Abstract
The diagnostic gap for rare neurodegenerative diseases is still considerable, despite continuous advances in gene identification. Many novel Mendelian genes have only been identified in a few families worldwide. Here we report the identification of an autosomal-dominant gene for hereditary spastic paraplegia (HSP) in 10 families that are of diverse geographic origin and whose affected members all carry unique truncating changes in a circumscript region of UBAP1 (ubiquitin-associated protein 1). HSP is a neurodegenerative disease characterized by progressive lower-limb spasticity and weakness, as well as frequent bladder dysfunction. At least 40% of affected persons are currently undiagnosed after exome sequencing. We identified pathological truncating variants in UBAP1 in affected persons from Iran, USA, Germany, Canada, Spain, and Bulgarian Roma. The genetic support ranges from linkage in the largest family (LOD = 8.3) to three confirmed de novo mutations. We show that mRNA in the fibroblasts of affected individuals escapes nonsense-mediated decay and thus leads to the expression of truncated proteins; in addition, concentrations of the full-length protein are reduced in comparison to those in controls. This suggests either a dominant-negative effect or haploinsufficiency. UBAP1 links endosomal trafficking to the ubiquitination machinery pathways that have been previously implicated in HSPs, and UBAP1 provides a bridge toward a more unified pathophysiology.
Keywords: hereditary spastic paraplegia, neurodegenerative diseases, endosomal trafficking, ubiquitination, zebrafish, animal model, genetic diseases, spasticity
Main Text
Hereditary spastic paraplegia (HSP) represents a group of genetically highly heterogeneous rare inherited neurodegenerative diseases, which are characterized by the pathological hallmark of a length-dependent degeneration of corticospinal-tract axons (see GeneReviews in Web Resources).1 Clinically, HSPs are marked by progressive spastic paraparesis, although the clinical presentation encompasses a wide spectrum of phenotypes. In pure forms of HSP, progressive spasticity and weakness in the lower extremities are the main features. In complex forms of HSP, additional clinical symptoms include cataracts, ataxia, epilepsy, cognitive impairment, peripheral neuropathy, optic neuropathy, and deafness (see GeneReviews in Web Resources).1 The prevalence of HSP has been estimated to be 1.3–9.6 in 100,000 (see GeneReviews in Web Resources).1, 2 Thus far, at least 58 genes have been reported to cause HSP in a Mendelian fashion.3 Yet approximately 40% of affected persons are still not diagnosed even after whole-exome sequencing (WES). Furthermore, many of the genes reported in recent years have only been described in a few families.4
In an effort to further close this diagnostic gap in HSP, we have gathered a highly diverse sample of 10 families from six countries (Iran, 1; USA, 1; Germany, 4; Canada, 1; Bulgaria, 2; and Spain, 1). Prior to the initiation of this study, all participating affected individuals gave informed consent in agreement with each institutional review board. In one family of Persian origin, family 1, we were able to genetically ascertain a total of 14 affected individuals from three generations (Figure 1). Sequencing of an HSP gene panel and CNV analysis at the SPG4 locus were unremarkable. Subsequently, WES was performed in two affected individuals (V.1 and V.15). Bioinformatics analysis of the sequencing data used standard tools, including BWA aligner,5 FreeBayes,5 GATK,6 and GENESIS.7 Only non-synonymous variants with a minor-allele frequency of less than 0.0001 in gnomAD and in our in-house Iranian variant database (BayanGene; http://www.bayangene.com) of 1,500 exomes were further considered. Two heterozygous variants remained in SVEP1 (chr9: 113137668; rs373655861; p.Thr3527Met hg19 [c.10580C>T]; GenBank: NM_153366.3) and UBAP1 (chr9: 34241270; GenBank: NM_016525.4: c.436_437insTGAG [p.Ser146Metfs∗14]), respectively. The SVEP1 variant was ruled out by segregation studies involving Sanger sequencing of the entire pedigree. Thus, after confirmation of complete segregation, the truncating frameshift variant in ubiquitin-associated protein 1 (UBAP1) was considered as the causative allele in family 1 (Figure 1). This frameshift variant was not present in ExAC, gnomAD, GENESIS, nor in 1,500 Iranian genomes. It is predicted to truncate the protein at residue 158 out of 502 amino acids (GenBank: NM_016525.4). By including the Sanger-confirmed affected and unaffected participants (Figure 1), we performed parametric two-point linkage analysis by using the LINKAGE program, which rendered a two-point LOD score of 8.25 at the position of the UBAP1 insertion.
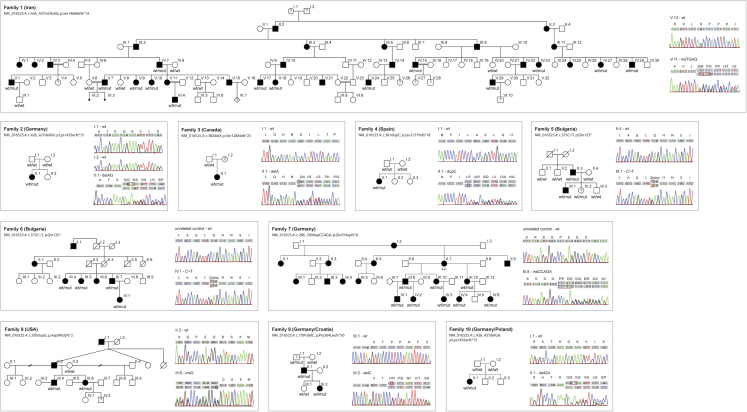
Figure 1.
Pedigrees of HSP-Affected Families with UBAP1 Truncations
All pedigrees suggest an autosomal-dominant or a de novo Mendelian trait. HSP-affected individuals are marked by filled symbols; individuals with unclear affection status are marked by a question mark. “mut” depicts the presence of a causative allele. Sanger traces exemplify the confirmation of variants detected via next-generation sequencing. The penetrance of truncating UBAP1 variants is reduced: individual F5-III.2 was subjectively unaffected at age 14 but showed brisk reflexes of lower limbs, indicating potential dysfunction of the corticospinal tract. The 80-year-old grandfather of the index case in family 9 (F9-III.1) was unfortunately not available for a neurological examination but was reported to be in good health and without any indication of a gait disturbance.
We then searched the GENESIS database for additional families with UBAP1 variants. GENESIS contains more than 3,000 exomes and genomes from affected persons with HSP and related disorders.7 We filtered for non-synonymous and truncating variants under an autosomal-dominant model with minor-allele frequency in gnomAD < 0.0001 and a minimum sequencing depth of 10 reads. We identified seven additional HSP families, all carrying truncating variants in UBAP1 (Table 1). In addition, predictively truncating UBAP1 variants were prioritized in two families (9 and 10) who underwent diagnostic exome sequencing at the University of Tuebingen. The detection of truncating alleles in all families is especially remarkable when one considers the almost complete constraint of UBAP1 for loss-of-function (truncating) variation in the ∼120,000 chromosomes in both ExAC and gnomAD, pLi = 0.95 and 0.92, respectively.8 We calculated the probability of significant enrichment of truncating variations in UBAP1 in our HSP dataset compared to ExAC. In the GENESIS dataset we found seven such variants in a cohort of 567 HSP samples versus 0 truncating variants in 60,000 ExAC samples (p = 6.187 × 10−15 by Fisher test. Odds ratio = infinity). Five truncating variants were reported in the ∼246,000 chromosomes in gnomAD, but none fell within the specific gene region containing the variants reported in this study.
Table 1.
Detailed Genomic Locations of Detected Pathogenic Variants
|
Isoform 1 (GenBank:NM_016525.4) Expressed in Neurons |
Isoform 4 (GenBank:NM_001171201.1) Canonical According to NCBI |
|||
|---|---|---|---|---|
| Family ID | Genome Assembly (hg19) | cDNA | Protein | Protein |
| 1 | chr9: 34241459–34241460 | c.436_437insTGAG | p.Ser146Metfs∗14 | p.Ser210Metfs∗14 |
| 2 | chr9: 34241449–34241450 | c.426_427delGA | p.Lys143Serfs∗15 | p.Lys207Serfs∗15 |
| 3 | chr9: 34241405–34241405 | c.382del | p.Ser128Alafs∗23 | p.Ser192Alafs∗23 |
| 4 | chr9: 34241384–34241384 | c.361dupC | p.Leu121Profs∗18 | p.Leu185Profs∗18 |
| 5 | chr9: 34241396–34241396 | c.373C > T | p.Gln125∗ | p.Gln161∗ |
| 6 | chr9: 34241396–34241396 | c.373C > T | p.Gln125∗ | p.Gln161∗ |
| 7 | chr9: 34241309–34241313 | c.286_290dupCCAGA | p.Glu97Aspfs∗8 | p.Glu161Aspfs∗8 |
| 8 | chr9: 34241318–34241318 | c.295dupG | p.Asp99Glyfs∗2 | p.Asp163Glyfs∗2 |
| 9 | chr9: 34249784–34249784 | c.1091delC | p.Pro364Leufs∗50 | p.Pro428Leufs∗50 |
| 10 | chr9: 34241449–34241450 | c.426_427delGA | p.Lys143Serfs∗15 | p.Lys207Serfs∗15 |
We refer to isoform 1 throughout the text.
All additional variants and their segregation with disease in the additional families were confirmed by Sanger sequencing. On the basis of transcript GenBank: NM_016525.4, the identified variants were as follows (Table 1 and Figure 1): families 2 and 10 from Germany, c.426_427delGA ( p.Lys143Serfs∗15); family 3 from Canada, c.382del (p.Ser128Alafs∗23); family 4 from Spain, c.361dupC (p.Leu121Profs∗18); families 5 and 6 from Bulgaria (Roma ethnicity), c.373C>T (p.Gln125∗); family 7 from Germany, c.286_290dupCCAGA (p.Glu97Aspfs∗8); family 8 from the United States, c.295dupG (p.Asp99Glyfs∗2), and family 9 from Germany c.1091delC (p.Pro364Leufs∗50).
In families 2 and 4, a de novo occurrence of the truncating variant was confirmed (Figure 1). Families 5 and 6 were of self-declared Bulgarian Roma ethnicity and carried the same p.Gln125∗ variant, although the two index participants are from reportedly unrelated families. Evaluating the prevalence of this allele in the European Roma population and in Gypsy HSP-affected persons will require further studies.
The first manifesting symptom in all 30 UBAP1 mutation carriers from 10 families for whom detailed clinical data were available was a progressive spastic-gait disorder with a median age at onset of 8 years (interquartile range 4–9 years; oldest onset age 26 years; one asymptomatic mutation carrier (F5-III.2) aged 14 years (detailed clinical information in Table S1). At the time of examination (median disease duration 28 years; interquartile range 15–36 years), lower-limb spastic paraparesis was still the most prominent clinical feature in all affected mutation carriers; this was accompanied by brisk lower-limb tendon reflexes (all carriers, including asymptomatic carrier F5-III.2) and extensor plantar response in all but the youngest affected individual (F7-IV.6). Although brisk tendon reflexes of the upper limbs were frequently present (26 of 30; 87%) significant upper-limb spasticity was seen only in a single case (1/30; 3%; F4-II.1), consistent with a length-dependent axonopathy of the corticospinal tract. Urinary urgency was reported in some cases (11 of 30; 37%), sensory deficits were absent or mild, and there was no evidence of peripheral neuropathy. In the majority of families (8/10; 80%), no additional signs or symptoms indicating affection of neuronal systems other than the corticospinal tract were seen, and the disease was accordingly classified as pure HSP. In family 7, however, seven out of nine family members had features of cerebellar involvement (such features included saccadic pursuit, gaze-evoked nystagmus, dysmetric saccades, and limb ataxia), features also present in family 9 (F9-II.1), indicating that the cerebellum is vulnerable to UBAP1 dysfunction at least in some cases.
Overall, truncating UBAP1 mutations are associated with a predominantly pure early-onset HSP phenotype; cerebellar involvement seems to be clustered in families and was observed in 2/10 families. Although there is thus minimal variation in terms of system involvement across families carrying UBAP1 mutations, phenotypic variability exists regarding the progression rate; for example, the disease progressed rather rapidly and led to early wheelchair dependency in families 2, 6, and 7 but was almost non-progressive in family 9 (F9-II.1 is still able to run and walk unlimited distances after 38 years of disease duration). Both intrafamilial as well as interfamilial variability are common or even the norm in HSP.3 A complete understanding of the phenotypic spectrum associated with UBAP1 mutations will require careful clinical evaluation of additional families carrying UBAP1 mutations.
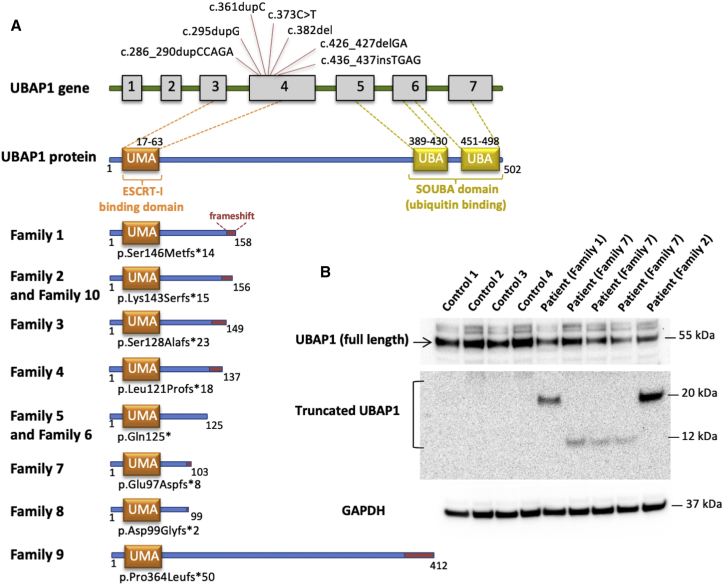
UBAP1 is a member of the endosomal sorting complex required for transport -1 (ESCRT-I) complex and a regulator of vesicular trafficking processes, binds to ubiquitinated cargo proteins, and is essential for sorting endocytic ubiquitinated cargos into multivesicular bodies (MVBs).9 It also plays an important role in proteasomal degradation of ubiquitinated cell-surface proteins, including EGFR (epidermal growth factor receptor) and BST2 (bone marrow stromal cell antigen 2).9 UBAP1 has two main domains: The UMA (UBAP1-MVB12-associated) domain in the N-terminal region (17–63 aa), which mediates the association with the ESCRT-I complex, and a SOUBA (solenoid of overlapping ubiquitin-associated domains) domain in the C-terminal region (389–498 aa).9, 10 Both domains allow UBAP1 to act as a molecular bridge connecting the endosomal trafficking pathways to the ubiquitination machinery. In an effort to decipher the pathophysiology of UBAP1 in HSP, we noted that all but one of the identified changes fall within a circumscript area of the protein between Asp 99 and Ser 146; the change in family 9 at Pro364 was the only outlier (GenBank: NM_016525.4). Interestingly, disease progression in this family has been dramatically slower than in the other families: the disease has been almost stationary over decades (see above), pointing toward a possible genotype-phenotype correlation. Yet, all changes preserve the UMA domain but cause a loss of the SOUBA domain.10 It has been shown that mutagenesis of the SOUBA domain in UBAP1 strongly reduces its interaction with ubiquitinated proteins (Figure 2).10 To determine whether the observed truncating variants would lead to nonsense-medicated mRNA decay and haploinsufficiency, we evaluated both the RNA and protein expression of mutant alleles. RT-PCR was performed on RNA extracted from the fibroblasts of an affected individual, and the RNA was sequenced by the Sanger method. Surprisingly, the c.436_437insTGAG was detected in the affected person’s cDNA, indicating escape of nonsense-medicated mRNA decay (Figure S1). Next, we performed immunoblot analysis to evaluate both wild-type and potential truncated mutant UBAP1. Total protein extracts were probed with an antibody raised against the N-terminal region of UBAP1 (amino acids 25–75), a part of the protein preserved in mutant UBAP1 proteins. The protein levels measured in affected individuals were compared with those in four control fibroblasts and normalized to GAPDH levels. Immunoblots showed decreased protein levels of full-length UBAP1 in fibroblasts from affected individuals; in addition, the truncated protein was detected (Figure 2). The reduced concentrations of the full-length protein in fibroblasts from affected individuals compared to controls along with the presence of the truncated protein could potentially lead to haploinsufficiency and/or a dominant-negative effect. To evaluate the effects of the truncated protein, we performed site-direct mutagenesis and generated a plasmid encoding the truncated protein fused to an HA tag at the N-terminal region. U2OS cells were co-transfected with either wild-type (HA-WT-UBAP1) or a truncated mutant (HA-Fs-UBAP1; p.Leu121Profs∗18) together with its known binding partner VPS28-Myc. Both the wild-type and the truncated mutant co-localize with VPS28 (Figure 3A). This suggests that interaction with the ESCRT-I complex is preserved; however, the lack of the SOUBA domain, essential for ubiquitin binding, would be detrimental. Interestingly, overexpression of truncated protein containing the UMA domain has been shown to result in a dominant-negative effect by inhibiting HIV-1 budding.10 It is thus possible that expression of the truncated protein in affected persons could cause a dominant-negative effect due to arrest of the ESCRT-complex without acquiring the ubiquitinated protein cargo.
Figure 2.
The Structure of UBAP1 and Mutations Carried by Affected Individuals
(A) Schematic diagram showing all exons and UTRs of UBAP1 on the basis of gene model GenBank: NM_016525.4. The gray boxes represent the coding sequence of UBAP1. All variants occurred in exon 4 of UBAP1. The UMA protein domain includes amino acids 17–63, and two UBAs include amino acids 389–430 and 451–489. All truncations are listed below; the preserving of the UMA domain is clearly depicted, but there is loss of the two SOUBA domains.
(B) Immunoblot analysis with an antibody recognizing amino acids 25–75 of UBAP1 shows a notable decrease in the amount of full-length UBAP1 in fibroblasts of the affected individuals from three different families when these cells are compared to control fibroblasts. Truncated UBAP1 of the predicted sizes was detected in fibroblasts from affected individuals but not in control fibroblasts.
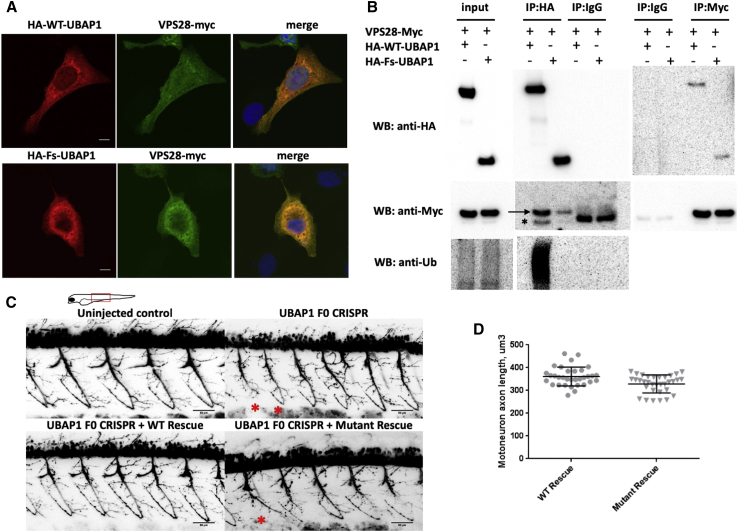
Figure 3.
Functional In Vitro and In Vivo Studies of Truncated UBAP1
(A) Immunostaining of U2OS cells transfected with HA-WT-UBAP1 HA-Fs-UBAP1 (p.Leu121Profs∗18) and VPS-28-Myc. Both wild-type and mutant UBAP1 co-localize with VPS28-Myc.
(B) A co-immunoprecipitation assay shows protein-protein interaction between VPS28-Myc and both HA-WT-UBAP1 and HA-Fs-UBAP1. Ubiquitinated proteins co-immunoprecipitated with HA-WT-UBAP1 but not with HA-Fs-UBAP1. The arrow points to VPS28-Myc, and the asterisk bellow the arrow indicates the IgG band.
(C) Motor-neuron axons in Tg(olig2∷DsRed) zebrafish embryos at 48 hpf. Embryos were injected with CRISPR Cas9 and sgRNAs against UBAP1; injection was supplemented with human RNA rescue of wild-type or truncated mutant UBAP1. Truncated and misshaped axons were more commonly observed with mutant hRNA rescue (indicated by asterisks). Scale bars represent 50 μm. The phenotypic difference between treated groups was evaluated by a Fisher exact test. Samples were assigned to either normal or affected categories on the basis of the presence of truncated and misshaped axons. The Fisher exact statistic value was determined to be 0.003; the result is significant at p < 0.005. Statistics describing normal versus affected phenotypes were calculated on the basis of the following sample sizes (number of embryos observed as having a phenotype). F0 CRISPR + wild-type hRNA rescue: normal = 11, affected = 1. F0 CRISPR + mutant hRNA rescue (family 4, p.Leu121Profs∗18), normal = 9, affected = 15.
(D) Quantification of the individual motor-axon lengths. p values were calculated with a one-tailed Student’s t test.: p = 0.0008 and n = 9 (number of embryos in each experimental group; four axons were measured per embryo).
We performed a co-immunoprecipitation (co-IP) assay to confirm the interaction between HA-Fs-UBAP1 and VPS28-Myc. HEK293T cells were co-transfected with VPS28-Myc and with either HA-WT-UBAP1 or HA-Fs-UBAP1 and immunoprecipitated with an anti-HA or an anti-Myc antibody and analyzed by immunoblot. Our results show that both wild-type and truncated UBAP1 co-immunoprecipitated with VPS28, confirming protein-protein interaction (Figure 3B). However, ubiquitinated proteins were co-immunoprecipitated with the HA-WT-UBAP1 but not with HA-Fs-UBAP1. It has previously been shown that siRNA depletion of UBAP1 in HeLa cells causes clustering of early-endosome accumulation of ubiquitinated proteins and enlargement and clustering of LAMP1-positive late endosomes and lysosomes.9 In fibroblasts of affected persons carrying UBAP1 mutations, however, none of these changes could be observed (Figure S2), even after exposure of cells to stress conditions. It therefore appears unlikely that loss of one UBAP1 allele results in the gross failure of multivesicular body sorting.
To investigate the effects of the truncated protein in vivo, we generated a zebrafish model with UBAP1 knockdown. We used a transgenic fish with fluorescently labeled motoneuron Tg(olig2∷DsRed).11 Embryos were injected with CRISPR Cas9 and sgRNAs against UBAP1 supplemented with human RNA rescue of either wild-type or truncated mutant UBAP1. At 48 hours post-fertilization (hpf), embryos were imaged in vivo with a confocal microscope. We observed significantly more truncated and misshaped axons in the mutant rescued embryos than in the wild-type rescued embryos (Figure 3C). Motoneuron axon lengths in the truncated mutant were significantly (p = 0.0008) shorter than those of the wild-type (Figure 3D). This result supports the pathogenic effects of the truncated protein in vivo.
In summary, we present strong genetic evidence that truncating mutations in UBAP1 cause a relatively frequent form of HSP. UBAP1 mutations were identified in a large Iranian kindred as well as in nine additional families with different ancestral backgrounds, including Bulgarian Roma, North American of European descent, German, Spanish, and Quebecois. All available affected persons in these families carried the respective mutation in UBAP1, although UBAP1 has a strong loss-of-function constraint in the 60,000 individuals studied in the ExAC dataset. In two families we were also able to show a de novo occurrence of the variants. In our dataset of 567 families affected by dominant HSP, UBAP1 accounts for 1.2% of cases. Evaluating the full allelic and clinical spectrum in this gene will require further studies. Because UBAP1 links two cellular pathways previously involved in HSP, this finding consolidates our current understanding of the pathophysiology of HSP and points to potential novel drug targets.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are thankful to the families who participated in this research. We thank Els De Vriendt for the excellent technical assistance. S.Z. and R.S. are supported by National Institutes of Health grant R01NS072248. The project was further supported by the European Union’s Horizon 2020 research and innovation program under grant 779257 (Solve-RD; R.S.) and in the context of the ERA-NET Cofund action N° 643578 by the German Bundesministerium für Bildung und Forschung (BMBF) (01GM1607) for the E-Rare-3 network PREPARE (M.S., S.R., and associated partner S.Z.). M.J.S. was supported by Instituto de Salud Carlos III grant PS09/01830 and FEDER (Fonds Européen de Développement Économique et Régional). A.J. is supported by an Excellence (TOP) grant of the University of Antwerp. The study was partly supported by NIMAD research grant (940714) awarded to MAF. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health. T.B.H. was supported by the BMBF through the “Juniorverbund in der Systemmedizin” “mitOmics” (FKZ 01ZX1405C to T.B.H.).
Published: March 28, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.001.
Web Resources
OMIM, www.omim.org
GENESIS platform, https://www.tgp-foundation.org/
GeneReviews, Fink, J.K. (1993). Hereditary Spastic Paraplegia Overview, https://www.ncbi.nlm.nih.gov/books/NBK1509/
Supplemental Data
References
- 1.Fink J.K. Hereditary spastic paraplegia: Clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126:307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parodi L., Fenu S., Stevanin G., Durr A. Hereditary spastic paraplegia: More than an upper motor neuron disease. Rev. Neurol. (Paris) 2017;173:352–360. doi: 10.1016/j.neurol.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Schüle R., Wiethoff S., Martus P., Karle K.N., Otto S., Klebe S., Klimpe S., Gallenmüller C., Kurzwelly D., Henkel D. Hereditary spastic paraplegia: Clinicogenetic lessons from 608 patients. Ann. Neurol. 2016;79:646–658. doi: 10.1002/ana.24611. [DOI] [PubMed] [Google Scholar]
- 4.Bis-Brewer D.M., Züchner S. Perspectives on the genomics of HSP beyond Mendelian inheritance. Front. Neurol. 2018;9:958. doi: 10.3389/fneur.2018.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez M., Falk M.J., Gai X., Postrel R., Schüle R., Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum. Mutat. 2015;36:950–956. doi: 10.1002/humu.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. ,2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefani F., Zhang L., Taylor S., Donovan J., Rollinson S., Doyotte A., Brownhill K., Bennion J., Pickering-Brown S., Woodman P. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr. Biol. 2011;21:1245–1250. doi: 10.1016/j.cub.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 10.Agromayor M., Soler N., Caballe A., Kueck T., Freund S.M., Allen M.D., Bycroft M., Perisic O., Ye Y., McDonald B. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–428. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucenas S., Snell H., Appel B. nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 2008;4:71–81. doi: 10.1017/S1740925X09990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.