Abstract
Background:
Macrophage plasticity allows cells to adopt different phenotypes, a property with important implications in disorders such as cystic fibrosis (CF) and asthma.
Objective:
To examine the transcriptional and functional significance of macrophage repolarization from an “M1” towards an “M2” phenotype, and assess the role of a common human genetic disorder (CF) and a prototypical allergic disease (asthma) in this transformation.
Methods:
Monocyte-derived macrophages were collected from healthy and CF subjects and polarized to an M2 state using IL-4, IL-10, glucocorticoids, apoptotic PMNs, or azithromycin. We performed transcriptional profiling and pathway analysis for each stimulus. We assessed the ability of M2-repolarized macrophages to respond to LPS re-challenge and clear apoptotic neutrophils, and used murine models to determine conserved functional responses to IL-4 and IL-10. We investigated whether M2 signatures were associated with alveolar macrophage phenotypes in asthma.
Results:
We found that macrophages exhibit highly diverse responses to distinct M2-polarizing stimuli. Specifically, IL-10 activated pro-inflammatory pathways and abrogated LPS-tolerance allowing for rapid restoration of LPS responsiveness. In contrast, IL-4 enhanced LPS-tolerance, dampening pro-inflammatory responses after repeat LPS challenge. A common theme observed across all M2 stimuli was suppression of interferon-associated pathways. We found that CF macrophages had intact reparative and transcriptional responses, suggesting that macrophage contributions to CF lung disease are primarily shaped by their environment. Finally, we leveraged in vitro-derived signatures to show that allergen provocation induces distinct M2-state transcriptional patterns in alveolar macrophages.
Conclusion:
Our findings highlight the diversity of macrophage polarization, attribute functional consequences to different “M2” stimuli, and provide a framework to phenotype macrophages in disease states.
Keywords: Macrophage, polarization, pathway, microarray, cystic fibrosis, asthma, efferocytosis, tolerance
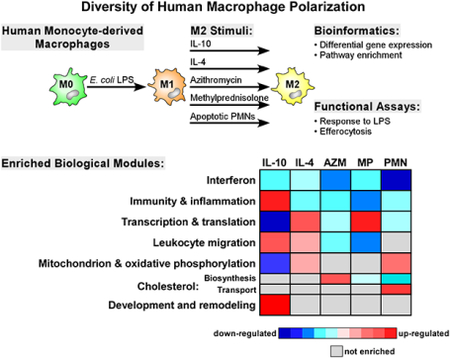
Graphical Abstract
CAPSULE SUMMARY
Macrophage polarization may influence the pathogenesis of many human diseases. In this study, we surveyed the effect of multiple polarizing stimuli on human macrophages in the context of cystic fibrosis and asthma.
INTRODUCTION
Macrophages are key effector cells of the immune system and in the lung contribute to both acute injury and its resolution via their functional heterogeneity and plasticity1, 2. Macrophages have been broadly classified into classically activated (M1) and alternatively activated (M2) states. The M1 phenotype is induced by pro-inflammatory T helper 1 (Th1) cytokines and factors, such as IFNγ or LPS. These cells release pro-inflammatory cytokines and are effective at killing bacteria3, 4. The M2 phenotype can be induced by TH2 cytokines, IL-4 and IL-13, as well as other factors, including apoptotic neutrophils, IL-10, and glucocorticoids5, 6. More recent evidence indicates that some drugs, such as azithromycin, can induce an M2 phenotype both in vitro and in cystic fibrosis (CF) patients7, 8. M2 macrophages dampen inflammatory gene expression, release anti-inflammatory factors, and promote wound repair1. The M1/M2 classification is based on in vitro studies and while it provides a framework to describe extremes of polarization, it does not recapitulate the complex spectrum and overlap of macrophage phenotypes in immuno-inflammatory disorders. We therefore sought to initially derive transcriptional signatures of macrophage polarization using in vitro exposures and then to assess the role of a common human genetic disorder (CF) in this transformation, and determine whether these signatures can be leveraged to understand the functional state macrophages in asthma.
Cystic fibrosis (CF), the most common fatal genetic disorder in Caucasians, is characterized by chronic inflammation in the lungs, suggesting an important role for macrophage phenotypes in disease progression. However, the contribution of the most common mutation in CF (F508del within the cystic fibrosis transmembrane conductance regulator [CFTR] gene) to macrophage repolarization is unknown. Human monocytes, alveolar macrophages, and monocyte-derived macrophages (MDMs) express CFTR protein9–11, and loss of functional CFTR in myeloid cells is associated with increased inflammatory responses12. Understanding whether CFTR alters other macrophage activation states may have important consequences both in CF lung disease and development of therapeutic approaches.
Macrophages have been implicated in the immuno-pathogenesis of asthma13, 14, and there is increasing evidence that disease severity is associated with abundance of M2-polarized cells15. Although environmental allergen exposure is the most common cause of asthma, large-scale phenotyping of alveolar macrophages following acute allergen exposure in asthmatics has not been previously explored.
To investigate these fundamental questions, we initially surveyed the stimulus-specific transcriptional and functional reprogramming of human MDMs during M1 to M2 transition with validation of specific IL-4 and IL-10 dependent pathways using murine derived bone marrow-derived macrophages. To assess the contribution of CFTR to macrophage repolarization, we studied the biological responses and gene expression profiles of macrophages from CF donors homozygous for F508del16. To examine the utility of these in vitro-derived polarization signatures in an in vivo setting, we investigated whether allergen-exposed alveolar macrophages in asthma were characterized by distinct M2 transcriptional patterns.
METHODS (please see supplementary Methods for details)
Human subjects, monocyte isolation and macrophage polarization
Human subjects provided informed written consent for blood donation at the University of Washington Medical Center as approved by the Institutional Review Boards of the University of Washington and Seattle Children’s Research Institute. Blood from 6 CF patients (4 males, 2 females) ages 27–32 (28.50±1.97 years) and from 6 healthy volunteers (4 males, 2 females) ages 25–31 (26.33±3.32 years) was drawn and peripheral blood mononuclear cells (PBMCs) were isolated, enriched, and differentiated for 7 days to generate M0 MDMs. M1 polarization was achieved by treating M0 MDMs with culture media containing E. Coli O111:B4 LPS for 24 h. M1 MDMs were then cultured in MDM media alone (control) or supplemented with IL-4 (10 ng/ml), IL-10 (50 ng/ml), methylprednisolone (100 nM), azithromycin (30μm), or apoptotic PMNs (5:1) for 24 h to repolarize towards an M2 state. The M2 stimuli were selected for their ability to promote an anti-inflammatory or M2 phenotype.5, 6,8,17
Microarray experiments
Total RNA from macrophages was isolated, labeled, and hybridized to Illumina HumanHT-12 v4 BeadChips (Illumina, Inc., San Diego, CA). All 72 experiments (6 conditions for 6 CF and 6 non-CF subjects) were normalized using lumi18. Detailed microarray information and raw data have been deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, GSE100521). For each M2 stimulus, differential gene expression between M1 (baseline) vs. M2 polarized cells of a subject was determined using a Bayesian paired t-test and a false discovery rate (FDR) threshold < 0.0119. Hierarchical clustering was performed on all differentially expressed genes that reached statistical significance (FDR < 0.01) in at least one experimental condition. Gene expression values were normalized across conditions using z-score: (xi − xm)/s, where xi refers to the expression value of gene x in condition i, xm is the mean expression value of gene x across all samples, and s indicates the standard deviation. Principal Component Analysis (PCA) was performed on these differentially expressed genes based on the covariance matrix.
Pathway analysis
To identify MDM transcriptional programs activated in response to M2-promoting stimuli, we applied Gene Set Enrichment Analysis (GSEA) using well-curated gene sets comprised of canonical pathways (Hallmark, KEGG, Reactome, Biocarta, etc.) and Gene Ontology (GO) annotations20. We used 31,401 unique transcripts for GSEA and applied an FDR < 0.05 for statistical significance of pathway enrichment. Network visualization of GSEA results was performed using Enrichment Map21 in Cytoscape22.
qPCR
qPCR was performed as described23 for II6, Tnf, II12b, CxcI1, CxcI2, CxcI8, II1b, and Hprt. The data were expressed as relative quantification (RQ) or fold change, which was calculated as 2−ΔΔCt.
Neutrophil isolation, induction of apoptosis and efferocytosis assay
Neutrophils (PMNs) were isolated from whole blood, labeled using the CellTrace CFSE Cell Proliferation Kit (Life Technologies), and underwent sterile, age-induced apoptosis. After 24 h treatment with M2 stimuli or media, culture media was replaced with fresh MDM media containing CFSE-labeled apoptotic neutrophils for 2 h incubation. The proportion of MDMs that ingested or bound at least one PMN (CD14+CFSE+) was determined by analysis on a BD FACSCanto RUO flow cytometer (BD Biosciences, San Jose, CA) using Flowjo software (Tree Star Inc., Ashland, OR). MFI represented the mean fluorescent intensity of the CFSE signal within the CD14+CFSE+ population.
Murine bone marrow-derived macrophages (BMDM)
Animal procedures were approved by the Institutional Animal Care and Use Committees at University of Washington. Stat6−/− mice on a BALB/c background, BALB/c mice, II10r−/− mice on a C57BI/6 background and C57Bl/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME). Bone marrow was collected from femurs and tibias of mice as described24. On day 7, cells received stimulation with E. coli LPS (10 ng/ml for 24 h) or control media. After 24 h, cells were washed and recombinant murine IL-4, IL-10, (10 ng/ml; Life Technologies) or control media was added to cultures for 24 h. At this time, cells were washed and received re-challenge with LPS or control media. At 4–6 h, cells were collected and processed for mRNA.
Statistical analysis
Statistical significance was determined using Student’s t-test (two-sided) after passing normality assumption (Shapiro-Wilk normality test), or Mann Whitney test for non-normally distributed data (GraphPad Software, La Jolla, CA).
Transcriptional analysis of macrophage polarization in allergen-induced asthma
We obtained publicly available RNA-sequencing data (http://www.ncbi.nlm.nih.gov/geo, GSE65039) on alveolar macrophages isolated from bronchoalveolar lavages performed at baseline and 48 hours following bronchial provocation with allergen from a subject with mild asthma who was on no asthma-controlling medications. This carefully designed allergen exposure protocol has been previously published25. Alveolar macrophages were purified by using Percall gradient method and adhesion purification. Total RNA was harvested using PureLink RNA micro Scale kit (Life Technologies), libraries were constructed using PrepX RNA-seq library kit (Wafergen, Fremont, CA), and sequencing was performed with Illumina HiSeq 2000 sequencing system. We performed differential gene expression analysis using DESeq226 statistical method. We next created 10 gene sets corresponding to differentially up- and down- regulated genes (FDR < 0.01 and absolute fold-change > 1.5) from the five M2-polarizing stimuli based on our microarray experiments on non-CF subjects. We combined these “M2” gene sets with canonical pathways and performed GSEA on statistically rank-ordered gene list from pre vs. post allergen-exposed alveolar macrophages. An FDR < 0.05 was used to determine statistical significance.
RESULTS
Different M2 stimuli elicit highly distinct transcriptional profiles in human macrophages independent of CF status
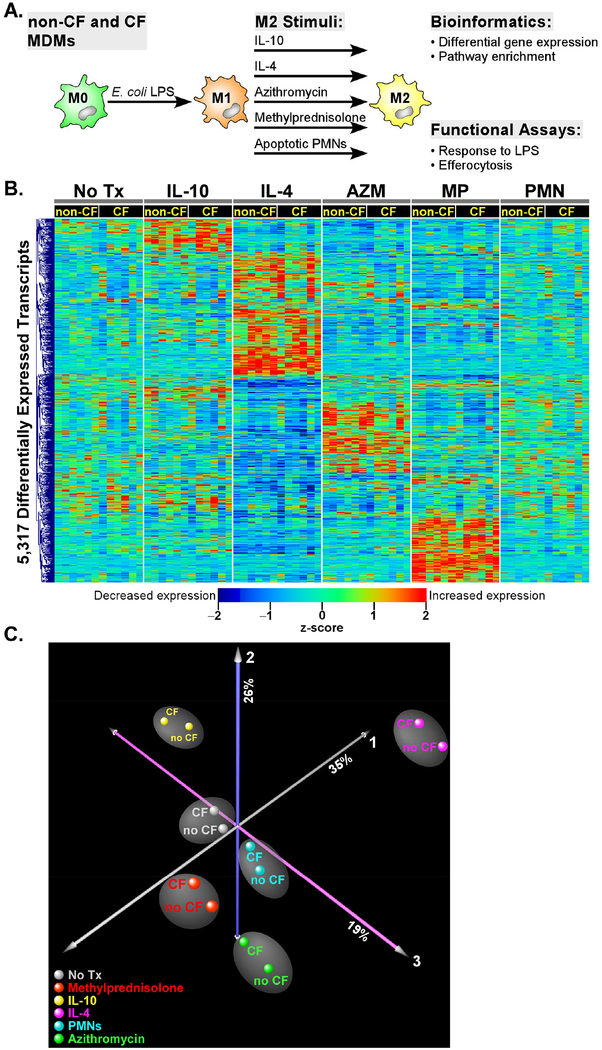
Blood monocytes were isolated from 6 healthy and 6 CF donors and differentiated to MDMs in culture. To reprogram these cells into an M1 phenotype and erase any bias based on prior in vivo activation states or as a result of in vitro differentiation, they were exposed to E. coli LPS for 24 h and repolarized using 5 different M2 stimuli (IL-4, IL-10, methylprednisolone, azithromycin, apoptotic neutrophils) for an additional 24 h (Figure 1A). Initially, we compared the differential transcriptional responses of MDMs to each of the five M2-polarizing stimuli relative to baseline (i.e., no treatment) in non-CF and CF subjects. Our experimental design allowed implementation of paired analysis for each subject (i.e., pre vs. post exposure) in each condition, thereby reducing inter-individual variability and enhancing the transcriptional signal. We identified a total of 4,882 transcripts that were significantly altered in the non-CF subjects in response to at least one of the five exposure conditions (stimulus-specific gene lists in Supplemental Table E1). In the CF patients, a total of 3,537 transcripts were differentially expressed in at least one of the five M2-polarizing stimuli (stimulus-specific gene lists in Supplemental Table E2). Cumulatively, 5,317 transcripts were differentially expressed across all subjects and conditions. Hierarchical clustering of these transcripts demonstrated highly distinct and stimulus-specific expression patterns (Figure 1B). A majority (~88%) of the differentially regulated genes were common to both genotypes implying similar responses to M2 stimuli in CF and non-CF MDMs. To further explore this observation, Principal Component Analysis (PCA), a statistical method for reducing complex high dimensional data, was applied to these differentially expressed genes, revealing that the primary driver of gene expression variability was the response to the M2 stimuli and not genotype status (Figure 1C).
Figure 1. Overview of experimental design.
(A) Blood monocytes from 6 non-CF and 6 CF donors were cultured with M-CSF over 7 days to produce MDMs. Cells were stimulated with E. coli LPS for 24h and then repolarized with IL-10, IL-4, azithromycin, methylprednisolone, apoptotic PMNs, or control media for 24 h. Cells were then processed for gene expression and functional assays. (B) Hierarchical cluster analysis of a total of 5,317 differentially expressed genes in non-CF and CF subjects across the five M2 stimuli. Note the distinct transcriptional pattern associated with each exposure, and the overall similar expression profile between non-CF and CF subjects. An FDR < 0.01 cutoff was used to designate significant differential expression in each M2 exposure relative to baseline (no treatment). (C) Principal Component Analysis. To simplify visualization of the 72 experiments, differentially expressed transcriptional profiles were averaged in CF (n = 6) and non-CF (n = 6) subjects for each M2 stimulus. Each orthogonal axis captures a percentage of the total gene expression variability, with ~80% of the variance captured by the three axes. Note that the non-CF and CF samples segregate based on M2 stimuli, implying that the primary determinant of separation between groups is exposure, not genotype
A spectrum of common and divergent transcriptional programs characterize macrophage polarization
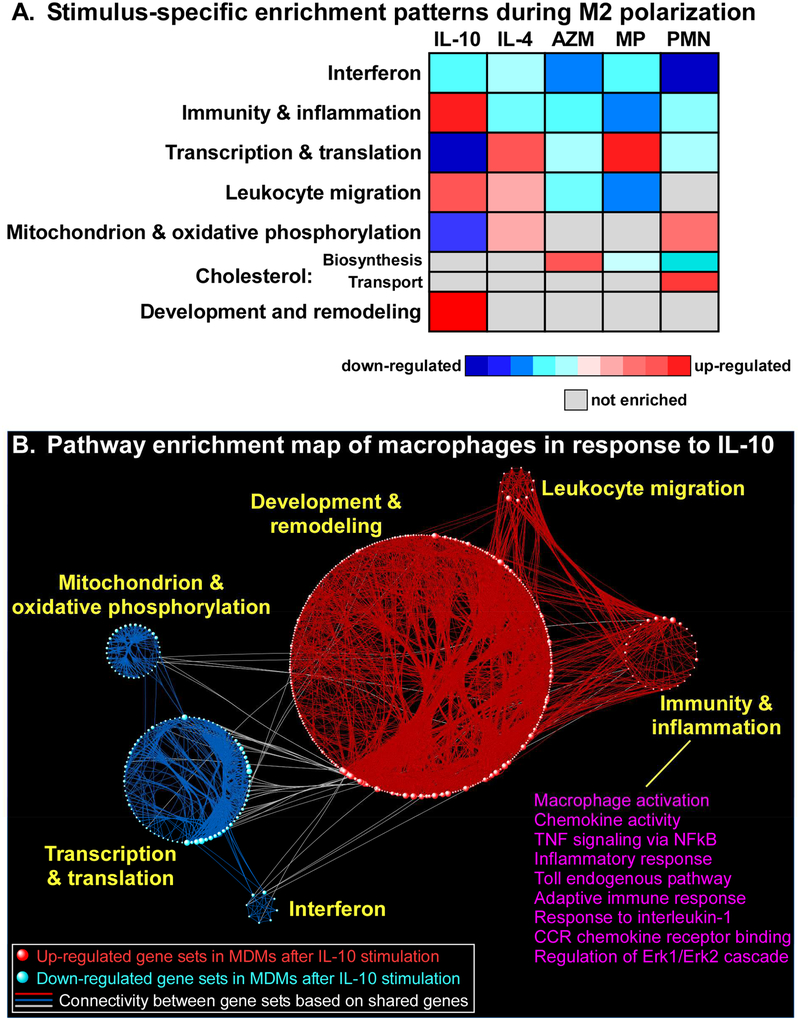
To further investigate the diverse transcriptional response of MDMs to different M2-promoting conditions, we applied gene set enrichment analysis (GSEA) to systematically identify up and down-regulated pathways under each polarizing state. A visual summary of our results is depicted in Figure 2A (see also Supplemental Table E3). We found that suppression of interferon signaling in MDMs was the most prominent signal shared by all M2-polarizing stimuli across subjects, with IL-10 down-regulating IFIT3 and STAT1; and apoptotic PMNs down-regulating IFIT1–3, and IRF7. However, we observed substantial variability in the gene members of enriched interferon-associated pathways depending on the M2 stimulus (see Supplemental Table E4 and Supplemental Figure E3). Interestingly, the suppression of interferon-stimulated genes and pathways was recently reported in human asthma.27 These findings are consistent with the widely accepted paradigm that M2 macrophage phenotypes are associated with a dampened immune state. Methylprednisolone is a well-established anti-inflammatory, and our findings also highlight strong up-regulation of the decoy receptor, IL1R2. Nevertheless, the overall pathway profile of MDMs during M2 polarization was quite heterogeneous and stimulus-dependent, with many processes displaying diametrical enrichment patterns.
Figure 2. Visual summary of pathway analysis.
(A) GSEA of the transcriptional response to M2-polarizing stimuli identified many differentially up and down-regulated pathways. Network analysis was used to group these pathways into seven larger aggregates known as modules. Note the divergent enrichment pattern across conditions, although common themes such as suppression of interferon-associated pathways are also revealed from this analysis. Complete list of enriched gene sets is provided in Supplemental Table E3. (B) Network-based visualization of the GSEA of MDM response to IL-10 treatment illustrates the emergence of modules comprised of interconnected pathways (shown as spheres). Unlike other M2 stimuli, exposure to IL-10 activated multiple immune-related programs, several of which are listed.
One striking example of the transcriptional heterogeneity observed in MDM’s response to M2 stimuli was the unexpected up-regulation of multiple pro-inflammatory gene sets after exposure to IL-10 (Figure 2B), including “Toll-like receptor signaling”, “Macrophage activation”, “TNF signaling via NF-kB”, “Inflammatory response”, and “Cytokine/chemokine signaling” pathways. These processes were enriched in both CF and non-CF MDMs stimulated with IL-10, but not under any other M2-polarizing condition. In fact, the other M2 stimuli were associated with down-regulation of inflammatory pathways; for example, IL-4 treatment resulted in suppression of “Toll-like receptor signaling”.
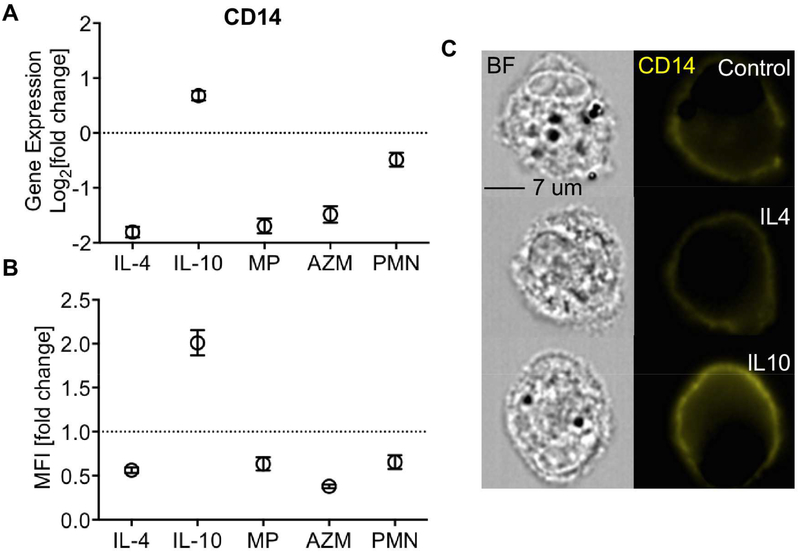
Since TLR signaling plays a critical role in LPS tolerance and our results revealed an opposite enrichment profile for this pathway in response to IL-10 versus IL-4, we assessed how its constituents were altered during M2 polarization. We found that the expression of CD14, the key co-receptor for LPS binding to TLR4, was significantly reduced by IL-4 but increased by IL-10 (Fig 3A). Furthermore, these distinct gene expression patterns were corroborated at the protein level by FACS measurement of CD14 protein expression on the surface of macrophages (Figure 3B, 3C).
Figure 3. CD14 is differentially regulated by IL-4 and IL-10.
Expression of the LPS-binding protein and TLR4 co-receptor, CD14 was significantly suppressed by all M2-stimuli with the exception of IL-10, which significantly increased its expression as shown by (A) mRNA from gene expression arrays, and (B) protein as measured by flow cytometry. (C) CD14 staining (yellow) on the cell membrane of non-CF MDMs as imaged using Image Stream. MFI=median fluorescent intensity. BF= bright field. (Mean ± SEM; n=6/group).
IL-10 and IL-4 have opposing effects on LPS tolerance
The distinctive transcriptional signature produced by IL-10 suggested that despite its M2-polarizing attribution, this stimulus may also “prime” MDMs towards a pro-inflammatory phenotype. Since macrophages develop a period of unresponsiveness following exposure to LPS, we hypothesized that MDMs treated with IL-10 would exhibit diminished LPS tolerance. We therefore assessed the functional consequences of various M2 stimuli on repeated LPS challenge by initially exposing non-polarized MDMs to LPS (inducing an M1 state) followed by M2-specific polarization, and then a re-challenge with LPS. Gene expression for multiple immuno-inflammatory genes (II6, Il8, II1b, Tnfa, CxcI1, CxcI2) was determined using qPCR.
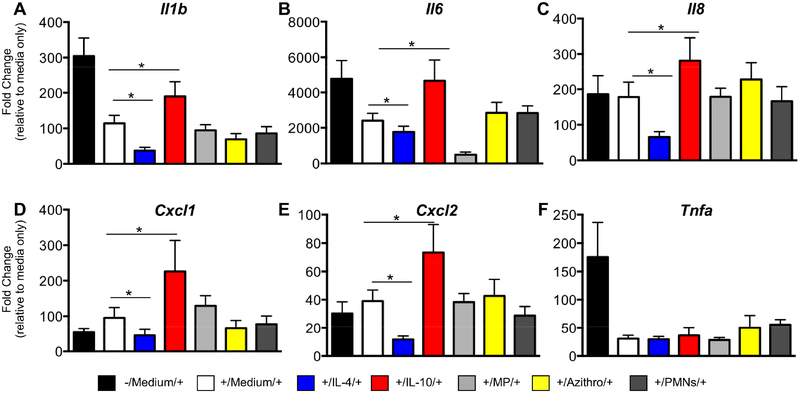
We found that IL-4 treatment decreased expression of pro-inflammatory cytokines (II1b, II8, CxcI1 and CxcI2) with the second LPS challenge, implying prolongation of LPS tolerance. In contrast, IL-10 exposure had the opposite effect by promoting a robust cytokine response to repeat LPS challenge and thereby reducing LPS tolerance of MDMs (Figure 4A–G). Methylprednisolone suppressed II6 expression but not the other inflammatory genes, and azithromycin and apoptotic PMNs did not alter LPS responsiveness of the macrophage. Together, these findings highlight the diverse functional ramifications of specific M2 stimuli and demonstrate the opposing effects of IL-4 and IL-10 on macrophage immune tolerance.
Figure 4. Consequences of M2 repolarization on LPS rechallenge.
Monocyte-derived macrophages from 6 healthy donors were stimulated with LPS (+) followed by M2 stimuli or control medium and then rechallenged with LPS. Additional control samples did not receive the first LPS challenge (-/Medium/+). Each donor’s samples were normalized to HPRT and expressed as fold change over their respective non-LPS treated samples. (A-F) For II1b, II6, II8, CxcI1, CxcI2 but not Tnfa, IL-4 exposure decreased the ability of the macrophage to produce pro-inflammatory cytokines, and IL-10 reversed LPS tolerance. (Mean ± SEM; n=6/group). *P-value <0.05.
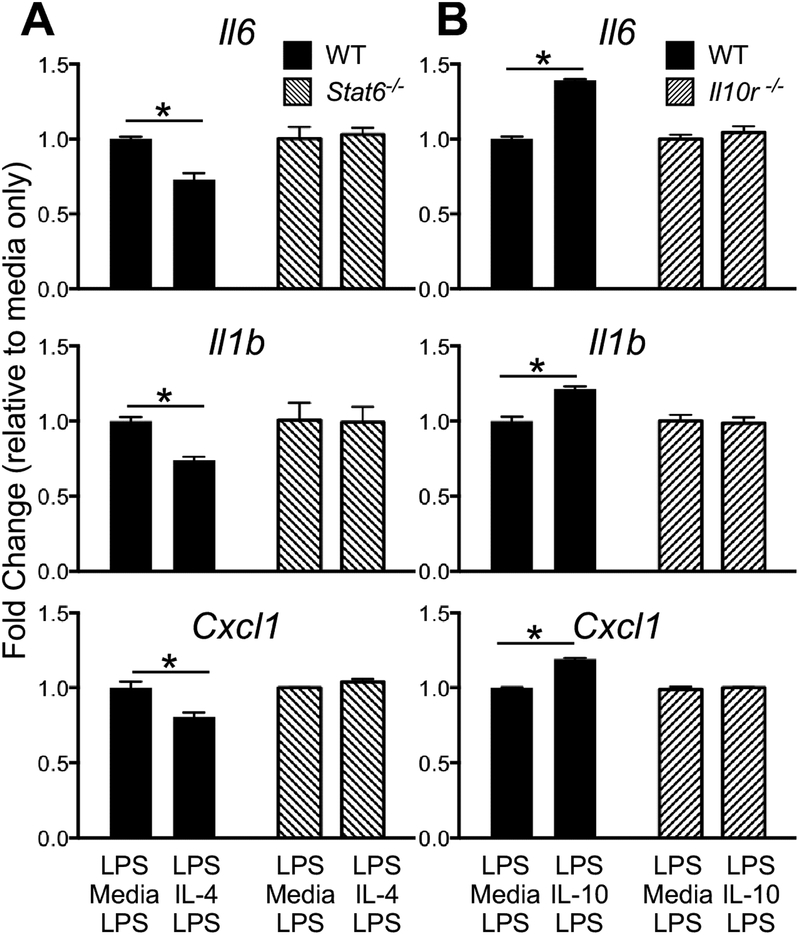
To further corroborate the IL-4 and IL-10 findings above, we performed similar repolarization experiments using murine bone marrow derived macrophages (BMDMs) with impaired IL-4 (Stat6−/−) or IL-10 (II10r−/−) pathways. Consistent with our observations in human macrophages, IL-4 impaired pro-inflammatory gene expression to the second LPS challenge, and this effect was abrogated in Stat6−/− cells (Figure 5A). Conversely, IL-10 increased pro-inflammatory gene expression to the second LPS challenge, an effect that was abolished in II10r−/− cells (Figure 5B). These experiments confirmed the dependency of LPS tolerance to critical signaling components of distinct M2 stimuli.
Figure 5. Murine macrophage inflammatory response to LPS rechallenge is modified by IL-4 and IL-10.
Stat6−/−, II10r−/−, and WT bone marrow-derived macrophages were stimulated with LPS followed by IL-4, IL-10, or control media and then rechallenged with LPS. Samples were normalized to HPRT and expressed as fold change over their respective control media-treated samples. (A) Experiments performed in BALB-C wildtype (WT) and Stat6−/− BMDM demonstrating a Stat6-dependent suppression of LPS-rechallenge by IL-4. (I) Experiments performed in C57BL/6 WT and II10r−/− BMDM demonstrating an IL-10r-dependent augmentation of LPS-rechallenge by IL-10. Each condition was performed in triplicate and 2 experiment replicates performed. *P-value < 0.05.
IL-10-repolarized macrophages have the greatest ability to clear apoptotic neutrophils
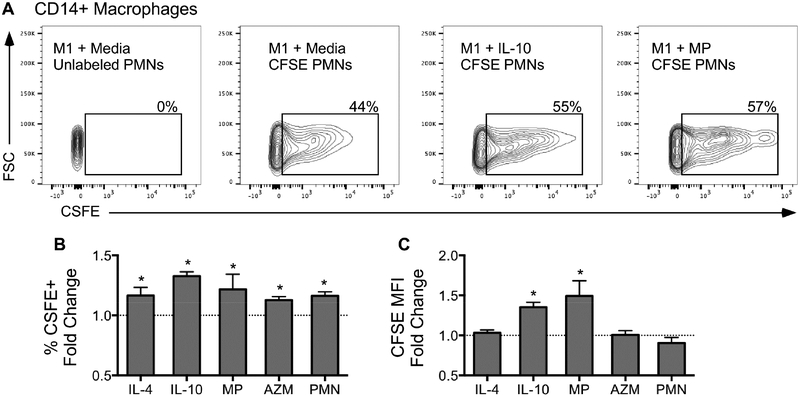
We next asked whether different M2-polarizing stimuli alter MDM’s ability to clear apoptotic cells. Purified apoptotic neutrophils were collected from non-CF donors, labeled with CFSE and allowed to undergo age-induced apoptosis for 48h prior to co-incubation with M1 cells or M1 cells treated with M2 stimuli. Efferocytosis was measured using flow cytometry for percentages of CD14+ macrophages with CFSE staining as well as mean fluorescence intensity (MFI) for CFSE (Figure 6A).
Figure 6. M2 stimuli have varying ability to promote macrophage efferocytosis and are independent of functional CFTR.
(A) Representative gating strategy to quantify efferocytosis of CD14+ macrophages using CFSE+ PMNs. The CFSE+ gate was drawn using control macrophages incubated with unlabeled PMNs. The percentage of CD14+ macrophages positive for ingested apoptotic PMNs (% CFSE+) and quantity of ingested apoptotic PMNs (CFSE MFI) was determined across stimuli. (B) Fold change in percentage of macrophages positive for ingested apoptotic PMNs (PMN) relative to their respective media-only controls. (C) Fold change in CFSE MFI in macrophages relative to their respective media-only controls. *P-value < 0.05 M2 stimuli vs. media control. AZM=azithromycin, MP=methylprednisolone, PMN=apoptotic PMNs, MFI=Mean fluorescence intensity. (Mean ± SEM; n=6/group).
We found that all M2 stimuli significantly enhanced macrophage efferocytosis as measured by percentage of macrophages staining for intracellular PMNs (Figure 6B), but IL-10 treatment had the greatest effect. IL-10 and methylprednisolone also increased the number of PMNs ingested by macrophages (Figure 6C). Interestingly, our pathway analysis identified several phagocytosis-associated processes (“phagocytosis”, “phagocytic cup”, “FCγR-mediated phagocytosis”) as being highly enriched in MDMs stimulated with IL-10, whose up-regulated member genes included well-known drivers of macrophage phagocytosis such as FCγRs, MARCO (macrophage receptor with collagenous structure), and CD3628, 29.
Apoptotic PMNs and azithromycin modulate the macrophage transcriptome by altering processes involved in lipid metabolism and transport
Whereas macrophage responses to well-characterized M2 stimuli such as IL-4 and IL-10 have been studied, there are no published reports on transcriptional programs induced by exposure of human MDMs to apoptotic PMNs or azithromycin. Our gene set analysis of MDMs stimulated by apoptotic neutrophils revealed widespread suppression of lipid and cholesterol biosynthesis pathways, with highly significant down-regulation of essential genes including HMGCR (HMG-CoA reductase), LDLR (low density lipoprotein receptor), and FDFT1 (farnesyl-diphosphate farnesyltransferase 1). Concomitantly, processes involved in lipid transport, cholesterol efflux, as well as fat storage and catabolism were activated (Supplemental Figure E4). Taken together, these results indicate that exposure and subsequent phagocytosis of apoptotic PMNs by macrophages initiates distinct lipid-processing pathways aimed at reducing the burden associated with accumulation of cellular debris and fat contents. We also found alterations in lipid-related transcriptional programs after stimulation of MDMs with azithromycin, however in contrast to apoptotic PMNs, we observed an opposite enrichment pattern whereby lipid biosynthetic processes were activated with up-regulation of several of the same key genes (e.g., LDLR, FDFT1) that were suppressed in response to apoptotic neutrophils (Figure 2A).
Functional changes to M2 stimuli are CFTR-independent
Given the potential role of CFTR in macrophage activation, we also examined the ability of M2 repolarized macrophages from CF donors to respond to LPS re-challenge and clear apoptotic PMNs, two critical macrophage functions that may modulate the inflammatory response in CF lung disease. As suggested by a similar transcriptional response between non-CF and CF macrophages (Figure 1B, C), we also observed a similar functional response to LPS rechallenge (Supplemental Figure E1A–D) and efferocytosis (Supplemental Figure E2A, B) across all M2 stimuli. These findings suggest that these LPS and M2 response patterns are broadly CFTR-independent.
Fingerprinting Macrophage Polarization States in Acute Allergic Asthma
Since the environmental milieu is a critical factor in macrophage polarization, we explored the utility of using our M2 transcriptional signatures to describe dynamic states of alveolar macrophages during acute allergen-induced asthma. We defined 10 new gene sets based on differentially up- and down-regulated genes in response to the five M2 stimuli (Supplemental Table E5) and included them with available canonical pathways. We performed GSEA to identify enriched gene sets associated with the transcriptional response of alveolar macrophages 48 hours after allergen exposure (Table 1). We found that “Induced in response to IL-4”, “Induced in response to IL-10”, “Suppressed in response to AZM” and “Suppressed in response to MP” gene sets were enriched among alveolar macrophage genes up-regulated post allergen exposure. This observation indicates that polarized alveolar macrophages in asthma share concordant transcriptional signatures with MDMs stimulated with IL-4 and IL-10, while having discordant expression signals compared to MDMs treated with azithromycin and methylprednisolone. In contrast, down-regulated alveolar macrophage genes following allergen challenge were enriched in “Suppressed in response to IL-4”, “Suppressed in response to IL-10”, “Induced in response to AZM” and “Induced in response to MP” gene sets. Collectively, these findings imply that acutely polarized alveolar macrophages in asthma are characterized by a complex phenotype that overlaps with IL-4 and IL-10 transcriptional signatures while displaying an expression pattern that is opposite of AZM and methylprednisolone-induced responses.
Table 1.
Top 60 most enriched gene sets in alveolar macrophages after allergen provocation.
| Pathways Up-regulated in Alveolar Macrophages Post Challenge (n = 30) | FDR |
| Class a1 rhodopsin like receptors | 0 |
| G-protein coupled receptor (GPCR) ligand binding | 0 |
| Cytokine cytokine receptor interaction | 0 |
| Allograft rejection | 0 |
| Inflammatory response | 0 |
| Hematopoietic cell lineage | 0 |
| Suppressed in response to AZM | 0 |
| Induced in response to IL-4 | 0 |
| Signaling by GPCR | 0 |
| Neuroactive ligand receptor interaction | 0 |
| GPCR downstream signaling | 0 |
| Suppressed in response to MP | 0 |
| IL-2 Stat5 signaling | 0 |
| Chemokine receptors bind chemokines | 0 |
| TNFa signaling via NF-kB | 0 |
| Immunoregulatory interactions between a lymphoid and a non lymphoid cell | 0 |
| Peptide ligand binding receptors | 0 |
| Gastrin creb signaling pathway via PKC and MAPK | 0 |
| Induced in response to IL-10 | 0 |
| G alpha (q) signaling events | 0 |
| CDB T-cell receptor pathway | 3.68×10−5 |
| Srp-dependent cotranslational protein targeting to membrane | 3.51×10−5 |
| Core matrisome | 1.01×10−4 |
| G2M checkpoint | 1.28×10−4 |
| CDB T-cell receptor downstream pathway | 2.18×10−4 |
| T-cell receptor signaling pathway | 2.71×10−4 |
| IL-12 pathway | 5.54×10−4 |
| T cytotoxic pathway | 1.06×10−3 |
| T-cell signal transduction | 1.12×10−3 |
| IL-17 pathway | 1.73×10−3 |
| Pathways Down-regulated in Alveolar Macrophages Post Challenge (n = 30) | FDR |
| Generic transcription pathway | 0 |
| Interferon alpha response | 0 |
| Oxidative phosphorylation | 0 |
| Metabolism of lipids and lipoproteins | 0 |
| Suppressed in response to IL-4 | 0 |
| Fatty acid metabolism | 0 |
| Adipogenesis | 0 |
| Peroxisome | 0 |
| Induced in response to MP | 0 |
| Interferon gamma response | 0 |
| Cholesterol homeostasis | 0 |
| Cholesterol biosynthesis | 0 |
| Induced in response to AZM | 0 |
| Pyruvate metabolism | 0 |
| Transcription | 0 |
| Aminoacyl tRNA biosynthesis | 0 |
| Phospholimetabolism | 0 |
| DNA repair | 6.40×10−5 |
| p53 pathway | 6.06×10−5 |
| Coagulation | 5.76×10−5 |
| Spliceosome | 5.48×10−5 |
| Valine leucine and isoleucine degradation | 5.23×10−5 |
| Recruitment of mitotic centrosome proteins and complexes | 5.01×10−5 |
| Interferon alpha beta signaling | 9.89×10−5 |
| Platelet sensitization by LDL | 2.38×10−4 |
| mRNA processing | 5.07×10−4 |
| Antigen processing ubiquitination proteasome degradation | 4.88×10−4 |
| Interferon gamma signaling | 5.13×10−4 |
| Suppressed in response to IL-10 | 6.79×10−4 |
| Glycan degradation | 9.73×10−4 |
DISCUSSION
Although macrophages exist in various polarized states, the concept of macrophage plasticity has important clinical implications in chronic inflammatory conditions such as cystic fibrosis lung disease and asthma. In our studies, we examined the functional significance of macrophage repolarization from an “M1” (LPS-stimulated) toward an “M2” phenotype with stimuli based on their known immune-dampening properties. Although the M1 and M2 nomenclature is a gross oversimplification of a dynamic process that may be better described as a “color wheel” of activation states30, we use the terms M1 and M2 to highlight a transition from a state that is pro-inflammatory to one that is anti-inflammatory. While the recommended terminology for reporting macrophages activation states by stimuli is useful for defining in vitro conditions31, markers for these stimuli are also necessary to help determine macrophage phenotypes in vivo. Hence, defining surrogate signatures based on a simplified in vitro protocol can provide an informative tool to discern the environmental factors driving in vivo activation states. Our study advances this concept by highlighting the diversity of individual M2 stimuli on macrophage programming at both the transcriptional level and macrophage effector function. It provides new insights into how these events may contribute to resolution of injury via down-regulation of inflammatory cytokines and clearance of apoptotic debris as well as restoration of macrophage responses to LPS, a key factor influencing susceptibility to recurrent infection.
By applying gene set enrichment analysis (GSEA) to systematically identify up and down-regulated pathways under each polarizing state, we found that the overall pathway profile of MDMs during M2 polarization was quite heterogeneous and stimulus-dependent. Suppression multiple interferon-associated pathways was one of the most prominent signals shared by all M2-polarizing stimuli across subjects, although even here, substantial variability among individual genes members was observed. These findings are consistent with the widely accepted paradigm that M2 macrophage phenotypes are associated with a dampened immune state and suggest a shared feature involving interferon pathways.
We previously demonstrated that alveolar macrophage repolarization from “M1” to “M2” occurs in vivo during the resolution of lung injury in a P. aeruginosa pneumonia murine model32, however the drivers of these changes in human pulmonary disorders remain uncertain. IL-4 is released during tissue injury by both innate and adaptive immune responses33, 34. Other important drivers of macrophage repolarization include clearance of apoptotic PMNs, IL-10, and immune-modulating drugs, including azithromycin and methylprednisolone among other factors35. Our study systematically identified distinct markers and pathways activated by each of these stimuli, thereby highlighting the transcriptional and functional diversity of macrophage polarization. For example, the most activated pathways induced by apoptotic PMNs were processes required for handling the lipid load that occurs with ingestion of cells. Genes regulating cholesterol efflux were upregulated, including ABCG1 and ABCA1, whereas genes controlling cholesterol synthesis were suppressed. The selective shift between the activation/suppression state of these processes is likely critical to macrophage survival, as reports have shown that ABCG1-null and ABCA1-null macrophages have increased susceptibility for programmed cell death following efferocytosis36, 37.
We also investigated the consequence of M2 repolarization in terms of restoring LPS responsiveness and clearing apoptotic cells. Although all M2 stimuli were associated with down-regulation of pro-inflammatory interferon-associated genes, IL-10 repolarization had the greatest effect on promoting efferocytosis and, unexpectedly, also activated pathways involved in restoring LPS responsiveness. Consistent with these observations, IL-10-repolarized cells responded more robustly to LPS rechallenge, whereas IL-4-stimulated macrophages displayed an immune-suppressive phenotype. These findings were conserved across species since murine macrophages showed the same distinctive LPS response patterns when repolarized by IL-4 versus IL-10. Hence, we demonstrated that the type of stimulus can dramatically alter key immune functions in polarizing macrophages and may contribute to non-resolving inflammation or susceptibility to new infection.
Translating our findings to understand the temporal dynamics of macrophage phenotypes in vivo will be necessary to determine how genetic and environmental stimuli contribute to acute and chronic inflammatory disorders in humans. As a proof of concept, we leveraged our MDM polarization signatures to perform exploratory pathway analysis on an established human model of allergen-induced asthma25. We found that the transcriptional response of alveolar macrophages 48 hours after allergen provocation was concordant with IL-4 and IL-10 induced programs in MDMs and discordant with azithromycin and methylprednisolone stimulated expression profiles. This observation highlights the heterogeneous activation state of airway macrophages in asthma that is, in part, likely due to the presence of different subpopulations38 but also reflective of the complexity of TH2 immune responses in human disease. Indeed, up-regulated gene sets in allergen-activated alveolar macrophages included several pro-inflammatory pathways enriched in response to IL-10 stimulation of MDMs such as, “TNF signaling via NF-kB”, “Inflammatory response”, and “Cytokine/chemokine signaling” (Table 1). Intriguingly, the discordant signal between allergen-challenged alveolar macrophage programs and methylprednisolone and azithromycin-induced signatures implies that these two stimuli counteract the pathogenic response in asthma. Since the treatment of asthma with glucocorticoids is well established and recent evidence supports a role for azithromycin39, our analytical Since the functional state of macrophages is influenced by both genetic macrophage polarization.
Since the functional state of macrophages is influenced by both genetic and environmental factors, we asked whether the most common mutation in cystic fibrosis (F508del) alters macrophage repolarization. There are conflicting data on the inflammatory responsiveness of peripheral monocytes from CF donors, with one small study demonstrating increased LPS responsiveness40, and another study demonstrating impaired LPS responsiveness41. The direct contribution of CFTR on monocyte function in these studies is uncertain, as the inflammatory milieu from which these cells are isolated is likely contributory.
Although our microarray experiments were not designed to examine LPS responses between CF vs. non-CF macrophages, we did examine individual gene expression changes pre- and post-LPS using qPCR. Using a set of inflammatory markers, we did not observe increased LPS responsiveness in CF macrophages. However, our sample size was small, and CFTR-dependent responses may not overcome that of other genetic modifiers of LPS-responsiveness. Alternatively, the primary driver of macrophage polarization may be the inflammatory milieu in CF, and this effect was likely abrogated by our ex vivo culture model where blood monocytes were differentiated to macrophages over 7 days. Importantly, in our study, CF and non-CF macrophages responded similarly to all M2 stimuli at the gene expression, pathway enrichment, and functional levels. Hence, our data suggest that in CF lung disease, environmental factors are likely to be the key determinants of macrophage biology and polarization in a CFTR-independent manner.
Our study has several limitations. The sample sizes were small and substantial inter-individual variability exists in macrophage polarization responses. We attempted to address this issue by designing our experiments such that for each stimulus, paired analysis (i.e., comparing pre vs. post exposure) could be performed in the same individual. A shortcoming of pathway-based approaches such as GSEA is their reliance on predefined gene sets, which may not accurately capture the complexity of human disease state and are prone to biases in selection of gene members. A distinctive feature of our study is its utilization of human MDMs shared across multiple donors, including those with cystic fibrosis. A limitation of this approach is that MDMs are different from tissue macrophages, and extension of our findings to resident macrophages will need to be confirmed within the appropriate organ. However, our exploratory analysis integrating in vivo MDM signatures with alveolar macrophage responses in asthma demonstrates the feasibility and promise of this approach. Nevertheless, defining the role of macrophage polarization in complex human disorders such as CF and asthma need to be confirmed by larger studies.
In conclusion, our findings highlight the transcriptional diversity of macrophage polarization across various “M2” stimuli, attribute functional consequences to these activation states, and provide a resource and methodology to leverage this information to phenotype macrophage subtypes in human diseases.
Supplementary Material
KEY MESSAGES.
Macrophage polarization from a classical (M1) to an alternative (M2) activation state is characterized by highly distinct functional and transcriptional responses based on the types of TH2 stimuli.
The most common mutation in the cystic fibrosis transmembrane conductance regulator does not significantly alter the effector function or gene expression of monocyte-derived macrophages to various M2-polarizing factors.
Stimulus-specific transcriptional signatures of monocyte-derived macrophages may be leveraged for molecular phenotyping of alveolar macrophages in asthma.
Acknowledgments
Funding: This study was supported by NIH/NIDDK P30 DK089507 (AMM, RSM, SAG), NIH/NHLBI R01 HL116514 (AMM), NIH/NIAID R01 AI137111 (SAG), NIH/NHLBI T32 HL007828 (MEL), and the Cystic Fibrosis Foundation Research Development Program CFF SINGH15R0 (MEL).
ABBREVIATIONS
- CF
cystic fibrosis
- MDM
monocyte-derived macrophages
- CFTR
cystic fibrosis transmembrane conductance regulator
- PBMC
peripheral blood mononuclear cell
- FDR
false discovery rate
- GO
gene ontology
- GSEA
gene set enrichment analysis
- LPS
lipopolysaccharide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
- 1.Gordon S Alternative activation of macrophages. Nat Rev Immunol 2003; 3:23–35. [DOI] [PubMed] [Google Scholar]
- 2.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol 2008; 181:3733–9. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005; 5:953–64. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity 2005; 23:344–6. [DOI] [PubMed] [Google Scholar]
- 5.Vallelian F, Schaer CA, Kaempfer T, Gehrig P, Duerst E, Schoedon G, et al. Glucocorticoid treatment skews human monocyte differentiation into a hemoglobin-clearance phenotype with enhanced heme-iron recycling and antioxidant capacity. Blood 2010; 116:5347–56. [DOI] [PubMed] [Google Scholar]
- 6.Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol 2012; 189:3508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cory TJ, Birket SE, Murphy BS, Hayes D Jr., Anstead MI, Kanga JF, et al. Impact of azithromycin treatment on macrophage gene expression in subjects with cystic fibrosis. J Cyst Fibros 2014; 13:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy BS, Sundareshan V, Cory TJ, Hayes D Jr., Anstead MI, Feola DJ. Azithromycin alters macrophage phenotype. J Antimicrob Chemother 2008; 61:554–60. [DOI] [PubMed] [Google Scholar]
- 9.Di A, Brown ME, Deriy LV, Li C, Szeto FL, Chen Y, et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol 2006; 8:933–44. [DOI] [PubMed] [Google Scholar]
- 10.Sorio C, Buffelli M, Angiari C, Ettorre M, Johansson J, Vezzalini M, et al. Defective CFTR expression and function are detectable in blood monocytes: development of a new blood test for cystic fibrosis. PLoS One 2011; 6:e22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Porto P, Cifani N, Guarnieri S, Di Domenico EG, Mariggio MA, Spadaro F, et al. Dysfunctional CFTR alters the bactericidal activity of human macrophages against Pseudomonas aeruginosa. PLoS One 2011; 6:e19970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruscia EM, Zhang PX, Satoh A, Caputo C, Medzhitov R, Shenoy A, et al. Abnormal trafficking and degradation of TLR4 underlie the elevated inflammatory response in cystic fibrosis. J Immunol 2011; 186:6990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balhara J, Gounni AS. The alveolar macrophages in asthma: a double-edged sword. Mucosal Immunol 2012; 5:605–9. [DOI] [PubMed] [Google Scholar]
- 14.Draijer C, Peters-Golden M. Alveolar Macrophages in Allergic Asthma: the Forgotten Cell Awakes. Curr Allergy Asthma Rep 2017; 17:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girodet PO, Nguyen D, Mancini JD, Hundal M, Zhou X, Israel E, et al. Alternative Macrophage Activation Is Increased in Asthma. Am J Respir Cell Mol Biol 2016; 55:467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal 2014; 26:192–7. [DOI] [PubMed] [Google Scholar]
- 17.Long ME, Eddy WE, Gong KQ, Lovelace-Macon LL, McMahan RS, Charron J, et al. MEK1/2 Inhibition Promotes Macrophage Reparative Properties. J Immunol 2017; 198:862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008; 24:1547–8. [DOI] [PubMed] [Google Scholar]
- 19.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res 2012; 40:W553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isserlin R, Merico D, Voisin V, Bader GD. Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res 2014; 3:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2:2366–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gharib SA, Johnston LK, Huizar I, Birkland TP, Hanson J, Wang Y, et al. MMP28 promotes macrophage polarization toward M2 cells and augments pulmonary fibrosis. J Leukoc Biol 2014; 95:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manicone AM, Birkland TP, Lin M, Betsuyaku T, van Rooijen N, Lohi J, et al. Epilysin (MMP-28) restrains early macrophage recruitment in Pseudomonas aeruginosa pneumonia. J Immunol 2009; 182:3866–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park GY, Lee YG, Berdyshev E, Nyenhuis S, Du J, Fu P, et al. Autotaxin production of lysophosphatidic acid mediates allergic asthmatic inflammation. Am J Respir Crit Care Med 2013; 188:928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhakta NR, Christenson SA, Nerella S, Solberg OD, Nguyen CP, Choy DF, et al. IFN-stimulated Gene Expression, Type 2 Inflammation, and Endoplasmic Reticulum Stress in Asthma. Am J Respir Crit Care Med 2018; 197:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 1999; 17:593–623. [DOI] [PubMed] [Google Scholar]
- 29.Jing J, Yang IV, Hui L, Patel JA, Evans CM, Prikeris R, et al. Role of macrophage receptor with collagenous structure in innate immune tolerance. J Immunol 2013; 190:6360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014; 41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston LK, Rims CR, Gill SE, McGuire JK, Manicone AM. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. Am J Respir Cell Mol Biol 2012; 47:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loke P, Gallagher I, Nair MG, Zang X, Brombacher F, Mohrs M, et al. Alternative activation is an innate response to injury that requires CD4+ T cells to be sustained during chronic infection. J Immunol 2007; 179:3926–36. [DOI] [PubMed] [Google Scholar]
- 34.Brandt E, Woerly G, Younes AB, Loiseau S, Capron M. IL-4 production by human polymorphonuclear neutrophils. J Leukoc Biol 2000; 68:125–30. [PubMed] [Google Scholar]
- 35.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012; 122:787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yvan-Charvet L, Pagler TA, Seimon TA, Thorp E, Welch CL, Witztum JL, et al. ABCA1 and ABCG1 protect against oxidative stress-induced macrophage apoptosis during efferocytosis. Circ Res 2010; 106:1861–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wojcik AJ, Skaflen MD, Srinivasan S, Hedrick CC. A critical role for ABCG1 in macrophage inflammation and lung homeostasis. J Immunol 2008; 180:4273–82. [DOI] [PubMed] [Google Scholar]
- 38.Draijer C, Boorsma CE, Robbe P, Timens W, Hylkema MN, Ten Hacken NH, et al. Human asthma is characterized by more IRF5+ M1 and CD206+ M2 macrophages and less IL-10+ M2-like macrophages around airways compared with healthy airways. J Allergy Clin Immunol 2017; 140:280–3 e3. [DOI] [PubMed] [Google Scholar]
- 39.Gibson PG, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, et al. Effect of azithromycin on asthma exacerbations and quality of life in adults with persistent uncontrolled asthma (AMAZES): a randomised, double-blind, placebo-controlled trial. Lancet 2017; 390:659–68. [DOI] [PubMed] [Google Scholar]
- 40.Jaresova I, Rozkova D, Spisek R, Janda A, Brazova J, Sediva A. Kinetics of Toll-like receptor-4 splice variants expression in lipopolysaccharide-stimulated antigen presenting cells of healthy donors and patients with cystic fibrosis. Microbes Infect 2007; 9:1359–67. [DOI] [PubMed] [Google Scholar]
- 41.del Fresno C, Gomez-Pina V, Lores V, Soares-Schanoski A, Fernandez-Ruiz I, Rojo B, et al. Monocytes from cystic fibrosis patients are locked in an LPS tolerance state: down-regulation of TREM-1 as putative underlying mechanism. PLoS One 2008; 3:e2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.