Identification of factors associated with the size of the Human Immunodeficiency Virus reservoir remains a priority for HIV cure research. We report that the activity of Indoleamine 2,3-Dioxygenase, an immunoregulatory enzyme that metabolizes tryptophan to kynurenines, is associated with the size of the HIV reservoir.
Keywords: HIV reservoir; indoleamine 2,3-dioxygenase; kynurenine; microbial translocation; ART
Abstract
Background
Indoleamine 2,3-dioxygenase (IDO) is an immunoregulatory enzyme that metabolizes tryptophan to immunosuppressive kynurenines. We investigated whether IDO activity is associated with the size of HIV reservoir.
Methods
Total human immunodeficiency virus (HIV) DNA in peripheral blood mononuclear cells (PBMCs) from 127 HIV-infected patients receiving antiretroviral therapy (ART) was quantified. Tryptophan and kynurenine concentrations, as well as microbial translocation markers, were measured in plasma samples. T-cell activation and exhaustion in PBMCs were assessed by flow cytometry.
Results
Elevated IDO activity prior to ART correlated with on-ART HIV DNA (r = 0.35, P = .004), but was not associated with pre-ART HIV DNA. A median duration of 15 months of ART significantly decreased IDO activity; however, these levels were still higher than those observed in HIV-uninfected controls. Among treated participants, IDO activity positively correlated with their concurrent HIV DNA (r = 0.36, P < .0001). Multivariate model showed an independent association of pre-ART CD4/CD8 ratio (adjusted odds ratio [aOR], 0.75 per 0.1 increase [95% confidence interval {CI}, .62–.91]) and on-ART IDO activity (aOR, 1.09 per nM/μM increase [95% CI, 1.04–1.14]) with higher levels of HIV DNA on-ART. A lack of association of the microbial translocation markers was observed with the size of HIV reservoir. HIV DNA positively correlated with the proportions of activated CD4 T and CD8 T cells and exhausted CD4 T cells.
Conclusions
We observed a positive correlation between IDO activity and total HIV DNA in blood, highlighting the important role of immunometabolic aberrations in HIV persistence.
Human immunodeficiency virus (HIV) reservoirs in resting CD4 T cells represent the major obstacle to an HIV cure owing to their persistence on effective antiretroviral therapy (ART) [1–3]. The reservoir is established early in HIV infection, which allows it to undergo rapid viral replication upon ART interruption. Such viral rebound has also been witnessed in patients initiating ART at a very early stage [4, 5]. Furthermore, Siliciano et al have calculated that the reservoir is stable with an estimated half-life of 44 months and will require >70 years of ART to eradicate [6]. Following ART discontinuation, HIV rebounds within 4 weeks in majority of patients and such time to rebound can be predicted by the size of the HIV reservoir on ART [7, 8]. To develop a cure, identifying factors that influence the size of HIV reservoir remains a research priority [9].
We and others have reported that indoleamine 2,3-dioxygenase (IDO) activity is elevated during acute HIV infection and that such elevation persists during the chronic phase and is not normalized in patients receiving long-term suppressive ART [10–12]. IDO is a rate-limiting enzyme in the kynurenine pathway that is induced by interferon-γ. IDO metabolizes tryptophan to immunosuppressive kynurenines, and thereby is implicated in the pathogenesis of cancers and infectious diseases [13]. During HIV infection, elevated expression of IDO skews CD4 T-cell differentiation into regulatory T cells instead of T-helper 17 cells and directly impairs T-cell immune responses, thus contributing to HIV disease progression [14–16]. As an immune checkpoint, IDO and its downstream kynurenine pathway are increasingly receiving attention for being immunotherapeutic targets along with programmed cell death 1 (PD-1) blockade. Therefore, IDO activity may play a role in HIV persistence and deserves further investigation.
While most previous studies suggest that chronic inflammation and immune activation contribute to HIV persistence, there have also been conflicting reports [17–21]. Chronic inflammation might lead to HIV persistence by generating new target cells and increasing the proliferation of infected cells [22]. In HIV-infected individuals, viral replication and persistently elevated levels of inflammation and T-cell activation may be consequences of gut microbial translocation [23]. The pre-ART elevated IDO activity was also associated with plasma levels of microbial translocation markers [11]. Whether microbial translocation contributes to HIV persistence is not yet established. Several soluble markers of microbial translocation, such as gram-negative bacterial cell wall component lipopolysaccharide (LPS) and the host acute response proteins LPS-binding protein (LBP) and soluble CD14 (sCD14), have been studied mostly in cross-sectional studies that reported divergent results [21]. Therefore, more comprehensive study of markers of immune dysfunction and microbial translocation is warranted to determine their role in HIV persistence.
Herein, we quantified total HIV type 1 (HIV-1) DNA levels in relation to IDO activity, microbial translocation, immune activation, and T-cell exhaustion in a cohort of HIV-infected participants receiving first-line ART.
METHODS
Study Population and Sample Size
The study population consisted of 127 HIV-1–infected patients without any history of opportunistic infections or cancers, who initiated first-line ART for a median duration of 15 (interquartile range [IQR], 12–27) months. The study population did not include any subjects with known autoimmune disease and/or pregnant women. Whole blood samples (10 mL) were collected from all participants to quantify total HIV DNA. Pre-ART plasma samples were available in 64 of the participants, which were quantified together with their on-ART samples to investigate the changes in IDO activity and the markers of microbial translocation. In addition, 25 HIV-uninfected controls were studied as a comparison group for the levels of microbial translocation and IDO activity. Clinical records of these patients were reviewed for the past medical history and ART regimens. Ethics approval was obtained from the Shanghai Public Health Clinical Center Ethic Committee, China. Informed consent was obtained from all study participants.
Quantification of HIV RNA and CD4 and CD8 T-Cell Counts
CD4 and CD8 T-cell counts were assessed by flow cytometry (BD Biosciences, Franklin Lakes, New Jersey) and HIV-1 RNA loads measured by polymerase chain reaction (PCR) (Cobas Amplicor, Roche, Basel, Switzerland) at the clinical laboratory of the Shanghai Public Health Clinical Center.
Quantification of Tryptophan and Kynurenine Levels in Plasma as a Measure of IDO Activity
Plasma levels of tryptophan and kynurenine were quantified using ultra-performance liquid chromatography–mass spectrometry as previously described [24]. IDO activity was calculated as the plasma kynurenine/tryptophan ratio (K/T ratio).
Quantification of Soluble Markers of Microbial Translocation and Immune Activation
Plasma levels of markers of microbial translocation including LPS (CUSABIO Life Science, Wuhan, China), endogenous endotoxin-core antibody (EndoCAb) (Hycult Biotech, the Netherlands), and LBP (Hycult Biotech), sCD14 (R&D Systems, Minneapolis, Minnesota), and soluble CD163 (sCD163) (R&D Systems) were determined using enzyme-linked immunosorbent assays according to the manufacturers’ instructions.
Flow Cytometry
Peripheral blood mononuclear cells (PBMCs) were isolated and stained with the following antibodies: CD3 APC-H7, CD4 FITC, CD8 APC, CD38 PE-Cy7, HLA-DR PerCP-Cy5.5, PD-1 PE, and 7AAD PE-Cy5 (all from BD Biosciences). Cells were fixed in 1% paraformaldehyde and analyzed within 24 hours of staining. Data were analyzed using FlowJo software version 10 (FlowJo, LLC, Ashland, Oregon).
Determination of HIV DNA
HIV-1 DNA was amplified and quantified using a fluorescence-based, real-time HIV quantitative detection kit (SUPBIO, Guangzhou, China) after extraction of total cellular DNA from peripheral blood using a DNA isolation kit (Qiagen, Valencia, California). The quantification range of this assay was 10–5 × 106 copies/106 PBMCs. Levels of total HIV DNA below the lower limit of detection were deemed as 10 copies/106 PBMCs.
Statistical Analyses
The normality of the data was assessed using the Shapiro-Wilk test. Normally distributed data were described as the mean ± standard deviation while nonnormally distributed variables were described as median with IQR. For comparisons between baseline and follow-up samples, paired t test or Wilcoxon signed-rank test was performed depending on the distribution of the variable. Spearman rank correlation test was used to measure the association between HIV DNA and other variables. All the associations were corrected for false discovery rate <0.05 using the Benjamini-Hochberg method (reported as q value). HIV DNA levels were categorized into lower HIV DNA group and higher HIV DNA group using the median as a cutoff. Binary logistic regression was used to determine the independent association of clinical covariates with HIV DNA. All analyses were performed using Stata version 12.0 (StataCorp, College Station, Texas) and GraphPad Prism 7.0 (GraphPad Software, La Jolla, California) software.
RESULTS
Demographic and Clinical Characteristics of the Study Participants
Among the 127 enrolled HIV-infected patients, 92.1% (117/127) were male, with a median age of 32 (IQR, 27–44) years (Table 1). The majority of the participants received 2 nucleoside reverse transcriptase inhibitors plus 1 nonnucleoside reverse transcriptase inhibitor–based regimen. HIV RNA was below the limit of detection (<20 copies/mL) in all the participants after a median of 15 (IQR, 12–27) months of ART. The on-ART total HIV DNA level in this cohort was 2.27 (IQR, 1.92–2.62) log10 copies/106 PBMCs.
Table 1.
Study Participants
| Characteristics | Total (N = 127) |
|---|---|
| Age at ART initiation, y, median (IQR) | 32 (27–44) |
| Male sex, No. (%) | 117 (92.1) |
| Pre-ART CD4 T-cell count, cells/μL, median (IQR) | 348 (258–459) |
| Pre-ART CD4/CD8 ratio, median (IQR) | 0.36 (0.25–0.49) |
| Pre-ART HIV RNAa, log10 copies/mL, median (IQR) | 4.61 (4.15–5.05) |
| ART regimen, No. (%) | |
| TDF + 3TC + EFV | 116 (91.3) |
| ZDV + 3TC + EFV/NVP | 7 (5.5) |
| TDF + 3TC + RAL | 2 (1.6) |
| TDF + 3TC + LPV/r | 2 (1.6) |
| ART duration, mo, median (IQR) | 15 (12–27) |
| On-ART CD4 T-cell count, cells/μL, median (IQR) | 403 (332–560) |
| On-ART CD4/CD8 ratio, median (IQR) | 0.62 (0.45–0.87) |
Abbreviations: 3TC, lamivudine; ART, antiretroviral therapy, EFV, efavirenz; HIV, human immunodeficiency virus; IQR, interquartile range; LPV/r, lopinavir/ritonavir; NVP, nevirapine; RAL, raltegravir; TDF, tenofovir disoproxil fumarate; ZDV, zidovudine.
aData from 99 participants.
Pre-ART CD4/CD8 Ratio Predicted On-ART HIV DNA Level
The age and sex did not influence the size of the HIV reservoir on-ART (Table 2). Pre-ART HIV RNA was marginally correlated with on-ART HIV DNA (r = 0.19, P = .059). Pre-ART CD4 T-cell counts and pre-ART CD4/CD8 ratio inversely predicted, while pre-ART CD8 T-cell counts positively predicted, on-ART HIV DNA (r = –0.27, P = .003; r = –0.41, P < .0001; and r = 0.22, P = .013, respectively). On-ART CD4 T-cell counts and CD4/CD8 ratio increased substantially, the latter of which was associated with the duration of ART. As expected, on-ART HIV DNA inversely correlated with the duration of ART (r = –0.38, P < .0001). In addition, on-ART CD8 T-cell counts and on-ART CD4/CD8 ratio correlated with their concurrent HIV DNA levels (r = 0.32, P = .0003 and r = –0.41, P < .0001, respectively). After adjusting for false discovery rate, all of these factors were still significantly associated with on-ART HIV DNA.
Table 2.
False Discovery Rate Adjusted P Value (q Value) for Each Association Calculated
| Characteristic | P Value | q Value |
|---|---|---|
| Association with on-ART HIV DNA | ||
| Age | .996 | .798 |
| Sex | .783 | .681 |
| Pre-ART HIV RNA | .059 | .112 |
| Pre-ART CD4 T-cell count | .003 | .011 |
| Pre-ART CD8 T-cell count | .013 | .040 |
| Pre-ART CD4/CD8 | .000002 | .00003 |
| Duration of ART | .00001 | .0001 |
| On-ART CD4 T-cell count | .039 | .079 |
| On-ART CD8 T-cell count | .0003 | .002 |
| On-ART CD4/CD8 | .000001 | .00003 |
| Pre-ART IDO activity | .004 | .016 |
| On-ART IDO activity | .00003 | .0003 |
| On-ART IDO activity (CD4 T-cell count <200 cells/μL) | .011 | .037 |
| Pre-ART sCD14 | .556 | .546 |
| Pre-ART LPS | .537 | .546 |
| Pre-ART LBP | .897 | .738 |
| Pre-ART EndoCAb | .546 | .546 |
| Pre-ART sCD163 | .398 | .520 |
| On-ART sCD14 | .348 | .481 |
| On-ART LPS | .461 | .520 |
| On-ART LBP | .783 | .681 |
| On-ART EndoCAb | .897 | .738 |
| On-ART sCD163 | .033 | .072 |
| Percentage of HLA-DR+CD38+ CD4 | .028 | .065 |
| Percentage of HLA-DR+CD38+ CD8 | .026 | .065 |
| Percentage of PD-1+ CD4 | .026 | .065 |
| Percentage of PD-1+ CD8 | .455 | .520 |
| Association with pre-ART IDO activity | ||
| Pre-ART HIV DNA | .601 | .571 |
| Pre-ART HIV RNA | .0002 | .001 |
| Association with on-ART IDO activity | ||
| On-ART sCD14 | .263 | .397 |
| On-ART LPS | .429 | .520 |
| On-ART LBP | .457 | .520 |
| On-ART EndoCAb | .757 | .681 |
| On-ART sCD163 | .274 | .397 |
| Percentage of HLA-DR+CD38+ CD4 | .211 | .337 |
| Percentage of HLA-DR+CD38+ CD8 | .519 | .546 |
| Percentage of PD-1+ CD4 | .083 | .148 |
| Percentage of PD-1+ CD8 | .116 | .196 |
Data in bold indicate significant after adjusted for false discovery rate. False discovery rate <0.05.
Abbreviations: ART, antiretroviral therapy; EndoCAb, endogenous endotoxin-core antibody; HIV, human immunodeficiency virus; HLA-DR, human leukocyte antigen – DR isotype; IDO, indoleamine 2,3-dioxygenase; LBP, lipopolysaccharide-binding protein; LPS, lipopolysaccharide; PD-1, programmed cell death 1; sCD14, soluble CD14; sCD163, soluble CD163.
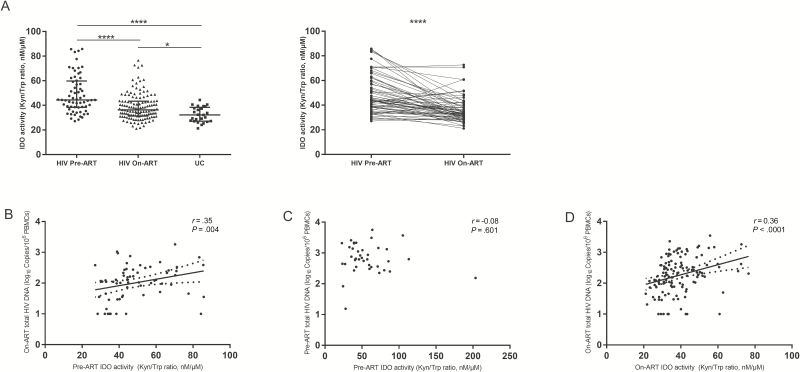
IDO Activity Was Correlated With On-ART HIV DNA
IDO activity was significantly elevated in HIV-infected patients before ART as compared to HIV-uninfected controls (Figure 1A). The elevated pre-ART IDO activity positively correlated with pre-ART HIV RNA (r = 0.46, P = .0002) and positively predicted on-ART HIV DNA level (r = 0.35, P = .004; Figure 1B and Table 2). To explore the association between pre-ART IDO activity and pre-ART HIV DNA, another 41 patients with plasma and PBMCs collected at the time of ART initiation were studied (Supplementary Table 1). Strikingly, pre-ART IDO activity was not associated with pre-ART HIV DNA in this group (r = –0.08, P = .601; Figure 1C).
Figure 1.
Indoleamine 2,3-dioxygenase (IDO) activity and its association with total human immunodeficiency virus (HIV) DNA. A, Elevated IDO activity in HIV-infected patients and longitudinal changes in IDO activity pre–antiretroviral therapy (ART) and on-ART. Pre-ART IDO activity predicted on-ART HIV DNA (B) but was not associated with total pre-ART HIV DNA (C). On-ART IDO activity was positively correlated with on-ART HIV DNA (D). *P < .05, ****P < .0001. Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus; IDO, indoleamine 2,3-dioxygenase; Kyn/Trp, kynurenine/tryptophan; PBMC, peripheral blood mononuclear cell; UC, human immunodeficiency virus–uninfected controls.
IDO activity decreased significantly on ART. However, IDO activity in HIV-infected participants was still higher than that in HIV-uninfected controls. On-ART IDO activity was not associated with any of the concurrent microbial translocation markers (data not shown). The on-ART IDO activity strongly correlated with on-ART HIV DNA (r = 0.36, P < .0001; Figure 1D). The correlation was stronger among participants with pre-ART CD4 T-cell counts <200 cells/μL (r = 0.51, P = .011; Table 2). After adjustment for other factors that were associated with on-ART HIV DNA, low pre-ART CD4/CD8 ratio and high on-ART IDO activity were independently associated with high on-ART HIV DNA (higher than the median level) in a multivariate binary logistic regression model (adjusted odds ratio [aOR], 0.75 per 0.1 increase [95% confidence interval {CI}, .62–.91]; aOR, 1.09 per nM/μM increase [95% CI, 1.04–1.14], respectively; Table 3).
Table 3.
Factors Associated With High Total Human Immunodeficiency Virus DNA on Antiretroviral Therapy
| Factor | Univariate Logistic Regression | Multivariate Logistic Regression | ||||
|---|---|---|---|---|---|---|
| OR | (95% CI) | P Value | OR | (95% CI) | P Value | |
| Agea | 1.00 | (.97–1.03) | .994 | |||
| Male sex | 1.02 | (.28–3.70) | .979 | |||
| CD4 T-cell countb | 0.74 | (.59–.93) | .011 | |||
| CD8 T-cell countb | 1.07 | (.99–1.16) | .075 | |||
| CD4/CD8 ratioc | 0.75 | (.63–.90) | .002 | 0.75 | (.62–.91) | .003 |
| HIV RNAd | 1.46 | (.80–2.69) | .215 | |||
| ART duratione | 0.92 | (.88–.97) | .001 | |||
| On-ART CD4 T-cell countb | 0.77 | (.62–.95) | .015 | |||
| On-ART CD8 T-cell countb | 1.17 | (1.04–1.32) | .011 | |||
| On-ART CD4/CD8 ratioc | 0.80 | (.71–.91) | .001 | |||
| On-ART IDO activityf | 1.09 | (1.04–1.13) | <.0001 | 1.09 | (1.04–1.14) | <.0001 |
Abbreviations: ART, antiretroviral therapy; CI, confidence interval; HIV, human immunodeficiency virus; IDO, indoleamine 2,3-dioxygenase; OR, odds ratio.
aShown per 1 year increase.
bShown per 100 cells/μL increase.
cShown per 0.1 increase.
dShown per 1 log10 copies/mL increase, data from 99 patients.
eShown per 1 month increase.
fShown per 1 nM/μM increase.
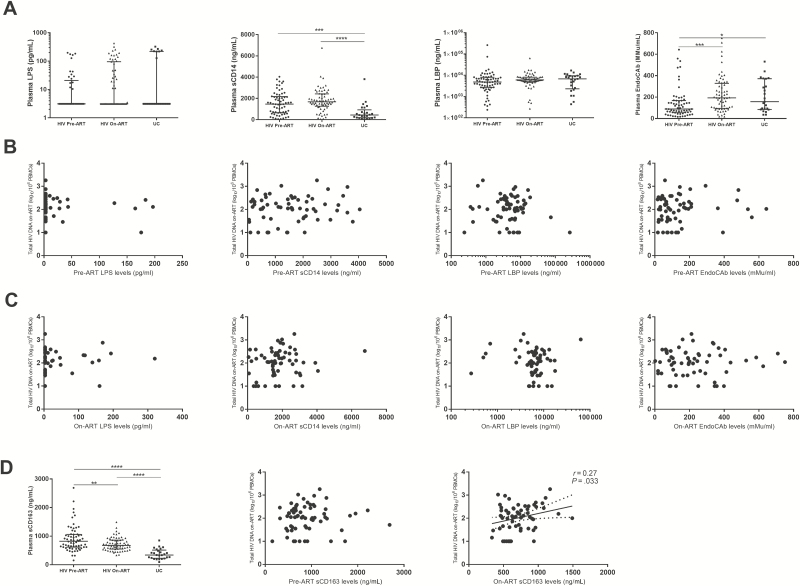
Microbial Translocation Persists on ART but Was Not Associated With HIV Reservoir Size
Plasma levels of sCD14 were elevated while LPS and LBP were comparable and EndoCAb was lower among untreated participants compared with HIV-uninfected controls (Figure 2A and Table 2). However, none of these markers predicted on-ART HIV DNA (Figure 2B). After effective ART, plasma EndoCAb increased to a level similar to that of HIV-uninfected controls. Levels of on-ART plasma sCD14 were still higher than those in HIV-uninfected controls. There was no correlation between on-ART HIV DNA and any of these plasma markers among treated participants (Figure 2C).
Figure 2.
No association between microbial translocation and human immunodeficiency virus (HIV) persistence. A, Changes of microbial translocation markers. B, Pre– antiretroviral therapy (ART) levels of microbial translocation could not predict on-ART total HIV DNA. C, Microbial translocation markers were not associated with on-ART total HIV DNA. D, On-ART but not pre-ART sCD163 was associated with on-ART total HIV DNA. Microbial translocation markers were quantified in 64 patients with paired samples available both before and after ART. *P < .05, **P < .01, ***P < .001, ****P < .0001. Abbreviations: ART, antiretroviral therapy; EndoCAb, endogenous endotoxin-core antibody; HIV, human immunodeficiency virus; LBP, lipopolysaccharide-binding protein; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cell; sCD14, soluble CD14; sCD163, soluble CD163; UC, human immunodeficiency virus–uninfected controls.
Plasma levels of sCD163 were elevated in untreated participants, and decreased but did not normalize on ART (Figure 2D). The pre-ART level of plasma sCD163 was not associated with on-ART HIV DNA, whereas the on-ART level of sCD163 almost positively correlated with HIV DNA (r = 0.11, P = .398 and r = 0.27, P = .033, q = .072, respectively; Figure 2D).
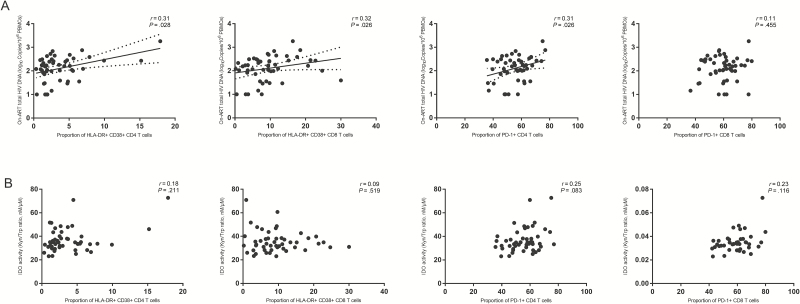
T-Cell Activation and CD4 T-Cell Exhaustion Were Positively Correlated With HIV DNA
The proportions of HLA-DR+ CD38+ cells in both CD4 and CD8 T-cell compartments marginally correlated with total HIV DNA levels in treated participants (r = 0.31, P = .028, q = .065 and r = 0.32, P = .026, q = .065, respectively; Figure 3A). In addition, the proportion of PD-1–expressing CD4 T cells but not PD-1+ CD8 T cells marginally significantly correlated with HIV DNA levels (r = 0.31, P = .026, q = .065 and r = 0 .11, P = .455, respectively; Figure 3A). Proportions of HLA-DR+CD38+ CD4 T cells, HLA-DR+CD38+ CD8 T cells, PD-1+ CD4 T cells, and PD-1+ CD8 T cells did not correlate with concurrent IDO activity (Figure 3B).
Figure 3.
T-cell activation and exhaustion were associated with human immunodeficiency virus (HIV) persistence (A) but not indoleamine 2,3-dioxygenase activity (B). HLA-DR, CD38, and PD-1 were assessed in samples from 50 of the participants. Abbreviations: ART, antiretroviral therapy; CD38, cluster of differentiation 38; HIV, human immunodeficiency virus; HLA-DR, human leukocyte antigen – DR isotype; IDO, indoleamine 2,3-dioxygenase; Kyn/Trp, kynurenine/tryptophan; PBMC, peripheral blood mononuclear cell; PD-1, programmed cell death 1.
DISCUSSION
Identification of factors that are linked with the size of HIV reservoir is essential to HIV eradication. In the current study, we assessed associations between IDO activity, microbial translocation, immune activation, T-cell exhaustion, and total HIV DNA in peripheral blood in a cohort of well-treated HIV-infected patients. We observed a positive correlation of total HIV DNA with IDO activity, with immune activation as well as with T-cell exhaustion, but not with microbial translocation.
To the best of our knowledge, this is the first study to report a correlation between IDO activity and total HIV DNA. During HIV infection, IDO activity has been attributed to pathogenesis. Elevated IDO activity has been associated with HIV disease progression, Kaposi sarcoma, active tuberculosis, poor CD4 T-cell recovery, cardiovascular disease, HIV-associated neurocognitive disorder, and mortality [12, 16, 25]. Alongside the persistence of HIV reservoirs on long-term suppressive ART, the elevated IDO activity is also not normalized even after >7 years of ART [11, 12, 14]. Interestingly, IDO activity was associated with total HIV DNA in our study and not with microbial translocation and immune activation. Along these lines, it is reasonable to speculate that the elevated IDO activity is driven by the persistent HIV infection or vice versa, suggesting it may be one of the strategies that HIV uses to escape eradication. Clonal expansion of the latently infected memory CD4 T cells, which can be driven by foreign antigens and cytokines, have been attributed to HIV persistence [19]. Recent studies suggested that clonal expansion may be driven by antigens from tumors and pathogens (eg, cytomegalovirus and Epstein-Barr virus) [26, 27]. Given the immunosuppressive role of IDO, elevated IDO activity may impede the clearance of these antigens, which in turn expand the HIV reservoir [28]. Although it is still controversial, another mechanism for HIV persistence involves ongoing low-level viral replication on ART, especially in anatomical sites such as lymph nodes where drug concentrations are insufficient to completely suppress HIV replication [1, 29, 30]. Owing to its immunosuppressive role, it is also possible that the up-regulated IDO activity in these sites facilitates HIV replication. In fact, IDO and its downstream kynurenine pathway play a critical role in maintaining immune privilege in brain, eyes, and testes, which are also sites of HIV persistence during suppressive ART [28]. Testes from HIV-infected patients show elevated IDO activity with an increase in the frequency of regulatory T cells as compared to blood, which may facilitate HIV persistence [31]. In an animal model of HIV-1 encephalitis, inhibition of IDO has been shown to enhance elimination of virus-infected macrophages [32]. Combined ART and blockade of IDO has also been shown to reduce the viral load significantly in simian immunodeficiency virus–infected rhesus macaques with unsuccessful ART [33]. Taken together, IDO activity may be involved in HIV persistence and might be a potential immunotherapeutic target to reduce HIV reservoir size. Therefore, further research, both in vitro and in vivo, needs to be done to examine the effects of IDO inhibitors on reducing the size of HIV reservoir.
In contrast to on-ART IDO activity, no association between pre-ART IDO activity and pre-ART total HIV DNA was observed. This can be explained by factors that influence IDO activity, for example, pre-ART microbial translocation and HIV RNA levels. However, it may also be attributed to the different distribution of the integrated HIV, as HIV DNA is more likely to be detected in active CD4 T cells in untreated participants and more in resting T cells after ART initiation [1].
Expression of PD-1 on T cells has been previously associated with HIV persistence on ART, which we also observed in this study [17, 19, 34]. PD-1 is an immune checkpoint marker that suppresses immune responses, high expression of which on T cells represents an immunomodulatory microenvironment that may contribute to HIV persistence. Our study did not find evidence to support this mechanism as no significant association between proportion of PD-1+ CD8 T cells and total HIV DNA was observed, which is in line with previous studies that assessed blood and gut tissues [18, 35]. However, a recent study reported correlation between CD8 T-cell exhaustion and integrated HIV DNA in lymph nodes [35]. Blockade of PD-1 also leads to reversal of HIV latency [36]. On the other hand, PD-1 itself could be a potential surrogate marker of HIV reservoir. In HIV-infected individuals, central and transitional memory CD4 T cells that express high levels of PD-1 have higher HIV DNA than those cells that express low levels of PD-1 [19, 34]. In lymph nodes, CD4 T cells expressing PD-1, which are mainly T follicular helper cells, are the major source of replication-competent HIV-1 [37]. In this context, blocking PD-1 might be a promising strategy to decrease HIV reservoir size. Interestingly, a case with drastic and sustained decrease of the HIV reservoir under anti–PD-1 therapy has been recently reported, although its reproducibility needs to be established [38].
In our study, the proportion of PD-1–expressing T cells was not associated with IDO activity. Interestingly, a landmark study recently revealed that increased IDO activity in tumor-repopulating cells could induce PD-1 expression on T cells [39]. Whether this strategy is adopted by HIV in the microenvironment still needs to be studied. If that is the case, a combination of anti–PD-1/PD-L1 and IDO inhibitors may be more effective than anti–PD-1/PD-L1 alone in eradicating HIV reservoirs. This combination strategy is promising in treating cancers, although a recent clinical trial failed to report additive effects of IDO inhibitor among patients with melanoma receiving anti–PD-1 therapy [36, 40].
Several groups have demonstrated the positive correlation between immune activation and HIV persistence, while others reported the lack of an association [17, 18, 20, 21, 35]. Such inconsistent results may be explained by differences in the study population with variable duration of ART, distinct sites of sample collection, and different methods for HIV quantification used. We observed that persistent microbial translocation after ART does not correlate, or at least does not directly contribute to HIV persistence, as we failed to find any association between microbial translocation markers and total HIV DNA.
Our results need to be interpreted with caution owing to some study limitations. First, we quantified HIV persistence and tryptophan and kynurenine concentrations in peripheral blood, which may not represent the microenvironments in tissues, where a significant portion of HIV reservoir is located. Another limitation is the use of PCR-based methods to measure HIV persistence, which could overestimate the HIV reservoir size as most of the viral genomes detected are defective or replication incompetent [2]. Third, IDO activity was measured at pre-ART and on-ART time points in 64 of 127 participants owing to the limited availability of samples. Finally, our study did not enroll patients with long-term (eg, >10 years) successive ART. The decay slopes of HIV reservoir among patients on variable durations of ART are different, which is greatest during the first 4 years of ART [21]. Additional studies of populations with such attributes are therefore required to increase the generalizability of the findings.
In summary, our study found a positive correlation between IDO activity and total HIV DNA in blood, suggesting its involvement in HIV persistence. Further studies are warranted to clarify underlying mechanisms and explore whether interventions targeting this pathway may help reduce HIV reservoir size.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all of the participants in this study. We also thank Jiangrong Wang, Wei Song, Yang Tang, Dan Yin, Rong Chen, Qi Tang, Jiadang Gu, Haijuan Wu, and Dongdong Luo for their coordination.
Financial support. This study was sponsored by the National Natural Science Foundation of China (grant numbers 81571977 and 81301420) and the Shanghai Municipal Commission of Health and Family Planning (grant numbers 20164Y0015 and 15GWZK0103).
Potential conflicts of interest. H. L. has received grants from the National Natural Science Foundation of China and the Shanghai Municipal Commission of Health and Family Planning. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Rosenbloom DIS, Hill AL, Laskey SB, Siliciano RF. Re-evaluating evolution in the HIV reservoir. Nature 2017; 551:E6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ho YC, Shan L, Hosmane NN, et al. . Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finzi D, Hermankova M, Pierson T, et al. . Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 4. Henrich TJ, Hatano H, Bacon O, et al. . HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: an observational study. PLoS Med 2017; 14:e1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colby DJ, Trautmann L, Pinyakorn S, et al. . Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 2018; 24: 923–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siliciano JD, Kajdas J, Finzi D, et al. . Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 7. Li JZ, Etemad B, Ahmed H, et al. . The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 2016; 30:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yerly S, Günthard HF, Fagard C, et al. . Swiss HIV Cohort Study Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 2004; 18:1951–3. [DOI] [PubMed] [Google Scholar]
- 9. Deeks SG, Lewin SR, Ross AL, et al. . International AIDS Society Towards a Cure Working Group International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 2016; 22:839–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jenabian MA, El-Far M, Vyboh K, et al. . Montreal Primary Infection and Slow Progressor Study Groups Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J Infect Dis 2015; 212:355–66. [DOI] [PubMed] [Google Scholar]
- 11. Chen J, Shao J, Cai R, et al. . Anti-retroviral therapy decreases but does not normalize indoleamine 2,3-dioxygenase activity in HIV-infected patients. PLoS One 2014; 9:e100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi Q, Hua S, Clish CB, et al. . Plasma tryptophan-kynurenine metabolites are altered in HIV infection and associated with progression of carotid artery atherosclerosis. Clin Infect Dis 2018; 67:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katz JB, Muller AJ, Prendergast GC. Indoleamine 2,3-dioxygenase in T-cell tolerance and tumoral immune escape. Immunol Rev 2008; 222:206–21. [DOI] [PubMed] [Google Scholar]
- 14. Jenabian MA, Patel M, Kema I, et al. . Distinct tryptophan catabolism and Th17/Treg balance in HIV progressors and elite controllers. PLoS One 2013; 8:e78146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Favre D, Mold J, Hunt PW, et al. . Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2010; 2:32ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byakwaga H, Boum Y 2nd, Huang Y, et al. . The kynurenine pathway of tryptophan catabolism, CD4+ T-cell recovery, and mortality among HIV-infected Ugandans initiating antiretroviral therapy. J Infect Dis 2014; 210:383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatano H, Jain V, Hunt PW, et al. . Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis 2013; 208:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cockerham LR, Siliciano JD, Sinclair E, et al. . CD4+ and CD8+ T cell activation are associated with HIV DNA in resting CD4+ T cells. PLoS One 2014; 9:e110731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chomont N, El-Far M, Ancuta P, et al. . HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Besson GJ, Lalama CM, Bosch RJ, et al. . HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 2014; 59:1312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gandhi RT, McMahon DK, Bosch RJ, et al. . ACTG A5321 Team Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13:e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev 2013; 254:326–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brenchley JM, Price DA, Schacker TW, et al. . Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12:1365–71. [DOI] [PubMed] [Google Scholar]
- 24. Cheng J, Jin H, Hou X, Lv J, Gao X, Zheng G. Disturbed tryptophan metabolism correlating to progression and metastasis of esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2017; 486:781–7. [DOI] [PubMed] [Google Scholar]
- 25. Hunt PW, Sinclair E, Rodriguez B, et al. . Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 2014; 210:1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simonetti FR, Sobolewski MD, Fyne E, et al. . Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016; 113:1883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henrich TJ, Hobbs KS, Hanhauser E, et al. . Human immunodeficiency virus type 1 persistence following systemic chemotherapy for malignancy. J Infect Dis 2017; 216:254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Routy JP, Routy B, Graziani GM, Mehraj V. The kynurenine pathway is a double-edged sword in immune-privileged sites and in cancer: implications for immunotherapy. Int J Tryptophan Res 2016; 9:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palmer S, Maldarelli F, Wiegand A, et al. . Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105:3879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lorenzo-Redondo R, Fryer HR, Bedford T, et al. . Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016; 530:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jenabian MA, Costiniuk CT, Mehraj V, et al. . Orchid Study Group Immune tolerance properties of the testicular tissue as a viral sanctuary site in ART-treated HIV-infected adults. AIDS 2016; 30:2777–86. [DOI] [PubMed] [Google Scholar]
- 32. Potula R, Poluektova L, Knipe B, et al. . Inhibition of indoleamine 2,3-dioxygenase (IDO) enhances elimination of virus-infected macrophages in an animal model of HIV-1 encephalitis. Blood 2005; 106:2382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boasso A, Vaccari M, Fuchs D, et al. . Combined effect of antiretroviral therapy and blockade of IDO in SIV-infected rhesus macaques. J Immunol 2009; 182:4313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fromentin R, Bakeman W, Lawani MB, et al. . CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 2016; 12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khoury G, Fromentin R, Solomon A, et al. . Human immunodeficiency virus persistence and T-cell activation in blood, rectal, and lymph node tissue in human immunodeficiency virus-infected individuals receiving suppressive antiretroviral therapy. J Infect Dis 2017; 215:911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evans VA, van der Sluis RM, Solomon A, et al. . Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 2018; 32:1491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Banga R, Procopio FA, Noto A, et al. . PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016; 22:754–61. [DOI] [PubMed] [Google Scholar]
- 38. Guihot A, Marcelin AG, Massiani MA, et al. . Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann Oncol 2018; 29:517–8. [DOI] [PubMed] [Google Scholar]
- 39. Liu Y, Liang X, Dong W, et al. . Tumor-repopulating cells induce PD-1 expression in CD8(+) T cells by transferring kynurenine and AhR activation. Cancer Cell 2018; 33: 480–94.e7. [DOI] [PubMed] [Google Scholar]
- 40. Long GV, Dummer R, Hamid O, et al. . Epacadostat (E) plus pembrolizumab (P) versus pembrolizumab alone in patients (pts) with unresectable or metastatic melanoma: Results of the phase 3 ECHO-301/KEYNOTE-252 study. J Clin Oncol 2018; 36(15 Suppl):108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.