and it was suggested that certain unknown genes could phenocopy MAX mutant tumors. Our findings suggest that DLST might be one of these genes and that the gene-expression alterations caused by UPD in paternal chromosomal region 14q are the cause of this characteristic expression profile, underscoring the relevance of this mechanism for PPGL pathogenesis. Finally, the absence of somatic rearrangements in other PPGL-susceptibility loci and the finding of 14q UPD as the only common chromosomal alteration in p.Gly374Glu-DLST mutated tumors further support the role of DLST in tumor development.
Previous methylome analyses on PPGLs identified stable clusters associated with distinct clinical features and mutational status.21, 23, 54 The homogeneous methylation profile exhibited by the PPGLs carrying the p.Gly374Glu-DLST variant rules out a CIMP profile, and it further points to a driver role of this DLST mutation in these tumors. Moreover, the expression profile shared by DLST-mutated tumors further suggests that they carry a similar driver genetic alteration. Depletion of DLST leads to the accumulation of endogenous HIF1A through L-2HG-mediated inhibition of enzymatic prolyl hydroxylase activities.33 The expression profile similar to that of EPAS1-mutated tumors and the consistent upregulation of HIF3A mRNA observed in p.Gly374Glu-DLST tumors further strengthen the connection of tumors harboring TCA mutations to the induction of a pseudo-hypoxic state.24, 55 Moreover, it is known that HIF3A overexpression in pancreatic cancer tissues is correlated with a shorter survival time and increased local invasion and distant metastasis.56 Further studies of HIF3A in this context are warranted.
Finally, the strong DLST immunostaining observed in DLST-mutated tumors might suggest that, as occurs with SDHD and the SDH complex,57 the presence of mutations in DLST disrupts OGDH complex assembly and make the DLST epitope more accessible. This could also explain the high intensity of DLST immunostaining found in PPGLs carrying other mutations in TCA-cycle-related genes because these mutations also lead to disruption of the cycle. The positive DLST staining observed in EPAS1-mutated tumors suggests that the activation of this pseudohypoxic pathway could also provoke changes in the assembly of TCA-cycle enzymes. This is in agreement with the proposed regulatory network between the TCA cycle and the hypoxia response, in which each of the pathways reciprocally affects the other.55 Although the mechanism remains unexplained, our data suggest that DLST immunohistochemistry might be useful in the classification of variants of unknown significance in genes related to the TCA cycle.
In summary, we have identified a recurrent germline variant (p.Gly374Glu) that functionally compromises DLST function. Tumors harboring this alteration display altered methylation and transcriptional profiles similar to those observed in EPAS1-mutated tumors, suggesting a connection between DLST functional abrogation and pseudohypoxia. Therefore, we propose DLST as a PPGL susceptibility gene.
Declaration of Interests
David Pirman, Christopher E. Mahoney, Giovanni Cianchetta, and Gromoslaw A. Smolen are employees of and have ownership interest in Agios Pharmaceuticals. The other authors declare no competing interests.
Acknowledgments
This work was supported by the Instituto de Salud Carlos III (ISCIII), through the “Acción Estratégica en Salud” (AES) (projects PI15/00783 and PI18/00454 to A.C. and PI17/01796 to M.R.); cofounded by the European Regional Development Fund [ERDF]), and the Deutsche Forschungsgemeinschaft (DFG RI2684/1-1 to S.R.). The Human Genotyping Unit is a member of the Centro Nacional de Genotipado – Plataforma de Recursos Biomoleculares (CeGen-PRB3) and is supported by grant PT17/0019 of the PE I+D+i 2013–2016, funded by ISCIII and ERDF.
Published: March 28, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.02.017.
Accession Numbers
The transcriptome from tumor #5A and the methylomes used in the analysis have been deposited in the National Center for Biotechnology Information GEO database under the accession numbers GEO: GSE123344, GSE111336, and GSE123185.
Web Resources
Centro Nacional de Genotipado (CEGEN-ISCIII), www.cegen.org
Genome Aggregation Consortium Database, https://gnomad.broadinstitute.org
MultAlin Software, http://multalin.toulouse.inra.fr
National Center for Biotechnology Information GEO Database, https://www.ncbi.nlm.nih.gov/geo/
Online Mendelian Inheritance in Man, http://www.omim.org
Phyre2 Server:, http://www.sbg.bio.ic.ac.uk/phyre2
RNAi Consortium from the Broad Institute, https://portals.broadinstitute.org/gpp/public/resources/protocols
Universal ProbeLibrary Set, https://www.roche-applied-science.com
Supplemental Data
References
- 1.Toledo R.A., Burnichon N., Cascon A., Benn D.E., Bayley J.-P., Welander J., Tops C.M., Firth H., Dwight T., Ercolino T., NGS in PPGL (NGSnPPGL) Study Group Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat. Rev. Endocrinol. 2017;13:233–247. doi: 10.1038/nrendo.2016.185. [DOI] [PubMed] [Google Scholar]
- 2.Buffet A., Morin A., Castro-Vega L.-J., Habarou F., Lussey-Lepoutre C., Letouzé E., Lefebvre H., Guilhem I., Haissaguerre M., Raingeard I. Germline mutations in the mitochondrial 2-oxoglutarate/malate carrier SLC25A11 gene confer a predisposition to metastatic paragangliomas. Cancer Res. 2018;78:1914–1922. doi: 10.1158/0008-5472.CAN-17-2463. [DOI] [PubMed] [Google Scholar]
- 3.Dahia P.L.M. Pheochromocytoma and paraganglioma pathogenesis: Learning from genetic heterogeneity. Nat. Rev. Cancer. 2014;14:108–119. doi: 10.1038/nrc3648. [DOI] [PubMed] [Google Scholar]
- 4.Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W.M., Lu C., Ward P.S. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linehan W.M., Spellman P.T., Ricketts C.J., Creighton C.J., Fei S.S., Davis C., Wheeler D.A., Murray B.A., Schmidt L., Vocke C.D., Cancer Genome Atlas Research Network Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remacha L., Comino-Méndez I., Richter S., Contreras L., Currás-Freixes M., Pita G., Letón R., Galarreta A., Torres-Pérez R., Honrado E. Targeted exome sequencing of Krebs cycle genes reveals candidate cancer-predisposing mutations in pheochromocytomas and paragangliomas. Clin. Cancer Res. 2017;23:6315–6324. doi: 10.1158/1078-0432.CCR-16-2250. [DOI] [PubMed] [Google Scholar]
- 7.Björklund P., Pacak K., Crona J. Precision medicine in pheochromocytoma and paraganglioma: current and future concepts. J. Intern. Med. 2016;280:559–573. doi: 10.1111/joim.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lussey-Lepoutre C., Buffet A., Gimenez-Roqueplo A.P., Favier J. Mitochondrial deficiencies in the predisposition to paraganglioma. Metabolites. 2017;7:1–13. doi: 10.3390/metabo7020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amar L., Baudin E., Burnichon N., Peyrard S., Silvera S., Bertherat J., Bertagna X., Schlumberger M., Jeunemaitre X., Gimenez-Roqueplo A.-P., Plouin P.F. Succinate dehydrogenase B gene mutations predict survival in patients with malignant pheochromocytomas or paragangliomas. J. Clin. Endocrinol. Metab. 2007;92:3822–3828. doi: 10.1210/jc.2007-0709. [DOI] [PubMed] [Google Scholar]
- 10.van Nederveen F.H., Gaal J., Favier J., Korpershoek E., Oldenburg R.A., de Bruyn E.M., Sleddens H.F., Derkx P., Rivière J., Dannenberg H. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: A retrospective and prospective analysis. Lancet Oncol. 2009;10:764–771. doi: 10.1016/S1470-2045(09)70164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z.H., McCarthy D.B., O’Connor C.M., Reed L.J., Stoops J.K. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA. 2001;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran S., Kota P., Ding F., Dokholyan N.V. Automated minimization of steric clashes in protein structures. Proteins. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrödinger. The PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, LLC.)
- 16.Simon-Sanchez J., Scholz S., Fung H.C., Matarin M., Hernandez D., Gibbs J.R., Britton A., de Vrieze F.W., Peckham E., Gwinn-Hardy K. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum. Mol. Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 17.Comino-Méndez I., Gracia-Aznárez F.J., Schiavi F., Landa I., Leandro-García L.J., Letón R., Honrado E., Ramos-Medina R., Caronia D., Pita G. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 18.Allen E.L., Ulanet D.B., Pirman D., Mahoney C.E., Coco J., Si Y., Chen Y., Huang L., Ren J., Choe S. Differential aspartate usage identifies a subset of cancer cells particularly dependent on OGDH. Cell Rep. 2016;17:876–890. doi: 10.1016/j.celrep.2016.09.052. [DOI] [PubMed] [Google Scholar]
- 19.Cascón A., Comino-Méndez I., Currás-Freixes M., de Cubas A.A., Contreras L., Richter S., Peitzsch M., Mancikova V., Inglada-Pérez L., Pérez-Barrios A. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. J. Natl. Cancer Inst. 2015;107:1–5. doi: 10.1093/jnci/djv053. [DOI] [PubMed] [Google Scholar]
- 20.Bibikova M., Le J., Barnes B., Saedinia-Melnyk S., Zhou L., Shen R., Gunderson K.L. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 21.de Cubas A.A., Korpershoek E., Inglada-Pérez L., Letouzé E., Currás-Freixes M., Fernández A.F., Comino-Méndez I., Schiavi F., Mancikova V., Eisenhofer G. DNA methylation profiling in pheochromocytoma and paraganglioma reveals diagnostic and prognostic markers. Clin. Cancer Res. 2015;21:3020–3030. doi: 10.1158/1078-0432.CCR-14-2804. [DOI] [PubMed] [Google Scholar]
- 22.Reich M., Ohm K., Angelo M., Tamayo P., Mesirov J.P. GeneCluster 2.0: An advanced toolset for bioarray analysis. Bioinformatics. 2004;20:1797–1798. doi: 10.1093/bioinformatics/bth138. [DOI] [PubMed] [Google Scholar]
- 23.Letouzé E., Martinelli C., Loriot C., Burnichon N., Abermil N., Ottolenghi C., Janin M., Menara M., Nguyen A.T., Benit P. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 24.López-Jiménez E., Gómez-López G., Leandro-García L.J., Muñoz I., Schiavi F., Montero-Conde C., de Cubas A.A., Ramires R., Landa I., Leskelä S. Research resource: Transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol. Endocrinol. 2010;24:2382–2391. doi: 10.1210/me.2010-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnichon N., Vescovo L., Amar L., Libé R., de Reynies A., Venisse A., Jouanno E., Laurendeau I., Parfait B., Bertherat J. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum. Mol. Genet. 2011;20:3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey E.R., Diaz-Uriarte R. Pomelo II: Finding differentially expressed genes. Nucleic Acids Res. 2009;37:W581-6. doi: 10.1093/nar/gkp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Knapp J.E., Mitchell D.T., Yazdi M.A., Ernst S.R., Reed L.J., Hackert M.L. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 1998;280:655–668. doi: 10.1006/jmbi.1998.1924. [DOI] [PubMed] [Google Scholar]
- 30.Ruderfer D.M., Hamamsy T., Lek M., Karczewski K.J., Kavanagh D., Samocha K.E., Daly M.J., MacArthur D.G., Fromer M., Purcell S.M., Exome Aggregation Consortium Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 2016;48:1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Nemeria N.S., Chandrasekhar K., Kumaran S., Arjunan P., Reynolds S., Calero G., Brukh R., Kakalis L., Furey W., Jordan F. Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. J. Biol. Chem. 2014;289:15215–15230. doi: 10.1074/jbc.M113.544080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter S., Gieldon L., Pang Y., Peitzsch M., Huynh T., Leton R., Viana B., Ercolino T., Mangelis A., Rapizzi E. Metabolome-guided genomics to identify pathogenic variants in isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase genes in pheochromocytoma and paraganglioma. Genet. Med. 2018;27 doi: 10.1038/s41436-018-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burr S.P., Costa A.S.H., Grice G.L., Timms R.T., Lobb I.T., Freisinger P., Dodd R.B., Dougan G., Lehner P.J., Frezza C., Nathan J.A. Mitochondrial protein lipoylation and the 2-oxoglutarate dehydrogenase complex controls HIF1α stability in aerobic conditions. Cell Metab. 2016;24:740–752. doi: 10.1016/j.cmet.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L., Shi Q., Ho D.J., Starkov A.A., Wille E.J., Xu H., Chen H.L., Zhang S., Stack C.M., Calingasan N.Y. Mice deficient in dihydrolipoyl succinyl transferase show increased vulnerability to mitochondrial toxins. Neurobiol. Dis. 2009;36:320–330. doi: 10.1016/j.nbd.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki J., Yamada T., Inoue K., Nabe S., Kuwahara M., Takemori N., Takemori A., Matsuda S., Kanoh M., Imai Y. The tumor suppressor menin prevents effector CD8 T-cell dysfunction by targeting mTORC1-dependent metabolic activation. Nat. Commun. 2018;9:3296. doi: 10.1038/s41467-018-05854-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vatrinet R., Leone G., De Luise M., Girolimetti G., Vidone M., Gasparre G., Porcelli A.M. The α-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metab. 2017;5:3. doi: 10.1186/s40170-017-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon W.H., Sandoval H., Nagarkar-Jaiswal S., Jaiswal M., Yamamoto S., Haelterman N.A., Putluri N., Putluri V., Sreekumar A., Tos T. Loss of nardilysin, a mitochondrial co-chaperone for α-ketoglutarate dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Guo Y.R., Liu K., Yin Z., Liu R., Xia Y., Tan L., Yang P., Lee J.H., Li X.J. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552:273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldham W.M., Clish C.B., Yang Y., Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Intlekofer A.M., Wang B., Liu H., Shah H., Carmona-Fontaine C., Rustenburg A.S., Salah S., Gunner M.R., Chodera J.D., Cross J.R., Thompson C.B. L-2-hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 2017;13:494–500. doi: 10.1038/nchembio.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanamori T., Nishimaki K., Asoh S., Ishibashi Y., Takata I., Kuwabara T., Taira K., Yamaguchi H., Sugihara S., Yamazaki T. Truncated product of the bifunctional DLST gene involved in biogenesis of the respiratory chain. EMBO J. 2003;22:2913–2923. doi: 10.1093/emboj/cdg299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiavi F., Milne R.L., Anda E., Blay P., Castellano M., Opocher G., Robledo M., Cascón A. Are we overestimating the penetrance of mutations in SDHB? Hum. Mutat. 2010;31:761–762. doi: 10.1002/humu.21269. [DOI] [PubMed] [Google Scholar]
- 44.Dénes J., Swords F., Rattenberry E., Stals K., Owens M., Cranston T., Xekouki P., Moran L., Kumar A., Wassif C. Heterogeneous genetic background of the association of pheochromocytoma/paraganglioma and pituitary adenoma: Results from a large patient cohort. J. Clin. Endocrinol. Metab. 2015;100:E531–E541. doi: 10.1210/jc.2014-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odièvre M.-H., Chretien D., Munnich A., Robinson B.H., Dumoulin R., Masmoudi S., Kadhom N., Rötig A., Rustin P., Bonnefont J.-P. A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of α-ketoglutarate dehydrogenase deficiency. Hum. Mutat. 2005;25:323–324. doi: 10.1002/humu.9319. [DOI] [PubMed] [Google Scholar]
- 46.Danhauser K., Sauer S.W., Haack T.B., Wieland T., Staufner C., Graf E., Zschocke J., Strom T.M., Traub T., Okun J.G. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am. J. Hum. Genet. 2012;91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson G.E., Hirsch J.A., Cirio R.T., Jordan B.D., Fonzetti P., Elder J. Abnormal thiamine-dependent processes in Alzheimer’s disease. Lessons from diabetes. Mol. Cell. Neurosci. 2013;55:17–25. doi: 10.1016/j.mcn.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourgeron T., Chretien D., Poggi-Bach J., Doonan S., Rabier D., Letouzé P., Munnich A., Rötig A., Landrieu P., Rustin P. Mutation of the fumarase gene in two siblings with progressive encephalopathy and fumarase deficiency. J. Clin. Invest. 1994;93:2514–2518. doi: 10.1172/JCI117261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., Rötig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 50.Fattal-Valevski A., Eliyahu H., Fraenkel N.D., Elmaliach G., Hausman-Kedem M., Shaag A., Mandel D., Pines O., Elpeleg O. Homozygous mutation, p.Pro304His, in IDH3A, encoding isocitrate dehydrogenase subunit is associated with severe encephalopathy in infancy. Neurogenetics. 2017;18:57–61. doi: 10.1007/s10048-016-0507-z. [DOI] [PubMed] [Google Scholar]
- 51.Spiegel R., Pines O., Ta-Shma A., Burak E., Shaag A., Halvardson J., Edvardson S., Mahajna M., Zenvirt S., Saada A. Infantile cerebellar-retinal degeneration associated with a mutation in mitochondrial aconitase, ACO2. Am. J. Hum. Genet. 2012;90:518–523. doi: 10.1016/j.ajhg.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punzi G., Porcelli V., Ruggiu M., Hossain M.F., Menga A., Scarcia P., Castegna A., Gorgoglione R., Pierri C.L., Laera L. SLC25A10 biallelic mutations in intractable epileptic encephalopathy with complex I deficiency. Hum. Mol. Genet. 2018;27:499–504. doi: 10.1093/hmg/ddx419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flynn A., Benn D., Clifton-Bligh R., Robinson B., Trainer A.H., James P., Hogg A., Waldeck K., George J., Li J. The genomic landscape of phaeochromocytoma. J. Pathol. 2015;236:78–89. doi: 10.1002/path.4503. [DOI] [PubMed] [Google Scholar]
- 54.Fishbein L., Leshchiner I., Walter V., Danilova L., Robertson A.G., Johnson A.R., Lichtenberg T.M., Murray B.A., Ghayee H.K., Else T., Cancer Genome Atlas Research Network Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31:181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raimundo N., Baysal B.E., Shadel G.S. Revisiting the TCA cycle: Signaling to tumor formation. Trends Mol. Med. 2011;17:641–649. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X., Guo X., Chen M., Xie C., Jiang J. HIF-3α promotes metastatic phenotypes in pancreatic cancer by transcriptional regulation of the RhoC-ROCK1 signaling pathway. Mol. Cancer Res. 2018;16:124–134. doi: 10.1158/1541-7786.MCR-17-0256. [DOI] [PubMed] [Google Scholar]
- 57.Menara M., Oudijk L., Badoual C., Bertherat J., Lepoutre-Lussey C., Amar L., Iturrioz X., Sibony M., Zinzindohoué F., de Krijger R. SDHD immunohistochemistry: A new tool to validate SDHx mutations in pheochromocytoma/paraganglioma. J. Clin. Endocrinol. Metab. 2015;100:E287–E291. doi: 10.1210/jc.2014-1870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.