Abstract
Purpose
To make the artificial intelligence (AI) classifiers of the image of the blastocyst implanted later in order to predict the probability of achieving live birth.
Methods
A system for using the machine learning approaches, which are logistic regression, naive Bayes, nearest neighbors, random forest, neural network, and support vector machine, of artificial intelligence to predict the probability of live birth from a blastocyst image was developed. Eighty images of blastocysts that led to live births and 80 images of blastocysts that led to aneuploid miscarriages were used to create an AI‐based method with 5‐fold cross‐validation retrospectively for classifying embryos.
Results
The logistic regression method showed the best results. The accuracy, sensitivity, specificity, positive predictive value, and negative predictive value were 0.65, 0.60, 0.70, 0.67, and 0.64, respectively. Area under the curve was 0.65 ± 0.04 (mean ± SE). Estimated probability of belonging to the live birth category was found significantly related to the probability of live birth (P < 0.005).
Conclusions
Classifiers using artificial intelligence applied toward a blastocyst image have a potential to show the probability of live birth being the outcome.
Keywords: artificial intelligence, blastocyst, live birth, machine learning
1. INTRODUCTION
Securing live birth with no chromosomal abnormality is considered to be the preferable ultimate goal in assisted reproductive technology. Failure of embryo development, or miscarriage, results in loss of time and cost. Moreover, in order to avoid ongoing aneuploid gestations, it is preferable to distinguish euploid embryos from aneuploid or mosaic embryos when possible. Reliable selection of embryos prior to embryo transfer has been investigated.
Morphological structures such as meiotic spindles, zona pellucidae, vacuoles or refractile bodies, and polar body shapes have been investigated, but none of these features have been conclusively assessed as having prognostic value for the further developmental competence of oocytes.1 Conventional morphological evaluation has had limited success at identifying aneuploid embryos .2, 3, 4, 5, 6 Several observational studies have proposed time‐lapse parameters as predictive of aneuploidy, though these have had diverging conclusions. Researchers have generally concluded that aneuploidy is reflected in cell cycle parameters up to day 2 of development because euploid embryos have been found to display more tightly clustered timings compared with aneuploid embryos .2, 7, 8, 9 However, it is well‐documented that embryos of good morphological quality may well be aneuploid and suboptimal embryos may be euploid .2, 10, 11 The morphological classification of aneuploidy or euploid, however, is not established. Strict evidence may still be too weak to justify introducing time lapse in routine clinical settings .2
Preimplantation genetic testing for aneuploidy (PGT‐A) 12, 13 is another method for examining chromosomal profiles. Performed on a small embryo biopsy prior to transfer, PGT‐A is an invasive technique for the embryo, which brings with it considerable ethical arguments. Transfer of the embryo after the biopsy is prohibited in some countries, including Japan. Moreover, the chromosomal profiles are not always uniform and differ at the blastocyst sites. The chromosomal profile of the biopsy specimen does not always represent the profile of the rest of the embryo because of this genetic heterogeneity. A mosaicism in the trophectoderm (TE) is observed much more than has been appreciated, and a single TE biopsy apparently may not be representative of the complete TE .14 A global Internet‐based survey indicated more randomized controlled trials are needed to legitimize use of PGT‐A .15
There is now clear need for a means of noninvasively predicting live birth of euploid or not. We therefore created a system applying the machine learning approach of artificial intelligence (AI) and applied it to blastocyst images to seek a solution for this challenge as a pilot study. Our image analysis via AI was able to detect and recognize information that the conventional methods could not. We studied comparisons of six types of machine learning methods on blastocyst images and then made a classifier program that retrospectively shows the probability of live birth. Confidence score that is the estimated probability of belonging to the live birth category can be viewed in terms of ranking of blastocysts; thus, it will be easier for embryologist to select superior blastocysts for transfer. Herein, we present the feasibility of using the classifier in our system for predicting the probability of live birth.
2. MATERIAL AND METHODS
2.1. Blastocyst images
This study used fully de‐identified data and was approved by the Institutional Review Board (IRB) at Okayama Couples’ Clinic (IRB no. 18000128‐04). This study carried out with explanation to the patients and a Web site with additional information and including an opt‐out option was set up for the study. After blastocyst formation followed by fertilization from May 2008 to January 2017 in Okayama Couples’ Clinic, an image of the incubated blastocyst was captured approximately 115 hours after insemination and saved on a USB storage device in JPEG format containing no data that could be used to identify the individual. The de‐identified image data were transferred to the AI system off‐line. Eighty images of blastocysts that led to live births and 80 images of blastocysts that led to aneuploid miscarriages were used to create a machine learning based method for classifying embryos. These were respectively defined as in the normal category or abortion category. In other words, blastocysts in the normal category in this study would become live birth with unknown chromosomal status and those in the abortion category would result in abortion due to chromosomal abnormalities confirmed with genetic testing of chorionic villus samples. Then, this algorithm was applied to a random sampling of embryos that led either to live birth or to aneuploid miscarriage.
2.2. Preparation for AI
All de‐identified images stored off‐line were transferred to our AI‐based system. Each image that had the artifacts outside the blastocyst deleted to the greatest possible extent was cropped as a square and then saved in 100 × 100 pixel size. One‐fifth of images from each category were randomly selected as test dataset and the rest used as training dataset. In this way, the test dataset could potentially be used for validation, as the training dataset contained no test data. The number of training dataset items could be augmented as often seen in computer science because the blastocyst image processing of the arbitrary degrees’ rotation led to being in the same category of the different vector data. For example, if an image was rotated at intervals of 90 degrees, four images with different vectors were generated.
2.3. AI classifier
We developed a classifier program using machine learning with L2regularization 16, 17 to categorize blastocyst images as either in the normal or abortion category and to obtain the mathematical probability for predicting normal category (as well as abnormal category). The analysis used supervised machine learning methods: logistic regression ,18 naive Bayes ,19 nearest neighbors ,20 random forest ,21 support vector machine ,22 and neural network 23 with automatic option for hyper‐parameters to find best classifier in each method. The automatic option of the neural network included activation function to use between layers, the parameter multiplying the L2 term, the type of network module to use, the depth of the network, the total number of the network's trainable parameters, and the optimization method such as ordinary stochastic gradient descent with momentum or stochastic gradient descent using an adaptive learning rate that is invariant to diagonal rescaling of the gradients. We applied cross‐validation ,24, 25, 26 a powerful method for model selection, to identify the optimal method of machine learning. The suitable number of images for the training data was investigated by evaluating accuracy and variances using the 5‐fold cross‐validation method, in which test data were analyzed using the training data, as follows. First, the test data were the initial one‐fifth of the images of each category's collection. Then, the test data were changed to the next one‐fifth of the images. This procedure was repeated five times to encompass all images as potential test data. The number of augmented training images was analyzed until the accuracy and variance were likely to show the maximum and minimum value, respectively (Figure 1). When the optimal number of training data was obtained, the histogram of the probability for prediction was investigated.
Figure 1.
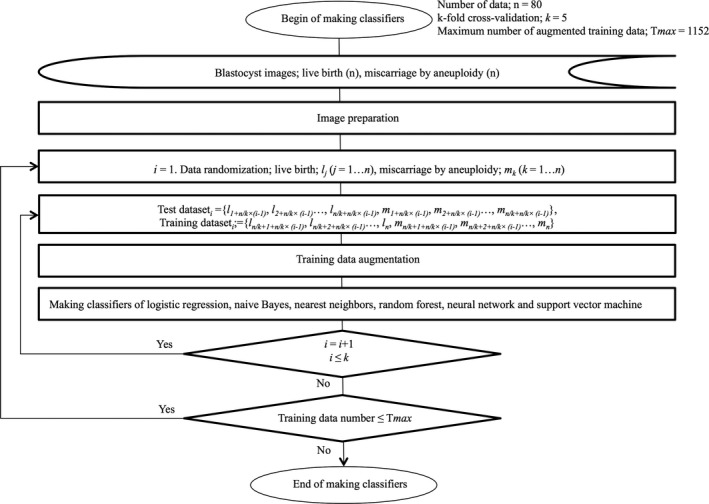
The flow chart of making classifiers for blastocysts that were implanted and led to live birth or aneuploid miscarriages
2.4. Development environment
The development environment used in the present study was as follows: a Mac (CPU; 2 x 2.8 GHz Quad‐Core Intel Xeon, RAM; 20 GB) running OS X 10.11.6 (Apple, Inc, Cupertino, CA, USA); Mathematica 11.3.0.0 (Wolfram Research, Champaign, IL).
2.5. Statistics
Mathematica 11.3.0.0 (Wolfram Research, Champaign, IL, USA) was used for statistical analysis.
3. RESULTS
3.1. Logistic regression as machine learning
The preliminary calculations revealed logistic regression was the best among the abovementioned machine learning methods. The accuracy by six types of machine learning methods (logistic regression, naive Bayes, nearest neighbors, random forest, neural network, and support vector machine) is shown in Figure 2, respectively. As the number of the training data became larger, the average of the accuracies showed the maximum and the standard deviation (SD) of the accuracies showed the minimum simultaneously in each method. The best accuracy of all with small SD was 0.650 ± 0.075 (mean ± SD) when the number of the training data was 640 in logistic regression. Therefore, the logistic regression should be selected as the best method among the machine learning methods and the results for both accuracy and SD suggested 640 as the suitable number of training images and selected as the number for this study. Consequently, further investigations were carried out with logistic regression.
Figure 2.
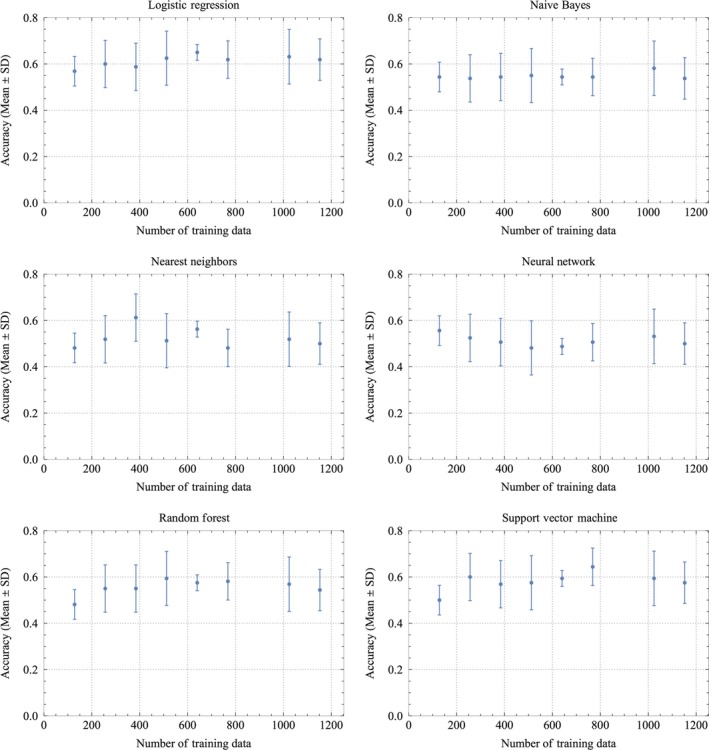
The relationship of the number of training files and accuracy in regard to resulting in live birth or abortion by machine learning with logistic regression method, naive Bayes method, nearest neighbors method, neural network method, random forest method, and support vector machine method as shown in each panels. Mean ± SD of accuracy at the number of the training data is shown in each panel. Accuracy is likely to approach a maximum of 0.65 with the minimum of the standard deviation of 0.075 when there are 640 files in the logistic regression method
3.2. AI classifier
Table 1 shows the results of the best classifier. The accuracy, sensitivity, specificity, positive predictive value, and negative predictive value were 0.650 ± 0.075, 0.600 ± 0.105, 0.700 ± 0.103, 0.669 ± 0.085, and 0.638 ± 0.069 (mean ± SD), respectively. Classifying an image took only less than one second on average. A probability of 0 or 1 meant the image was categorized in the abortion category or normal category, respectively. When the probability was ≥0.5, the image was put in the normal category; otherwise, it was in the abortion category. The histogram of the confidence score is shown in Figure 3. The confidence score increased, while the incidence of becoming live birth increased. A significant relationship was seen between the confidence score by the AI and the probability of incidence derived from the histogram (P < 0.005 by Cochran‐Armitage test). The linear regression model that fits the incidence are constructed as follows: y = 0.822 (±0.224) x + 0.128 (±0.129) (mean ± SE), x; the confidence score by the AI, y; the probability of incidence of becoming live birth. The area under the curve (AUC) of the receiver operating curve was 0.6594 ± 0.043 (mean ± SE), and the 95% confidence interval ranged 0.575‐0.743 as shown in Figure 4.
Table 1.
Discrimination ability of the best classifier. A classifier using a logistic regression method with L2 regularization with 640 selected images as the training data showed the best results
| Mean ± SD | Median | |
|---|---|---|
| Accuracy | 0.650 ± 0.075 | 0.656 |
| Sensitivity | 0.600 ± 0.105 | 0.625 |
| Specificity | 0.700 ± 0.103 | 0.750 |
| Positive predictive value | 0.669 ± 0.085 | 0.647 |
| Negative predictive value | 0.638 ± 0.069 | 0.667 |
Figure 3.
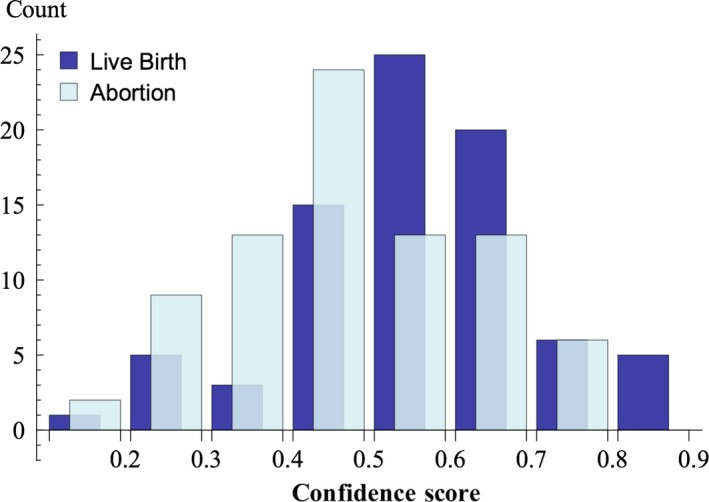
The histogram of confidence score; the probability that is likely to be live birth class of blastocyst images of which the outcome had reveled as either live birth or abortion with aneuploid. The probability of 1 and 0 means that the image is very likely to belong to the live birth class and the abortion class, respectively
Figure 4.
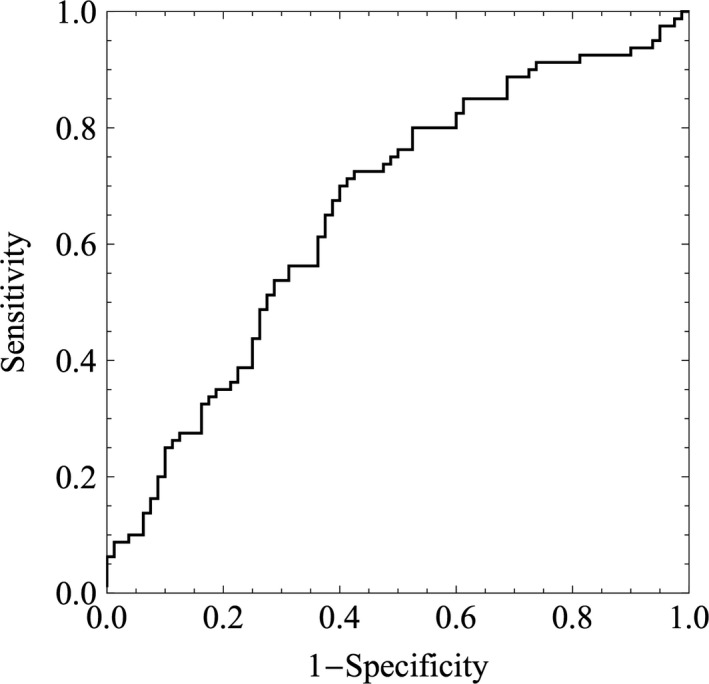
The area under the curve of the best classifier for predicting live birth by the logistic regression method. The value of the curve is 0.659 ± 0.043 (mean ± SE), and the 95% confidence interval ranged 0.575‐0.743
4. DISCUSSION
We obtained the classifier of the machine learning program using logistic regression with L2 regularization to classify blastocyststhat were implanted and led to live birth or aneuploid miscarriages. The best classifier with 640 selected training images showed the accuracy, sensitivity, specificity, positive predictive value, and negative predictive value for predicting live birth were 0.650 ± 0.075, 0.600 ± 0.105, 0.700 ± 0.103, 0.669 ± 0.085, and 0.638 ± 0.069 (mean ± SD), respectively (Table 1). One study found the live birth rate per transfer was 0.668, by using clinical factors such as age and body mass index .27 Another study reported that the grading of the TE was the only statistically significant independent predictor of live birth outcome, and the live birth probabilities of grade A, B, or C of the TE were 0.499, 0.339, and 0.080, respectively .28 These reports cannot be directly compared with our results because all of the miscarriage causes were not aneuploidy. But they suggest a classifier with no harm inflicted, owing to the use of AI in this study, is superior to those clinical factors and the examinations of conventional morphology.
There are several reports of AI29 with using machine learning, which is mainly deep learning 30 with the convolutional neural networks for medicine .31 The deep learning required a lot of data such as more than several thousands to make a classifier. Because there are 160 images in this study, we studied machine learning except deep learning. The accuracies with the deep learning have been reported such as 0.997 for histopathological diagnosis of breast cancer ,32 0.90‐0.83 for the early diagnosis of the Alzheimer's disease ,33 0.83 for urological dysfunctions ,34 0.72 ,35 and 0.50 36 for the colposcopy, 0.83 for the diagnostic imaging of orthopedic trauma .37 In practical sterility, there are some clinical disincentives for the embryo to achieve live birth; the uterine factors 38—(intrauterine adhesions ,39, 40 uterine myomas ,41 and endometrial polyps),42 endometriosis ,43 ovarian function ,44 oviduct obstruction ,45, 46 female diseases such as diabetes mellitus ,47 immune disorder ,48, 49 and uterine microbiota .50 Because these factors cannot be detected by the AI classifier from the image of the blastocyst, the accuracy, the sensitivity, and the specificity for live birth cannot reach to 1. These clinical characteristics of the blastocyst prevent the accuracy to predict live birth by any means from as close to 1. But we think the accuracy of 0.650 by the AI for predicting live birth distinguishing from abortion or miscarriage is feasible.
In our repeated calculations, the logistic regression continually showed the best result among the six machine learning methods examined. In short, the logistic regression models the log probabilities of each category with logistic functions of a linear combination of numerical features. However, further investigation in the future may prove other machine learning methods superior. Though we preliminarily tested the deep learning method of the convolutional neural networks with training data increased to 20 000, this did not show better accuracy than logistic regression. However, the deep learning might be able to provide better accuracy if a greater number of training images can be obtained.
We found 640 to be a suitable number for the training data because the maximum value of accuracy was obtained, while the error was close to the minimum value, as shown in Figure 2. L2 regularization seemed to sufficiently control over‐fitting for reducing the error. This was a key process in that the smaller size of both error and bias may yield a good classifier. Only a few seconds were needed to finish calculation for an image once the classifier was established. We therefore can envision the classifier being applied for clinical use.
In one report, embryos categorized using a model in which the two morphokinetic variables are applied to predict chromosomal content by blastomere biopsy on day 3, followed by array‐comprehensive genomic hybridization, showed a significant decrease in the percentage of chromosomally normal embryos for each decreasing category (A, 35.9%; B, 26.4%; C, 12.1%; D, 9.8%; P < 0.001) .51 Our method appears more viable than this because the classifier in this study induces no harm, and it shows a wider range of percentage for category prediction.
The receiver operating curve of the classifier shows the AUC value is 0.659 ± 0.043 (mean ± SE). Regarding the AUC of preimplantation genetic screening, a study reported a prediction model to classify embryos into high‐, medium‐, or low‐risk categories, with an AUC of 0.72 .52 The model could be useful for ranking embryos and for prioritizing them for PGT‐A. However, it does have limited predictive value for patients undergoing IVF in general .53 In terms of the discriminative ability of an examination, the classifier using AI seems inferior to PGT‐A; therefore, the classifier in this study may need to be improved. Though the ability of the classifier in this study shows viability for classifying blastocysts, the histogram of the probability of live birth and abortion categories suggests correct assigning to the classifier was difficult for some images. More precise recognition of atypical images would improve our classifier and increase accuracy.
There are further potential ways of improving the classifier. First, more images for both the normal and abortion categories may be needed. In this pilot study, the analysis is restricted to simply evaluating a pregnancy once it has been detected. As the blastocysts images are required prior to transfer, a further study with blastocyst images including images failed to implant is required in order to classify all embryos suitable for transfer by AI following this pilot study. It may also be important to statistically investigate the relationship of the probability given by the classifier with other clinical and morphological information, such as maternal age, body mass index, and TE grading. The data in this pilot study were not big and not stratified by such as maternal age or cycle history. The further investigation including the statistical stratification with more number of data might result in better results. Though we used images of blastocyst on only day 5 in order to reduce bias in this pilot study, the further investigation might be considered to prepare dataset by adding images of day 6 blastocyst that seem to grow late. Predictive ability could be improved for when the image and certain information are combined, and the outcome is investigated by statistical analysis. The proper conditions of capturing images should also be investigated and a standard determined so the classifier can be easily implemented at any institute. Blastocyst images showing chromosomal mosaicism or aneuploids confirmed by the PGT‐A were not included in this study. In further work, we would be able to develop an advanced classifier that can classify blastocyst images into three or more categories such as live birth, aneuploid, mosaic, and abortion.
Biologically, this method shows capabilities at least as good as other modes of examination. Ethically speaking, it inflicts no harm on the blastocyst. It offers economic savings for patients and/or clinical institutes, gives quick and efficient diagnosis of the classification, and permits examination over distances when the image is transferred via the Internet.
We applied machine learning in the realm of AI to develop a classifier for predicting from an image of the blastocyst that was implanted. The classifier can predict the probability of becoming live birth with the accuracy of 0.65. The further validation to test the feasibility of a classifier by machine learning using the data of the images and conventional methods might be conducted. Only less than one second are needed to complete analysis of each image. This method does not harm the embryo, which can be transferred after establishing the prediction. Though further study may be required to validate the classifier, the system showed a possibility that the AI would be feasible for clinical use and may bring benefits to both patients and medical personnel.
The contents in this manuscript were approved as a patent in Japan; patent 6422142.
DISCLOSURES
Conflict of interest: Yasunari Miyagi, Toshihiro Habara, Rei Hirata, and Nobuyoshi Hayashi declare that they have no conflict of interest. Human rights statements and informed consent:Human rights statements and informed consent: All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for which identifying information is included in this article. A Web site with additional information including an “opt‐out” option was set up for the study. Animal studies:No animals were used in this study. Approval by ethics committee:The protocol for the research project including human subjects has been approved by the Institutional Review Board at Okayama Couples’ Clinic (IRB no. 18000128‐04).
ACKNOWLEDGEMENTS
Daniel Bodri MD MSc PhD for advice of proofs.
Miyagi Y, Habara T, Hirata R, Hayashi N. Feasibility of artificial intelligence for predicting live birth without aneuploidy from a blastocyst image. Reprod Med Biol. 2019;18:204–211. 10.1002/rmb2.12267
Clinical Trial Registry: This study is not a clinical trial.
REFERENCES
- 1. Rienzi L, Vajta G, Ubaldi F. Predictive value of oocyte morphology in human IVF: a systematic review of the literature. Hum Reprod. 2011;17:34‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirkegaard K, Ahlström A, Ingerslev HJ, Hardarson T. Choosing the best embryo by time lapse versus standard morphology. Fertil Steril. 2015;103:323‐332. [DOI] [PubMed] [Google Scholar]
- 3. Finn A, Scott L, O'Leary T, Davies D, Hill J. Sequential embryo scoring as a predictor of aneuploidy in poor‐prognosis patients. Reprod Biomed Online. 2010;21:381‐390. [DOI] [PubMed] [Google Scholar]
- 4. Eaton JL, Hacker MR, Barrett CB, Thornton KL, Penzias AS. Influence of patient age on the association between euploidy and day‐3 embryo morphology. Fertil Steril. 2010;94:365‐367. [DOI] [PubMed] [Google Scholar]
- 5. Eaton JL, Hacker MR, Harris D, Thornton KL, Penzias AS. Assessment of day‐3 morphology and euploidy for individual chromosomes in embryos that develop to the blastocyst stage. Fertil Steril. 2009;91:2432‐2436. [DOI] [PubMed] [Google Scholar]
- 6. Wells D. Embryo aneuploidy and the role of morphological and genetic screening. Reprod Biomed Online. 2010;21:274‐277. [DOI] [PubMed] [Google Scholar]
- 7. Chavez SL, Loewke KE, Han J, et al. Dynamic blastomere behaviour reflects human embryo ploidy by the four‐cell stage. Nat Commun. 2012;3:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong CC, Loewke KE, Bossert NL, et al. Non‐invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115‐1121. [DOI] [PubMed] [Google Scholar]
- 9. Conaghan J, Chen AA, Willman SP, et al. Improving embryo selection using a computer‐automated time‐lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100:412‐419.e5. [DOI] [PubMed] [Google Scholar]
- 10. Alfarawati S, Fragouli E, Colls P, et al. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil Steril. 2011;95:520‐524. [DOI] [PubMed] [Google Scholar]
- 11. Ziebe S, Lundin K, Loft A, et al. FISH analysis for chromosomes 13, 16, 18, 21, 22, X and Y in all blastomeres of IVF pre‐embryos from 144 randomly selected donated human oocytes and impact on pre‐embryo morphology. Hum Reprod. 2003;18:2575‐2581. [DOI] [PubMed] [Google Scholar]
- 12. Dahdouh EM, Balayla J, Audibert F, et al. Technical update: preimplantation genetic diagnosis and screening. J Obstet Gynaecol Can. 2015;37:451‐463. [DOI] [PubMed] [Google Scholar]
- 13. Brezina PR, Kutteh WH. Clinical applications of preimplantation genetic testing. BMJ. 2015;350:g7611‐g7611. [DOI] [PubMed] [Google Scholar]
- 14. Gleicher N, Metzger J, Croft G, Kushnir VA, Albertini DF, Barad DH. A single trophectoderm biopsy at blastocyst stage is mathematically unable to determine embryo ploidy accurately enough for clinical use. Reprod Biol Endocrinol. 2017;15:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weissman A, Shoham G, Shoham Z, Fishel S, Leong M, Yaron Y. Preimplantation genetic screening: results of a worldwide web‐based survey. Reprod Biomed Online. 2017;35(6):693‐700. [DOI] [PubMed] [Google Scholar]
- 16. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Lean Res. 2014;15:1929‐1958. [Google Scholar]
- 17. Nowlan SJ, Hinton GE. Simplifying neural networks by soft weight‐sharing. Neural Comput. 1992;4:473‐493. [Google Scholar]
- 18. Dreiseitl S, Ohno‐Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35:352‐359. [DOI] [PubMed] [Google Scholar]
- 19. Ben‐Bassat M, Klove KL, Weil MH. Sensitivity analysis in Bayesian classification models: multiplicative deviations. IEEE Trans Pattern Anal Mach Intell. 1980;3:261‐266. [DOI] [PubMed] [Google Scholar]
- 20. Friedman J, Baskett F, Shustek L. An algorithm for finding nearest neighbors. IEEE Trans Comput. 1975;100:1000‐1006. [Google Scholar]
- 21. Breiman L. Random forests. Mach Learn. 2001;45:5‐32. [Google Scholar]
- 22. Tsochantaridis I, Joachims T, Hofmann T, Altun Y. Large margin methods for structured and interdependent output variables. J Mach Learn Res. 2005;6:1453‐1484. [Google Scholar]
- 23. Rumelhart D, Hinton G, Williams R. Learning representations by back‐propagating errors. Nature. 1986;323:533‐536. [Google Scholar]
- 24. Kohavi R.A Study of Cross‐Validation and Bootstrap for Accuracy Estimation and Model Selection. Proceedings of the 14th International Joint Conference on Artificial Intelligence. 1995 Aug 20–25; Montreal. Montreal: International Joint Conference on Artificial Intelligence, 1995;2:1137‐43. [Google Scholar]
- 25. Schaffer C. Selecting a classification method by cross‐validation. Mach Learn. 1993;13:135‐143. [Google Scholar]
- 26. Refaeilzadeh P, Tang L. Cross‐Validation In: Liu H, Özsu MT, editors. Encyclopedia of database systems. New York, NY: Springer; 2009:532‐538. [Google Scholar]
- 27. Kresowik JD, Sparks AE, Van Voorhis BJ. Clinical factors associated with live birth after single embryo transfer. Fertil Steril. 2012;98:1152‐1156. [DOI] [PubMed] [Google Scholar]
- 28. Ahlström A, Westin C, Reismer E, Wikland M, Hardarson T. Trophectoderm morphology: an important parameter for predicting live birth after single blastocyst transfer. Hum Reprod. 2011;26:3289‐3296. [DOI] [PubMed] [Google Scholar]
- 29. Miyagi Y, Fujiwara K, Oda T, Miyake T, Coleman RL. Development of new method for the prediction of clinical trial results using compressive sensing of artificial intelligence. J Biostat Biometric. 2018;3:202. [Google Scholar]
- 30. Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798‐1828. [DOI] [PubMed] [Google Scholar]
- 31. Abbod MF, Catto JW, Linkens DA, Hamdy FC. Application of artificial intelligence to the management of urological cancer. J Urol. 2007;178:1150‐1156. [DOI] [PubMed] [Google Scholar]
- 32. Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortiz A, Munilla J, Gorriz JM, Ramirez J. Ensembles of deep learning architectures for the early diagnosis of the Alzheimer’s disease. Int J neural systems. 2016;26:1650025. [DOI] [PubMed] [Google Scholar]
- 34. Gil D, Johnsson M, Chamizo J, Paya AS, Fernandez DR. Application of artificial neural networks in the diagnosis of urological disfunctions. Expert Syst Appl. 2009;36:5754‐5760. [Google Scholar]
- 35. Simões PW, Izumi NB, Casagrande RS, Venson R, Veronezi CD, Moretti GP. Classification of images acquired with colposcopy using artificial neural networks. Cancer Inform. 2014;13:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato M, Horie K, Hara A, et al. Application of deep learning to the classification of images from colposcopy. Oncol lett. 2018;15:3518–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olczak J, Fahlberg N, Maki A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 2017;88:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanders B. Uterine factors and infertility. J Reprod Med. 2006;51:169–176. [PubMed] [Google Scholar]
- 39. Pabuccu R, Atay V, Orhon E, Urman B, Ergün A. Hysteroscopic treatment of intrauterine adhesions is safe and effective in the restoration of normal menstruation and fertility. Fertil Steril. 1997;68:1141–1143. [DOI] [PubMed] [Google Scholar]
- 40. Liu L, Huang X, Xia E, Zhang X, Li TC, Liu Y. A cohort study comparing 4 mg and 10 mg daily doses of postoperative oestradiol therapy to prevent adhesion reformation after hysteroscopic adhesiolysis. Hum Fertil. 10.1080/14647273.2018.1444798 [DOI] [PubMed] [Google Scholar]
- 41. Ikhena DE, Bulun SE. Literature review on the role of uterine fibroids in endometrial function. Reprod Sci. 2018;25:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor E, Gomel V. The uterus and fertility. Fertil Steril. 2008;89:204–16. [DOI] [PubMed] [Google Scholar]
- 43. Tomassett C, D’Hooghe T. Endometriosis and infertility: insights into the causal link and management strategies. Best Pract Res Clin Obstet Gynaecol. 2018;51:25–33. [DOI] [PubMed] [Google Scholar]
- 44. Christ JP, Gunning MN, Palla G, et al. Estrogen deprivation and cardiovascular disease risk in primary ovarian insufficiency. Fertil Steril. 2018;109:594–600. [DOI] [PubMed] [Google Scholar]
- 45. Arronet GH, Eduljee SY, O'Brien JR. A nine‐year surgery of fallopian tube dysfunction in human infertility: diagnosis and therapy. Fertil Steril. 1969;20:903–918. [DOI] [PubMed] [Google Scholar]
- 46. Segars JH, Hill GA, Herbert CM, Colston A, Moore DE, Winfield AC. Selective fallopian tube cannulation: initial experience in an infertile population. Fertil Steril. 1990;53:357–359. [DOI] [PubMed] [Google Scholar]
- 47. Ota K, Ohta H, Yamagishi S. Diabetes and Female Sterility/Infertility: Diabetes and Aging‐related Complications. Singapore: Springer; 2018. [Google Scholar]
- 48. Practice Committee of the American Society for Reproductive Medicine . The role of immunotherapy in in vitro fertilization: a guideline. Fertil Steril. 2018;110:387–400. [DOI] [PubMed] [Google Scholar]
- 49. Hong YH, Kim SJ, Moon KY, et al. Impact of presence of antiphospholipid antibodies on in vitro fertilization outcome. Obstet Gynecol Sci. 2018;61:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moreno I, Simon C. Relevance of assessing the uterine microbiota in infertility. Fertil Steril. 2018;110:337–343. [DOI] [PubMed] [Google Scholar]
- 51. Basile N, Nogales Mdel C, Bronet F, et al. Increasing the probability of selecting chromosomally normal embryos by time‐lapse morphokinetics analysis. Fertil Steril. 2014;101:699–704. [DOI] [PubMed] [Google Scholar]
- 52. Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non‐invasive morphokinetics. Reprod Biomed Online. 2013;26:477–485. [DOI] [PubMed] [Google Scholar]
- 53. Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time‐lapse imaging without PGS. Reprod Biomed Online. 2013;27:140–146. [DOI] [PubMed] [Google Scholar]


